Published online Jun 15, 2002. doi: 10.3748/wjg.v8.i3.499
Revised: December 23, 2001
Accepted: January 23, 2002
Published online: June 15, 2002
AIM: To study HCV polyprotein processing is important for the understanding of the natural history of HCV and the design of vaccines against HCV. The purpose of this study is to investigate the affection of context sequences on hepatitis C virus (HCV) E2 processing.
METHODS: HCV genes of different lengths were expressed and compared in vaccinia virus/T7 system with homologous patient serum S94 and mouse anti-serum ME2116 raised against E. coli-derived E2 peptide, respectively. Deglycosylation analysis and GNA (Galanthus nivalus) lectin binding assay were performed to study the post-translational processing of the expressed products.
RESULTS: E2 glycoproteins with different molecular weights (~75 kDa and ~60 kDa) were detected using S94 and ME2116, respectively. Deglycosylation analysis showed that this difference was mainly due to different glycosylation. Endo H resistance and its failure to bind to GNA lectin demonstrated that the higher molecular weight form (75 kDa) of E2 was complex-type glycosylated, which was readily recognized by homologous patient serum S94. Expression of complex-type glycosylated E2 could not be detected in all of the core-truncated constructs tested, but readily detected in constructs encoding full-length core sequences.
CONCLUSION: The upstream conserved full-length core coding sequence was required for the production of E2 glycoproteins carrying complex-type N-glycans which reacted strongly with homologous patient serum and therefore possibly represented more mature forms of E2. As complex-type N-glycans indicated modification by Golgi enzymes, the results suggest that the presence of full-length core might be critical for E1/E2 complex to leave ER. Our data may contribute to a better understanding of the processing of HCV structural proteins as well as HCV morphogenesis.
- Citation: Zhu LX, Liu J, Li YC, Kong YY, Staib C, Sutter G, Wang Y, Li GD. Full-length core sequence dependent complex-type glycosylation of hepatitis C virus E2 glycoprotein. World J Gastroenterol 2002; 8(3): 499-504
- URL: https://www.wjgnet.com/1007-9327/full/v8/i3/499.htm
- DOI: https://dx.doi.org/10.3748/wjg.v8.i3.499
Hepatitis C virus (HCV), the major cause of post-transfusion and community-acquired non-A, non-B hepatitis[1,2], is a member of the Flaviviridae family[3]. This virus has a positive-sense, single stranded RNA genome of about 9.6 kb, which encodes a polyprotein precursor of about 3000 amino acids. The polyprotein is further processed into various precursors and mature viral proteins[4,5]. The structural proteins are encoded in the order NH2-core-E1-E2-P7, which are processed into core (C), E1, E2, and P7 by host membrane-associated signal peptidase (s)[6-11]. The downstream nonstructural region is processed by a viral metalloprotease and a viral serine protease located at the N-terminus of NS3[12-17]. The core protein is thought to constitute the viral capsid with E1 and E2 being the virus envelope proteins. Numerous studies have shown that E1 and E2 are heavily glycosylated and associate to form a noncovalent heterodimeric complex[9,10,18,19]. E1 and E2 are believed to be type I transmembrane proteins with an N-terminal glycosylated ectodomain and a C-terminal hydrophobic anchor.
The lack of an efficient in vitro cell culture system for productive HCV propagation[20-27] and low levels of HCV particles in the liver tissues or blood of infected patients[28,29] have hampered the study of native viral proteins. Fortunately, a variety of prokaryotic and eukaryotic expression systems have proved useful for the production and characterization of HCV encoded proteins[8,9,11,30-32]. However, diverse findings have been reported, regarding the molecular weights of E2, which most likely is a reflection of the differences in efficiency of HCV polyprotein processing and post-translational modification achieved in the particular systems[7-9,11,17-19,30]. In this study, the E2 expression of recombinant plasmids carrying various length of HCV C-E1-E2 coding sequences was analyzed in the vaccinia virus/bacteriophage T7 RNA polymerase expression system[33]. The results suggest that the upstream conserved core coding sequence is required for the production of E2 glycoproteins carrying complex-type N-glycans which react strongly with homologous patient serum and therefore possibly represent more mature forms of E2.
Human HeLa (ATCC #CCL-2) and monkey BS-C-1 (ATCC #CCL-26) cells were maintained in Dulbecco’s modified essential medium (DMEM/HG) supplemented with 5% heat inactivated fetal calf serum (FCS) at 37 °C in a 5% CO2 atmosphere. Recombinant vaccinia virus vTT7 that expresses the bacteriophage T7 RNA polymerase gene under the control of vaccinia virus early/late promoter P7.5 was generated and propagated as previously described[34]. PFU (plaque forming unit) titration was performed on BS-C-1 cell monolayers.
