Published online Sep 21, 2024. doi: 10.3748/wjg.v30.i35.3932
Revised: August 12, 2024
Accepted: August 26, 2024
Published online: September 21, 2024
Processing time: 190 Days and 10.6 Hours
In this editorial, we comment on an article published in the recent issue of the World Journal of Gastroenterology. Celiac disease (CeD) is a disease occurring in genetically susceptible individuals, which is mainly characterized by gluten intolerance in the small intestine and clinical symptoms such as abdominal pain, diarrhea, and malnutrition. Therefore, patients often need a lifelong gluten-free diet, which greatly affects the quality of life and expenses of patients. The gold standard for diagnosis is intestinal mucosal biopsy, combined with serological and genetic tests. At present, the lack of safe, effective, and satisfactory drugs for CeD is mainly due to the complexity of its pathogenesis, and it is difficult to find a perfect target to solve the multi-level needs of patients. In this editorial, we mainly review the pathological mechanism of CeD and describe the current experimental and improved drugs for various pathological aspects.
Core Tip: This editorial provides a comprehensive review of the pathophysiological mechanisms underlying celiac disease and explores emerging therapeutic strategies, including the potential role of Aspergillus niger-derived prolyl endopeptidase. The discussion emphasizes the need for new treatments that address the multifaceted nature of the disease, aiming to improve patient quality of life beyond the limitations of a strict gluten-free diet.
- Citation: Ge HJ, Chen XL. Advances in understanding and managing celiac disease: Pathophysiology and treatment strategies. World J Gastroenterol 2024; 30(35): 3932-3941
- URL: https://www.wjgnet.com/1007-9327/full/v30/i35/3932.htm
- DOI: https://dx.doi.org/10.3748/wjg.v30.i35.3932
Celiac disease (CeD) is mainly characterized by gastrointestinal symptoms, including abdominal pain, diarrhea (fatty diarrhea), and weight loss, as well as more and more attention to extra-intestinal manifestations, such as anemia, osteoporosis, liver damage, and epilepsy[1]. Some patients are complicated with other diseases, such as type 1 diabetes and thyroiditis[2,3]. The main manifestations of CeD are intestinal villus atrophy and secondary nutritional malab
At present, genetic factors are considered to be the main cause of CeD[10]. More than 90% of the patients were positive for human leukocyte antigen (HLA)-DQ2 or HLA-DQ8[6,11]. However, the positive target gene does not indicate that the patient must have CeD, and the significance of negative gene test is much clear. It is widely proved that HLA-DQ2 or HLA-DQ8 negativity can basically exclude CeD[7]. The gold standard for diagnosis of CeD is multiple intestinal mucosal biopsies under gastroenteroscopy[12,13]. Serological antibody detection is an important means for screening and diagnosis of CeD. The antibodies with diagnostic value mainly include anti-endomyocardial antibody (EMA), anti-tissue transglutaminase antibody (anti-tTG), anti-deacylated glial protein peptide, and anti-gliadin antibody (AGA)[14,15]. Previous research showed that serum anti-tTG IgA (tTG-IgA) had a sensitivity of 98% and specificity of 75% for CeD diagnosis[16]. As a result, in adults with a strong suspicion of CeD and elevated serum tTG-IgA levels, it may be reason
Gluten-free diet (GFD) is the first and main choice for CeD treatment, and is currently the most effective way[15,17]. However, due to the different proportions of gluten in the diet structure of different countries and regional cultures and the fact that the risk of involuntary intake could not be assessed everywhere, GFD would increase the food cost of patients' daily life. Therefore, completely GFD is difficult to achieve and affects the daily life and social interaction of patients[18]. Therefore, there is an urgent need to clarify the comprehensive and accurate pathological mechanisms of CeD and develop new drugs or treatments on this basis to improve the quality of life and reduce the cost of life of CeD patients. This editorial reviews the pathological mechanisms of CeD and the possible effective drugs in the future.
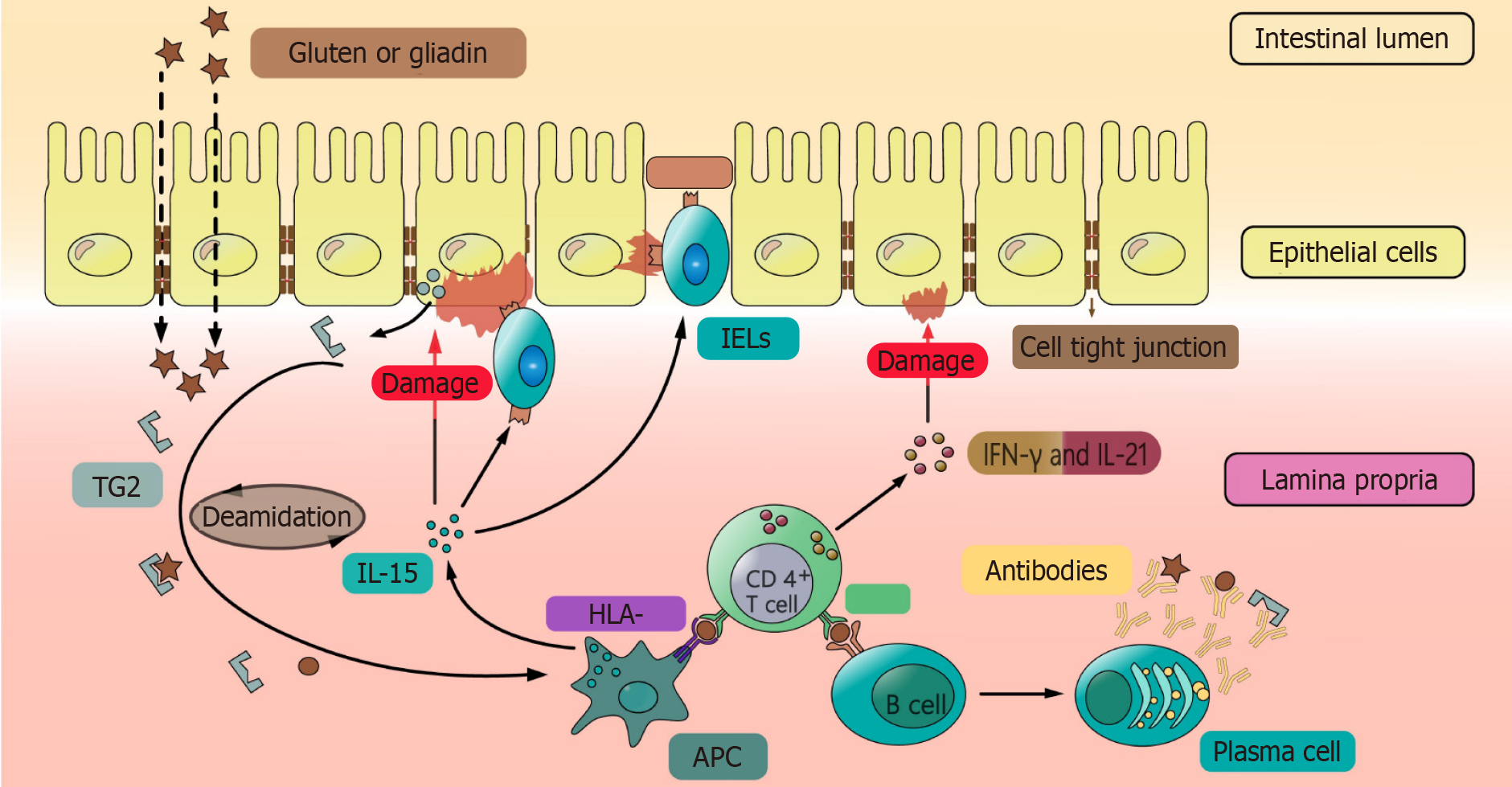
The majority of CeD patients are positive for the susceptibility gene HLA-DQ2 or HLA-DQ8, which is the most important part of the pathogenesis of most CeD[6,11]. HLA class I and class II genes are located on the 6th of chromosomes of major histocompatibility complexes[19]. We summarize the information of possible HLA genes and single nucleotide polymorphisms associated with CeD in Table 1 and Table 2. These genes encode HLA-DQ2/DQ8 on the surface of antigen-presenting cells (APCs), and they bind to gluten and are degraded in the small intestine to form HLA polypeptide complexes, which are presented to CD4+ T cells[20,21]. APCs secrete interleukin (IL)-15 to promote the proliferation of intraepithelial lymphocytes (IELs) interspersed between or beneath epithelial cells, express natural killer cell receptor (NKG2D) on the cell surface, and induce epithelial cells to express the NKG2D ligand major histocompatibility complex related gene A (MICA). NK cells that express NKG2D recognize MICA and kill the intestinal epithelial cells (IECs) that express MICA on the surface, thus increasing intestinal permeability[22-24]. On the one hand, T cells activate lymphoid B cells to differentiate into plasma cells, which secrete antibodies against gluten and transglutaminase 2[25,26]. On the other hand, T cells secrete pro-inflammatory factors such as interferon-γ (IFN-γ) and IL-21, which cause damage to the intestinal epithelium and enhance its permeability[27].
