Published online Dec 15, 1999. doi: 10.3748/wjg.v5.i6.535
Revised: May 12, 1999
Accepted: July 13, 1999
Published online: December 15, 1999
- Citation: Feng DY, Zheng H, Fu CY, Cheng RX. An improvement method for the detection of in situ telomerase activity: in situ telomerase activity labeling. World J Gastroenterol 1999; 5(6): 535-537
- URL: https://www.wjgnet.com/1007-9327/full/v5/i6/535.htm
- DOI: https://dx.doi.org/10.3748/wjg.v5.i6.535
Telomerase is a special reverse transcriptase which consists of a template RNA and protein, and uses the RNA template to catalyze the addition of telomeric DNA to chromosome ends[1-5]. The enzyme is active in adult male germ-line cells, embryonic cells and hemopoietic stems, but is undetectable in normal somatic cells except for proliferative cells of renewal tissue, e.g., activated lymphocytes, basal cells of the epidermis and intestinal crypt cells, whereas in almost all malignant tumor cells and immortal cell lines, telomerase activity has been measured. Thus, it is believed that activation of telomerase is closely as sociated with genesis and development of malignant tumors. Most studies to date have measured telomerase activity by telomeric repeat amplification protocol ( TRAP ) in heterogeneous tissue extracts[1,3-5]. With the introduction of sensitive TRAP, telomerase has been reported to be detectable in small tissue samples from almost all tumors and tumors-derived cell lines. It is not known if all cells within a tumor have telomerase activity or if only a subset does. It is necessary to develop an in situ assay for detecting telomerase activity levels in cytological and tissue samples. Ohyashiki[2]reported an in situ assay for telomerase activity by in situ PCR with fluorescence TS and CX telomerase primers to detect telomerase activity of cultured cells and free cells, but the attempts to use in situ PCR telomerase assay on frozen sections of pathological materials were unsuccessful. Now we report an improved in situ telomerase activity assay on the basis of TRAP, which is very easy to be performed. A stable result can be gained on frozen sections of tumor tissue.
Twenty-four hepatocellular carcinoma (HCC), 20 colorectal carcinoma (CRC) and 18 nasopharyngeal carcinoma ( NPC ) tissues were obtained from patients in Xiangya Hospital and the Second Affiliated Hospital of Hunan Medical University, Changs ha, People’s Republic of China. Each tissue was divided into 2 pieces. One piece was fixed in 100 mL/L formalin and embedded in paraffin for pathological diagnosis, the other was soaked in liquid nitrogen, and preserved at -76 °C.
In situ TRAP kit was purchased from Department of Pathology, Beijing Medical University (P.R.China). SP detection kit and development kit were from Maixing Comp (Fuzhou, P.R.China). RNase was the product of Sino-American Ltd (Shanghai, P.R.China).
Six ìm frozen tissue sections were cut by freezing microtome (Leica, Germany) and the slides treated by poly-lysine were mounted, and detected for in situ telomerase activity.Briefly, each section was added 20 μL telomerase reaction mixture containing up-stream primer TS (5’-AATCCGTCGAGCAGAGTT-3’) 0.5 μL, down-stream primer CX (5’-CCCTTACCCTTACCCTTACCCTTA-3’) 0.5 μL, 2 mM biotin-dNTP 2 μL, 2 × buffer A 5 μL, 10 × buffer B μL and H2O 11 μL, coverslips were sealed. They were then incubated at 30 °C for 60 min rinsed with 0.01 M PBS for 3 min, fixed by 40 g/L paraformaldehyde for 15 min, rinsed with 0.01 M PBS for 2 × 3 min, submerged into 800 mL/L ethanol for 2 min and dried, dealt with 2% H2O2 for 20 min to block endoperoxidase, rinsed with 0.01 M PBS for 2 × 3 min, blocked by normal goat serum for 60 min, incubated by peroxidase-streptavidin at 37 °C for 60 min, rinsed with 0.01 M PBS for 3 × 5 min, developed with DAB, and then counterstained with hematoxylin.