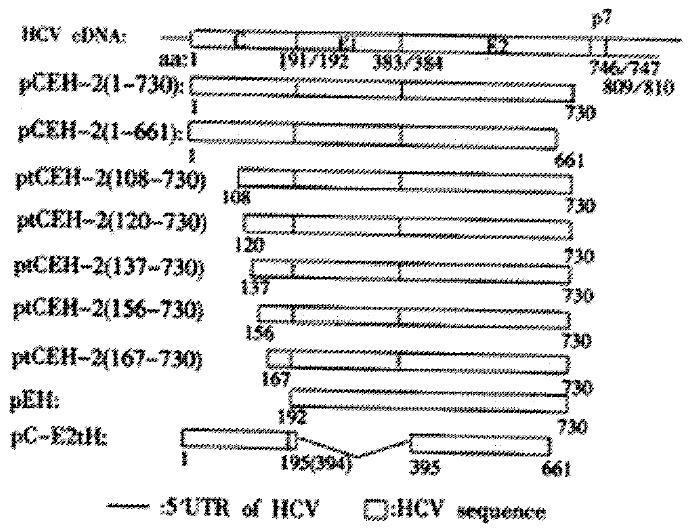
The vaccinia virus/T7 promoter expression vector pTM1 was kindly provided by Bernard Moss (NIH, Bethesda, USA) and all of the expression plasmids carrying HCV cDNA encoding structural proteins described below were derived from pTM1. Figure 1 depicts the HCV gene fragment in the expression plasmids. Plasmids pCEH-2 (1-730) and pEH containing HCV C, E1 and E2 gene of subtype 1b[35] (GENBANK accession #D10934) were described previously[34]. Briefly, cDNA sequences encoding HCV polyprotein amino acids 192 to 730 were inserted into pTM1 to obtain pEH. HCV sequences encoding complete C were fused with the E1/E2 sequences of pEH to result in plasmid pCEH-2 (1-730). The latter plasmid served as basis for PCR cloning to generate plasmids pCE1 (1-341), ptCEH-2 (108-730), ptCEH-2 (120-730), ptCEH-2 (137-730), ptCEH-2 (156-730), ptCEH-2 (167-730), pCEH-2 (1-661) and pTM1/EH (192-661) for the expression of 5’ or/and 3’ truncated HCV sequences encoding HCV polyprotein amino acids as indicated by numbers in parenthesis. Plasmid pC-E2tH (1-195/394-661) was generated by deleting E1 coding sequences from pCEH-2 (1-661) and linking polyprotein amino acids aa194 and aa394 together. Authenticity of HCV cDNA sequences in all constructed plasmids was confirmed by automatic sequencing.
HeLa cells grown to 80% confluency were infected with recombinant vaccinia virus vTT7 at a multiplicity of infection (MOI) of 10 PFU per cell to allow for production of recombinant T7 RNA polymerase. At 2 h post-infection, the inoculum was removed and DNA of pTM1 based expression plasmids was transfected using DOTAP liposomal transfection reagent as described by the manufacturer (Roche Molecular Biochemicals, Mannheim). After 24 hours of incubation, infected/transfected cells were washed twice with PBS, harvested by scraping, pelleted upon brief centrifugation, and resuspended in a small volume of PBS. Cell lysates were prepared by adding SDS-PAGE sample loading buffer and stored at -80 °C until further analysis.
Cell lysates were separated by reductive SDS-PAGE and then transferred onto nitrocellulose membranes (Schleicher & Schuell). Blocking was done using 5% fat-free milk powder. For immunodetection of HCV proteins, blots were incubated with primary antibodies, washed, and incubated with 1000 fold diluted HRP-protein A (Sigma). The membranes were then washed again and reactive proteins were detected using the ECL system (Amersham Phamacia Biotech) according to the manufacturers’ instructions. The primary antibodies used in this study include: anti-HCV human serum S94 at a dilution of 1:500 (kindly provided by Wang, Y., Beijing University, China), anti-HCV human serum S268 at a dilution of 1: 500 (kindly provided by Lu Z., Shanghai Ruijin Hospital, China), anti-E2 mouse polyclonal antibody ME2116[36] at a dilution of 1:300 (raised against E. coli-derived HCV E2 polypeptide aa450 to 565), and anti-E1 rabbit polyclonal antibody RE1135-C at a dilution of 1:250 (raised against an E. coli-derived C-terminally truncated HCV E1 fragment).
For deglycosylation analysis, cell pellets were directly lysed in denaturing buffer provided by the manufacturer and digested with PNGase F (NEB) or Endo H (NEB) for 2 hours at 37 °C.
