Published online Mar 14, 2025. doi: 10.3748/wjg.v31.i10.102289
Revised: November 28, 2024
Accepted: January 17, 2025
Published online: March 14, 2025
Processing time: 135 Days and 7.4 Hours
Recent studies have shown a noticeable increase in global Helicobacter pylori
Core Tip: The rise of antibiotic resistance in Helicobacter pylori (H. pylori) has become a global concern and led to the reduction of efficacy of conventional therapies. Resistant H. pylori strains, often inadequately mapped by regional surveillance, frequently demand multiple eradication attempts, imposing considerable financial burdens and adverse effects and contributing to secondary resistance development. This study aimed to provide a comprehensive review of the current landscape of mechanisms and prevalence of H. pylori resistance and to summarize promising therapeutic alternatives under evaluation. These strategies might improve treatment efficacy, enhancing patient outcomes in this challenging scenario.
- Citation: Rocha GR, Lemos FFB, Silva LGO, Luz MS, Correa Santos GL, Rocha Pinheiro SL, Calmon MS, de Melo FF. Overcoming antibiotic-resistant Helicobacter pylori infection: Current challenges and emerging approaches. World J Gastroenterol 2025; 31(10): 102289
- URL: https://www.wjgnet.com/1007-9327/full/v31/i10/102289.htm
- DOI: https://dx.doi.org/10.3748/wjg.v31.i10.102289
Helicobacter pylori (H. pylori) is a microaerophilic, gram-negative bacterium that commonly colonizes the human gastric mucosa with an estimated prevalence of approximately 44% of adults and 35% of children and adolescents worldwide[1]. The discovery of H. pylori in 1983 by Warren and Marshall[2] and their subsequent research revealing its association with chronic gastritis and peptic ulcer disease[3] profoundly impacted the gastroduodenal pathology field and revolutionized clinical management of these conditions.
Although most H. pylori-positive individuals are asymptomatic, the presence of the bacterium invariably leads to persistent inflammation of the gastric mucosa (chronic gastritis), which raises the risk of developing functional and structural abnormalities as well as neoplasms[4]. Thus, H. pylori infection can culminate in conditions such as peptic ulcer disease, atrophic gastritis[5], gastric adenocarcinoma[6,7], and gastric mucosa-associated lymphoid tissue lymphoma[8]. Since the eradication of H. pylori can reduce dyspeptic symptoms[9], halt the progression of preneoplastic lesions[10], lower the risk of cancer[11,12], and induce regression of early-stage mucosa-associated lymphoid tissue lymphoma[13], treatment of the infection is beneficial and therefore recommended for both symptomatic and asymptomatic individuals[4].
The activity of hydrogenase, catalase, superoxide dismutase, and urease enzymes allow the bacteria to survive and thrive in the stomach[14,15]. While H. pylori can elicit a robust immune response with lymphocytes, eosinophils, macrophages, and dendritic cells being the most identified cell types[16], the bacterium has also developed mechanisms to evade immune detection and maintain its colonization. For instance, vacuolating cytotoxin A, a pore-forming toxin present in many bacterial strains and a major determinant of H. pylori pathogenicity[17], has been demonstrated to inhibit macrophage maturation, T cell proliferation, and the antigen presentation process[18,19]. This immunomodulatory activity combined with a significant arsenal of enzymes essential for its successful colonization of the gastric mucosa, may contribute significantly to the evasion of host defenses and long-term persistence of the infection.
Thus, once adults are H. pylori-positive, the infection usually persists throughout life unless approached with specific therapy or until severe atrophic gastritis[20-22]. Indeed, the capacity of H. pylori to not only be in gastric mucus attached to gastric epithelial cells but also inhabit the intracellular environment (and being inaccessible to many antibiotics) provides the bacteria an intrinsic mechanism of relative antimicrobial resistance[23,24]. Thus, eradicating H. pylori generally demands the combination of different antibiotics and adjunctive drugs that are conventionally administered in 14 days[4]. Additionally, while its high genetic plasticity is often examined to explain the ability of H. pylori to successfully infect hosts and cause disease[25,26], it also plays a critical role in the development of highly resistant strains[27]. This adaptability enables the bacteria to efficiently acquire developing resistance to antimicrobials, even when other factors such as horizontal gene transferring are considered[28].
Clarithromycin, amoxicillin, tetracycline, nitroimidazoles, levofloxacin, rifabutin, and furazolidone are among the antibiotics that may be effective against H. pylori. To prevent excessive use of antibiotics, strategies for empirically eradicating H. pylori are categorized into first-line, second-line, and rescue approaches, which must consider the local antimicrobial resistance profile[4,29]. When aiming for rational medication usage, antimicrobial susceptibility testing (AST) is a reasonable strategy that enables tailored therapy when feasible[4]. However, the limited availability of these methods hinders their widespread adoption, necessitating cost-effectiveness evaluations in different areas.
Indeed, from 2006 to 2016, resistance rates to clarithromycin, metronidazole, and levofloxacin have increased across all World Health Organization (WHO) regions[30]. Several studies are currently being carried out to evaluate different approaches for optimizing empirical treatment in this context. For example, alternative options may include the use of new antisecretory drugs to enhance treatment efficacy and reduce the need for multiple antibiotic usage[31,32], shortening the duration of well-established therapy regimens[33,34] and decreasing the concomitant administration of drugs[35]. Ultimately, the objectives are to improve effectiveness, enhance patient adherence (by streamlining therapeutic regimens or minimizing adverse events), and mitigate the selection pressure of antibiotics.
Therapeutic regimens for H. pylori infection consist of a combination of antibiotics with a strong acid suppressant[20]. To ensure the bactericidal efficacy of antimicrobials, it is important to raise gastric pH with antisecretory drugs, such as proton pump inhibitors (PPIs), like omeprazole and lansoprazole, or potassium-competitive acid blockers (P-CABs), like vonoprazan and tegoprazan[24]. Additionally, first-line therapy often includes bismuth. Although the mechanisms of bismuth salts are not fully understood, they offer advantages including antibacterial properties through the inhibition of different enzymes (like urease, fumarase, alcohol dehydrogenase, and phospholipase) and a cytoprotective effect on the gastric mucosa, facilitating ulcer healing and impeding H. pylori from attaching to gastric epithelial cells[36].
According to the Maastricht VI/Florence consensus (2022)[4], the bismuth-containing quadruple therapy (BQT), which includes a PPI, bismuth, tetracycline, and metronidazole, is recommended as a first-line empirical therapy globally. In regions where clarithromycin resistance is below 15%, another first-line option is the clarithromycin triple therapy, consisting of a PPI, clarithromycin, and amoxicillin. However, in areas with high (≥ 15%) or unknown clarithromycin resistance, if BQT is not available, the preferred first-line treatment is the non-bismuth concomitant quadruple therapy (non-BQT), which includes a PPI, clarithromycin, amoxicillin, and metronidazole administered concomitantly. After the failure of first-line therapies, second-line and rescue treatments should be guided by local resistance patterns. Alternative options include levofloxacin-based therapies (comprising a PPI, levofloxacin, amoxicillin, with or without bismuth), high-dose PPI-amoxicillin dual therapy, and rifabutin-based therapies[4].
Regarding AST-based therapy, traditional culture-based methods are limited by technical constraints, subjective interpretation, prolonged execution time, and the necessity for invasive procedures (prior endoscopy), which is not always required for diagnosis except in high cancer-risk cases[4]. Molecular methods, such as PCR, provide reliability in identifying particular mutations that cause resistance, but they usually rely on endoscopic procedures. Emerging non-invasive AST methods, such as real-time PCR on fecal samples, are being investigated as alternatives[37,38]. Further evaluation is needed to determine the overall advantages and cost-effectiveness of AST-guided tailored treatment compared to empirical treatments before issuing widespread clinical advice.
H. pylori inherently possesses a highly dynamic genome, characterized by extensively repetitive chromosomal sequences that facilitate frequent mutations, even in the absence of multiple coexisting lineages within a single host[28]. Addi
Resistance to clarithromycin in H. pylori is linked to the structures through which macrolides operate, namely the ribosomal 50S subunit[42]. A2142G, A2143G, and A2142C are the main nucleotide substitutions in the 23S rRNA molecule, accounting for more than 90% of clarithromycin-resistant H. pylori[43]. These point mutations are located in the peptidyl transferase loop, a critical area for the binding of macrolide antibiotics, and their alterations compromise the ability of the drug to operate effectively, leading to a less effective treatment of the bacteria[44]. Despite this, A2115G, G2212A, G2141A, A2144T, and T2289C point mutations are also currently known to impede clarithromycin action against the bacteria[45]. Furthermore, it is believed that efflux pump systems, specifically the HP0605-HP0607 gene cluster, also may worsen H. pylori resistance when the bacteria already present 23S rRNA alterations[46].
Regarding fluoroquinolones, H. pylori primarily develops resistance through mutations in the genes encoding bacterial type II topoisomerases, specifically DNA gyrase, which is crucial for DNA replication and serves as the target of these drugs[47]. Resistance typically arises from alterations in the A subunits of DNA gyrase, encoded by the gyrA gene[48]. The quinolone resistance-determining region within this gene is particularly susceptible to mutations that alter the target protein structure and binding affinity to the enzyme, especially in codons 91 and 87[49]. Nevertheless, quinolone resistance can also develop through mutations in the B subunits of DNA gyrase, encoded by the gyrB gene[50].
Resistance to metronidazole in H. pylori primarily arises from disruptions in the activation of the drug, which occurs through redox reactions inside the bacteria carried out by enzymes, such as oxygen-insensitive NADPH nitroreductase[51]. This interaction triggers the production of reactive oxygen species and other reductive intermediates that lead to severe DNA damage and cytotoxicity. Despite impairing the survival of the bacteria, it also heightens the mutation rate, resulting in higher resistance of H. pylori to metronidazole compared to other antibiotics[52,53]. RdxA is one of the genes responsible for encoding reductase enzymes and is a primary site for mutations, which can lead to the inactivation of these enzymes and the disruption of metronidazole activation within the bacterial cell[54]. However, several mutations in rdxA are developmental signals rather than being associated with resistance, limiting the reliability of molecular methods for detecting metronidazole resistance[53]. Another gene, frxA, also plays a similar role in metronidazole resistance, particularly when combined with other mutations, although its specific contribution to resistance is not as clearly agreed upon as rdxA[55,56].
Concerning beta-lactams, H. pylori resistance mechanisms hinge on mutations in a few critical genes. Amoxicillin and other drugs in this class target penicillin-binding proteins in the bacterial periplasm, forming a stable complex that disrupts the cross-linking of peptidoglycan in the cell wall[57]. Unlike many other gram-negative pathogens where beta-lactamase production is central to resistance development, H. pylori primarily relies on mutations in the pbp1A gene that may occur at multiple sites[58]. These mutations reduce the affinity of penicillin-binding proteins for beta-lactams, significantly diminishing antibiotic effectiveness[59]. Additionally, genetic alterations in porin proteins, particularly hopB and hopC, are also believed to contribute to resistance to multiple beta-lactams, especially when they occur in synergy with each other and with pbp1A mutations[60,61].
Tetracycline is a broad-spectrum antibiotic that inhibits bacterial protein synthesis by binding to the 30S subunit of ribosomes and blocking aminoacyl-tRNA attachment[62]. Resistance mechanisms to tetracycline have been documented in other bacteria, typically involving specific antibiotic efflux pumps[63]. In H. pylori, however, efflux pumps specific to tetracycline are rarely reported, with references only to a homolog of the tetA(P) efflux pump and other multidrug efflux systems as potential resistance mechanisms[64,65]. Tetracycline resistance in H. pylori is more commonly associated with mutations in the 16S rRNA-encoding genes, particularly at positions 926-928 and 965-967, which disrupt the binding site of the antibiotic and decrease its efficiency[63,66].
Lastly, furazolidone and rifampicin are considered alternative antibiotics for rescue treatment[67]. Furazolidone is a nitrofuran and shares similarities with metronidazole, dealing damage to the DNA of the bacteria but also requiring activation by reductase reactions within the bacteria[68,69], such as flavodoxin pyruvate oxidoreductase, encoded by the porD gene[70]. Mutations in this gene are thought to be the main contributors to increased resistance to furazolidone by H. pylori, although most research on this mechanism is dated and scarce[71]. Rifampicin functions by binding to the β subunit of DNA-dependent RNA polymerase, encoded by the rpoB gene, thereby severely disrupting RNA transcription and protein synthesis[72]. Resistance to rifampicin is typically associated with mutations in the rifampicin resistance-determining region of the rpoB gene, though resistance can also arise from mutations outside this region[73,74].
Multidrug resistance (MDR) (i.e. resistance to three or more antibiotics of different classes) in H. pylori presents a significant challenge in clinical management. Boyanova et al[75] reported that the most common patterns of MDR in H. pylori may be resistance to clarithromycin, metronidazole, and fluoroquinolone. MDR can be developed through the accumulation of gene mutations that confer single-drug resistance. However, other direct contributors are intrinsically related to MDR manifestation, including the upregulation of multidrug efflux pump systems and biofilm formation.
The expression of several genes is associated with active efflux phenotypes that lead to MDR in H. pylori by protecting the bacteria from toxic antibiotic effects. Key genes involved include HP0605 (hefA), HP1174 (gluP), HP1181, and HP1184[76-79]. Specifically, the upregulation of gluP by the activity of spoT enzyme can stimulate biofilm formation in H. pylori[78]. Notably, when compared to planktonic (freely existing) cells, biofilm-forming cells also exhibit significantly higher expression of other efflux pump proteins genes, including hefA, HP1181 (both related to MDR), and HP1165 (related to tetracycline resistance) and those coding for transmembrane ABC transporters[76,78,80].
Certain H. pylori strains can form biofilms on gastric mucosa, consisting of dead cells and extracellular polymeric substances, including mannose-related proteoglycans and extracellular DNA[81-83]. In various pathogens and situations, the biofilm matrix serves as a robust, non-specific barrier that shields bacterial communities from direct antimicrobial effects, aiding horizontal gene transfer and promoting the overexpression of resistance mechanisms[84,85]. Conceivably, studies have shown that biofilm-forming H. pylori exhibit increased levels of minimum bactericidal concentration for amoxicillin, metronidazole, tetracycline, and erythromycin than planktonic counterparts[76,86]. Moreover, when biofilm cells are subjected to the same antimicrobial concentration as planktonic cells, they present decreased susceptibility to clarithromycin[86]. Although the mechanisms are not fully understood, biofilm formation appears to significantly contribute to the MDR observed in H. pylori strains. Hence, targeting biofilm inhibition could be a promising strategy in combating these resistant infections.
The coexistence of subpopulations with different levels of antibiotic resistance within a single patient is termed heteroresistance[87]. In H. pylori infection, this phenomenon can be perceived within a single biopsy (intraniche heteroresistance) or across different biopsy sites (interniche heteroresistance)[88]. Heteroresistance can arise from simultaneous infection with multiple H. pylori strains with distinct resistance profiles or within a single monoclonal strain due to antibiotic pressure or spontaneous mutations[89].
Due to variations in heteroresistance frequency among diverse populations and geographic areas[90], the overall prevalence of this phenomenon in H. pylori is not well defined, with reported rates ranging from 7%-60% for clarithromycin and 14%-61% for metronidazole[91,92]. Given the significance of these frequencies, it is both cost-effective and recommended that endoscopic procedures aimed at AST include the collection of combined samples from both the antrum and corpus as well as the retrieval of multiple isolates from each biopsy site[90,93]. Consequently, a more precise drug-susceptibility profile is achieved, leading to more precise antibiotic regimen adjustments[90].
Sustaining intragastric acid suppression in H. pylori-positive patients is essential for relieving peptic ulcer symptoms and enhancing antibiotic efficacy. Since cytochrome P450 2C19 (CYP2C19) is responsible for around 80% of the biotransformation of first-generation PPIs such as omeprazole, lansoprazole, and pantoprazole[94], CYP2C19 polymorphisms significantly impact the pharmacokinetics, bioavailability, and clinical efficacy of these drugs[95]. The phenotype manifestations of CYP2C19 polymorphisms include ultrarapid metabolizer (UM), normal (NM) (previously referred to as extensive), intermediate, and poor metabolizers (PM)[96].
The distribution of CYP2C19 phenotypes shows significant ethnic and geographic differences, with nearly one-third of the global population exhibiting a significant variation, i.e. being either PM or UM[96,97]. Notably, a recent meta-analysis concluded that patients with CYP2C19 UM or NM phenotypes undergoing eradication regimens containing first-generation PPIs (omeprazole, pantoprazole, or lansoprazole) have a 2.14-fold significantly higher likelihood of H. pylori eradication failure compared to those with intermediate or PM phenotypes[98]. Thus, pharmacogenetic guidelines recommend considering a 50%-100% increase in the dosage of these PPIs to optimize therapeutic efficacy in both CYP2C19 UM and NM[99]. Regarding the impact of CYP2C19 polymorphisms on the H. pylori eradication rate using new-generation PPIs, such as rabeprazole and esomeprazole, some studies indicate less interference, but the available evidence is inconsistent and graded as moderate or weak[99].
There are no well-established international guidelines regarding the use of CYP2C19 genetic testing in the pretreatment setting. CYP2C19 genetic tests have significant limitations in terms of sensitivity, as they are not designed to detect all possible variants of the CYP2C19 gene, including rare alleles with deletions in the gene locus[99]. Additionally, according to randomized controlled trials (RCTs), CYP2C19 phenotypes do not impact the acid suppression effect of P-CABs like vonoprazan and tegoprazan[100,101], which are predominantly metabolized by CYP3A4 according to in vitro evaluation[102]. Consequently, given that these drugs are currently recommended as alternatives to PPIs in first-line pharmacological regimens, the clinical utility of CYP2C19 testing before treatment is uncertain, requiring further cost-effectiveness evaluation across diverse patient populations.
