Published online Jun 15, 2000. doi: 10.3748/wjg.v6.i3.344
Revised: April 13, 2000
Accepted: April 28, 2000
Published online: June 15, 2000
AIM: To investigate the application of confocal laser scanning microscopy (CLSM) in tumor pathology and three-dimensional (3-D) reconstruction by CLSM in pathologic specimens of hepatocellular carcinoma (HCC).
METHODS: The 30 μm thick sections were cut from the paraffin-embedded tissues of HCC, hyperplasia and normal liver, stained with DNA fluorescent probe YOYO-1 iodide and examined by CLSM to collect optical sections of nuclei and 3-D images reconstructed.
RESULTS: HCC displayed chaotic arrangement of carcinoma cell nuclei, marked pleomorphism, indented and irregular nuclear surface, and irregular and coarse chromatin texture.
CONCLUSION: The serial optical tomograms of CLSM can be used to create 3-D reconstruction of cancer cell nuclei. Such 3-D impressions might be helpful or even essential in making an accurate diagnosis.
- Citation: Zhang WH, Zhu SN, Lu SL, Huang YL, Zhao P. Three-dimensional image of hepatocellular carcinoma under confocal laser scanning microscope. World J Gastroenterol 2000; 6(3): 344-347
- URL: https://www.wjgnet.com/1007-9327/full/v6/i3/344.htm
- DOI: https://dx.doi.org/10.3748/wjg.v6.i3.344
Under conventional light microscope, histopath-ologists often use plane image to evaluate the three-dimensional (3-D) cellular characteristics. Three-D configuration may be reconstructed by using serial mechanical sectioning, but it s axis definition is not good, image is blur and tiny structure can't be sh own clearly, and the specimen might be damaged. Using the serial optical tomograms and 3-D reconstruction function, confocal laser scanning microscopy (CLSM) can provide a much better quality 3-D image than conventional light microscope, and lead the observer into a brand-new 3-D world. Although CLSM has been used extensively in cell biology[1], few applications were reported in routine clinical pathology such as three-dimensional DNA image cytometry by CLSM in thick tissue blocks of prostatic lesions and 3-D reconstruction by CLSM in routine pathologic specimens of benign and malignant lesions of human breast[2-4]. In this study, 3-D reconstruction was performed on routine formalin-fixed, paraffin-embedded tissues of normal, and hyperplastic tissues of liver and hepatocellular carcinoma by using computer- assisted CLSM together with 3-D reconstruction. The goals of our study were to present 3-D morphologic characteristics of benign and malignant specimens of the liver and to attempt to demonst rate the usefulness of CLSM in routinely obtained surgical pathologic tissues.
Two cases of normal liver tissue were selected from autopsy specimens collected in the Department of Pathology of Shanghai Medical University, six cases of hepatocellular carcinoma including adjacent liver tissue were routine clinical specimens collected in 1996 from the Liver Cancer Institute of Zhongshan Hospital affiliated to Shanghai Medical University.
All tissues were fixed in 10% formalin, embedded in paraffin, serial sect ions were cut at 5 μm and 30 μm. The 5 μm slices were stained with hematoxylin and eosin for conventional light microscopic observation. The 30 μm slices were stained with DNA fluorescent probe, YOYO-1 iodide (Molecular Probes, Eugene, Ore., USA). The sections were deparaffinized with xylene (10 min × 2) and dehydrated with 100%, 95%, and 70% ethanol (5 min × 2) and rinsed in distilled water for 2 min × 5. The specimens were then fixed with 10% neutral buffered formalin for 30 min and washed with tap water. After rinsing with distilled water and 0.01 M phosphate buffer 5 min × 2, nuclear RNA was removed by incubating the sections for 30 min at 37 °C in 200 μL of ribonuclease A (RNAase; Sigma, USA) at a concentration of 160 g/L in PBS. DNA was next hydrolyzed with 2N HCl for 25 min at 27.5 °C. After rinsing with distilled water for 2 min × 5, the sections were covered with 200 μL of YOYO-1 iodide diluted into 1:2000 with PBS. The PBS was diluted 1:5 with distilled water to reduce the salt concentration. To this 200 μL working solution of YOYO-1 iodide, 20 μL of 0.1N HCl was added and the final solution was stored in the dark at 4 °C for use. Homogeneous fluorescence intensity of nuclei at different depths of the confocal slices was obtained by agitating the YOYO-1 iodide for 1 h in the dark. Afterwards, the sections were rinsed with distilled water, covered with buffered glycerol, and the glass cover slip were scaled with finger nail polish. Sections were stored at 4 °C in the dark until CLSM examinati on.
