Published online May 14, 2024. doi: 10.3748/wjg.v30.i18.2454
Revised: April 5, 2024
Accepted: April 18, 2024
Published online: May 14, 2024
Processing time: 81 Days and 4.5 Hours
Drug-induced liver injury (DILI) is one of the most common adverse events of medication use, and its incidence is increasing. However, early detection of DILI is a crucial challenge due to a lack of biomarkers and noninvasive tests.
To identify salivary metabolic biomarkers of DILI for the future development of noninvasive diagnostic tools.
Saliva samples from 31 DILI patients and 35 healthy controls (HCs) were sub
We found 247 differentially expressed salivary metabolites between the DILI group and the HC group. Using WMCNA, we identified a set of 8 DEMs closely related to liver injury for further prediction testing. Interestingly, the distinct separation of DILI patients and HCs was achieved with five metabolites, namely, 12-hydroxydodecanoic acid, 3-hydroxydecanoic acid, tetradecanedioic acid, hypoxanthine, and inosine (area under the curve: 0.733-1).
Salivary metabolomics revealed previously unreported metabolic alterations and diagnostic biomarkers in the saliva of DILI patients. Our study may provide a potentially feasible and noninvasive diagnostic method for DILI, but further validation is needed.
Core Tip: Drug-induced liver injury (DILI) is one of the most common and serious adverse reactions to drugs. Conventional biomarkers are not specific and there is an urgent need for a non-invasive DILI marker. Our study has revealed a significant difference in salivary metabolites between patients with DILI and healthy individuals, and identified five metabolites that can distinguish DILI from healthy control, namely 12-hydroxydodecanoic acid, 3-hydroxydecanoic acid, tetradecanedioic acid, hypoxanthine, and inosine. Our study may provide a potential feasible non-invasive diagnostic method for DILI.
- Citation: Yu SM, Zheng HC, Wang SC, Rong WY, Li P, Jing J, He TT, Li JH, Ding X, Wang RL. Salivary metabolites are promising noninvasive biomarkers of drug-induced liver injury. World J Gastroenterol 2024; 30(18): 2454-2466
- URL: https://www.wjgnet.com/1007-9327/full/v30/i18/2454.htm
- DOI: https://dx.doi.org/10.3748/wjg.v30.i18.2454
Drug-induced liver injury (DILI) refers to liver damage caused by the consumption of prescription or nonprescription drugs, health products, natural remedies, biological agents, and herbal and dietary supplements (HDSs)[1]. According to recent epidemiological studies, the incidence of DILI in Western countries is approximately 14-19 cases per 100000 people, with an increasing trend[2,3]. While liver injury can improve after the discontinuation of the suspected drugs in most patients with DILI, more than 10% of patients with DILI may progress to chronic hepatitis, acute liver failure, or even death[4]. It has been reported that DILI is the most common cause of acute liver failure, with a 180-day case fatality rate of 8% in the United States[5,6]. Unlike metabolic liver diseases, which have a well-defined histology, a liver biopsy from a patient with DILI can reveal a variety of histological features, such as inflammation, fibrosis, vascular injury, cholestasis, necrosis, nodular regeneration, and ductal destruction[7]. Thus, establishing a diagnosis of DILI can be challenging, as it relies mainly on excluding other common causes of liver injury. As there are currently no diagnostic tests or biomarkers available for idiosyncratic DILI, diagnosis typically depends on a thorough medical history that includes a detailed medication history, exclusion of other potential causes of liver disease, and a series of liver biochemical tests before and after cessation of the medication[8]. Several clinical tools have been developed to assess the causality of DILI, with the Roussel Uclaf Causality Assessment Method (RUCAM) being the most widely used. However, importantly, RUCAM is far from perfect in terms of its accuracy and effectiveness[9]. It is therefore extremely important to develop noninvasive biomarkers with high sensitivity and specificity for the early detection and therapeutic evaluation of DILI.
Metabolomics involves profiling small-molecule metabolites and offers the potential to identify and characterize specific metabolic phenotypes associated with a given disease[10]. To date, metabolite biomarkers for DILI have been identified in plasma, serum and urine[11-14]. Saliva is a highly promising biological fluid for the detection of disease biomarkers owing to its noninvasive and painless collection procedures as well as its simple storage and handling requirements, which require minimal training[15]. Several recent studies have highlighted the potential use of metabolite biomarkers in saliva for identifying patients with oral cancer[16], breast cancer[17], periodontitis[18], diabetes[19], schizophrenia[20], Alzheimer’s disease[21], hepatocellular carcinoma and chronic liver disease[22], but there are no related reports in patients with DILI. This study aimed to identify novel biomarkers for DILI and represents the first demonstration of the utility of salivary metabolites for discriminating patients with DILI from healthy individuals.
This study included patients with DILI who were hospitalized at the Fifth Medical Center of PLA General Hospital between July 2020 and June 2021. The study was approved by the Ethics Committee of the Fifth Medical Center of PLA General Hospital (2020050D). Prior to their inclusion in the study, all research subjects or their representatives were required to sign a written informed consent form. Patient demographic information and laboratory data were obtained through electronic medical records and questionnaires.
The inclusion criteria for this study were as follows: (1) Aged between 18 and 70 years; (2) newly diagnosed with DILI; and (3) voluntary participation in the study and signing of the informed consent form upon inclusion.
The exclusion criteria for this study were as follows: (1) Patients with other concomitant causes of liver injury (such as viral, alcoholism, autoimmune, metabolic, tumor, genetic, or biliary diseases); and (2) patients with autoimmune diseases, malignant tumors, or severe heart, lung, or renal insufficiency.
