Published online Mar 14, 2025. doi: 10.3748/wjg.v31.i10.99583
Revised: December 26, 2024
Accepted: January 23, 2025
Published online: March 14, 2025
Processing time: 125 Days and 18.6 Hours
As a non-coding RNA molecule, circular RNAs (circRNAs) have significant specificity, and existing data suggest a close relationship between them and the prognosis of patients with gastric cancer (GC). However, this mechanism has no evidence yet. This article explores the functions of hsa_circRNA_102415 in the malignant behavior and potential downstream signaling of GC cells. The chosen approach is loss of signal and functional gain.
To investigate and analyze the relationship between hsa_circRNA_102415 and GC and explore its specific role. Results provide reference for other researchers to develop targeted treatment plans.
The gene expression omnibus (GEO) database can be used to obtain the microarray dataset GSE83521. Data were analyzed using the GEO2R tool to identify differences in circRNAs between normal and GC samples. Quantitative real-time polymerase chain reaction was used to detect differentially expressed genes in GC tissue samples and adjacent cancer tissue samples. GC cells were transfected with small interfering-hsa_circRNA_104415 and plasmid DNA (pcDNA)-hsa_ircRNA_102415. Multiple detection methods, such as Transwell and cell counting kit 8, were used to evaluate cellular physiological activities, including cell invasion and proliferation. The relationship between Wnt family members 2B, microRNA (miR)-4529-5p, etc., including argonaute 2-RNA immunoprecipitation and luciferase reporter genes was analyzed. Rescue experiments were conducted to analyze and explore the relationship between the malignant behavior of GC cells and hsa_circRNA_102415.
GEO2R analysis confirmed that hsa_circRNA_102415 had significantly higher expression levels in disease tissues. hsa_circRNA_102415 and miR-4529-5p showed a negative correlation in disease cells,
miR-4529-5p was used to successfully activate the potential of WNT2B, clarify the role of hsa_circRNA_102415 in GC cells, and provide reference for other researchers to develop targeted treatment plans.
Core Tip: This article explores the functions of circular RNAs (hsa_circRNA_102415) in the malignant behavior and potential downstream signaling of gastric cancer (GC) cells. The experimental content of this article utilizes sponge like microRNA-4529-5p to successfully activate the potential of WNT2B, clarify the role of hsa_circRNA_102415 in GC cells, and provide reference for other researchers to develop targeted treatment plans.
- Citation: Bai W, Guo ZL, Guo JH, Li F, Bu P, Liu J. Circular RNA contributes to gastric cancer by targeting Wnt family member 2B as a competing endogenous RNA. World J Gastroenterol 2025; 31(10): 99583
- URL: https://www.wjgnet.com/1007-9327/full/v31/i10/99583.htm
- DOI: https://dx.doi.org/10.3748/wjg.v31.i10.99583
According to the global cancer statistics published by the International Agency for Research on Cancer in 2020, gastric cancer (GC) is among the most commonly diagnosed cancers worldwide, ranking fourth in incidence among malignant tumors. There is a notable variation in GC rates across different regions, with East Asia experiencing higher incidence rates attributed to a combination of dietary practices, Helicobacter pylori infection, and genetic predispositions[1]. In China, for instance, regions such as Liaoning, Fujian, Gansu, and Shandong report higher-than-average rates of GC, potentially linked to a preference for salt-preserved foods, elevated Helicobacter pylori infection rates, and familial inheritance[2]. Over recent years, the incidence and mortality of GC have shown a gradual decline in some regions due to the westernization of lifestyles, the implementation of early detection technologies, and advancements in treatment methods[3]. Despite these improvements, the overall outlook remains challenging, and the prevention and control of GC continue to pose a significant challenge in the public health domain. Therefore, an in-depth research of the disease should be conducted, and its mechanisms should be identified to provide effective targets for the treatment of GC.
Eukaryotic cells should present circular RNA (circRNA) in eukaryotic cells, which exists in a covalent closed loop form. Further research shows that this substance does not exist or has a polyadenylation tail. CircRNA has better conservation and stability than linear RNA[3]. High-throughput technology reveals that a number of circRNAs exist in living organisms[4]. CircRNA is related to the physiological activities of many malignant tumors, including breast cancer[5], cervical cancer[6], hepatocellular carcinoma[7], colorectal cancer[8], esophageal cancer[9], etc. Scholars have discovered many circRNAs in GC tissues, which may be the reasons for changes in genetic function[10]. For example, circRNA can adsorb with microRNA (miR)-331-3p and promote the development of GC by regulating TGFBR1[11]. CircFAT1 (e2) can act on the miR-548g/RUNX1 axis to exert inhibitory effects on disease cells[12]. Although many researchers have conducted studies, few have investigated the role of a single circRNA in the development of diseases.
A large amount of research data shows the high expression of miR in malignant tumors, and it can regulate the occurrence and development of tumors[13]. circRNAs can act as a sponge to protect target genes and prevent abnormal mRNA function, thereby regulating the occurrence and development of diseases[14]. Research indicates that hsa_circ_0009172 suppresses tumor cell proliferation by targeting the miR-485-3p/NTRK3 axis. Hsa_circ_0009172 specifically binds to miR-485-3p, mitigating its suppressive impact on NTRK3. With the upregulation of NTRK3, it subsequently inhibits proliferation-associated signaling pathways in tumor cells, including the mitogen-activated protein kinase/extracellular regulated protein kinases and phosphatidylin-ositol-3-kinase/Akt cascades, thus impeding tumor cell proliferation[15]. Liu et al[16] pointed out that circRNA circ-PVT1 can regulate ZEB1 and affect its expression, thereby making GC cells resistant to paclitaxel[16]. In the context of GC research, miR-4529-5p has garnered attention. Studies have begun to reveal its involvement in the modulation of physiological activities in GC cells, potentially impacting cellular behavior through interactions with certain target genes. For instance, the research by Sun et al[17] suggests a link between miR-4529-5p and ROCK2, which subsequently affects the proliferation and migration of GC cells. Yet, these studies merely offer preliminary insights into the role of miR-4529-5p in GC, with its specific regulatory network, interactions with other molecules, and dynamic changes across different stages of GC remaining obscure. Consequently, our study aims to delve deeper into the mechanisms of miR-4529-5p in GC, particularly its interplay with hsa_circRNA_102415, to provide a more nuanced understanding of the pathogenesis of GC. Thus far, researchers have not elucidated the relationship between GC progression and miR-4529-5p.
In this study, the gene expression omnibus (GEO) dataset was used to analyze the differential expression of circRNAs in GC. hsa_circRNA_102415 had significantly higher expression levels in patients than in controls. A correlation analysis was conducted between miR-4529-5p and hsa_circRNA_102415, and the results showed their negative correlation. In this regard, we assume an interaction between the two, which can have an impact on the occurrence and development of GC. Cell analysis experiments were also conducted to explore the relationship between the malignant behavior of GC and hsa_circRNA_102415. This study aims to investigate and analyze the relationship between hsa_circRNA_102415 and GC and explore its specific role. Results provide reference for other researchers to develop targeted treatment plans.
Seventy-five tumor specimens and matched normal samples were obtained from Shanxi Province Cancer Hospital from March 2021 to February 2023. All subjects signed the informed consent form. The research content of this article had been approved by the relevant committee. The inclusion criteria for this article were as follows: No history of cancer diagnosis, first-time diagnosis, and no evidence of disease within 30 days after the first surgery. During the follow-up, testing was conducted every 3-6 months for the first five years, followed by annual testing thereafter. The number of months from surgery to death was set as the overall survival time, and the number of months from surgery to recurrence date was set as the recurrence free survival time. All research subjects did not receive relevant preoperative treatment, and the diagnosis was independently made by two experienced experts. The sample was stored at -80 °C.
