Published online Mar 21, 2025. doi: 10.3748/wjg.v31.i11.101124
Revised: January 24, 2025
Accepted: February 17, 2025
Published online: March 21, 2025
Processing time: 189 Days and 1.4 Hours
Gastric cancer (GC) is one of the most aggressive malignancies worldwide and is characterized by its poor prognosis and resistance to conventional therapies. Autophagy and long non-coding RNAs (lncRNAs) play critical yet complex roles in GC, functioning as both tumor suppressors and promoters depending on the disease stage and context. Autophagy influences cellular homeostasis and metabolism, whereas lncRNAs regulate gene expression through epigenetic modifications, RNA sponging, and protein interactions. Notably, the interplay between lncRNAs and autophagy modulates tumor progression, metastasis, chemoresistance, and the tumor microenvironment. This study explored the intricate relationship between lncRNAs and autophagy in GC, highlighting their roles in pathogenesis and treatment resistance. By addressing current knowledge gaps and proposing innovative therapeutic strategies, we have emphasized the potential of targeting this dynamic interplay for improved diagnostic and therapeutic outcomes.
Core Tip: Autophagy and long non-coding RNAs play pivotal roles in the progression of gastric cancer (GC), driving key processes such as tumor growth, metastasis, chemoresistance, and immune evasion. While each functions distinctly, their intricate interplay amplifies their impact, reshaping the gastric tumor microenvironment and accelerating disease progression. Understanding these mechanisms and their connections not only deepens our knowledge of GC pathogenesis but also unlocks new opportunities for innovative, targeted therapeutic strategies to combat this challenging disease.
- Citation: Luong TV, Cao MTT, Nguyen NVD, Dang HNN, Nguyen TT. Roles of autophagy and long non-coding RNAs in gastric cancer. World J Gastroenterol 2025; 31(11): 101124
- URL: https://www.wjgnet.com/1007-9327/full/v31/i11/101124.htm
- DOI: https://dx.doi.org/10.3748/wjg.v31.i11.101124
Gastric cancer (GC) is one of the most aggressive malignancies worldwide and is characterized by rapid progression, high metastatic potential, and resistance to conventional therapies, resulting in dismal patient outcomes[1-3]. Current treatment strategies, including surgery and chemotherapy, are not highly effective in the advanced stages of the disease[3,4]. This pressing challenge highlights the need for a deeper understanding of GC pathophysiology to uncover novel therapeutic approaches. Two key molecular mechanisms that have gained significant attention in recent years are autophagy and long non-coding RNAs (lncRNAs), both of which play critical roles in the progression and treatment resistance of GC.
Autophagy is the principal mechanism that mediates the delivery of various cellular cargoes to lysosomes for degradation and recycling. Debnath et al[5] highlighted the essential protective functions of autophagy in various diseases. However, in the context of cancer, autophagy has contrasting roles. It acts to prevent early tumor development but also facilitates the maintenance and metabolic adaptation of advanced and metastasizing tumors. Research has shown that autophagy plays a key role in the occurrence, development, treatment, prognosis, and drug resistance of GC[6-8].
In a recent study, Chang et al[9] explored the role of autophagy in gastrointestinal diseases, shedding light on its complex functions. Their study revealed that autophagy acted as a prosurvival mechanism in benign gastrointestinal conditions, such as intestinal ischemia-reperfusion injury, inflammatory bowel disease, and motility disorders. We agree with the conclusion of Chang et al[9] that the dual nature of autophagy under pathological conditions depends on the extent of autophagic activity and the influence of concurrent factors.
Another layer of complexity in GC biology involves lncRNAs, coding RNA molecules that regulate critical cellular processes such as proliferation, apoptosis, metastasis, and chemoresistance[10]. Emerging evidence suggests that autophagy and lncRNAs are not independent mechanisms; instead, they are intimately linked, forming a regulatory network that significantly influences GC progression[11]. For example, specific lncRNAs can either activate or inhibit autophagy through signaling pathways, while autophagy reciprocally regulates lncRNA stability and activity[12,13]. This dynamic interplay not only drives tumor growth and survival but also offers a promising avenue for therapeutic intervention[14].
In this study, we investigated the individual roles of autophagy and lncRNAs in GC as well as their interconnected functions. By elucidating their crosstalk, we aimed to bridge critical knowledge gaps and propose new perspectives for targeted therapies and biomarker development. Expanding upon the foundational insights provided by Chang et al[9], our work highlighted the potential of these molecular pathways to transform the diagnosis and treatment of GC.
Autophagy is a ubiquitous biological phenomenon in eukaryotic cells that plays a pivotal role in maintaining cellular homeostasis and influencing pathophysiological processes, including cancer development[15]. In GC, autophagy has a context-dependent function. It suppresses tumor initiation in the early stages by preserving cellular integrity but supports tumor growth and survival in advanced stages under stress conditions. The following sections explore its mechanisms, contrasting roles, and implications for GC treatment.
