Published online Oct 15, 2000. doi: 10.3748/wjg.v6.i5.626
Revised: August 23, 2000
Accepted: August 25, 2000
Published online: October 15, 2000
- Citation: Caselmann WH, Serwe M, Lehmann T, Ludwig J, Sproat BS, Engels JW. Design, delivery and efficacy testing of therapeutic nucleic acids used to inhibit hepatitis C virus gene expression in vitro and in vivo. World J Gastroenterol 2000; 6(5): 626-629
- URL: https://www.wjgnet.com/1007-9327/full/v6/i5/626.htm
- DOI: https://dx.doi.org/10.3748/wjg.v6.i5.626
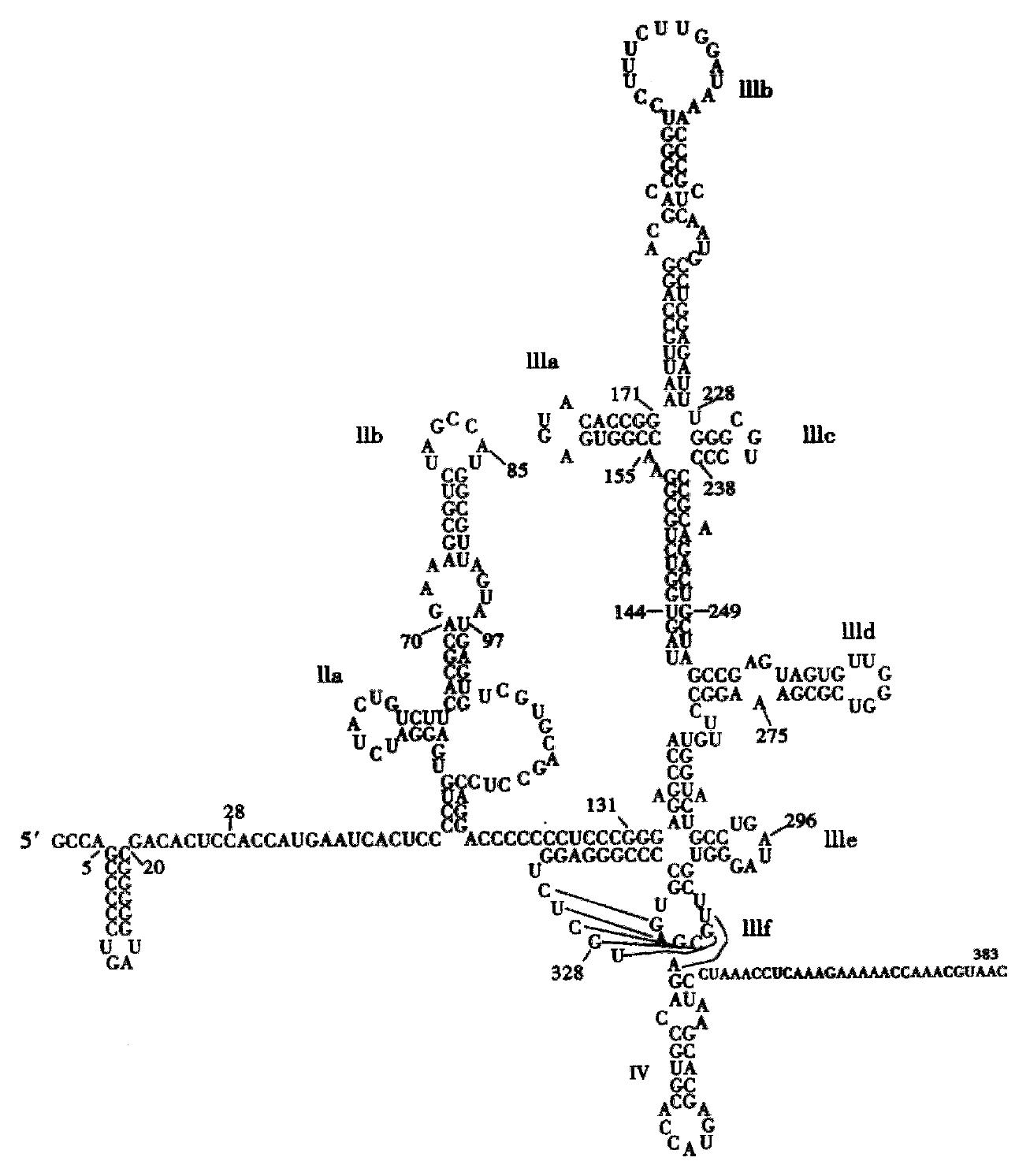
Despite major achievements in the treatment of chronic hepatitis C with the combination of interferons and the nucleoside analog ribavirin[1] the majority of patients with chronic hepatitis C virus (HCV) infection cannot be treated effectively. To improve this response rate we used antis ense technologies to inhibit HCV translation as possible additional option for experimental treatment. Antisense oligodeoxynucleotides (ODN) are short nucleic acid oligomers, which bind complementary to the single-stranded HCV-RNA with plus (+) strand polarity and can induce hybrid arrest