Published online Apr 14, 2025. doi: 10.3748/wjg.v31.i14.104397
Revised: February 1, 2025
Accepted: March 21, 2025
Published online: April 14, 2025
Processing time: 113 Days and 9.4 Hours
Celiac disease (CD) is a systemic autoimmune disorder triggered by gluten ingestion ingenetically predisposed individuals. It is characterized by intestinal histological damage and the production of specific autoantibodies. The latest European Society for Paediatric Gastroenterology, Hepatology, and Nutrition (ESPGHAN) 2020 guidelines have excluded human leukocyte antigen (HLA) genotyping from the no-biopsy diagnostic approach due to its weak positive predictive value, limited availability, and high cost in some countries. However, HLA genetic testing remains valuable in certain clinical contexts. This study provided practical indications for when to request and how to interpret HLA genotyping, emphasizing its continued relevance for CD diagnosis in specific cases. We also proposed a strategy for monitoring the risk of developing type 1 diabetes (T1D) in patients with CD, based on the risk stratification carried by different HLA genotypes. A retrospective analysis of 746 patients with CD and 627 controls was conducted at our hospital starting in 2012, when HLA geno
Core Tip: This guide explained how to interpret human leukocyte antigens (HLA) genetics associated with celiac disease (CD) and the different clinical situations where HLA genotyping can be useful in the diagnosis of CD. It also provided a strategy based on HLA genotyping for monitoring patients with CD at risk for the future development of type 1 diabetes (T1D). Only a subset of HLA genotypes linked to CD is associated with the development of T1D. Interestingly, some HLA genotypes that carry a high risk for CD may offer protection against T1D. Therefore, HLA genotyping in patients with CD could help in identifying those at high risk for T1D, enabling proactive interventions and therapies to preserve beta cell function.
- Citation: Schirru E, Rossino R, Jores RD, Corpino M, Muntoni S, Cucca F, Congia M. Clinical settings in which human leukocyte antigen typing is still useful in the diagnosis of celiac disease. World J Gastroenterol 2025; 31(14): 104397
- URL: https://www.wjgnet.com/1007-9327/full/v31/i14/104397.htm
- DOI: https://dx.doi.org/10.3748/wjg.v31.i14.104397
Celiac disease (CD) is a systemic autoimmune disease characterized by a specific serological antibody profile and a peculiar intestinal histological damage. It is triggered and sustained by the ingestion of gluten and related prolamins in genetically predisposed individuals[1]. CD has a strong hereditary component[2]; 7.5%-15.0% of first-degree relatives and dizygotic twins are affected by CD, while CD concordance is estimated to range between 50%-80% in monozygotic twins[3]. CD is polygenic and multifactorial due to the interaction of multiple genes with a series of environmental factors and lifestyles that can vary rapidly depending on the historical periods in which these interactions occur.
A series of data indicated that the prevalence of CD appears to be increasing[4-9] and that this does not seem to be influenced by either the age at which gluten is introduced or the duration of breastfeeding[10,11]. It is likely that the cause of such an increase in prevalence is attributable not to a single factor but to a combination of elements, including the constant increase, starting from the mid-twentieth century, in the consumption of gluten-containing foods[12] and a modification of the intestinal microbiota due to widespread use of antibiotics and a Western-style diet[13]. In the coming years, the real estimates of CD prevalence will probably be more difficult to establish due to the increasingly high number of individuals who undertake a gluten-free diet (GFD) for health reasons without first excluding CD[14]. In this category of individuals, help for the clinician can come from the human leukocyte antigen (HLA) genetic test of susceptibility for CD.
The heritability of CD is due to 56% of genes outside the major histocompatibility complex (MHC) system, and as much as 44% of genes in the MHC family. Numerous genome-wide association studies have identified that 57 non-HLA susceptibility genes for CD would explain only about 6% of heritability, while 50% would consist of multiple common variants as observed in other polygenic diseases[15]. Recently, five susceptibility variants in the MHC that act independently of the HLA-DQA1 and HLA-DQB1 loci have been identified and explain 18% of heritability[16]. Finally, 26% of heritability is made up of the strong association with some alleles at the HLA-DQA1 and HLA-DQB1 loci that encode for HLA-DQ2 and HLA-DQ8 molecules[17]. Therefore, to date, the strongest single genetic association with CD is carried by the HLA-DQ loci.
It should be immediately noted that by adding up the frequencies of HLA-DQ2 and HLA-DQ8 in Caucasian populations, their positivity reaches 30%-40% or even about 50% in Sardinians[18] and Sahrawis[19], but the disease manifests itself only in a fraction of them. In addition, disease prevalence does not statistically correlate with the frequency of such HLA molecules in different populations[20]. Overall, these observations confirm the polygenic nature and the need for various environmental factors for the development of CD.
In Table 1 the genetic terminology used in the manuscript is explained, allowing a better understanding of the HLA complex located on the short arm of chromosome 6 (occupying about 3 Mbp in 6p21.3). The region contains more than 220 genes, mostly with immunological functions. The loci that compose it in the center-telomeric direction are HLA-DP, HLA-DM, HLA-DQ, and HLA-DR (belonging to class II) and HLA-B, HLA-C, HLA-E, and HLA-A (belonging to class I). The class III, including some genes that encode for complement fractions, is located between class I and class II. The system has a high degree of polymorphism[21], and for this reason numerous alleles exist at the same locus. For example, 10273 alleles have been reported at the HLA-B locus of class I and 2782 at the HLA-DQB1 locus of class II[22]. For updated data, see the Immuno Polymorphism Database central platform (https://www.ebi.ac.uk/ipd/imgt/hla/allele.html).
| Terminology | Explanation |