| HLA gene name | Encoding alleles | Chromosome location | Association with celiac disease |
| HLA-DQ2.5[78] | DQA1*05, DQB1*02 | 6p21.3 | Strongly associated with celiac disease; present in 90%-95% of patients |
| HLA-DQ8[79] | DQA1*03, DQB1*03: 02 | 6p21.3 | Associated with celiac disease; present in 5%-10% of patients |
| HLA-DQ7[80] | DQA1*05, DQB1*03: 01 | 6p21.3 | Possibly associated with celiac disease; lower risk, often requires combination with other high-risk genes |
| HLA-DR3[81] | DRB1*03 | 6p21.3 | Typically coexists with HLA-DQ2.5, indirectly increases celiac disease risk |
| HLA-DR4[82] | DRB1*04 | 6p21.3 | Risk with celiac disease is low, but in some cases may lead to the disease |
| Single nucleotide polymorphism | Gene | Location | Variation(s) | Association with celiac disease |
| rs2187668[83] | HLA-DQA1 | chr6: 32638107 | C>T | Strongly associated with celiac disease; often used as a marker for the HLA-DQ2.5 haplotype |
| rs7775228[84] | HLA-DQB1 | chr6: 32690302 | T>A, T>C | Associated with the HLA-DQ8 haplotype, another strong genetic risk factor for celiac disease |
| rs3135388[85] | HLA-DRB1 | chr6: 32445274 | A>C, A>G, A>T | Linked to the HLA-DR3 haplotype, which may indirectly increase celiac disease risk when co-inherited with DQ2.5 |
| rs13015714[86] | IL18R1 | chr2: 102355405 | G>A, G>T | Associated with increased risk of celiac disease and severe symptoms due to its role in immune response regulation |
| rs1738074[87] | TAGAP | chr6: 159044945 | T>C | Linked to immune function and associated with increased susceptibility to celiac disease |
| rs3087243[88] | CTLA4 | chr2: 203874196 | G>A, G>T | Rare variant associated with autoimmune conditions; may exacerbate celiac disease symptoms when present |
| rs3184504[85] | SH2B3 | chr12: 111446804 | T>A, T>C, T>G | Involved in immune response, linked to increased risk of autoimmune diseases, including celiac disease |
| rs13031237[89] | REL | chr2: 60908994 | G>T | Associated with celiac disease and other autoimmune disorders, may affect the severity of the disease |
Gluten is the main protein component in a variety of grains (such as wheat, barley, and rye)[28]. It can be broken down into amino acids and peptides by enzymes in the lumen and on the brush edge of the small intestine. Gluten mainly includes gliadin and glutenin, in which gliadin rich in glutamine and proline, is the main antigen protein leading to CeD, and its decomposition products are difficult to be digested by digestive enzymes in intestinal lumen[13,29].
The activation of intestinal immune response by gluten is mainly through two aspects: Innate immunity and adaptive immunity. Regarding innate immunity, the non-immunogen dominant peptide (P31-P43) in gluten could directly promote the proliferation and differentiation of epithelial lymphocytes, the proliferation of CD8+ T cells and NK cells, and the production of IFN-γ[29]. In addition, it induces the expression of NKG2D receptor ligands in the intestinal epithelium and enhances the killing effect of NK cells on normal IECs, resulting in increased intestinal epithelial permeability[22-24]. With regard to adaptive immunity, due to the increase of intestinal mucosal epithelial permeability caused by infection or innate immunity, some larger peptides in gluten (such as 33mer) could enter the intestinal lamina propria through the intestinal mucosal barrier[30]. After deamidation by tTG, it is easier to be captured by APCs and presented to CD4+ T cells[31]. 33-mer peptide could promote the proliferation and differentiation of B cells to produce antibodies such as AGA, anti-EMA, and anti-tTG actibody[32,33]. Because the damaged intestinal cells can express CD71 transporter on the apical side, the secretory IgA-gliadin complex enhances the transport of gluten from the lumen to the lamina propria by reversing endocytosis[34,35], and that is also the reason why gluten immunogenic peptides (GIPs) could cross the epithelial cells into blood stream and be tested out in urine[36,37]. Finally, the interaction between CD4+ T cells and gliadin in the lamina propria of the intestinal tract induces their activation and proliferation, and induces crypt proliferation by producing pro-inflammatory cytokines which also causes chemotaxis of immune cells[38], and villi would become blunt after IEC death[39].