In situ telomerase activity was also detected by in situ TRAP. The proce dure was as follows: 6 μm frozen sections were dripped with 20 μL mixture (primer TS 0.5 μL, 2 × buffer A 5ìL, H2O 14.5 μL), incubated at 30 °C for 60 min, rinsed with 0.01 M PBS for 3 min, fixed by 40 g/L paraformaldehyde for 10 min, rinsed with 0.01 M PBS for 3 min, digested with 25 mg/L proteinase K at 37 °C for 10 min, washed with 0.01 M PBS for 2 × 3 min, soaked in 800 mL/L ethanol for 2 min and dried, dripped with PCR mixture (primer TS and CX 0.5 μL respectively, 2 mM biotin-dNTPs 2 μL, 10 × buffer B 1 μL, Taq polymerase 2 U and H2O 15 μL), added coverglasses and sealed with liquid paraffin, and amplified by PCR (PCR conditions were 35 cycles at 95 °C for 1 min, at 50 °C for 1min, at 72 °C for 1 min) on Ericomp Thermocycler (Ericomp Ltd Comp. America), washed by chloroform and absolute ethanol for 5 min respectively to scavenge the liquid paraffin and the coverglasses, washed with 0.01 M PBS for 4 × 5 min, blocked by normal goat serum at 37 °C for 60 min, dripped with ABC reagent and incubated at 37 °C for 1 h, rinsed with 0.01 M PBS for 3 × 5 min, developed by DAB, and counterstained with hematoxylin.
Distilled water replaced primer TS and CX in reaction mixture or biotin-dNTPs in PCR miture. The sections were digested by RNase at 37 °C for 60 min to remove te lomerase RNA templates. The sections were heated at 95 °C for 10min to inactivate telomerase.
Positive sections in the kit were used for positive control.
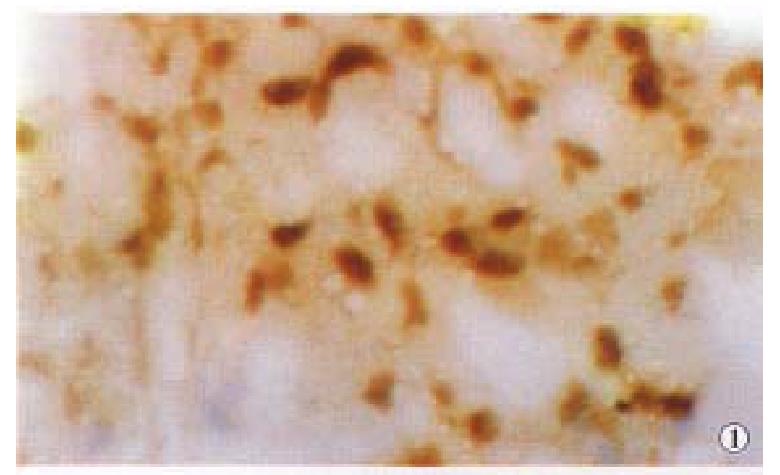
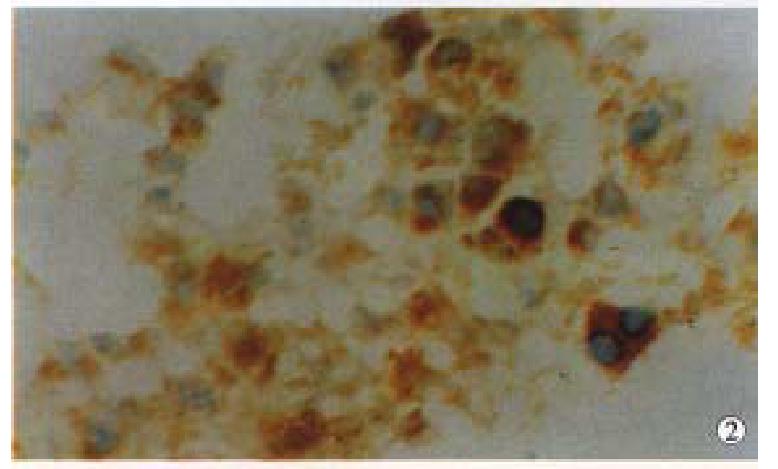
We first tested in situ labeling to detect telomerase activity in HCC, CRC and NPC tissues. Cancer cells contained detectable levels of telomerase activity. Positive signals of telomerase activity in HCC were almost in nuclei (Figure 1), a few in cytoplasm. Distributions of the positive cells were clustered or diffused in HCC. Weaker telomerase activity was also detected in cytoplasm and nuclei in liver tissues surrounding HCC, and mainly localized in nuclei of hepatocytes near cancer tissues, and positive cells were small patchy or clustered (Figure 2). In CRC, positive signals of telomerase activity were principally localized in nuclei, and much weaker in cytoplasm. In NPC, the telomerase activity was detected in nuclei (Figure 3). Distributions of positive cells were small nested or patchy in shape. Positive results for telomerase activity were obtained from positive control group, and negative results from negative control groups.