Furthermore, poor compliance with therapeutic regimens conceivably increases the risk of unsuccessful H. pylori eradication. Adverse drug events, such as nausea, diarrhea, fatigue, and abdominal pain, although generally mild, are common, reported by 26%-74% of patients[103,104], and must be considered due to their impact on therapeutic adherence and patient well-being. Reinforcing medication adherence might importantly improve H. pylori eradication rates in developing countries[105]. Lastly, treatment compliance is impacted by cost and complexity[104], emphasizing the critical need for more affordable and streamlined therapeutic regimens.
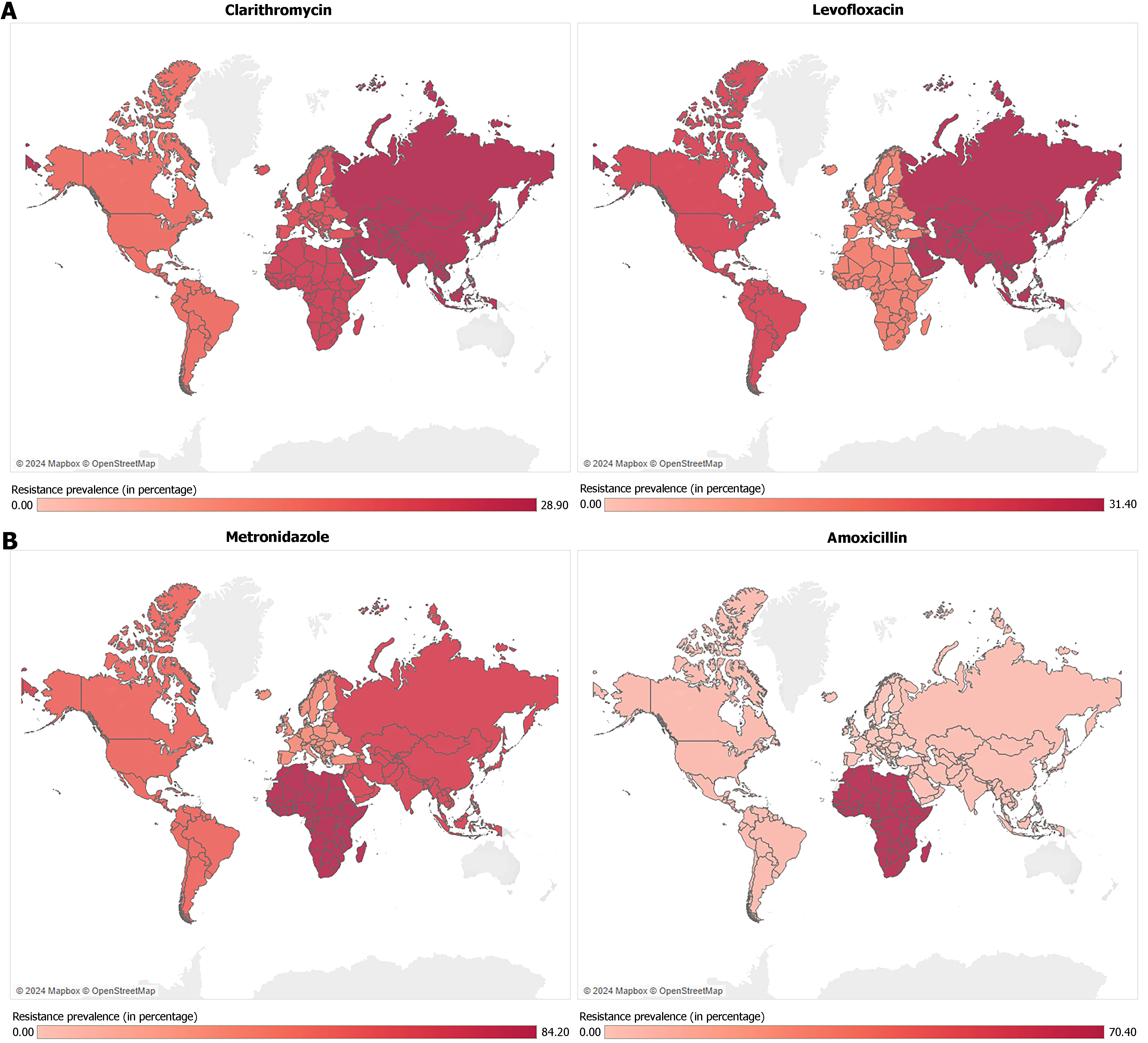
The rising resistance of H. pylori to antibiotics poses a significant global health challenge, with resistance patterns differing markedly across regions[106]. Meta-analyses have documented a substantial increase in antibiotic resistance worldwide. A 2018 meta-analysis[30] indicated metronidazole resistance as the most common resistance pattern, surpassing 15% in all regions WHO regions, while clarithromycin resistance exceeded 15% in most WHO regions, except in the Americas. A more recent meta-analysis from 2024[107] indicated that pooled primary resistance to clarithromycin (from 2013 to 2023) surpassed 15% in all regions, ranging from 16.0% [95% confidence interval (CI): 11.7%-20.8%)] in the Americas to 28.9% (95%CI: 26.6%-31.2%) in Asia (Figure 1A). Remarkably, in children, clarithromycin resistance is the predominant pattern of resistance with a global prevalence of 38.2%, which is significantly higher than that observed in adults (25.6%)[107]. Nonetheless, recent findings show substantial variability regarding resistance in H. pylori-infected children[107-110].
Amoxicillin and tetracycline generally show lower resistance rates (< 10%) across most regions, making them more reliable options in treatment protocols within these areas[30]. However, amoxicillin resistance is considerably higher in Africa at 70.4% (95%CI: 64%-76.4%), where metronidazole resistance is also remarkably frequent at 84.2% (95%CI: 78.9%-88.9%)[107] (Figure 1B). Despite being highly heterogeneous, these findings align with past research[111] and highlight the ongoing need to improve efforts in antibiotic stewardship and monitoring antibiotic resistance trends in Africa.
In India, a recent meta-analysis encompassing studies from 2000 to 2023 identified the prevalence of resistance to clarithromycin at 35.6% (with a downward trend), to metronidazole at 77.7% (stable), and to levofloxacin at 32.8% of patients (with an upward trend)[112]. Thus, metronidazole-containing regimens are inadequate as a first-line treatment in most regions of India, and amoxicillin, tetracycline, and furazolidone are being considered as options[112]. Similarly, in China, antimicrobial resistance is 36.7%, 69%, 29.4%, and 1.4% for clarithromycin, metronidazole, levofloxacin and amoxicillin, respectively. This shows that amoxicillin may be important for treatment[113] and that bismuth is essential for regimens containing metronidazole and clarithromycin in China, given its ability to overcome resistance[114].
In Australia, the prevalence of clarithromycin resistance has been over 20.0% since 2010, with an average increase of 3.7% per year, while stable trends have been seen for metronidazole resistance (35.3%)[115]. In Portugal, a 2018 meta-analysis identified resistance to clarithromycin in 42% and metronidazole in 25% of patients[116]. Resistance rates to clarithromycin and levofloxacin are higher than 25% in Turkey[117] and 30% in the United States[118]. In the studies cited, resistance to tetracycline and amoxicillin showed low rates[115-118].
In the Americas, steady upward trends in resistance to clarithromycin (1.85%-32.20%) and levofloxacin (9.20%-58.10%) were seen in Mexico between 1997 and 2017[119]. In Brazil, a multicenter study identified resistance to clarithromycin in 16.9% and metronidazole in 13.5% of patients admitted between 2012 and 2015[120]. In subsequent studies in northeastern Brazil, the prevalence of resistance to clarithromycin detected was 14.4%-14.5%[120,121], while in the southern region resistance to clarithromycin ranged from 8.7%-19.1% and to levofloxacin from 16.4%-22.5%[120,122]. However, the recent evaluation of resistance in H. pylori is scarce in Latin America[107], which limits a representative determination of the profile in these territories. For example, resistance in H. pylori has not been investigated in Brazil since 2020, a period marked by the severe acute respiratory syndrome coronavirus 2 pandemic, in which there was indiscriminate consumption of antibiotics, including macrolides[123]. In 2022, a study in Ecuador found resistance to clarithromycin in 33.6% of cases[124]. Furthermore, on the African continent, studies conducted in Egypt reported rates of resistance to clarithromycin ranging from 28.7% to 52.8% among H. pylori-positive patients between 2021 and 2022[125,126]. Still, there are significant gaps in the profile of recent resistance in other African countries.
The economic unavailability of resources for detecting resistance profiles and underlying mechanisms leads to several limitations regarding epidemiological surveillance in developing countries. Additionally, the lack of standardized testing protocols and follow-up data in some regions further complicates the accurate assessment of antibiotic resistance, leading to potential underestimation or regional disparities in reported resistance levels[116]. Moreover, most studies present high levels of heterogeneity, complicating the accurate assessment of antibiotic resistance and potentially contributing to the underestimation of regional disparities in resistance rates[30,107,111]. The currently available data concerning the mechanisms and the prevalence of H. pylori resistance to clarithromycin, metronidazole, levofloxacin, tetracycline, amoxicillin, rifampicin, and MDR patterns, at global and continental levels, are summarized in Table 1.
| Resistance pattern | Main resistance mechanisms | Resistance prevalence by region (2013-2023) | ||||
| Africa | Americas | Asia | Europe | General | ||
| Clarithromycin | Nucleotide substitutions disrupting the antibiotic binding site in the 23S rRNA (A2142G, A2143G, A2142C, A2115G, G2212A, G2141A, A2144T, and T2289C mutations); efflux pump systems (HP0605-HP0607) | 24.6% (16.4-33.9)1 | 16.0% (11.7-20.8)1 | 28.9% (26.6-31.2)1 | 21.3% (18.1-24.6)1 | 26.7% (24.7-28.8)1 |
| Metronidazole | Partial inactivation of reductase enzymes required for metronidazole activity (mutations mainly in rdxA and possibly in frxA gene) | 84.2% (78.9-88.9)1 | 48.1% (43.2-53.1)1 | 66.1% (62.1-69.9)1 | 29.9% (24.0-36.1)1 | 59.6% (55.2-63.9)1 |
| Levofloxacin | Disrupted antibiotic binding site in the DNA gyrase (mainly gyrA but also gyrB gene mutations) | 14.3% (0-49.9)1 | 25.0% (5.2-52.8)1 | 31.4% (28.3-34.6)1 | 13.3% (10.4-16.4)1 | 26.2% (23.5-28.9)1 |
| Amoxicillin | Reduced affinity to PBPs (pbp1A gene mutations); porin protein alterations | 70.4% (64.0-76.4)1 | 4.8% (2.8-7.3)1 | 2.8% (2.0-3.7)1 | 0% (0-0.2)1 | 2.6% (1.8-3.5)1 |
| Tetracycline | Disrupted antibiotic binding site in rRNA (mutations in 16S rRNA-encoding genes); efflux pump tetA[P] (HP1165), and other multidrug efflux systems | 1% (0.1-2.5)1 | 1.1% (0-4.2)1 | 2.2% (1.3-3.2)1 | 0% (0-0)1 | 1.5% (0.9-2.3)1 |
| Rifampicin | Amino acid exchanges disrupting antibiotic binding site in the β subunit of DNA-dependent RNA polymerase (mutations in rpoB gene) | 0%3 (Algeria)[234,235] | 0%-23.0%3 (Colombia)[236,237], 7.0%-19.0%3 (New York, United States)[238] | 14.4%3 (Iran)[239], 5.4%-73.2%3 (China)[240] | 11.4%3 (Belgium)[241], 8.3%3 (Bulgaria)[242], 1.2%3 (France)[243], 33.3%3 (Spain)[244] | Unavailable |
| Multi-drug resistance patterns | Accumulation of single-drug resistance genes; upregulation of multidrug efflux pump systems (hefA, gluP, HP1181 and HP1184); biofilm formation | 1.6%3 (Egypt)[245], 15.7%3 (Cameroon)[246] | 12.5%3 (Chile)[247] | 10.0% (7-14)2 (children in East Asia)[248], 14.7%2 (India)[112], 24.9%3 (China)[249] | 1.4%2 (Portugal)[116], 0.8%3 (Austria)[250], 2.4%3 (Spain)[251] | 6.0%2 (children)[252] |
With increasing antibiotic resistance leading to H. pylori eradication failure, evaluating the most effective treatment approaches is essential. According to the Taipei global consensus[127], a highly effective empiric regimen is generally preferred over a susceptibility-guided approach due to cost and convenience. However, AST-guided therapy can be used as a first-line option when available[20]. This approach might be particularly beneficial in settings where data on local resistance patterns lacks an update. Hence, a more in-depth evaluation is required to compare the efficacy and cost-effectiveness of AST-guided therapies vs empirical approaches from a broader public health perspective. Furthermore, to prevent eradication failure it is essential to consider including medications that improve effectiveness and adherence if they are accessible.
PPIs have been employed for the treatment of dyspepsia since the 1980s, with the first-generation including omeprazole, lansoprazole, and pantoprazole[128]. However, limitations in the effects of PPIs on H. pylori eradication highlighted the need for more efficacious therapeutic approaches, driving the development of newer generations of PPIs and additional pharmaceutical agents aimed at improving therapeutic outcomes[128].
Subsequent generations of PPIs, such as rabeprazole, esomeprazole, ilaprazole, anaprazole, tenatoprazole, and dexlansoprazole, were introduced as alternative options for the treatment of H. pylori[128,129]. McNicholl et al[130] performed a meta-analysis to compare the efficacy of esomeprazole and rabeprazole to first-generation PPIs (omeprazole, lansoprazole, and pantoprazole) in H. pylori treatment effectiveness. The analysis indicated that regimens containing esomeprazole and rabeprazole achieved higher eradication rates compared to those with first-generation PPIs[130]. However, the authors cautioned that these findings should not be generalized due to variations in therapeutic regimens and differences in patient metabolism related to CYP2C19 polymorphisms. Notably, the efficacy differences were minimal in poor metabolizers and significant in normal metabolizers[130].
These findings underscore that identifying a single PPI as the most effective for eradication regimens may not be appropriate, and the choice of PPI should be tailored to individual clinical circumstances[130,131]. It is important to note that the overall efficacy of newer generations of PPIs remains comparable in H. pylori eradication, as demonstrated by Zhu et al[132] and Jin et al[133], that found no significant differences between anaprazole and rabeprazole and ilaprazole and esomeprazole, respectively.
Advances in pharmacotherapy have also introduced vonoprazan, a P-CAB, as a new option in H. pylori eradication therapy[134]. Vonoprazan acts by ionically, reversibly, and competitively binding to the H+/K+-ATPase pump, which is responsible for gastric acid secretion[134]. This mechanism inhibits acid secretion more efficiently and for a longer time than PPIs, due to the high pKa of vonoprazan, which promotes increased drug accumulation in the gastric environment[134,135].
In this context, an RCT conducted by Bunchorntavakul and Buranathawornsom[136], involving 118 patients, evaluated two treatment groups: One received vonoprazan 20 mg, amoxicillin 1 g, and clarithromycin 500 mg, while the other received omeprazole 20 mg, amoxicillin 1 g, and clarithromycin 500 mg. The study concluded that the vonoprazan group was not inferior to the omeprazole group and demonstrated slightly superior eradication rates[136].
Further, Chey et al[137] conducted a RCT involving 1046 patients and concluded that vonoprazan-containing regimens, including both triple (vonoprazan/amoxicillin/clarithromycin) and dual (vonoprazan/amoxicillin) therapies, achieved superior outcomes in the overall study population compared to the PPI-containing triple therapy, regardless of clarithromycin resistance. The intention-to-treat (ITT) eradication rates were 80.8% for vonoprazan in triple therapy, 77.2% for vonoprazan in dual therapy, and 68.5% for lansoprazole in triple therapy[137].