The diagnostic criteria utilized in this study to identify DILI were as follows[8]: (1) Recent liver biochemistry indices displaying clinically significant abnormalities; (2) a comprehensive history of medication and HDS use within the 180 d prior to presentation; and (3) exclusion of alternative causes of liver injury. Clinically significant abnormalities in liver biochemistry commonly met at least one of the following criteria[8]: (1) Serum aspartate aminotransferase (AST) or alanine aminotransferase (ALT) levels greater than 5 times the upper limit of normal (ULN) or alkaline phosphatase (ALP) levels greater than 2 times the ULN on at least two separate occasions, each 24 hours apart; (2) total serum bilirubin (TBIL) levels greater than 2.5 mg/dL in conjunction with elevated serum AST, ALT, or ALP levels; or (3) international normalized ratio (INR) levels greater than 1.5 with elevated serum AST, ALT, or ALP levels. According to the updated RUCAM[23], patients with definite or probable DILI (score 6 or higher) were included in this study.
Saliva was collected in Salivette tubes (Sarstedt AG and Co., Numbrecht, Germany) as described previously[24]. All samples were collected between 9 a.m. and 12 a.m. Each subject was asked to refrain from smoking, eating, drinking and tooth brush procedures for at least 1 h before saliva collection. Additionally, the subjects were required to gently gargle with water prior to saliva collection to remove food debris. To stimulate salivation, subjects were instructed to gently chew and roll the cotton swab for 60-90 seconds and then to spit the cotton swab back into the collection tube of the kit. The tubes were immediately kept on ice and centrifuged within 1 h at 10000 × g for 10 min at 4 °C. The collected supernatants were aliquoted (100 μL) without any further processing and stored at -80 °C until sample analysis.
This study was conducted according to previously described methods[25,26]. The saliva samples (100 μL) were placed in EP tubes and resuspended in prechilled 80% methanol by thorough vortexing. The samples were then incubated on ice for 5 minutes and centrifuged at 15000 g and 4 °C for 20 min. A portion of the supernatant was diluted with liquid chromatography (LC)/mass spectrometry (MS)-grade water to obtain a final concentration of 53% methanol. The resulting mixture was transferred to a fresh Eppendorf tube and centrifuged at 15000 × g and 4 °C for 20 min. Finally, the supernatant was injected into the LC-MS/MS system for analysis.