A Leica TCS-NT confocal laser scanning microscope equipped with epifluoresce nce optics and an appropriate combination of filters to visualize and digitize t he images of the different specimens. An argon laser with an excitation wavelength of 488 nm was used to activate the green fluorescence of the YOYO-1 iodide-stained nuclear DNA (maximal absorption 491 nm and emission 509 nm). A 16 × objective (numerical aperture of 1.30) was used to observe the specificity of t he staining, and a 100 × water objective (numerical aperture of 1.30) was use d to study the details of chromatin pattern. Optical sections were collected throughout the entire stained thickness of the paraffin sections with a Z-step interval of 0.3 μm or 0.6 μm. The 3-D image processing was performed on a Leica computer with the original 3-D interactive visualization software. The collected confocal optical sections and the 3-D reconstructed images were printed with a Panasonic color vide o dye-sublimation copy processor.
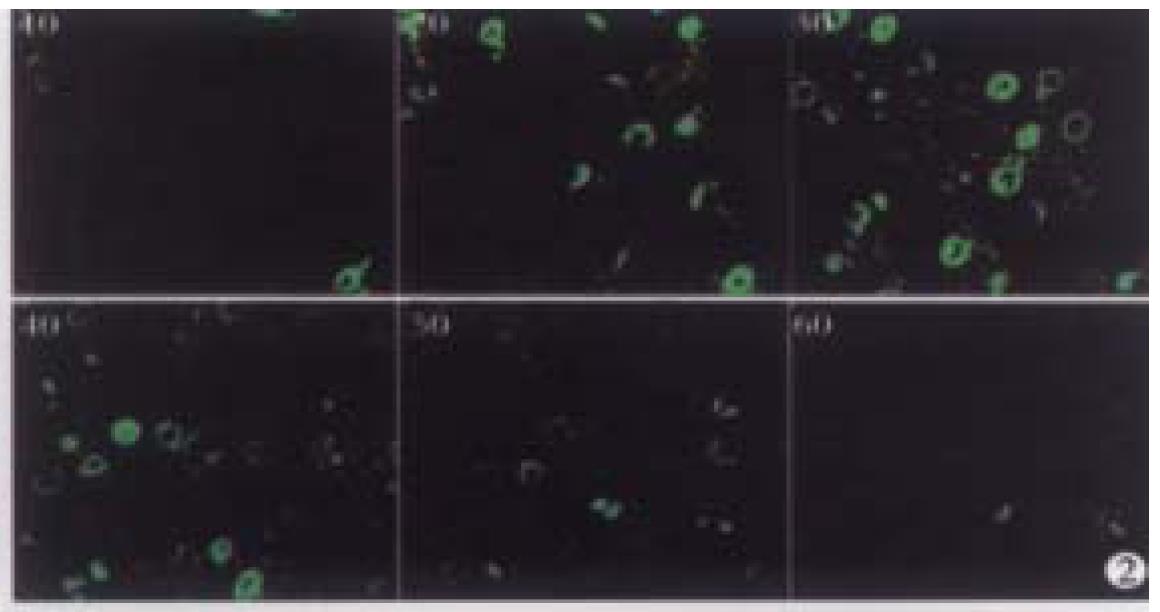
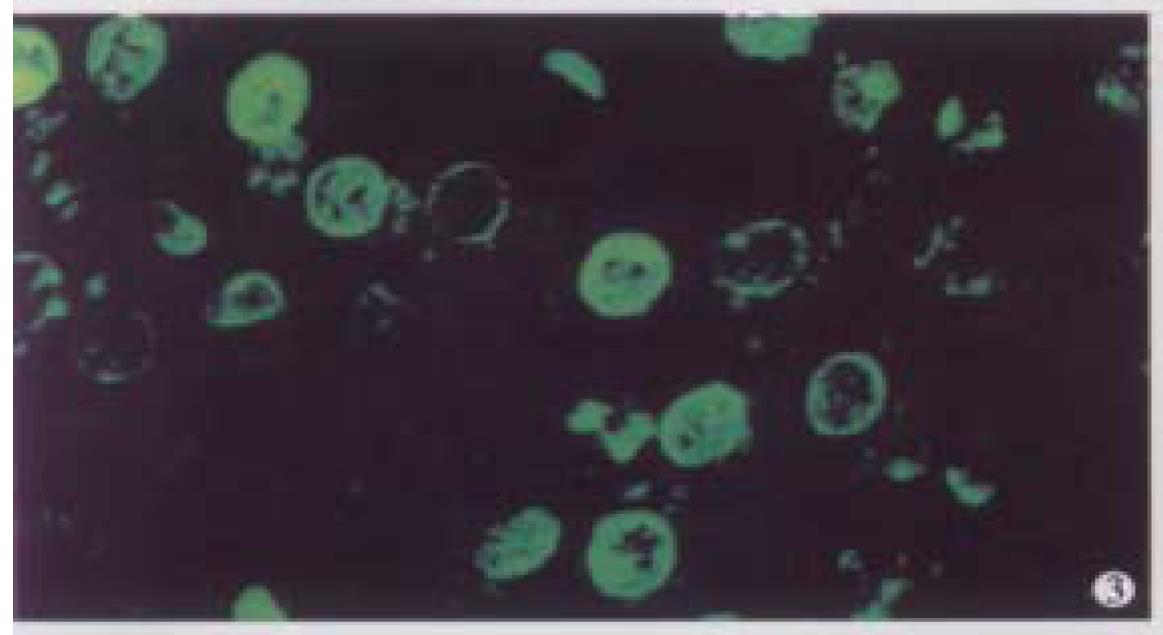
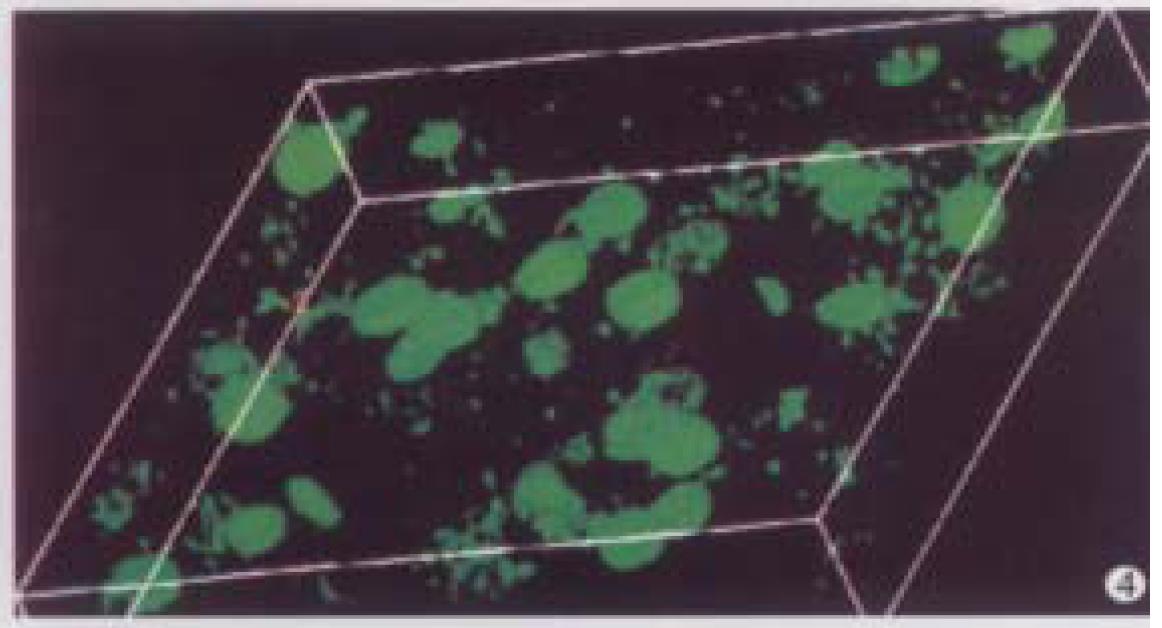
A microscopic field of view was selected from the normal liver specimen and comparison of the images taken under transmission conventional light microscope and CLSM is illustrated in Figure 1. The transmission conventional light microscopic image was shown on the right side and the out-of-focus signals were visible. The out-of-focus blur reduced the contrast and sharpness of the final image. In the confocal image (left) the out-of-focus signals were cut off an d only signals in focus were clearly visible. Optical sections (planes 1-60) were taken from the surface to the bottom of normal liver specimen with a Z-step interval of 0.5 μm. Figure 2 shows the 10th, 20th, 30th, 40th, 50th and 60th plane digital images of Z-series. The confocal images w ere taken at 5, 10, 15, 20, 25 and 30 μm depths, respectively. In the confocal images some nuclei appeared or disappeared depending on their orientation in space. The 60 2-D optical sections were computer focused on a plane (deep-focusing) to analyze the fine structure of chromatin patterns inside the nucleus and reconstructed 3-D images to display the 3-D detailed a rrangement of nucleus. Figure 3 shows normal liver cells with similar round or ovoid nuclei, homogeneous intensity of YOYO-1 iodide fluorescence as well. Three-dimensional view was shown in Figure 4. The nuclear surface appeared smooth with homogeneous fluorescence intensity.