Human gastric mucosal cell lines GES-1 and human GC cell lines AGS, HGC-27, SNU-1, BGC823, SGC7901, MGC803, and MKN-45 were obtained from the National Type Culture Collection Center. When implementing cell culture, dulbecco’s modified eagle medium was chosen and added with 100 U/mL penicillin and 100 μg/mL streptomycin as well as 10% fetal bovine serum. During cell culture, the cells were placed in a carbon dioxide incubator for cultivation.
The overexpression vector and the control of hsa_cirRNA_102415 were obtained from Gene Pharmaceutical Co., Ltd. Specific small interfering RNAs, such as WNT2B (si-WNT2B), as well as miR-4529-5p inhibitors and miR-4529-5p mimetics, were purchased from Genechem, Shanghai, China. Before carrying out transfection, the cells were first digested using trypsin, counted, and seeded in a six-well plate. The cells were cultured for 24 hours, and cell status was observed. When the fusion rate was 40%-60%, the cells were transfected, diluted, and mixed with Lipofectamine®3000 reagent in Opti minimal essential medium. Diluted DNA with Opti minimal essential medium and add p3000™ reagent to it. The sample was placed at room temperature for 5 minutes and added with DNA liposome complexes. Transfection was performed according to the instructions of the reagent kit. After 48 hours, the cells were collected for later use.
RNA was extracted from the cells by using Trizol reagent by strictly following the instructions of the reagent kit. RNA was reverse transcribed using the Prime Script RT kit. The sample was subjected to polymerase chain reaction (PCR) analysis using Applied Biosystems 7500 with PCR parameters of 95 °C for 60 seconds, 95 °C for 20 seconds, 56 °C for 10 seconds, 72 °C for 15 seconds, for a total of 35 cycles. The PCR reaction system was prepared by adding 6 μL double distilled water, 0.4 μL of Taq polymerase, 10 μL of 2 × SYBR green PCR main mixture, and 2 μL of complementary DNA. The primers were designed as follows: U6 small nuclear RNA forward: 5’-GCT TCG GCA GCA CAT ATA CTA AAA T-3’, and reverse: 5’-CGC TTC ACG AAT TTG CGT GTC AT-3’; Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) forward: 5’-TGC AGT GGC AAA GTG GAG ATT-3’, and reverse: 5’-TCG CTC CTG GAA GAT GGT GAT-3’; MiR-497-5p forward: 5’-CCT TCA GCA GCA CAC TGT GG-3’, and reverse: 5’-CAG TGC AGG GTC CGA GGT AT-3’; Hsa_circRNA_102415 forward: 5’-TTG GCG TAC CTC CGA GAG AAG CA-3’; and reverse: 5’-TGC TTC TCT CGT CGG AGG TAC GCC AA-3’. Each sample was extracted three times. When calculating the expression of water products, the 2-ΔΔCT method was chosen, and U6 small nuclear RNA and GAPDH were used as endogenous controls.
AGS cells were washed with pre-cooled phosphate-buffered saline (PBS) and added an appropriate amount of radio-immunoprecipitation assay lysis buffer containing protease inhibitors. Protein concentration was detected using the bicinchoninic acid assay protein assay kit (Thermo Fisher Scientific, Waltham, Massachusetts, United States). Sodium dodecyl sulfate polyacrylamide gel electrophoresis electrophoresis was performed on the protein. The voltage was set at 70 V for 0.5 hour and adjusted to 120 V for 1.5 hours. The protein bands were subjected to membrane transfer treatment, allowing the protein to transfer to the polyvinylidene difluoride membrane for 120 minutes. The membrane was placed in 5% skim milk and sealed for 2 hours. Primary antibodies, including rabbit monoclonal anti-WNT2B antibody and rabbit polyclonal anti-β-actin antibody, were diluted appropriately and added to the cells. The primary antibody was removed, and the secondary antibody was appropriately diluted to obtain goat anti rabbit IgG. The secondary antibody was added, and the sample was placed at room temperature for 60 minutes. β-Actin was chosen as the endogenous control. The bands were visualized using enhanced chemiluminescence plus chemiluminescence assay kit, and images of the samples were taken using an imaging system. Band density was analyzed using Image J software.
The cells were inoculated into a 96-well plate, with 2000 cells per well. The cells were incubated and added with 10 μL of cell counting kit (CCK)-8 solution per well at different times. A microplate reader was used to detect absorbance at
Apoptosis was detected using the annexin V-fluorescein isothiocyanate/propidium iodide apoptosis detection kit by strictly following the kit instructions. At 48 hours after completing the transfection operation, the cells were centrifuged at 1000 rpm and 4 °C for 5 minutes. The precipitate was collected and suspended in annexin V-fluorescein isothiocyanate and binding buffer at room temperature for 0.5 hours. Flow cytometry analysis was performed on a Gallios Flow Cytometer (Beckman Coulter, United States). Data were analyzed using software Kaluza analysis.
Cell invasion ability was detected using an 8.0-micron chamber plate. Matrigel was applied onto the surface of the Transwell filter. The cells were planted in a culture dish. Serum-free medium was added to the upper part of the dish, and an appropriate amount of fetal bovine serum containing medium was added to the lower part. The cells were cultured for 2 days. The cells measured on the chamber were suspended using cotton swabs, and invasive cells were fixed with 4% paraformaldehyde. The cells were stained with crystal violet. The stained infiltrating cells were imaged with Olympus IX70 inverted microscope under randomly selected optimal field of view. The experiment was repeated three times.
Cell cycle was analyzed using relevant reagent kits. The cells were cultivated in serum-free medium, with 5 × 102 cells per well, and treated with GM or SC79 at 37 °C for 24 hours. Eagle’s minimal essential medium was used to treat cells in the control group. The cells were washed using PBS solution, followed by enzymatic treatment, and then centrifuged at 300 and 4 °C for 5 minutes. The cells were fixed with 70% ethanol 4 °C for 24 hours, washed with PBS solution, and stained using propidium iodide from RNase A staining solution. Cell cycle was evaluated using flow cytometry with relevant software.
Computer-aided algorithms were used to predict circRNA and miR targets.
In brief, hsa_circRNA_102415 was amplified, and the sequence was transferred into the pmirGLO vector to construct wild-type (WT) reporter and hsa_circRNA-102415-WT containing the mutation binding site.
WNT2B 3’ untranslated region (UTR) was amplified, and the sequence was transferred into pmirGLO vector to construct WNT2B 3’-UTR WNT2B-WT and WNT2B-MUT containing mutation binding sites. AGS cells were then transfected with miR-4529-5p mimic and luciferase plasmids WNT2B-WT and WNT2B-MUT. At 48 hours after transfection, luciferase activity was detected and analyzed using a dual-luciferase reporter assay system.
For the synthesis of bio-miR-4529-5p-WT and biological probe negative control, a mixture of Sp6 RNA polymerase and biotin RNA labeling produced by Roche was used for labeling. The cells were lysed using cell lysis buffer to lyse cells. The lysate was collected, mixed with streptavidin agarose beads and labeled RNA, and incubated at 4 °C for 60 minutes. The combined sediment was analyzed.