Statistical analyses were conducted using R software (version R-4.2.2) and Python (version 3.11.1). In cases where the data were not normally distributed, the area normalization method was employed to attempt normal transformations. Principal component analysis (PCA) was carried out in an unsupervised manner to provide a broad view of the data distribution. Partial least squares-discriminant analysis (PLS-DA) was performed as a supervised model to evaluate metabolic alterations among groups, and a permutation test was carried out 200 times to check for overfitting risks in the PLS-DA model. Pathway enrichment analysis was conducted using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. The least absolute shrinkage and selection operator (LASSO) and random forest (RF) methods were employed for optimization and modeling. Weighted metabolite coexpression network analysis (WMCNA) was used to identify sets of metabolites with similar expression profiles across samples. The Pearson correlation coefficient was used to determine the correlation between metabolites and liver function indices. The χ2 test or Fisher’s exact test was used for categorical parameters, while student’s t test or the Mann-Whitney U test was used for two-group comparisons, as appropriate. A P value < 0.05 was considered to indicate statistical significance.
A total of 31 patients diagnosed with DILI and 35 healthy controls (HCs) were included in this study. Table 1 presents the clinical characteristics of both groups. Compared with the HCs, the patients in the DILI group exhibited significantly greater ALT, AST, TBIL, ALP, INR, gamma-glutamyl transpeptidase (GGT), and total bile acid levels and significantly lower levels of albumin (P < 0.001). There were no significant differences in other baseline characteristics, including age, sex, weight, or body mass index, between the two groups (P > 0.05).
| Clinical characteristic | DILI (n = 31) | HC (n = 35) | P value |
| Age, yr, median (IQR) | 50.0 (46.0-60.0) | 52.0 (45.0-61.0) | 0.585 |
| Female | 19 (61.3) | 19 (54.3) | 0.566 |
| Weight, kg, median (IQR) | 60.0 (53.0-74.0) | 63.0 (58.0-70.0) | 0.310 |
| BMI, kg/m2, median (IQR) | 21.5 (20.2-25.0) | 22.9 (20.8-25.4) | 0.676 |
| Laboratory examination | |||
| ALT, U/L, median (IQR) | 238.0 (91.0-526.0) | 22.0 (12.0-27.0) | < 0.001 |
| AST, U/L, median (IQR) | 182.0 (74.0-270.0) | 25.0 (16.0-30.0) | < 0.001 |
| TBIL, μmol/L, median (IQR) | 72.7 (32.1-117.4) | 8.5 (5.7-13.1) | < 0.001 |
| ALP, U/L, median (IQR) | 137.0 (92.0-215.0) | 86.0 (77.0-114.0) | < 0.001 |
| GGT, U/L, median (IQR) | 140.0 (82.0-213.0) | 32.0 (17.0-41.0) | < 0.001 |