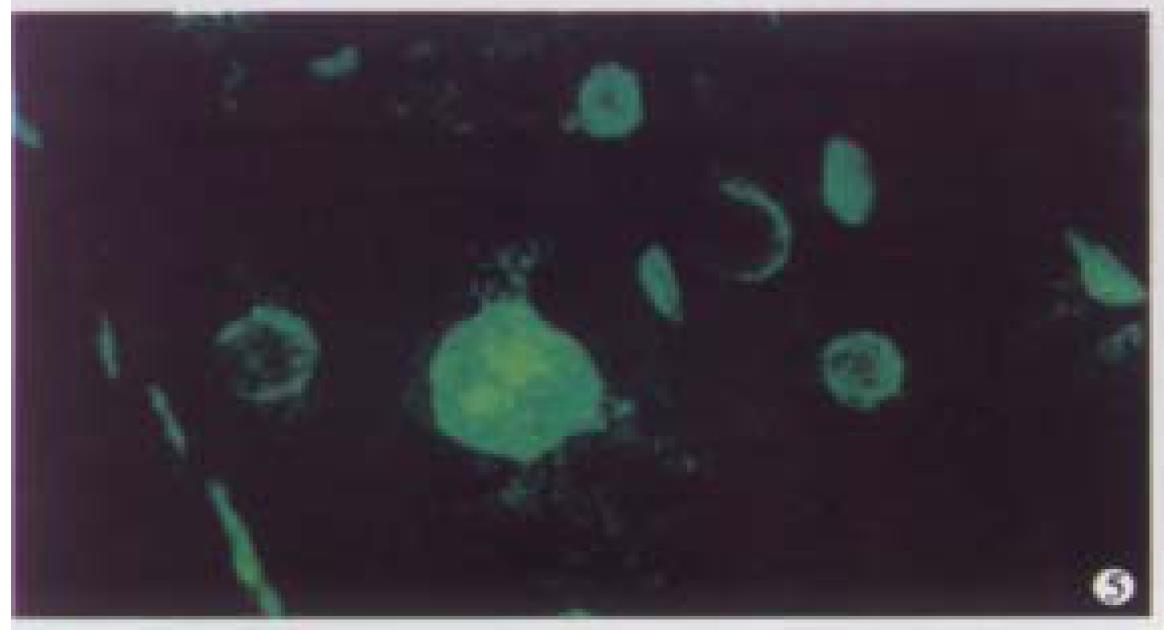
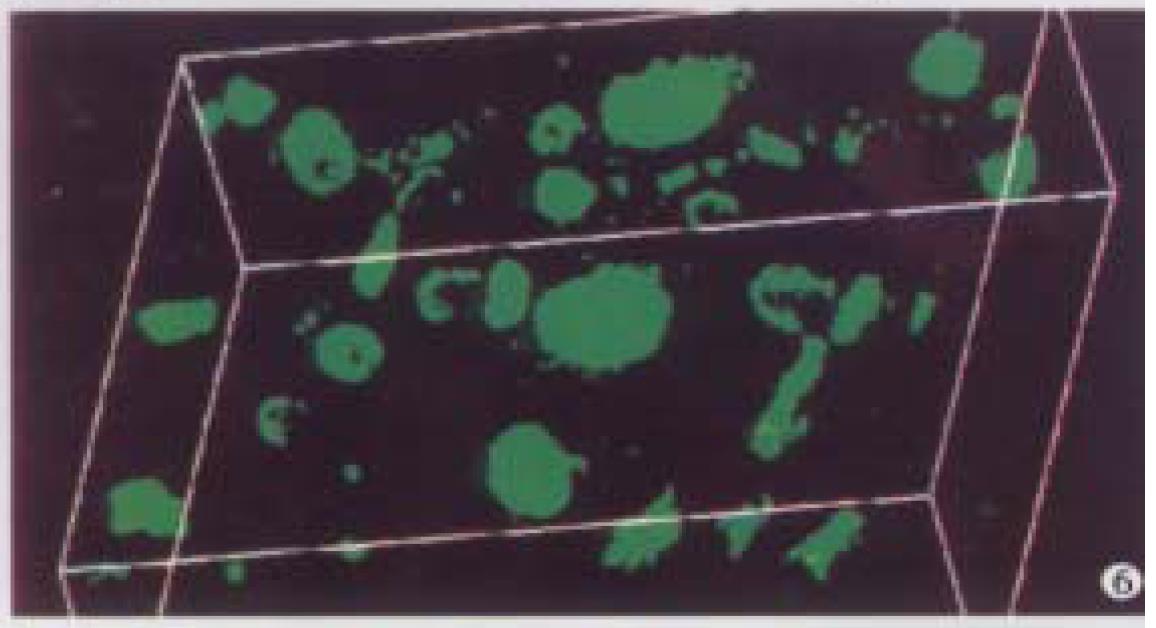
Optical sections (planes 1-50) were taken from the surface to the bottom of atypical hyperplasia of liver specimen with a Z-step interval of 0.6 μm. The structure of chromatin patterns inside the nucleus with homogeneous fluorescence intensity is shown with