In the context of quadruple therapy, a recent RCT conducted by Yang et al[32], including 600 patients, found that vonoprazan-dual therapy, either administered for 14 or 10 days, achieved better eradication rates than a 14-day qua
| No. of patients | Group with vonoprazan (days) | Eradication | Control group (days) | Eradication | Identification | Ref. |
| 1046 | Vonoprazan 20 mg + amoxicillin 1 g + clarithromycin 500 mg (14 days) | 80.8%1 | Lansoprazole 30 mg + amoxicillin 1 g + clarithromycin 500 mg (14 days) | 68.5%1 | NCT04167670 | |
| 118 | Vonoprazan 20 mg + amoxicillin 1 g + clarithromycin 500 mg (7 days) | 96.7%1 | Omeprazole 20 mg + amoxicillin 1 g + clarithromycin 500 mg (14 days) | 88.5%1 | TCTR20210219007 | [134] |
| 600 | Vonoprazan 20 mg + amoxicillin 1 g (14 days) | 92.5%2 | Rabeprazole 20 mg + bismuth potassium citrate/tinidazole/clarithromycin, combined packet 4.2 g (14 days) | 81.5%2 | NCT05469685 | [32] |
| 234 | Vonoprazan 20 mg + amoxicillin 1 g + furazolidone 100 mg + bismuth 200 mg (10 days) | 96.2%1 | Esomeprazole 20 mg + amoxicillin 1000 mg + furazolidone 100 mg + bismuth 200 mg (14 days) | 93.6%1 | NCT04907747 | [136] |
Meta-analyses also highlight the promise of vonoprazan as a first-line treatment for H. pylori compared to PPIs. A recent meta-analysis conducted by Liu et al[140], which included 27 studies comparing vonoprazan-containing therapies among themselves or with PPI-containing therapies, indicated that vonoprazan-based BQT achieved the highest pooled eradication rate among the therapies. Also, vonoprazan dual therapy (vonoprazan-amoxicillin) did not present a significant difference in eradication rates vs PPI-containing BQT in both ITT [odds ratio (OR) = 1.02, 95%CI: 0.70-1.47] and PP analyses (OR = 0.82, 95%CI: 0.41-1.62), but it did demonstrate a significantly reduced risk of adverse events (OR = 0.40, 95%CI: 0.24-0.68)[140]. Similarly, previous meta-analyses have indicated the general superiority of vonoprazan-based regimens compared to those based on PPIs[139,141] and that vonoprazan dual therapy is as effective as PPI-containing BQT[142]. Thus, RCTs and meta-analyses performed so far underscore the promise of P-CABs in H. pylori treatment, positioning it as a feasible option with both efficacy and safety. Once more evidence is collected from various regions, these drugs may be included in future guidelines in first-line regimens, as they appear to enable less use of antimicrobials without compromising effectiveness.
Enhancing first-line empirical therapy is essential to prevent the development of secondary MDR in H. pylori strains, as first-line treatment failure plays a significant role in the emergence of MDR in H. pylori[143,144]. However, the rising levels of antibiotic resistance hinder the selection of therapies that effectively balance efficacy, a low incidence of adverse effects, and high patient adherence. In this context, greater efforts are required to develop treatment options that cost-effectively meet these criteria[145].
Since evidence suggests that clarithromycin resistance has likely exceeded 15% on all continents[30,107,146], BQT is widely recommended as the preferred empirical treatment on a global scale. Thus, clarithromycin triple therapy should be reserved for areas with reliably updated surveillance confirming resistance rates below 15%, as outdated data risks compromising treatment efficacy. In a meta-analysis from 2021, Rokkas et al[147] found that vonoprazan triple therapy and reverse-hybrid therapies consisting of a PPI and amoxicillin for 14 days, with clarithromycin plus metronidazole in the initial 7 days were the most effective among eight first-line regimens, while standard (PPI) triple therapy was the least effective[147].
Additionally, more recent RCTs following the Maastricht VI/Florence Consensus Report have provided stronger evidence for different therapeutic approaches and alternatives in light of the high levels of current antibiotic resistance. In China, a recent RCT found that 10-day vonoprazan-amoxicillin dual therapy with vonoprazan 20 mg twice/day provided lower adverse events and a non-inferior efficacy compared to 14-day BQT[31]. Further, poor compliance (less than 80% of prescribed drugs taken) was significantly linked to treatment failure in the vonoprazan-amoxicillin dual therapy group but not in the BQT group, which indicates that the extended 14-day duration of BQT may be redundant[31]. Indeed, recent research in China and Taiwan has found that 10-day BQT has non-inferior efficacy to the 14-day BQT as a first-line therapy and presents a lower incidence and severity of adverse effects, such as dizziness and vomiting[34,148,149].
Recently, the LEGACy consortium, which included patients from European and Latin American countries, demonstrated that BQT was the only regimen to achieve a cure rate exceeding 90.0% (compared to 88.7% for non-BQT regimens and 75.2% for triple therapy)[150]. Additionally, the 2024-Hp-EuReg trial, involving 49690 patients, provided a comprehensive analysis further confirming that BQT consistently achieves eradication rates above 90% across all European regions[151]. Notably, the 10-day single-capsule BQT proved to be the regimen most reliably associated with optimal effectiveness[151].
Regarding second-line therapy for H. pylori infection after failure of clarithromycin triple therapy[152], an RCT conducted in Taiwan reported that both levofloxacin-based quadruple therapy and BQT have shown comparable rates of effectiveness (88%) in a scenario with increased trends in resistance[153]. In the context of rescue treatment regimens, rifabutin-containing triple therapy and high-dose amoxicillin dual therapy demonstrate similar efficacy to BQT but with fewer side effects[154,155]. Furthermore, a recent multicenter trial[156] found that a BQT regimen containing amoxicillin and tetracycline achieved optimal eradication rates as a rescue therapy[156]. Notably, administering tetracycline three times daily instead of four reduced adverse events without compromising efficacy[156].
AST is essential to ensure effective tailored bacterial eradication therapies. AST can be conducted through both culture-dependent methods, mostly performed to determine the minimum inhibitory concentration (MIC) of antibiotics, and molecular assays[157-159].
Culture-dependent AST: Given the possibility of interniche heteroresistance, culture-dependent techniques require the collection of at least two biopsy samples from both the antrum and the corpus and involve time-consuming and labor-intensive H. pylori isolation[93]. Among these, phenotypical methods [e.g., Agar dilution (AD), disk diffusion (DD), broth microdilution (BD), and gradient E-tests (GET)] are commonly used for AST, though each method has its own limitations[160].
AD comprises the incorporation of different antibiotic concentrations into agar plates, inoculation of H. pylori strains, and observation of the growth for MIC determination[161,162]. Despite being fastidious, this technique is regarded as the golden standard to assess the MIC of antimicrobial agents and as the reference method to compare other AST approaches[4]. However, widely accepted clinical MIC breakpoints have not yet been established for antibiotics other than clarithromycin, including amoxicillin, metronidazole, levofloxacin, and furazolidone[163]. The current cutoff points established under the European Committee on Antimicrobial Susceptibility Testing are based on epidemiological thresholds, which constrain their applicability in clinical practice[163]. This limitation affects the interpretation of AD results as well as other MIC-oriented techniques, such as BD and GET.
Notwithstanding, BD and GET offer less fastidious methods for MIC determination. BD consists in the inoculation of H. pylori into a range of serially diluted antibiotics within a liquid medium[164]. Despite the challenges associated with the growth of H. pylori in broth, the development of supplemented media, automation, and the possibility of simul
Although quantitative methods such as AD, BD, and GET are valuable for assessing H. pylori susceptibility, DD remains a widely used qualitative AST method for H. pylori[170]. In this technique, paper disks saturated with antibiotics are placed on an agar plate inoculated with the target bacteria. Upon incubation, the zone of inhibition around each disk is measured to assess the effectiveness of the antibiotic in impeding bacterial growth[171,172]. Despite its simplicity and ease of execution, DD does not provide precise MIC values, thereby limiting its ability to offer detailed information about bacterial sensitivity[173]. Collectively, all these techniques highlight the need for continued development and standardization in susceptibility testing methods to improve their clinical utility and accessibility.
Molecular-based AST: In contrast to phenotypic techniques, molecular-based antibiotic susceptibility tests are designed to detect specific genetic mutations or markers in the H. pylori genome that are associated with resistance to particular antibacterial agents[174]. Currently, the most commonly employed molecular-based techniques are PCR and next-generation sequencing[163]. PCR is particularly effective for detecting specific mutations within the H. pylori genome, whereas next-generation sequencing offers a more comprehensive overview of resistance determinants and is increasingly used to profile antibiotic resistance in clinical isolates of the bacterium[175-178]. Although these techniques usually require gastric biopsy, isolation of viable specimens is not necessary, which streamlines the process and reduces the time and labor involved in susceptibility testing. Furthermore, results from molecular-based ASTs using fecal samples have shown a close correlation with those obtained from gastric biopsies, suggesting that non-invasive testing is a viable alternative[179,180].
Despite these advantages, several factors still limit the widespread use of molecular-based methods. Notably, there is variability in the concordance between genotype and phenotype across different antibiotics[181]. For example, determining susceptibility to metronidazole can be highly complex, whereas studies have shown excellent concordance for clarithromycin and levofloxacin[39,182,183]. Additionally, the high costs and the need for advanced technological infrastructure, which is often unavailable in certain regions, further restrict the widespread adoption of these methods. Consequently, there are still constraints preventing molecular-based AST from reaching its full potential.
The extensive variation in susceptibility among H. pylori strains and the differing availability of AST in different regions make it difficult to achieve universally applicable results from AST-based treatments, which demand specific analysis under different populations and clinical contexts. Previous meta-analyses indicated that AST-guided therapy could provide higher eradication rates when compared to unspecified first-line empirical therapies[184,185]. While some analyses suggested the superior effectiveness of AST-guided therapy over BQT[185,186], the substantial heterogeneity identified precluded a significant conclusion. Still, no significant differences were found in second-line or third-line treatment scenarios[185], which might occur since subsequent empirical therapies culminate in progressively broader spectrum coverage of resistant strains.
Presently, there is a specific focus on the effectiveness and feasibility of genotypic (molecular-based) tailored therapies in eradicating H. pylori. They have shown similar eradication outcomes to culture (phenotypic)-guided therapies in first-line treatment and non-inferiority in third-line therapy, as reported by a recent multicenter RCT[187]. While molecular-guided therapies consistently have better eradication rates than standard triple therapy[188,189], a meta-analysis by Li et al[188] showed lower efficacy when compared to empirical quadruple therapy (either BQT or non-BQT), but the authors emphasized the limited number of RCTs (only five) focusing on this comparison at the time, avoiding unwarranted conclusions[188]. Yet, the pooled eradication rates of genotypic-tailored therapy reported in the study were notably lower than those presented in a single-arm meta-analysis (79.0% vs 86.9% and 86.0% vs 91.5% by ITT and PP analyses, res
Aside from effectiveness, choosing between tailored or empirical therapy includes considering local factors such as the background rate of antimicrobial resistance, availability of resources (especially ASTs and bismuth), and cost-effectiveness of ASTs[186,192]. Due to the unavoidable variability in cost-effectiveness evaluation across different periods, locations, and sample constraints, it is not feasible to make broad generalizations.
In France, a recent multicenter RCT (presenting an 18.7% rate of clarithromycin resistance) reported that 14-day PCR-guided triple therapy (consisting of esomeprazole, amoxicillin plus clarithromycin or levofloxacin) was non-inferior and less expensive than 14-day non-BQT in ITT analysis[193]. In South Korea, numerous studies have been carried out to assess the cost-effectiveness of dual priming oligonucleotide-based multiplex PCR, which can be used to detect point 23S ribosomal RNA gene mutations related to clarithromycin resistance. Although these studies were confined to a single country and focused on the same analytical tool, the results were inconsistent, with some reporting that tailored therapy reduced average costs for successful eradication[194,195], while others showed higher average medical costs[196,197].
Nevertheless, as H. pylori infection can be accurately diagnosed with non-invasive methods (such as stool antigen test and urea breath test), the need for endoscopy and biopsy, generally required for AST, is often avoidable in a “test-and-treat” approach that employs empirical regimens, ultimately preventing additional costs. Changing this paradigm, the development of non-invasive methods that accurately identify antibiotic resistance can be convenient alternatives in tailored treatment compared to conventional biopsy-ASTs. Genotypic testing of clarithromycin resistance in stool samples is a promising alternative, presenting a pooled sensitivity of 93% (95%CI: 90%-96%) and specificity of 98% (95%CI: 93%-100%), according to a meta-analysis by Ren et al[41].
Despite often showing enhanced outcomes, according to current evidence, tailored treatments achieve high cure rates (> 90%) in only 40% to 63% of cases[198,199]. While robust trials have demonstrated the consistency of BQT in achieving > 90% eradication rates, suggesting that routine AST-guided therapy may not significantly improve overall therapeutic success[151]. However, the unavailability of bismuth in some regions and the complexity of its traditional regimen, requiring multiple pills daily, pose challenges to adherence in real-world settings. Furthermore, a retrospective cohort study in Thailand involving 1080 patients concluded that AST-guided therapy offered higher efficacy and could be a cost-effective strategy if initiated immediately after first-line treatment failure[200]. Notably, this approach was linked to reduced costs for subsequent medication, post-treatment urea breath test, and hospital visits in a real-world scenario[200]. In the context of pediatric H. pylori infection, recent clinical guidelines recommend AST to assess clarithromycin susceptibility as a first-line strategy, highlighting its importance in improving treatment outcomes and reducing antimicrobial resistance[201].
Due to growing concerns about antimicrobial resistance, there is an increased emphasis on developing new treatments for H. pylori. Adjuvant therapies are designed to boost the effectiveness of antibiotic treatments, either by countering bacterial resistance mechanisms or by altering the response of the host[202]. While ongoing enhancements to antibacterial drug combinations may offer short-term effectiveness, their impact often diminishes over time[203]. Thus, research into complementary approaches, including antibiofilm agents and the use of probiotics, is still in progress.
The formation of H. pylori biofilms decreases the efficacy of conventional treatments[81]. In this sense, several studies are being developed to investigate the use of antibiofilm agents as adjuvants[204]. Most antibiofilm agents originate from natural products, with many being secondary metabolites produced by microorganisms, including phytochemicals, biosurfactants, antimicrobial peptides, and microbial enzymes[205]. Additionally, certain quorum-sensing inhibitors and probiotics have been identified to exhibit anti-biofilm properties[206,207]. These natural products demonstrate strong anti-biofilm and antibacterial properties in vitro. Notably, some, like Pistacia vera L. oleoresin, Casearia sylvestris leaf derivatives, Amu-ru 7, and dihydrotanshinone I, have shown effectiveness against H. pylori-resistant strains in both in vitro and in vivo studies[208-212], suggesting the potential to mitigate H. pylori MDR and creating a complementary effect against it. For instance, Pistacia vera L. oleoresin boosts the effectiveness of levofloxacin, aiding in the suppression of drug resistance in H. pylori strains. Armeniaspirol A is another antibiofilm agent that may exhibit significant antibacterial activity against H. pylori, including strain resistant to multiple drugs[204]. Furthermore, the combination of armeniaspirol A with omeprazole was more successful in eliminating H. pylori in vivo than standard triple therapy in a mouse model of MDR infection[213].
In recent years, nanomaterials have been utilized to eliminate H. pylori biofilms and reduce drug resistance[214-216]. A recent study reported that combining antibiotics with rhamnolipid, a glycolipid biosurfactant that can disrupt biofilms and potentially inhibit bacterial adhesion, effectively prevented biofilm formation in vitro[217]. Also, nanodrugs formulated with berberine derivatives and rhamnolipids successfully penetrated the mucus layer and effectively eradicated H. pylori biofilms in both in vitro and in vivo studies[218]. Moreover, the data presented in a study demonstrated that New Synthesized Silver Ultra-NanoClusters could represent a novel strategy for the treatment of H. pylori infections either alone or in combination with metronidazole[219]. Previous studies have indicated that N-acetylcysteine, an antioxidant that helps to break down mucus, decreases bacterial load and improves eradication rates[220,221]. It is currently the only molecule in clinical trials that has shown effectiveness against H. pylori biofilms[222-224].
Recent research is focused on the impact of adding probiotics to H. pylori eradication therapy. Probiotics are believed to potentially influence treatment primarily by reducing drug-associated side effects, competing at microbial adhesion sites, and enhancing the immune response[225]. Clinical study results, however, have been inconsistent[225].
For instance, a double-blind RCT by Ismail et al[226] found that the use of Lactobacillus reuteri after standard triple therapy was significantly associated with a higher eradication rate (22.2% and 24.3% differences in ITT and PP analyses, respectively) and mitigation of adverse effects. However, regarding the quadruple therapy scenario, while certain research reported enhanced treatment efficacy by probiotics supplementation[227], several double-blind RCTs[228,229] did not find a statistically significant improvement in eradication rates with Lactobacillus reuteri strains, despite a general agreement in their ability to reduce the frequency of side effects.
Some meta-analyses have suggested that the addition of probiotics in H. pylori treatment may be beneficial in standard triple therapy since they are associated with enhanced eradication rates and reduced risk of treatment adverse effects[230,231]. Notably, Lau et al[230] indicated possible benefits through Lactobacillus and Saccharomyces incorporation in standard triple therapy. However, these findings have not been universally accepted, as other studies did not find an improvement in the eradication rate with probiotics[232].
