Published online Oct 15, 2001. doi: 10.3748/wjg.v7.i5.726
Revised: August 6, 2001
Accepted: August 16, 2001
Published online: October 15, 2001
AIM: To construct subtracted cDNA libraries and further identify differentially expressed genes that are related to the development of colorectal carcinoma (CRC).
METHODS: Suppression subtractive hybridization (SSH) was done on cDNAs of normal mucosa, adenoma and adenocarcinoma tissues from the same patient. Three subtracted cDNA libraries were constructed and then hybridized with forward and backward subtracted probes for differential screening. Positive clones from each subtracted cDNA library were selected for sequencing and BLAST analysis. Finally, virtual Northern Blot confirmed such differential expression.
RESULTS: By this way, there were about 3-4 × 102 clones identified in each subtracted cDNA library, in which about 85% positive clones were differentially screened. Sequencing and BLAST homology search revealed some clones containing sequences of known gene fragments and several possibly novel genes showing few or no sequence homologies with any known sequences in the database.
CONCLUSION: All results confirmed the effectiveness and sensitivity of SSH. The differentially expressed genes during the development of CRC can be used to shed light on the pathogenesis of CRC and be useful genetic markers for early diagnosis and therapy.
- Citation: Luo MJ, Lai MD. Identification of differentially expressed genes in normal mucosa, adenoma and adenocarcinoma of colon by SSH. World J Gastroenterol 2001; 7(5): 726-731
- URL: https://www.wjgnet.com/1007-9327/full/v7/i5/726.htm
- DOI: https://dx.doi.org/10.3748/wjg.v7.i5.726
Colorectal carcinoma (CRC) is one of the most common forms of malignancy and the major cause of mortality in the population. The adenoma-carcinoma sequence has been accepted as the main pathway involving a series of molecular changes, which include activation of oncogenes, inactivation of tumor suppressor genes, etc. in colorectal carcinogenesis for several years[1]. However, due to the complexity of carcinogenesis, there are still many problems unsolved for the model of CRC. Not only are all the known genetic alterations not enough to explain fully the progression toward malignancy, but also there is no one genetic marker which is entirely tissue and/or cancer specific. Hence, an attempt was made to find new genes that are related to the development of CRC. Our goal was to clone and identify new cancer-predisposing genes.
In almost any area of biology or medicine, questions arise from the differential gene expression. Transformations in the normal mucosa-adenoma-carcinoma sequence result from the expression changes of specific gene (s) too. The identification and characterization of human genes expressed exclusively or preferentially in tumor tissue will hopefully shed light on the mechanisms of CRC development and provide useful genetic markers for screening, diagnosis, prognosis, therapeutic monitoring and even follow-up development of therapeutic vaccines.
There are many techniques that AIM at producing an inventory of differential transcripts between two populations of mRNAs. Although all these methods have proven successfully isolating differentially expressed genes, they all had some intrinsic drawbacks: need for plenty of initiating materials, difficulty in obtaining rare transcripts, significant incidence of false positives, labor intensive, etc. Suppression subtractive hybridization (SSH), a PCR-based cDNA subtracted method[2], combines normalization and subtraction in a single procedure and generates subtracted cDNA libraries. This method overcomes many drawbacks mentioned above.
In this research, we conducted SSH to identify genes differentially expressed among cDNAs of normal mucosa, adenoma and adenocarcinoma tissues from the same patient. Three subtracted cDNA libraries were constructed and numerous sequences were isolated.
All normal (N), adenoma (A) and adenocarcinoma (T) tissue samples obtained from the same patient were histologically confirmed by a pathologist. The tissues were frozen in liquid nitrogen and homogenized with mortar. Total RNA was prepared using Trizol reagent (Gibco BRL, USA). RNA integrity was checked on 1% formaldehyde agarose gel. Poly (A) + RNA was then purified using PolyATract mRNA cDNA system kit (Promega, USA) as per manufacturer’s instructions.