| Gene | A particular nucleotide DNA sequence at a specific locus |
| Allele | Polymorphisms in the sequence of a DNA at the same locus. In a diploid individual, at most two different alleles can be present |
| MHC and HLA | Major histocompatibility complex. A large region of vertebrate DNA containing a set of closely linked polymorphic genes encoding immune cell surface proteins called MHC molecules. In humans the region is called HLA |
| Center-telomeric | Refers to the known order of the HLA class II (DP, DM, DQ, and DR loci), class III (containing the C4 and TNF genes), and class I (B, C, E and A) genes, from centromere to telomere along the chromosome. Centromer is the constricted region of chromosome connecting the sister chromatids and creating a short arm (p) and a long arm (q) on the chromatids. The telomere is a region of repetitive nucleotide sequences associated with specialized proteins at the ends of chromosomes |
| Polymorphism | Variations in the nucleotide sequence of a given locus (determines more than one allele at that locus) |
| Homozygous | Presence of alleles with the same nucleotide sequence at the same locus on homologous chromosomes (HLA-DQB1 02:01/HLA-DQB1 02:01) |
| Heterozygous | Presence of alleles with different nucleotide sequences at the same locus on homologous chromosomes (HLA-DQB1 02:01/HLA-DQB1 03:02) |
| Haplotype | Alleles inherited together on a certain chromosomal segment (HLA-DQA1 05:01/HLA-DQB1 02:01). In the MHC, due to a strong linkage disequilibrium, haplotypes sometimes extending from the A locus to the DQB1 locus are found quite frequently. A1, Cw7, B8, DR3, and DQ2 are present in Northern European populations, and A30, Cw5, B18, DR3, and DQ2 are present in the Sardinian population |
| Genotype | Genetic constitution of an individual (e.g., of an HLA genotype -A1, Cw7, B8, DR3, DQ2/-A30, Cw5, B18, DR3, DQ2) |
| Cis | Alleles at different loci located on the same chromosome (e.g., HLA-DQA1 03:01 HLA-DQB1 03:02, coding for the molecule HLA-DQ8) |
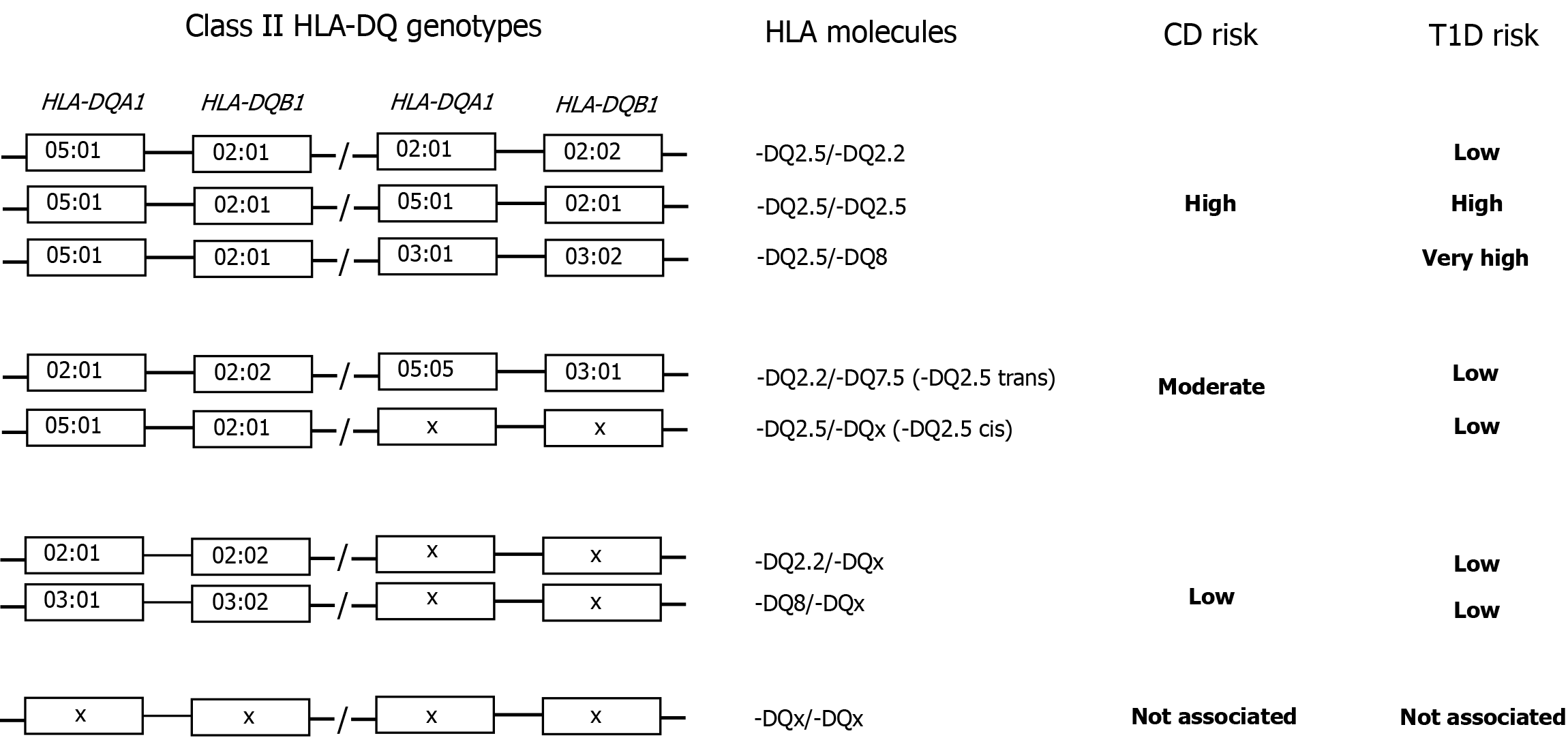
| Trans | Alleles located at different loci on opposite chromosomes (e.g., HLA-DQA1 05:05 HLA-DQB1 03:01/HLA-DQA1 02:01 HLA-DQB1 02:02; see also Figure 1) |
| Heterodimer | Protein complex consisting of two different subunits (e.g., α chain and β chain of HLA-DQ2), also named HLA-DQ molecule |
| Codominance | Both maternal and paternal alleles are expressed. This allows the formation of the HLA-DQ2 heterodimer in trans |
| Linkage disequilibrium | The nonrandom association of alleles of different loci within a population |
HLA is characterized by the presence of strong linkage disequilibrium (LD). Two or more loci are in LD when the frequency of association of their different alleles is higher or lower than expected if the loci were independent and therefore randomly associated. The presence of LD and interdependent immunological function genes suggest a possible selective advantage of particular configurations of alleles, sometimes arranged in extended haplotypes from the class I HLA-A locus to the class II HLA-DQB1 locus[23]. Numerous studies, including ours on the Sardinian population[18,24] and their particular genetic makeup, have shown that HLA genes involved in CD are located at the HLA-DQA1 and HLA-DQB1 loci of class II[24-26]. Among the many alleles encoded by these loci, those most frequently associated with CD are HLA-DQA1 05:01 and HLA-DQB1 02:01. These two alleles are in strong LD and encode the alpha chain and beta chain of the HLA-DQ heterodimer (DQα1 05:01, DQβ1 02:01) called HLA-DQ2. This heterodimer can be encoded in cis when the two alleles HLA-DQA1 05:01 and HLA-DQB1 02:01 are located on the same chromosome (almost always in LD with the HLA-DRB1 03:01 allele of the HLA-DRB1 locus to form the HLA-DRB1 03:01, HLA-DQA1 05:01, HLA-DQB1 02:01 haplotype) or in trans in the HLA-DQA1 05:05, HLA-DQB1 03:01/HLA-DQA1 02:01, HLA-DQB1 02:02 genotype[25].
The alleles HLA-DQA1 02:01 and HLA-DQB1 02:02 also encode a molecule that has been named HLA-DQ2, even though it is composed of a different alpha chain (the polymorphism that differentiates the DQβ1 0201 and DQβ1 0202 chains is negligible from a functional point of view). To differentiate these two HLA-DQ2 molecules at the protein level, the HLA-DQ2 molecule encoded by the HLA-DQA1 05:01 and HLA-DQB1 02:01 alleles is called HLA-DQ2.5, while the HLA-DQ molecule encoded by the HLA-DQA1 02:01 and HLA-DQB1 02:02 alleles is called HLA-DQ2.2 (Figure 1 and Table 2).
| HLA (genotype) | HLA (heterodimer) | CD (n = 746) | Controls (n = 627) | OR | P value | CI |
| HLA-DQA1 05 HLA-DQB1 02:01/HLA-DQA1 02:01 HLA-DQB1 02:02 | -DQ2.5/-DQ2.2 | 111 | 17 | 6.3 | 1.1 × 10-14 | 3.7-10.6 |
| HLA-DQA1 05 HLA-DQB1 02:01/HLA-DQA1 05 HLA-DQB1 02:01 | -DQ2.5/-DQ2.5 | 153 | 33 | 4.6 | 2 × 10-16 | 3.1-6.9 |
| HLA-DQA1 05 HLA-DQB1 02:01/HLA-DQA1 03:01 HLA-DQB1 03:02 | -DQ2.5/-DQ8 | 57 | 21 | 2.4 | 6.2 × 10-4 | 1.4-4.0 |
| HLA-DQA1 05 HLA-DQB1 03:01/HLA-DQA1 02:01 HLA-DQB1 02:02 | -DQ2.5 trans | 41 | 20 | 1.8 | 3.9 × 10-2 | 1.0-3.0 |
| HLA-DQA1 05 HLA-DQB1 02:01/HLA-DQA1 X HLA-DQB1X | -DQ2.5 cis | 271 | 165 | 1.6 | 7.2 × 10-5 | 1.3-2.0 |
| HLA-DQA1 02:01 HLA-DQB1 02:02/HLA-DQA1 X HLA-DQB1 X | -DQ2.2/X | 17 | 28 | 0.5 | 2.3 × 10-2 | 0.3-0.9 |
| HLA-DQA1 03:01 HLA-DQB1 03:02/HLA-DQA1 X HLA-DQB1 X | -DQ8/X | 15 | 38 | 0.3 | 1 × 10-4 | 0.2-0.6 |
Approximately 90%-95% of patients with CD are positive for alleles that encode for HLA-DQ2 and more specifically for HLA-DQ2.5 in cis or trans (Figure 1 and Table 2). The other HLA molecule associated with CD in about 5%-7% of patients is HLA-DQ8 encoded by the HLA-DQA1 03:01 and HLA-DQB1 03:02 alleles, while the remaining 3%-5% is composed of HLA-DQ2.2[27] or very rarely of half heterodimers such as in the HLA-DQ2.3molecule[28] or in the HLA-DQ7.5 molecule[29].
Table 2 shows the HLA genotypes and heterodimers of susceptibility for CD. HLA genotypes were ordered by decreasing odds ratio (OR) using the cases of patients with CD from our center[30]. These associations are similar to those observed by other authors[31] and by meta-analysis studies[32]. A brief discussion of some of these high-frequency heterodimers with higher OR will allow us to better understand the HLA association and how to interpret the results provided by the genotyping laboratory.