Intestinal bacterial populations can be categorized into three groups based on their interaction with the host: Probiotics (e.g., Lactobacillus and Bifidobacterium), pathogenic bacteria (e.g., Clostridium and Enterococcus faecalis), and opportunistic pathogens. There was a disorder of digestive tract bacterial populations in patients with CeD, and the abundance and diversity of beneficial bacteria decreased while those of pathogenic bacteria increased[40]. The human body lacks proteases that could fully digest gluten, and many genera of bacteria in the intestine have been found to be related to gluten metabolism (such as Lactobacillus, Streptococcus, Staphylococcus, Clostridium, and Bifidobacterium). These bacteria are mainly colonized in the large intestine and partly distributed in the small intestine[41,42]. The sensitivity of different CeD patients to different intake of gluten may also be related to the disorder of intestinal bacterial populations[43,44].
Aspergillus niger-derived prolyl endoprotease: Aspergillus niger-derived prolyl endoprotease (AN-PEP), a kind of PEP extracted from Aspergillus niger, can effectively degrade the antigen protein in gluten, thus reducing its immunogenicity and avoiding intestinal damage[37]. The advantage of this kind of PEP is that it is an oral enzyme with activity between pH 2-8 and 4-5, so it can resist pepsin digestion without any special treatment[45]. Previous studies have shown that oral AN-PEP can enhance the degradation and digestion of gluten in healthy people, and similar result has been obtained in patients with gluten sensitivity[46]. Oral AN-PEP has also been found to degrade most of the gluten collagen before the chyme reaches the small intestine[47,48]. In in vitro model experiments, it was found that dietary composition affected the degradation efficiency of AN-PEP for gluten. For example, the addition of carbonated beverage strongly enhanced the activity of AN-PEP due to its acidification, fat did not affect the degradation of glutenin by AN-PEP, but the presence of food protein slowed down the detoxification of glutenin[49]. In this study, compared with the placebo group, AN-PEP treatment did not significantly reduce the total fecal concentration of GIPs, and there was no statistical difference in serological tests, but the clinical information system questionnaire showed that the rate of patients with severe symptoms in the AN-PEP group was significantly lower, suggesting that the auxiliary effect of AN-PEP on the GFD is still worthy of further exploration[50].
ALV003: ALV003, also known as latiglutinase, is an experimental drug composed of two recombinant proteases admi
AGA: AGA is a polyclonal IgY antibody against gluten. Because of its cross-reactivity, it can neutralize all CeD-inducing prolamins. IgY was collected from the yolks of highly immunized hens, sprayed and dried, and then encapsulated with 50% mannitol[55]. In a clinical trial of patients with a GFD, 1 g of AGA was given per meal, and 10 patients had fewer celiac symptoms (especially tiredness, headache, and bloating), improved quality of life, lowered antibodies, and lactu
TAK-062: TAK-062, derived from the enzyme kumamolisin of Alicyclobacillus sendaiensis and engineered to enhance its proteolytic activity, specifically targets proline-glutamine (P-Q) dipeptide motifs and is designed to maintain its proteolytic function even in the varying pH levels and presence of proteases in the gastrointestinal tract[57,58]. TAK-062 is expected to work irrespectively of meal composition. In vitro, TAK-062 at a dose of 900 mg was well-tolerated and degraded more than 99% of gluten (3 g and 9 g) within 10 min[58].
TG2 inhibition: ZED1227 is a small molecule tTG2 inhibitor. The compound selectively binds to the active state of tTG2, forming a stable covalent bond with the cysteine in its catalytic center[59]. Clinical trials have confirmed that ZED1227 has high biological safety, and it is still safe and effective when the dose is up to 500 mg. In a proof-of-concept trial, it was confirmed by intestinal mucosal biopsy that ZED1227 could effectively reduce intestinal mucosal damage with the increase of drug dose (10 mg, 50 mg, and 100 mg)[60].
IL-15 antibody: The pro-inflammatory cytokine IL-15 has been identified as the main pathophysiological mediator of CeD[61]. IL-15 is produced by APCs and epithelial cells in the small intestine and is a necessary factor for the activation and proliferation of IELs[61,62]. IELs, mainly CD8+ T cells, destroy IECs and cause villus atrophy, which is characteristic of CeD. AMG714 is the first anti-IL-15 monoclonal antibody to be investigated for the treatment of CeD. In a clinical trial, patients with CeD who received a long-term GFD were treated with subcutaneous injection of AMG714 twice every 2 wk for 10 wk (a total of 6 doses) and gluten stimulation (2-4 g per day) from 2 to 12 wk. Compared with the placebo group, AMG714 did not prevent intestinal mucosal damage caused by gluten exposure at 150 mg and 300 mg doses[63]. However, at 300 mg, the increase of IELs in the treatment group was less significant and the symptoms were milder. In addition, another clinical trial found no difference in the main end points of abnormal IELs between patients with refractory type 2 CeD who received AMG714 or placebo for 10 wk[64].
Larazotide acetate (also known as AT-1001) is a synthesis of occlusive zone toxin produced by Vibrio cholerae[65]. Zonulin is the main regulatory element of paracellular junction. The main role of AT-1001 is to act as an anti-zonulin receptor inhibitor to reduce the increase of intestinal barrier permeability mediated by zonulin[66]. Existing clinical trials have confirmed its good biological safety, and the improvement of intestinal symptoms is better in patients treated with AT-1001 combined with a GFD than the GFD alone, but there is no statistical difference in lactulose-to-mannitol ratio when compared with the placebo group[67,68]. Whether there are other mechanisms for the effect of AT-1001 on intestinal permeability and how to apply it in clinical practice need further research.
Gluten enters the lamina propria of the mucosa and is deacylated by TG2 and then presented to CD4+ T cells by APCs, which then activates a series of immune responses, resulting in intestinal mucosal damage. Therefore, it is expected to become a target for the treatment of CeD by interfering with antigen presentation and activating T cells, inducing immune tolerance, and reducing the activation of CD4+ T cells.
TAK-101: TAK-101 is a drug in which gluten (a complex subcomponent of natural wheat gliadin) is encapsulated in nega
KAN-101: KAN-101 is also a drug that induces immune tolerance in the liver by intravenous injection. It consists of a synthetic liver-targeted glycosylated polymer that is connected to a synthetic immunodominant deaminated gliadin peptide (KAN0009) that binds to HLA-DQ2.5[68]. The possible mechanism is to inhibit the immune response of CD4+ T cells by activating Treg cells[72]. Its biosafety has been confirmed in clinical trials but needs further clinical verification.
Nexvax2: Nexvax2 is composed of three synthetic peptides and contains six HLA-DQ2.5 restricted immune dominant T cell epitopes, which is similar to the induction of bystander inhibition to play the role of immune tolerance[73]. Unfortunately, the follow-up clinical trial was terminated because the symptom improvement and serological tests did not meet the requirements.