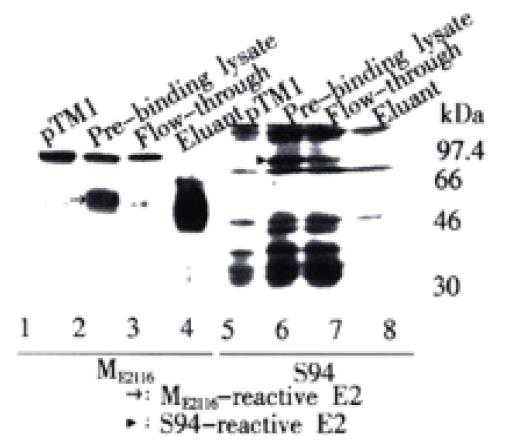
The type of N-glycans on expressed E2 glycoproteins was also analyzed by testing its ability to bind to GNA (Galanthus nivalus) lectin. HeLa cells infected with vTT7 and transfected with pCEH-2 (1-730) were collected by scraping, washed in cold PBS, and then lysed with lysis buffer (50 mM Tris-HCl [pH8.0], 150 mM NaCl, 0.5% Nonidet P-40, 1 mM PMSF). After centrifugation at 10000 g, the supernatant was allowed to bind to GNA-agarose (Sigma). The flow-through fraction was collected and the gel matrices were washed with lysis buffer. Bound proteins were eluted with 1M α-D-mannopyranoside (Sigma) in lysis buffer. Samples were then analyzed by Western-blotting.
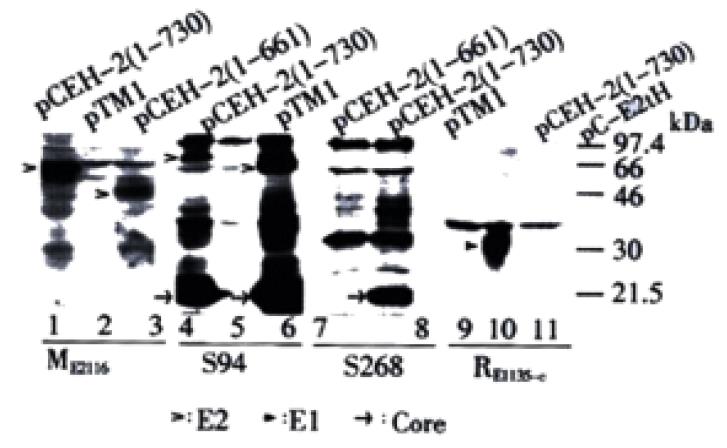
The hybrid vaccinia virus/T7 bacteriophage RNA polymerase expression system was used to study the expression of the HCV structural proteins. The transient expression products of plasmids pCEH-2 (1-730) and pCEH-2 (1-661), which contain HCV cDNA encoding the structural region terminating at amino acid 730 and 661 of the polyprotein respectively, were analyzed by Western blot using polyclonal mouse serum ME2116 raised against E. coli-derived E2 protein[36]. E2 products with apparent molecular weights (MWs) of ~60 kDa and ~50 kDa were detected for pCEH-2 (1-730) and pCEH-2 (1-661) respectively (Figure 2, lane 1, 3). The apparent molecular weights were higher than calculated values, which, along with the heterogeneous appearance of the detected bands, suggested that these were glycosylated expression products. The lower molecular weight of the E2 species obtained from expression of pCEH-2 (1-661) in comparison to pCEH-2 (1-730) was consistent with the introduced truncation at the 3’ end of the E2 coding sequences leading to the loss of 70 amino acids in the recombinant polypeptide backbone.
The expression products were also analyzed with HCV patient serum S94. Multiple prominent bands were detected for pCEH-2 (1-730) and pCEH-2 (1-661), but not for vector plasmid pTM1, representing expressed HCV structure proteins and possibly some precursors. The bands of ~75 kDa and ~66 kDa (Figure 2, lane 4, 6), which again consistent with the different length of E2 coding sequence in both plasmids, should represented the E2 proteins, although their MWs were higher than that detected by ME2116. It is worth noting that the analyzed HCV cDNA originated from the same patient from whom S94 was collected. The core antigen and some precursors of expression products could also be detected by another HCV patient serum S268, but we could not detect the E2-specific 75 kDa band (Figure 2, lane 8), which could be attributed to the high variability of the E2 glycoprotein[37,38].
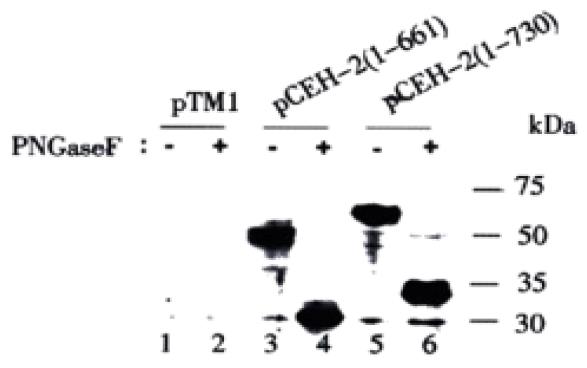
The antibody dependent detection of different E2 glycoprotein species was surprising. To rule out that the E2 bands of higher MW is the uncleaved E1-E2 precursors, the expression products from pCEH-2 (1-730) were analyzed with anti-E1 rabbit sera. A heavy band of about 30 kDa was detected, possibly representing multiple forms of E1 proteins (Figure 2, lane 10), while no E1-specific band with higher MW was detected. Another possible explanation could be that E2 polypeptides of varying sizes were synthesized due to incomplete or irregular processing of the polyprotein. This hypothesis was abandoned when we subjected recombinant proteins to deglycosylation with PNGase F prior to Western blot analysis with mouse polyclonal antibody ME2116. After PNGase F treatment, only one E2-specific band with apparent molecular weight of 30 kDa was detected for pCEH-2 (1-661) expression products, while an E2-specific doublet band of 33/34 kDa was detected for pCEH-2 (1-730) expression products (Figure 3), which is consistent with the MWs of calculated E2 polypeptide backbones. It suggests that difference of the MWs of E2 species detected by ME2116 and S94 from the same transient expression products (pCEH-2 (1-730): ~60 kDa and ~75 kDa, pCEH-2 (1-661): ~50 kDa and ~66 kDa) is mainly due to different N-glycosylation. The detection of a E2 peptide doublet with pCEH-2 (1-730) but not with pCEH-2 (1-661) is in agreement with the fact that E2 contains a PKR-eIF2a phosphorylation site (PKR: RNA-activated protein kinase) at aa659-670[39], which is largely deleted in the pCEH-2 (1-661) construct.