Autophagy is a highly conserved biological process that involves the engulfment of unfolded proteins or damaged organelles by double-membrane cytosolic vesicles known as autophagosomes, which are then delivered to the lysosome for breakdown. This multistep process ensures cellular homeostasis through recycling macromolecules and removing potentially toxic components[5,16]. In mammals, autophagy is tightly controlled by core autophagy-related genes and complex signaling pathways. The process consists of four key stages: (1) Initiation; (2) Elongation; (3) Autophagosome formation; and (4) Lysosomal fusion and degradation[17,18]. For example, autophagy associated gene-5 (ATG5) and ATG12 play crucial roles in autophagosome expansion and elongation, whereas the Rab7 and SNARE proteins regulate fusion with lysosomes. Dysregulation of these pathways and genes has been implicated in various diseases, including GC, where autophagy can contribute to tumor suppression or progression depending on the context[19].
Autophagy regulates multiple processes involved in cancer development, including apoptosis, ferroptosis, metastasis, and cell cycle regulation. In GC, autophagy plays opposing roles depending on the tumor stage, acting as a double-edged sword. In the early stages, autophagy suppresses tumor initiation by maintaining genomic stability and eliminating damaged organelles and proteins. However, during tumor progression, autophagy supports cancer cell survival by mitigating metabolic stress and facilitating adaptation to the tumor microenvironment (TME), thereby promoting tumor growth and metastasis[20,21].
Autophagy is tightly regulated by ATG genes and various signaling pathways. Key oncogenic regulators, such as Bcl-2/Bcl-XL, protein kinase B (AKT), and mammalian target of rapamycin-1 (mTORC1), often increase autophagy in advanced cancer stages, whereas tumor-suppressive proteins, such as Beclin-1, p53, and phosphatase and tensin homolog deleted on chromosome ten, modulate its inhibitory effects on tumor initiation[22,23]. Non-coding RNAs, including microRNAs, lncRNAs and circular RNAs, also play pivotal roles in regulating autophagy[24]. For example, miR-183 promotes GC development by modulating autophagy through the sirtuin 1 (SIRT1) and phosphoinositide 3-kinase (PI3K)/AKT/mTOR pathways, whereas the overexpression of miR-543 inhibits autophagy by targeting SIRT1, enhancing the proliferation, migration, and invasion of GC cells[25,26]. Similarly, Lnc-SNHG11 has been shown to drive oncogenic autophagy, facilitating epithelial-to-mesenchymal transition (EMT), proliferation, and metastasis via activation of the Wnt/β-catenin pathway[6].
Autophagy plays a complex role in the treatment of GC, acting as both a potential therapeutic target and a mechanism of drug resistance. On the one hand, enhancing autophagy has been proposed as a strategy to induce cancer cell death, with some anticancer drugs designed to activate autophagy for this purpose[27,28]. On the other hand, autophagy is often activated in tumor cells as a stress response, enabling them to survive chemotherapy and radiotherapy by mitigating cellular damage and metabolic stress[27,29,30].
Drug resistance remains a major challenge in GC therapy, and understanding the mechanisms by which autophagy contributes to this phenomenon is crucial for developing more effective treatments. ATG lncRNAs have been shown to play critical regulatory roles in this process. For example, YiRen et al[31] demonstrated that long non-coding metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) enhances autophagy-mediated drug resistance by sponging miR-23b-3p, leading to increased autophagic activity in drug-resistant GC cells. Similarly, the lncRNA Colorectal Neoplasia Differentially Expressed reduces chemoresistance by modulating alternative splicing of PICALM via SRSF6, while circCUL2 promotes cisplatin resistance through autophagy activation mediated by the miR-142-3p/Rho-associated coiled-coil-containing protein kinase 2 axis[32,33]. These findings highlight the intricate interplay between autophagy and lncRNAs in regulating GC cell survival under therapeutic stress.
LncRNAs are RNA molecules longer than 200 nucleotides that do not encode proteins but strongly regulate gene expression and various critical biological processes[10]. These functions are achieved through three main mechanisms. First, through epigenetic regulation, lncRNAs interact with DNA or histone proteins to modify chromatin structure and regulate gene expression. For example, the lncRNA HOTAIR has been shown to activate methyltransferase enzymes to modulate gene methylation, thereby promoting the expression of oncogenic genes[10]. Second, lncRNAs act as sponges for microRNAs, competing with them to prevent mRNA degradation and maintain the expression of key proteins. An example is MALAT1, which sponges miR-23b-3p, enhancing autophagy and supporting cancer cell survival in adverse environments[34]. Third, lncRNAs interact with proteins to regulate the activity of oncogenic signaling pathways, such as the mTOR pathway, which controls cell growth and proliferation[12]. Dysregulation of these mechanisms has significant implications in GC, where lncRNAs contribute to tumor biology, including uncontrolled proliferation, metastasis, and resistance to chemotherapy.
LncRNAs play a central role in various biological processes associated with GC, including proliferation, metastasis, chemoresistance, and interactions with the TME. The diverse functions of these genes underscore their potential as key regulators in GC pathogenesis.
Proliferation and metastasis: Proliferation and metastasis are hallmarks of GC, and lncRNAs are key regulators of these processes[35]. For example, AC093818.1, an oncogenic lncRNA, is markedly upregulated during the advanced stages of GC. Studies have shown that AC093818.1 epigenetically activates the expression of phosphoinositide-dependent kinase 1, a crucial regulator of energy metabolism and cell growth, thereby promoting the metastatic potential of cancer cells[35]. Additionally, MAGI2-AS3, another oncogenic lncRNA, supports EMT (a critical step in metastasis) by competing with miR-141/200a to sustain the expression of zinc finger E-box binding homology box 1, a key transcription factor that drives cell motility and invasiveness[36].