In this context, Yang et al[233] recently conducted an umbrella review of systematic reviews with meta-analyses, which suggested that therapies incorporating probiotics were significantly associated with improved eradication rates and a lower risk of side effects compared to standard therapy alone. Nevertheless, as reported by the authors, methodological aspects regarding low-quality studies and heterogeneity constrain the applicability of these findings as general clinical recommendations[233]. Despite the promising potential of probiotic supplementation, there is still a critical need for high-quality, multicenter RCTs that focus on specific formulations to comprehensively evaluate their effects in clinical settings. Therefore, reliable and evidence-based data can be used to update treatment guidelines and recommendations concerning the use of probiotics in H. pylori therapy. Table 3 provides an integrated summary of the Helicobacter pylori treatment strategies discussed, encompassing empirical and AST-guided antimicrobial regimens, probiotics, and antibiofilm agents, while outlining emerging alternatives and future research directions for optimizing therapeutic outcomes.
| AST-guided therapy | Empirical therapy | Antibiofilm agents | Probiotics | |||||||
| By phenotypic determination | By molecular determination | BQT | Non-BQT | CLA-TT | LEV-TT or LEV-QT | AMX-DT | RIF-TT | |||
| Advantages | Antimicrobial stewardship; AST can be done for all recommended antibiotics (vs molecular-AST) | Antimicrobial stewardship; reliable to detect CLA resistance; does not require the isolation of viable species; non-inferior efficacy vs BQT; less cost vs non-BQT in some RCTs | Optimal reliability (> 90% cure rate) regardless of resistance profile in Europe and some Latin American countries | Possible option when bismuth is not available | Reduced costs; fewer antibiotics | In second-line therapy, LEV-QT is comparable to BQT in areas with increased trends in resistance | Good reliability in general | Good reliability in general | May improve eradication rates in biofilm-forming multidrug-resistant strains | May improve eradication rates and lower the risk of side effects |
| Limitations | Highly time-consuming; affected by collection, transport, and techniques; availability | Limited number of antimicrobial agents; limited correlation with MET resistance; availability | Complex dosing in conventional regimens; availability | Not indicated in regions with dual resistance > 15% | Indicated only if CLA resistance < 15% in updated data | High trends in resistance compromise LEV-TT efficacy | High resistance rates in Africa and some areas in Asia | Potential adverse events; costs | Currently, only NAC has been shown effective in clinical trials | Methodological aspects regarding low-quality studies; heterogeneity of strains |
| Emerging approaches | Vonoprazan-containing AST-guided therapies; non-invasive AST, including CLA resistance in stool samples | Sc-BQT; 10-day BQT (non-inferior efficacy and lower adverse events vs 14-day BQT); vonoprazan-containing BQT and TT; vonoprazan-amoxicillin; BQT with amoxicillin-tetracycline | NAC; rhamnolipids; SUNCs; Pistacia vera L. oleoresin; Casearia sylvestris leaf derivative; ARM1 | Lactobacillus reuteri; Saccharomyces spp. | ||||||
| Future steps | Update regional and local resistance surveillance, especially in developing countries; validation of PCR-AST in diverse populations by correlating detected mutations with actual resistance profiles; identification of novel mutations and determinants of resistance through WGS; development and validation of non-invasive molecular-AST | Cost-effectiveness evaluation between empirical and AST-guided therapies in both RCTs and real-world data; evaluation of vonoprazan-containing therapies through multicenter RCTs in different regions and populations; evaluation of the impact of different antimicrobial therapies on the gut microbiota resistome through multicenter RCTs | Further evaluation of potential agents in high-quality clinical trials | Determination of specific strains and formulations through high-quality and multicenter RCTs | ||||||
Given the increasing global trends in H. pylori resistance, particularly against clarithromycin, metronidazole, and levofloxacin, it is essential to enhance regional and local resistance surveillance efforts, especially in developing countries where data are sparse and empirical regimens might be inappropriate. Furthermore, the validation of current PCR-based AST methods across diverse populations should be prioritized by establishing correlations between identified mutations and confirmed resistance phenotypes. Whole-genome sequencing can also play a pivotal role in identifying novel mutations and resistance determinants, particularly in regions outside Europe and Asia, where research remains limited and robust data are lacking. While BQT is generally recommended as the first-line treatment, except in regions with clarithromycin resistance rates below 15%, an evaluation of the cost-effectiveness of empirical vs AST-guided strategies should be conducted through RCTs and prospective real-world studies.
In parallel, improved treatments can be potentially attained by introducing emergent drugs, including P-CABs. Region-specific studies assessing their efficacy and cost-effectiveness will be essential to determine their applicability and future recommendations in guidelines, as they seem to allow for reduced antimicrobial usage without sacrificing efficacy. Additionally, P-CAB-containing regimens could be further explored within the context of AST-guided therapies. Furthermore, ongoing research is assessing the potential benefits of anti-biofilm agents and probiotics in clinical settings, with an emphasis on identifying specific beneficial strains and formulations.
Post-treatment evaluation remains crucial in clinical management, and confirming H. pylori eradication can be achieved using non-invasive tests like the urea breath test and stool antigen test. In light of the challenges associated with the cost and availability of AST and P-CABs, alongside the significant limitations of antibiotic resistance data due to insufficient surveillance, routine post-treatment assessment is indispensable for informed clinical decision-making.
| 1. | Chen YC, Malfertheiner P, Yu HT, Kuo CL, Chang YY, Meng FT, Wu YX, Hsiao JL, Chen MJ, Lin KP, Wu CY, Lin JT, O'Morain C, Megraud F, Lee WC, El-Omar EM, Wu MS, Liou JM. Global Prevalence of Helicobacter pylori Infection and Incidence of Gastric Cancer Between 1980 and 2022. Gastroenterology. 2024;166:605-619. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 166] [Article Influence: 166.0] [Reference Citation Analysis (0)] |
| 2. | Warren JR, Marshall B. Unidentified curved bacilli on gastric epithelium in active chronic gastritis. Lancet. 1983;1:1273-1275. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 515] [Cited by in RCA: 454] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 3. | Marshall BJ, Warren JR. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet. 1984;1:1311-1315. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3302] [Cited by in RCA: 3268] [Article Influence: 79.7] [Reference Citation Analysis (1)] |
| 4. | Malfertheiner P, Megraud F, Rokkas T, Gisbert JP, Liou JM, Schulz C, Gasbarrini A, Hunt RH, Leja M, O'Morain C, Rugge M, Suerbaum S, Tilg H, Sugano K, El-Omar EM; European Helicobacter and Microbiota Study group. Management of Helicobacter pylori infection: the Maastricht VI/Florence consensus report. Gut. 2022;gutjnl-2022. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 744] [Cited by in RCA: 667] [Article Influence: 222.3] [Reference Citation Analysis (0)] |
| 5. | Kuipers EJ, Klinkenberg-Knol EC, Vandenbroucke-Grauls CM, Appelmelk BJ, Schenk BE, Meuwissen SG. Role of Helicobacter pylori in the pathogenesis of atrophic gastritis. Scand J Gastroenterol Suppl. 1997;223:28-34. [PubMed] |
| 6. | Herrera V, Parsonnet J. Helicobacter pylori and gastric adenocarcinoma. Clin Microbiol Infect. 2009;15:971-976. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 154] [Cited by in RCA: 171] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 7. | Laird-Fick HS, Saini S, Hillard JR. Gastric adenocarcinoma: the role of Helicobacter pylori in pathogenesis and prevention efforts. Postgrad Med J. 2016;92:471-477. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 6] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 8. | Floch P, Mégraud F, Lehours P. Helicobacter pylori Strains and Gastric MALT Lymphoma. Toxins (Basel). 2017;9:132. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 58] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 9. | Ford AC, Tsipotis E, Yuan Y, Leontiadis GI, Moayyedi P. Efficacy of Helicobacter pylori eradication therapy for functional dyspepsia: updated systematic review and meta-analysis. Gut. 2022;gutjnl-2021. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 44] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 10. | Zhu F, Zhang X, Li P, Zhu Y. Effect of Helicobacter pylori eradication on gastric precancerous lesions: A systematic review and meta-analysis. Helicobacter. 2023;28:e13013. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 20] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 11. | Ford AC, Yuan Y, Forman D, Hunt R, Moayyedi P. Helicobacter pylori eradication for the prevention of gastric neoplasia. Cochrane Database Syst Rev. 2020;7:CD005583. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 39] [Article Influence: 7.8] [Reference Citation Analysis (1)] |
| 12. | Lee YC, Chiang TH, Chou CK, Tu YK, Liao WC, Wu MS, Graham DY. Association Between Helicobacter pylori Eradication and Gastric Cancer Incidence: A Systematic Review and Meta-analysis. Gastroenterology. 2016;150:1113-1124.e5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 737] [Cited by in RCA: 674] [Article Influence: 74.9] [Reference Citation Analysis (0)] |
| 13. | Lemos FFB, de Castro CT, Calmon MS, Silva Luz M, Pinheiro SLR, Faria Souza Mendes Dos Santos C, Correa Santos GL, Marques HS, Delgado HA, Teixeira KN, Souza CL, Oliveira MV, Freire de Melo F. Effectiveness of Helicobacter pylori eradication in the treatment of early-stage gastric mucosa-associated lymphoid tissue lymphoma: An up-to-date meta-analysis. World J Gastroenterol. 2023;29:2202-2221. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 21] [Cited by in RCA: 16] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 14. | Karkhah A, Ebrahimpour S, Rostamtabar M, Koppolu V, Darvish S, Vasigala VKR, Validi M, Nouri HR. Helicobacter pylori evasion strategies of the host innate and adaptive immune responses to survive and develop gastrointestinal diseases. Microbiol Res. 2019;218:49-57. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 68] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 15. | de Brito BB, da Silva FAF, Soares AS, Pereira VA, Santos MLC, Sampaio MM, Neves PHM, de Melo FF. Pathogenesis and clinical management of Helicobacter pylori gastric infection. World J Gastroenterol. 2019;25:5578-5589. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 198] [Cited by in RCA: 169] [Article Influence: 28.2] [Reference Citation Analysis (15)] |
| 16. | Suzuki T, Kato K, Ohara S, Noguchi K, Sekine H, Nagura H, Shimosegawa T. Localization of antigen-presenting cells in Helicobacter pylori-infected gastric mucosa. Pathol Int. 2002;52:265-271. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 34] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 17. | Atherton JC, Peek RM Jr, Tham KT, Cover TL, Blaser MJ. Clinical and pathological importance of heterogeneity in vacA, the vacuolating cytotoxin gene of Helicobacter pylori. Gastroenterology. 1997;112:92-99. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 424] [Cited by in RCA: 406] [Article Influence: 14.5] [Reference Citation Analysis (1)] |
| 18. | Gebert B, Fischer W, Weiss E, Hoffmann R, Haas R. Helicobacter pylori vacuolating cytotoxin inhibits T lymphocyte activation. Science. 2003;301:1099-1102. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 404] [Cited by in RCA: 409] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 19. | Zheng PY, Jones NL. Helicobacter pylori strains expressing the vacuolating cytotoxin interrupt phagosome maturation in macrophages by recruiting and retaining TACO (coronin 1) protein. Cell Microbiol. 2003;5:25-40. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 141] [Cited by in RCA: 134] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 20. | Malfertheiner P, Camargo MC, El-Omar E, Liou JM, Peek R, Schulz C, Smith SI, Suerbaum S. Helicobacter pylori infection. Nat Rev Dis Primers. 2023;9:19. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 229] [Cited by in RCA: 404] [Article Influence: 202.0] [Reference Citation Analysis (1)] |
| 21. | Gao L, Weck MN, Nieters A, Brenner H. Inverse association between a pro-inflammatory genetic profile and Helicobacter pylori seropositivity among patients with chronic atrophic gastritis: enhanced elimination of the infection during disease progression? Eur J Cancer. 2009;45:2860-2866. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 14] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 22. | Suerbaum S, Michetti P. Helicobacter pylori infection. N Engl J Med. 2002;347:1175-1186. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1848] [Cited by in RCA: 1907] [Article Influence: 82.9] [Reference Citation Analysis (3)] |
| 23. | Graham DY, Shiotani A. New concepts of resistance in the treatment of Helicobacter pylori infections. Nat Clin Pract Gastroenterol Hepatol. 2008;5:321-331. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 270] [Cited by in RCA: 281] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 24. | Shiotani A, Lu H, Dore MP, Graham DY. Treating Helicobacter pylori effectively while minimizing misuse of antibiotics. Cleve Clin J Med. 2017;84:310-318. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 56] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 25. | Jolaiya TF, Fowora MA, Onyekwere C, Ugiagbe R, Agbo II, Lesi O, Ndububa DA, Adekanle O, Njom HA, Idowu A, Adeleye IA, Bamidele M, Ngoka FN, Palamides PF, Smith SI. Duodenal ulcer promoting gene (DupA), plasticity region genes and sigma factors in H. pyloristrains from Nigeria. J Infect Dev Ctries. 2020;14:162-168. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 3] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 26. | Ganguly M, Sarkar S, Ghosh P, Sarkar A, Alam J, Karmakar BC, De R, Saha DR, Mukhopadhyay AK. Helicobacter pylori plasticity region genes are associated with the gastroduodenal diseases manifestation in India. Gut Pathog. 2016;8:10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 27. | Mehrotra T, Devi TB, Kumar S, Talukdar D, Karmakar SP, Kothidar A, Verma J, Kumari S, Alexander SM, Retnakumar RJ, Devadas K, Ray A, Mutreja A, Nair GB, Chattopadhyay S, Das B. Antimicrobial resistance and virulence in Helicobacter pylori: Genomic insights. Genomics. 2021;113:3951-3966. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 28. | Aras RA, Kang J, Tschumi AI, Harasaki Y, Blaser MJ. Extensive repetitive DNA facilitates prokaryotic genome plasticity. Proc Natl Acad Sci U S A. 2003;100:13579-13584. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 100] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 29. | Fallone CA, Chiba N, van Zanten SV, Fischbach L, Gisbert JP, Hunt RH, Jones NL, Render C, Leontiadis GI, Moayyedi P, Marshall JK. The Toronto Consensus for the Treatment of Helicobacter pylori Infection in Adults. Gastroenterology. 2016;151:51-69.e14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 695] [Cited by in RCA: 636] [Article Influence: 70.7] [Reference Citation Analysis (0)] |
| 30. | Savoldi A, Carrara E, Graham DY, Conti M, Tacconelli E. Prevalence of Antibiotic Resistance in Helicobacter pylori: A Systematic Review and Meta-analysis in World Health Organization Regions. Gastroenterology. 2018;155:1372-1382.e17. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 617] [Cited by in RCA: 825] [Article Influence: 117.9] [Reference Citation Analysis (0)] |
| 31. | Yan TL, Wang JH, He XJ, Zhu YB, Lu LJ, Wang YJ, Wang ZW, Gao JG, Xu CF, Ma H, Luan SM, Li L, Chen Y. Ten-Day Vonoprazan-Amoxicillin Dual Therapy vs Standard 14-Day Bismuth-Based Quadruple Therapy for First-Line Helicobacter pylori Eradication: A Multicenter Randomized Clinical Trial. Am J Gastroenterol. 2024;119:655-661. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 7] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 32. | Yang F, Yu B, Qin L, Dai X. A randomized clinical study on the efficacy of vonoprazan combined with amoxicillin duo regimen for the eradication of Helicobacter pylori. Medicine (Baltimore). 2023;102:e35610. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 13] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 33. | Apostolopoulos P, Ekmektzoglou K, Georgopoulos S, Chounta E, Theofanopoulou A, Kalantzis C, Vlachou E, Tsibouris P, Alexandrakis G. 10-Day Versus 14-Day Quadruple Concomitant Nonbismuth Therapy for the Treatment of Helicobacter pylori Infection: Results From a Randomized Prospective Study in a High Clarithromycin Resistance Country. J Clin Gastroenterol. 2020;54:522-527. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 12] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 34. | Ding YM, Duan M, Han ZX, Song XH, Zhang FL, Wang Z, Ning Z, Zeng SY, Kong QZ, Zhang WL, Liu J, Wan M, Lin MJ, Lin BS, Nan XP, Wang H, Li YY, Zuo XL, Li YQ. Bismuth-Containing Quadruple Therapy for Helicobacter pylori Eradication: A Randomized Clinical Trial of 10 and 14 Days. Dig Dis Sci. 2024;69:2540-2547. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 35. | Georgopoulos SD, Xirouchakis E, Liatsos C, Apostolopoulos P, Kasapidis P, Martinez-Gonzalez B, Laoudi F, Stoupaki M, Axiaris G, Sgouras D, Mentis A, Michopoulos S. Equivalence Trial of the Non-Bismuth 10-Day Concomitant and 14-Day Hybrid Therapies for Helicobacter pylori Eradication in High Clarithromycin Resistance Areas. Antibiotics (Basel). 2024;13:280. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 36. | Alkim H, Koksal AR, Boga S, Sen I, Alkim C. Role of Bismuth in the Eradication of Helicobacter pylori. Am J Ther. 2017;24:e751-e757. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 67] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 37. | Iannone A, Giorgio F, Russo F, Riezzo G, Girardi B, Pricci M, Palmer SC, Barone M, Principi M, Strippoli GF, Di Leo A, Ierardi E. New fecal test for non-invasive Helicobacter pylori detection: A diagnostic accuracy study. World J Gastroenterol. 2018;24:3021-3029. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 36] [Cited by in RCA: 28] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 38. | Mommersteeg MC, Nieuwenburg SAV, Wolters LMM, Roovers BHCM, van Vuuren HAJ, Verhaar AP, Bruno MJ, Kuipers EJ, Peppelenbosch MP, Spaander MCW, Fuhler GM. The use of non-invasive stool tests for verification of Helicobacter pylori eradication and clarithromycin resistance. United European Gastroenterol J. 2023;11:894-903. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 39. | Zhou Y, Zhong Z, Hu S, Wang J, Deng Y, Li X, Chen X, Li X, Tang Y, Li X, Hao Q, Liu J, Sang T, Bo Y, Bai F. A Survey of Helicobacter pylori Antibiotic-Resistant Genotypes and Strain Lineages by Whole-Genome Sequencing in China. Antimicrob Agents Chemother. 2022;66:e0218821. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 13] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 40. | Feng Y, Hu W, Wang Y, Lu J, Zhang Y, Tang Z, Miao S, Zhou Y, Huang Y. Efficacy of Phenotype-vs. Genotype-Guided Therapy Based on Clarithromycin Resistance for Helicobacter pylori Infection in Children. Front Pediatr. 2022;10:854519. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Reference Citation Analysis (0)] |