Altogether, the above results suggest that the 75 kDa and 66 kDa bands detected by S94 are HCV E2 glycoproteins of heavier glycosylation and thus higher molecular weight.
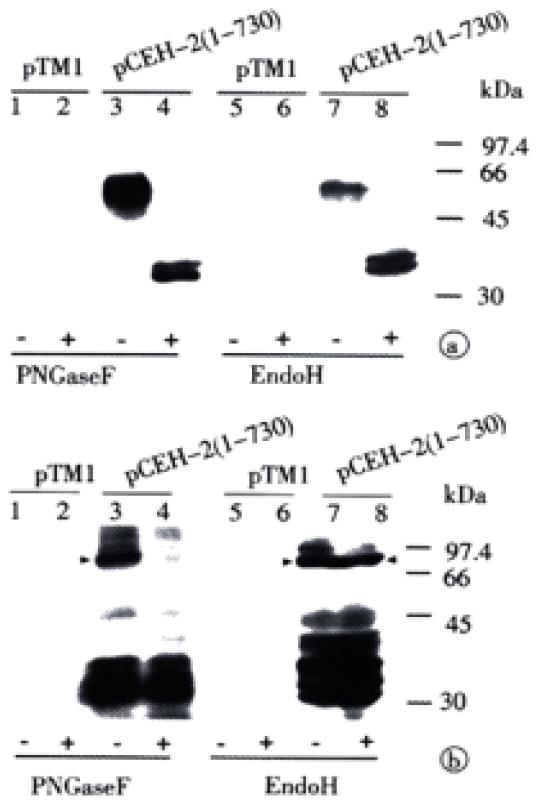
Since ME2116- and S94-reactive E2 species had polypeptide backbones of the same size, the difference in apparent molecular weight and antibody reactivity could only be attributed to differences in the degree and/or type of glycosylation. The glycan type on different E2 species expressed from pCEH-2 (1-730) was then analyzed by testing their sensitivity to PNGase F and Endo H. PNGase F hydrolyzes all types of N-glycan chains from glycopeptides and glycoproteins unless they carry α-1-3 linked core fucose residues present in insect and plant glycoproteins[40], while Endo H cleaves only high mannose structures and hybrid structures on N-linked oligosaccharides of glycoproteins[41]. Figure 4A shows that the ME2116-reactive E2 species was sensitive to both PNGase F and Endo H digestion. The S94-reactive E2 species disappeared after PNGase F digestion (Figure 4B). It was difficult to detect the deglycosylated E2 with S94 after PNGase F treatment, because there were multiple HCV polyprotein precursor proteins of about 30000 reacted strongly with S94 and unglycosylated E2 seemed to react weakly with S94 (unpublished data). However, the highly glycosylated, S94-reactive E2 band remained after Endo H digestion (Figure 4B). The glycan type of different E2 species expressed from pCEH-2 (1-730) were also analyzed by testing their ability to bind to GNA lectin. GNA is specific for the non-reducing end of α-D-mannosyl residue of glycoconjugate and therefore can be used to probe the presence of high mannose type or hybrid type glycans on glycoproteins[42,43]. The ME2116-reactive E2 species could quantitatively bind to and be eluted from GNA-agarose, whereas no obvious binding could be demonstrated for S94-reactive E2 species (Figure 5).
The resistance of S94-reactive E2 glycoprotein species to Endo H digestion together with the fact that it could not bind to GNA indicates that the S94-reactive E2 protein carries complex-type glycans.