The HLA-DQ2.5 heterodimer, a variant of HLA-DQ2, can be defined as the most permissive for CD since it is present in more than 90% of patients with CD[25]. It can be encoded in cis in the HLA-DRB1 03:01, HLA-DQA1 05:01, HLA-DRB1 02:01 haplotype or in trans in the HLA-DQA1 05:05, HLA-DQB1 03:01/HLA-DQA1 02:01, HLA-DQB1 02:02 genotype, conferring a very similar disease risk (Figure 1 and Table 2). The beta chains (DQβ1 02:01 and DQβ1 02:02) encoded by HLA-DQB1 02:01 and HLA-DQB1 02:02 differ by one amino acid residue at position 135, which is distant from the peptide-binding region and therefore functionally irrelevant. Instead, homozygosity for HLA-DQ2.5 confers a high risk for the development of celiac autoimmunity[4,33].
According to some authors this “double dose” of HLA-DQ2.5 allows for the presentation of a broader repertoire of gluten peptides in greater quantities[34] and stability[35] to T cells than other molecules associated with CD, such as HLA-DQ2.2 and HLA-DQ8. This quantitative model of intestinal damage in CD, where HLA-DQ2.5 expression and the number of gluten-specific T cells available represent the main limiting factors, has been indirectly strengthened by several studies conducted on patients with CD[36,37] and a recent meta-analysis[38].
HLA-DQ8 is encoded by the HLA-DQA1 03 and HLA-DQB1 03:02 alleles. When HLA-DQA1 03 is associated with HLA-DQB1 03:01, the molecule is called HLA-DQ7.3. When it is associated with HLA-DQB1 03:03, it is called HLA-DQ9. Only HLA-DQ8 is permissive for CD (Figure 1 and Table 2). HLA-DQ8, when present in heterozygosity with HLA-DQ2.5, confers a high risk for CD. In addition, it carries a strong risk for the development of autoimmune type 1 diabetes (T1D). One possible explanation may be the formation of HLA trans-dimers between the alpha chain encoded by HLA-DQA1 05:01 and the beta chain encoded by HLA-DQB1 03:02[39-41]. On the other hand, when HLA-DQ8 is not in combination with HLA-DQ2.5 it confers a low risk for CD.
The HLA-DQ2.2 heterodimer differs functionally from HLA-DQ2.5 due to the alpha chain encoded by the HLA-DQA1 02:01 allele. The differences between the two alpha chains make the HLA-DQ2.2 molecule less suited than HLA-DQ2.5 to present gluten-derived peptides to cluster of differentiation 4 positive T cells[35,42]. However, when in association with the HLA-DQ2.5 haplotype (i.e. HLA-DQ2.5/HLA-DQ2.2 genotype) an OR of 6.3 with a frequency of 14.9% was observed[30], which was comparable to that observed in HLA-DQ2.5 homozygotes (Table 2), a finding consistent with other case studies[31]. On the contrary, when it is not associated with the HLA-DQ2.5 haplotype, it confers a low risk for CD (Table 2). When in association with HLA-DQA1 05:05 and HLA-DQB1 03:01 to form the HLA-DQ2.5 heterodimer in trans, it confers a similar risk of CD to that of HLA-DQ2.5 in cis (Table 2). Therefore, in both cases, HLA-DQ2.2, due to the genetic mechanism of codominance, behaves as a donor of beta chains for the constitution of the HLA-DQ2.5 heter
Although very rare, HLA genotypes other than HLA-DQ2 and HLA-DQ8 have been reported in CD. In a multicenter European study, such patients were detected in 2% of cases[33]. In another American study, the frequency of endomisial antibody (EMA)-positive individuals but negative for HLA-DQ2 and HLA-DQ8 was even lower (0.16%)[36]. In our case series only 10 patients were non-HLA-DQ2 and non-HLA-DQ8. All 10 of these patients received a diagnosis of CD in the late 1980s, when the anti-tissue transglutaminase type 2-IgA (t-TG2-IgA) and t-TG2-IgG determinations were not yet available. Unfortunately, we were unable to reverify these diagnoses to exclude that mucosal damage was due to other pathologies. On the other hand, none were found to be negative for HLA-DQ2 or HLA-DQ8 when the t-TG2-IgA determination was available. Therefore, we believe that the diagnosis of CD in patients with HLA genotypes other than HLA-DQ2 and HLA-DQ8 or with heterodimeric molecules such as in HLA-DQ2.3[27] or HLA-DQ7.5[28] should be entrusted to reference centers and should be based on unequivocal evidence that mucosal damage is gluten-dependent.
In the following paragraphs, we outlined the clinical situations for which HLA genetic testing for CD can be proposed to the patient or family. This practical guide should reduce the use of the test to appropriate cases, helping the clinicians make useful decisions for the diagnosis of the disease or the follow-up of individuals at risk (Table 3).
| Condition | HLA-DQ2 and HLA-DQ8 negativity |
| Before biopsy | |
| First-degree relatives of a patient affected by CD | Allows the exclusion of serological monitoring in individuals not carrying any genetic risk |
| Individuals who have started a GFD without performing t-TG2-IgA measurement | Allows the exclusion of CD as the cause of gastrointestinal symptoms regardless of the clinical response to the GFD |
| Individuals with persistent low t-TG2-IgA titer | Allows unequivocal definition of the false positives, including first-degree relatives of a proband with reduced gluten intake |
| Individuals affected by IgA deficiency | Allows the exclusion of serological monitoring in individuals not carrying any HLA genetic risk |
| Patients with chromosomal pathologies associated with increased CD risk (Down syndrome, Turner syndrome, Williams syndrome) | Allows the limit of periodic serological follow-up exclusively to positive patients |
| Patients affected by Hashimoto’s thyroiditis | Allows the limit of the periodic serological follow-up exclusively to positive patients |
| After biopsy | |
| Ineffectiveness of GFD in patients with CD | Allows exclusion of CD and suspect other pathologies. Could help in excluding refractory CD type II and enteropathy-associated T cell lymphoma |
| Dubious CD biopsy performed for other reasons | Allows exclusion of CD and suspect other pathologies |
Since the publication of the ESPGHAN guidelines in 2012 for the diagnosis of CD and up to 2020, the main use of HLA genotyping was directed towards formulating new diagnoses of CD in children and adolescents without performing intestinal biopsy[41]. In fact, these guidelines provided the option not to perform intestinal biopsy in case the patient presented with: (1) Symptoms suggestive of CD; (2) A value of t-TG2-IgA greater than 10 times the normal values and, in a second sample, positivity of EMA confirming the positivity of t-TG2-IgA; and (3) HLA genetics compatible with CD, i.e. positivity for HLA-DQ2 or HLA-DQ8[43].
These recommendations were revised in the new ESPGHAN 2020 guidelines[44], where neither HLA genotyping for the HLA-DQA1 and HLA-DQB1 loci nor the presence of symptoms associated with CD are mandatory to formulate the diagnosis of CD in a pediatric patient with high t-TG2-IgA greater than 10 times normal values confirmed by EMA positivity in a second sample. The reasons given in the new guidelines for this change are that HLA testing is not widely available, is expensive, and would not improve the ability of serology to approach diagnosis without biopsy[44]. However, it is important to underscore that the HLA genotyping expenses can be performed at very convenient costs by tagging single nucleotide polymorphisms as shown by Bastos et al[45].
Even in a retrospective study conducted on the Sardinian population in subjects attending the ambulatory of the Pediatric Gastroenterological Unit in Cagliari, Italy, between 2005 and 2012, we found that all symptomatic patients with CD with t-TG2-IgA greater than 10 times normal values were positive for HLA-DQ2 or HLA-DQ8, thus confirming the ESPGHAN 2020 recommendations, at least for symptomatic patients[46]. Therefore, it is likely that by adhering to the ESPGHAN 2020 guidelines that HLA genetic testing for diagnosing CD will no longer be used except in selected cases[44].
Table 3 provides a detailed list of scenarios in which HLA genotyping can be proposed as a useful analysis in the diagnostic process of CD to the patient or the patient’s parents. The request for the test has been divided into situations where it may be useful before or after performing an intestinal biopsy. It is always necessary to explain rigorously that the test has value only when negative for HLA-DQ2 or HLA-DQ8, while its positivity is of little clinical usefulness. In fact, negativity has a very high negative predictive value, avoiding the need for negative patients to undertake further serological, endoscopic, or possible gluten reintroduction tests[47].