VSL#3 probiotic preparation has the capacity to hydrolyze gliadin polypeptides responsible for celiac sprue, which was testified in vitro[74]. However, in a multicenter study, the probiotic preparation VSL#3 was tested with a GFD in 45 adults with symptoms of CeD, but there was no significant improvement in symptom severity and fecal microbiota[75]. A clinical trial showed that the treatment combined with lactic acid bacteria could improve gastrointestinal symptoms and increase the possible presence of lactic acid bacteria, staphylococci, and bifidobacteria in stool[76]. In addition, another trial explored the effects of Bifidobacterium on 22 untreated adult patients with active CeD. The results showed that the probiotics improved gastrointestinal symptoms and normalized immune markers, including a reduction in the number of α-defensins and Paneth cells in duodenal biopsy samples. At the end of the study, all patients were diagnosed with CeD by duodenal biopsy[68,77].
The pathological mechanism of CeD is complex (Figure 1). It is most important to find the key points in the pathogenesis of CeD and develop targeted drugs to interrupt the process of immune response. Glutenin and drugs that can degrade or bind gluten are expected to improve patients' quality of life and symptoms when their diet is contaminated with gluten. Immunosuppressants for pathological processes are expected to completely save patients from the suffering of a strict GFD. In addition, in many clinical trials, there is a phenomenon that symptoms are not completely consistent with serological examination and intestinal mucosal biopsy results. CeD patients often experience subjective symptoms such as abdominal pain and diarrhea. It is important to explore whether there is a temporal or spatial discrepancy between these symptoms and medical examination findings, and how they may be linked to the patients' emotional state, the brain-gut axis, and intestinal flora. Further research is needed to better understand these relationships.
| 1. | Volta U, Caio G, Stanghellini V, De Giorgio R. The changing clinical profile of celiac disease: a 15-year experience (1998-2012) in an Italian referral center. BMC Gastroenterol. 2014;14:194. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 90] [Cited by in RCA: 127] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 2. | Passali M, Josefsen K, Frederiksen JL, Antvorskov JC. Current Evidence on the Efficacy of Gluten-Free Diets in Multiple Sclerosis, Psoriasis, Type 1 Diabetes and Autoimmune Thyroid Diseases. Nutrients. 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 43] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 3. | Volta U, Tovoli F, Caio G. Clinical and immunological features of celiac disease in patients with Type 1 diabetes mellitus. Expert Rev Gastroenterol Hepatol. 2011;5:479-487. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 60] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 4. | Yuan J, Gao J, Li X, Liu F, Wijmenga C, Chen H, Gilissen LJ. The tip of the "celiac iceberg" in China: a systematic review and meta-analysis. PLoS One. 2013;8:e81151. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 54] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 5. | Cichewicz AB, Mearns ES, Taylor A, Boulanger T, Gerber M, Leffler DA, Drahos J, Sanders DS, Thomas Craig KJ, Lebwohl B. Diagnosis and Treatment Patterns in Celiac Disease. Dig Dis Sci. 2019;64:2095-2106. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 42] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 6. | Husby S, Koletzko S, Korponay-Szabó IR, Mearin ML, Phillips A, Shamir R, Troncone R, Giersiepen K, Branski D, Catassi C, Lelgeman M, Mäki M, Ribes-Koninckx C, Ventura A, Zimmer KP; ESPGHAN Working Group on Coeliac Disease Diagnosis; ESPGHAN Gastroenterology Committee; European Society for Pediatric Gastroenterology, Hepatology, and Nutrition. European Society for Pediatric Gastroenterology, Hepatology, and Nutrition guidelines for the diagnosis of coeliac disease. J Pediatr Gastroenterol Nutr. 2012;54:136-160. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1708] [Cited by in RCA: 1837] [Article Influence: 141.3] [Reference Citation Analysis (1)] |
| 7. | Bai JC, Ciacci C. World Gastroenterology Organisation Global Guidelines: Celiac Disease February 2017. J Clin Gastroenterol. 2017;51:755-768. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 96] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 8. | Singh P, Arora A, Strand TA, Leffler DA, Catassi C, Green PH, Kelly CP, Ahuja V, Makharia GK. Global Prevalence of Celiac Disease: Systematic Review and Meta-analysis. Clin Gastroenterol Hepatol. 2018;16:823-836.e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1076] [Cited by in RCA: 944] [Article Influence: 134.9] [Reference Citation Analysis (1)] |
| 9. | Theethira TG, Dennis M, Leffler DA. Nutritional consequences of celiac disease and the gluten-free diet. Expert Rev Gastroenterol Hepatol. 2014;8:123-129. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 102] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 10. | Breuning MH, van den Berg-Loonen EM, Bernini LF, Bijlsma JB, van Loghem E, Meera Khan P, Nijenhuis LE. Localization of HLA on the short arm of chromosome 6. Hum Genet. 1977;37:131-139. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 38] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 11. | Ludvigsson JF, Leffler DA, Bai JC, Biagi F, Fasano A, Green PH, Hadjivassiliou M, Kaukinen K, Kelly CP, Leonard JN, Lundin KE, Murray JA, Sanders DS, Walker MM, Zingone F, Ciacci C. The Oslo definitions for coeliac disease and related terms. Gut. 2013;62:43-52. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1254] [Cited by in RCA: 1159] [Article Influence: 96.6] [Reference Citation Analysis (1)] |
| 12. | Rubio-Tapia A, Hill ID, Kelly CP, Calderwood AH, Murray JA; American College of Gastroenterology. ACG clinical guidelines: diagnosis and management of celiac disease. Am J Gastroenterol. 2013;108:656-76; quiz 677. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1173] [Cited by in RCA: 1157] [Article Influence: 96.4] [Reference Citation Analysis (0)] |
| 13. | Kneepkens CM, von Blomberg BM. Clinical practice : coeliac disease. Eur J Pediatr. 2012;171:1011-1021. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 32] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 14. | Downey L, Houten R, Murch S, Longson D; Guideline Development Group. Recognition, assessment, and management of coeliac disease: summary of updated NICE guidance. BMJ. 2015;351:h4513. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 80] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 15. | Ludvigsson JF, Bai JC, Biagi F, Card TR, Ciacci C, Ciclitira PJ, Green PH, Hadjivassiliou M, Holdoway A, van Heel DA, Kaukinen K, Leffler DA, Leonard JN, Lundin KE, McGough N, Davidson M, Murray JA, Swift GL, Walker MM, Zingone F, Sanders DS; BSG Coeliac Disease Guidelines Development Group; British Society of Gastroenterology. Diagnosis and management of adult coeliac disease: guidelines from the British Society of Gastroenterology. Gut. 2014;63:1210-1228. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 689] [Cited by in RCA: 782] [Article Influence: 71.1] [Reference Citation Analysis (2)] |