| ALB, g/L, median (IQR) | 36.0 (32.0-39.0) | 41.0 (39.0-46.0) | < 0.001 |
| TBA, μmol/L, median (IQR) | 42.0 (11.0-73.0) | 9.0 (5.0-17.0) | < 0.001 |
| LDH, U/L, median (IQR) | 186.0 (158.0-202.0) | 188.0 (146.0-243.0) | 0.550 |
| INR, median (IQR) | 1.0 (1.0-1.1) | 0.9 (0.9-1.0) | < 0.001 |
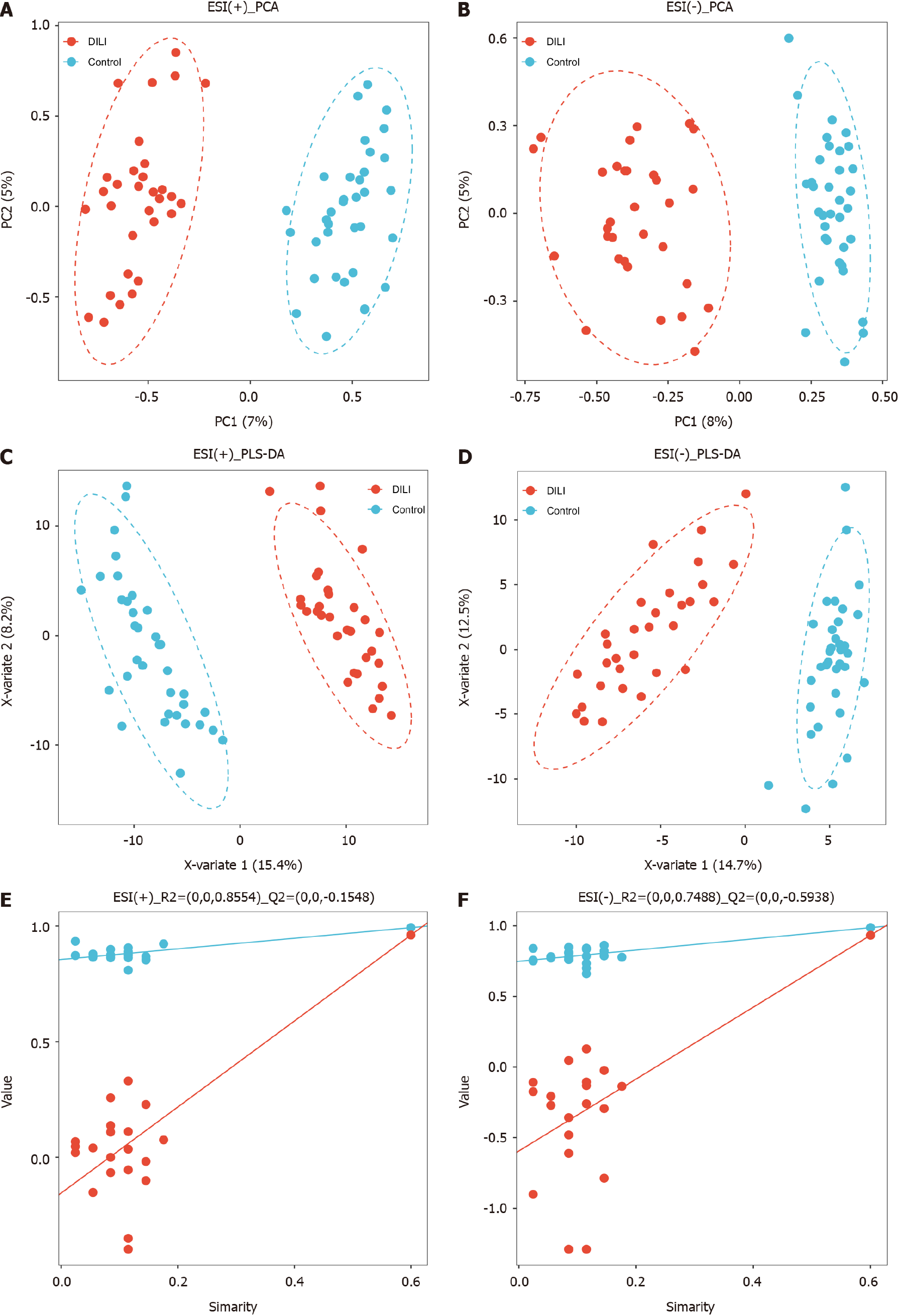
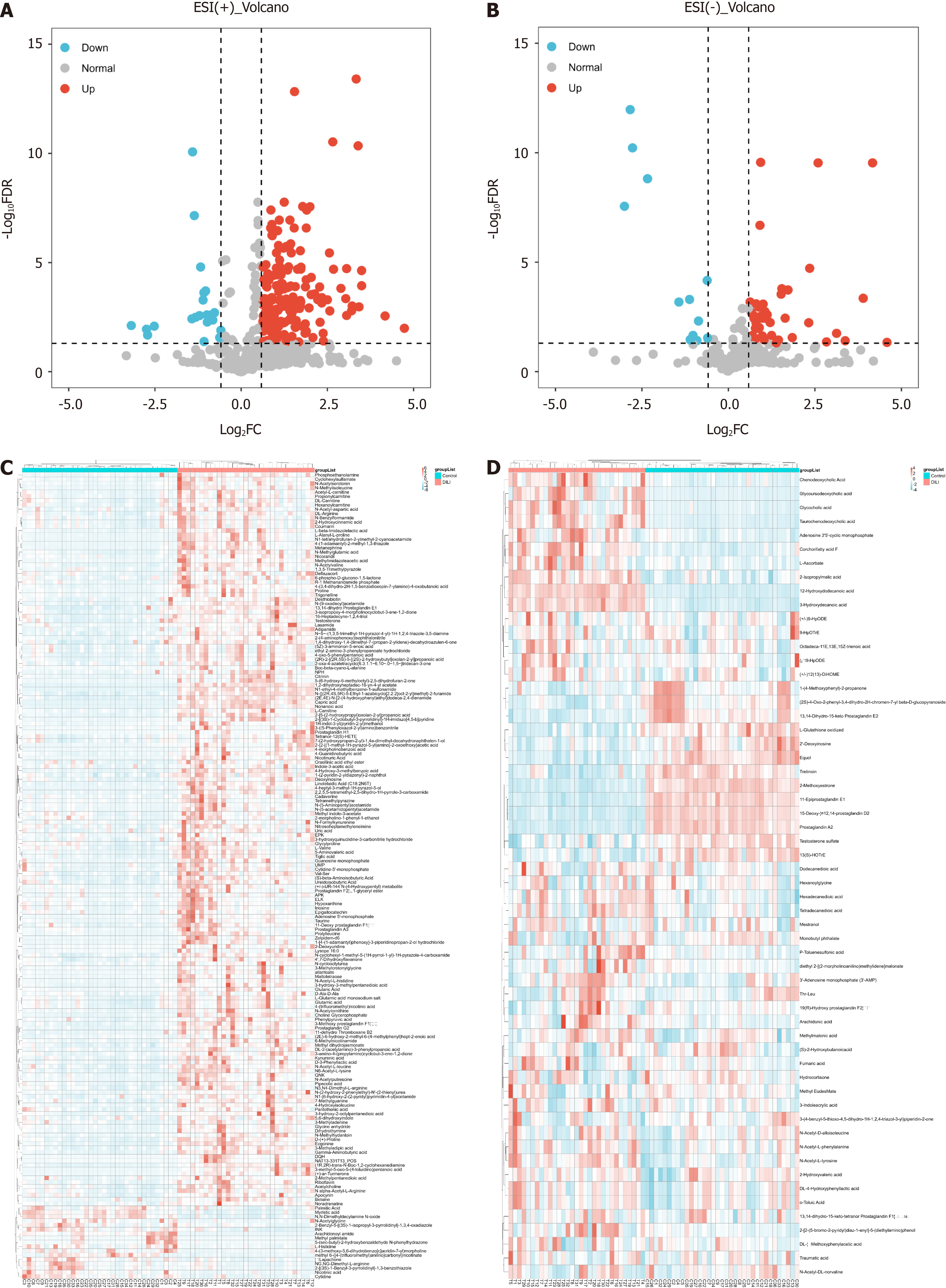
In total, 853 metabolites were detected by ultrahigh-pressure LC coupled with tandem MS. The HC and DILI groups were not well separated according to unsupervised PCA (Figure 1A and B). A PLS-DA was performed for both the positive and negative modes of MS, which clearly differentiated between the HC and DILI groups (Figure 1C and D). The 200-permutation test further validated the reliability and validity of the PLS-DA model (Figure 1E and F), indicating that there was no overfitting in the PLS-DA model. Interestingly, the salivary metabolite composition significantly differed between the control and DILI groups. The differentially expressed metabolites (DEMs) between the two groups were screened using an independent sample t test (variable importance in the projection ≥ 1, FC ≥ 1.5 or ≤ 0.67, and P < 0.05). We found 188 salivary DEMs screened in positive mode and 59 in negative mode (Figure 2) (Supplementary Table 1).
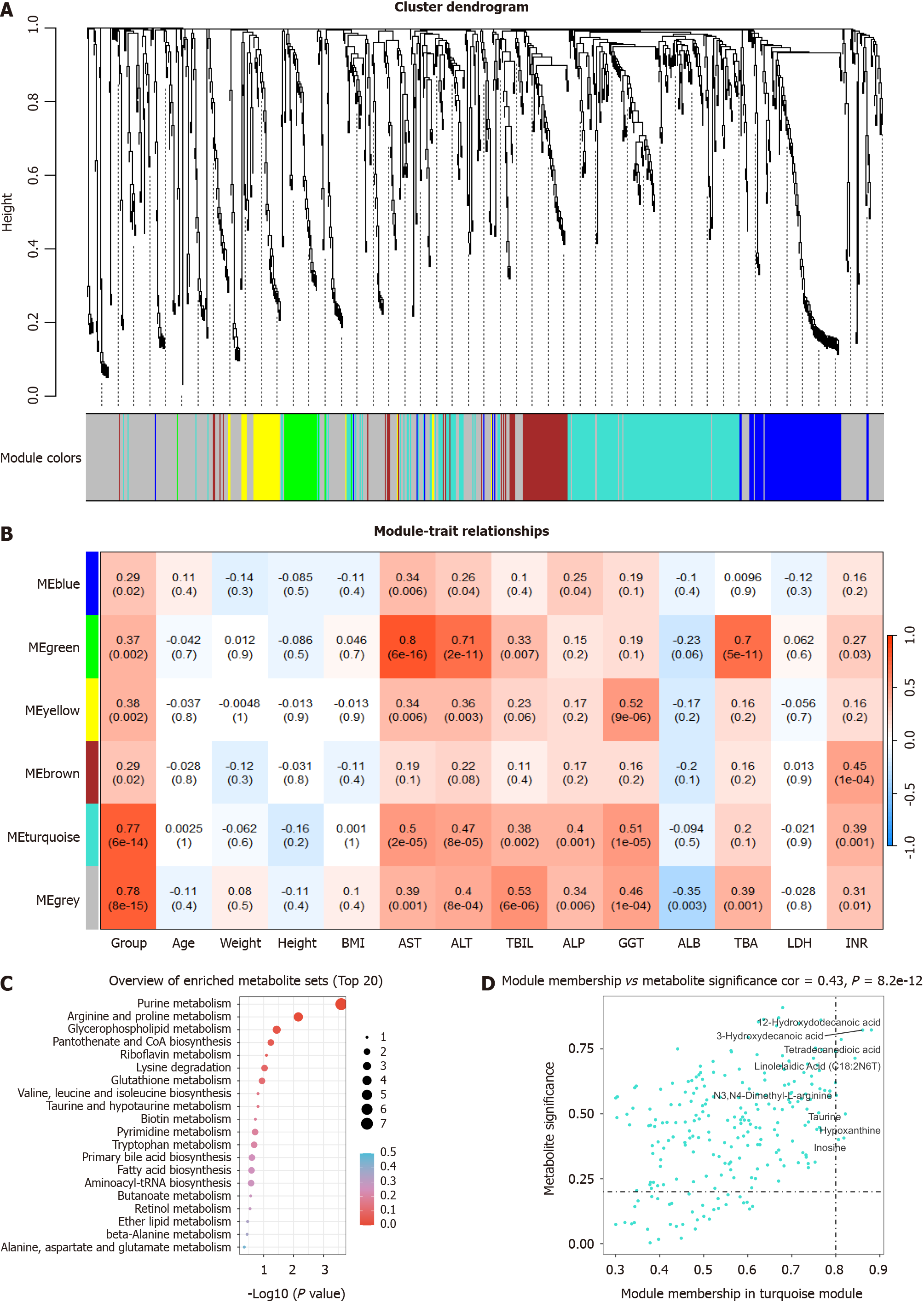
WMCNA is a powerful tool for identifying sets of metabolites that exhibit highly synergistic changes. Rather than focusing solely on differentially abundant metabolites, WMCNA leverages information from thousands or even tens of thousands of relevant metabolites and conducts significant association analyses of the metabolites with phenotypes. Therefore, we used WMCNA and constructed a network to further investigate the relationships between metabolites and DILI. The results of the WMCNA classified metabolites into five modules of closely associated metabolites, among which the turquoise consensus module was the most relevant to DILI (r = 0.77, P < 0.001) (Figure 3A and B) (Supplementary Table 2). Consequently, the turquoise module was selected as a clinically significant module for further analysis. The results of KEGG enrichment analysis showed that metabolites in the turquoise module were mainly involved in purine metabolism and beta-oxidation of very long-chain fatty acids (Figure 3C). Additionally, eight metabolites in the turquoise module with a metabolite significance of > 0.2 and a module membership of > 0.8 were selected (Figure 3D). Remarkably, these eight metabolites in the turquoise module overlapped with those previously identified in the PLS-DA.
