TO THE EDITOR
Hepatitis B virus (HBV) remains the leading cause of viral hepatitis globally[1]. According to the World Health Organization, as of 2022, approximately 254 million individuals worldwide are living with chronic hepatitis B, with 1.2 million new infections annual and around 1.1 million deaths due to HBV-related cirrhosis and hepatocellular carcinoma (HCC)[2]. Vietnam ranks among the top 20 countries most affected by hepatitis B, with an adult prevalence rate of approximately 10.5%[3]. Recently, Nguyen et al[4] employed multi-clustering analysis to provide a deeper insight into the intricate relationship between human leukocyte antigen (HLA) gene polymorphisms and the risk of HBV-associated cirrhosis and HCC within a Vietnamese population. The HLA system plays a pivotal role in modulating the immune response, governing how individuals recognize and combat viruses, bacteria, and other foreign agents[5,6]. Genome-wide association studies (GWAS) have identified specific single nucleotide polymorphisms within the HLA-DP and HLA-DQ regions that are associated with the risk of chronic HBV infection and HCC, particularly in regions such as Japan, China, and Saudi Arabia[7]. However, the complete range of genetic factors associated with achieving a functional cure remains unclear. Therefore, comprehensive analytical approaches are essential to deepen our understanding of genetic, clinical and immunological parameters. This study offers valuable insights that could contribute to the development of personalized medical treatments and prevention strategies, while also identifying potential molecular markers for future research and clinical applications.
Nguyen et al[4] firstly extracted genomic DNA from blood samples of 315 healthy controls and 721 patients with chronic HBV infection, all recruited from multiple hospitals in Vietnam. HLA-DP and HLA-DQ gene polymorphisms were analyzed using real-time quantitative polymerase chain reaction, focusing on the HLA-DPA1 rs3077, HLA-DPB1 rs9277535, and single nucleotide polymorphism rs2856718 variants. Subsequently, the genotype distributions were assessed for Hardy-Weinberg equilibrium and linkage disequilibrium, followed by hierarchical cluster analysis. Notably, the authors identified that the “A” allele in the combination of rs2856718, rs3077, and rs9277535 significantly reduced the risk of liver disease. In the cases of HCC, the A-A-A/G haplotype increased the risk of HCC development by 1.58-fold. The study confirmed the complex relationship between mutations in HLA genotype and disease susceptibility, suggesting that this relationship might be mediated through key markers such as bilirubin, gamma-glutamyl transferase, aspartate aminotransferase, alanine aminotransferase, and alpha-fetoprotein. The conclusion drawn from this study are commendable for the innovative methodology and comprehensive exploration of a Vietnamese population. This approach, integrating paraclinical analysis with bioinformatics, holds significant potential for further development in pre-machine learning research, paving the way for personalized treatment strategies in medicine.
The study revealed a clinically significant correlation between HLA-DP-DQ genotypes and the development of HCC and liver cirrhosis, there also exhibits visible considerations that need to be addressed. For example, the prevalence of HBV among Indigenous Australians is more than four times higher than in the non-Indigenous population, which is associated with elevated rates of liver disease and liver cancer[8,9]. This suggests that the same population residing in different environments may lead to varying incidences of HBV-related liver disease. Thus, it is crucial to consider other environmental or lifestyle elements that influence the progression of HBV infection, in addition to genetic factors, to fully understand their role in disease development. Moreover, as the authors noted, machine learning analysis combining clinical parameters, immune phenotyping, and genetic information could effectively predict the likelihood of a functional cure in patients with HBV. However, larger cohorts of the healthy controls and HBV-related patients are necessary in future research to enhance the reliability of these findings.