Relatives of patients diagnosed with CD have a higher risk of developing CD over time. A recent retrospective study by the Mayo Clinic found a frequency of 44.4% of CD in first-degree relatives of patients with CD[48], a finding confirmed by other authors[49]. This frequency is much higher than that found in previous studies[50,51]. The awareness that CD can occur at any time in an individual’s life often silently (in the Mayo Clinic study, 28% were asymptomatic)[48] seems to indicate a rational choice to exclude from long-term serological screening those first-degree relatives negative for HLA-DQ2 or HLA-DQ8.
This category of individuals is likely to increase in the coming years due to the growing popularity of GFD as a healthier option capable of alleviating chronic gastrointestinal symptoms[52,53]. In addition, gluten-free products are now more abundant, easier to purchase, and less expensive than in the past[54]. Finally, an increasing number of individuals are diagnosed or self-diagnosed with non-celiac gluten sensitivity without following a rigorous clinical path[55,56]. These individuals on GFD, in whom CD has not previously been excluded, may be offered the opportunity of HLA genetic testing for CD, which if negative for HLA-DQ2 or HLA-DQ8 allows the exclusion of CD as the cause of gastrointestinal symptoms.
It is not uncommon in clinical practice, including referral centers, to observe adult and pediatric patients with low t-TG2-IgA positivity who may be affected by CD or may be false positives. A series of clinical conditions are known to be associated with non-specific production of t-TG2-IgA antibodies, such as chronic inflammatory bowel diseases[57], autoimmune diseases[58], simple respiratory and gastrointestinal infections[59], chronic liver diseases[60], HIV infection[61], and other conditions[62]. Even the intestinal production of t-TG2-IgA does not seem to be completely specific for CD[63]. In these particular situations, it seems rational to propose HLA genotyping to identify patients negative for HLA-DQ2 or HLA-DQ8 before undertaking invasive examinations such as esophagogastroduodenoscopy in pediatric patients or in patients with chronic diseases in whom esophagogastroduodenoscopy may be contraindicated.
Selective IgA deficiency (IgAD) is a clinical condition that predisposes patients to autoimmune manifestations, the most frequently associated being CD[64]. IgAD also has a strong genetic component that maps to the MHC region[65], and although it tends to be associated with positive HLA-DQ2 haplotypes[66,67], a certain number of haplotypes are HLA-DQ2 negative. In a study of Swedish and Iranian patients with IgAD, about 38% were found to be negative for HLA-DQ2 or HLA-DQ8[68]. The ESPGHAN 2020 guidelines consider intestinal biopsy mandatory in patients with IgAD and positive t-TG2-IgG, as it was not possible to derive a safe t-TG2-IgG cutoff value from the literature capable of predicting CD in IgAD[44]. These guidelines do not mention the possible usefulness of HLA genotyping. We believe that HLA genotyping should be considered in patients with IgAD and positive t-TG2-IgG to spare individuals negative for HLA-DQ2 or HLA-DQ8 from biopsies.
The 2012 CD diagnostic guidelines clearly indicated HLA genotyping as the initial screening test in such risk groups for CD[43]. Only in individuals positive for HLA-DQ2 or HLA-DQ8 was periodic serological screening recommended using t-TG2-IgA. The 2020 guidelines have been modified, and diagnosis in symptomatic or asymptomatic patients without performing duodenal biopsy can be made with serology alone, even for these conditions[44]. However, in these patients, gastrointestinal symptoms can be frequent but not necessarily related to CD[69]. Furthermore, it is known that CD can run silently in the same patients[69], that some symptoms commonly found in CD can be mistakenly attributed to the syndrome for years[70], and that CD can also occur in adulthood[71]. This implies that such patients, whether symptomatic or asymptomatic, should undergo indefinite serological follow-up over time because they belong to at-risk groups, including those who do not require it. It seems, therefore, more rational to adhere to the ESPGHAN 2012 guidelines in these at-risk groups[43].
HLA genotyping in children with a diagnosis of CD can also establish the risk of future T1D development. The risk may be stratified by high to low risk according to different HLA genotypes. Indeed, we found that CD patients with HLA-DQ2.5/HLA-DQ8, and HLA-DQ2.5/HLA-DQ2.5 genotypes were strongly associated with concomitance of CD and T1D. Conversely, the HLA-DQ2.5/HLA-DQ2.2 genotype appeared to confer protection against T1D development (Table 2)[30]. Therefore, early screening for CD around 2-3 years of age with subsequent CD diagnosis with or without biopsy followed by HLA genotyping could help in identifying those patients at high risk of T1D[30]. This strategy may detect patients with CD at the very early stages of T1D (phase 1) who should undergo periodic pancreatic autoantibody monitoring and further immune, genetic, and metabolic tests to identify those patients susceptible to immunotherapies able to preserve endogenous beta cell function.
Recent meta-analysis studies have confirmed an increased prevalence of CD in patients with Hashimoto’s thyroiditis (HT), recommending screening for CD in such patients[72]. The association with HLA-DQ2 or HLA-DQ8 in HT is not as strong as in CD[73,74]. The HLA-DQ2.2 allele appears to confer protection, while the HLA-DQ2.5 allele has a weak association at the limits of significance and the strongly associated HLA-DRB1 04 haplotype is in LD with HLA-DQ7.3 and not with HLA-DQ8[73,74]. Therefore, it is crucial to be able to exclude patients who are negative for HLA-DQ2 or HLA-DQ8 from long-term serological screening. Therefore, we consider it rational to offer this option to pediatric and adult patients in follow-up for HT, as indicated in the 2012 guidelines[43].
An increased prevalence of CD is also observed in Graves’ disease[75]. Therefore, serological screening for CD is recommended in this autoimmune thyroid disease as well[75]. Regarding the possible use of genetic testing for CD, it is found that among the alleles most strongly associated with Graves’ disease, we find the same ones present in CD, in particular HLA-DRB1 03:01 (in LD with HLA-DQ2) and HLA-DQA1 05[76]. Therefore, HLA genotyping in patients with Graves’ disease will be of limited clinical utility in excluding CD. Therefore, its determination is not recommended to establish the risk of CD in patients with Graves’ disease.
Approximately 7%-30% of adult patients with CD and 25% of pediatric patients with CD appear to have an unsatisfactory response to a GFD[77-80]. In another retrospective study conducted in a single referral center in patients with different grades of intestinal damage, about 20% who were on a GFD were negative for HLA-DQ2 or HLA-DQ8[47]. Therefore, genetic testing for CD may be useful to identify misdiagnoses related to false positives of t-TG2-IgA and indicate a functional gastrointestinal disorder or other intestinal pathology as responsible for the symptoms.
Refractory celiac disease (RCD) is defined as CD that remains unresponsive to at least 12 months of a strict GFD[81,82]. RCD includes various subtypes with differing characteristics, making diagnosis and management challenging. The overlap between RCD and enteropathy-associated T-cell lymphoma (EATL) adds to this complexity. Genetic factors, such as HLA-DQ2 homozygosity, are strongly linked to RCD-II and EATL, occurring in 44%-65% of RCD-II cases and 53.3% of EATL cases compared with 25.5% in RCD-I and 20.7% in uncomplicated CD[83]. Identifying HLA-DQ2 homozygous patients early could help detect those at higher risk of severe complications. Single-cell analysis highlighted significant variability in abnormal cell populations in RCD-II[84], offering insights that could improve diagnostics and treatments, similar to advances seen in cancer research[85-87].
Sometimes when patients undergo endoscopic investigations and biopsies of the upper gastrointestinal tract for reasons unrelated to the diagnosis of CD, histological lesions may be found that could be part of the initial pathological picture of CD, such as increased intraepithelial T lymphocytes. Since such lesions can also appear in a large group of other pathologies including cases of villous atrophy and negative serology for CD, such as drug-induced mucosal damage[88-90], negativity for HLA-DQ2 or HLA-DQ8 allows CD to be excluded from the differential diagnosis of such pathologies.
Recently, a new therapy for autoimmune diseases, including CD, has been proposed. It involves creating nanoparticles coated with MHC-peptide complexes where both the MHC and peptide are disease-specific[91,92]. These nanoparticles are designed to bind specifically to T cell receptors on cluster of differentiation 4-positive cells, triggering the transformation of effector T cells into regulatory T cells. Regulatory T cells in turn could promote antigen-specific immune tolerance while preserving the overall integrity of the immune system[93,94].