RNA was extracted from AGS cells by using Trizol following strictly the instructions provided by the reagent kit company. Protein A/G magnetic beads, immunoprecipitation buffer, anti IgG antibody, and anti WNT2B antibody were mixed and incubated at 4 °C for 12 hours. The antibody and RNA were mixed in immunoprecipitation buffer and subjected to elution operation on the magnetic beads to allow RNA to precipitate on the impurity wall. The precipitate was washed and reverse transcribed. Relative fold enrichment was calculated using 2-ΔΔCT method.
GSE83521 (annotated probe files) was obtained from the GEO database. Analysis of GSE83521 revealed that it included a sample size of 6, including 6 tumor tissues and 6 normal tissues. GEO data were analyzed using GEO2R to compare and screen circRNAs with differential expression between normal and GC samples. When setting the threshold for expression differences, we set 1.5 as the P value.
Statistical product and service solutions software was used for statistical analysis. Results were expressed using mean ± SEM method, t-test was used to compare two data sets, and analysis of variance was applied to compare data between groups. Differences at P < 0.05 were considered significant.
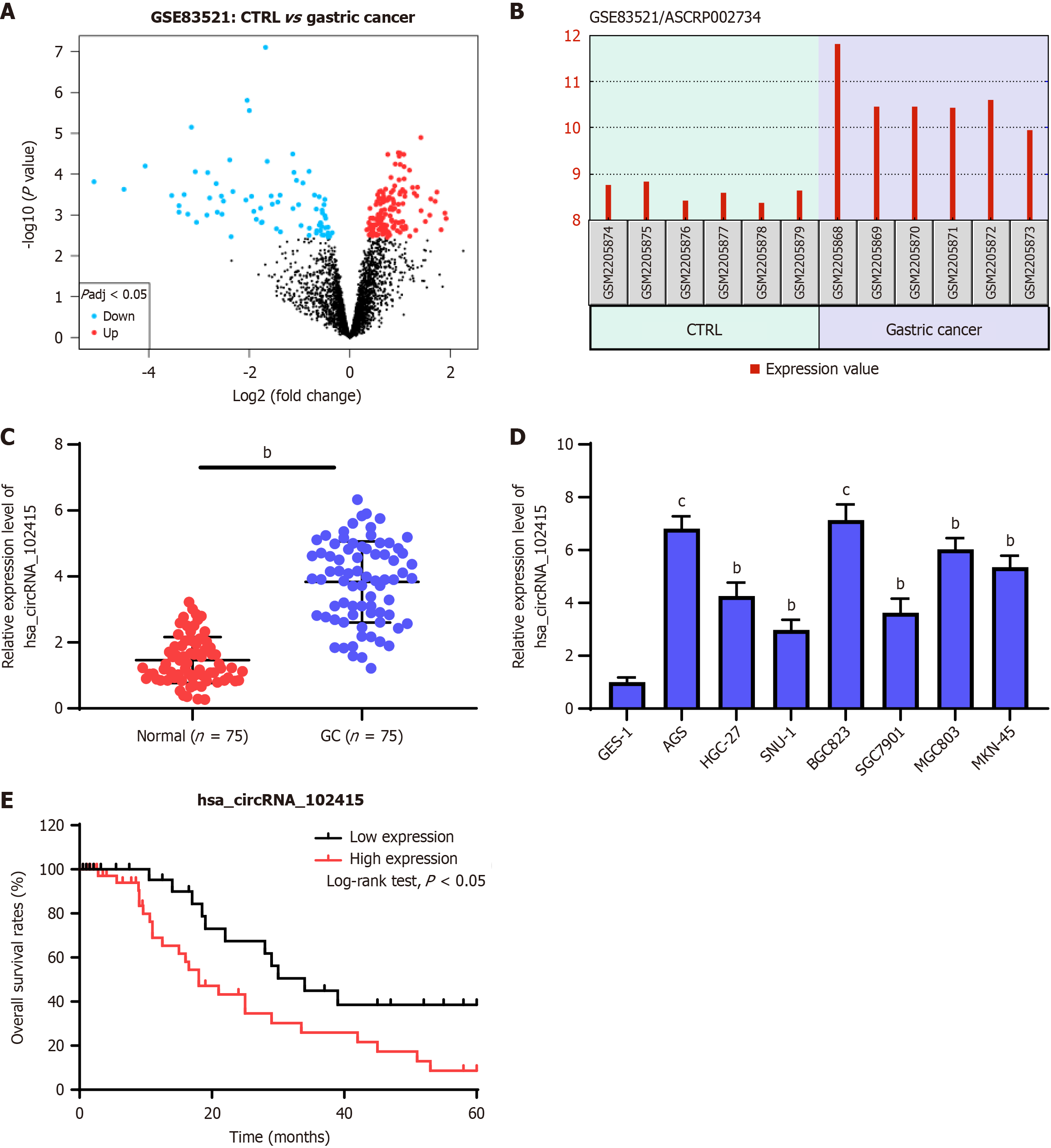
The GC dataset GSE83521 was downloaded from the GEO. The tumor tissue had significantly higher differential expression of circRNA than the normal tissue (Figure 1A and B). This study used Quantitative real-time PCR (RT-qPCR) to detect the expression levels of hsa_circRNA_102415 in normal and malignant tumor tissues. The tumor tissue had a significantly higher expression level of hsa_cirRNA_102415 than the normal tissue (Figure 1C, P < 0.01). The human gastric mucosal cell line had a significantly higher expression level of hsa_cirRNA_102415 than the human GC cell line (Figure 1D, P < 0.01). This study investigated and analyzed the relationship between the overall survival rate of patients with GC and the expression level of this gene. If patients had a high expression level of this gene, then they often did not have a good survival rate (Figure 1E, P < 0.01). Therefore, the GC may be have a better survival rate (Figure 1E, P < 0.01). Although the relationship between hsa_circRNA_102415 expression and patient survival rate was preliminarily explored in this study, due to the limited sample size and lack of further experimental verification, a definite conclusion cannot be drawn at present. Future studies with larger sample sizes are needed to more accurately analyze the correlation between them. Among them, hsa_circRNA_102415 had significantly induced expression, which may be closely related to the occurrence and development of GC.