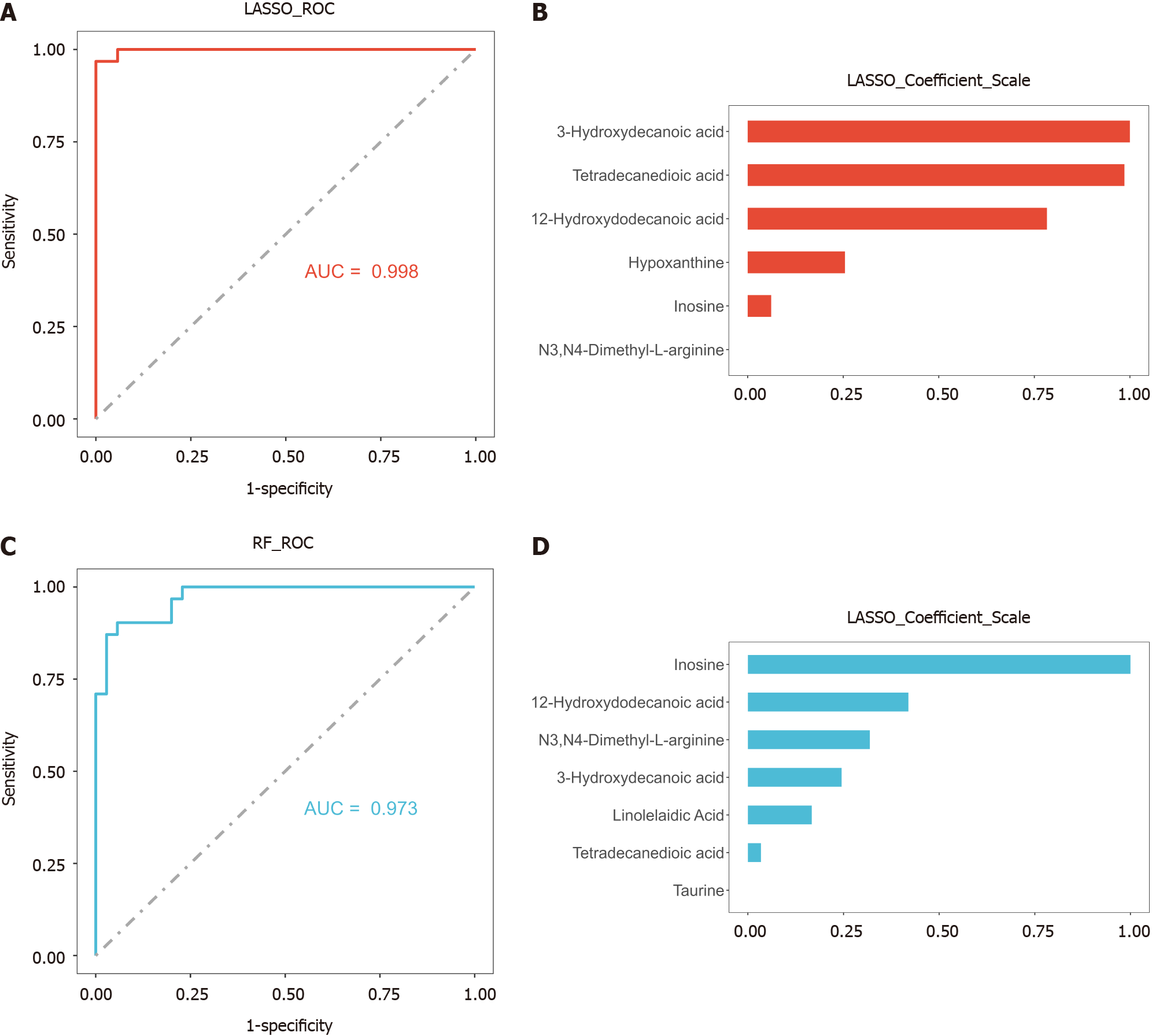
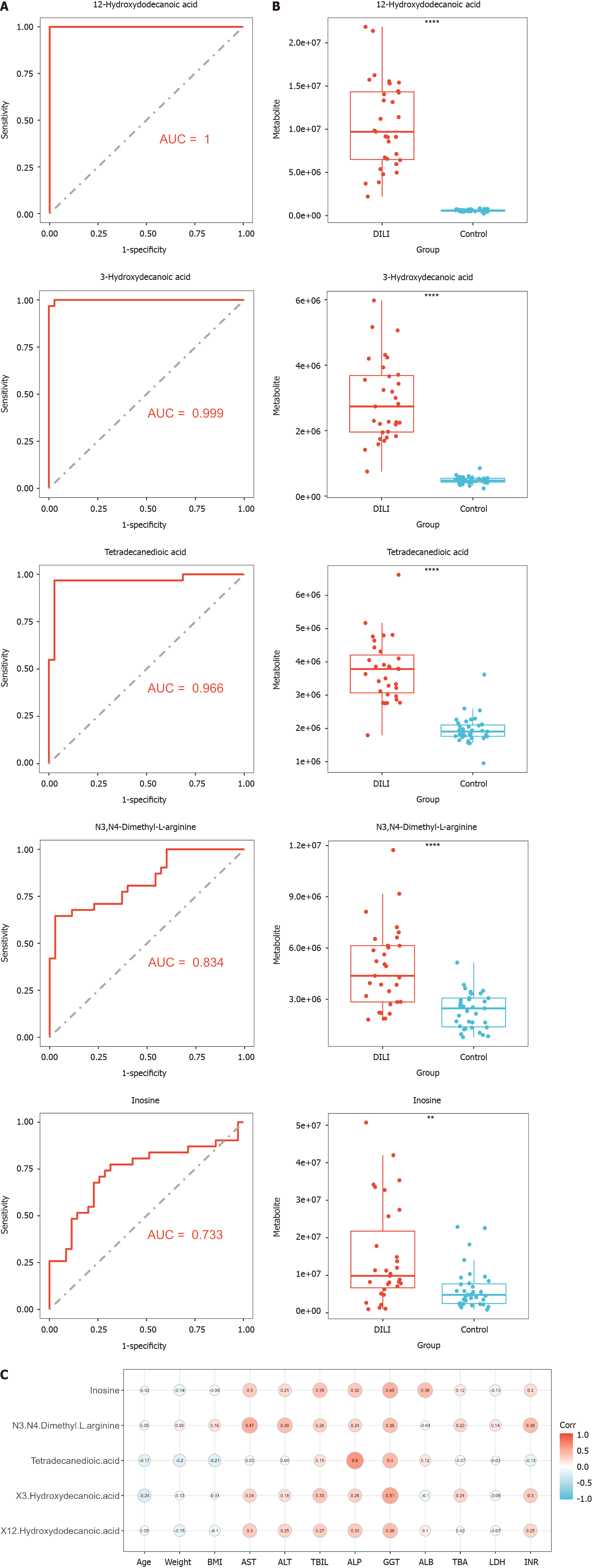
To establish a diagnostic model of DILI, LASSO and RF were performed on the above eight metabolites. There were five hub metabolites in LASSO area under the curve (AUC) = 0.998 and seven hub metabolites in RF (AUC = 0.969) (Figure 4). We further intersected the hub metabolites of the two models to obtain five common metabolites, namely, 12-hydroxydodecanoic acid, 3-hydroxydecanoic acid, tetradecanedioic acid, hypoxanthine, and inosine. These results suggest that the five hub metabolites may serve as potential diagnostic biomarkers for DILI. The AUC values of 12-hydroxydodecanoic acid, 3-hydroxydecanoic acid, tetradecanedioic acid, hypoxanthine, and inosine were 1.000, 0.999, 0.966, 0.834, and 0.733, respectively (Figure 5A). In particular, the levels of the five hub metabolites were significantly greater than those in HCs (Figure 5B). Additionally, distance correlation matrix plots revealed correlations between the five hub metabolites and crucial clinical indicators of patients with DILI, including the levels of ALT, AST, TBIL, ALP, and GGT (Figure 5C).
DILI is one of the most serious and common adverse reactions to drugs and is a major cause of clinically acute liver injury or failure[27,28]. It is crucial to detect DILI signal events early before they become symptomatic or severe to prevent clinically significant liver injury. The currently available biomarkers for DILI, namely, ALT, AST, and ALP, lack both specificity and sensitivity for detecting DILI at an early stage and are not reliable for predicting clinical outcomes[8]. Accordingly, a noninvasive marker for DILI and elucidation of the pathogenic mechanisms underlying DILI in humans are urgently needed. Using saliva for biomarker discovery is appealing because it can be collected noninvasively and is stable for long periods at room temperature[29]. To the best of our knowledge, this is the first study to investigate