Before testing these innovative immunomodulatory therapies in humans, researchers can utilize a mouse model for CD that was recently developed after years of research[95]. It is clear that future patients with CD or other autoimmune diseases, such as multiple sclerosis or T1D, who may benefit from these personalized therapies will need to undergo HLA genotyping for HLA-DRB1, HLA-DQA1, and HLA-DQB1.
HLA typing, while deemed non-essential by the 2020 ESPGHAN guidelines for the diagnosis of CD, continues to hold significant diagnostic value in specific clinical contexts. This guide provided clinicians with practical recommendations on when and how to effectively incorporate HLA genetic testing into the diagnostic pathway. In addition, a monitoring strategy in patients with CD for future development of T1D based on the risk stratification conferred by different HLA genotypes is also presented.
We thank the non-profit organization Associazione Diabete Zero ODV.
| 1. | Caio G, Volta U, Sapone A, Leffler DA, De Giorgio R, Catassi C, Fasano A. Celiac disease: a comprehensive current review. BMC Med. 2019;17:142. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 564] [Cited by in RCA: 563] [Article Influence: 93.8] [Reference Citation Analysis (0)] |
| 2. | Iversen R, Sollid LM. The Immunobiology and Pathogenesis of Celiac Disease. Annu Rev Pathol. 2023;18:47-70. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 103] [Article Influence: 51.5] [Reference Citation Analysis (0)] |
| 3. | Lundin KE, Wijmenga C. Coeliac disease and autoimmune disease-genetic overlap and screening. Nat Rev Gastroenterol Hepatol. 2015;12:507-515. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 109] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 4. | Liu E, Dong F, Barón AE, Taki I, Norris JM, Frohnert BI, Hoffenberg EJ, Rewers M. High Incidence of Celiac Disease in a Long-term Study of Adolescents With Susceptibility Genotypes. Gastroenterology. 2017;152:1329-1336.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 72] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 5. | Myléus A, Ivarsson A, Webb C, Danielsson L, Hernell O, Högberg L, Karlsson E, Lagerqvist C, Norström F, Rosén A, Sandström O, Stenhammar L, Stenlund H, Wall S, Carlsson A. Celiac disease revealed in 3% of Swedish 12-year-olds born during an epidemic. J Pediatr Gastroenterol Nutr. 2009;49:170-176. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 213] [Cited by in RCA: 222] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 6. | Rubio-Tapia A, Kyle RA, Kaplan EL, Johnson DR, Page W, Erdtmann F, Brantner TL, Kim WR, Phelps TK, Lahr BD, Zinsmeister AR, Melton LJ 3rd, Murray JA. Increased prevalence and mortality in undiagnosed celiac disease. Gastroenterology. 2009;137:88-93. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 583] [Cited by in RCA: 519] [Article Influence: 32.4] [Reference Citation Analysis (1)] |
| 7. | Lionetti E, Pjetraj D, Gatti S, Catassi G, Bellantoni A, Boffardi M, Cananzi M, Cinquetti M, Francavilla R, Malamisura B, Montuori M, Zuccotti G, Cristofori F, Gaio P, Passaro T, Penagini F, Testa A, Trovato CM, Catassi C. Prevalence and detection rate of celiac disease in Italy: Results of a SIGENP multicenter screening in school-age children. Dig Liver Dis. 2023;55:608-613. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 27] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 8. | Gatti S, Lionetti E, Balanzoni L, Verma AK, Galeazzi T, Gesuita R, Scattolo N, Cinquetti M, Fasano A, Catassi C; Celiac Screening Team. Increased Prevalence of Celiac Disease in School-age Children in Italy. Clin Gastroenterol Hepatol. 2020;18:596-603. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 70] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 9. | Gatti S, Rubio-Tapia A, Makharia G, Catassi C. Patient and Community Health Global Burden in a World With More Celiac Disease. Gastroenterology. 2024;167:23-33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Reference Citation Analysis (0)] |
| 10. | Lionetti E, Castellaneta S, Francavilla R, Pulvirenti A, Tonutti E, Amarri S, Barbato M, Barbera C, Barera G, Bellantoni A, Castellano E, Guariso G, Limongelli MG, Pellegrino S, Polloni C, Ughi C, Zuin G, Fasano A, Catassi C; SIGENP (Italian Society of Pediatric Gastroenterology, Hepatology, and Nutrition) Working Group on Weaning and CD Risk. Introduction of gluten, HLA status, and the risk of celiac disease in children. N Engl J Med. 2014;371:1295-1303. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 386] [Cited by in RCA: 320] [Article Influence: 29.1] [Reference Citation Analysis (0)] |
| 11. | Vriezinga SL, Auricchio R, Bravi E, Castillejo G, Chmielewska A, Crespo Escobar P, Kolaček S, Koletzko S, Korponay-Szabo IR, Mummert E, Polanco I, Putter H, Ribes-Koninckx C, Shamir R, Szajewska H, Werkstetter K, Greco L, Gyimesi J, Hartman C, Hogen Esch C, Hopman E, Ivarsson A, Koltai T, Koning F, Martinez-Ojinaga E, te Marvelde C, Pavic A, Romanos J, Stoopman E, Villanacci V, Wijmenga C, Troncone R, Mearin ML. Randomized feeding intervention in infants at high risk for celiac disease. N Engl J Med. 2014;371:1304-1315. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 317] [Cited by in RCA: 306] [Article Influence: 27.8] [Reference Citation Analysis (0)] |
| 12. | Andrén Aronsson C, Lee HS, Koletzko S, Uusitalo U, Yang J, Virtanen SM, Liu E, Lernmark Å, Norris JM, Agardh D; TEDDY Study Group. Effects of Gluten Intake on Risk of Celiac Disease: A Case-Control Study on a Swedish Birth Cohort. Clin Gastroenterol Hepatol. 2016;14:403-409.e3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 86] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 13. | Verdu EF, Galipeau HJ, Jabri B. Novel players in coeliac disease pathogenesis: role of the gut microbiota. Nat Rev Gastroenterol Hepatol. 2015;12:497-506. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 150] [Cited by in RCA: 169] [Article Influence: 16.9] [Reference Citation Analysis (0)] |
| 14. | Kim HS, Patel KG, Orosz E, Kothari N, Demyen MF, Pyrsopoulos N, Ahlawat SK. Time Trends in the Prevalence of Celiac Disease and Gluten-Free Diet in the US Population: Results From the National Health and Nutrition Examination Surveys 2009-2014. JAMA Intern Med. 2016;176:1716-1717. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 87] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 15. | Coleman C, Quinn EM, Ryan AW, Conroy J, Trimble V, Mahmud N, Kennedy N, Corvin AP, Morris DW, Donohoe G, O'Morain C, MacMathuna P, Byrnes V, Kiat C, Trynka G, Wijmenga C, Kelleher D, Ennis S, Anney RJ, McManus R. Common polygenic variation in coeliac disease and confirmation of ZNF335 and NIFA as disease susceptibility loci. Eur J Hum Genet. 2016;24:291-297. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 26] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 16. | Gutierrez-Achury J, Zhernakova A, Pulit SL, Trynka G, Hunt KA, Romanos J, Raychaudhuri S, van Heel DA, Wijmenga C, de Bakker PI. Fine mapping in the MHC region accounts for 18% additional genetic risk for celiac disease. Nat Genet. 2015;47:577-578. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 107] [Cited by in RCA: 109] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 17. | Dieli-Crimi R, Cénit MC, Núñez C. The genetics of celiac disease: A comprehensive review of clinical implications. J Autoimmun. 2015;64:26-41. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 101] [Article Influence: 10.1] [Reference Citation Analysis (1)] |
| 18. | Cucca F, Lampis R, Frau F, Macis D, Angius E, Masile P, Chessa M, Frongia P, Silvetti M, Cao A, De Virgiliis S, Congia M. The distribution of DR4 haplotypes in Sardinia suggests a primary association of type I diabetes with DRB1 and DQB1 loci. Hum Immunol. 1995;43:301-308. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 74] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 19. | Catassi C, Doloretta Macis M, Rätsch IM, De Virgiliis S, Cucca F. The distribution of DQ genes in the Saharawi population provides only a partial explanation for the high celiac disease prevalence. Tissue Antigens. 2001;58:402-406. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 43] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 20. | Lionetti E, Catassi C. Co-localization of gluten consumption and HLA-DQ2 and -DQ8 genotypes, a clue to the history of celiac disease. Dig Liver Dis. 2014;46:1057-1063. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 43] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 21. | Williams TM. Human leukocyte antigen gene polymorphism and the histocompatibility laboratory. J Mol Diagn. 2001;3:98-104. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 93] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 22. | Robinson J, Halliwell JA, Hayhurst JD, Flicek P, Parham P, Marsh SG. The IPD and IMGT/HLA database: allele variant databases. Nucleic Acids Res. 2015;43:D423-D431. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1479] [Cited by in RCA: 1499] [Article Influence: 136.3] [Reference Citation Analysis (0)] |