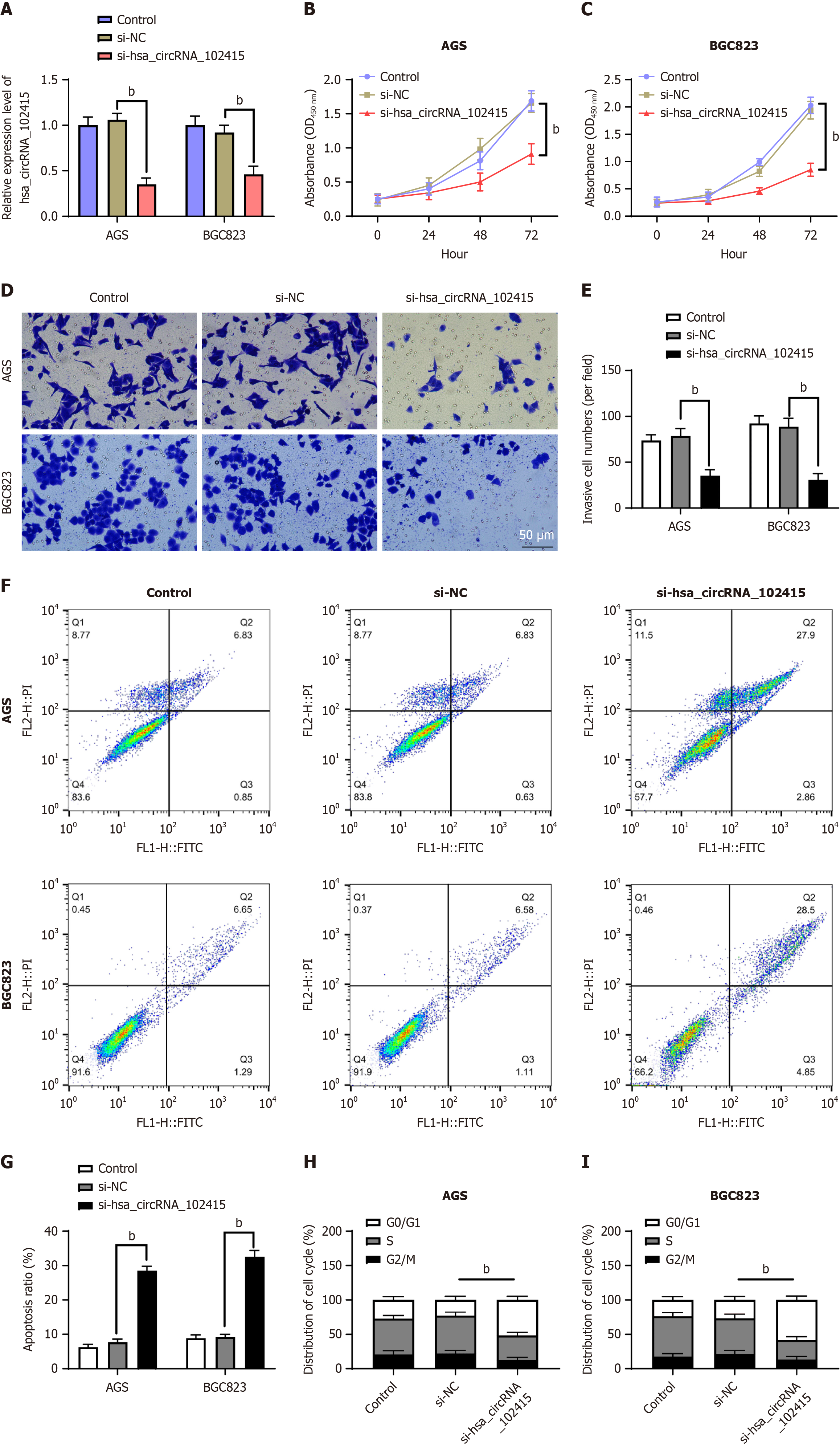
To further investigate and analyze the relationship between the two, this study chose to transfect si-hsa_circRNA_102451 in cells to silence hsa_circRNA_102451 (Figure 2A, P < 0.01). Silencing CCK-8 significantly reduced the proliferation of GC cells (Figure 2B and C, P < 0.01). When analyzing the invasion ability of the cells, Transwell was chosen. After the gene knockout, the invasion ability of GC cells changed (Figure 2D and E, P < 0.01). After the knock out, the apoptosis of GC cells studied in this study was significantly improved (Figure 2B and C, P < 0.01; Figure 2F and G, P < 0.01). Analysis of the cell cycle revealed that hsa_circRNA_102415 can have a certain impact on G0/G1 phase cells, by significantly reducing the proportion of G2/M phase cells. After knocking out the memory, the cell cycle process was significantly inhibited (Figure 2H and I, P < 0.01). Hence, hsa_circRNA_102415 silencing appeared to suppress key GC phenotypes.
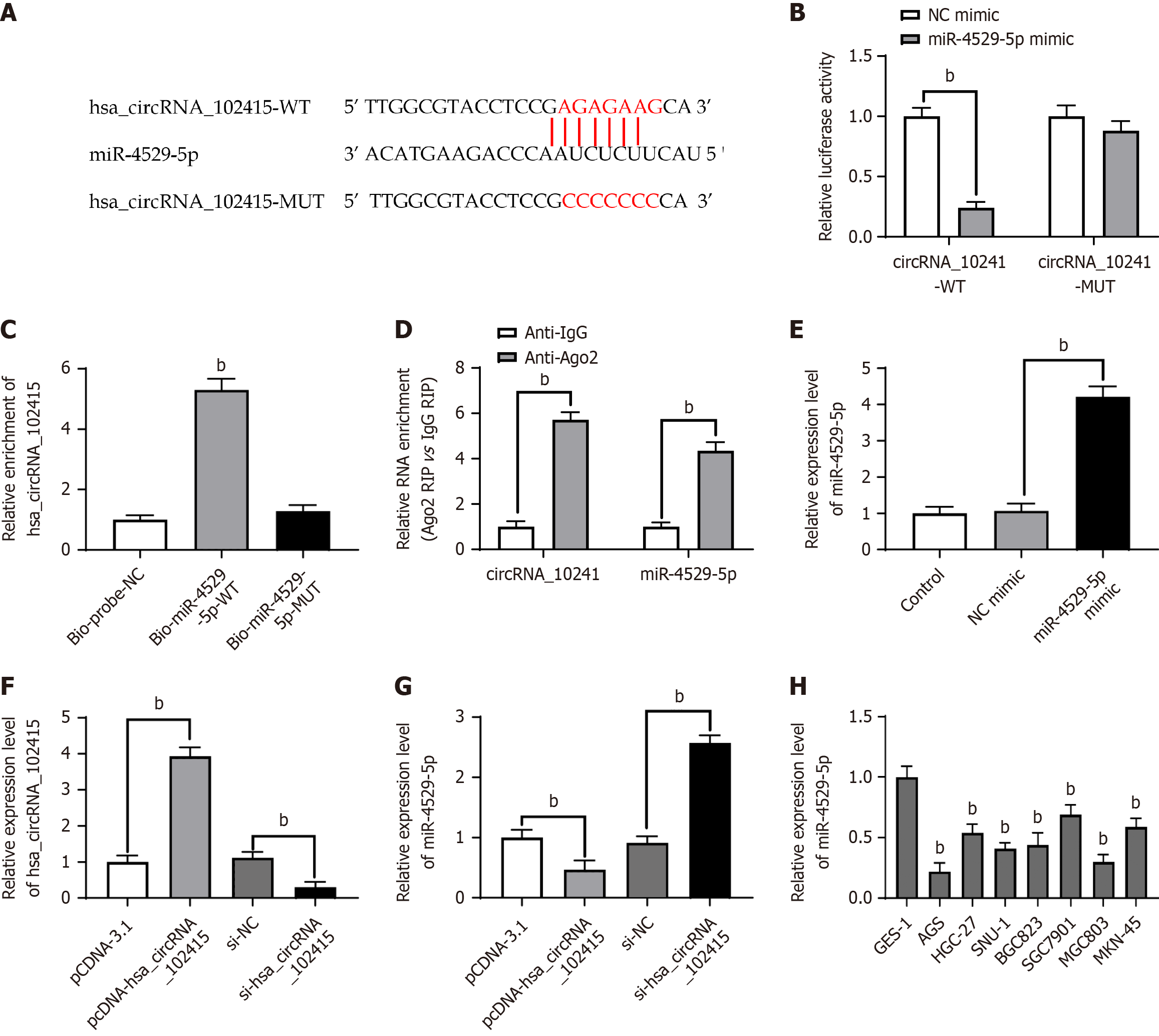
Accumulating evidence suggests that circRNAs act as sponges for miRs, which in turn affect downstream activities[18]. Therefore, we focused on investigating the potential mechanism by which hsa_circRNA_102415 participates in the progression of GC. Potential miRs for gene inactivation were investigated and analyzed using databases such as circBank. The results showed that hsa_circRNA_102415 and hsa-miR-4529-5p have complementary base pairing (Figure 3A). We then constructed hsa_circRNA_102415 WT and hsa_circRNA_102415 MUT luciferase vectors based on the results. miR-4529-5p was co-transfected with hsa_circRNA_102415-WT or hsa_circRNA_102415-MUT luciferase vectors. The luciferase activity dramatically decreased in the hsa_circRNA_102415-WT group. However, for the hsa_circRNA_102415-MUT group, changes in luciferase activity were not significant (Figure 3B, P < 0.01). To investigate the presence of a direct relationship between hsa_circRNA_102415 and miR-4529-5p, this study conducted biotin labeled pull-down analysis. For hsa_circRNA_102415, miR-4529-5p had significantly higher levels in its precipitate (Figure 3C, P < 0.01). miR can induce RNA, regulate its silencing complex, and thus enable silencing to take effect. For RNA-induced silencing complex (RISC), argonaute 2 (Ago2) is its core. To clarify whether the two are in the same RISC, we performed RNA immunoprecipitation (RIP) analysis was performed and compared with Ago2 immunoprecipitates with control IgG immunoprecipitates. Compared with the latter, the former had significantly higher RNA levels (Figure 3D, P < 0.01). Figure 3E shows the overexpression efficiency of miR-4529-5p mimics in AGS cells (Figure 3E, P < 0.01). To further investigate and analyze the regulatory relationship, this study chose to transfect short hairpin (sh)-hsa_circRNA_104415 