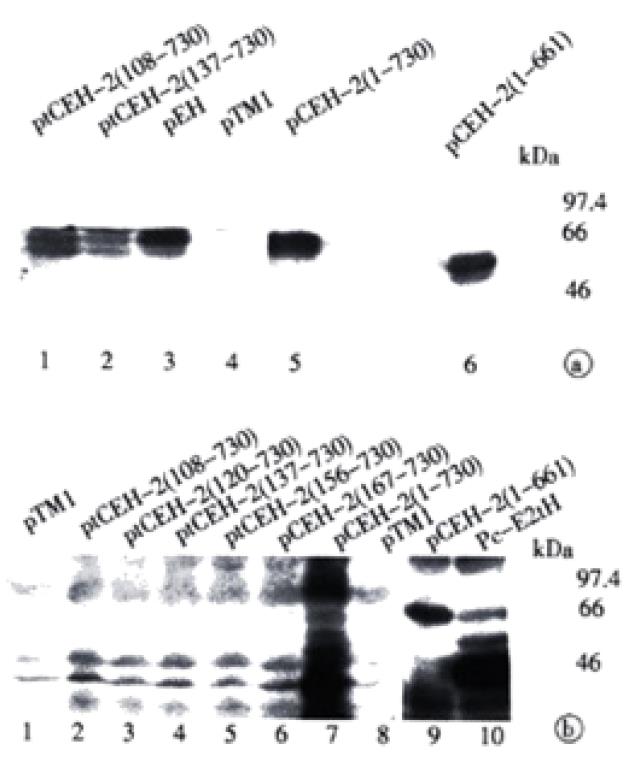
We also assessed if co-expression of core or E1 coding sequences had any effect on the production of the E2 proteins, which was readily recognized by homologous patient serum S94. A set of expression plasmids containing HCV cDNAs with various deletions in the core sequence (ORF starting at aa108, aa120, aa137, aa151, or aa167, respectively) were constructed. The lysates from vTT7-infected and plasmid DNA transfected HeLa cells were analyzed by Western blot with either the mouse anti-serum ME2116 or the patient serum S94 (Figure 6).
When using mouse antibody ME2116, recombinant E2 of ~60 kDa and ~50 kDa could be detected upon expression of all constructs tested (Figure 6A). Patient serum S94 allowed detection of E2 for pCEH-2 (1-730) and pCEH-2 (1-661) with full-length core coding sequences (Figure 6B, lanes 7, 9), similar to that described in Figure 2. In contrast, no E2 products could be visualized after expression of constructs containing no or only partial core sequences (Figure 6B, lanes 1-6). Interestingly, deletion of E1 coding sequences had no significant effect on the synthesis of S94 detectable E2 protein (Figure 6B, lane 10). These results suggests that the presence of complete HCV core sequence is crucial for the expression and/or post-translational processing of the complex-type glycosylated form of E2.
In this study, various constructs of HCV cDNAs placed under transcriptional control of the bacteriophage T7 promoter were transiently expressed using vaccinia virus/T7 system. Upon characterization of the HCV gene products with different antibodies, two species of E2 with different MWs were identified in the expression products of the same plasmid. The high molecular weight forms of E2 were readily recognized by a patient serum, but displayed weak reactivity with antibodies raised against E. coli derived E2. These high molecular weight forms of E2 were not likely produced from inefficient proteolytical processing at the E1/E2 boundary as these proteins were not stained with E1-specific antibodies. Efficient processing at E1/E2 was confirmed by deglycosylation analysis. The difference of the MWs of E2 species detected by S94 and by ME2116 was therefore mainly due to different N-glycosylation. The S94-reactive E2 glycoproteins, which were resistant to Endo H digestion and could not bind to GNA, carry complex-type glycans.
The specific recognition of the complex-type glycosylated E2 but not the high-mannose-type glycosylated E2 by homologous patient serum S94 suggested that the former could be a better representation of native E2 proteins on HCV virions. Similar results were also reported by Inudoh et al[44]. By comparing the reactivity of complex-type glycosylated E2 and the high-mannose-type glycosylated E2 with different patient sera, they demonstrated that the former is superior in diagnosing HCV infection. Their results and our results reported here are in concordance with the finding that E2 protein on patient derived virions contained complex-type sugars indicating Golgi-specific modification[45].
Expression of full-length or C-terminally truncated envelop proteins in eukaryotic cells has demonstrated that E1 and E2 are retained within the ER membrane system due to the presence of ER-retention signals in the C-termini of both envelope proteins[46-50]. However, recent study indicates that HCV E2 proteins could also present in the Golgi apparatus of the stably transfected cell line expressing HCV C-E1-E2-NS2 fragment. A possible explanation could be that Martire et al[51] used an HCV gene fragment including full-length core sequences in their study while structural protein sequences without full-length core sequences were used to study the localization of envelop proteins. The results reported here demonstrated that the complex-type glycosylated, possibly more mature form of E2 is only detectable upon co-expression of the complete HCV core coding sequence. Deletion of the first 107 N-terminal core amino acid residues was obviously sufficient to abrogate production of complex-glycosylated E2. This result suggest that the core protein might allow for targeting the envelope glycoproteins to Golgi-specific modification, which could be a key step in the morphogenesis of HCV virions. HCV-like particles were observed when HCV cDNA encoding whole core, E1 and E2 was expressed in baculovirus-insect expression system[52]. After binding of core to the E1-E2 complex statically located on the ER membrane, virus-like particles might be formed and the conformation of E1-E2 complex changed, which could result in the abrogation of the ER-retention signal for the E1-E2 complex. Then the virus-like particles might migrate along the secretion pathway, where E2 (and E1) proteins undergo more complex glycosylation by the Golgi enzymes.