| 23. | Alter I, Gragert L, Fingerson S, Maiers M, Louzoun Y. HLA class I haplotype diversity is consistent with selection for frequent existing haplotypes. PLoS Comput Biol. 2017;13:e1005693. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 36] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 24. | Congia M, Frau F, Lampis R, Frau R, Mele R, Cucca F, Muntoni F, Porcu S, Boi F, Contu L. A high frequency of the A30, B18, DR3, DRw52, DQw2 extended haplotype in Sardinian celiac disease patients: further evidence that disease susceptibility is conferred by DQ A1*0501, B1*0201. Tissue Antigens. 1992;39:78-83. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 48] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 25. | Louka AS, Sollid LM. HLA in coeliac disease: unravelling the complex genetics of a complex disorder. Tissue Antigens. 2003;61:105-117. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 137] [Cited by in RCA: 129] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 26. | Sollid LM. Coeliac disease: dissecting a complex inflammatory disorder. Nat Rev Immunol. 2002;2:647-655. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 686] [Cited by in RCA: 657] [Article Influence: 28.6] [Reference Citation Analysis (0)] |
| 27. | Spurkland A, Sollid LM, Polanco I, Vartdal F, Thorsby E. HLA-DR and -DQ genotypes of celiac disease patients serologically typed to be non-DR3 or non-DR5/7. Hum Immunol. 1992;35:188-192. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 91] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 28. | Karell K, Louka AS, Moodie SJ, Ascher H, Clot F, Greco L, Ciclitira PJ, Sollid LM, Partanen J; European Genetics Cluster on Celiac Disease. HLA types in celiac disease patients not carrying the DQA1*05-DQB1*02 (DQ2) heterodimer: results from the European Genetics Cluster on Celiac Disease. Hum Immunol. 2003;64:469-477. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 399] [Cited by in RCA: 411] [Article Influence: 18.7] [Reference Citation Analysis (0)] |
| 29. | Tinto N, Cola A, Piscopo C, Capuano M, Galatola M, Greco L, Sacchetti L. High Frequency of Haplotype HLA-DQ7 in Celiac Disease Patients from South Italy: Retrospective Evaluation of 5,535 Subjects at Risk of Celiac Disease. PLoS One. 2015;10:e0138324. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 28] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 30. | Schirru E, Rossino R, Diana D, Jores RD, Baldera D, Muntoni S, Spiga C, Ripoli C, Ricciardi MR, Cucca F, Congia M. HLA Genotyping in Children With Celiac Disease Allows to Establish the Risk of Developing Type 1 Diabetes. Clin Transl Gastroenterol. 2024;15:e00710. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 3] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 31. | Pietzak MM, Schofield TC, McGinniss MJ, Nakamura RM. Stratifying risk for celiac disease in a large at-risk United States population by using HLA alleles. Clin Gastroenterol Hepatol. 2009;7:966-971. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 62] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 32. | De Silvestri A, Capittini C, Poddighe D, Valsecchi C, Marseglia G, Tagliacarne SC, Scotti V, Rebuffi C, Pasi A, Martinetti M, Tinelli C. HLA-DQ genetics in children with celiac disease: a meta-analysis suggesting a two-step genetic screening procedure starting with HLA-DQ β chains. Pediatr Res. 2018;83:564-572. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 32] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 33. | Liu E, Lee HS, Aronsson CA, Hagopian WA, Koletzko S, Rewers MJ, Eisenbarth GS, Bingley PJ, Bonifacio E, Simell V, Agardh D; TEDDY Study Group. Risk of pediatric celiac disease according to HLA haplotype and country. N Engl J Med. 2014;371:42-49. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 253] [Cited by in RCA: 250] [Article Influence: 22.7] [Reference Citation Analysis (0)] |
| 34. | Vader W, Stepniak D, Kooy Y, Mearin L, Thompson A, van Rood JJ, Spaenij L, Koning F. The HLA-DQ2 gene dose effect in celiac disease is directly related to the magnitude and breadth of gluten-specific T cell responses. Proc Natl Acad Sci U S A. 2003;100:12390-12395. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 270] [Cited by in RCA: 278] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 35. | Bodd M, Kim CY, Lundin KE, Sollid LM. T-cell response to gluten in patients with HLA-DQ2.2 reveals requirement of peptide-MHC stability in celiac disease. Gastroenterology. 2012;142:552-561. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 70] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 36. | Karinen H, Kärkkäinen P, Pihlajamäki J, Janatuinen E, Heikkinen M, Julkunen R, Kosma VM, Naukkarinen A, Laakso M. Gene dose effect of the DQB1*0201 allele contributes to severity of coeliac disease. Scand J Gastroenterol. 2006;41:191-199. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 66] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 37. | Jores RD, Frau F, Cucca F, Grazia Clemente M, Orrù S, Rais M, De Virgiliis S, Congia M. HLA-DQB1*0201 homozygosis predisposes to severe intestinal damage in celiac disease. Scand J Gastroenterol. 2007;42:48-53. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 31] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 38. | Bajor J, Szakács Z, Farkas N, Hegyi P, Illés A, Solymár M, Pétervári E, Balaskó M, Pár G, Sarlós P, Szűcs Á, Czimmer J, Szemes K, Huszár O, Varjú P, Vincze Á. Classical celiac disease is more frequent with a double dose of HLA-DQB1*02: A systematic review with meta-analysis. PLoS One. 2019;14:e0212329. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 23] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 39. | Chow IT, Gates TJ, Papadopoulos GK, Moustakas AK, Kolawole EM, Notturno RJ, McGinty JW, Torres-Chinn N, James EA, Greenbaum C, Nepom GT, Evavold BD, Kwok WW. Discriminative T cell recognition of cross-reactive islet-antigens is associated with HLA-DQ8 transdimer-mediated autoimmune diabetes. Sci Adv. 2019;5:eaaw9336. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 18] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 40. | Gioia L, Holt M, Costanzo A, Sharma S, Abe B, Kain L, Nakayama M, Wan X, Su A, Mathews C, Chen YG, Unanue E, Teyton L. Position β57 of I-A(g7) controls early anti-insulin responses in NOD mice, linking an MHC susceptibility allele to type 1 diabetes onset. Sci Immunol. 2019;4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 45] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 41. | van Lummel M, van Veelen PA, Zaldumbide A, de Ru A, Janssen GM, Moustakas AK, Papadopoulos GK, Drijfhout JW, Roep BO, Koning F. Type 1 diabetes-associated HLA-DQ8 transdimer accommodates a unique peptide repertoire. J Biol Chem. 2012;287:9514-9524. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 62] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 42. | Fallang LE, Bergseng E, Hotta K, Berg-Larsen A, Kim CY, Sollid LM. Differences in the risk of celiac disease associated with HLA-DQ2.5 or HLA-DQ2.2 are related to sustained gluten antigen presentation. Nat Immunol. 2009;10:1096-1101. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 106] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 43. | Husby S, Koletzko S, Korponay-Szabó IR, Mearin ML, Phillips A, Shamir R, Troncone R, Giersiepen K, Branski D, Catassi C, Lelgeman M, Mäki M, Ribes-Koninckx C, Ventura A, Zimmer KP; ESPGHAN Working Group on Coeliac Disease Diagnosis; ESPGHAN Gastroenterology Committee; European Society for Pediatric Gastroenterology, Hepatology, and Nutrition. European Society for Pediatric Gastroenterology, Hepatology, and Nutrition guidelines for the diagnosis of coeliac disease. J Pediatr Gastroenterol Nutr. 2012;54:136-160. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1708] [Cited by in RCA: 1837] [Article Influence: 141.3] [Reference Citation Analysis (1)] |