0.5 μg mRNAs from each sample were reversely transcribed using MuLV SuperScript II reverse transcriptase (Gibco BRL, USA), SMART II oligonucleotides and CDS primers from SMART cDNA synthesis kit (Clontech, USA). The first strand cDNA mixtures were amplified by LD PCR. Before amplification, the number of PCR cycles was optimized (19 for N, 17 for both A and T) to ensure that cDNA products remain in the exponential phase of amplification to reduce nonspecific amplification.
After cDNAs were produced, they were purified by passing through Chroma spin-400 columns (Clontech, USA). Each purified tester cDNA was digested with RsaI to give an average insert length approximately longer than that of 600 base pairs. Then they were phenol chloroform-extracted, ethanol-precipitated, and dissolved in 6 μL ddH2O. The tester cDNAs were subdivided into two equal groups and then ligated to adaptor 1 and 2R in separate ligation reactions. Ligation efficiency was tested. Subtractive hybridization was performed by annealing an excess of driver cDNAs with each sample of adaptor-ligated tester cDNAs. The cDNAs were heat-denatured and incubated in 68 °C for 8 h. After the first hybridization, the two samples were mixed together and hybridized again with freshly heat-denatured driver cDNAs for 20 h at 68 °C. Two rounds of hybridization would generate a normalized population of tester-specific cDNAs with different adaptors on each end.
After filling in the ends, two rounds of PCR amplification were performed to enrich desired cDNAs containing both adaptors by exponential amplification of these products. The optimized cycles for the first and second PCR were 26 and 12 respectively to increase representation and reduce redundancy of subtracted cDNA libraries.
Secondary PCR products were used as templates for PCR amplification of human G3PDH at 18, 23, 28 and 33 cycles to assure subtraction efficiency. PCR products were run on 1.8% agarose gel.
Both forward and backward SSH were performed. The difference between them was that the tester sample in forward SSH was used as driver sample in backward SSH.
Products from the secondary PCR were T-A cloned into pGEM-T Easy Vector using pGEM-T Easy Vector System II kit (Promega, USA). The ligation products were transformed into DH5α competent cells. The transformed cells were plated on 2-YT agar plates containing ampicilin, X-Gal and IPTG allowed for color selection of colonies. Transformation efficiency was calculated and white clones were selected for further characterization.
Randomly selected individual clones were grown for 8 h in 96-well culture plates and then used for clone PCR amplification. The primers used were nested Primer1 and 2R of adaptors. After 27 cycles amplification, the PCR products were subjected to a differential screening process. Briefly, 3 μL of amplified products of each candidate gene was mixed with DNA dilution buffer (50 μg/mL herring sperm DNA, 10 mM Tris. Cl, pH8.0, 1 mM EDTA), spotted onto duplicate Hybond N+ Membranes (Boeheringer Mannheim, Germany) and UV cross-linked for 1 min. The forward and backward subtracted cDNAs were used as probes. These cDNAs were DIG-labeled by PCR amplification using PCR DIG probe synthesis kit (Boeheringer Mannheim, Germany), digested with RsaI, EagI and SmalI to remove the adaptors and purified with column. After prehybridization, the duplicate membranes were hybridized with forward and backward subtracted cDNA libraries probes respectively at 42 °C for 16 h-20 h in hybridization solution with 50% deionized formamide, 0.1% (w/v)-Lauroylsarcosinee, 2% blocking reagent, 0.02% (w/v) SDS, 2 μg/mL NP1, NP2R and CDS primer sequences. The membranes were then washed for 2 ± 5 min with 2 × SSC, 0.1% SDS at room temperature and for 2 ± 15 min in 0.5 × SSC, 0.1% SDS at 68 °C. Then, they were blocked by gently agitating in blocking solution at 37 °C for 1 h, incubated in buffer I with Anti-DIG (1∶4000) at 37 °C for 30 min, washed in buffer I for 2 ± 15 min and equilibrated in buffer III for 5 min. Finally, the membranes were incubated in mixture of NBT and BCIP for about 15 min. The clones giving at least 5-fold higher hybridization signals with the forward probes were selected for further analysis.
