Published online Jun 15, 2000. doi: 10.3748/wjg.v6.i3.356
Revised: January 8, 2000
Accepted: January 15, 2000
Published online: June 15, 2000
AIM: To detect the expression of caspase 3 gene in primary huma n hepatocellular carcinoma (HCC) and investigate its relationship to p21WAF1 gene expression and HCC apoptosis.
METHODS: In situ hybridization was employed to determine caspase 3 and p21WAF1 expression in HCC. In situ end-labeling was used to detect hepatocytic apoptosis in HCC.
RESULTS: Twenty-one of 39 (53.8%) cases of HCC were found to express caspase 3 transcripts, while 46.2% of HCC failed to express caspase 3. Non-cancerous adjacent liver tissues showed more positive caspase 3 (87.5%, 7/8) as compared with HCC (P < 0.05). The expression of caspase 3 is correlated with HCC differentiation, 72.2% (13/18) of moderately to highly differentiated HCC showed caspase 3 transcripts positive, while only 38.1% of poorly differentiated HCC harbored caspase 3 transcripts (P < 0.05). No relationship was found between caspase 3 expression and tumor size or grade or metastasis, although 62.5% (5/8) of HCC with metastasis were caspase 3 positive and a little higher than that with no metastasis (51.6%, P > 0.05). Expression of caspase 3 alone did not affect the apoptosis index (AI) of HCC. The AI was 7.12‰ in caspase 3-positive tumors (n = 21), while in caspase 3-negative cases (n = 18) 6.59‰ (P > 0.05). Expression of caspase 3 clearly segregated with p21WAF1 positive tumors as compared with p21WAF1 negative cases (16 of 23, 69.6% vs 5 of 16, 31.3%) with statistical significance(P = 0.017). In the cases with positive caspase 3 and negative p21WAF1, the AI was found slightly higher, but with no statistical significance, than that with expression of p21WAF1 and caspase 3 (7.21‰vs 6.98‰, P > 0.05).
CONCLUSION: Loss of caspase 3 expression may contribute to HCC carcinogenesis, although the expression of caspase 3 does not correlate well with cell apoptosis in HCC. p21WAF1 may be merely one of the inhibitors which can reduce caspase 3 mediated cell apoptosis in HCCs.
- Citation: Sun BH, Zhang J, Wang BJ, Zhao XP, Wang YK, Yu ZQ, Hao LJ, Yang DL. Analysis of in vivo patterns of caspase 3 gene expression in primary hepatocellular carcinoma and its relationship to p21WAF1 expression and hepatic apoptosis. World J Gastroenterol 2000; 6(3): 356-360
- URL: https://www.wjgnet.com/1007-9327/full/v6/i3/356.htm
- DOI: https://dx.doi.org/10.3748/wjg.v6.i3.356
Hepatocellular carcinoma(HCC), represent 80%-90% of the primary liver cancer, is one of the leading causes of cancer morbidity and mortality on a global scale. More than 80% of liver cancer cases occurr in the developing world, especially in China, where HCC is the second cause of cancer death and responsible for 130000 deaths every year[1]. Despite dramatic advances in basic and clinical research in the past decades, the exact molecular mechanism for hepatocarcinogenesis is unclear. Gene expression changes have been demonstrated in accordance with cell growth, differentiation and carcinogenesis[2]. Tumor formation can result from a decrease in cell death, as well a s an increase in cell proliferation. In addition to altered expression of cell cycle-related gene, dysregulation of apoptosis (programmed cell death) is thought to contribute to cancer by aberrantly extending cell viability and favoring the accumulation of transforming mutations[3].