In summary, upon expression of recombinant HCV core, E1, and E2 sequences, the E2 proteins of different glycosylations could be identified. The complex-glycosylated E2 protein might represent a more mature form of E2 and its formation required the conserved core coding sequences. Our data may contribute to a better understanding of the processing of HCV structural proteins as well as HCV morphogenesis.
We thank Wang, Y. and Lu, Z. for providing the HCV clones and the HCV specific patient sera.
Edited by Schmid R and Pang LH
| 1. | Farci P. Choo QL, Kuo G, Weiner AJ, Overby LR, Bradley DW, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome [Science 1989; 244: 359-362]. J Hepatol. 2002;36:582-585. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4996] [Cited by in RCA: 4656] [Article Influence: 129.3] [Reference Citation Analysis (0)] |
| 2. | Kuo G, Choo QL, Alter HJ, Gitnick GL, Redeker AG, Purcell RH, Miyamura T, Dienstag JL, Alter MJ, Stevens CE. An assay for circulating antibodies to a major etiologic virus of human non-A, non-B hepatitis. Science. 1989;244:362-364. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2495] [Cited by in RCA: 2343] [Article Influence: 65.1] [Reference Citation Analysis (0)] |
| 3. | Major ME, Feinstone SM. The molecular virology of hepatitis C. Hepatology. 1997;25:1527-1538. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 226] [Cited by in RCA: 212] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 4. | Choo QL, Richman KH, Han JH, Berger K, Lee C, Dong C, Gallegos C, Coit D, Medina-Selby R, Barr PJ. Genetic organization and diversity of the hepatitis C virus. Proc Natl Acad Sci USA. 1991;88:2451-2455. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1133] [Cited by in RCA: 1132] [Article Influence: 33.3] [Reference Citation Analysis (0)] |
| 5. | Houghton M, Weiner A, Han J, Kuo G, Choo QL. Molecular biology of the hepatitis C viruses: implications for diagnosis, development and control of viral disease. Hepatology. 1991;14:381-388. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 586] [Cited by in RCA: 500] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 6. | Harada S, Watanabe Y, Takeuchi K, Suzuki T, Katayama T, Takebe Y, Saito I, Miyamura T. Expression of processed core protein of hepatitis C virus in mammalian cells. J Virol. 1991;65:3015-3021. [PubMed] |
| 7. | Hijikata M, Kato N, Ootsuyama Y, Nakagawa M, Shimotohno K. Gene mapping of the putative structural region of the hepatitis C virus genome by in vitro processing analysis. Proc Natl Acad Sci USA. 1991;88:5547-5551. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 465] [Cited by in RCA: 460] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 8. | Lanford RE, Notvall L, Chavez D, White R, Frenzel G, Simonsen C, Kim J. Analysis of hepatitis C virus capsid, E1, and E2/NS1 proteins expressed in insect cells. Virology. 1993;197:225-235. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 88] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 9. | Ralston R, Thudium K, Berger K, Kuo C, Gervase B, Hall J, Selby M, Kuo G, Houghton M, Choo QL. Characterization of hepatitis C virus envelope glycoprotein complexes expressed by recombinant vaccinia viruses. J Virol. 1993;67:6753-6761. [PubMed] |
| 10. | Selby MJ, Choo QL, Berger K, Kuo G, Glazer E, Eckart M, Lee C, Chien D, Kuo C, Houghton M. Expression, identification and subcellular localization of the proteins encoded by the hepatitis C viral genome. J Gen Virol. 1993;74:1103-1113. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 169] [Cited by in RCA: 163] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 11. | Spaete RR, Alexander D, Rugroden ME, Choo QL, Berger K, Crawford K, Kuo C, Leng S, Lee C, Ralston R. Characterization of the hepatitis C virus E2/NS1 gene product expressed in mammalian cells. Virology. 1992;188:819-830. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 90] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 12. | Grakoui A, McCourt DW, Wychowski C, Feinstone SM, Rice CM. A second hepatitis C virus-encoded proteinase. Proc Natl Acad Sci USA. 1993;90:10583-10587. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 305] [Cited by in RCA: 283] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 13. | Hijikata M, Mizushima H, Akagi T, Mori S, Kakiuchi N, Kato N, Tanaka T, Kimura K, Shimotohno K. Two distinct proteinase activities required for the processing of a putative nonstructural precursor protein of hepatitis C virus. J Virol. 1993;67:4665-4675. [PubMed] |
| 14. | Eckart MR, Selby M, Masiarz F, Lee C, Berger K, Crawford K, Kuo C, Kuo G, Houghton M, Choo QL. The hepatitis C virus encodes a serine protease involved in processing of the putative nonstructural proteins from the viral polyprotein precursor. Biochem Biophys Res Commun. 1993;192:399-406. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 145] [Cited by in RCA: 133] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 15. | Grakoui A, McCourt DW, Wychowski C, Feinstone SM, Rice CM. Characterization of the hepatitis C virus-encoded serine proteinase: determination of proteinase-dependent polyprotein cleavage sites. J Virol. 1993;67:2832-2843. [PubMed] |