at the ribosome as well as cellular RNase H activity, which cleaves the RNA template in DNA-RNA hybrids. The 5’-non-coding region (NCR) is highly conserved among the 6 main genotypes and exerts the function of an internal ribosomal entry site (IRES), which is important for translation[2] and possibly also for viral replication (Figure 1).
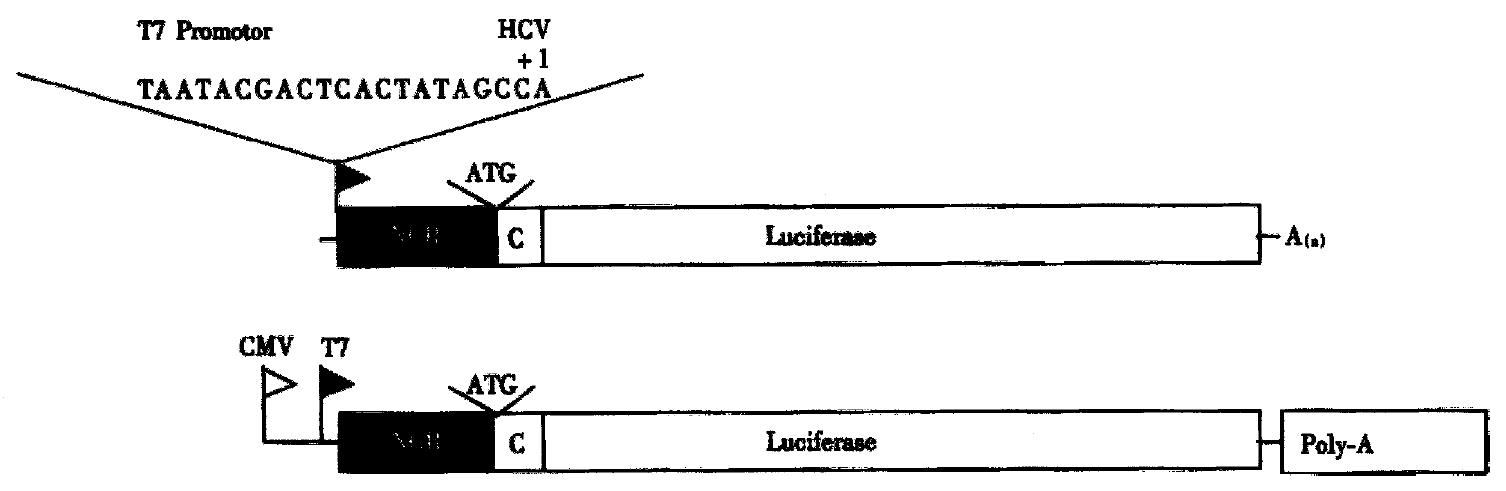
Various antisense oligodeoxynucleotides varying from 8 to 23-mers and covering stem-loop structures within the 5’-NCR and the adjacent core gene were synthesized[3]. ODN4 comprises HCV 5’-NCR nucleotides (nts.) 326-348 including the start AUG of the polyprotein coding sequence. Since easy-to-handle and generally a vailable infection or replication systems for HCV are lacking. We used model sys tems (Figure 2), in which the HCV 5’-NCR and 66 nts. Of the core gene were fused to the firefly luciferase gene for in vitro transcription/translation from the bacteriophage T7 promoter. A similar construct with the cytomegalovirus (CMV) immediate early promoter was constructed for transfection experiments of hepatoma and non-liver cell lines[4].