| 41. | Ren X, Shi Y, Suo B, Yao X, Lu H, Li C, Zhang Y, Zhou L, Tian X, Song Z. Individualized diagnosis and eradication therapy for Helicobacter pylori infection based on gene detection of clarithromycin resistance in stool specimens: A systematic review and meta-analysis. Helicobacter. 2023;28:e12958. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 12] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 42. | Kocsmár É, Buzás GM, Szirtes I, Kocsmár I, Kramer Z, Szijártó A, Fadgyas-Freyler P, Szénás K, Rugge M, Fassan M, Kiss A, Schaff Z, Röst G, Lotz G. Primary and secondary clarithromycin resistance in Helicobacter pylori and mathematical modeling of the role of macrolides. Nat Commun. 2021;12:2255. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 43] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 43. | Pourakbari B, Mahmoudi S, Parhiz J, Sadeghi RH, Monajemzadeh M, Mamishi S. High frequency of metronidazole and clarithromycin-resistant Helicobacter pylori in formalin-fixed, paraffin-embedded gastric biopsies. Br J Biomed Sci. 2018;75:61-65. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 18] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 44. | Mohammad K, Maryam B, Abbas D, Sadegh GD. Clarithromycin resistance and 23S rRNA mutations in Helicobacter pylori isolates in Iran. Afr J Microbiol Res. 2011;5:853-856. [RCA] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 11] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 45. | Marques AT, Vítor JMB, Santos A, Oleastro M, Vale FF. Trends in Helicobacter pylori resistance to clarithromycin: from phenotypic to genomic approaches. Microb Genom. 2020;6:e000344. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 32] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 46. | Hirata K, Suzuki H, Nishizawa T, Tsugawa H, Muraoka H, Saito Y, Matsuzaki J, Hibi T. Contribution of efflux pumps to clarithromycin resistance in Helicobacter pylori. J Gastroenterol Hepatol. 2010;25 Suppl 1:S75-S79. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 91] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 47. | Mori H, Suzuki H. Update on quinolone-containing rescue therapies for Helicobacter pylori infection. World J Gastroenterol. 2020;26:1733-1744. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 17] [Cited by in RCA: 19] [Article Influence: 3.8] [Reference Citation Analysis (1)] |
| 48. | Redgrave LS, Sutton SB, Webber MA, Piddock LJ. Fluoroquinolone resistance: mechanisms, impact on bacteria, and role in evolutionary success. Trends Microbiol. 2014;22:438-445. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 515] [Cited by in RCA: 648] [Article Influence: 58.9] [Reference Citation Analysis (0)] |
| 49. | Haumaier F, Schneider-Fuchs A, Backert S, Vieth M, Sterlacci W, Wöhrl BM. Rapid Detection of Quinolone Resistance Mutations in gyrA of Helicobacter pylori by Real-Time PCR. Pathogens. 2022;11:59. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 8] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 50. | Ziver-Sarp T, Yuksel-Mayda P, Saribas S, Demiryas S, Gareayaghi N, Ergin S, Tasci I, Ozbey D, Bal K, Erzin Y, Akkus S, Bahar-Tokman H, Demirci M, Tufan-Kocak B, Kocazeybek B. Point Mutations at gyrA and gyrB Genes of Levofloxacin Resistant Helicobacter pylori Strains and Dual Resistance with Clarithromycin. Clin Lab. 2021;67. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 2] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 51. | Martínez-Júlvez M, Rojas AL, Olekhnovich I, Espinosa Angarica V, Hoffman PS, Sancho J. Structure of RdxA--an oxygen-insensitive nitroreductase essential for metronidazole activation in Helicobacter pylori. FEBS J. 2012;279:4306-4317. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 49] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 52. | Huang J, Li Z, Ge F, Sun C, Deng Z, Yao W, He X. Functional determination of site-mutations in rdxA involved in metronidazole resistance of Helicobacter pylori. Front Cell Dev Biol. 2024;12:1435064. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 53. | Zhang S, Wang X, Wise MJ, He Y, Chen H, Liu A, Huang H, Young S, Tay CY, Marshall BJ, Li X, Chua EG. Mutations of Helicobacter pylori RdxA are mainly related to the phylogenetic origin of the strain and not to metronidazole resistance. J Antimicrob Chemother. 2020;75:3152-3155. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 17] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 54. | Kargar M, Baghernejad M, Doosti A. Role of NADPH-insensitive nitroreductase gene to metronidazole resistance of Helicobacter pylori strains. Daru. 2010;18:137-140. [PubMed] |
| 55. | Lee SM, Kim N, Kwon YH, Nam RH, Kim JM, Park JY, Lee YS, Lee DH. rdxA, frxA, and efflux pump in metronidazole-resistant Helicobacter pylori: Their relation to clinical outcomes. J Gastroenterol Hepatol. 2018;33:681-688. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 35] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 56. | Marques B, Donato MM, Cardoso O, Luxo C, Martinho A, Almeida N. Study of rdxA and frxA genes mutations in metronidazole-resistant and -susceptible Helicobacter pylori clinical isolates from the central region of Portugal. J Glob Antimicrob Resist. 2019;17:300-304. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 20] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 57. | Matta AJ, Zambrano DC, Martínez YC, Fernández FF. Point mutations in the glycosyltransferase domain of the pbp1a gene in amoxicillin-resistant Helicobacter pylori isolates. Rev Gastroenterol Mex (Engl Ed). 2023;88:100-106. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 3] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 58. | Kuo CJ, Ke JN, Kuo T, Lin CY, Hsieh SY, Chiu YF, Wu HY, Huang MZ, Bui NN, Chiu CH, Chiu CT, Lai CH. Multiple amino acid substitutions in penicillin-binding protein-1A confer amoxicillin resistance in refractory Helicobacter pylori infection. J Microbiol Immunol Infect. 2023;56:40-47. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 12] [Reference Citation Analysis (0)] |
| 59. | Tran TT, Nguyen AT, Quach DT, Pham DT, Cao NM, Nguyen UT, Dang AN, Tran MA, Quach LH, Tran KT, Le NQ, Ung VV, Vo MN, Nguyen DT, Ngo KD, Tran TL, Nguyen VT. Emergence of amoxicillin resistance and identification of novel mutations of the pbp1A gene in Helicobacter pylori in Vietnam. BMC Microbiol. 2022;22:41. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 15] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 60. | Co EM, Schiller NL. Resistance mechanisms in an in vitro-selected amoxicillin-resistant strain of Helicobacter pylori. Antimicrob Agents Chemother. 2006;50:4174-4176. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 27] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 61. | Matteo MJ, Granados G, Olmos M, Wonaga A, Catalano M. Helicobacter pylori amoxicillin heteroresistance due to point mutations in PBP-1A in isogenic isolates. J Antimicrob Chemother. 2008;61:474-477. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 42] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 62. | Trieber CA, Taylor DE. Mutations in the 16S rRNA genes of Helicobacter pylori mediate resistance to tetracycline. J Bacteriol. 2002;184:2131-2140. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 106] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 63. | Wu JY, Kim JJ, Reddy R, Wang WM, Graham DY, Kwon DH. Tetracycline-resistant clinical Helicobacter pylori isolates with and without mutations in 16S rRNA-encoding genes. Antimicrob Agents Chemother. 2005;49:578-583. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 50] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 64. | Khan R, Nahar S, Mukhopadhyay AK, Berg DE, Ahmad MM, Okamoto K, Nair GB, Rahman M. Isolation of tetracycline-resistant clinical Helicobacter pylori without mutations in 16S rRNA gene in Bangladesh. Microbiol Immunol. 2008;52:508-511. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 3] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 65. | Raj DS, Kumar Kesavan D, Muthusamy N, Umamaheswari S. Efflux pumps potential drug targets to circumvent drug Resistance – Multi drug efflux pumps of Helicobacter pylori. Mater Today Proc. 2021;45:2976-2981. [DOI] [Full Text] |
| 66. | Seriki AT, Smith SI, Adeleye AI, Fowora MA. Molecular analysis of low-level tetracycline resistance in clinical isolates of Helicobacter pylori among dyspeptic patients in South West Nigeria. J Glob Antimicrob Resist. 2018;13:143-145. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 10] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 67. | Ji CR, Liu J, Li YY, Guo CG, Qu JY, Zhang Y, Zuo X. Safety of furazolidone-containing regimen in Helicobacter pylori infection: a systematic review and meta-analysis. BMJ Open. 2020;10:e037375. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 17] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 68. | Guzman KA, Hidalgo A, Pazos AJ. Point Mutations in Furazolidone and Rifampicin Resistance Genes in Helicobacter pylori Strains from Colombia. Antibiotics (Basel). 2024;13:643. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 69. | Zullo A, Ierardi E, Hassan C, De Francesco V. Furazolidone-based therapies for Helicobacter pylori infection: a pooled-data analysis. Saudi J Gastroenterol. 2012;18:11-17. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 30] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 70. | Kwon DH, Lee M, Kim JJ, Kim JG, El-Zaatari FA, Osato MS, Graham DY. Furazolidone- and nitrofurantoin-resistant Helicobacter pylori: prevalence and role of genes involved in metronidazole resistance. Antimicrob Agents Chemother. 2001;45:306-308. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 49] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 71. | Hu Y, Zhu Y, Lu NH. Primary Antibiotic Resistance of Helicobacter pylori in China. Dig Dis Sci. 2017;62:1146-1154. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 83] [Article Influence: 10.4] [Reference Citation Analysis (2)] |
| 72. | Glocker E, Bogdan C, Kist M. Characterization of rifampicin-resistant clinical Helicobacter pylori isolates from Germany. J Antimicrob Chemother. 2007;59:874-879. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 48] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 73. | Hays C, Burucoa C, Lehours P, Tran CT, Leleu A, Raymond J. Molecular characterization of Helicobacter pylori resistance to rifamycins. Helicobacter. 2018;23. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 35] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 74. | Yang T, Liu B, Zhou J, Shen Y, Song X, Tang X, Benghezal M, Marshall BJ, Tang H, Li H. The Inappropriateness of Using Rifampicin E-Test to Predict Rifabutin Resistance in Helicobacter pylori. J Infect Dis. 2022;226:S479-S485. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 75. | Boyanova L, Hadzhiyski P, Kandilarov N, Markovska R, Mitov I. Multidrug resistance in Helicobacter pylori: current state and future directions. Expert Rev Clin Pharmacol. 2019;12:909-915. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 71] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 76. | Attaran B, Falsafi T, Ghorbanmehr N. Effect of biofilm formation by clinical isolates of Helicobacter pylori on the efflux-mediated resistance to commonly used antibiotics. World J Gastroenterol. 2017;23:1163-1170. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 40] [Cited by in RCA: 41] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 77. | Falsafi T, Ehsani A, Attaran B, Niknam V. Association of hp1181 and hp1184 Genes With the Active Efflux Phenotype in Multidrug-Resistant Isolates of Helicobacter pylori. Jundishapur J Microbiol. 2016;9:e30726. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 11] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 78. | Ge X, Cai Y, Chen Z, Gao S, Geng X, Li Y, Li Y, Jia J, Sun Y. Bifunctional Enzyme SpoT Is Involved in Biofilm Formation of Helicobacter pylori with Multidrug Resistance by Upregulating Efflux Pump Hp1174 (gluP). Antimicrob Agents Chemother. 2018;62:e00957-18. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 54] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 79. | Liu ZQ, Zheng PY, Yang PC. Efflux pump gene hefA of Helicobacter pylori plays an important role in multidrug resistance. World J Gastroenterol. 2008;14:5217-5222. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 56] [Cited by in RCA: 68] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 80. | Kazakos EI, Dorrell N, Polyzos SA, Deretzi G, Kountouras J. Comment on "Effect of biofilm formation by clinical isolates of Helicobacter pylori on the efflux-mediated resistance to commonly used antibiotics". World J Gastroenterol. 2017;23:6194-6196. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 15] [Cited by in RCA: 16] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 81. | Yonezawa H, Osaki T, Kamiya S. Biofilm Formation by Helicobacter pylori and Its Involvement for Antibiotic Resistance. Biomed Res Int. 2015;2015:914791. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 82] [Cited by in RCA: 105] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 82. | Grande R, Di Giulio M, Bessa LJ, Di Campli E, Baffoni M, Guarnieri S, Cellini L. Extracellular DNA in Helicobacter pylori biofilm: a backstairs rumour. J Appl Microbiol. 2011;110:490-498. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 74] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 83. | Yang FL, Hassanbhai AM, Chen HY, Huang ZY, Lin TL, Wu SH, Ho B. Proteomannans in biofilm of Helicobacter pylori ATCC 43504. Helicobacter. 2011;16:89-98. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 31] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 84. | Madsen JS, Burmølle M, Hansen LH, Sørensen SJ. The interconnection between biofilm formation and horizontal gene transfer. FEMS Immunol Med Microbiol. 2012;65:183-195. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 364] [Cited by in RCA: 420] [Article Influence: 32.3] [Reference Citation Analysis (0)] |
| 85. | Tshibangu-Kabamba E, Yamaoka Y. Helicobacter pylori infection and antibiotic resistance - from biology to clinical implications. Nat Rev Gastroenterol Hepatol. 2021;18:613-629. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 292] [Article Influence: 73.0] [Reference Citation Analysis (0)] |
| 86. | Yonezawa H, Osaki T, Hojo F, Kamiya S. Effect of Helicobacter pylori biofilm formation on susceptibility to amoxicillin, metronidazole and clarithromycin. Microb Pathog. 2019;132:100-108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 47] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 87. | Rizvanov AA, Haertlé T, Bogomolnaya L, Talebi Bezmin Abadi A. Helicobacter pylori and Its Antibiotic Heteroresistance: A Neglected Issue in Published Guidelines. Front Microbiol. 2019;10:1796. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 31] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 88. | Kocsmár É, Kocsmár I, Buzás GM, Szirtes I, Wacha J, Takáts A, Hritz I, Schaff Z, Rugge M, Fassan M, Kiss A, Lotz G. Helicobacter pylori heteroresistance to clarithromycin in adults-New data by in situ detection and improved concept. Helicobacter. 2020;25:e12670. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 21] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 89. | Spagnuolo R, Scarlata GGM, Paravati MR, Abenavoli L, Luzza F. Change in Diagnosis of Helicobacter pylori Infection in the Treatment-Failure Era. Antibiotics (Basel). 2024;13:357. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 90. | Islam JM, Yano Y, Okamoto A, Matsuda R, Shiraishi M, Hashimoto Y, Morita N, Takeuchi H, Suganuma N, Takeuchi H. Evidence of Helicobacter pylori heterogeneity in human stomachs by susceptibility testing and characterization of mutations in drug-resistant isolates. Sci Rep. 2024;14:12066. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 91. | Keikha M, Karbalaei M. Prevalence of antibiotic heteroresistance associated with Helicobacter pylori infection: A systematic review and meta-analysis. Microb Pathog. 2022;170:105720. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 17] [Reference Citation Analysis (0)] |