deep-focusing in Figure 5. Three-dimensional view is shown in Figure 6. The nuclear surface appeared smooth with homogeneous fluorescence intensity. But the volume of nucleus of atypical hyperplasia liver cell was bigger than that of normal liver cell.
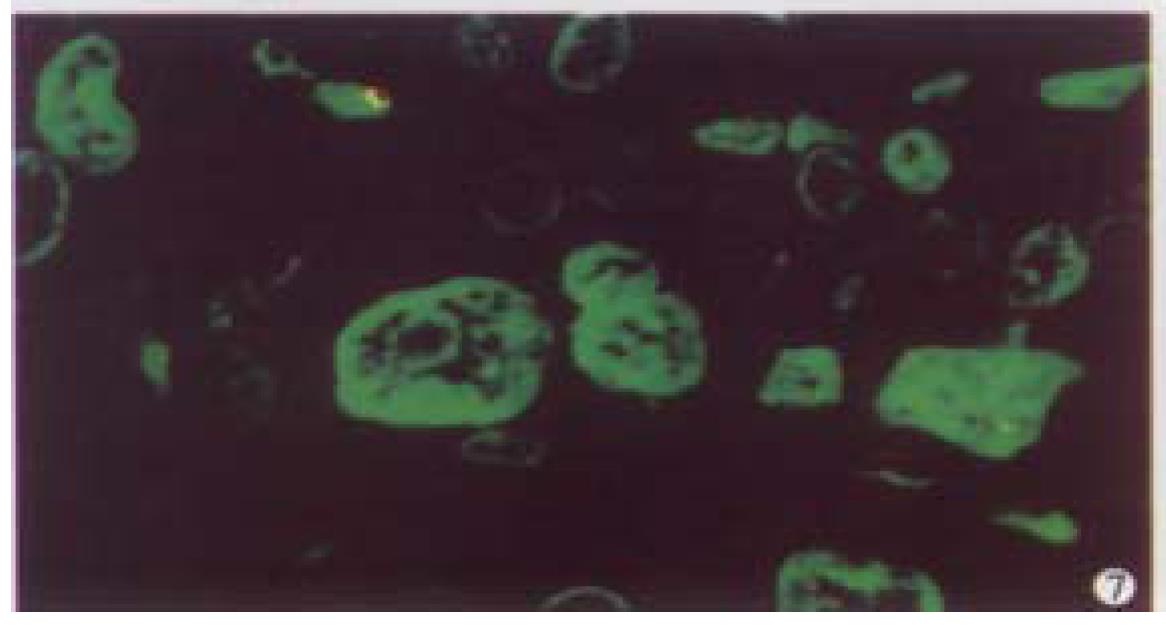
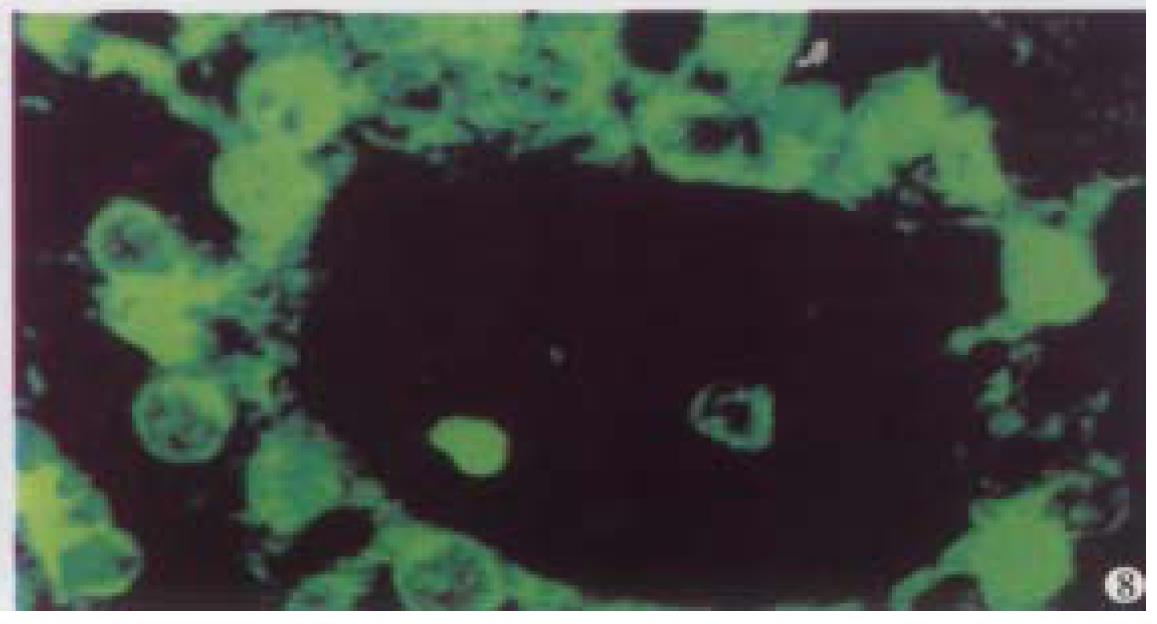