Conversely, FENDRR is known as a tumor-suppressive lncRNA that inhibits metastasis. It downregulates the expression of fibronectin-1, a protein that enhances cancer cell invasiveness, thereby reducing the migratory and invasive capabilities of GC cells[37]. These findings underscore the role of lncRNAs in regulating proliferation and metastasis, highlighting their potential as therapeutic targets to inhibit GC progression.
Chemoresistance: Chemoresistance poses a significant challenge in GC treatment, with lncRNAs playing pivotal roles[34]. MALAT1, an oncogenic lncRNA, enhances chemoresistance through autophagy activation. MALAT1 sponges miR-23b-3p, leading to increased expression of ATG12, a key gene in autophagy, allowing cancer cells to survive under harsh chemotherapeutic conditions[34]. Furthermore, DS cell adhesion molecule antisense RNA 1 (DSCAM-AS1) has been shown to confer resistance to taxane-based therapies by regulating the miR-204/SOX4 axis[38]. These mechanisms demonstrate that lncRNAs not only enable GC cells to endure treatment-induced stress but also diminish the efficacy of standard chemotherapies. Targeting lncRNAs such as MALAT1 could offer a promising approach to enhancing che
Energy metabolism and the TME: Energy metabolism and TME adaptation are critical in GC progression, with lncRNAs playing key regulatory roles. H19 modulates glucose and lipid metabolism by upregulating hexokinase 2, providing energy for tumor growth and enhancing adaptability under adverse conditions. Single nucleotide polymorphisms (SNPs) in the promoter region of H19 further increase its expression, promoting cancer cell invasiveness and metastasis[39]. Similarly, SOX2-overlapping transcript (OT) supports lipid metabolism while impairing immune cell function in the TME, weakening the activity of T cells and macrophages and thus allowing cancer cells to evade immune surveillance[40]. Additionally, HAGLROS enhances cancer cell survival by activating mTORC1, regulating autophagy, and facilitating adaptation under metabolic stress[12]. These findings suggest that lncRNAs not only regulate energy metabolism but also reshape the TME to promote cancer progression, opening new avenues for therapeutic intervention by targeting these key lncRNAs.
Collectively, these insights illustrate the multifaceted roles of lncRNAs in GC pathogenesis, providing a strong rationale for their exploration as therapeutic targets to mitigate disease progression and improve patient outcomes.
SNPs in lncRNAs significantly influence GC risk and prognosis by altering lncRNA expression and function. For example, the SNP rs2795025 in MALAT1 enhances ATG12 expression by sponging miR-23b-3p, leading to autophagy activation and chemoresistance. This SNP is associated with poor prognosis and increased recurrence risk[41]. Similarly, SNPs in HOXD-AS1 promote tumor invasiveness and poor treatment response by activating the Janus kinase/signal transducer and activator of transcription 3 (STAT3) axis[42]. Furthermore, the SNP rs2839698 in the promoter region of H19 supports glucose metabolism and increases metastatic potential, further aggravating GC progression[39]. These SNPs highlight the intricate relationship between genetic variants and GC pathogenesis. They not only serve as effective prognostic biomarkers but also open new avenues for personalized therapies, with the potential to improve survival outcomes and reduce recurrence in GC patients.