| 16. | Ciacci C, Bai JC, Holmes G, Al-Toma A, Biagi F, Carroccio A, Ciccocioppo R, Di Sabatino A, Gingold-Belfer R, Jinga M, Makharia G, Niveloni S, Norman GL, Rostami K, Sanders DS, Smecuol E, Villanacci V, Vivas S, Zingone F; Bi. A.CeD study group. Serum anti-tissue transglutaminase IgA and prediction of duodenal villous atrophy in adults with suspected coeliac disease without IgA deficiency (Bi.A.CeD): a multicentre, prospective cohort study. Lancet Gastroenterol Hepatol. 2023;8:1005-1014. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 25] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 17. | Allen B, Orfila C. The Availability and Nutritional Adequacy of Gluten-Free Bread and Pasta. Nutrients. 2018;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 61] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 18. | Lee AR, Wolf RL, Lebwohl B, Ciaccio EJ, Green PHR. Persistent Economic Burden of the Gluten Free Diet. Nutrients. 2019;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 88] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 19. | Sollid LM, Thorsby E. HLA susceptibility genes in celiac disease: genetic mapping and role in pathogenesis. Gastroenterology. 1993;105:910-922. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 409] [Cited by in RCA: 402] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 20. | Kim CY, Quarsten H, Bergseng E, Khosla C, Sollid LM. Structural basis for HLA-DQ2-mediated presentation of gluten epitopes in celiac disease. Proc Natl Acad Sci U S A. 2004;101:4175-4179. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 319] [Cited by in RCA: 307] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 21. | Ráki M, Tollefsen S, Molberg Ø, Lundin KE, Sollid LM, Jahnsen FL. A unique dendritic cell subset accumulates in the celiac lesion and efficiently activates gluten-reactive T cells. Gastroenterology. 2006;131:428-438. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 89] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 22. | Meresse B, Chen Z, Ciszewski C, Tretiakova M, Bhagat G, Krausz TN, Raulet DH, Lanier LL, Groh V, Spies T, Ebert EC, Green PH, Jabri B. Coordinated induction by IL15 of a TCR-independent NKG2D signaling pathway converts CTL into lymphokine-activated killer cells in celiac disease. Immunity. 2004;21:357-366. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 583] [Cited by in RCA: 618] [Article Influence: 29.4] [Reference Citation Analysis (0)] |
| 23. | Hüe S, Mention JJ, Monteiro RC, Zhang S, Cellier C, Schmitz J, Verkarre V, Fodil N, Bahram S, Cerf-Bensussan N, Caillat-Zucman S. A direct role for NKG2D/MICA interaction in villous atrophy during celiac disease. Immunity. 2004;21:367-377. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 500] [Cited by in RCA: 515] [Article Influence: 24.5] [Reference Citation Analysis (0)] |
| 24. | Setty M, Discepolo V, Abadie V, Kamhawi S, Mayassi T, Kent A, Ciszewski C, Maglio M, Kistner E, Bhagat G, Semrad C, Kupfer SS, Green PH, Guandalini S, Troncone R, Murray JA, Turner JR, Jabri B. Distinct and Synergistic Contributions of Epithelial Stress and Adaptive Immunity to Functions of Intraepithelial Killer Cells and Active Celiac Disease. Gastroenterology. 2015;149:681-91.e10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 89] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 25. | du Pré MF, Sollid LM. T-cell and B-cell immunity in celiac disease. Best Pract Res Clin Gastroenterol. 2015;29:413-423. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 55] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 26. | Iversen R, Amundsen SF, Kleppa L, du Pré MF, Stamnaes J, Sollid LM. Evidence That Pathogenic Transglutaminase 2 in Celiac Disease Derives From Enterocytes. Gastroenterology. 2020;159:788-790. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 45] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 27. | Bodd M, Ráki M, Tollefsen S, Fallang LE, Bergseng E, Lundin KE, Sollid LM. HLA-DQ2-restricted gluten-reactive T cells produce IL-21 but not IL-17 or IL-22. Mucosal Immunol. 2010;3:594-601. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 115] [Cited by in RCA: 114] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 28. | Malalgoda M, Meinhardt SW, Simsek S. Detection and quantitation of immunogenic epitopes related to celiac disease in historical and modern hard red spring wheat cultivars. Food Chem. 2018;264:101-107. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 32] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 29. | Ribeiro M, Picascia S, Rhazi L, Gianfrani C, Carrillo JM, Rodriguez-Quijano M, Branlard G, Nunes FM. In Situ Gluten-Chitosan Interlocked Self-Assembled Supramolecular Architecture Reduces T-Cell-Mediated Immune Response to Gluten in Celiac Disease. Mol Nutr Food Res. 2018;62:e1800646. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 12] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 30. | Nanayakkara M, Kosova R, Lania G, Sarno M, Gaito A, Galatola M, Greco L, Cuomo M, Troncone R, Auricchio S, Auricchio R, Barone MV. A celiac cellular phenotype, with altered LPP sub-cellular distribution, is inducible in controls by the toxic gliadin peptide P31-43. PLoS One. 2013;8:e79763. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 22] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 31. | Herrán AR, Pérez-Andrés J, Caminero A, Nistal E, Vivas S, Ruiz de Morales JM, Casqueiro J. Gluten-degrading bacteria are present in the human small intestine of healthy volunteers and celiac patients. Res Microbiol. 2017;168:673-684. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 63] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 32. | Green PH, Cellier C. Celiac disease. N Engl J Med. 2007;357:1731-1743. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1321] [Cited by in RCA: 1271] [Article Influence: 70.6] [Reference Citation Analysis (0)] |
| 33. | Caio G, Volta U, Sapone A, Leffler DA, De Giorgio R, Catassi C, Fasano A. Celiac disease: a comprehensive current review. BMC Med. 2019;17:142. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 564] [Cited by in RCA: 563] [Article Influence: 93.8] [Reference Citation Analysis (0)] |