and plasmid DNA (pcDNA)-hsa_circRNA_102415 vectors in AGS cells. By using RT-qPCR to detect the expression level of the vector in cells, the expression efficiency was determined (Figure 3F, P < 0.01). If hsa_circRNA_102415 was overexpressed, then it will significantly inhibit the expression of miR-4529-5p. If it was silenced, then it will significantly promote the expression of miR-4529-5p (Figure 3G, P < 0.01). This study found that miR-4529-5p also had a low expression level in GC cell lines (Figure 3H, P < 0.01). Hence, hsa_circRNA_102415 may play a sponge role in the study of GC cells.
For hsa_circRNA_102415, as miR-4529-5p acts as a target downstream, we conducted further research on whether a functional relationship exists between the two and performed rescue experiments. From the cellular behavior, overexpression of hsa_circRNA_102415 significantly aggravated the proliferation (Figure 4A, P < 0.01), invasion (Figure 4B and C, P < 0.01) and G2/M phase cell number (Figure 4D, P < 0.01) of AGS cells and inhibited apoptosis (Figure 4E and F, P < 0.01) and G0/G1 phase cell number. However, upregulation of miR-4529-5p prominently prevented the malignant behavior of AGS cells, as reflected by suppressed cell proliferation (Figure 4A, P < 0.01), invasion (Figure 4B and C, P < 0.01) and G2/M phase cell number (Figure 4D, P < 0.01) and accelerated apoptosis (Figure 4E and F, P < 0.01) and G0/G1 phase cell number. Interestingly, the overexpression of miR-4529-5p obviously reversed the promoting effect of hsa_circRNA_102415 upregulation on the malignant behavior of AGS cells.
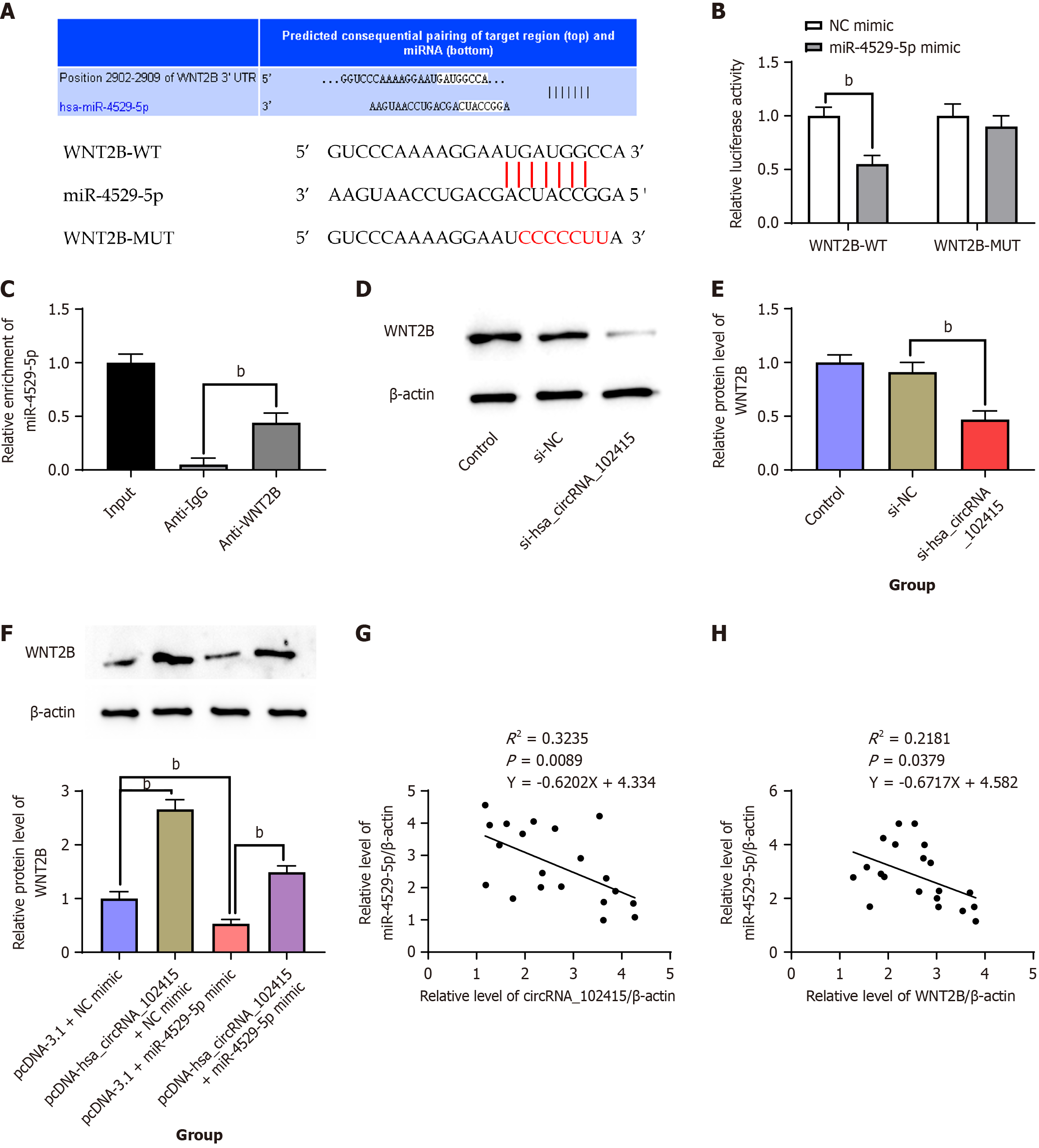
We determined whether WNT2B is a functional target by using the Target scan data site. Figure 5A reveals potential binding sites between WNT2B 3’-UTR and miR-4529-5p. When the latter was overexpressed, it had an impact on the WNT2B 3’-UTR-MUT group, resulting in a significant decrease in its luciferase activity level (Figure 5B, P < 0.01). RIP results revealed that miR-4529-5p was significantly enriched in WNT2B antibody compared with that in IgG, which further validated the binding relationship of miR-4529-5p on WNT2B (Figure 5C, P < 0.01). Experiments were grouped in reference to sections 3.2 and 3.4. The Western blot data displayed that hsa_circRNA_102415 knockdown evidently suppressed WNT2B protein expression levels in AGS cells, whereas hsa_circRNA_102415 overexpression distinctly increased WNT2B protein levels, which were reversed by transfection of miR-4529-5p mimic (Figure 5D-F, P < 0.01). According to the correlation analysis results, the expression of miR-4529-5p was negatively correlated with the levels of WNT2B and hsa_circRNA_102415 in GC tissues (Figure 5G and H, P < 0.01), suggesting that hsa_circRNA_102415 upregulated WNT2B expression in GC cells as a competing endogenous RNA (ceRNA) for miR-4529-5p.