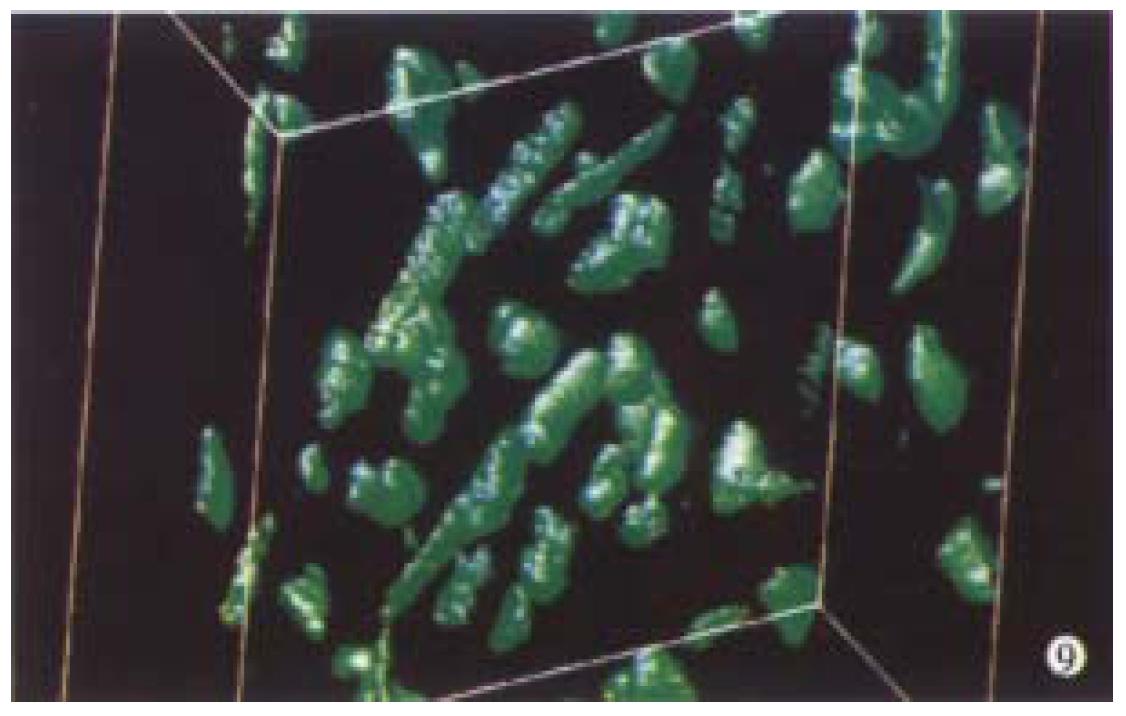
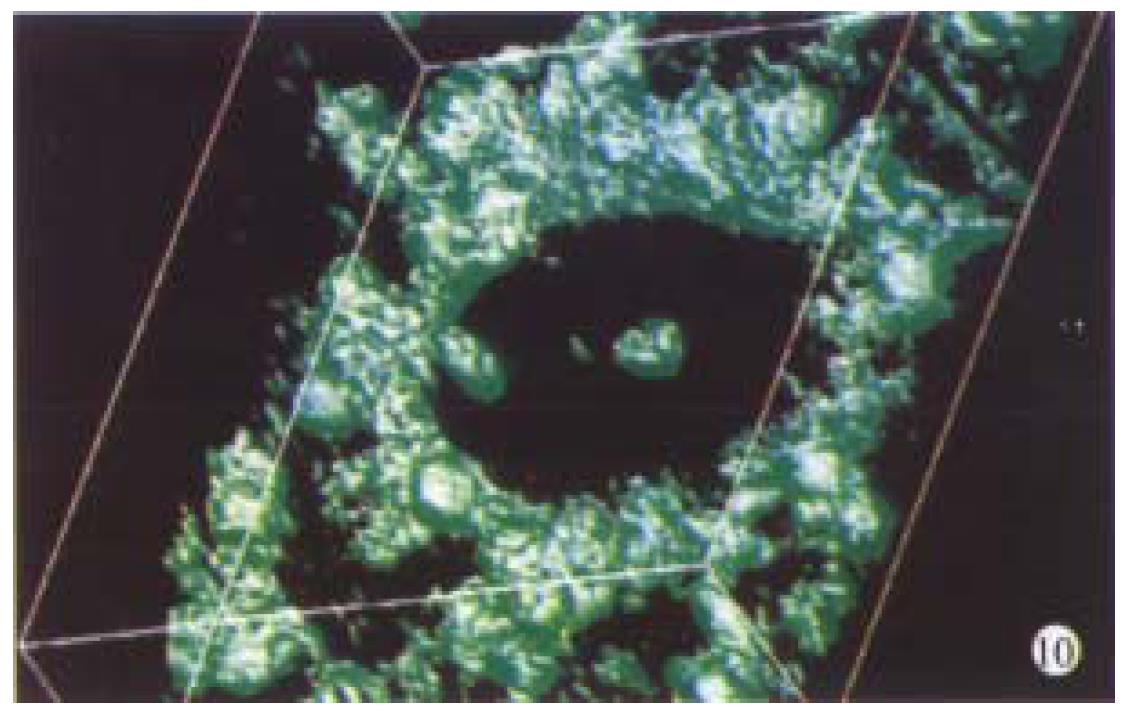
Optical sections (planes 1-50) were taken from the surface to the bottom of hepatocellular carcinoma specimen with a Z-step interval of 0.6 μm. The deep-focusing images were shown in Figure 7 and Figure 8. The structure of chromatin patterns inside the nucleus with heterogeneous karyotheca thickness, irregular and coarse chromatin texture, chromatin underside the karyotheca mainly. Three-dimensional view is shown in Figure 9 and Figure 10. HCC displayed remarkably different features in 3-D morphology, including: indented, molding, and irregular nuclear surface; marked pleomorphism; chaotic arrangement of tumor cell nuclei.