salivary metabolites in patients with DILI. Our study revealed a significant difference in salivary metabolites between patients with DILI and healthy individuals. Subsequently, we employed machine learning to identify combinations of salivary metabolites with predictive power to serve as biomarkers for DILI diagnosis.
In this study, WMCNA yielded five modules, of which the turquoise module was deemed clinically relevant. Metabolite pathway analysis of the turquoise module suggested that metabolic disorders associated with DILI might be linked to purine metabolism and beta-oxidation of very long-chain fatty acids. Additionally, eight hub metabolites in the turquoise module showed significant associations with DILI and were also found to exhibit changes in the PLS-DA model. Notably, 12-hydroxydodecanoic acid, 3-hydroxydecanoic acid, tetradecanedioic acid, hypoxanthine, and inosine were identified as having diagnostic value.
Based on research, disruptions in purine metabolism can cause the liver to release ATP, which can modulate immune responses and lead to the death of hepatocytes[30-32]. Purine metabolism may also affect DILI by interfering with drug metabolism or cell death processes[33-35]. Thus, purine metabolism is considered an important metabolic change in DILI, with hypoxanthine and inosine identified as key metabolites that impact this pathway[36,37]. Hypoxanthine and inosine are intermediates in the purine degradation pathway, and their levels increase during hypoxia, which is when adenine nucleotides are rapidly degraded[38-40]. When oxygen becomes available during tissue reperfusion, xanthine oxidase oxidizes hypoxanthine and inosine, generating reactive oxygen species that can trigger liver injury[41,42]. Our findings suggest that hypoxanthine and inosine may serve as potential indicators for diagnosing DILI, and efforts to repair impaired purine metabolism could benefit patients with this condition. Further research is needed to confirm these results.