Candidate positive clones from each subtracted cDNA library were selected for sequencing. Cycle sequencing reactions were conducted with Thermo Sequenase cycle sequencing core kit (Amersham Life Science, USA). Each clone was sequenced on both 5’ and 3’ end. The BLASTN program was used to search for the DNA sequence homology of the isolated clones in the database of Genbank.
High yields of full-length cDNAs were generated from 1 μg total RNA of normal mucosa, adenoma and adenocarcinoma using SMART PCR cDNA Synthesis kit (Clontech, USA). 0.2 μg cDNAs were electrop hosed on a 1.0% agarose gel, transferred onto Hybond N+ Membrane (Boeheringer Mannheim, Germany) and hybridized with DIG-labeled clone probes. These probes were generated by PCR amplification of the relevant inserts, adaptor-cutting and column-purification. Hybridization and washing conditions were the same as described above in screening of cDNA libraries. DIG-labeled G3PDH probes were used as control.
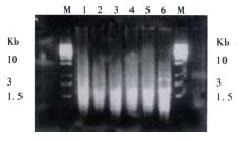
In this research, SSH was done to identify differentially expressed genes among cDNAs of normal mucosa, adenoma and adenocarcinoma tissues from the same patient. Three subtracted cDNA libraries were constructed. The subtracted A-N cDNA library used normal mucosa as tester cDNA and adenoma tissue as driver cDNA; the subtracted T-N cDNA library used adenocarcinoma tissue as tester cDNA and normal mucosa as driver cDNA; the subtracted T-A cDNA library used adenocarcinoma tissue as tester cDNA and adenoma tissue as driver cDNA. Figure 1 shows subtracted cDNA libraries after secondary PCR amplification. They usually looked like smears with or without several discrete bands. However, the patterns among them were different.
Subtraction efficiency analysis showed the effectively reduced abundance of non -differentially expressed genes. In nonsubtracted cDNA libraries, housekeeping gene G3PDH PCR products were visible after 18 cycles of amplification and became saturated after 23-28 cycles (Figure 2). However, subtracted libraries required a higher number of amplification cycles for G3PDH to be detected. Depletion of G3PDH was more significant in the subtracted A-N and T-A cDNA libraries, suggesting that these two subtractions were more complete.
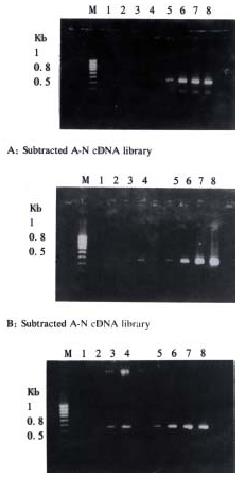
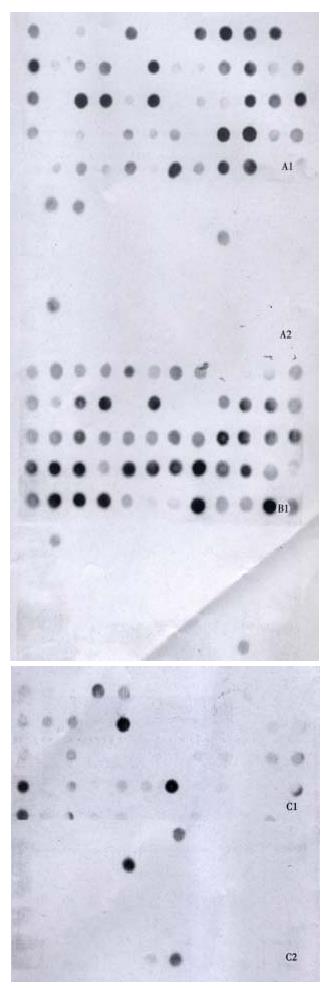
About 4 × 102, 3 × 102 and 3.5 × 102 clones were respectively identified in subtracted A-N, T-N and T-A cDNA libraries with average insert size of 0.2-1 kb (Figure 3). These clones were screened by hybridization with both forward and backward subtracted cDNA library probes. Clones corresponding to truly differentially expressed genes would mainly hybridize with the forward subtracted cDNA probe, but not, or only faintly, with the backward subtracted cDNA probe, asshown by signal comparison between two blots. By this way, more than 85% positive clones in three subtracted cDNA libraries were differentially screened.