Caspase is a large family which contains at least 14 members. It has been shown that caspase play an important role in regulating cancer cell death both induced by activated lymphocytes through Fas/FasL pathway and by chemotherapy agents[4]. Caspase 3 or cpp32 is the key member of effector caspases. Caspase 3 had overexpression in B-cell chronic lymphocytic leukemia (B-CLL)[5], acute myelogenous leukemia (AML)[6], follicular small cleaved cell non-Hodgkin's B-cell lymphomas[7,8], human breast cancer cell lines and primary breast tumors[9]and neurob lastomas[10]. Caspase 3 is a potent protease which can cleave a large scale of substrate, including cell cycle related genes such as p21WAF1[4]. p21WAF1 is a cyclin-dependent kinase inhibitor and plays an important role in DNA damage induced growth arrest. p21WAF1 overexpression can cause G1 cell cycle arrest and further interrupt the apoptotic process at a point upstream from caspase 3 activation[11]. In this study, we investigated the expression of caspase 3 in primary human HCC and its potential impact on tumor cell apoptosis and its relationship to p21WAF1 expression.
The surgically resected specimens employed in this study were obtained from consecutive patients with primary HCC who had undergone potentially curative tumor resection at the Department of General and Hepato-Biliary Surgery, Tongji Hospi tal during 1996-1997. A cohort of 39 cases was involved in this study. All cases were selected on the basis of availability of frozen material for study and on the absence of extensive chemotherapy-induced tumor necrosis. Materials were co mposed of 3 cases of grade I, 18 cases of grade II, 11 cases of grade III, the remaining 7 cases were grade IV according to TNM system (1987). The tumor lesions analyzed here included 21 poor, 9 moderate and 9 well differentiations. There were 34 males and 5 females, and the age ranged from 24 to 71 years with an average of 46.1 (SD, 12.5). Eight cases of non -cancerous adjacent liver tissues were also included in the study. Routinely processed 40 g/L paraformaldehyde-fixed, paraffin-embedded blocks containing principal tumor were selected. Serial sections of 5 μm were prepared from the cut surface of blocks at the maximum cross-section of the tumor.
The plasmid pET21b-cpp32 containing caspase 3 (cpp32) cDNA probe was kindly provided by Dr. JC. Reed(La Jolla, USA). After digestion with Xho I and Nde I, the fragment was separated by electrophoresis through an agar ose gel and recovered by QIA quick gel extraction kit (QIAGEN) using a micro-centrifuge according to the manufacturer's protocol. The p21WAF1 cDNA probe was kindly provided by Dr. SJ Elledge (Houston, USA). Preparation of p21WAF1 probe was described previously[12]. The probes were labeled and detected using a Dig DNA labeling and detection kit (Boehringer Mannheim Biochemica, Germany). Briefly, 40 g/L paraformaldehyde-fixed paraffin embedded samples were cut at 5 μm and adhered to APES-treated slides. After deparaffinized and rehydrated through a graded series of ethanol, the sections were immersed in a 0.01 mol/L DEPC-treated PBS (pH7.4) two times each for 5 min, and then, in PBS containing 100 mmol/L glycine and PBS containing 3 mL/L Triton X-100 for 5 min in turns. Sections were permeabilized for 30 min at 37 °C with TE buffer (100 mmol/L Tris-HCl, 50 mmol/L EDTA, pH8.0) containing 10 mg/L RNase-free proteinase K and washed with DEPC-treated PBS, then incubated at 42 °C for 2 h with pre-hybridization buffer. Hybridization solution (400 mL/L deionized formamide, 500 g/L dextra sulfate, 1 × Dehardt's reagent, 4 × SSC, 10 mmol/L DTT, 1 g/L yeast tRNA, 1 g/L denatured salmon sperm DNA) containing 2 mg/L probe overlay each section after deprive prehybr idization buffer from slides and hybridize at 42 °C for 36 h in a humid chamber. The sections were washed in a shaking water bath at 37 °C in 2 × SSC, 1 × SSC, 0.1 × SSC for 15 min each, then washed with buffer I (100 mmol/L Tris-HCl, pH7.5, 150 mmol/L NaCl) for 20 min and with blocking solution (buffer I containing 20 mL/L normal sheep serum) for 30 min, and added sheep anti-Dig-alkaline phosphates (diluted at 1:800 in buffer I) and incubated for another 1 h before development by NBT at 37 °C for 3 h in the dark. Hybridization buffer containing no probe was used for negativ e control for each staining. Scoring method for caspase 3 and p21WAF1 expression was described by Kawasaki[13]. Positive tumor cells were quantified by two independent observers, and the average percentage of positive tumor cells was determined in at least 5 areas at × 400 and assigned to one of five categories: (a) 0, < 1%; (b)1, 1%-25%; (c)2, 25%-50%; (d) 3, 50%-75% and (e) 4, > 75%. The ISH staining intensity was scored as (a) weak 1+; (b) moderate, 2+; and intense, 3+. For tumors showing heterogeneous staining, the predominant pattern was taken into account for scoring. The percentage of positive tumor cells and staining intensity were multiplied to produce a weighted score for each case. Cases with weighted scores < 1 were defined as negative, otherwise were defined as positive.