| 16. | Manabe S, Fuke I, Tanishita O, Kaji C, Gomi Y, Yoshida S, Mori C, Takamizawa A, Yosida I, Okayama H. Production of nonstructural proteins of hepatitis C virus requires a putative viral protease encoded by NS3. Virology. 1994;198:636-644. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 86] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 17. | Tomei L, Failla C, Santolini E, De Francesco R, La Monica N. NS3 is a serine protease required for processing of hepatitis C virus polyprotein. J Virol. 1993;67:4017-4026. [PubMed] |
| 18. | Grakoui A, Wychowski C, Lin C, Feinstone SM, Rice CM. Expression and identification of hepatitis C virus polyprotein cleavage products. J Virol. 1993;67:1385-1395. [PubMed] |
| 19. | Matsuura Y, Suzuki T, Suzuki R, Sato M, Aizaki H, Saito I, Miyamura T. Processing of E1 and E2 glycoproteins of hepatitis C virus expressed in mammalian and insect cells. Virology. 1994;205:141-150. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 76] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 20. | Hiramatsu N, Dash S, Gerber MA. HCV cDNA transfection to HepG2 cells. J Viral Hepat. 1997;4 Suppl 1:61-67. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 15] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 21. | Seipp S, Mueller HM, Pfaff E, Stremmel W, Theilmann L, Goeser T. Establishment of persistent hepatitis C virus infection and replication in vitro. J Gen Virol. 1997;78:2467-2476. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 86] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 22. | Rumin S, Berthillon P, Tanaka E, Kiyosawa K, Trabaud MA, Bizollon T, Gouillat C, Gripon P, Guguen-Guillouzo C, Inchauspé G. Dynamic analysis of hepatitis C virus replication and quasispecies selection in long-term cultures of adult human hepatocytes infected in vitro. J Gen Virol. 1999;80:3007-3018. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 81] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 23. | Lohmann V, Körner F, Koch J, Herian U, Theilmann L, Bartenschlager R. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science. 1999;285:110-113. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2294] [Cited by in RCA: 2251] [Article Influence: 86.6] [Reference Citation Analysis (0)] |
| 24. | Song ZQ, Hao F, Min F, Ma QY, Liu GD. Hepatitis C virus infection of human hepatoma cell line 7721 in vitro. World J Gastroenterol. 2001;7:685-689. [PubMed] |
| 25. | Bartenschlager R, Lohmann V. Novel cell culture systems for the hepatitis C virus. Antiviral Res. 2001;52:1-17. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 152] [Cited by in RCA: 146] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 26. | Lohmann V, Körner F, Dobierzewska A, Bartenschlager R. Mutations in hepatitis C virus RNAs conferring cell culture adaptation. J Virol. 2001;75:1437-1449. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 370] [Cited by in RCA: 368] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 27. | Pietschmann T, Lohmann V, Rutter G, Kurpanek K, Bartenschlager R. Characterization of cell lines carrying self-replicating hepatitis C virus RNAs. J Virol. 2001;75:1252-1264. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 290] [Cited by in RCA: 288] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 28. | Ni YH, Chang MH, Lue HC, Hsu HY, Wang MJ, Chen PJ, Chen DS. Posttransfusion hepatitis C virus infection in children. J Pediatr. 1994;124:709-713. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 44] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 29. | Luengrojanakul P, Vareesangthip K, Chainuvati T, Murata K, Tsuda F, Tokita H, Okamoto H, Miyakawa Y, Mayumi M. Hepatitis C virus infection in patients with chronic liver disease or chronic renal failure and blood donors in Thailand. J Med Virol. 1994;44:287-292. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 42] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 30. | Hsu HH, Donets M, Greenberg HB, Feinstone SM. Characterization of hepatitis C virus structural proteins with a recombinant baculovirus expression system. Hepatology. 1993;17:763-771. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.0] [Reference Citation Analysis (0)] |
| 31. | Yan BS, Liao LY, Leou K, Chang YC, Syu WJ. Truncating the putative membrane association region circumvents the difficulty of expressing hepatitis C virus protein E1 in Escherichia coli. J Virol Methods. 1994;49:343-351. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 14] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 32. | Zhu LX, Kong YY, Wang Y, Li GD. Effect of Downstream Sequence on the Cleavage of Envelop Protein 1 Signal Sequence in Hepatitis C Virus. Sheng Wu Hua Xue Yu Sheng Wu Wu Li Xue Bao (Shanghai). 2001;33:682-686. [PubMed] |