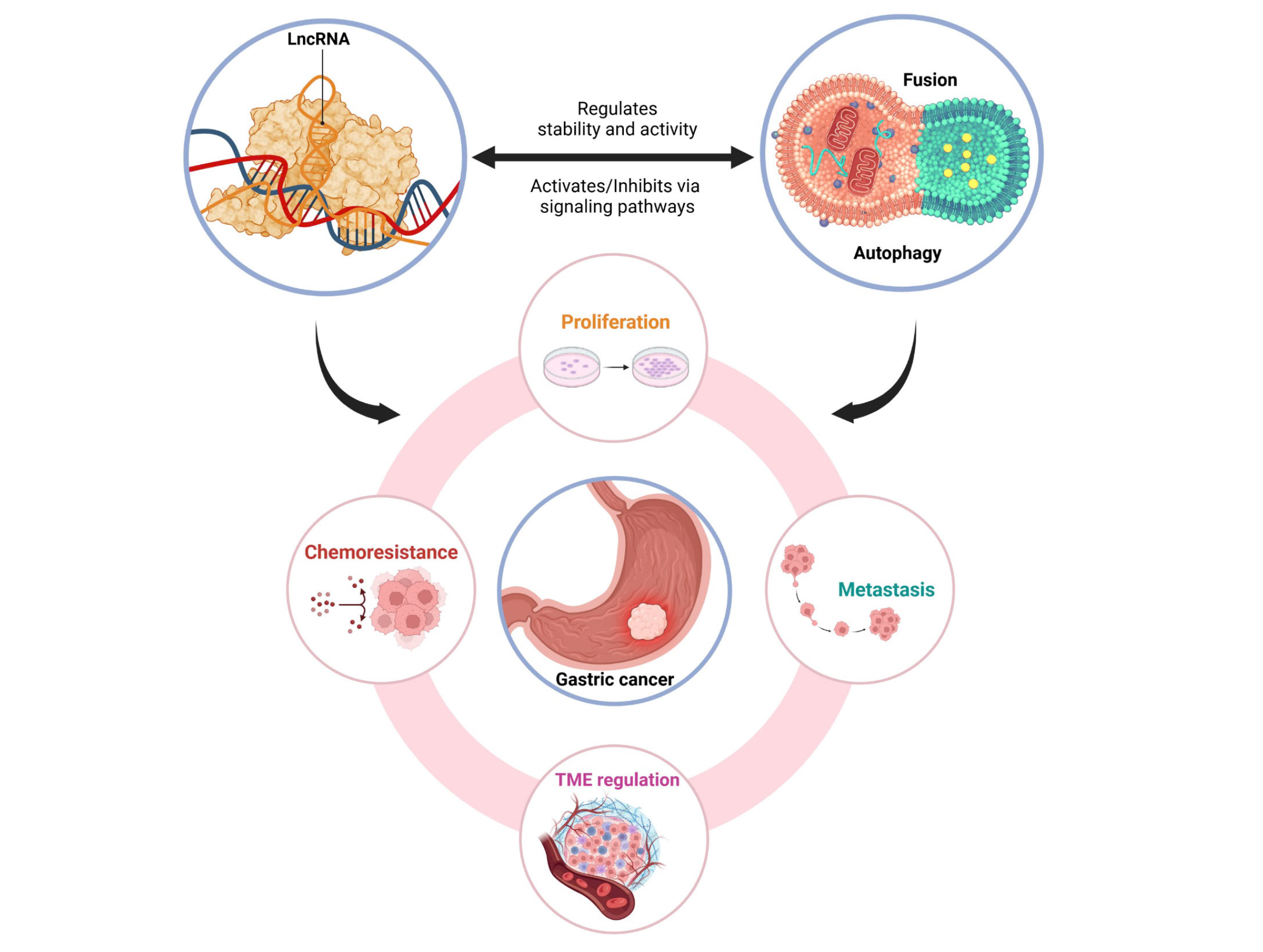
The interaction between lncRNAs and autophagy in GC is a complex and emerging area of research. LncRNAs not only regulate autophagy through diverse mechanisms but are also influenced by it, forming a dynamic feedback loop that impacts GC progression and therapeutic response. Figure 1 visually illustrates this relationship by highlighting the key regulatory mechanisms and biological impacts they exert in the context of GC.
Activation of autophagy: LncRNAs can modulate autophagy by activating or inhibiting critical pathways. For example, MALAT1 enhances autophagy by sponging miR-23b-3p and upregulating ATG12, enabling cancer cells to survive under stress conditions[12]. According to Lu et al[13], H19 also participates in autophagy activation by regulating energy metabolism signaling pathways, aiding cancer cells in adapting to the harsh TME. Additionally, GBCDRlnc1, although more extensively studied in gallbladder cancer, has been shown to activate autophagy via the mTOR pathway, a mechanism likely conserved in GC, thereby enhancing chemoresistance[8]. Wang et al[11] further emphasized that lncRNAs such as LINC00963 and HULC activate autophagy, playing critical roles in maintaining cancer cell survival under hypoxic or chemotherapeutic stress. These findings underscore the central role of lncRNAs in activating auto
Inhibition of autophagy: Some lncRNAs inhibit autophagy, thereby promoting tumor growth and progression. HAGLROS, which is regulated by STAT3 signaling, activates the mTORC1 pathway to suppress autophagy, subsequently driving GC cell proliferation and invasion[12]. ARHGAP5-AS1 similarly inhibits autophagy by stabilizing oncogenic mRNAs through m6A modification, thereby enhancing chemoresistance[43]. Wang et al[11] noted that autophagy suppression can lead to the accumulation of oncogenic factors and increased metastasis potential. They highlighted the roles of lncRNAs such as NEAT1 and MALAT1 in these processes[11]. These findings suggest that lncRNAs not only flexibly control autophagy but also exploit this mechanism to drive tumor progression and therapeutic resistance in GC.
Autophagy is not only regulated by lncRNAs but also affects their expression and stability reciprocally. It can degrade oncogenic lncRNAs, indirectly reducing tumorigenic activity. Conversely, autophagy also maintains homeostasis to protect essential lncRNAs required for cancer cell survival and proliferation[14]. This bidirectional relationship between lncRNAs and autophagy underscores their pivotal role in the biological regulation of cancer cells (Figure 1).
Proliferation and metastasis: Autophagy and lncRNAs collaborate to promote proliferation and metastasis through complex regulatory mechanisms. MAGI2-AS3 supports EMT by modulating autophagy, providing the energy and nutrients essential for the migration and invasion of GC cells[36]. Additionally, LINC00963 has been reported to increase autophagy via the miR-4458/ATG16L1 axis, thereby facilitating tumor growth and invasion[44]. According to Lu et al[13], lncRNAs such as H19 play pivotal roles in supporting autophagy, thereby promoting cancer cell proliferation and adaptation under adverse conditions such as hypoxia or nutrients. Conversely, FENDRR, a tumor-suppressive lncRNA, inhibits autophagy and reduces the activity of metastasis-promoting factors, thereby limiting GC progression[37].