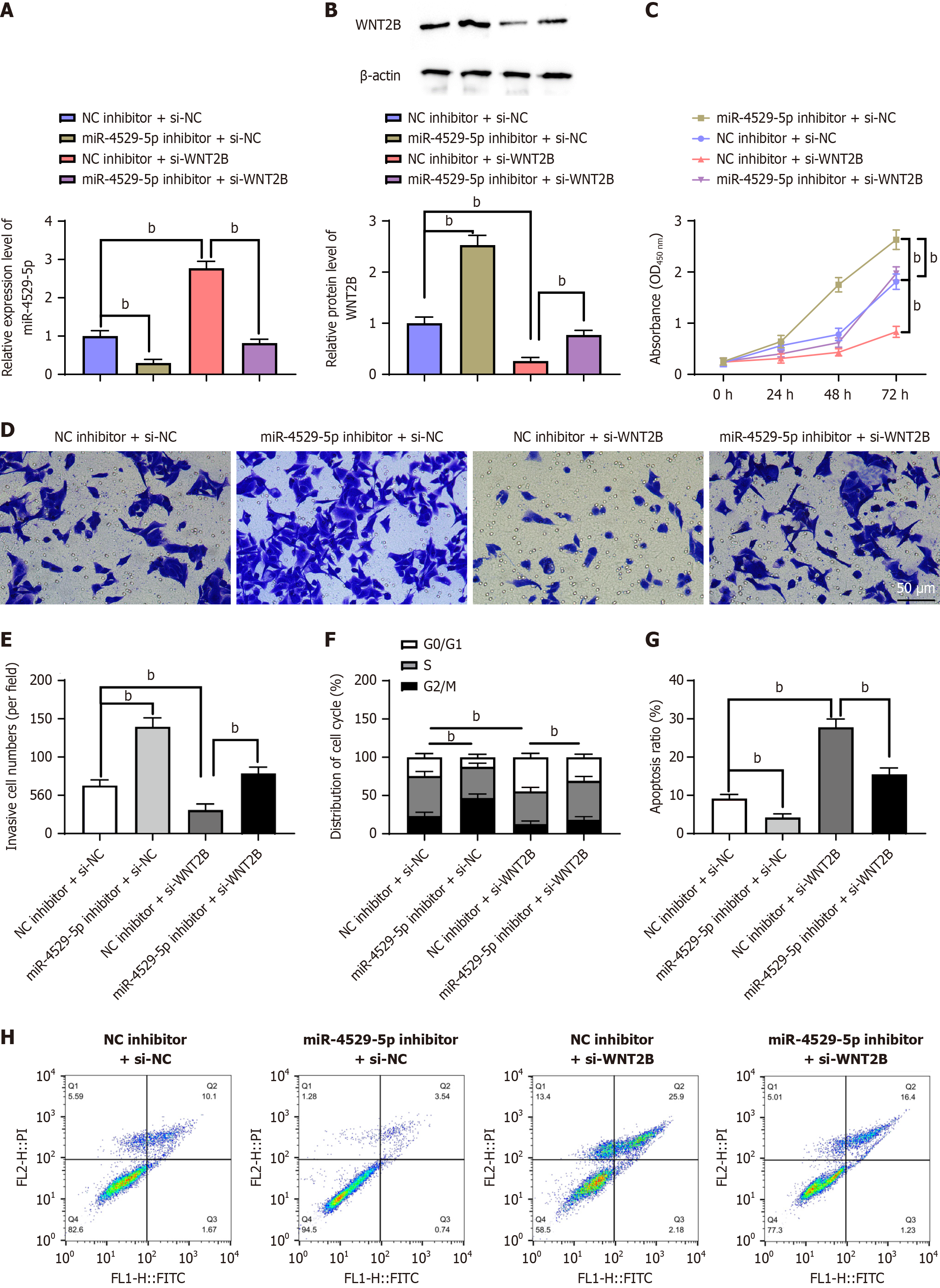
To explore whether hsa_circRNA_102415 regulates WNT2B expression as a ceRNA of miR-4529-5p, which in turn is involved in GC malignant behavior, we investigated the functional correlation between miR-4529-5p and WNT2B and transfected sh-WNT2B and miR-4529-5p inhibitors in AGS cells. In terms of expression level efficiency, multiple methods including RT-qPCR were used for validation (Figure 6A and B, P < 0.01). The CCK-8 assay showed that the inhibition of miR-4529-5p expression apparently promoted AGS cell proliferation (Figure 6C, P < 0.01), invasion (Figure 6D and E, P < 0.01), and G2/M phase cell number (Figure 6F, P < 0.01) and impeded cell apoptosis (Figure 6G and H, P < 0.01) and G0/G1 phase cell number. WNT2B knockdown showed the opposite results and partially reversed the promoting effects of miR-4529-5p expression inhibition on cell behavior.
Researchers often focus on non-coding RNAs when studying malignant tumors[19]. Among RNA aggregates, circRNAs are disease specific and tissue-specific, so they may play a role as biomass markers for cancer specialization[20]. CircRNA has significantly better stability than linear RNA, which leads to its ability to function as a transcription regulator[21]. However, researchers have not yet clarified the mechanism of action between malignant tumors and circRNA; however, in the process of treatment and diagnosis, the mechanism of action of circRNA has become increasingly important. CircRNA can play a certain regulatory role in its development process. Yuan et al[22] pointed out that in GC, circRNAs_102231 has significantly higher expression levels and can be considered a new oncogene that can serve as a potential biomarker for therapeutic and diagnosis. Zhang et al[23] registered four GC associated circRNA expression profiles from the GEO database and identified hsa_circRNA_00067997 as an oncogene in GC. In addition, a study screened for differentially expressed circRNA_0043691 from the GEO GSE141977 dataset. After knocking out the gene, it could regulate the relevant axis, thereby significantly slowing down the development of GC[24]. In this study, the circRNA expression profile was screened, differentially expressed circRNAs were identified, and significance thresholds were set and adjusted. GEO2R analysis showed that it had significantly higher expression levels of hsa_circRNA_102415. If the gene is in a silent state, then it can inhibit the physiological processes of GC cells and effectively promote cell apoptosis. Bioinformatics tools were used to predict whether miR-4529-5p could bind to hsa_circRNA_102415, which was speculated to be the molecular basis of GC regulation by hsa_circRNA_102415. While previous research has uncovered several circRNAs linked to GC, the precise roles and regulatory mechanisms of hsa_circRNA_102415 in this context are not yet fully understood. Our study is designed to address this knowledge gap and contribute to the foundation for precision therapy in GC.