| 33. | Moss B, Elroy-Stein O, Mizukami T, Alexander WA, Fuerst TR. Product review. New mammalian expression vectors. Nature. 1990;348:91-92. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 434] [Cited by in RCA: 458] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 34. | Li Y, Li G, Kong Y, Wang Y, Wang Y, Wen Y. Expression of structural proteins of hepatitis C virus (HCV) in mammalian cells. Sci China C Life Sci. 1998;41:47-55. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 35. | Wang Y, Okamoto H, Tsuda F, Nagayama R, Tao QM, Mishiro S. Prevalence, genotypes, and an isolate (HC-C2) of hepatitis C virus in Chinese patients with liver disease. J Med Virol. 1993;40:254-260. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 68] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 36. | Liu J, Zhu L, Zhang X, Lu M, Kong Y, Wang Y, Li G. Expression, purification, immunological characterization and application of Escherichia coli-derived hepatitis C virus E2 proteins. Biotechnol Appl Biochem. 2001;34:109-119. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 16] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 37. | Hijikata M, Kato N, Ootsuyama Y, Nakagawa M, Ohkoshi S, Shimotohno K. Hypervariable regions in the putative glycoprotein of hepatitis C virus. Biochem Biophys Res Commun. 1991;175:220-228. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 252] [Cited by in RCA: 236] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 38. | Weiner AJ, Brauer MJ, Rosenblatt J, Richman KH, Tung J, Crawford K, Bonino F, Saracco G, Choo QL, Houghton M. Variable and hypervariable domains are found in the regions of HCV corresponding to the flavivirus envelope and NS1 proteins and the pestivirus envelope glycoproteins. Virology. 1991;180:842-848. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 433] [Cited by in RCA: 415] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 39. | Taylor DR, Shi ST, Romano PR, Barber GN, Lai MM. Inhibition of the interferon-inducible protein kinase PKR by HCV E2 protein. Science. 1999;285:107-110. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 548] [Cited by in RCA: 530] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 40. | Tarentino AL, Gómez CM, Plummer TH. Deglycosylation of asparagine-linked glycans by peptide: N-glycosidase F. Biochemistry. 1985;24:4665-4671. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 882] [Cited by in RCA: 901] [Article Influence: 22.5] [Reference Citation Analysis (0)] |
| 41. | Tai T, Yamashita K, Ogata-Arakawa M, Koide N, Muramatsu T, Iwashita S, Inoue Y, Kobata A. Structural studies of two ovalbumin glycopeptides in relation to the endo-beta-N-acetylglucosaminidase specificity. J Biol Chem. 1975;250:8569-8575. [PubMed] |
| 42. | Kaku H, Goldstein IJ. Snowdrop lectin. Methods Enzymol. 1989;179:327-331. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 19] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 43. | Shibuya N, Berry JE, Goldstein IJ. One-step purification of murine IgM and human alpha 2-macroglobulin by affinity chromatography on immobilized snowdrop bulb lectin. Arch Biochem Biophys. 1988;267:676-680. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 46] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 44. | Inudoh M, Nyunoya H, Tanaka T, Hijikata M, Kato N, Shimotohno K. Antigenicity of hepatitis C virus envelope proteins expressed in Chinese hamster ovary cells. Vaccine. 1996;14:1590-1596. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 10] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 45. | Sato K, Okamoto H, Aihara S, Hoshi Y, Tanaka T, Mishiro S. Demonstration of sugar moiety on the surface of hepatitis C virions recovered from the circulation of infected humans. Virology. 1993;196:354-357. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 37] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 46. | Cocquerel L, Duvet S, Meunier JC, Pillez A, Cacan R, Wychowski C, Dubuisson J. The transmembrane domain of hepatitis C virus glycoprotein E1 is a signal for static retention in the endoplasmic reticulum. J Virol. 1999;73:2641-2649. [PubMed] |
| 47. | Flint M, McKeating JA. The C-terminal region of the hepatitis C virus E1 glycoprotein confers localization within the endoplasmic reticulum. J Gen Virol. 1999;80:1943-1947. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 37] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 48. | Duvet S, Cocquerel L, Pillez A, Cacan R, Verbert A, Moradpour D, Wychowski C, Dubuisson J. Hepatitis C virus glycoprotein complex localization in the endoplasmic reticulum involves a determinant for retention and not retrieval. J Biol Chem. 1998;273:32088-32095. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117] [Cited by in RCA: 116] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 49. | Cocquerel L, Meunier JC, Pillez A, Wychowski C, Dubuisson J. A retention signal necessary and sufficient for endoplasmic reticulum localization maps to the transmembrane domain of hepatitis C virus glycoprotein E2. J Virol. 1998;72:2183-2191. [PubMed] |
| 50. | Mottola G, Jourdan N, Castaldo G, Malagolini N, Lahm A, Serafini-Cessi F, Migliaccio G, Bonatti S. A new determinant of endoplasmic reticulum localization is contained in the juxtamembrane region of the ectodomain of hepatitis C virus glycoprotein E1. J Biol Chem. 2000;275:24070-24079. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 26] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 51. | Martire G, Viola A, Iodice L, Lotti LV, Gradini R, Bonatti S. Hepatitis C virus structural proteins reside in the endoplasmic reticulum as well as in the intermediate compartment/cis-Golgi complex region of stably transfected cells. Virology. 2001;280:176-182. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 32] [Article Influence: 1.3] [Reference Citation Analysis (0)] |