Chemoresistance: Autophagy and lncRNAs are critical in modulating chemoresistance in GC cells. MALAT1 activates autophagy to remove the toxic byproducts of chemotherapy, reducing treatment efficacy and increasing cancer cell survival[12]. DSCAM-AS1 regulates autophagy through the miR-204/SOX4 axis, protecting cancer cells from taxane-based chemotherapy and increasing drug resistance[38]. Wang et al[11] reported that lncRNAs such as GBCDRlnc1 and HULC support cancer cell survival under chemotherapy-induced stress by modulating autophagy. Furthermore, ARHGAP5-AS1 hinders autophagic degradation, leading to the accumulation of protective factors against chemotherapeutic agents[43]. Notably, the circular RNA multiple C2 domain containing transmembrane protein (MCTP2) has been shown to reduce cisplatin resistance in GC by regulating Myotubularin-related phosphatase 3 expression via miR-99a-5p, thereby limiting autophagy and enhancing chemotherapeutic efficacy[45].
While immunotherapy has transformed the landscape of cancer treatment, its effectiveness in GC is often hampered by immunosuppressive TMEs[46]. LncRNAs and autophagy collaborate to influence key components of the TME, supporting cancer cell survival and therapy resistance. LncRNAs such as SOX2-OT modulate autophagy in immune cells, enabling the TME to evade immune surveillance[40]. Moreover, autophagy stabilizes oncogenic lncRNAs such as H19, enhancing cancer cell adaptability under hypoxic or nutrient-deficient conditions[39]. This bidirectional relationship ensures that cancer cells can thrive despite environmental stressors. Additionally, Wang et al[11] reported that autophagy protects key lncRNAs such as MCTP2, sustaining cellular homeostasis and enabling cancer cells to overcome chemotherapeutic stress.
The relationship between lncRNAs and autophagy in GC involves complex interactions, which play central roles in proliferation, metastasis, chemoresistance, and interactions with the TME. These insights not only shed light on the fundamental biological mechanisms of cancer but also suggest the potential application of lncRNAs and autophagy as novel therapeutic targets.
While substantial progress has been made in understanding the roles of autophagy and lncRNAs in GC, several critical gaps remain. First, the dynamic interplay between autophagy and lncRNAs is not fully understood, particularly how lncRNAs regulate autophagy through signaling pathways such as the PI3K/AKT/mTOR pathway and how autophagy reciprocally influences lncRNA stability and function. These interactions remain poorly defined across different stages of GC progression.
Second, heterogeneity in lncRNA and ATG gene expression among GC subtypes and patient populations limits the development of universal biomarkers. SNPs in lncRNAs, which may affect GC risk and therapy response, have not been systematically studied, particularly in diverse populations. This lack of data hinders personalized treatment strategies.
Third, while lncRNAs such as SOX2-OT and H19 are implicated in reshaping the TME, the role of autophagy in mediating these effects remains unclear. This limits the potential for developing therapies that target both tumor cells and their microenvironments.
Finally, translational research on leveraging ATG lncRNAs for therapeutic purposes is limited. Although preclinical studies suggest that these molecules are promising targets, clinical trials evaluating their efficacy and safety are scarce, and the absence of precise tools for selective modulation presents additional challenges.
To address these gaps, integrating advanced technologies is essential. Multiomics approaches, including genomics and single-cell RNA sequencing, can map interactions between autophagy and lncRNAs, offering insights into their roles across tumor stages and subtypes. These tools can also help identify dual biomarkers, such as MALAT1 and H19, that combine autophagy and lncRNA profiles to improve early detection and prognosis.
Therapeutic innovations should focus on nanotechnology-based delivery systems to selectively target ATG lncRNAs. For example, inhibiting the expression of oncogenic lncRNAs such as MALAT1 or increasing the expression of tumor-suppressive lncRNAs such as FENDRR could increase treatment specificity while minimizing off-target effects. Combining autophagy modulators with chemotherapy or immunotherapy could also overcome drug resistance, a major challenge in GC treatment.
Personalized medicine, which involves the stratification of patients on the basis of their unique lncRNA and autophagy profiles, is critical. Clinical trials targeting specific SNPs in lncRNAs, such as H19, can refine therapeutic approaches. Additionally, robust in vivo models, such as patient-derived xenografts, should be employed to validate these strategies before clinical application.
Exploring the immunomodulatory roles of ATG lncRNAs within the TME is another promising avenue. Targeting lncRNAs that regulate immune evasion, and metabolic reprogramming could increase the effectiveness of immunotherapies. Understanding these mechanisms will be key to designing combination therapies that address both tumor cells and their microenvironment.
In conclusion, addressing the interplay between autophagy and lncRNAs in GC through innovative technologies and personalized approaches has the potential to revolutionize diagnosis and treatment. By bridging these knowledge gaps, future research can pave the way for targeted therapies and improved outcomes for patients with GC.