Furthermore, we observed a significant increase in the levels of certain lipid metabolism metabolites, including 12-hydroxydodecanoic acid, 3-hydroxydecanoic acid, and tetradecanedioic acid, in DILI patients compared to HCs. Fatty acids, which are synthesized and metabolized primarily in the liver, are heavily impacted by liver injury[43]. The excessive buildup of medium- and long-chain fatty acids, such as 12-hydroxydodecanoic acid, 3-hydroxydecanoic acid, and tetradecanedioic acid, may hinder beta-oxidation in DILI patients, leading to anomalous energy metabolism and fat storage[44-46]. Treating DILI may involve preventing the accumulation of these toxic fatty acids in the liver, but very little research has been conducted in this field, which makes it an interesting area of study. Taken together, these findings indicate that abnormal fatty acid accumulation could act as a biomarker for DILI and contribute to its development, which is consistent with the literature[47].
To the best of our knowledge, this study provides initial evidence showing the ability of salivary metabolites to distinguish patients with DILI from healthy individuals. Our approach to sampling DILI biomarkers was not only less invasive but also less expensive than prior diagnostic methods. Moreover, additional investigations are currently underway to substantiate these findings across separate cohorts and ascertain the role of these metabolites in DILI patient outcomes.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report’s classification
Scientific Quality: Grade A
Novelty: Grade A
Creativity or Innovation: Grade A
Scientific Significance: Grade A
P-Reviewer: Stan FG, Romania S-Editor: Qu XL L-Editor: A P-Editor: Yuan YY
| 1. | Yu YC, Mao YM, Chen CW, Chen JJ, Chen J, Cong WM, Ding Y, Duan ZP, Fu QC, Guo XY, Hu P, Hu XQ, Jia JD, Lai RT, Li DL, Liu YX, Lu LG, Ma SW, Ma X, Nan YM, Ren H, Shen T, Wang H, Wang JY, Wang TL, Wang XJ, Wei L, Xie Q, Xie W, Yang CQ, Yang DL, Yu YY, Zeng MD, Zhang L, Zhao XY, Zhuang H; Drug-induced Liver Injury (DILI) Study Group; Chinese Society of Hepatology (CSH); Chinese Medical Association (CMA). CSH guidelines for the diagnosis and treatment of drug-induced liver injury. Hepatol Int. 2017;11:221-241. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 221] [Cited by in RCA: 207] [Article Influence: 25.9] [Reference Citation Analysis (1)] |
| 2. | Björnsson ES, Bergmann OM, Björnsson HK, Kvaran RB, Olafsson S. Incidence, presentation, and outcomes in patients with drug-induced liver injury in the general population of Iceland. Gastroenterology. 2013;144:1419-1425, 1425.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 535] [Cited by in RCA: 582] [Article Influence: 48.5] [Reference Citation Analysis (0)] |
| 3. | Sgro C, Clinard F, Ouazir K, Chanay H, Allard C, Guilleminet C, Lenoir C, Lemoine A, Hillon P. Incidence of drug-induced hepatic injuries: a French population-based study. Hepatology. 2002;36:451-455. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 555] [Cited by in RCA: 535] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 4. | Chalasani N, Bonkovsky HL, Fontana R, Lee W, Stolz A, Talwalkar J, Reddy KR, Watkins PB, Navarro V, Barnhart H, Gu J, Serrano J; United States Drug Induced Liver Injury Network. Features and Outcomes of 899 Patients With Drug-Induced Liver Injury: The DILIN Prospective Study. Gastroenterology. 2015;148:1340-52.e7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 635] [Cited by in RCA: 640] [Article Influence: 64.0] [Reference Citation Analysis (0)] |
| 5. | Chalasani N, Fontana RJ, Bonkovsky HL, Watkins PB, Davern T, Serrano J, Yang H, Rochon J; Drug Induced Liver Injury Network (DILIN). Causes, clinical features, and outcomes from a prospective study of drug-induced liver injury in the United States. Gastroenterology. 2008;135:1924-1934, 1934.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 624] [Cited by in RCA: 611] [Article Influence: 35.9] [Reference Citation Analysis (0)] |
| 6. | Ostapowicz G, Fontana RJ, Schiødt FV, Larson A, Davern TJ, Han SH, McCashland TM, Shakil AO, Hay JE, Hynan L, Crippin JS, Blei AT, Samuel G, Reisch J, Lee WM; U. S. Acute Liver Failure Study Group. Results of a prospective study of acute liver failure at 17 tertiary care centers in the United States. Ann Intern Med. 2002;137:947-954. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1562] [Cited by in RCA: 1462] [Article Influence: 63.6] [Reference Citation Analysis (0)] |
| 7. | Kleiner DE. Drug-induced Liver Injury: The Hepatic Pathologist's Approach. Gastroenterol Clin North Am. 2017;46:273-296. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 53] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 8. | Fontana RJ, Liou I, Reuben A, Suzuki A, Fiel MI, Lee W, Navarro V. AASLD practice guidance on drug, herbal, and dietary supplement-induced liver injury. Hepatology. 2023;77:1036-1065. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 91] [Article Influence: 45.5] [Reference Citation Analysis (0)] |
| 9. | Katarey D, Verma S. Drug-induced liver injury. Clin Med (Lond). 2016;16:s104-s109. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 182] [Cited by in RCA: 199] [Article Influence: 22.1] [Reference Citation Analysis (0)] |
| 10. | Schrimpe-Rutledge AC, Codreanu SG, Sherrod SD, McLean JA. Untargeted Metabolomics Strategies-Challenges and Emerging Directions. J Am Soc Mass Spectrom. 2016;27:1897-1905. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 780] [Cited by in RCA: 859] [Article Influence: 95.4] [Reference Citation Analysis (0)] |