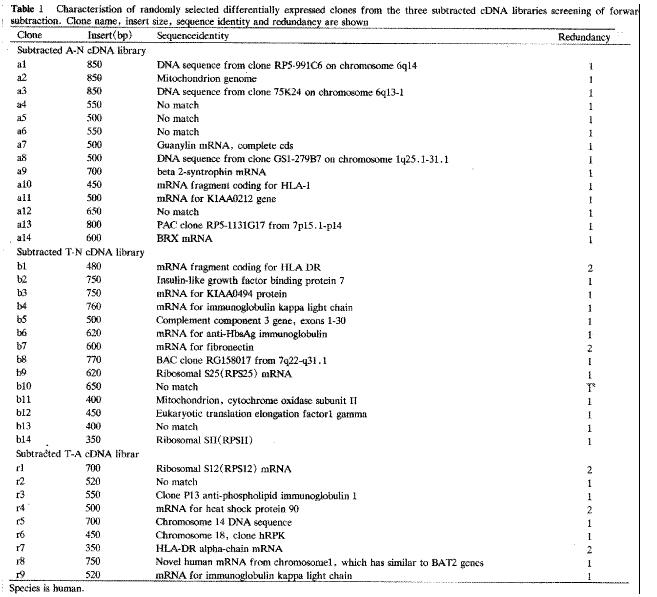
Automated sequencing proved that most clones were isolated no more than two times. BLASTN homology search revealed that some clones contained substantial sequence homologies with known gene fragments and several possibly novel genes, showed few sequence homologies with any known sequences in the GenBank database. The results of homology search are shown in Figure 4.
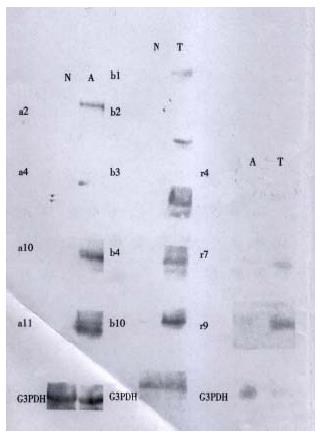
The cDNAs of selected clones were mainly expressed in tester tissue but not, or to a lesser extent, in driver tissue as Figure 5 shows. Virtual Northern Blot showed such differential expression in most clones. These results were also confirmed by RNA Dot blot (data not shown) (Figure 6).
It was learned in the last two decades that genetic changes lie at the root of all cancers. In response, the Cancer Genome Anatomy Project (CGAP) will unite the newest technologies to identify all the genes responsible for the initiation and growth of cancer. In this research, we used SSH to identify candidate genes that were differentially expressed during CRC development.
The novel technique called SSH is an ideal subtractive system that combines high subtraction efficiency with normalized representation of differentially expressed genes, and is based on suppression PCR that permits exponential amplification of genes differing in abundance, at the same time that amplification of sequences of identical abundance genes in two populations is suppressed. Therefore, differentially expressed genes of low abundance can be cloned, while differentially expressed genes of high abundance are not excessively isolated[3,4]. It was reported that SSH achieved more than 1000-fold enrichment of low-abundance genes but only required 2 μg mRNA instead of the 10-20 μg required in other METHODS. This method has been successfully used to isolate significant genes in many rese arches. Here we report the efficiency of SSH technique in identifying differentially expressed genes in normal mucosa, adenoma and adenocarcinoma from the same patient. All of the results confirmed the effectiveness and sensitivity of this method.