| 92. | Kouhsari E, Sadeghifard N, Khadiv A, Sayadi H, Amiriani T, Ghafourian S, Valadbeigi H, Krutova M. Heteroresistance to clarithromycin and metronidazole in patients with a Helicobacter pylori infection: a systematic review and meta-analysis. Ann Clin Microbiol Antimicrob. 2022;21:19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 20] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 93. | Graham DY, Moss SF. Antimicrobial Susceptibility Testing for Helicobacter pylori Is Now Widely Available: When, How, Why. Am J Gastroenterol. 2022;117:524-528. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 55] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 94. | Andersson T, Holmberg J, Röhss K, Walan A. Pharmacokinetics and effect on caffeine metabolism of the proton pump inhibitors, omeprazole, lansoprazole, and pantoprazole. Br J Clin Pharmacol. 1998;45:369-375. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 84] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 95. | Tomalik-Scharte D, Lazar A, Fuhr U, Kirchheiner J. The clinical role of genetic polymorphisms in drug-metabolizing enzymes. Pharmacogenomics J. 2008;8:4-15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 86] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 96. | Koopmans AB, Braakman MH, Vinkers DJ, Hoek HW, van Harten PN. Meta-analysis of probability estimates of worldwide variation of CYP2D6 and CYP2C19. Transl Psychiatry. 2021;11:141. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 67] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 97. | Biswas M. Global distribution of CYP2C19 risk phenotypes affecting safety and effectiveness of medications. Pharmacogenomics J. 2021;21:190-199. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 17] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 98. | Shah SC, Tepler A, Chung CP, Suarez G, Peek RM Jr, Hung A, Roumie C, Narula N. Host Genetic Determinants Associated With Helicobacter pylori Eradication Treatment Failure: A Systematic Review and Meta-analysis. Gastroenterology. 2021;161:1443-1459. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 22] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 99. | Lima JJ, Thomas CD, Barbarino J, Desta Z, Van Driest SL, El Rouby N, Johnson JA, Cavallari LH, Shakhnovich V, Thacker DL, Scott SA, Schwab M, Uppugunduri CRS, Formea CM, Franciosi JP, Sangkuhl K, Gaedigk A, Klein TE, Gammal RS, Furuta T. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for CYP2C19 and Proton Pump Inhibitor Dosing. Clin Pharmacol Ther. 2021;109:1417-1423. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 166] [Cited by in RCA: 184] [Article Influence: 46.0] [Reference Citation Analysis (0)] |
| 100. | Yang E, Kim S, Kim B, Kim B, Kim Y, Park SS, Song GS, Yu KS, Jang IJ, Lee S. Night-time gastric acid suppression by tegoprazan compared to vonoprazan or esomeprazole. Br J Clin Pharmacol. 2022;88:3288-3296. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 51] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 101. | Ozaki H, Harada S, Takeuchi T, Kawaguchi S, Takahashi Y, Kojima Y, Ota K, Hongo Y, Ashida K, Sakaguchi M, Tokioka S, Sakamoto H, Furuta T, Tominaga K, Higuchi K. Vonoprazan, a Novel Potassium-Competitive Acid Blocker, Should Be Used for the Helicobacter pylori Eradication Therapy as First Choice: A Large Sample Study of Vonoprazan in Real World Compared with Our Randomized Control Trial Using Second-Generation Proton Pump Inhibitors for Helicobacter pylori Eradication Therapy. Digestion. 2018;97:212-218. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 30] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 102. | Nishihara M, Yamasaki H, Czerniak R, Jenkins H. In Vitro Assessment of Potential for CYP-Inhibition-Based Drug-Drug Interaction Between Vonoprazan and Clopidogrel. Eur J Drug Metab Pharmacokinet. 2019;44:217-227. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 103. | Ford AC, Malfertheiner P, Giguere M, Santana J, Khan M, Moayyedi P. Adverse events with bismuth salts for Helicobacter pylori eradication: systematic review and meta-analysis. World J Gastroenterol. 2008;14:7361-7370. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 85] [Cited by in RCA: 97] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 104. | Mesquita A, Rocha-Castro C, Guimarães D, Costa J, Soutinho J, Taveira-Gomes T. Multicentric Study to Assess Helicobacter pylori Incidence, Patient Reported Adverse Events, Compliance and Effectiveness, in Real-World Setting. Int J Environ Res Public Health. 2022;19:12847. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 5] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 105. | Zeng R, Li X, Wang F, Xie J, Song C, Xie Y. Reinforced medication adherence improves Helicobacter pylori eradication rate in developing countries: A systematic review and meta-analysis of randomized controlled trials. Helicobacter. 2023;28:e12989. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Reference Citation Analysis (0)] |
| 106. | Thung I, Aramin H, Vavinskaya V, Gupta S, Park JY, Crowe SE, Valasek MA. Review article: the global emergence of Helicobacter pylori antibiotic resistance. Aliment Pharmacol Ther. 2016;43:514-533. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 410] [Cited by in RCA: 553] [Article Influence: 61.4] [Reference Citation Analysis (2)] |
| 107. | Yu Y, Xue J, Lin F, Liu D, Zhang W, Ru S, Jiang F. Global Primary Antibiotic Resistance Rate of Helicobacter pylori in Recent 10 years: A Systematic Review and Meta-Analysis. Helicobacter. 2024;29:e13103. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 27] [Article Influence: 27.0] [Reference Citation Analysis (1)] |
| 108. | Xu W, Yang B, Lin L, Lin Q, Wang H, Yang L, Li Z, Lamm S, Chen Y, Yang N, Chen Y, Yu C, Li L. Antibiotic resistance of Helicobacter pylori in Chinese children: A multicenter study from 2016 to 2023. Helicobacter. 2024;29:e13038. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 109. | Li L, Ke Y, Yu C, Li G, Yang N, Zhang J, Li Y. Antibiotic resistance of Helicobacter pylori in Chinese children: A multicenter retrospective study over 7 years. Helicobacter. 2017;22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 27] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 110. | Cabrera C, Torres J, Serrano CA, Gallardo P, Orellana V, George S, O'Ryan M, Lucero Y. Antimicrobial Resistance of Helicobacter pylori Isolated From Latin American Children and Adolescents (2008-2023): A Systematic Review. Helicobacter. 2024;29:e13101. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 111. | Jaka H, Rhee JA, Östlundh L, Smart L, Peck R, Mueller A, Kasang C, Mshana SE. The magnitude of antibiotic resistance to Helicobacter pylori in Africa and identified mutations which confer resistance to antibiotics: systematic review and meta-analysis. BMC Infect Dis. 2018;18:193. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 52] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 112. | Dutta S, Jain S, Das K, Verma P, Som A, Das R. Primary antibiotic resistance of Helicobacter pylori in India over the past two decades: A systematic review. Helicobacter. 2024;29:e13057. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 7] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 113. | Wang Y, Du J, Zhang D, Jin C, Chen J, Wang Z, Mei T, Fu K, Qian Q, Pang T. Primary antibiotic resistance in Helicobacter pylori in China: a systematic review and meta-analysis. J Glob Antimicrob Resist. 2023;34:30-38. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 14] [Reference Citation Analysis (0)] |
| 114. | Long X, Chen Q, Yu L, Liang X, Liu W, Lu H. Bismuth improves efficacy of proton-pump inhibitor clarithromycin, metronidazole triple Helicobacter pylori therapy despite a high prevalence of antimicrobial resistance. Helicobacter. 2018;23:e12485. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 44] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 115. | Schubert JP, Warner MS, Rayner CK, Roberts-Thomson IC, Mangoni AA, Costello S, Bryant RV. Increasing Helicobacter pylori clarithromycin resistance in Australia over 20 years. Intern Med J. 2022;52:1554-1560. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 15] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 116. | Sarıkaya B, Çetinkaya RA, Özyiğitoğlu D, Işık SA, Kaplan M, Kırkık D, Görenek L. High antibiotic resistance rates in Helicobacter pylori strains in Turkey over 20 years: implications for gastric disease treatment. Eur J Gastroenterol Hepatol. 2024;36:545-553. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Reference Citation Analysis (0)] |
| 117. | Ho JJC, Navarro M, Sawyer K, Elfanagely Y, Moss SF. Helicobacter pylori Antibiotic Resistance in the United States Between 2011 and 2021: A Systematic Review and Meta-Analysis. Am J Gastroenterol. 2022;117:1221-1230. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 69] [Article Influence: 23.0] [Reference Citation Analysis (0)] |
| 118. | Lopo I, Libânio D, Pita I, Dinis-Ribeiro M, Pimentel-Nunes P. Helicobacter pylori antibiotic resistance in Portugal: Systematic review and meta-analysis. Helicobacter. 2018;23:e12493. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 38] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 119. | Camorlinga-Ponce M, Gómez-Delgado A, Aguilar-Zamora E, Torres RC, Giono-Cerezo S, Escobar-Ogaz A, Torres J. Phenotypic and Genotypic Antibiotic Resistance Patterns in Helicobacter pylori Strains From Ethnically Diverse Population in México. Front Cell Infect Microbiol. 2020;10:539115. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 17] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 120. | Sanches BS, Martins GM, Lima K, Cota B, Moretzsohn LD, Ribeiro LT, Breyer HP, Maguilnik I, Maia AB, Rezende-Filho J, Meira AC, Pinto H, Alves E, Mascarenhas R, Passos R, de Souza JD, Trindade OR, Coelho LG. Detection of Helicobacter pylori resistance to clarithromycin and fluoroquinolones in Brazil: A national survey. World J Gastroenterol. 2016;22:7587-7594. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 31] [Cited by in RCA: 31] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 121. | Benigno TGDS, Ribeiro Junior HL, Azevedo OGR, Pinheiro RF, Oliveira RTG, Maciel FS, Oliveira EL, Queiroz DMM, Braga LLBC. Clarithromycin-resistant H. pylori primary strains and virulence genotypes in the Northeastern region of Brazil. Rev Inst Med Trop Sao Paulo. 2022;64:e47. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 122. | Vianna JS, Ramis IB, Ramos DF, Gastal OL, Silva RAD, Gonçalves CV, Silva PEAD. The interplay between mutations in cagA, 23S rRNA, gyrA and drug resistance in Helicobacter pylori. Rev Inst Med Trop Sao Paulo. 2018;60:e25. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 6] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 123. | Rodrigues SO, Santiago FR, Silva MS, Lima ASG, Godoy LE, De Waard M, Fouad D, Batiha GE, Santos TL, Pagnossa JP. Macrolide resistance outcomes after the Covid-19 pandemic: A one health approach investigation. Biomed Pharmacother. 2024;180:117437. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 124. | Zurita J, Sevillano G, Paz Y Miño A, Zurita-Salinas C, Peñaherrera V, Echeverría M; Helicobacter pylori Research Group, Navarrete H. Mutations associated with Helicobacter pylori antimicrobial resistance in the Ecuadorian population. J Appl Microbiol. 2022;132:2694-2704. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 125. | Asaad AM, El-Azab G, Abdelsameea E, Elbahr O, Kamal A, Abdel-Samiee M, Abdelfattah A, Abdallah H, Maher D, El-Refaie A, Ghanem SE, Ansari S, Awad SM. Susceptibility patterns and virulence genotypes of Helicobacter pylori affecting eradication therapy outcomes among Egyptian patients with gastroduodenal diseases. World J Gastroenterol. 2023;29:2950-2960. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 2] [Cited by in RCA: 5] [Article Influence: 2.5] [Reference Citation Analysis (2)] |
| 126. | Abdulrahman MS, Mansy MS, Al-Ghreib KA, Johar D, Zaky S. PCR-based RFLP and ERIC-PCR patterns of Helicobacter pylori strains linked to multidrug resistance in Egypt. Sci Rep. 2024;14:22273. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 127. | Liou JM, Malfertheiner P, Lee YC, Sheu BS, Sugano K, Cheng HC, Yeoh KG, Hsu PI, Goh KL, Mahachai V, Gotoda T, Chang WL, Chen MJ, Chiang TH, Chen CC, Wu CY, Leow AH, Wu JY, Wu DC, Hong TC, Lu H, Yamaoka Y, Megraud F, Chan FKL, Sung JJ, Lin JT, Graham DY, Wu MS, El-Omar EM; Asian Pacific Alliance on Helicobacter and Microbiota (APAHAM). Screening and eradication of Helicobacter pylori for gastric cancer prevention: the Taipei global consensus. Gut. 2020;69:2093-2112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 335] [Cited by in RCA: 306] [Article Influence: 61.2] [Reference Citation Analysis (0)] |
| 128. | Shin JM, Sachs G. Pharmacology of proton pump inhibitors. Curr Gastroenterol Rep. 2008;10:528-534. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 370] [Cited by in RCA: 323] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 129. | Ali A, AlHussaini KI. Helicobacter pylori: A Contemporary Perspective on Pathogenesis, Diagnosis and Treatment Strategies. Microorganisms. 2024;12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 48] [Article Influence: 48.0] [Reference Citation Analysis (0)] |
| 130. | McNicholl AG, Linares PM, Nyssen OP, Calvet X, Gisbert JP. Meta-analysis: esomeprazole or rabeprazole vs. first-generation pump inhibitors in the treatment of Helicobacter pylori infection. Aliment Pharmacol Ther. 2012;36:414-425. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 125] [Cited by in RCA: 143] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 131. | Chen L, He J, Wang L, Ge Q, Chu H, Chen Y, Chen X, Long Y, Deng Y, He H, Li A, Chen S. Efficacies of different proton pump inhibitor-based 14-day bismuth-furazolidone quadruple regimens for the initial eradication of Helicobacter pylori in the southeast coastal region of China: an open-label, randomized clinical trial. Clin Exp Med. 2018;18:569-576. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 11] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 132. | Zhu H, Pan X, Zhang L, Sun H, Fan H, Pan Z, Huang C, Shi Z, Ding J, Wang Q, Du Y, Lyu N, Li Z. Effect and safety of anaprazole in the treatment of duodenal ulcers: a randomized, rabeprazole-controlled, phase III non-inferiority study. Chin Med J (Engl). 2022;135:2941-2949. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 9] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 133. | Jin Y, Zhang S, Pan J, Yue M, Zhang G, Yao D, Wang Q. Comparison of efficacy and safety of ilaprazole and esomeprazole both in initial treatment regimen and retreatment regimen of Helicobacter pylori infection in chronic gastritis. Pharmazie. 2019;74:432-438. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 134. | Shirley M. Vonoprazan: A Review in Helicobacter pylori Infection. Drugs. 2024;84:319-327. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 15] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 135. | Kiyotoki S, Nishikawa J, Sakaida I. Efficacy of Vonoprazan for Helicobacter pylori Eradication. Intern Med. 2020;59:153-161. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 68] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 136. | Bunchorntavakul C, Buranathawornsom A. Randomized clinical trial: 7-day vonoprazan-based versus 14-day omeprazole-based triple therapy for Helicobacter pylori. J Gastroenterol Hepatol. 2021;36:3308-3313. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 46] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 137. | Chey WD, Mégraud F, Laine L, López LJ, Hunt BJ, Howden CW. Vonoprazan Triple and Dual Therapy for Helicobacter pylori Infection in the United States and Europe: Randomized Clinical Trial. Gastroenterology. 2022;163:608-619. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 172] [Article Influence: 57.3] [Reference Citation Analysis (2)] |
| 138. | Lu L, Wang Y, Ye J, Han Y, Lou G, Li Y, Yan H, Du Q. Quadruple therapy with vonoprazan 20 mg daily as a first-line treatment for Helicobacter pylori infection: A single-center, open-label, noninferiority, randomized controlled trial. Helicobacter. 2023;28:e12940. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 33] [Reference Citation Analysis (0)] |
| 139. | Sun Y, Yue L, Hu W. Effectiveness and safety of vonoprazan-based regimens compared with those of proton pump inhibitor (PPI)-based regimens as first-line agents for Helicobacter pylori: a meta-analysis of randomized clinical trials. Eur J Clin Pharmacol. 2023;79:279-288. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 14] [Reference Citation Analysis (0)] |
| 140. | Liu L, Shi H, Shi Y, Wang A, Guo N, Li F, Nahata MC. Vonoprazan-based therapies versus PPI-based therapies in patients with H. pylori infection: Systematic review and meta-analyses of randomized controlled trials. Helicobacter. 2024;29:e13094. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 13] [Reference Citation Analysis (1)] |
| 141. | Simadibrata DM, Syam AF, Lee YY. A comparison of efficacy and safety of potassium-competitive acid blocker and proton pump inhibitor in gastric acid-related diseases: A systematic review and meta-analysis. J Gastroenterol Hepatol. 2022;37:2217-2228. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 51] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 142. | Zhou BG, Jiang X, Ding YB, She Q, Li YY. Vonoprazan-amoxicillin dual therapy versus bismuth-containing quadruple therapy for Helicobacter pylori eradication: A systematic review and meta-analysis. Helicobacter. 2024;29:e13040. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 7] [Article Influence: 7.0] [Reference Citation Analysis (1)] |
| 143. | Li SY, Li J, Dong XH, Teng GG, Zhang W, Cheng H, Gao W, Dai Y, Zhang XH, Wang WH. The effect of previous eradication failure on antibiotic resistance of Helicobacter pylori: A retrospective study over 8 years in Beijing. Helicobacter. 2021;26:e12804. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 22] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 144. | Xie J, Peng J, Liu D, Zeng R, Qiu J, Shen L, Gong X, Liu D, Xie Y. Treatment failure is a key factor in the development of Helicobacter pylori resistance. Helicobacter. 2024;29:e13091. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 145. | Garrido-Treviño LF, López-Martínez M, Flores-Hinojosa JA, Tijerina-Rodríguez L, Bosques-Padilla F. Empiric treatment vs susceptibility-guided treatment for eradicating H. pylori: Is it possible to change that paradigm using modern molecular methods? Rev Gastroenterol Mex (Engl Ed). 2022;87:330-341. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 146. | Schubert JP, Tay A, Lee KHC, Leong LEX, Rayner CK, Warner MS, Roberts-Thomson IC, Costello SP, Bryant RV. Genomic analysis of Helicobacter pylori in Australia: Antimicrobial resistance, phylogenetic patterns, and virulence factors. J Gastroenterol Hepatol. 2024;39:1869-1875. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 147. | Rokkas T, Gisbert JP, Malfertheiner P, Niv Y, Gasbarrini A, Leja M, Megraud F, O'Morain C, Graham DY. Comparative Effectiveness of Multiple Different First-Line Treatment Regimens for Helicobacter pylori Infection: A Network Meta-analysis. Gastroenterology. 2021;161:495-507.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 120] [Article Influence: 30.0] [Reference Citation Analysis (0)] |
| 148. | Yang EH, Chen WY, Chiang HC, Li CH, Wu IH, Chen PJ, Wu CT, Tsai YC, Cheng WC, Huang CJ, Sheu BS, Cheng HC. 10-Day versus 14-day bismuth quadruple therapy for first-line eradication of Helicobacter pylori infection: a randomised, open-label, non-inferiority trial. EClinicalMedicine. 2024;70:102529. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 9] [Reference Citation Analysis (0)] |