When our improved method was used to detect in situ telomerase activity, its positive signal localization, intensity and character, and distribution of positive cells were very similar to the results by in situ PCR-TRAP with much better morphosis preservation and background of tissues.
Telomerase is a specific reverse transcriptase which utilizes its own RNA as a template to catalyze synthesis of telomere[1-5]. According to molecular biological theory, if the telomerase is active, it can synthesize cDNA by adding the primers and dNTPs under suitable condition. Because dNTPs used to synthesize cDNA are labeled with biotin which has an affinity for streptavidin-peroxidae, and when developed by DAB, the telomerase activity can be detected. Affinity reaction of streptavidin with biotin can amplify positive signals, therefore PCR amplif ication procedure may be omitted for in situ detection of telomerase activit y. Thus, the method for in situ detection of telomerase activity is very simple and particularly successful on frozen sections of pathological materials. Morphological preservation and background of tissue sections are much better.
According to the data, in order to achieve satisfactory results during experiment. The following points are worth notice: (1) Coverglasses, tips, forceps and distilled water must be sterilized by high temperature; (2) After sections were incu bated with reaction mixture, the sections should not be rinsed too long, and intensity should not be too strong, otherwise, synthetic cDNA by reverse transcription is easy to be washed away, resulting in false negative, because cDNA is not fixed and fast adhered to the sections; (3) The slides must be treated with poly-lysine to avoid separation of the sections from slides.
Project supported by the Health Ministry Science Fund of C hina, No. 98-1-110
Edited by Xian-Lin Wang
Proofread by Jing-Yun Ma
| 1. | Nakayama J, Tahara H, Tahara E, Saito M, Ito K, Nakamura H, Nakanishi T, Tahara E, Ide T, Ishikawa F. Telomerase activation by hTRT in human normal fibroblasts and hepatocellular carcinomas. Nat Genet. 1998;18:65-68. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 443] [Cited by in RCA: 437] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 2. | Ohyashiki K, Ohyashiki JH, Nishimaki J, Toyama K, Ebihara Y, Kato H, Wright WE, Shay JW. Cytological detection of telomerase activity using an in situ telomeric repeat amplification protocol assay. Cancer Res. 1997;57:2100-2103. [PubMed] |
| 3. | Nouso K, Urabe Y, Higashi T, Nakatsukasa H, Hino N, Ashida K, Kinugasa N, Yoshida K, Uematsu S, Tsuji T. Telomerase as a tool for the differential diagnosis of human hepatocellular carcinoma. Cancer. 1996;78:232-236. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 4. | Miura N, Horikawa I, Nishimoto A, Ohmura H, Ito H, Hirohashi S, Shay JW, Oshimura M. Progressive telomere shortening and telomerase reactivation during hepatocellular carcinogenesis. Cancer Genet Cytogenet. 1997;93:56-62. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 135] [Cited by in RCA: 124] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 5. | Wu S, Liew CT, Li X. [Study on telomerase activity in hepatocellular carcinoma and chronic hepatitic disease]. Zhonghua Binglixue Zazhi. 1998;27:91-93. [PubMed] |