| 44. | Husby S, Koletzko S, Korponay-Szabó I, Kurppa K, Mearin ML, Ribes-Koninckx C, Shamir R, Troncone R, Auricchio R, Castillejo G, Christensen R, Dolinsek J, Gillett P, Hróbjartsson A, Koltai T, Maki M, Nielsen SM, Popp A, Størdal K, Werkstetter K, Wessels M. European Society Paediatric Gastroenterology, Hepatology and Nutrition Guidelines for Diagnosing Coeliac Disease 2020. J Pediatr Gastroenterol Nutr. 2020;70:141-156. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 363] [Cited by in RCA: 690] [Article Influence: 138.0] [Reference Citation Analysis (0)] |
| 45. | Bastos MD, Kowalski TW, Puñales M, Tschiedel B, Mariath LM, Pires ALG, Faccini LS, Silveira TR. Search for DQ2.5 and DQ8 alleles using a lower cost technique in patients with type 1 diabetes and celiac disease in a population of southern Brazil. Arch Endocrinol Metab. 2017;61:550-555. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 2] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 46. | Schirru E, Danjou F, Cicotto L, Rossino R, Macis MD, Lampis R, Jores RD, Congia M. Anti-actin IgA antibodies identify celiac disease patients with a Marsh 3 intestinal damage among subjects with moderate anti-TG2 levels. Biomed Res Int. 2013;2013:630463. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 9] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 47. | Pallav K, Kabbani T, Tariq S, Vanga R, Kelly CP, Leffler DA. Clinical utility of celiac disease-associated HLA testing. Dig Dis Sci. 2014;59:2199-2206. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 35] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 48. | Nellikkal SS, Hafed Y, Larson JJ, Murray JA, Absah I. High Prevalence of Celiac Disease Among Screened First-Degree Relatives. Mayo Clin Proc. 2019;94:1807-1813. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 20] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 49. | Meijer CR, Auricchio R, Putter H, Castillejo G, Crespo P, Gyimesi J, Hartman C, Kolacek S, Koletzko S, Korponay-Szabo I, Ojinaga EM, Polanco I, Ribes-Koninckx C, Shamir R, Szajewska H, Troncone R, Villanacci V, Werkstetter K, Mearin ML. Prediction Models for Celiac Disease Development in Children From High-Risk Families: Data From the PreventCD Cohort. Gastroenterology. 2022;163:426-436. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 21] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 50. | Karinen H, Kärkkäinen P, Pihlajamäki J, Janatuinen E, Heikkinen M, Julkunen R, Kosma VM, Naukkarinen A, Laakso M. HLA genotyping is useful in the evaluation of the risk for coeliac disease in the 1st-degree relatives of patients with coeliac disease. Scand J Gastroenterol. 2006;41:1299-1304. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 23] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 51. | Rubio-Tapia A, Van Dyke CT, Lahr BD, Zinsmeister AR, El-Youssef M, Moore SB, Bowman M, Burgart LJ, Melton LJ 3rd, Murray JA. Predictors of family risk for celiac disease: a population-based study. Clin Gastroenterol Hepatol. 2008;6:983-987. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 114] [Cited by in RCA: 102] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 52. | Niland B, Cash BD. Health Benefits and Adverse Effects of a Gluten-Free Diet in Non-Celiac Disease Patients. Gastroenterol Hepatol (N Y). 2018;14:82-91. [PubMed] |
| 53. | Choung RS, Unalp-Arida A, Ruhl CE, Brantner TL, Everhart JE, Murray JA. Less Hidden Celiac Disease But Increased Gluten Avoidance Without a Diagnosis in the United States: Findings From the National Health and Nutrition Examination Surveys From 2009 to 2014. Mayo Clin Proc. 2016;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 89] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 54. | Lee AR, Wolf RL, Lebwohl B, Ciaccio EJ, Green PHR. Persistent Economic Burden of the Gluten Free Diet. Nutrients. 2019;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 88] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 55. | Catassi C, Elli L, Bonaz B, Bouma G, Carroccio A, Castillejo G, Cellier C, Cristofori F, de Magistris L, Dolinsek J, Dieterich W, Francavilla R, Hadjivassiliou M, Holtmeier W, Körner U, Leffler DA, Lundin KE, Mazzarella G, Mulder CJ, Pellegrini N, Rostami K, Sanders D, Skodje GI, Schuppan D, Ullrich R, Volta U, Williams M, Zevallos VF, Zopf Y, Fasano A. Diagnosis of Non-Celiac Gluten Sensitivity (NCGS): The Salerno Experts' Criteria. Nutrients. 2015;7:4966-4977. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 422] [Cited by in RCA: 380] [Article Influence: 38.0] [Reference Citation Analysis (0)] |
| 56. | Reese I, Schäfer C, Kleine-Tebbe J, Ahrens B, Bachmann O, Ballmer-Weber B, Beyer K, Bischoff SC, Blümchen K, Dölle S, Enck P, Enninger A, Huttegger I, Lämmel S, Lange L, Lepp U, Mahler V, Mönnikes H, Ockenga J, Otto B, Schnadt S, Szepfalusi Z, Treudler R, Wassmann-Otto A, Zuberbier T, Werfel T, Worm M. Non-celiac gluten/wheat sensitivity (NCGS)-a currently undefined disorder without validated diagnostic criteria and of unknown prevalence: Position statement of the task force on food allergy of the German Society of Allergology and Clinical Immunology (DGAKI). Allergo J Int. 2018;27:147-151. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 26] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 57. | Di Tola M, Sabbatella L, Anania MC, Viscido A, Caprilli R, Pica R, Paoluzi P, Picarelli A. Anti-tissue transglutaminase antibodies in inflammatory bowel disease: new evidence. Clin Chem Lab Med. 2004;42:1092-1097. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 22] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 58. | Lerner A, Ramesh A, Matthias T. Serologic Diagnosis of Celiac Disease: New Biomarkers. Gastroenterol Clin North Am. 2019;48:307-317. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 59. | Ferrara F, Quaglia S, Caputo I, Esposito C, Lepretti M, Pastore S, Giorgi R, Martelossi S, Dal Molin G, Di Toro N, Ventura A, Not T. Anti-transglutaminase antibodies in non-coeliac children suffering from infectious diseases. Clin Exp Immunol. 2010;159:217-223. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 50] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 60. | Villalta D, Crovatto M, Stella S, Tonutti E, Tozzoli R, Bizzaro N. False positive reactions for IgA and IgG anti-tissue transglutaminase antibodies in liver cirrhosis are common and method-dependent. Clin Chim Acta. 2005;356:102-109. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 40] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 61. | Kurien M, Chalkiadakis I, Evans K, Sanders DS. False-positive tissue transglutaminase antibody levels occur in HIV-positive patients: HLA typing is essential. J Clin Gastroenterol. 2012;46:346. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 2] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 62. | Freeman HJ. Strongly positive tissue transglutaminase antibody assays without celiac disease. Can J Gastroenterol. 2004;18:25-28. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 35] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 63. | Maglio M, Ziberna F, Aitoro R, Discepolo V, Lania G, Bassi V, Miele E, Not T, Troncone R, Auricchio R. Intestinal Production of Anti-Tissue Transglutaminase 2 Antibodies in Patients with Diagnosis Other Than Celiac Disease. Nutrients. 2017;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 22] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 64. | Ludvigsson JF, Neovius M, Hammarström L. Association between IgA deficiency & other autoimmune conditions: a population-based matched cohort study. J Clin Immunol. 2014;34:444-451. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 90] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 65. | Ferreira RC, Pan-Hammarström Q, Graham RR, Fontán G, Lee AT, Ortmann W, Wang N, Urcelay E, Fernández-Arquero M, Núñez C, Jorgensen G, Ludviksson BR, Koskinen S, Haimila K, Padyukov L, Gregersen PK, Hammarström L, Behrens TW. High-density SNP mapping of the HLA region identifies multiple independent susceptibility loci associated with selective IgA deficiency. PLoS Genet. 2012;8:e1002476. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 49] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 66. | Olerup O, Smith CI, Hammarström L. Different amino acids at position 57 of the HLA-DQ beta chain associated with susceptibility and resistance to IgA deficiency. Nature. 1990;347:289-290. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 86] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 67. | Olerup O, Smith CI, Björkander J, Hammarström L. Shared HLA class II-associated genetic susceptibility and resistance, related to the HLA-DQB1 gene, in IgA deficiency and common variable immunodeficiency. Proc Natl Acad Sci U S A. 1992;89:10653-10657. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 74] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 68. | Mohammadi J, Pourpak Z, Jarefors S, Saghafi S, Zendehdel K, Pourfathollah AA, Amirzargar AA, Aghamohammadi A, Moin M, Hammarstrom L. Human leukocyte antigens (HLA) associated with selective IgA deficiency in Iran and Sweden. Iran J Allergy Asthma Immunol. 2008;7:209-214. [PubMed] |