Tumor cell apoptosis was identified by DNA fragmentation detection kit (QIA33-kit, Calbiochem). Briefly, deparaffinized and rehydrated sections were permeated with proteinase K (20 mg/L in 10 mmol/L Tris, pH8.0) for 20 min at room temperature and washed with 1 × TBS (20 mmol/L Tris pH7.6, 140 mmol/L NaCl). After endogenous peroxidases were inactivated by using 30 mL/L hydrogen peroxide for 5 min and washed with 1 × TBS, equilibration buffer was added to each section and incubated at room temperature for 20 min. Terminal deoxynucleotidyl transferase (TDT) enzyme in TDT labeling reaction mix at a 1:20 dilution was pipetted onto the sections, followed by 1.5 h incubation at 37 °C. After the reaction was terminated by immersing sections into stop solution and washed with blocking buffer for 10 min at room temperature, the anti-digoxingenin-peroxidase was added to t he sections. DAB solution was used for color development. Sections were counterstained by methyl green. A positive control generated covering specimen with DNase I (1 mg/L) for the first procedure. Specific positive tissue sections were used for negative control by substituting distilled water for the TDT in the reaction mixture. The AI was expressed as the ratio of positively stained tumor cells and bodies to all tumor cells, given a percentage for each case. A minimum of 1000 cells was counted under a 400-fold magnification. Positively staining tumor cells with morphological characteristics of apoptosis were identified using standard criteria, including chromatin condensation, nuclear disintegration and formation of crescentic caps of condensed chromatin at the nuclear periphery.
Variables associated with caspase 3 expression as well as the relationship between caspase 3 and p21WAF1 were analyzed by χ2 test. Differences in the tumor cell AI for groups dichotomized according to caspase 3 expression were checked by independent t test.
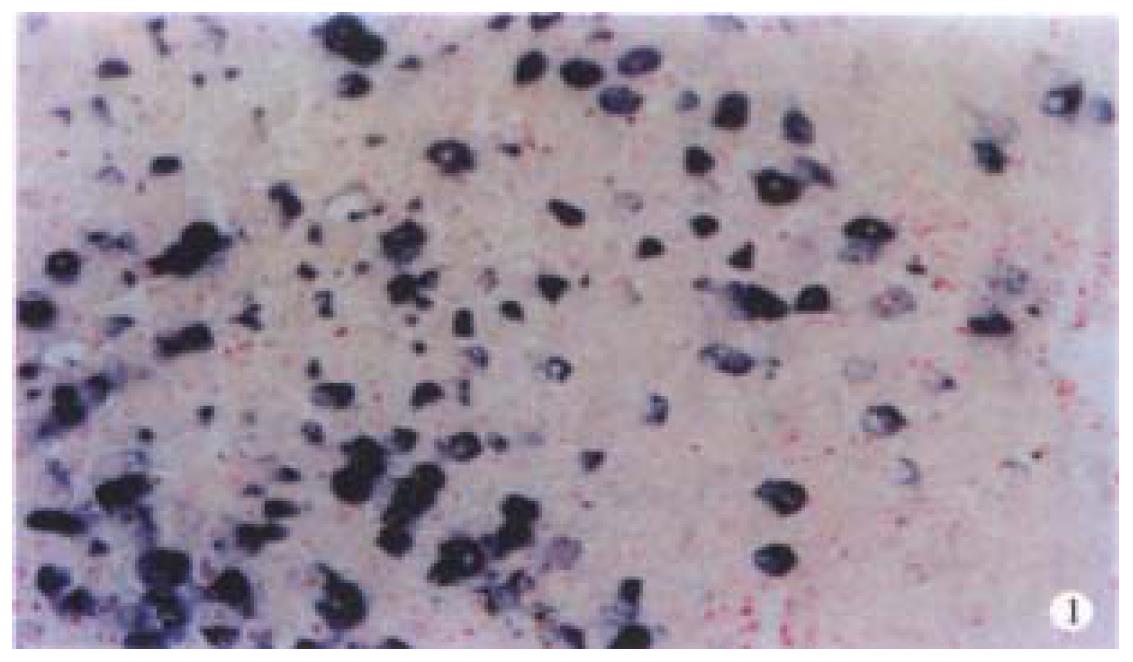