CircRNAs can play multiple functions, including regulatory factors, sponges, etc[25,26]. For RNA, ceRNAs are its transcripts that can utilize miR response elements to achieve regulatory effects[27]. A large amount of data shows that circRNA can act on GC cells and regulate their development process. For example, studies have shown that hsa_circ_0017639 can regulate relevant axes, thereby affecting the physiological activity of GC cells and significantly inhibiting apoptosis. Hence, it can be considered a potential target for therapeutic effects[28]. A new circRNA can regulate the physiological activity of GC cells by using related axis pairs, which in turn accelerated tumorigenesis[29]. Moreover, some researchers have pointed out that circRNA can play a sponge like role, which can regulate the development process of GC[30]. According to some data, miR-4529-5p can serve as a target expression for long non-coding RNA MHRT in this study. Functional experiments have shown that this gene can regulate the apoptosis process mediated by the latter[17]. On the basis of clarifying the relevant information, this article conducts research on the relationship between hsa_circRNA_102415 and miRs, explores whether there is any functional interaction between them, and verifies their expression relationship in GC tissues studied in this article by using multiple detection methods. In GC tissues, there is an opposite expression trend, and in the GC cell lines studied in this article, the two can directly interact with each other. Subsequently, this article elaborates on the biological functions of relevant biological axes in GC cell lines. The results found that hsa_circRNA_102415 interacts with miR-4529-5p through complementary binding, thereby neutralizing the suppressive effect of miR-4529-5p on its downstream target genes. This interaction influences critical biological processes in GC cells, including proliferation, apoptosis, invasion, and cell cycle regulation. The ceRNA regulatory network, such as this one, may be pivotal in the initiation and progression of GC, offering a novel perspective on the mechanisms underlying the disease.
Numerous researchers have demonstrated that miRs can act as regulatory factors, regulating the expression of target genes in the post transcriptional stage[31]. When Wnt signaling molecules undergo changes, they can have an impact on related signaling pathways, significantly increasing the likelihood of malignant tumors occurring[32]. Researchers have cloned and identified the human WNT2B/WNT13 gene, and found that it shares homology with the oncogene WNT2. The peptides encoded by the WNT2B gene have high similarity, with differences mainly at the N-terminus, because of the alternative splicing promoter[33]. WNT2B is the target gene for miR-4529-5p. According to data, WNT2B has a high expression level in patients with GC in this article. Knocking it out can significantly slow down the growth rate of tumors[34]. Therefore, this article conducted research and analysis on the role of hsa_circRNA_102415 in the development of GC. Functional experiments further found that WNT2B knockdown memorably inhibited GC cell proliferation, invasion and G2/M phase cell number as well as inhibited apoptosis and G0/G1 phase cell number, as well as was able to partially reverse the promoting effects of miR-4529-5p expression inhibition on cell behavior.
This study revealed that hsa_circRNA_102415 exacerbated GC cell malignant behavior as a ceRNA of miR-4529-5p by upregulating WNT2B expression. While this study sheds light on the significant role and regulatory mechanisms of hsa_circRNA_102415 in GC, it acknowledges certain limitations. Firstly, the relatively small sample size may not capture the full spectrum of the GC patient population, potentially limiting the generalizability of our findings. Secondly, the absence of in vivo experimental validation prevents a conclusive understanding of hsa_circRNA_102415’s specific impact on GC growth and metastasis within a biological context. Furthermore, although the regulatory axis involving hsa_circRNA_102415/miR-4529-5p/WNT2B in GC has been identified, there may be additional, yet undiscovered, molecules or signaling pathways that also play a role. Future research can be advanced in several directions. Firstly, expanding the clinical sample size will further validate the diagnostic and prognostic value of hsa_circRNA_102415 in GC, thereby enhancing the reliability and precision of our research outcomes. Secondly, initiating animal model experiments will allow for a deeper investigation into the influence of hsa_circRNA_102415 on GC development in vivo, as well as its potential as a therapeutic target. Thirdly, a more profound exploration of the interactions between hsa_circRNA_102415 and other signaling pathways or molecules will help construct a more holistic regulatory network. This comprehensive understanding of its complex role in GC pathogenesis will provide a theoretical foundation for the development of more effective treatment strategies for GC.
| 1. | Suzuki S, Takahashi A, Ishikawa T, Akazawa K, Katai H, Isobe Y, Miyashiro I, Ono H, Tanabe S, Fukagawa T, Muro K, Nunobe S, Kadowaki S, Suzuki H, Irino T, Usune S, Miyata H, Kakeji Y; Registration Committee of the Japanese Gastric Cancer Association. Surgically treated gastric cancer in Japan: 2011 annual report of the national clinical database gastric cancer registry. Gastric Cancer. 2021;24:545-566. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 22] [Article Influence: 5.5] [Reference Citation Analysis (1)] |
| 2. | Wang Z, Han W, Xue F, Zhao Y, Wu P, Chen Y, Yang C, Gu W, Jiang J. Nationwide gastric cancer prevention in China, 2021-2035: a decision analysis on effect, affordability and cost-effectiveness optimisation. Gut. 2022;71:2391-2400. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 44] [Article Influence: 14.7] [Reference Citation Analysis (1)] |
| 3. | Kano Y, Ohashi M, Ida S, Kumagai K, Makuuchi R, Sano T, Hiki N, Nunobe S. Therapeutic value of splenectomy to dissect splenic hilar lymph nodes for type 4 gastric cancer involving the greater curvature, compared with other types. Gastric Cancer. 2020;23:927-936. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 13] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 4. | Wang Y, Wang H, Xi F, Wang H, Han X, Wei W, Zhang H, Zhang Q, Zheng Y, Zhu Q, Kohnen MV, Reddy ASN, Gu L. Profiling of circular RNA N(6) -methyladenosine in moso bamboo (Phyllostachys edulis) using nanopore-based direct RNA sequencing. J Integr Plant Biol. 2020;62:1823-1838. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 37] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 5. | Liu T, Ye P, Ye Y, Lu S, Han B. Circular RNA hsa_circRNA_002178 silencing retards breast cancer progression via microRNA-328-3p-mediated inhibition of COL1A1. J Cell Mol Med. 2020;24:2189-2201. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 82] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 6. | Xu T, Song X, Wang Y, Fu S, Han P. Genome-Wide Analysis of the Expression of Circular RNA Full-Length Transcripts and Construction of the circRNA-miRNA-mRNA Network in Cervical Cancer. Front Cell Dev Biol. 2020;8:603516. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 19] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 7. | Dong W, Dai ZH, Liu FC, Guo XG, Ge CM, Ding J, Liu H, Yang F. The RNA-binding protein RBM3 promotes cell proliferation in hepatocellular carcinoma by regulating circular RNA SCD-circRNA 2 production. EBioMedicine. 2019;45:155-167. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 85] [Cited by in RCA: 132] [Article Influence: 22.0] [Reference Citation Analysis (0)] |