GC remains a formidable clinical challenge because of its aggressive progression, late detection, and therapeutic resistance. This review underscored the multifaceted roles of autophagy and lncRNAs in GC, demonstrating their significant contributions to tumor biology, from proliferation and metastasis to chemoresistance and immune modulation. Despite substantial progress, critical gaps in understanding of the dynamic interplay between these mechanisms persist, particularly in diverse patient populations and tumor contexts. Bridging these gaps through advanced research and personalized approaches holds transformative potential for GC management. By targeting ATG lncRNAs and leveraging innovations such as nanotechnology and multiomics, future therapies could achieve greater specificity and effectiveness, ultimately improving outcomes for patients worldwide.
| 1. | Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75126] [Cited by in RCA: 64587] [Article Influence: 16146.8] [Reference Citation Analysis (176)] |
| 2. | Arnold M, Abnet CC, Neale RE, Vignat J, Giovannucci EL, McGlynn KA, Bray F. Global Burden of 5 Major Types of Gastrointestinal Cancer. Gastroenterology. 2020;159:335-349. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 857] [Cited by in RCA: 1229] [Article Influence: 245.8] [Reference Citation Analysis (0)] |
| 3. | Voeten DM, van Berge Henegouwen MI. ASO Author Reflections: Failure to Cure in Patients Undergoing Surgery for Gastric Cancer: A Nationwide Cohort Study. Ann Surg Oncol. 2021;28:4497-4498. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (1)] |
| 4. | Wöhrer SS, Raderer M, Hejna M. Palliative chemotherapy for advanced gastric cancer. Ann Oncol. 2004;15:1585-1595. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 138] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 5. | Debnath J, Gammoh N, Ryan KM. Autophagy and autophagy-related pathways in cancer. Nat Rev Mol Cell Biol. 2023;24:560-575. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 105] [Cited by in RCA: 679] [Article Influence: 339.5] [Reference Citation Analysis (0)] |
| 6. | Wu Q, Ma J, Wei J, Meng W, Wang Y, Shi M. lncRNA SNHG11 Promotes Gastric Cancer Progression by Activating the Wnt/β-Catenin Pathway and Oncogenic Autophagy. Mol Ther. 2021;29:1258-1278. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 145] [Cited by in RCA: 142] [Article Influence: 35.5] [Reference Citation Analysis (0)] |
| 7. | Li GM, Li L, Li MQ, Chen X, Su Q, Deng ZJ, Liu HB, Li B, Zhang WH, Jia YX, Wang WJ, Ma JY, Zhang HL, Xie D, Zhu XF, He YL, Guan XY, Bi J. DAPK3 inhibits gastric cancer progression via activation of ULK1-dependent autophagy. Cell Death Differ. 2021;28:952-967. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 54] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 8. | Cao Y, Luo Y, Zou J, Ouyang J, Cai Z, Zeng X, Ling H, Zeng T. Autophagy and its role in gastric cancer. Clin Chim Acta. 2019;489:10-20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 123] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 9. | Chang YF, Li JJ, Liu T, Wei CQ, Ma LW, Nikolenko VN, Chang WL. Morphological and biochemical characteristics associated with autophagy in gastrointestinal diseases. World J Gastroenterol. 2024;30:1524-1532. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 3] [Reference Citation Analysis (0)] |
| 10. | Deng K, Wang H, Guo X, Xia J. The cross talk between long, non-coding RNAs and microRNAs in gastric cancer. Acta Biochim Biophys Sin (Shanghai). 2016;48:111-116. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 41] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 11. | Wang Z, Liu J, Xie J, Yuan X, Wang B, Shen W, Zhang Y. Regulation of autophagy by non-coding RNAs in gastric cancer. Front Oncol. 2022;12:947332. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 12. | Chen JF, Wu P, Xia R, Yang J, Huo XY, Gu DY, Tang CJ, De W, Yang F. STAT3-induced lncRNA HAGLROS overexpression contributes to the malignant progression of gastric cancer cells via mTOR signal-mediated inhibition of autophagy. Mol Cancer. 2018;17:6. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 130] [Cited by in RCA: 193] [Article Influence: 27.6] [Reference Citation Analysis (0)] |
| 13. | Lu L, Liang Q, Zhang X, Xu Y, Meng D, Liang Z. Autophagy Related Noncoding RNAs: Emerging Regulatory Factors of Gastric Cancer. Cancer Manag Res. 2022;14:2215-2224. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Reference Citation Analysis (0)] |
| 14. | Saha S, Panigrahi DP, Patil S, Bhutia SK. Autophagy in health and disease: A comprehensive review. Biomed Pharmacother. 2018;104:485-495. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 280] [Cited by in RCA: 384] [Article Influence: 54.9] [Reference Citation Analysis (0)] |