| 34. | Schumann M, Richter JF, Wedell I, Moos V, Zimmermann-Kordmann M, Schneider T, Daum S, Zeitz M, Fromm M, Schulzke JD. Mechanisms of epithelial translocation of the alpha(2)-gliadin-33mer in coeliac sprue. Gut. 2008;57:747-754. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 108] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 35. | Matysiak-Budnik T, Moura IC, Arcos-Fajardo M, Lebreton C, Ménard S, Candalh C, Ben-Khalifa K, Dugave C, Tamouza H, van Niel G, Bouhnik Y, Lamarque D, Chaussade S, Malamut G, Cellier C, Cerf-Bensussan N, Monteiro RC, Heyman M. Secretory IgA mediates retrotranscytosis of intact gliadin peptides via the transferrin receptor in celiac disease. J Exp Med. 2008;205:143-154. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 210] [Cited by in RCA: 212] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 36. | Moreno ML, Cebolla Á, Muñoz-Suano A, Carrillo-Carrion C, Comino I, Pizarro Á, León F, Rodríguez-Herrera A, Sousa C. Detection of gluten immunogenic peptides in the urine of patients with coeliac disease reveals transgressions in the gluten-free diet and incomplete mucosal healing. Gut. 2017;66:250-257. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 181] [Cited by in RCA: 235] [Article Influence: 29.4] [Reference Citation Analysis (0)] |
| 37. | Palanski BA, Weng N, Zhang L, Hilmer AJ, Fall LA, Swaminathan K, Jabri B, Sousa C, Fernandez-Becker NQ, Khosla C, Elias JE. An efficient urine peptidomics workflow identifies chemically defined dietary gluten peptides from patients with celiac disease. Nat Commun. 2022;13:888. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 31] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 38. | Lammers KM, Chieppa M, Liu L, Liu S, Omatsu T, Janka-Junttila M, Casolaro V, Reinecker HC, Parent CA, Fasano A. Gliadin Induces Neutrophil Migration via Engagement of the Formyl Peptide Receptor, FPR1. PLoS One. 2015;10:e0138338. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 37] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 39. | Kupfer SS, Jabri B. Pathophysiology of celiac disease. Gastrointest Endosc Clin N Am. 2012;22:639-660. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 96] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 40. | Nadal I, Donant E, Ribes-Koninckx C, Calabuig M, Sanz Y. Imbalance in the composition of the duodenal microbiota of children with coeliac disease. J Med Microbiol. 2007;56:1669-1674. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 271] [Cited by in RCA: 287] [Article Influence: 16.9] [Reference Citation Analysis (0)] |
| 41. | Chibbar R, Dieleman LA. The Gut Microbiota in Celiac Disease and probiotics. Nutrients. 2019;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 77] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 42. | Bodkhe R, Shetty SA, Dhotre DP, Verma AK, Bhatia K, Mishra A, Kaur G, Pande P, Bangarusamy DK, Santosh BP, Perumal RC, Ahuja V, Shouche YS, Makharia GK. Comparison of Small Gut and Whole Gut Microbiota of First-Degree Relatives With Adult Celiac Disease Patients and Controls. Front Microbiol. 2019;10:164. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 81] [Cited by in RCA: 64] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 43. | Caio G, Lungaro L, Segata N, Guarino M, Zoli G, Volta U, De Giorgio R. Effect of Gluten-Free Diet on Gut Microbiota Composition in Patients with Celiac Disease and Non-Celiac Gluten/Wheat Sensitivity. Nutrients. 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 83] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 44. | Elli L, Branchi F, Tomba C, Villalta D, Norsa L, Ferretti F, Roncoroni L, Bardella MT. Diagnosis of gluten related disorders: Celiac disease, wheat allergy and non-celiac gluten sensitivity. World J Gastroenterol. 2015;21:7110-7119. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 156] [Cited by in RCA: 152] [Article Influence: 15.2] [Reference Citation Analysis (4)] |
| 45. | Stepniak D, Spaenij-Dekking L, Mitea C, Moester M, de Ru A, Baak-Pablo R, van Veelen P, Edens L, Koning F. Highly efficient gluten degradation with a newly identified prolyl endoprotease: implications for celiac disease. Am J Physiol Gastrointest Liver Physiol. 2006;291:G621-G629. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 190] [Cited by in RCA: 186] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 46. | Salden BN, Monserrat V, Troost FJ, Bruins MJ, Edens L, Bartholomé R, Haenen GR, Winkens B, Koning F, Masclee AA. Randomised clinical study: Aspergillus niger-derived enzyme digests gluten in the stomach of healthy volunteers. Aliment Pharmacol Ther. 2015;42:273-285. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 55] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 47. | Mitea C, Havenaar R, Drijfhout JW, Edens L, Dekking L, Koning F. Efficient degradation of gluten by a prolyl endoprotease in a gastrointestinal model: implications for coeliac disease. Gut. 2008;57:25-32. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 183] [Cited by in RCA: 182] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 48. | König J, Holster S, Bruins MJ, Brummer RJ. Randomized clinical trial: Effective gluten degradation by Aspergillus niger-derived enzyme in a complex meal setting. Sci Rep. 2017;7:13100. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 47] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 49. | Montserrat V, Bruins MJ, Edens L, Koning F. Influence of dietary components on Aspergillus niger prolyl endoprotease mediated gluten degradation. Food Chem. 2015;174:440-445. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 28] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 50. | Stefanolo JP, Segura V, Grizzuti M, Heredia A, Comino I, Costa AF, Puebla R, Temprano MP, Niveloni SI, de Diego G, Oregui ME, Smecuol EG, de Marzi MC, Verdú EF, Sousa C, Bai JC. Effect of Aspergillus niger prolyl endopeptidase in patients with celiac disease on a long-term gluten-free diet. World J Gastroenterol. 2024;30:1545-1555. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 6] [Cited by in RCA: 12] [Article Influence: 12.0] [Reference Citation Analysis (3)] |
| 51. | Gass J, Bethune MT, Siegel M, Spencer A, Khosla C. Combination enzyme therapy for gastric digestion of dietary gluten in patients with celiac sprue. Gastroenterology. 2007;133:472-480. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 169] [Cited by in RCA: 168] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 52. | Siegel M, Garber ME, Spencer AG, Botwick W, Kumar P, Williams RN, Kozuka K, Shreeniwas R, Pratha V, Adelman DC. Safety, tolerability, and activity of ALV003: results from two phase 1 single, escalating-dose clinical trials. Dig Dis Sci. 2012;57:440-450. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 84] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 53. | Murray JA, Syage JA, Wu TT, Dickason MA, Ramos AG, Van Dyke C, Horwath I, Lavin PT, Mäki M, Hujoel I, Papadakis KA, Bledsoe AC, Khosla C, Sealey-Voyksner JA; CeliacShield Study Group. Latiglutenase Protects the Mucosa and Attenuates Symptom Severity in Patients With Celiac Disease Exposed to a Gluten Challenge. Gastroenterology. 2022;163:1510-1521.e6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 43] [Article Influence: 14.3] [Reference Citation Analysis (1)] |