| 8. | Liu Y, Chen L, Liu T, Su X, Peng L, Chen J, Tan F, Xing P, Wang Z, Di J, Jiang B, Qu H. Genome-wide circular RNA (circRNA) and mRNA profiling identify a circMET-miR-410-3p regulatory motif for cell growth in colorectal cancer. Genomics. 2022;114:351-360. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 11] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 9. | Shi Y, Fang N, Li Y, Guo Z, Jiang W, He Y, Ma Z, Chen Y. Circular RNA LPAR3 sponges microRNA-198 to facilitate esophageal cancer migration, invasion, and metastasis. Cancer Sci. 2020;111:2824-2836. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 97] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 10. | Shao Y, Li J, Lu R, Li T, Yang Y, Xiao B, Guo J. Global circular RNA expression profile of human gastric cancer and its clinical significance. Cancer Med. 2017;6:1173-1180. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 154] [Cited by in RCA: 216] [Article Influence: 27.0] [Reference Citation Analysis (0)] |
| 11. | Zhang L, Song X, Chen X, Wang Q, Zheng X, Wu C, Jiang J. Circular RNA CircCACTIN Promotes Gastric Cancer Progression by Sponging MiR-331-3p and Regulating TGFBR1 Expression. Int J Biol Sci. 2019;15:1091-1103. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 81] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 12. | Fang J, Hong H, Xue X, Zhu X, Jiang L, Qin M, Liang H, Gao L. A novel circular RNA, circFAT1(e2), inhibits gastric cancer progression by targeting miR-548g in the cytoplasm and interacting with YBX1 in the nucleus. Cancer Lett. 2019;442:222-232. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 128] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 13. | Urbanek-Trzeciak MO, Galka-Marciniak P, Nawrocka PM, Kowal E, Szwec S, Giefing M, Kozlowski P. Pan-cancer analysis of somatic mutations in miRNA genes. EBioMedicine. 2020;61:103051. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 34] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 14. | Liu J, Li Z, Teng W, Ye X. Identification of downregulated circRNAs from tissue and plasma of patients with gastric cancer and construction of a circRNA-miRNA-mRNA network. J Cell Biochem. 2020;121:4590-4600. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 12] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 15. | Wang H, Wang N, Zheng X, Wu L, Fan C, Li X, Ye K, Han S. Circular RNA hsa_circ_0009172 suppresses gastric cancer by regulation of microRNA-485-3p-mediated NTRK3. Cancer Gene Ther. 2021;28:1312-1324. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 17] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 16. | Liu YY, Zhang LY, Du WZ. Circular RNA circ-PVT1 contributes to paclitaxel resistance of gastric cancer cells through the regulation of ZEB1 expression by sponging miR-124-3p. Biosci Rep. 2019;39. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 69] [Cited by in RCA: 104] [Article Influence: 20.8] [Reference Citation Analysis (0)] |
| 17. | Sun X, Zhang X, Chen S, Fan M, Ma S, Zhai H. Myosin Heavy Chain-Associated RNA Transcripts Promotes Gastric Cancer Progression Through the miR-4529-5p/ROCK2 Axis. Dig Dis Sci. 2019;64:3539-3548. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 18. | Shu X, Cheng L, Dong Z, Shu S. Identification of circular RNA-associated competing endogenous RNA network in the development of cleft palate. J Cell Biochem. 2019;120:16062-16074. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 11] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 19. | Liao Y, Jung SH, Kim T. A-to-I RNA editing as a tuner of noncoding RNAs in cancer. Cancer Lett. 2020;494:88-93. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 27] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 20. | Han B, Chao J, Yao H. Circular RNA and its mechanisms in disease: From the bench to the clinic. Pharmacol Ther. 2018;187:31-44. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 403] [Cited by in RCA: 607] [Article Influence: 86.7] [Reference Citation Analysis (0)] |
| 21. | Xiao MS, Wilusz JE. An improved method for circular RNA purification using RNase R that efficiently removes linear RNAs containing G-quadruplexes or structured 3' ends. Nucleic Acids Res. 2019;47:8755-8769. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 76] [Cited by in RCA: 159] [Article Influence: 26.5] [Reference Citation Analysis (0)] |
| 22. | Yuan G, Ding W, Sun B, Zhu L, Gao Y, Chen L. Upregulated circRNA_102231 promotes gastric cancer progression and its clinical significance. Bioengineered. 2021;12:4936-4945. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 52] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 23. | Zhang H, Wang X, Huang H, Wang Y, Zhang F, Wang S. Hsa_circ_0067997 promotes the progression of gastric cancer by inhibition of miR-515-5p and activation of X chromosome-linked inhibitor of apoptosis (XIAP). Artif Cells Nanomed Biotechnol. 2019;47:308-318. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 36] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 24. | Zhang Y, Hu G, Zhang Z, Jing Y, Tao F, Ye M. CircRNA_0043691 sponges miR-873-3p to promote metastasis of gastric cancer. Mamm Genome. 2021;32:476-487. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 4] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 25. | Zhao B, Li Z, Qin C, Li T, Wang Y, Cao H, Yang X, Wang W. Mobius strip in pancreatic cancer: biogenesis, function and clinical significance of circular RNAs. Cell Mol Life Sci. 2021;78:6201-6213. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 16] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 26. | Zhang Y, Yu F, Bao S, Sun J. Systematic Characterization of Circular RNA-Associated CeRNA Network Identified Novel circRNA Biomarkers in Alzheimer's Disease. Front Bioeng Biotechnol. 2019;7:222. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 54] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 27. | Cardenas J, Balaji U, Gu J. Cerina: systematic circRNA functional annotation based on integrative analysis of ceRNA interactions. Sci Rep. 2020;10:22165. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 12] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 28. | Li B, Jin M, Cao F, Li J, Wu J, Xu L, Liu X, Shi Y, Chen W. Hsa_circ_0017639 expression promotes gastric cancer proliferation and metastasis by sponging miR-224-5p and upregulating USP3. Gene. 2020;750:144753. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 31] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 29. | Huang S, Zhang X, Guan B, Sun P, Hong CT, Peng J, Tang S, Yang J. A novel circular RNA hsa_circ_0008035 contributes to gastric cancer tumorigenesis through targeting the miR-375/YBX1 axis. Am J Transl Res. 2019;11:2455-2462. [PubMed] |
| 30. | Lai Z, Yang Y, Wang C, Yang W, Yan Y, Wang Z, Xu J, Jiang K. Circular RNA 0047905 acts as a sponge for microRNA4516 and microRNA1227-5p, initiating gastric cancer progression. Cell Cycle. 2019;18:1560-1572. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 15] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 31. | Yang M, Woolfenden HC, Zhang Y, Fang X, Liu Q, Vigh ML, Cheema J, Yang X, Norris M, Yu S, Carbonell A, Brodersen P, Wang J, Ding Y. Intact RNA structurome reveals mRNA structure-mediated regulation of miRNA cleavage in vivo. Nucleic Acids Res. 2020;48:8767-8781. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 37] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 32. | Koni M, Pinnarò V, Brizzi MF. The Wnt Signalling Pathway: A Tailored Target in Cancer. Int J Mol Sci. 2020;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 106] [Cited by in RCA: 119] [Article Influence: 23.8] [Reference Citation Analysis (0)] |
| 33. | Katoh M, Kirikoshi H, Terasaki H, Shiokawa K. WNT2B2 mRNA, up-regulated in primary gastric cancer, is a positive regulator of the WNT- beta-catenin-TCF signaling pathway. Biochem Biophys Res Commun. 2001;289:1093-1098. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 64] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 34. | Zhang L, Liu F, Meng Z, Luo Q, Pan D, Qian Y. Inhibited HDAC3 promotes microRNA-376c-3p to suppress malignant phenotypes of gastric cancer cells by reducing WNT2b. Genomics. 2021;113:3512-3522. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 3] [Article Influence: 0.8] [Reference Citation Analysis (0)] |