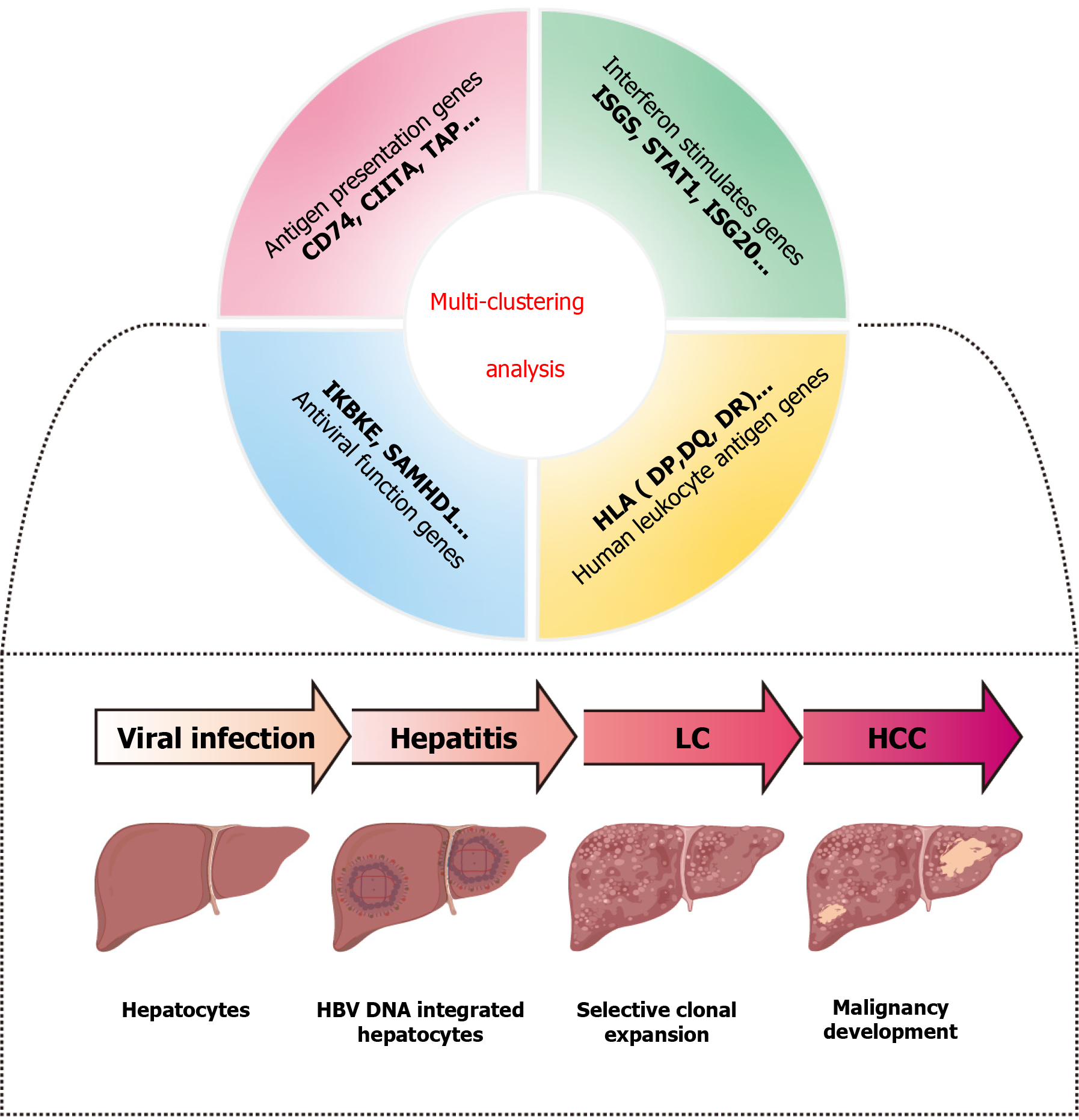
During the immune-active and immune escape (ENEG) phases of chronic HBV infection, distinct inflammatory profiles and notable differences in intrahepatic gene expression were observed. Genes involved in antigen presentation genes (leukocyte differentiation antigen 74, class II transactivator, transporters associated with antigen processing), interferon-stimulated genes (interferon-stimulated genes; signal transducer and activator of transcription 1, interferon-stimulated gene 20), and antiviral function-related genes (inhibitor of nuclear factor kappa-B kinase subunit epsilon, sterile alpha motif and HD domain-containing protein 1) were significantly induced[10] besides HLA genes highlighted in this study of Nguyen et al[4]. While HLA genes have been identified as important genetic factors in HBV persistence through GWAS[11], future research could explorer the intricate interactions between HLA and other genes in relation to HBV-associated liver disease (Figure 1). Additionally, the HLA system is divided into two main classes: The HLA class I group, which is primarily associated with the incidence of rejection[12], and the HLA-II genes, which include HLA-DP, HLA-DQ, and HLA-DR, are primarily link to immune response[13]. A GWAS identified HLA-DR as the major locus for susceptibility to HBV-related acute-on-chronic liver failure[14]. Moreover, genetic polymorphisms in HLA-DR have also been suggested to influence HCC risks, possibly through interactions with HBV mutations[15]. Therefore, expanding the scope of research to include other HLA gene regions, such as HLA-DR, and their relationship with immune responses and liver disease progression would provide deeper insights into disease mechanisms. In future clinical practice, genetic markers recognized through multi-clustering analysis could be used to identify high-risk individuals. By integrating biochemical indicators, such as alanine aminotransferase and alpha-fetoprotein, more precise risk assessment tools could be developed. These tools may facilitate the design of personalized vaccines or immunotherapies for individuals with specific high-risk genotypes. Furthermore, for patients already infected with HBV, personalized follow-up plans could be developed by combining genetic markers with virological and immunological data, thereby reducing the risk of liver disease progression.
Figure 1 Machine learning revealed the complex relationship between different genotypes and hepatitis B virus liver diseases.
CD74: Leukocyte differentiation antigen 74; CIITA: Class II transactivator; TAP: Transporters associated with antigen processing; ISGS: Interferon-stimulated genes; STAT1: Signal transducer and activator of transcription 1; ISG20: Interferon-stimulated gene 20; IKBKE: Inhibitor of nuclear factor kappa-B kinase subunit epsilon; SAMHD1: Sterile alpha motif and HD domain-containing protein 1; HLA: Human leukocyte antigen; LC: Liver cirrhosis; HCC: Hepatocellular carcinoma; HBV: Hepatitis B virus.