PCR analysis of the subtraction efficiency after subtractive hybridization was showed that the abundance of the non-differentially expressed gene G3PDH was effectively reduced. The difference of subtraction efficiency among the three subtracted libraries might be due to the diversity of starting materials for comparison. As we know, no subtraction technique is 100% effective. Although SSH greatly improved the subtraction efficiency, some false positive clones were still present in the studied material. Therefore, non-radioactive hybridization with forward and backward subtracted cDNA library probes was used to differentially screen these subtracted libraries before Virtual Northern blot confirmation[5]. About more than 85% positive clones in each subtracted cDNA library were differentially screened. von Stein OD et al[3] found about 94% positive rate in their research, so they considered confirmation of differential expression by Northern blot analysis for each clone obtained was probably unnecessary. In our research, Virtual Northern Blot and RNA Dot Blot were still performed to con firm such differential expression before further analysis of the clones.
By this way, we constructed three subtracted cDNA libraries, A-N, T-N and T-A, as mentioned above. About 3-4 × 102 clones were identified in each subtracted cDNA library through differential screening. After sequencing and BLASTN homology search, these clones could be divided into three groups: known genes preciously described as differentially expressed genes in cancer and closely related with carcinogenesis of CRC; known genes never before associated with carcinogenesis; and unknown genes that remain to be fully characterized. The identification of the first group of known genes attested to the validity of this method. In addition, no more than only two times isolation of most clones also proved the high efficiency of SSH.
HLA-DR, identified in both subtracted T-N and T-A cDNA libraries, was reported to be abundantly expressed in CRC but less so in adenoma and normal mucosa [6,7]. However, the relationship of HLA-DR expression with prognosis is still disputed[6,8]. Other molecules such as invariant chain may take part in this mechanism. So it is very important to conduct comprehensive research on the role of these molecules.
Heat shock protein (Hsps) is thought to play an important role in the cell cycles and various processes of carcinogenesis. It can be divided into two types: Hsp90α and Hsp90β[9]. The former may play a role in cell proliferation; the later is inversely related to differentiation. Both of them are present in normal tissue but are more highly expressed in neoplasm. More important, Hsp90 was recently reported to interact with HLA-DR. In this study, Hsp90 was screened in the subtracted T-A cDNA library two times, which showed its high expression in carcinoma.
IGFBP-rp1 belongs to the IGFBP family with low affinity with IGFs. It had been shown to decrease expression with disease progression in breast carcinomas[10] and in the worsening from the benign to malignant prostate epithelial cells[11,12]. Methylation with down-regulated expression was found to be associated with liver tumorigenesis[13]. So it is inferred that IGFBP-rp1 may have a suppressive effect on cancer development. Surprisingly, in this study, IGFBP-rp1 was overexpressed in CRC in contrast to their down-regulation in carcinoma, which was also shown in prostate cancer tissue[14]. It remains to be further investigated what the role of IGFBP-rp1 is and whether such different expression is individual- specific, tissue- specific or even cell-specific.
Two cDNA clones were shown to have more than 90% homology with Ig κ gene, one screened in the subtracted T-N cDNA library and the other in T-A. Hu et al[15] reported a new transforming gene Tx showed very high (99.5%) homology with the C region of Ig κ. A literature survey revealed Ig or Ig-like materials expression in carcinoma[16]. It is generally considered that Ig is mainly secreted by B-lympho cytes, so the relationship between Ig κ and carcinogenesis of CRC should be further confirmed.
Guanylin (GN) is the endogenous peptide ligand for guanylyl cyclase C (GCC) and can regulate intestinal Cl-secretion. It is robustly expressed in normal intestinal epithelium of colon but is absent in CRC[17]. So, it is possible that loss of GN activity leads to or is a result of adenocarcinoma formation. In this study, GN was expressed at higher levels in adenoma than in normal mucosa but not in CRC. It is uncertain if such increase of GN activity in adenoma means a kind of reaction to carcinogen stimulation.