By ISH staining, caspase 3 transcripts was detected predominantly in cytoplasm (Figure 1). Consistent with the presence of caspase 3 protein in human biopsy liver samples, expression of caspase 3 in non-cancerous adjacent liver tissue was also observed in 87.5% (7/8) of cases. The intensity of caspase 3 staining was heterogeneous within a case detected. The tumor cells positively stained by ISH range from 10% to 90%, depending on the cases examine d. After multiplying the weighted caspase 3 score, 21 cases of HCC in the present study were defined as positive (53.8%), with weighted caspase 3 score from 1 to 12.
A clinicopathological analysis of caspase 3 positive cases is shown in Table 1. No statistical significance was observed in the prognostic parameters, including tumor size, metastasis, TNM grade, analyzed in the present study except for differentiation. The expression of caspase 3 is correlated with HCC differ entiation. It was found that as high as 72.2% (13/18) of moderately to highly differentiated HCC showed caspase 3 transcripts positive, while only 38.1% of poorly differentiated HCC harbored caspase 3 transcripts (P < 0.05).
| No. | Caspase 3 expression (%) | P | |
| Samples | |||
| Non-cancerous adjacent liver | 8 | 7 (87.5) | < 0.05 |
| HCC | 39 | 21 (53.8) | |
| Age (year) | |||
| < 60 | 28 | 15 (53.6) | NS |
| > 60 | 11 | 6 (54.5) | |
| Sex | |||
| Male | 34 | 18 (52.9) | NS |
| Female | 5 | 3 (60.0) | |
| Tumor size (cm) | |||
| > 5.0 | 27 | 15 (55.6) | NS |
| < 5.0 | 12 | 6 (50.0) | |
| Differentiation | |||
| Well-moderate | 18 | 13 (72.2) | < 0.05 |
| Poor | 21 | 8 (38.1) | |
| TNM grade | |||
| I-II | 21 | 12 (57.1) | NS |
| III-IV | 18 | 9 (50.0) | |
| Metastasis | |||
| Negative | 31 | 16 (51.6) | NS |
| Positive | 8 | 5 (62.5) | |
| p21WAF1 | |||
| Positive | 23 | 16 (69.6) | < 0.05 |
| Negative | 16 | 5 (31.3) |
Twenty-three cases were detected expression p21WAF1 transcripts. Positive signal was predominantly located in cytoplasm with a heterogeneous distribution of positive tumor cells. The significance of p21WAF1 gene expression was discussed in our previous study[12]. Expression of caspase 3 clearly segregated with p21WAF1 positive tumors as compared with p21WAF1 negative cases (16 of 23, 69.6% vs 5 of 16, 31.3%) with statistical significance (P < 0.05).