The area, volume, shape, DNA content, and chromatin pattern of nuclei may be important for the diagnosis and prognosis of cancer. Histopath ologists often use 4 μm to 6 μm thick paraffin sections to obtain representative and diagnostically relevant images. Due to the very limited section thickness in comparison with the size of the tissue, and the images are nearly two-dimensions, focusing up and down at high magnification provides a rough idea of the 3-D cellular characteristics, and such 3-D impression may be helpful or even essential in arriving at a certain diagnosis, especially for borderline lesions or tumors. However, in spite of the usefulness of such 3-D information about nuclei, conventional light microscopy is not always the ideal tool for 3-D evaluation due to the interference of out-of-focus structures with the images of the focus plane studied. Much of the light emitted from the regions of specimen above and below the focal plane contributes to the out-of-focus blur, which seriously reduces the contrast and sharpness of the final image. Confocal laser scanning microscope allows the acquisition of optical sections from a thick specimen and out-of-focus blur can be reduced considerably and thus much sharper and clearer images will be obtained. CLSM has become an exciting new instrument in biomedical research because of its increased resolution over conventional light microscope and its utility for subsequent 3-D-reconstructi on analysis[5,6].
In this paper, the 3-D reconstruction have demonstrated 3-D contour of representative characteristics of normal liver cells, atypical hyperplasia liver cells, and HCC cells and the spatial relationship of nuclei, as well as the subtle structure of chromatin texture inside nuclei. This paper emphasized the practical feasibility of CLSM and 3-D reconstruction from routine surgical histopathologic materials. To obtain desirable quality 3-D image from formalin-fixed, paraffin-embedded specimens, YOYO-1 iodide, a highly specific and sensitive (picogram sensitivity)[7] DNA probe was utilized. Intense and homogeneous fluorescence was obtained by incubating the YOYO-1 iodide for 1 h with agitation in the dark. To reveal subtle details of nuclear structure, RNA was removed by RNase predigestion, since nuclear RNA was stained by YOYO-1 iodide as well.
CLSM combines the three most advanced and important elements of our era, the microscope, the laser and the computer in one, moreover it is non-invasive and can be used on archival paraffin blocks. We anticipate that, in the future, pathologists may utilize these new techniques to make more precise diagnosis.
Dr. Wang-Hai Zhang, graduated from Shanghai Medical University as a PhD in 1999, majoring in surgical pathology and molecular pathology, having 10 papers published.
Edited by WANG Xian-Lin
proofread by Sun SM
| 1. | Minsky M. Memoir on inventing the confocal scanning microscope. Scanning. 1988;10:128-138. [DOI] [Full Text] |
| 2. | Boon ME, Schut JJ, Suurmeijer AJ, Benita EM, Hut PK, Kok LP. Confocal microscopy of false-negative breast aspirates. Diagn Cytopathol. 1995;12:42-48. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 3. | Liu S, Weaver DL, Taatjes DJ. Three-dimensional reconstruction by confocal laser scanning microscopy in routine pathologic specimens of benign and malignant lesions of the human breast. Histochem Cell Biol. 1997;107:267-278. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 19] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 4. | Beil M, Irinopoulou T, Vassy J, Rigaut JP. Chromatin texture analysis in three-dimensional images from confocal scanning laser microscopy. Anal Quant Cytol Histol. 1995;17:323-331. [PubMed] |
| 5. | White JG, Amos WB, Fordham M. An evaluation of confocal versus conventional imaging of biological structures by fluorescence light microscopy. J Cell Biol. 1987;105:41-48. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 576] [Cited by in RCA: 482] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 6. | Shotton DM. Robert Feulgen Prize Lecture 1995. Electronic light microscopy: present capabilities and future prospects. Histochem Cell Biol. 1995;104:97-137. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 36] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 7. | Tekola P, Baak JP, Beliën JA, Brugghe J. Highly sensitive, specific, and stable new fluorescent DNA stains for confocal laser microscopy and image processing of normal paraffin sections. Cytometry. 1994;17:191-195. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 20] [Article Influence: 0.6] [Reference Citation Analysis (0)] |