Several genes shown to be dysregulated in neoplasm cells such as EF1-r[18], mitochondrion, RPS12, RPS 22/25, RPSII[19] were also identified. All of them were increased in cancer cells, as previously reported and some of them were related to the robust growth characteristics.
Besides the known genes, a very important finding in this study was the identification of several new genes that were previously undescribed in Genbank. Both Virtual Northern blot and RNA dot blot hybridization has showed such different expression. Among three subtracted cDNA libraries, A-N showed the highest rate of novel genes isolation. Although it remains to be explored the full-length sequence of these new genes by RACE or other METHODS and their function in carcinogenesis of CRC, these results really provide clues for us to understand newly proposed mechanisms of cancer formation.
In the past years, much of cancer research laid particular stress on analysis of single gene expressed differentially in tumor cells as compared with their normal counterparts, but little stress on comprehensive study of gene expression in cancer. It was interesting that in this study several genes including anti-HbsAg and IGFBP-rp1 were also differentially expressed in carcinoma. Based on available reports, anti-HbsAg gene was mainly related with the individual character, while IGFBP-rp1 was even supposed to be an anti-oncogene. So we propose that such diversity and specificity of gene expression may contribute to the heterogeneity in biological properties of cancer, which may be individual-specific, tumor-specific, process-specific or even cell-specific. Literature also indicated that there were differences of gene expression among cancers. Therefore, two novel concepts can be brought forward on carcinigenesis: one is the model formed by a total of individual gene expression in cancer, the other is the module formed by interaction among molecules[20]. Cellular functions, such as signal transduction, are carried out by modules. Connecting different levels of analysis from molecules, through modules, to models is essential for an understanding of carcinogenesis. Differentially expressed genes between tissues comprise the molecular basis for understanding model and module. So, despite the limitations of tissue from the same patient, this approach has provided interesting results for further research.
This is the first time that differentially expressed genes in colorectal adenoma and adenocarcinoma were isolated by the new technique SSH. Now, microarray detection of these differentially expressed genes as model and module to hybridize with other tumor samples is being investigated[21]. Comprehensive study of these genes will be helpful for us to create theoretical models of some systems and verify that such predictions match reality, which, in turn, will be useful for screening, detecting, classifying and therapy of CRC.
Edited by Pan BR
| 1. | Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759-767. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8087] [Cited by in RCA: 7997] [Article Influence: 228.5] [Reference Citation Analysis (1)] |
| 2. | Diatchenko L, Lau YF, Campbell AP, Chenchik A, Moqadam F, Huang B, Lukyanov S, Lukyanov K, Gurskaya N, Sverdlov ED. Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc Natl Acad Sci USA. 1996;93:6025-6030. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2262] [Cited by in RCA: 2003] [Article Influence: 69.1] [Reference Citation Analysis (0)] |
| 3. | von Stein OD, Thies WG, Hofmann M. A high throughput screening for rarely transcribed differentially expressed genes. Nucleic Acids Res. 1997;25:2598-2602. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 117] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 4. | Kuang WW, Thompson DA, Hoch RV, Weigel RJ. Differential screening and suppression subtractive hybridization identified genes differentially expressed in an estrogen receptor-positive breast carcinoma cell line. Nucleic Acids Res. 1998;26:1116-1123. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 85] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 5. | Girard JP, Baekkevold ES, Yamanaka T, Haraldsen G, Brandtzaeg P, Amalric F. Heterogeneity of endothelial cells: the specialized phenotype of human high endothelial venules characterized by suppression subtractive hybridization. Am J Pathol. 