| 15. | Klionsky DJ, Petroni G, Amaravadi RK, Baehrecke EH, Ballabio A, Boya P, Bravo-San Pedro JM, Cadwell K, Cecconi F, Choi AMK, Choi ME, Chu CT, Codogno P, Colombo MI, Cuervo AM, Deretic V, Dikic I, Elazar Z, Eskelinen EL, Fimia GM, Gewirtz DA, Green DR, Hansen M, Jäättelä M, Johansen T, Juhász G, Karantza V, Kraft C, Kroemer G, Ktistakis NT, Kumar S, Lopez-Otin C, Macleod KF, Madeo F, Martinez J, Meléndez A, Mizushima N, Münz C, Penninger JM, Perera RM, Piacentini M, Reggiori F, Rubinsztein DC, Ryan KM, Sadoshima J, Santambrogio L, Scorrano L, Simon HU, Simon AK, Simonsen A, Stolz A, Tavernarakis N, Tooze SA, Yoshimori T, Yuan J, Yue Z, Zhong Q, Galluzzi L, Pietrocola F. Autophagy in major human diseases. EMBO J. 2021;40:e108863. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 555] [Cited by in RCA: 981] [Article Influence: 245.3] [Reference Citation Analysis (0)] |
| 16. | Beilankouhi EAV, Valilo M, Dastmalchi N, Teimourian S, Safaralizadeh R. The Function of Autophagy in the Initiation, and Development of Breast Cancer. Curr Med Chem. 2024;31:2974-2990. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 5] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 17. | Nair U, Klionsky DJ. Molecular mechanisms and regulation of specific and nonspecific autophagy pathways in yeast. J Biol Chem. 2005;280:41785-41788. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 113] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 18. | Levy JMM, Towers CG, Thorburn A. Targeting autophagy in cancer. Nat Rev Cancer. 2017;17:528-542. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1382] [Cited by in RCA: 1946] [Article Influence: 243.3] [Reference Citation Analysis (0)] |
| 19. | Zhang J, Xiang Q, Wu M, Lao YZ, Xian YF, Xu HX, Lin ZX. Autophagy Regulators in Cancer. Int J Mol Sci. 2023;24:10944. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 17] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 20. | Cao Z, Zhang H, Cai X, Fang W, Chai D, Wen Y, Chen H, Chu F, Zhang Y. Luteolin Promotes Cell Apoptosis by Inducing Autophagy in Hepatocellular Carcinoma. Cell Physiol Biochem. 2017;43:1803-1812. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 100] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 21. | Maiuri MC, Tasdemir E, Criollo A, Morselli E, Vicencio JM, Carnuccio R, Kroemer G. Control of autophagy by oncogenes and tumor suppressor genes. Cell Death Differ. 2009;16:87-93. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 305] [Cited by in RCA: 344] [Article Influence: 20.2] [Reference Citation Analysis (0)] |
| 22. | Ávalos Y, Canales J, Bravo-Sagua R, Criollo A, Lavandero S, Quest AF. Tumor suppression and promotion by autophagy. Biomed Res Int. 2014;2014:603980. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 118] [Cited by in RCA: 138] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 23. | Rakesh R, PriyaDharshini LC, Sakthivel KM, Rasmi RR. Role and regulation of autophagy in cancer. Biochim Biophys Acta Mol Basis Dis. 2022;1868:166400. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 107] [Article Influence: 35.7] [Reference Citation Analysis (0)] |
| 24. | Yan H, Bu P. Non-coding RNA in cancer. Essays Biochem. 2021;65:625-639. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 386] [Article Influence: 96.5] [Reference Citation Analysis (0)] |
| 25. | Li H, He C, Wang X, Wang H, Nan G, Fang L. MicroRNA-183 affects the development of gastric cancer by regulating autophagy via MALAT1-miR-183-SIRT1 axis and PI3K/AKT/mTOR signals. Artif Cells Nanomed Biotechnol. 2019;47:3163-3171. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 45] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 26. | Shi Y, Yang Z, Zhang T, Shen L, Li Y, Ding S. SIRT1-targeted miR-543 autophagy inhibition and epithelial-mesenchymal transition promotion in Helicobacter pylori CagA-associated gastric cancer. Cell Death Dis. 2019;10:625. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 45] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 27. | Ahmadi-Dehlaghi F, Mohammadi P, Valipour E, Pournaghi P, Kiani S, Mansouri K. Autophagy: A challengeable paradox in cancer treatment. Cancer Med. 2023;12:11542-11569. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 30] [Reference Citation Analysis (0)] |
| 28. | Yun CW, Lee SH. The Roles of Autophagy in Cancer. Int J Mol Sci. 2018;19:3466. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 377] [Cited by in RCA: 684] [Article Influence: 97.7] [Reference Citation Analysis (0)] |
| 29. | Huang T, Song X, Yang Y, Wan X, Alvarez AA, Sastry N, Feng H, Hu B, Cheng SY. Autophagy and Hallmarks of Cancer. Crit Rev Oncog. 2018;23:247-267. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 82] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 30. | Carew JS, Kelly KR, Nawrocki ST. Autophagy as a target for cancer therapy: new developments. Cancer Manag Res. 2012;4:357-365. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 81] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 31. | YiRen H, YingCong Y, Sunwu Y, Keqin L, Xiaochun T, Senrui C, Ende C, XiZhou L, Yanfan C. Long noncoding RNA MALAT1 regulates autophagy associated chemoresistance via miR-23b-3p sequestration in gastric cancer. Mol Cancer. 2017;16:174. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 189] [Cited by in RCA: 277] [Article Influence: 34.6] [Reference Citation Analysis (0)] |