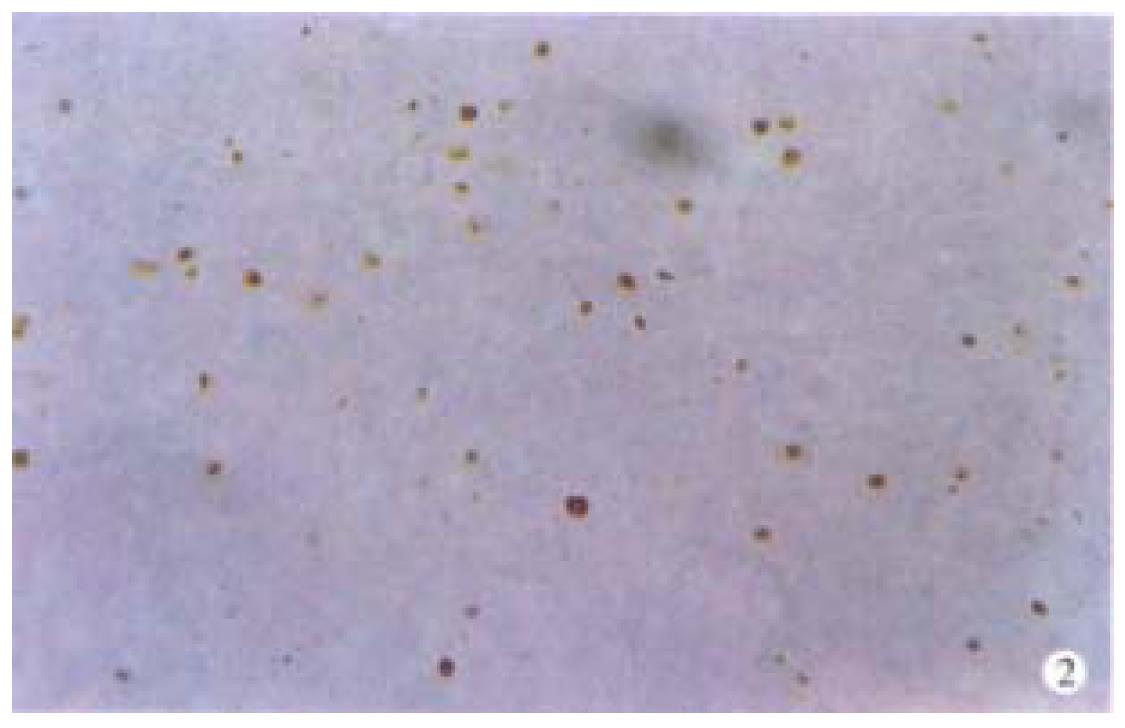
Apoptotic cells and apoptotic bodies were found in all cases of HCCs examined by in situ end-labeling (Figure 2). The mean AI of all tumors cases was 6.82‰ (s, 3.36‰; range 0.87‰-17.3‰). No significant association was observed between AI and tumor stage. The mean AI for caspase 3-positive tumors (n = 21) was 7.12‰ (s, 3.75‰), while in caspase 3-negative cases (n = 18) the AI was 6.59‰ (s, 2.98‰), no statistical significance was found between the two groups (P > 0.05). In the cases with co-expression of p21WAF1 and caspase 3, the AI was found lower, but with no significance, than that of cases with positive caspase 3 and negative p21WAF1 (6.98‰vs 7.21‰).
In this study, we have shown that caspase 3 was expressed in most of HCC cases. In our opinion, we are the first to describe the cpp32 expression in human primary HCCs. Like the result of human biopsy and autopsy liver materials[14], the non-cancerous adjacent liver tissue showed strong caspase 3 expression. A s high as 46.2% of human primary HCCs failed to show caspase 3 expression. In t he 21 cases which express caspase 3, the expression showed heterogeneous pattern with a weighted score from 1 to 12. Because caspase 3 is the effector caspase in the apoptosis pathways, we think that loss of caspase 3 expression may play an important role in HCC carcinogenesis. Caspase 3 expression had no relationsh ip with clinicopathological features except for tumor differentiation (Table 1). The result suggest that the expression of caspase 3 is correlated with tumor differentiation in HCC. It was reported that 33.1% cases of gastric carcinoma were caspase 3 positive[7]. Two of 3 breast carcinoma tissues expressed caspase 3, the immunointensity was generally higher in invasive cancers[8]. It raised the possibility that expression of casapse 3 in tumors showed tissue specificity. It was observed in this study that the apoptosis index (AI) was not associated with the expression of caspase 3 in HCC (7.12‰ in cpp32-positive cases vs 6.59‰ in cpp32-negative group). In the non-cancerous adjacent liver tissues more than 50% of the cells showed positive caspase 3, the AI was not increased as compared with HCC, no matter caspase 3 was positive or negative. This suggests that other factor(s) may exist in regulating normal cell apoptosis.