1999;155:2043-2055. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 77] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 6. | Jiang Z, Xu M, Savas L, LeClair P, Banner BF. Invariant chain expression in colon neoplasms. Virchows Arch. 1999;435:32-36. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 33] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 7. | Norheim Andersen S, Breivik J, Løvig T, Meling GI, Gaudernack G, Clausen OP, Schjölberg A, Fausa O, Langmark F, Lund E. K-ras mutations and HLA-DR expression in large bowel adenomas. Br J Cancer. 1996;74:99-108. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 28] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 8. | Teh M, Wee A, Raju GC. HLA-DR antigen expression in colonic adenomas and adenocarcinomas. Pathology. 1994;26:123-126. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 4] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 9. | Yano M, Naito Z, Tanaka S, Asano G. Expression and roles of heat shock proteins in human breast cancer. Jpn J Cancer Res. 1996;87:908-915. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 79] [Cited by in RCA: 80] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 10. | Burger AM, Zhang X, Li H, Ostrowski JL, Beatty B, Venanzoni M, Papas T, Seth A. Down-regulation of T1A12/mac25, a novel insulin-like growth factor binding protein related gene, is associated with disease progression in breast carcinomas. Oncogene. 1998;16:2459-2467. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 98] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 11. | Hwa V, Tomasini-Sprenger C, Bermejo AL, Rosenfeld RG, Plymate SR. Characterization of insulin-like growth factor-binding protein-related protein-1 in prostate cells. J Clin Endocrinol Metab. 1998;83:4355-4362. [PubMed] |
| 12. | Sprenger CC, Damon SE, Hwa V, Rosenfeld RG, Plymate SR. Insulin-like growth factor binding protein-related protein 1 (IGFBP-rP1) is a potential tumor suppressor protein for prostate cancer. Cancer Res. 1999;59:2370-2375. [PubMed] |
| 13. | Komatsu S, Okazaki Y, Tateno M, Kawai J, Konno H, Kusakabe M, Yoshiki A, Muramatsu M, Held WA, Hayashizaki Y. Methylation and downregulated expression of mac25/insulin-like growth factor binding protein-7 is associated with liver tumorigenesis in SV40T/t antigen transgenic mice, screened by restriction landmark genomic scanning for methylation (RLGS-M). Biochem Biophys Res Commun. 2000;267:109-117. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 54] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 14. | Degeorges A, Wang F, Frierson HF, Seth A, Chung LW, Sikes RA. Human prostate cancer expresses the low affinity insulin-like growth factor binding protein IGFBP-rP1. Cancer Res. 1999;59:2787-2790. [PubMed] |
| 15. | Hu WX, Cao Y, Li XY, Lee LM, Yao KT. Comparison of homology of Tx gene with Ig kappa gene and its expression in different cell lines. Acta Biochimi caet Biophysica Sinica. 1995;27:215-221. |
| 16. | Takemura K, Hirokawa K, Esaki Y, Mishima Y. Distribution of immunoglobulins and secretory component in gastric cancer of the aged. Cancer. 1990;66:2168-2173. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 17. | Cohen MB, Hawkins JA, Witte DP. Guanylin mRNA expression in human intestine and colorectal adenocarcinoma. Lab Invest. 1998;78:101-108. [PubMed] |
| 18. | Mathur S, Cleary KR, Inamdar N, Kim YH, Steck P, Frazier ML. Overexpression of elongation factor-1gamma protein in colorectal carcinoma. Cancer. 1998;82:816-821. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 19. | Zhang L, Zhou W, Velculescu VE, Kern SE, Hruban RH, Hamilton SR, Vogelstein B, Kinzler KW. Gene expression profiles in normal and cancer cells. Science. 1997;276:1268-1272. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 985] [Cited by in RCA: 957] [Article Influence: 34.2] [Reference Citation Analysis (0)] |
| 20. | Hartwell LH, Hopfield JJ, Leibler S, Murray AW. From molecular to modular cell biology. Nature. 1999;402:C47-C52. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2566] [Cited by in RCA: 2102] [Article Influence: 80.8] [Reference Citation Analysis (0)] |
| 21. | Berns A. Gene expression in diagnosis. Nature. 2000;403:491-492. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 38] [Article Influence: 1.5] [Reference Citation Analysis (0)] |