| 54. | Lähdeaho ML, Kaukinen K, Laurila K, Vuotikka P, Koivurova OP, Kärjä-Lahdensuu T, Marcantonio A, Adelman DC, Mäki M. Glutenase ALV003 attenuates gluten-induced mucosal injury in patients with celiac disease. Gastroenterology. 2014;146:1649-1658. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 160] [Cited by in RCA: 181] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 55. | Gujral N, Löbenberg R, Suresh M, Sunwoo H. In-vitro and in-vivo binding activity of chicken egg yolk immunoglobulin Y (IgY) against gliadin in food matrix. J Agric Food Chem. 2012;60:3166-3172. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 28] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 56. | Sample DA, Sunwoo HH, Huynh HQ, Rylance HL, Robert CL, Xu BW, Kang SH, Gujral N, Dieleman LA. AGY, a Novel Egg Yolk-Derived Anti-gliadin Antibody, Is Safe for Patients with Celiac Disease. Dig Dis Sci. 2017;62:1277-1285. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 26] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 57. | Wolf C, Siegel JB, Tinberg C, Camarca A, Gianfrani C, Paski S, Guan R, Montelione G, Baker D, Pultz IS. Engineering of Kuma030: A Gliadin Peptidase That Rapidly Degrades Immunogenic Gliadin Peptides in Gastric Conditions. J Am Chem Soc. 2015;137:13106-13113. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 95] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 58. | Pultz IS, Hill M, Vitanza JM, Wolf C, Saaby L, Liu T, Winkle P, Leffler DA. Gluten Degradation, Pharmacokinetics, Safety, and Tolerability of TAK-062, an Engineered Enzyme to Treat Celiac Disease. Gastroenterology. 2021;161:81-93.e3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 54] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 59. | Büchold C, Hils M, Gerlach U, Weber J, Pelzer C, Heil A, Aeschlimann D, Pasternack R. Features of ZED1227: The First-In-Class Tissue Transglutaminase Inhibitor Undergoing Clinical Evaluation for the Treatment of Celiac Disease. Cells. 2022;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 30] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 60. | Schuppan D, Mäki M, Lundin KEA, Isola J, Friesing-Sosnik T, Taavela J, Popp A, Koskenpato J, Langhorst J, Hovde Ø, Lähdeaho ML, Fusco S, Schumann M, Török HP, Kupcinskas J, Zopf Y, Lohse AW, Scheinin M, Kull K, Biedermann L, Byrnes V, Stallmach A, Jahnsen J, Zeitz J, Mohrbacher R, Greinwald R; CEC-3 Trial Group. A Randomized Trial of a Transglutaminase 2 Inhibitor for Celiac Disease. N Engl J Med. 2021;385:35-45. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 119] [Article Influence: 29.8] [Reference Citation Analysis (0)] |
| 61. | Mayassi T, Ladell K, Gudjonson H, McLaren JE, Shaw DG, Tran MT, Rokicka JJ, Lawrence I, Grenier JC, van Unen V, Ciszewski C, Dimaano M, Sayegh HE, Kumar V, Wijmenga C, Green PHR, Gokhale R, Jericho H, Semrad CE, Guandalini S, Dinner AR, Kupfer SS, Reid HH, Barreiro LB, Rossjohn J, Price DA, Jabri B. Chronic Inflammation Permanently Reshapes Tissue-Resident Immunity in Celiac Disease. Cell. 2019;176:967-981.e19. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 132] [Article Influence: 22.0] [Reference Citation Analysis (0)] |
| 62. | Fehniger TA, Caligiuri MA. Interleukin 15: biology and relevance to human disease. Blood. 2001;97:14-32. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 714] [Cited by in RCA: 758] [Article Influence: 31.6] [Reference Citation Analysis (0)] |
| 63. | Lähdeaho ML, Scheinin M, Vuotikka P, Taavela J, Popp A, Laukkarinen J, Koffert J, Koivurova OP, Pesu M, Kivelä L, Lovró Z, Keisala J, Isola J, Parnes JR, Leon F, Mäki M. Safety and efficacy of AMG 714 in adults with coeliac disease exposed to gluten challenge: a phase 2a, randomised, double-blind, placebo-controlled study. Lancet Gastroenterol Hepatol. 2019;4:948-959. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 86] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 64. | Cellier C, Bouma G, van Gils T, Khater S, Malamut G, Crespo L, Collin P, Green PHR, Crowe SE, Tsuji W, Butz E, Cerf-Bensussan N, Macintyre E, Parnes JR, Leon F, Hermine O, Mulder CJ; RCD-II Study Group Investigators. Safety and efficacy of AMG 714 in patients with type 2 refractory coeliac disease: a phase 2a, randomised, double-blind, placebo-controlled, parallel-group study. Lancet Gastroenterol Hepatol. 2019;4:960-970. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 59] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 65. | Fasano A, Baudry B, Pumplin DW, Wasserman SS, Tall BD, Ketley JM, Kaper JB. Vibrio cholerae produces a second enterotoxin, which affects intestinal tight junctions. Proc Natl Acad Sci U S A. 1991;88:5242-5246. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 378] [Cited by in RCA: 359] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 66. | Slifer ZM, Krishnan BR, Madan J, Blikslager AT. Larazotide acetate: a pharmacological peptide approach to tight junction regulation. Am J Physiol Gastrointest Liver Physiol. 2021;320:G983-G989. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 24] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 67. | Paterson BM, Lammers KM, Arrieta MC, Fasano A, Meddings JB. The safety, tolerance, pharmacokinetic and pharmacodynamic effects of single doses of AT-1001 in coeliac disease subjects: a proof of concept study. Aliment Pharmacol Ther. 2007;26:757-766. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 189] [Cited by in RCA: 193] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 68. | Kivelä L, Caminero A, Leffler DA, Pinto-Sanchez MI, Tye-Din JA, Lindfors K. Current and emerging therapies for coeliac disease. Nat Rev Gastroenterol Hepatol. 2021;18:181-195. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 79] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 69. | Getts DR, Shea LD, Miller SD, King NJ. Harnessing nanoparticles for immune modulation. Trends Immunol. 2015;36:419-427. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 151] [Cited by in RCA: 177] [Article Influence: 17.7] [Reference Citation Analysis (0)] |
| 70. | Casey LM, Hughes KR, Saunders MN, Miller SD, Pearson RM, Shea LD. Mechanistic contributions of Kupffer cells and liver sinusoidal endothelial cells in nanoparticle-induced antigen-specific immune tolerance. Biomaterials. 2022;283:121457. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 33] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 71. | Kelly CP, Murray JA, Leffler DA, Getts DR, Bledsoe AC, Smithson G, First MR, Morris A, Boyne M, Elhofy A, Wu TT, Podojil JR, Miller SD; TAK-101 Study Group. TAK-101 Nanoparticles Induce Gluten-Specific Tolerance in Celiac Disease: A Randomized, Double-Blind, Placebo-Controlled Study. Gastroenterology. 2021;161:66-80.e8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 122] [Article Influence: 30.5] [Reference Citation Analysis (0)] |