| 32. | Peng L, Sang H, Wei S, Li Y, Jin D, Zhu X, Li X, Dang Y, Zhang G. circCUL2 regulates gastric cancer malignant transformation and cisplatin resistance by modulating autophagy activation via miR-142-3p/ROCK2. Mol Cancer. 2020;19:156. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 70] [Cited by in RCA: 175] [Article Influence: 35.0] [Reference Citation Analysis (0)] |
| 33. | Zhang F, Wang H, Yu J, Yao X, Yang S, Li W, Xu L, Zhao L. LncRNA CRNDE attenuates chemoresistance in gastric cancer via SRSF6-regulated alternative splicing of PICALM. Mol Cancer. 2021;20:6. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 141] [Article Influence: 35.3] [Reference Citation Analysis (0)] |
| 34. | Zhou J, Wang M, Mao A, Zhao Y, Wang L, Xu Y, Jia H, Wang L. Long noncoding RNA MALAT1 sponging miR-26a-5p to modulate Smad1 contributes to colorectal cancer progression by regulating autophagy. Carcinogenesis. 2021;42:1370-1379. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 27] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 35. | Ba MC, Ba Z, Long H, Cui SZ, Gong YF, Yan ZF, Lin KP, Wu YB, Tu YN. LncRNA AC093818.1 accelerates gastric cancer metastasis by epigenetically promoting PDK1 expression. Cell Death Dis. 2020;11:64. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 35] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 36. | Li D, Wang J, Zhang M, Hu X, She J, Qiu X, Zhang X, Xu L, Liu Y, Qin S. LncRNA MAGI2-AS3 Is Regulated by BRD4 and Promotes Gastric Cancer Progression via Maintaining ZEB1 Overexpression by Sponging miR-141/200a. Mol Ther Nucleic Acids. 2020;19:109-123. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 76] [Cited by in RCA: 101] [Article Influence: 20.2] [Reference Citation Analysis (0)] |
| 37. | Xu TP, Huang MD, Xia R, Liu XX, Sun M, Yin L, Chen WM, Han L, Zhang EB, Kong R, De W, Shu YQ. Decreased expression of the long non-coding RNA FENDRR is associated with poor prognosis in gastric cancer and FENDRR regulates gastric cancer cell metastasis by affecting fibronectin1 expression. J Hematol Oncol. 2014;7:63. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 181] [Cited by in RCA: 226] [Article Influence: 20.5] [Reference Citation Analysis (0)] |
| 38. | Lu C, Xie T, Guo X, Wu D, Li S, Li X, Lu Y, Wang X. LncRNA DSCAM-AS1 Promotes Colon Cancer Cells Proliferation and Migration via Regulating the miR-204/SOX4 Axis. Cancer Manag Res. 2020;12:4347-4356. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 13] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 39. | Yang J, Tan F, Chen Y, Li X, Yuan C. The emerging role of long non-coding RNA SOX2-OT in cancers and non-malignant diseases. J Physiol Biochem. 2024;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Reference Citation Analysis (0)] |
| 40. | Tang SY, Zhou PJ, Meng Y, Zeng FR, Deng GT. Gastric cancer: An epigenetic view. World J Gastrointest Oncol. 2022;14:90-109. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 2] [Cited by in RCA: 14] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 41. | Jiang J, Jia ZF, Cao DH, Wu YH, Sun ZW, Cao XY. Association of the miR-146a rs2910164 polymorphism with gastric cancer susceptibility and prognosis. Future Oncol. 2016;12:2215-2226. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 37] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 42. | Zheng L, Chen J, Zhou Z, He Z. Knockdown of long non-coding RNA HOXD-AS1 inhibits gastric cancer cell growth via inactivating the JAK2/STAT3 pathway. Tumour Biol. 2017;39:1010428317705335. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 45] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 43. | Zhu L, Zhu Y, Han S, Chen M, Song P, Dai D, Xu W, Jiang T, Feng L, Shin VY, Wang X, Jin H. Impaired autophagic degradation of lncRNA ARHGAP5-AS1 promotes chemoresistance in gastric cancer. Cell Death Dis. 2019;10:383. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 74] [Cited by in RCA: 143] [Article Influence: 23.8] [Reference Citation Analysis (0)] |
| 44. | Hou M, Li C, Dong S. LINC00963/miR-4458 regulates the effect of oxaliplatin in gastric cancer by mediating autophagic flux through targeting of ATG16L1. Sci Rep. 2021;11:20951. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 45. | Sun G, Li Z, He Z, Wang W, Wang S, Zhang X, Cao J, Xu P, Wang H, Huang X, Xia Y, Lv J, Xuan Z, Jiang T, Fang L, Yang J, Zhang D, Xu H, Xu Z. Circular RNA MCTP2 inhibits cisplatin resistance in gastric cancer by miR-99a-5p-mediated induction of MTMR3 expression. J Exp Clin Cancer Res. 2020;39:246. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 70] [Cited by in RCA: 76] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 46. | Pi YN, Qi WC, Xia BR, Lou G, Jin WL. Long Non-Coding RNAs in the Tumor Immune Microenvironment: Biological Properties and Therapeutic Potential. Front Immunol. 2021;12:697083. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 49] [Article Influence: 12.3] [Reference Citation Analysis (0)] |