| 69. | Pavlovic M, Berenji K, Bukurov M. Screening of celiac disease in Down syndrome - Old and new dilemmas. World J Clin Cases. 2017;5:264-269. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 26] [Cited by in RCA: 17] [Article Influence: 2.1] [Reference Citation Analysis (2)] |
| 70. | Mårild K, Størdal K, Hagman A, Ludvigsson JF. Turner Syndrome and Celiac Disease: A Case-Control Study. Pediatrics. 2016;137:e20152232. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 34] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 71. | Henderson A, Lynch SA, Wilkinson S, Hunter M. Adults with Down's syndrome: the prevalence of complications and health care in the community. Br J Gen Pract. 2007;57:50-55. [PubMed] |
| 72. | Roy A, Laszkowska M, Sundström J, Lebwohl B, Green PH, Kämpe O, Ludvigsson JF. Prevalence of Celiac Disease in Patients with Autoimmune Thyroid Disease: A Meta-Analysis. Thyroid. 2016;26:880-890. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 61] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 73. | Jacobson EM, Huber A, Tomer Y. The HLA gene complex in thyroid autoimmunity: from epidemiology to etiology. J Autoimmun. 2008;30:58-62. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 201] [Cited by in RCA: 184] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 74. | Zeitlin AA, Heward JM, Newby PR, Carr-Smith JD, Franklyn JA, Gough SC, Simmonds MJ. Analysis of HLA class II genes in Hashimoto's thyroiditis reveals differences compared to Graves' disease. Genes Immun. 2008;9:358-363. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 75] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 75. | Ch'ng CL, Biswas M, Benton A, Jones MK, Kingham JG. Prospective screening for coeliac disease in patients with Graves' hyperthyroidism using anti-gliadin and tissue transglutaminase antibodies. Clin Endocrinol (Oxf). 2005;62:303-306. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 62] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 76. | Maciel LM, Rodrigues SS, Dibbern RS, Navarro PA, Donadi EA. Association of the HLA-DRB1*0301 and HLA-DQA1*0501 alleles with Graves' disease in a population representing the gene contribution from several ethnic backgrounds. Thyroid. 2001;11:31-35. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 40] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 77. | Kurada S, Yadav A, Leffler DA. Current and novel therapeutic strategies in celiac disease. Expert Rev Clin Pharmacol. 2016;9:1211-1223. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 22] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 78. | Leffler DA, Kelly CP. Update on the evaluation and diagnosis of celiac disease. Curr Opin Allergy Clin Immunol. 2006;6:191-196. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 10] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 79. | Veeraraghavan G, Leffler DA, Kaswala DH, Mukherjee R. Celiac disease 2015 update: new therapies. Expert Rev Gastroenterol Hepatol. 2015;9:913-927. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 17] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 80. | Leffler DA, Dennis M, Hyett B, Kelly E, Schuppan D, Kelly CP. Etiologies and predictors of diagnosis in nonresponsive celiac disease. Clin Gastroenterol Hepatol. 2007;5:445-450. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 231] [Cited by in RCA: 248] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 81. | Hujoel IA, Murray JA. Refractory Celiac Disease. Curr Gastroenterol Rep. 2020;22:18. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 50] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 82. | Ludvigsson JF, Leffler DA, Bai JC, Biagi F, Fasano A, Green PH, Hadjivassiliou M, Kaukinen K, Kelly CP, Leonard JN, Lundin KE, Murray JA, Sanders DS, Walker MM, Zingone F, Ciacci C. The Oslo definitions for coeliac disease and related terms. Gut. 2013;62:43-52. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1254] [Cited by in RCA: 1160] [Article Influence: 96.7] [Reference Citation Analysis (1)] |
| 83. | Al-Toma A, Goerres MS, Meijer JW, Peña AS, Crusius JB, Mulder CJ. Human leukocyte antigen-DQ2 homozygosity and the development of refractory celiac disease and enteropathy-associated T-cell lymphoma. Clin Gastroenterol Hepatol. 2006;4:315-319. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 143] [Cited by in RCA: 147] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 84. | Dieckman T, Schreurs M, Mahfouz A, Kooy-Winkelaar Y, Neefjes-Borst A, Bouma G, Koning F. Single-Cell Analysis of Refractory Celiac Disease Demonstrates Inter- and Intra-Patient Aberrant Cell Heterogeneity. Cell Mol Gastroenterol Hepatol. 2022;14:173-192. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 85. | Li Y, Liu H. Clinical powers of Aminoacyl tRNA Synthetase Complex Interacting Multifunctional Protein 1 (AIMP1) for head-neck squamous cell carcinoma. Cancer Biomark. 2022;34:359-374. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 32] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 86. | Liu H, Dong A, Rasteh AM, Wang P, Weng J. Identification of the novel exhausted T cell CD8 + markers in breast cancer. Sci Rep. 2024;14:19142. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 67] [Article Influence: 67.0] [Reference Citation Analysis (0)] |
| 87. | Liu H, Li Y. Potential roles of Cornichon Family AMPA Receptor Auxiliary Protein 4 (CNIH4) in head and neck squamous cell carcinoma. Cancer Biomark. 2022;35:439-450. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 40] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 88. | Brown I, Mino-Kenudson M, Deshpande V, Lauwers GY. Intraepithelial lymphocytosis in architecturally preserved proximal small intestinal mucosa: an increasing diagnostic problem with a wide differential diagnosis. Arch Pathol Lab Med. 2006;130:1020-1025. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 108] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 89. | Martins C, Teixeira C, Ribeiro S, Trabulo D, Cardoso C, Mangualde J, Freire R, Alves AL, Gamito É, Cremers I, Oliveira AP. Seronegative Intestinal Villous Atrophy: A Diagnostic Challenge. Case Rep Gastrointest Med. 2016;2016:6392028. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 3] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 90. | Oberhuber G. Histopathology of celiac disease. Biomed Pharmacother. 2000;54:368-372. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 181] [Cited by in RCA: 201] [Article Influence: 8.0] [Reference Citation Analysis (1)] |
| 91. | Serra P, Santamaria P. Nanoparticle-based approaches to immune tolerance for the treatment of autoimmune diseases. Eur J Immunol. 2018;48:751-756. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 44] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 92. | Singha S, Shao K, Yang Y, Clemente-Casares X, Solé P, Clemente A, Blanco J, Dai Q, Song F, Liu SW, Yamanouchi J, Umeshappa CS, Nanjundappa RH, Detampel P, Amrein M, Fandos C, Tanguay R, Newbigging S, Serra P, Khadra A, Chan WCW, Santamaria P. Peptide-MHC-based nanomedicines for autoimmunity function as T-cell receptor microclustering devices. Nat Nanotechnol. 2017;12:701-710. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 119] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 93. | Carballido JM, Santamaria P. Taming autoimmunity: Translating antigen-specific approaches to induce immune tolerance. J Exp Med. 2019;216:247-250. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 20] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 94. | Serra P, Santamaria P. Antigen-specific therapeutic approaches for autoimmunity. Nat Biotechnol. 2019;37:238-251. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 156] [Article Influence: 26.0] [Reference Citation Analysis (0)] |
| 95. | Abadie V, Kim SM, Lejeune T, Palanski BA, Ernest JD, Tastet O, Voisine J, Discepolo V, Marietta EV, Hawash MBF, Ciszewski C, Bouziat R, Panigrahi K, Horwath I, Zurenski MA, Lawrence I, Dumaine A, Yotova V, Grenier JC, Murray JA, Khosla C, Barreiro LB, Jabri B. IL-15, gluten and HLA-DQ8 drive tissue destruction in coeliac disease. Nature. 2020;578:600-604. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 127] [Cited by in RCA: 147] [Article Influence: 29.4] [Reference Citation Analysis (0)] |