As compared to mismatch or sense controls completely phosphorothioate-modified ODN 4 showed the best inhibitory effect of luciferase activity in the rabbit reticulocyte lysate, which ranged in the order 96% ± 1% at about 4 μM concentration. The effectiveness of ODN 4 is possibly due to its interference with a pseudonot structure (Figure 1) in HCV RNA which is disrupted, when ODN4 binds to the target RNA. In HepG2 tissue cultures comparable specific effects ware obtained. However, fluorescence labeling of this ODN demonstrated that only a small percentage of transfected cells did take up the therapeutic ODN.
In order to improve the capability of membrane permeation by variation of their lipophilicity chemically modified ODN such as methylphosphonate and benzylphosp honate ODN 4 were compared with phosphorothioate ODN 4, which had been used in completely modified form in the previously described experiments. For the following experimental setting partially modified ODN4, which carried 6 modifications over a length of 23 nts. either in terminal localization or scattered along the molecule (Table 1). Toxicological testing revealed acceptable IC50 concentrations between 0.2 and 5.8 μM and the best therapeutic indices of 3.8 for terminally modified benzylphosphonate (tB-) ODN 4 and terminally modified phosphorothioates (tS-) ODN4. While both scattered (s) and terminal (t) modifications inhibited HCV gene expression in case of phosphorothioate (S)-ODN 4, only terminally modified methylphosphonate (tM)-ODN 4 and benzylphosphonate (tB)-ODN 4 showed specific and effective inhibition. This is possibly correlated to an induction of RNaseH activity and the steric hindrance of this enzyme by differently modified molecules[5,6]. In cell culture, tB-ODN 4 showed the best inhibition of about 96%, leaving, however, 4% of reporter gene expression unaffected.
| 5’-TGCTCATGGTGCACGGTCTACA-3’ | 4 |
| 5’-TGCTGATGCTGCACGCTCTAGGA-3’ | K |
| 5’-TCGTAGACCGTGCACATGAGCA-3’ | S |
| 5’-TGCTCATGGTGCACGGTCTACGA-3’ | 4 |
| 5’-TGCTGATGCTGCACGCTCTAGGA-3’ | K |
| 5’-TCGTAGACCGTGCACCATGAGCA-3’ | S |
Therefore, katalyticly active antisense RNAs, so-called hammerhead ribozymes (RZ), which cleave at NUH recognition sites (N: any nt., U: uridine, H: any nt. except guanosine) of a RNA template, were synthesized. Taking these recognition motifs into account, one of the RZ was directed towards similar stem-loop struc tures as the previously tested ODN. After purification of the RZ by denaturing polyacrylamide gel electro-phoresis and reverse phase chromatography their katalytic activity was tested with short 14-mer substrates under defined cleaving conditions. Best cleavage was obtained for RZ A328-R which cleaves between HCV nts. positions 330 and 331. Its Michaelis Menton constant K-m was 69.1 ± 19.3 nM. The katalyt ic constant Kcat was 4, 3 min-1 indicating multiple turnover. The katalytic effectiveness Kcat/Km was 6.4 × 107 M-1× min-1. The cleavage with longer (452 nts.) HCV-luciferase fusion RNA substrates was best effective, if a high molar excess of RZ were used. In direct comparison with antisense ODN or not katalyticly active antisense RNAs targeting exactly identical HCV-RNA sequences RZ A328-R displayed the best about 90% inhibition of luciferase activity in vitro.