p21WAF1 was first reported as a universal inhibitor of cyclin-dependent kinase, which is required for G1 to S transition[15]. Previous studies demonstrated that p21WAF1 can interrupt the apoptotic process at a point upstream from caspase-3 activation, because serum starvation, which also synchronized cells in G1 but did not induce p21WAF1, did not protect cells from apoptosis, while restoring a late G1 checkpoint by inducing p21WAF1 expression can protect cells from DNA damage induced apoptosis[16]. p21WAF1 can bind procaspase-3 but not activate caspase 3. On the other hand, activa ted caspase 3 can cleave p21WAF1. P21 cleavage by the activated cpp32 specifically abolished its interaction with PCNA and may interfere with normal PCNA-dependent repair[15]. The presence of p21WAF1 in human HCCs was reported previously in our paper[12]. In this context, the expression of p21WAF1 was also determined together with caspase 3 in a n attempt to find whether there is relationship between them. We found that expression of caspase 3 was strongly associated with p21WAF1 in 16 cases of HCC. There was no significant difference in AI between p21WAF1(+)/cpp32(+) and p21WAF1(-)/cpp32(+) (6.98‰vs 7.21‰). The results indicated that p21WAF1 may be merely one of the inhibitors which can reduce caspase 3 mediated cell apoptosis in HCCs. In fact, some other caspase 3 inhibitors, for example survivin, were reported to have overexpression in human tumors, including gastric and colorectal cancer [17,18]. Survivin is believed to bind activated caspase 3 and further inhibit cell apoptosis. It was reported that XIAP can interrupt caspase 3 mediated apoptosis via the same way as p21WAF1. So, further investig ation on other caspase 3 regulating protein is needed to find the regulation mechanism of caspase 3 mediated apoptosis in human HCCs.
Dr. Bao-Hua Sun, graduated from the Department of Pathology of Tongji Medical University in 1997, Postdoctor, currently engaged in apoptosis research of hepatocellular carcinoma, having 17 papers published.
Edited by Ma JY
proofread by Sun SM
| 1. | Tang ZY, Yu YQ, Zhou XD, Ma ZC, Wu ZQ. Progress and prospects in hepatocellular carcinoma surgery. Ann Chir. 1998;52:558-563. [PubMed] |
| 2. | Kokura K, Nakadai T, Kishimoto T, Makino Y, Muramatsu M, Tamura T. Gene expression in hepatomas. J Gastroenterol Hepatol. 1998;13 Suppl:S132-S141. [PubMed] |
| 3. | Wyllie AH. Apoptosis and carcinogenesis. Eur J Cell Biol. 1997;73:189-197. [PubMed] |
| 4. | Kidd VJ. Proteolytic activities that mediate apoptosis. Annu Rev Physiol. 1998;60:533-573. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 202] [Cited by in RCA: 205] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 5. | Kitada S, Andersen J, Akar S, Zapata JM, Takayama S, Krajewski S, Wang HG, Zhang X, Bullrich F, Croce CM. Expression of apoptosis-regulating proteins in chronic lymphocytic leukemia: correlations with In vitro and In vivo chemoresponses. Blood. 1998;91:3379-3389. [PubMed] |
| 6. | Estrov Z, Thall PF, Talpaz M, Estey EH, Kantarjian HM, Andreeff M, Harris D, Van Q, Walterscheid M, Kornblau SM. Caspase 2 and caspase 3 protein levels as predictors of survival in acute myelogenous leukemia. Blood. 1998;92:3090-3097. [PubMed] |