| 72. | Murray JA, Wassaf D, Dunn K, Arora S, Winkle P, Stacey H, Cooper S, Goldstein KE, Manchanda R, Kontos S, Grebe KM. Safety and tolerability of KAN-101, a liver-targeted immune tolerance therapy, in patients with coeliac disease (ACeD): a phase 1 trial. Lancet Gastroenterol Hepatol. 2023;8:735-747. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 30] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 73. | Truitt KE, Daveson AJM, Ee HC, Goel G, MacDougall J, Neff K, Anderson RP. Randomised clinical trial: a placebo-controlled study of subcutaneous or intradermal NEXVAX2, an investigational immunomodulatory peptide therapy for coeliac disease. Aliment Pharmacol Ther. 2019;50:547-555. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 40] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 74. | De Angelis M, Rizzello CG, Fasano A, Clemente MG, De Simone C, Silano M, De Vincenzi M, Losito I, Gobbetti M. VSL#3 probiotic preparation has the capacity to hydrolyze gliadin polypeptides responsible for Celiac Sprue. Biochim Biophys Acta. 2006;1762:80-93. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 159] [Cited by in RCA: 162] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 75. | Harnett J, Myers SP, Rolfe M. Probiotics and the Microbiome in Celiac Disease: A Randomised Controlled Trial. Evid Based Complement Alternat Med. 2016;2016:9048574. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 45] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 76. | Francavilla R, Piccolo M, Francavilla A, Polimeno L, Semeraro F, Cristofori F, Castellaneta S, Barone M, Indrio F, Gobbetti M, De Angelis M. Clinical and Microbiological Effect of a Multispecies Probiotic Supplementation in Celiac Patients With Persistent IBS-type Symptoms: A Randomized, Double-Blind, Placebo-controlled, Multicenter Trial. J Clin Gastroenterol. 2019;53:e117-e125. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 114] [Cited by in RCA: 111] [Article Influence: 18.5] [Reference Citation Analysis (1)] |
| 77. | Smecuol E, Hwang HJ, Sugai E, Corso L, Cherñavsky AC, Bellavite FP, González A, Vodánovich F, Moreno ML, Vázquez H, Lozano G, Niveloni S, Mazure R, Meddings J, Mauriño E, Bai JC. Exploratory, randomized, double-blind, placebo-controlled study on the effects of Bifidobacterium infantis natren life start strain super strain in active celiac disease. J Clin Gastroenterol. 2013;47:139-147. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 127] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 78. | Margaritte-Jeannin P, Babron MC, Bourgey M, Louka AS, Clot F, Percopo S, Coto I, Hugot JP, Ascher H, Sollid LM, Greco L, Clerget-Darpoux F. HLA-DQ relative risks for coeliac disease in European populations: a study of the European Genetics Cluster on Coeliac Disease. Tissue Antigens. 2004;63:562-567. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 130] [Cited by in RCA: 138] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 79. | Hill ID, Dirks MH, Liptak GS, Colletti RB, Fasano A, Guandalini S, Hoffenberg EJ, Horvath K, Murray JA, Pivor M, Seidman EG; North American Society for Pediatric Gastroenterology, Hepatology and Nutrition. Guideline for the diagnosis and treatment of celiac disease in children: recommendations of the North American Society for Pediatric Gastroenterology, Hepatology and Nutrition. J Pediatr Gastroenterol Nutr. 2005;40:1-19. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 752] [Cited by in RCA: 694] [Article Influence: 34.7] [Reference Citation Analysis (0)] |
| 80. | Tinto N, Cola A, Piscopo C, Capuano M, Galatola M, Greco L, Sacchetti L. High Frequency of Haplotype HLA-DQ7 in Celiac Disease Patients from South Italy: Retrospective Evaluation of 5,535 Subjects at Risk of Celiac Disease. PLoS One. 2015;10:e0138324. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 28] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 81. | Demarchi M, Carbonara A, Ansaldi N, Santini B, Barbera C, Borelli I, Rossino P, Rendine S. HLA-DR3 and DR7 in coeliac disease: immunogenetic and clinical aspects. Gut. 1983;24:706-712. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 46] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 82. | Herrera M, Theiler G, Augustovski F, Chertkoff L, Fainboim L, DeRosa S, Cowan EP, Satz ML. Molecular characterization of HLA class II genes in celiac disease patients of Latin American Caucasian origin. Tissue Antigens. 1994;43:83-87. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 21] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 83. | Rostami-Nejad M, Romanos J, Rostami K, Ganji A, Ehsani-Ardakani MJ, Bakhshipour AR, Zojaji H, Mohebbi SR, Zali MR, Wijmenga C. Allele and haplotype frequencies for HLA-DQ in Iranian celiac disease patients. World J Gastroenterol. 2014;20:6302-6308. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 33] [Cited by in RCA: 45] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 84. | Baaqeel RH, Banaganapalli B, Al Mahdi HB, Salama MA, Alhussaini BH, Alaifan MA, Bin-Taleb Y, Shaik NA, Al-Aama JY, Elango R, Saadah OI. TagSNP approach for HLA risk allele genotyping of Saudi celiac disease patients: effectiveness and pitfalls. Biosci Rep. 2021;41. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 85. | Hagopian W, Lee HS, Liu E, Rewers M, She JX, Ziegler AG, Lernmark Å, Toppari J, Rich SS, Krischer JP, Erlich H, Akolkar B, Agardh D; TEDDY Study Group. Co-occurrence of Type 1 Diabetes and Celiac Disease Autoimmunity. Pediatrics. 2017;140. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 80] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 86. | Hunt KA, Zhernakova A, Turner G, Heap GA, Franke L, Bruinenberg M, Romanos J, Dinesen LC, Ryan AW, Panesar D, Gwilliam R, Takeuchi F, McLaren WM, Holmes GK, Howdle PD, Walters JR, Sanders DS, Playford RJ, Trynka G, Mulder CJ, Mearin ML, Verbeek WH, Trimble V, Stevens FM, O'Morain C, Kennedy NP, Kelleher D, Pennington DJ, Strachan DP, McArdle WL, Mein CA, Wapenaar MC, Deloukas P, McGinnis R, McManus R, Wijmenga C, van Heel DA. Newly identified genetic risk variants for celiac disease related to the immune response. Nat Genet. 2008;40:395-402. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 528] [Cited by in RCA: 488] [Article Influence: 28.7] [Reference Citation Analysis (0)] |
| 87. | Pehlivan M, Ayna TK, Baran M, Soyöz M, Koçyiğit AÖ, Çerçi B, Pirim İ. Investigation of TAGAP gene polymorphism (rs1738074) in Turkish pediatric celiac patients. Turk J Biochem. 2021;46:293-298. [DOI] [Full Text] |
| 88. | Haimila K, Einarsdottir E, de Kauwe A, Koskinen LL, Pan-Hammarström Q, Kaartinen T, Kurppa K, Ziberna F, Not T, Vatta S, Ventura A, Korponay-Szabo IR, Adány R, Pocsai Z, Széles G, Dukes E, Kaukinen K, Mäki M, Koskinen S, Partanen J, Hammarström L, Saavalainen P. The shared CTLA4-ICOS risk locus in celiac disease, IgA deficiency and common variable immunodeficiency. Genes Immun. 2009;10:151-161. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 36] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 89. | Trynka G, Zhernakova A, Romanos J, Franke L, Hunt KA, Turner G, Bruinenberg M, Heap GA, Platteel M, Ryan AW, de Kovel C, Holmes GK, Howdle PD, Walters JR, Sanders DS, Mulder CJ, Mearin ML, Verbeek WH, Trimble V, Stevens FM, Kelleher D, Barisani D, Bardella MT, McManus R, van Heel DA, Wijmenga C. Coeliac disease-associated risk variants in TNFAIP3 and REL implicate altered NF-kappaB signalling. Gut. 2009;58:1078-1083. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 159] [Cited by in RCA: 155] [Article Influence: 9.7] [Reference Citation Analysis (0)] |