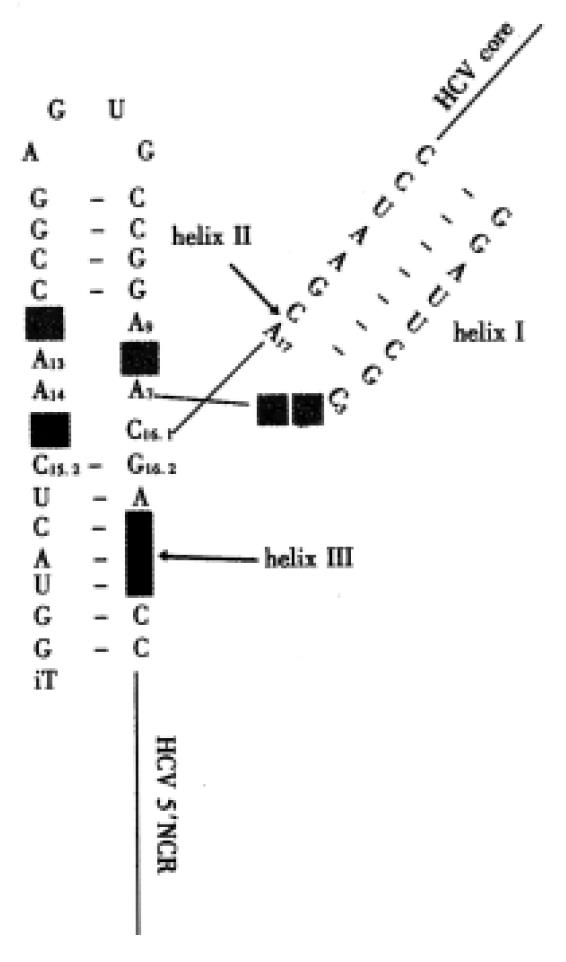
To further extend the amount of possible target regions on the HCV genome a A ≥ I exchange at nt. position 15.1 was introduced (Figure 3), which reverts the polarity of an important H-bond in the hammerhead structure and allows efficient cleavage at a non-natural NCH recognition site (N: any nt., C: cytidine, H: any nt. except guanosine). For stability reasons various 2’O-allyl or amino-ribonucleotide modifications had been introduced in these RZ[7], which cleave between HCV nts. 346 and 347 (Figure 3). A inactive cleavage antisense RZ and an unspecific RNA completely unrelated to HCV were used as controls. Regardless of prehybridisation of the RNA molecules with their target sequence, the best inhibitions of luciferase activity were obtained with the NCH recognizing RZ. The inhibition was less prono unced, when inactive cleavage antisense RZ were used. This data could be reproduced in HCV-5’-NCR/core-luciferase- recombinant CCl13 cell lines. which stably express the HCV core-luciferase fusion protein above from the CMV immediate early promoter. In vivo esting using CMV-HCV 5’-NCR/core-luciferase-transgenic mice are presently going on.