| 7. | Krajewski S, Gascoyne RD, Zapata JM, Krajewska M, Kitada S, Chhanabhai M, Horsman D, Berean K, Piro LD, Fugier-Vivier I. Immunolocalization of the ICE/Ced-3-family protease, CPP32 (Caspase-3), in non-Hodgkin's lymphomas, chronic lymphocytic leukemias, and reactive lymph nodes. Blood. 1997;89:3817-3825. [PubMed] |
| 8. | Chhanabhai M, Krajewski S, Krajewska M, Wang HG, Reed JC, Gascoyne RD. Immunohistochemical analysis of interleukin-1beta-converting enzyme/Ced-3 family protease, CPP32/Yama/Caspase-3, in Hodgkin's disease. Blood. 1997;90:2451-2455. [PubMed] |
| 9. | Zapata JM, Krajewska M, Krajewski S, Huang RP, Takayama S, Wang HG, Adamson E, Reed JC. Expression of multiple apoptosis-regulatory genes in human breast cancer cell lines and primary tumors. Breast Cancer Res Treat. 1998;47:129-140. [PubMed] |
| 10. | Nakagawara A, Nakamura Y, Ikeda H, Hiwasa T, Kuida K, Su MS, Zhao H, Cnaan A, Sakiyama S. High levels of expression and nuclear localization of interleukin-1 beta converting enzyme (ICE) and CPP32 in favorable human neuroblastomas. Cancer Res. 1997;57:4578-4584. [PubMed] |
| 11. | Suzuki A, Tsutomi Y, Akahane K, Araki T, Miura M. Resistance to Fas-mediated apoptosis: activation of caspase 3 is regulated by cell cycle regulator p21WAF1 and IAP gene family ILP. Oncogene. 1998;17:931-939. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 265] [Cited by in RCA: 268] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 12. | Sun B, Wu Z, Ruan Y, Yang M, Liu B. p21WAF1/Cip1 gene expression in primary human hepatocellular carcinoma and its relationship with p53 gene mutation. J Tongji Med Univ. 1999;19:1-5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 13. | Kawasaki H, Altieri DC, Lu CD, Toyoda M, Tenjo T, Tanigawa N. Inhibition of apoptosis by survivin predicts shorter survival rates in colorectal cancer. Cancer Res. 1998;58:5071-5074. [PubMed] |
| 14. | Krajewska M, Wang HG, Krajewski S, Zapata JM, Shabaik A, Gascoyne R, Reed JC. Immunohistochemical analysis of in vivo patterns of expression of CPP32 (Caspase-3), a cell death protease. Cancer Res. 1997;57:1605-1613. [PubMed] |
| 15. | Li R, Waga S, Hannon GJ, Beach D, Stillman B. Differential effects by the p21 CDK inhibitor on PCNA-dependent DNA replication and repair. Nature. 1994;371:534-537. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 463] [Cited by in RCA: 478] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 16. | Bissonnette N, Hunting DJ. p21-induced cycle arrest in G1 protects cells from apoptosis induced by UV-irradiation or RNA polymerase II blockage. Oncogene. 1998;16:3461-3469. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 108] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 17. | Lu CD, Altieri DC, Tanigawa N. Expression of a novel antiapoptosis gene, survivin, correlated with tumor cell apoptosis and p53 accumulation in gastric carcinomas. Cancer Res. 1998;58:1808-1812. [PubMed] |
| 18. | LaCasse EC, Baird S, Korneluk RG, MacKenzie AE. The inhibitors of apoptosis (IAPs) and their emerging role in cancer. Oncogene. 1998;17:3247-3259. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 728] [Cited by in RCA: 711] [Article Influence: 26.3] [Reference Citation Analysis (0)] |