| 149. | Ding YM, Li YY, Liu J, Wang J, Wan M, Lin MJ, Lin BS, Zhang WL, Kong QZ, Wang ST, Mu YJ, Duan M, Han ZX, Zuo XL, Li YQ. The cure rate of 10-day bismuth-containing quadruple therapy for Helicobacter pylori eradication is equivalent to 14-day: a systematic review and meta-analysis. Clin Exp Med. 2023;23:1033-1043. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 14] [Reference Citation Analysis (0)] |
| 150. | Medel-Jara P, Reyes Placencia D, Fuentes-López E, Corsi O, Latorre G, Antón R, Jiménez E, Miralles-Marco A, Caballero C, Boggino H, Cantero D, Barros R, Santos-Antunes J, Díez M, Quiñones LA, Riquelme E, Rollán A, Cerpa LC, Valdés I, Nyssen OP, Moreira L, Gisbert JP, Camargo MC, Ortiz-Olvera N, Leon-Takahashi AM, Ruiz-Garcia E, Fernández-Figueroa EA, Garrido M, Owen GI, Cervantes A, Fleitas T, Riquelme A. Quadruple therapies show a higher eradication rate compared to standard triple therapy for Helicobacter pylori infection within the LEGACy consortium. A multicenter observational study in European and Latin American countries. United European Gastroenterol J. 2024;12:1190-1199. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 151. | Olmedo L, Calvet X, Gené E, Bordin DS, Voynovan I, Castro-Fernandez M, Pabón-Carrasco M, Keco-Huerga A, Perez-Aisa Á, Lucendo AJ, Rodrigo L, Sarsenbaeva AS, Khlinov IB, Fadieienko G, Zaytsev O, Lanas Á, Martínez-Domínguez SJ, Alfaro E, Jonaitis L, Núñez Ó, Pellicano R, Hernández L, Gridnyev O, Kupcinskas J, Gasbarrini A, Boltin D, Niv Y, Babayeva G, Marcos-Pinto R, Tepes B, Venerito M, Papp V, Lerang F, Leja M, Phull PS, Marlicz W, Doulberis M, Smith SM, Milivojevic V, Kunovsky L, Mestrovic A, Matysiak-Budnik T, Simsek H, Cano-Català A, Puig I, Moreira L, Parra P, Nyssen OP, Megraud F, O'Morain C, Gisbert JP; Hp-EuReg investigators*. Evolution of the use, effectiveness and safety of bismuth-containing quadruple therapy for Helicobacter pylori infection between 2013 and 2021: results from the European registry on H. pylori management (Hp-EuReg). Gut. 2024;74:15-25. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 10] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 152. | Hong TC, El-Omar EM, Kuo YT, Wu JY, Chen MJ, Chen CC, Fang YJ, Leow AHR, Lu H, Lin JT, Tu YK, Yamaoka Y, Wu MS, Liou JM; Asian Pacific Alliance on Helicobacter and Microbiota. Primary antibiotic resistance of Helicobacter pylori in the Asia-Pacific region between 1990 and 2022: an updated systematic review and meta-analysis. Lancet Gastroenterol Hepatol. 2024;9:56-67. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 56] [Reference Citation Analysis (0)] |
| 153. | Liou JM, Jiang XT, Chen CC, Luo JC, Bair MJ, Chen PY, Chou CK, Fang YJ, Chen MJ, Chen CC, Lee JY, Yang TH, Yu CC, Kuo CC, Chiu MC, Chen CY, Shun CT, Hu WH, Tsai MH, Hsu YC, Tseng CH, Chang CY, Lin JT, El-Omar EM, Wu MS; Taiwan Gastrointestinal Disease and Helicobacter Consortium. Second-line levofloxacin-based quadruple therapy versus bismuth-based quadruple therapy for Helicobacter pylori eradication and long-term changes to the gut microbiota and antibiotic resistome: a multicentre, open-label, randomised controlled trial. Lancet Gastroenterol Hepatol. 2023;8:228-241. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 36] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 154. | Chen J, Guo Y, Huang Y, Ding Z, Wang J, Liang X, Xu P, Han Y, Lu H. Rifabutin-Containing Triple Therapy Versus Bismuth Quadruple Therapy for Helicobacter pylori Rescue Treatment: A Multicenter, Randomized Controlled Trial. J Infect Dis. 2023;228:511-518. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 11] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 155. | Bi H, Chen X, Chen Y, Zhao X, Wang S, Wang J, Lyu T, Han S, Lin T, Li M, Yuan D, Liu J, Shi Y. Efficacy and safety of high-dose esomeprazole-amoxicillin dual therapy for Helicobacter pylori rescue treatment: a multicenter, prospective, randomized, controlled trial. Chin Med J (Engl). 2022;135:1707-1715. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 24] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 156. | Han Z, Zhang Q, Mirza IA, Ding Y, Nan X, Zhao Q, Li R, Xu L, Zhang N, Duan M, Zeng S, Kong Q, Zhang W, Wang H, Wu X, Zuo X, Li Y, Li Y. Efficacy of Tetracycline Three Times Daily was Comparable to That of Four Times Daily for Helicobacter pylori Rescue Treatment: A Multicenter, Noninferiority, Randomized Controlled Trial. Helicobacter. 2024;29:e13102. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 157. | Francesco V, Zullo A, Manta R, Satriano A, Fiorini G, Pavoni M, Saracino IM, Giostra F, Monti G, Vaira D. Culture-based antibiotic susceptibility testing for Helicobacter pylori infection: a systematic review. Ann Gastroenterol. 2022;35:127-134. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 5] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 158. | Graham DY. Molecular-based Helicobacter pylori Susceptibility Testing Is Almost Ready for Prime Time. Gastroenterology. 2021;160:1936-1937. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 19] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 159. | O'Morain C, Smith SM. Antimicrobial susceptibility testing for Helicobacter pylori comes of age. Lancet Gastroenterol Hepatol. 2023;8:593-595. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Reference Citation Analysis (0)] |
| 160. | Ishibashi F, Suzuki S, Nagai M, Mochida K, Morishita T. Optimizing Helicobacter pylori Treatment: An Updated Review of Empirical and Susceptibility Test-Based Treatments. Gut Liver. 2023;17:684-697. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Reference Citation Analysis (0)] |
| 161. | Wiegand I, Hilpert K, Hancock RE. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat Protoc. 2008;3:163-175. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3102] [Cited by in RCA: 3855] [Article Influence: 226.8] [Reference Citation Analysis (0)] |
| 162. | Jorgensen JH, Hindler JF. New consensus guidelines from the Clinical and Laboratory Standards Institute for antimicrobial susceptibility testing of infrequently isolated or fastidious bacteria. Clin Infect Dis. 2007;44:280-286. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 128] [Cited by in RCA: 191] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 163. | Li H, Shen Y, Song X, Tang X, Hu R, Marshall BJ, Tang H, Benghezal M. Need for standardization and harmonization of Helicobacter pylori antimicrobial susceptibility testing. Helicobacter. 2022;27:e12873. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 19] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 164. | Mégraud F, Lehours P. Helicobacter pylori detection and antimicrobial susceptibility testing. Clin Microbiol Rev. 2007;20:280-322. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 476] [Cited by in RCA: 488] [Article Influence: 27.1] [Reference Citation Analysis (0)] |
| 165. | Tang X, Shen Y, Song X, Benghezal M, Marshall BJ, Tang H, Li H. Reassessment of the Broth Microdilution Method for Susceptibility Testing of Helicobacter pylori. J Infect Dis. 2022;226:S486-S492. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 11] [Reference Citation Analysis (0)] |
| 166. | Knezevic P, Aleksic Sabo V, Simin N, Lesjak M, Mimica-Dukic N. A colorimetric broth microdilution method for assessment of Helicobacter pylori sensitivity to antimicrobial agents. J Pharm Biomed Anal. 2018;152:271-278. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 21] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 167. | Miftahussurur M, Fauzia KA, Nusi IA, Setiawan PB, Syam AF, Waskito LA, Doohan D, Ratnasari N, Khomsan A, Adnyana IK, Akada J, Yamaoka Y. E-test versus agar dilution for antibiotic susceptibility testing of Helicobacter pylori: a comparison study. BMC Res Notes. 2020;13:22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 40] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 168. | Chen J, Huang Y, Ding Z, Liang X, Lu H. E-Test or Agar Dilution for Metronidazole Susceptibility Testing of Helicobacter pylori: Importance of the Prevalence of Metronidazole Resistance. Front Microbiol. 2022;13:801537. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 3] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 169. | Ng HY, Leung WK, Cheung KS. Antibiotic Resistance, Susceptibility Testing and Stewardship in Helicobacter pylori Infection. Int J Mol Sci. 2023;24:11708. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 16] [Reference Citation Analysis (0)] |
| 170. | Tang X, Shen Y, Hu R, Yang T, Benghezal M, Li H, Tang H. Re-assessment of the disk diffusion technique for routine antimicrobial susceptibility testing for Helicobacter pylori. Helicobacter. 2020;25:e12703. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 24] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 171. | Mansour-Ghanaei F, Poostizadeh G, Joukar F, Siavoshi F. Efficacy of Disc Diffusion and Agar Dilution Methods in Evaluating Helicobacter pylori Susceptibility to Antibiotics. Middle East J Dig Dis. 2022;14:207-213. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 172. | Ogata SK, Gales AC, Kawakami E. Antimicrobial susceptibility testing for Helicobacter pylori isolates from Brazilian children and adolescents: comparing agar dilution, E-test, and disk diffusion. Braz J Microbiol. 2014;45:1439-1448. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 42] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 173. | Yamin D, Uskoković V, Wakil AM, Goni MD, Shamsuddin SH, Mustafa FH, Alfouzan WA, Alissa M, Alshengeti A, Almaghrabi RH, Fares MAA, Garout M, Al Kaabi NA, Alshehri AA, Ali HM, Rabaan AA, Aldubisi FA, Yean CY, Yusof NY. Current and Future Technologies for the Detection of Antibiotic-Resistant Bacteria. Diagnostics (Basel). 2023;13:3246. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 46] [Reference Citation Analysis (0)] |
| 174. | Medakina I, Tsapkova L, Polyakova V, Nikolaev S, Yanova T, Dekhnich N, Khatkov I, Bordin D, Bodunova N. Helicobacter pylori Antibiotic Resistance: Molecular Basis and Diagnostic Methods. Int J Mol Sci. 2023;24:9433. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 18] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 175. | Hortelano I, Moreno MY, García-Hernández J, Ferrús MA. Optimization of pre- treatments with Propidium Monoazide and PEMAX™ before real-time quantitative PCR for detection and quantification of viable Helicobacter pylori cells. J Microbiol Methods. 2021;185:106223. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 5] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 176. | Redondo JJ, Keller PM, Zbinden R, Wagner K. A novel RT-PCR for the detection of Helicobacter pylori and identification of clarithromycin resistance mediated by mutations in the 23S rRNA gene. Diagn Microbiol Infect Dis. 2018;90:1-6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 44] [Article Influence: 5.5] [Reference Citation Analysis (2)] |
| 177. | Fernandez-Caso B, Miqueleiz A, Valdez VB, Alarcón T. Are molecular methods helpful for the diagnosis of Helicobacter pylori infection and for the prediction of its antimicrobial resistance? Front Microbiol. 2022;13:962063. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 11] [Reference Citation Analysis (0)] |
| 178. | Nezami BG, Jani M, Alouani D, Rhoads DD, Sadri N. Helicobacter pylori Mutations Detected by Next-Generation Sequencing in Formalin-Fixed, Paraffin-Embedded Gastric Biopsy Specimens Are Associated with Treatment Failure. J Clin Microbiol. 2019;57:e01834-18. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 40] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 179. | Gong RJ, Xu CX, Li H, Liu XM. Polymerase chain reaction-based tests for detecting Helicobacter pylori clarithromycin resistance in stool samples: A meta-analysis. World J Clin Cases. 2021;9:133-147. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 8] [Cited by in RCA: 13] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 180. | Beckman E, Saracino I, Fiorini G, Clark C, Slepnev V, Patel D, Gomez C, Ponaka R, Elagin V, Vaira D. A Novel Stool PCR Test for Helicobacter pylori May Predict Clarithromycin Resistance and Eradication of Infection at a High Rate. J Clin Microbiol. 2017;55:2400-2405. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 47] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 181. | Wang YH, Li Z, Wang L, Zhu-Ge LY, Zhao RL, Wu S, Wang Y, An Y, Xie Y. A systematic review and meta-analysis of genotypic methods for detecting antibiotic resistance in Helicobacter pylori. Helicobacter. 2018;23:e12467. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 35] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 182. | Tuan VP, Narith D, Tshibangu-Kabamba E, Dung HDQ, Viet PT, Sokomoth S, Binh TT, Sokhem S, Tri TD, Ngov S, Tung PH, Thuan NPM, Truc TC, Phuc BH, Matsumoto T, Fauzia KA, Akada J, Trang TTH, Yamaoka Y. A Next-Generation Sequencing-Based Approach to Identify Genetic Determinants of Antibiotic Resistance in Cambodian Helicobacter pylori Clinical Isolates. J Clin Med. 2019;8:858. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 59] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 183. | Yang T, Hu R, Tang X, Shen Y, Tay A, Pi X, Wang G, Debowski AW, Stubbs KA, Benghezal M, Marshall BJ, Li H, Tang H. Susceptibility-guided bismuth quadruple therapies for resistant Helicobacter pylori infections. Precis Clin Med. 2020;3:127-135. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 9] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 184. | Nyssen OP, Espada M, Gisbert JP. Empirical vs. Susceptibility-Guided Treatment of Helicobacter pylori Infection: A Systematic Review and Meta-Analysis. Front Microbiol. 2022;13:913436. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 33] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 185. | Ma Q, Li H, Liao J, Cai Z, Zhang B. Tailored therapy for Helicobacter pylori eradication: A systematic review and meta-analysis. Front Pharmacol. 2022;13:908202. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 19] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 186. | Ouyang Y, Zhang W, He C, Zhu Y, Lu N, Hu Y. Susceptibility-Guided Therapy vs. Bismuth-Containing Quadruple Therapy as the First-Line Treatment for Helicobacter pylori Infection: A Systematic Review and Meta-Analysis. Front Med (Lausanne). 2022;9:844915. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 187. | Chen MJ, Chen PY, Fang YJ, Bair MJ, Chen CC, Chen CC, Yang TH, Lee JY, Yu CC, Kuo CC, Chiu MC, Chou CK, Chen CY, Hu WH, Tsai MH, Hsu YC, Shun CT, Luo JC, Lin JT, El-Omar EM, Wu MS, Liou JM; Taiwan Gastrointestinal Disease and Helicobacter Consortium. Molecular testing-guided therapy versus susceptibility testing-guided therapy in first-line and third-line Helicobacter pylori eradication: two multicentre, open-label, randomised controlled, non-inferiority trials. Lancet Gastroenterol Hepatol. 2023;8:623-634. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 18] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 188. | Li M, Wang X, Meng W, Dai Y, Wang W. Empirical versus tailored therapy based on genotypic resistance detection for Helicobacter pylori eradication: a systematic review and meta-analysis. Therap Adv Gastroenterol. 2023;16:17562848231196357. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 5] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 189. | Kim JS, Kim BW, Kim JI, Chung WC, Jung SW, Bang CS, Kim GH, Jeon SW, Joo MK, Lee SH, Lim YJ, Sung JK, Seo SY, Park SY, Lee WS, Lee HL, Kim KB, Kim HU. Empirical Therapy Versus Tailored Therapy of Helicobacter pylori in Korea: Results of the K-CREATE Study. Helicobacter. 2024;29:e13126. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Reference Citation Analysis (0)] |
| 190. | Lin K, Huang L, Wang Y, Li K, Ye Y, Yang S, Li A. Efficacy of genotypic susceptibility-guided tailored therapy for Helicobacter pylori infection: A systematic review and single arm meta-analysis. Helicobacter. 2023;28:e13015. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 7] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 191. | Li CL, Zhou K, Zhang YX, Suo BJ, Tian XL, Zhang YX, Ren XL, Shi YY, Zhou LY, Song ZQ. Tailored therapy guided by genotypic resistance of clarithromycin and levofloxacin detected by polymerase chain reaction in the first-line treatment of Helicobacter pylori infection. J Dig Dis. 2024;25:36-43. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 192. | Lee JH, Min BH, Gong EJ, Kim JY, Na HK, Ahn JY, Kim DH, Choi KD, Min YW, Lee H, Lee JH, Jung HY, Kim JJ. Culture-based susceptibility-guided tailored versus empirical concomitant therapy as first-line Helicobacter pylori treatment: A randomized clinical trial. United European Gastroenterol J. 2024;12:941-950. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Reference Citation Analysis (0)] |
| 193. | Amiot A, Hacoon J, Heluwaert F, Mion F, Lamarque D, Moussata D, Mimouni M, Delchier JC, Durand-Zaleski I, Audureau E, Bastuji-Garin S; HEPYSE Study Group. 14-day tailored PCR-guided triple therapy versus 14-day non-Bismuth concomitant quadruple therapy for Helicobacter pylori eradication: A multicenter, open-label randomized noninferiority controlled trial. Helicobacter. 2024;29:e13076. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 194. | Cho JH, Jeon SR, Kim HG, Jin SY, Park S. Cost-effectiveness of a tailored Helicobacter pylori eradication strategy based on the presence of a 23S ribosomal RNA point mutation that causes clarithromycin resistance in Korean patients. J Gastroenterol Hepatol. 2019;34:700-706. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 52] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 195. | Choe AR, Shim KN, Park Y, Song EM, Tae CH, Jung SA. Cost-Effectiveness, Efficacy, and Safety Analysis of Tailored Therapy in Patients with Helicobacter pylori Infection. J Clin Med. 2021;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 15] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 196. | Cho JH, Jin SY, Park S. Comparison of tailored Helicobacter pylori eradication versus modified bismuth quadruple therapy in Korea: a randomized controlled trial. Expert Rev Anti Infect Ther. 2022;20:923-929. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 8] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 197. | Chang YW, Shin GY, Kim JW, Moon JC, Chang EJ, Oh CH, Jang JY. Cost-Effectiveness of Empirical Bismuth-Based Quadruple Therapy and Tailored Therapy After Clarithromycin Resistance Tests for Helicobacter pylori Eradication. Dig Dis Sci. 2022;67:1222-1230. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 5] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 198. | Rokkas T, Ekmektzoglou K, Graham DY. Current role of tailored therapy in treating Helicobacter pylori infections. A systematic review, meta-analysis and critical analysis. Helicobacter. 2023;28:e12936. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |