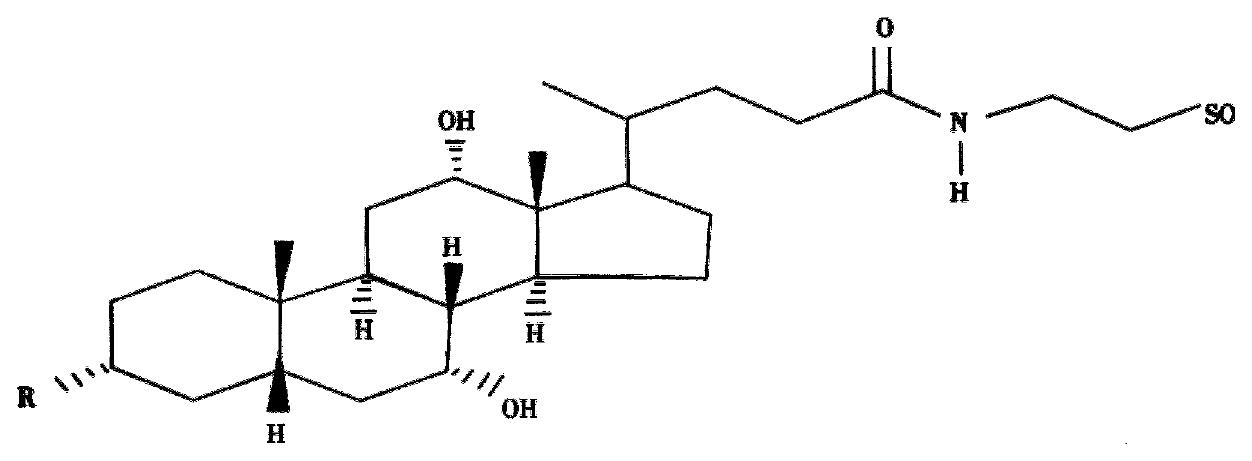
Coupling of effective antisense molecules to biomolecules such as bile acids which are specificly transported into the hepatocyte by bile acid transporters are in progress (Figure 4). The size of the antisense ODN coupled to cholic acid or taurocholic acid has been optimized with respect to efficient and specific inhibition of HCV-luciferase translation in vitro. Experiments with cell lines overexpressing the organic anion transporting polypeptide (OATP) and the sodium taurocholate cotransporting polypeptide (NTCP) are in progress.
Antisense oligodeoxynucleotides, RNAs and ribozymes can effectively inhibit HCV translation in model systems in vitro and in vivo. Chemical modifications such as phosphorothioate or benzylph-osphonate modifications of oligonucl eotides as well as O-allyl or amino-ribonucleotide modifications of ribozymes guarantee sufficient stability towards nuclease degradation. The specific exchange of single ribonucleotides in synthetic ribozymes permits to extend the ribozyme cleavage specificity to non-natural NCH motifs. If problems of hepatocyte-directed delivery and sufficient intracellular effector concentration can be solved, therapeutic nucleic acids may turn out to be a save and powerful treatment option in the multimodal therapy concept of chronic hepatitis C.
Edited by Ma JY
| 1. | Wedemeyer H, Caselmann WH, Manns MP. Combination therapy of chronic hepatitis C: an important step but not the final goal! J Hepatol. 1998;29:1010-1014. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 15] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 2. | Varaklioti A, Georgopoulou U, Kakkanas A, Psaridi L, Serwe M, Caselmann WH, Mavromara P. Mutational analysis of two unstructured domains of the 5' untranslated region of HCV RNA. Biochem Biophys Res Commun. 1998;253:678-685. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 13] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 3. | Alt M, Renz R, Hofschneider PH, Caselmann WH. Core specific antisense phosphorothioate oligodeoxynucleotides as potent and specific inhibitors of hepatitis C viral translation. Arch Virol. 1997;142:589-599. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 18] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 4. | Alt M, Renz R, Hofschneider PH, Paumgartner G, Caselmann WH. Specific inhibition of hepatitis C viral gene expression by antisense phosphorothioate oligodeoxynucleotides. Hepatology. 1995;22:707-717. [PubMed] |
| 5. | Alt M, Eisenhardt S, Serwe M, Renz R, Engels JW, Caselmann WH. Comparative inhibitory potential of differently modified antisense oligodeoxynucleotides on hepatitis C virus translation. Eur J Clin Invest. 1999;29:868-876. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 21] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 6. | Caselmann WH, Eisenhardt S, Alt M. Synthetic antisense oligodeoxynucleotides as potential drugs against hepatitis C. Intervirology. 1997;40:394-399. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 20] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 7. | Ludwig J, Blaschke M, Sproat BS. Extending the cleavage rules for the hammerhead ribozyme: mutating adenosine15.1 to inosine15.1 changes the cleavage site specificity from N16.2U16.1H17 to N16.2C16.1H17. Nucleic Acids Res. 1998;26:2279-2285. [PubMed] |