| 199. | Huang Y, Chen J, Ding Z, Liang X, Lu H. Susceptibility testing alone will not reliably achieve high Helicobacter pylori cure rates: A systematic review and meta-analysis. J Gastroenterol Hepatol. 2022;37:1212-1221. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 200. | Aumpan N, Gamnarai P, Wongcha-Um A, Miftahussurur M, Yamaoka Y, Vilaichone RK. The use of real-world evidence to generate cost analysis of antibiotic susceptibility testing (AST) in patients with Helicobacter pylori treatment failure in Thailand: A large population-based study. Heliyon. 2024;10:e39189. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 201. | Homan M, Jones NL, Bontems P, Carroll MW, Czinn SJ, Gold BD, Goodman K, Harris PR, Jerris R, Kalach N, Kori M, Megraud F, Rowland M, Tavares M; on behalf of ESPGHAN/NASPGHAN. Updated joint ESPGHAN/NASPGHAN guidelines for management of Helicobacter pylori infection in children and adolescents (2023). J Pediatr Gastroenterol Nutr. 2024;79:758-785. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 13] [Reference Citation Analysis (0)] |
| 202. | Hasanuzzaman M, Bang CS, Gong EJ. Antibiotic Resistance of Helicobacter pylori: Mechanisms and Clinical Implications. J Korean Med Sci. 2024;39:e44. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Reference Citation Analysis (0)] |
| 203. | Huang TT, Cao YX, Cao L. Novel therapeutic regimens against Helicobacter pylori: an updated systematic review. Front Microbiol. 2024;15:1418129. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 204. | Hou C, Yin F, Wang S, Zhao A, Li Y, Liu Y. Helicobacter pylori Biofilm-Related Drug Resistance and New Developments in Its Anti-Biofilm Agents. Infect Drug Resist. 2022;15:1561-1571. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 41] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 205. | Melander RJ, Basak AK, Melander C. Natural products as inspiration for the development of bacterial antibiofilm agents. Nat Prod Rep. 2020;37:1454-1477. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 86] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 206. | Carradori S, Di Giacomo N, Lobefalo M, Luisi G, Campestre C, Sisto F. Biofilm and Quorum Sensing inhibitors: the road so far. Expert Opin Ther Pat. 2020;30:917-930. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 35] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 207. | Maccelli A, Carradori S, Puca V, Sisto F, Lanuti P, Crestoni ME, Lasalvia A, Muraro R, Bysell H, Di Sotto A, Roos S, Grande R. Correlation between the Antimicrobial Activity and Metabolic Profiles of Cell Free Supernatants and Membrane Vesicles Produced by Lactobacillus reuteri DSM 17938. Microorganisms. 2020;8:1653. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 26] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 208. | Bai CL, Osaki T, Yonezawa H, Hanawa T, Zaman C, Kurata S, Kamiya S, Tanaka H. In vitro and in vivo effects of the Mongolian drug Amu-ru 7 on Helicobacter pylori growth and viability. Microbiol Immunol. 2010;54:508-515. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 5] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 209. | Cataldi V, Di Bartolomeo S, Di Campli E, Nostro A, Cellini L, Di Giulio M. In vitro activity of Aloe vera inner gel against microorganisms grown in planktonic and sessile phases. Int J Immunopathol Pharmacol. 2015;28:595-602. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 21] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 210. | Di Lodovico S, Napoli E, Di Campli E, Di Fermo P, Gentile D, Ruberto G, Nostro A, Marini E, Cellini L, Di Giulio M. Pistacia vera L. oleoresin and levofloxacin is a synergistic combination against resistant Helicobacter pylori strains. Sci Rep. 2019;9:4646. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 28] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 211. | Luo P, Huang Y, Hang X, Tong Q, Zeng L, Jia J, Zhang G, Bi H. Dihydrotanshinone I Is Effective against Drug-Resistant Helicobacter pylori In Vitro and In Vivo. Antimicrob Agents Chemother. 2021;65:e01921-20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 20] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 212. | Spósito L, Oda FB, Vieira JH, Carvalho FA, Dos Santos Ramos MA, de Castro RC, Crevelin EJ, Crotti AEM, Santos AG, da Silva PB, Chorilli M, Bauab TM. In vitro and in vivo anti-Helicobacter pylori activity of Casearia sylvestris leaf derivatives. J Ethnopharmacol. 2019;233:1-12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 45] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 213. | Jia J, Zhang C, Liu Y, Huang Y, Bai Y, Hang X, Zeng L, Zhu D, Bi H. Armeniaspirol A: a novel anti-Helicobacter pylori agent. Microb Biotechnol. 2022;15:442-454. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 23] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 214. |
Arif M, Sharaf M; |
| 215. | Cai J, Huang H, Song W, Hu H, Chen J, Zhang L, Li P, Wu R, Wu C. Preparation and evaluation of lipid polymer nanoparticles for eradicating H. pylori biofilm and impairing antibacterial resistance in vitro. Int J Pharm. 2015;495:728-737. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 59] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 216. | Puca V, Traini T, Guarnieri S, Carradori S, Sisto F, Macchione N, Muraro R, Mincione G, Grande R. The Antibiofilm Effect of a Medical Device Containing TIAB on Microorganisms Associated with Surgical Site Infection. Molecules. 2019;24:2280. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 20] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 217. | Chen X, Li P, Shen Y, Zou Y, Yuan G, Hu H. Rhamnolipid-involved antibiotics combinations improve the eradication of Helicobacter pylori biofilm in vitro: A comparison with conventional triple therapy. Microb Pathog. 2019;131:112-119. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 24] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 218. | Shen Y, Zou Y, Chen X, Li P, Rao Y, Yang X, Sun Y, Hu H. Antibacterial self-assembled nanodrugs composed of berberine derivatives and rhamnolipids against Helicobacter pylori. J Control Release. 2020;328:575-586. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 40] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 219. | Grande R, Sisto F, Puca V, Carradori S, Ronci M, Aceto A, Muraro R, Mincione G, Scotti L. Antimicrobial and Antibiofilm Activities of New Synthesized Silver Ultra-NanoClusters (SUNCs) Against Helicobacter pylori. Front Microbiol. 2020;11:1705. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 36] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 220. | Fontes LES, Martimbianco ALC, Zanin C, Riera R. N-acetylcysteine as an adjuvant therapy for Helicobacter pylori eradication. Cochrane Database Syst Rev. 2019;2:CD012357. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 18] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 221. | Yoon H, Lee DH, Jang ES, Kim J, Shin CM, Park YS, Hwang JH, Kim JW, Jeong SH, Kim N. Effects of N-acetylcysteine on First-Line Sequential Therapy for Helicobacter pylori Infection: A Randomized Controlled Pilot Trial. Gut Liver. 2016;10:520-525. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 222. | Cammarota G, Branca G, Ardito F, Sanguinetti M, Ianiro G, Cianci R, Torelli R, Masala G, Gasbarrini A, Fadda G, Landolfi R, Gasbarrini G. Biofilm demolition and antibiotic treatment to eradicate resistant Helicobacter pylori: a clinical trial. Clin Gastroenterol Hepatol. 2010;8:817-820.e3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 73] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 223. | Chen CC, Luo JC, Fang YJ, Lee JY, Kuo CC, Yang TH, Chiu MC, Yu JJ, Bair MJ, Chen PY, Chou CK, Chen CY, Chang CY, Hsu YC, Tseng CH, Hsu WF, Hu WH, Tsai MH, Hsieh CL, Chen MJ, Shun CT, Liu TY, Lee YC, Liou JM, Wu MS; and for the Taiwan Gastrointestinal Disease and Helicobacter Consortium. Comparison of the effect of clarithromycin triple therapy with or without N-acetylcysteine in the eradication of Helicobacter pylori: a randomized controlled trial. Therap Adv Gastroenterol. 2020;13:1756284820927306. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 6] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 224. | Gurbuz AK, Ozel AM, Ozturk R, Yildirim S, Yazgan Y, Demirturk L. Effect of N-acetyl cysteine on Helicobacter pylori. South Med J. 2005;98:1095-1097. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 34] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 225. | Mestre A, Sathiya Narayanan R, Rivas D, John J, Abdulqader MA, Khanna T, Chakinala RC, Gupta S. Role of Probiotics in the Management of Helicobacter pylori. Cureus. 2022;14:e26463. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 12] [Reference Citation Analysis (0)] |
| 226. | Ismail NI, Nawawi KNM, Hsin DCC, Hao KW, Mahmood NRKN, Chearn GLC, Wong Z, Tamil AM, Joseph H, Raja Ali RA. Probiotic containing Lactobacillus reuteri DSM 17648 as an adjunct treatment for Helicobacter pylori infection: A randomized, double-blind, placebo-controlled trial. Helicobacter. 2023;28:e13017. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 23] [Reference Citation Analysis (0)] |
| 227. | Viazis N, Argyriou K, Kotzampassi K, Christodoulou DK, Apostolopoulos P, Georgopoulos SD, Liatsos C, Giouleme O, Koustenis K, Veretanos C, Stogiannou D, Moutzoukis M, Poutakidis C, Mylonas II, Tseti I, Mantzaris GJ. A Four-Probiotics Regimen Combined with A Standard Helicobacter pylori-Eradication Treatment Reduces Side Effects and Increases Eradication Rates. Nutrients. 2022;14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 55] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 228. | Mohtasham M, Joukar F, Maroufizadeh S, Mojtahedi K, Asgharnezhad M, Mansour-Ghanaei F. Lactobacillus ruteri compared with placebo as an adjuvant in quadruple therapy for Helicobacter pylori eradication: A randomized, double-blind, controlled trial. Arab J Gastroenterol. 2023;24:40-44. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 9] [Reference Citation Analysis (0)] |
| 229. | Moreno Márquez C, Fernández Álvarez P, Valdés Delgado T, Castro Laria L, Argüelles Arias F, Caunedo Álvarez Á, Gómez Rodríguez BJ. Randomized, double-blind, placebo-controlled clinical trial on the usefulness of probiotic Lactobacillus reuteri in bismuth-containing quadruple eradication therapy for infection with Helicobacter pylori. Rev Esp Enferm Dig. 2022;114:89-95. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 12] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 230. | Lau CS, Ward A, Chamberlain RS. Probiotics improve the efficacy of standard triple therapy in the eradication of Helicobacter pylori: a meta-analysis. Infect Drug Resist. 2016;9:275-289. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 51] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 231. | Penumetcha SS, Ahluwalia S, Irfan R, Khan SA, Rohit Reddy S, Vasquez Lopez ME, Zahid M, Busmail A, Mohammed L. The Efficacy of Probiotics in the Management of Helicobacter Pylori: A Systematic Review. Cureus. 2021;13:e20483. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 13] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 232. | Lu C, Sang J, He H, Wan X, Lin Y, Li L, Li Y, Yu C. Probiotic supplementation does not improve eradication rate of Helicobacter pylori infection compared to placebo based on standard therapy: a meta-analysis. Sci Rep. 2016;6:23522. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 74] [Cited by in RCA: 69] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 233. | Yang Z, Zhou Y, Han Z, He K, Zhang Y, Wu D, Chen H. The effects of probiotics supplementation on Helicobacter pylori standard treatment: an umbrella review of systematic reviews with meta-analyses. Sci Rep. 2024;14:10069. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 234. | Bachir M, Allem R, Benejat L, Tifrit A, Medjekane M, Drici AE, Megraud F, Douidi KT. Molecular detection of mutations involved in Helicobacter pylori antibiotic resistance in Algeria. J Antimicrob Chemother. 2018;73:2034-2038. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 21] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 235. | Raaf N, Amhis W, Saoula H, Abid A, Nakmouche M, Balamane A, Ali Arous N, Ouar-Korichi M, Vale FF, Bénéjat L, Mégraud F. Prevalence, antibiotic resistance, and MLST typing of Helicobacter pylori in Algiers, Algeria. Helicobacter. 2017;22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 18] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 236. | Alvarez-Aldana A, Fernandez Uribe PA, Mejía Valencia T, Guaca-Gonzalez YM, Santacruz-Ibarra JJ, Arturo-Arias BL, Castañeda-Chavez LJ, Pacheco-López R, Londoño-Giraldo LM, Moncayo-Ortiz JI. Antimicrobial susceptibility of clinical Helicobacter pylori isolates and its eradication by standard triple therapy: a study in west central region of Colombia. Microbiol Spectr. 2024;12:e0040124. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 237. | Mannion A, Dzink-Fox J, Shen Z, Piazuelo MB, Wilson KT, Correa P, Peek RM Jr, Camargo MC, Fox JG. Helicobacter pylori Antimicrobial Resistance and Gene Variants in High- and Low-Gastric-Cancer-Risk Populations. J Clin Microbiol. 2021;59:e03203-20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 34] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 238. | Saranathan R, Levi MH, Wattam AR, Malek A, Asare E, Behin DS, Pan DH, Jacobs WR Jr, Szymczak WA. Helicobacter pylori Infections in the Bronx, New York: Surveying Antibiotic Susceptibility and Strain Lineage by Whole-Genome Sequencing. J Clin Microbiol. 2020;58:e01591-19. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 33] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 239. | Shokrzadeh L, Alebouyeh M, Mirzaei T, Farzi N, Zali MR. Prevalence of multiple drug-resistant Helicobacter pylori strains among patients with different gastric disorders in Iran. Microb Drug Resist. 2015;21:105-110. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 28] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 240. | Huang X, Wu B, Chen Q, Chen Y, Ji X, Zhou X, Suo B, Lin Z, Zheng X. Antibiotic resistance profile of Helicobacter pylori to 14 antibiotics: a multicenter study in Fujian, China. PeerJ. 2023;11:e15611. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 241. | Vanden Bulcke A, Waked B, Haems L, Lambrecht G, Hervent AS, Alliet G, Baert F, Vervaeke S. Antimicrobial resistance of Helicobacter pylori in West Flanders - Belgium: an observational cross-sectional study. Acta Clin Belg. 2022;77:360-367. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 6] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 242. | Boyanova L, Gergova G, Evstatiev I, Spassova Z, Kandilarov N, Yaneva P, Markovska R, Mitov I. Helicobacter pylori resistance to six antibiotics by two breakpoint systems and resistance evolution in Bulgaria. Infect Dis (Lond). 2016;48:56-62. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 25] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 243. | Mégraud F, Alix C, Charron P, Bénéjat L, Ducournau A, Bessède E, Lehours P. Survey of the antimicrobial resistance of Helicobacter pylori in France in 2018 and evolution during the previous 5 years. Helicobacter. 2021;26:e12767. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 17] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 244. | Macías-García F, Llovo-Taboada J, Díaz-López M, Bastón-Rey I, Domínguez-Muñoz JE. High primary antibiotic resistance of Helicobacter Pylori strains isolated from dyspeptic patients: A prevalence cross-sectional study in Spain. Helicobacter. 2017;22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 35] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 245. | Diab M, El-Shenawy A, El-Ghannam M, Salem D, Abdelnasser M, Shaheen M, Abdel-Hady M, El-Sherbini E, Saber M. Detection of antimicrobial resistance genes of Helicobacter pylori strains to clarithromycin, metronidazole, amoxicillin and tetracycline among Egyptian patients. Egypt J Med Hum Genet. 2018;19:417-423. [RCA] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 12] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 246. | Kouitcheu Mabeku LB, Eyoum Bille B, Tepap Zemnou C, Tali Nguefack LD, Leundji H. Broad spectrum resistance in Helicobacter pylori isolated from gastric biopsies of patients with dyspepsia in Cameroon and efflux-mediated multiresistance detection in MDR isolates. BMC Infect Dis. 2019;19:880. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 23] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 247. | Parra-Sepúlveda C, Merino JS, Sáez-Carrillo K, González C, García-Cancino A. Antibiotic resistance surveillance of Helicobacter pylori at the Biobío region (Chile) in a decade. Arq Gastroenterol. 2019;56:361-366. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 14] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 248. | Zhou Y, Zhang Y, Du S. Antibiotic resistance in Helicobacter pylori among children and adolescents in East Asia: A systematic review and meta-analysis. Chin Med J (Engl). 2024;137:1926-1938. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Reference Citation Analysis (0)] |
| 249. | Zhang YX, Zhou LY, Song ZQ, Zhang JZ, He LH, Ding Y. Primary antibiotic resistance of Helicobacter pylori strains isolated from patients with dyspeptic symptoms in Beijing: a prospective serial study. World J Gastroenterol. 2015;21:2786-2792. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 88] [Cited by in RCA: 104] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 250. | Zollner-Schwetz I, Leitner E, Plieschnegger W, Semlitsch G, Stepan V, Reiter L, Reicht G, Mörth E, Pavek J, Parsché P, Betterklieber C, Atzmüller D, Krause R, Högenauer C. Primary resistance of Helicobacter pylori is still low in Southern Austria. Int J Med Microbiol. 2016;306:206-211. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 11] [Article Influence: 1.2] [Reference Citation Analysis (1)] |
| 251. | Cosme A, Torrente Iranzo S, Montes Ros M, Fernández-Reyes Silvestre M, Alonso Galán H, Lizasoain J, Bujanda L. Helicobacter pylori antimicrobial resistance during a 5-year period (2013-2017) in northern Spain and its relationship with the eradication therapies. Helicobacter. 2019;24:e12557. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 17] [Article Influence: 2.8] [Reference Citation Analysis (1)] |
| 252. | Karbalaei M, Keikha M, Talebi Bezmin Abadi A. Prevalence of Primary Multidrug-resistant Helicobacter pylori in Children: A Systematic Review and Meta-analysis. Arch Med Res. 2022;53:634-640. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 2.7] [Reference Citation Analysis (0)] |