| 11. | Huang Y, Zhao X, Zhang ZT, Chen SS, Li SS, Shi Z, Jing J, Huang A, Guo YM, Bai ZF, Zou ZS, Xiao XH, Wang JB, Niu M. Metabolomics Profiling and Diagnosis Biomarkers Searching for Drug-Induced Liver Injury Implicated to Polygonum multiflorum: A Cross-Sectional Cohort Study. Front Med (Lausanne). 2020;7:592434. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 12] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 12. | Xie Z, Zhang L, Chen E, Lu J, Xiao L, Liu Q, Zhu D, Zhang F, Xu X, Li L. Targeted Metabolomics Analysis of Bile Acids in Patients with Idiosyncratic Drug-Induced Liver Injury. Metabolites. 2021;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 24] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 13. | Soga T, Sugimoto M, Honma M, Mori M, Igarashi K, Kashikura K, Ikeda S, Hirayama A, Yamamoto T, Yoshida H, Otsuka M, Tsuji S, Yatomi Y, Sakuragawa T, Watanabe H, Nihei K, Saito T, Kawata S, Suzuki H, Tomita M, Suematsu M. Serum metabolomics reveals γ-glutamyl dipeptides as biomarkers for discrimination among different forms of liver disease. J Hepatol. 2011;55:896-905. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 207] [Cited by in RCA: 192] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 14. | van Swelm RP, Kramers C, Masereeuw R, Russel FG. Application of urine proteomics for biomarker discovery in drug-induced liver injury. Crit Rev Toxicol. 2014;44:823-841. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 15] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 15. | Kaczor-Urbanowicz KE, Martin Carreras-Presas C, Aro K, Tu M, Garcia-Godoy F, Wong DT. Saliva diagnostics - Current views and directions. Exp Biol Med (Maywood). 2017;242:459-472. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 192] [Cited by in RCA: 268] [Article Influence: 29.8] [Reference Citation Analysis (0)] |
| 16. | Khurshid Z, Zafar MS, Khan RS, Najeeb S, Slowey PD, Rehman IU. Role of Salivary Biomarkers in Oral Cancer Detection. Adv Clin Chem. 2018;86:23-70. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 147] [Article Influence: 21.0] [Reference Citation Analysis (0)] |
| 17. | Murata T, Yanagisawa T, Kurihara T, Kaneko M, Ota S, Enomoto A, Tomita M, Sugimoto M, Sunamura M, Hayashida T, Kitagawa Y, Jinno H. Salivary metabolomics with alternative decision tree-based machine learning methods for breast cancer discrimination. Breast Cancer Res Treat. 2019;177:591-601. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 58] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 18. | Romano F, Meoni G, Manavella V, Baima G, Tenori L, Cacciatore S, Aimetti M. Analysis of salivary phenotypes of generalized aggressive and chronic periodontitis through nuclear magnetic resonance-based metabolomics. J Periodontol. 2018;89:1452-1460. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 46] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 19. | Bencharit S, Carlson J, Byrd WC, Howard-Williams EL, Seagroves JT, McRitchie S, Buse JB, Sumner S. Salivary Metabolomics of Well and Poorly Controlled Type 1 and Type 2 Diabetes. Int J Dent. 2022;2022:7544864. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 4] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 20. | Cui G, Qing Y, Li M, Sun L, Zhang J, Feng L, Li J, Chen T, Wang J, Wan C. Salivary Metabolomics Reveals that Metabolic Alterations Precede the Onset of Schizophrenia. J Proteome Res. 2021;20:5010-5023. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 23] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 21. | Yilmaz A, Geddes T, Han B, Bahado-Singh RO, Wilson GD, Imam K, Maddens M, Graham SF. Diagnostic Biomarkers of Alzheimer's Disease as Identified in Saliva using 1H NMR-Based Metabolomics. J Alzheimers Dis. 2017;58:355-359. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 93] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 22. | Hershberger CE, Rodarte AI, Siddiqi S, Moro A, Acevedo-Moreno LA, Brown JM, Allende DS, Aucejo F, Rotroff DM. Salivary Metabolites are Promising Non-Invasive Biomarkers of Hepatocellular Carcinoma and Chronic Liver Disease. Liver Cancer Int. 2021;2:33-44. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 21] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 23. | Danan G, Teschke R. RUCAM in Drug and Herb Induced Liver Injury: The Update. Int J Mol Sci. 2015;17. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 480] [Cited by in RCA: 509] [Article Influence: 50.9] [Reference Citation Analysis (0)] |
| 24. | Feng Y, Li Q, Chen J, Yi P, Xu X, Fan Y, Cui B, Yu Y, Li X, Du Y, Chen Q, Zhang L, Jiang J, Zhou X, Zhang P. Salivary protease spectrum biomarkers of oral cancer. Int J Oral Sci. 2019;11:7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51] [Cited by in RCA: 61] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 25. | Want EJ, O'Maille G, Smith CA, Brandon TR, Uritboonthai W, Qin C, Trauger SA, Siuzdak G. Solvent-dependent metabolite distribution, clustering, and protein extraction for serum profiling with mass spectrometry. Anal Chem. 2006;78:743-752. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 329] [Cited by in RCA: 407] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 26. | Barri T, Dragsted LO. UPLC-ESI-QTOF/MS and multivariate data analysis for blood plasma and serum metabolomics: effect of experimental artefacts and anticoagulant. Anal Chim Acta. 2013;768:118-128. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 164] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 27. | Navarro VJ, Senior JR. Drug-related hepatotoxicity. N Engl J Med. 2006;354:731-739. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 690] [Cited by in RCA: 644] [Article Influence: 33.9] [Reference Citation Analysis (0)] |
| 28. | Björnsson HK, Björnsson ES. Drug-induced liver injury: Pathogenesis, epidemiology, clinical features, and practical management. Eur J Intern Med. 2022;97:26-31. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 76] [Article Influence: 25.3] [Reference Citation Analysis (0)] |
| 29. | Hyvärinen E, Savolainen M, Mikkonen JJW, Kullaa AM. Salivary Metabolomics for Diagnosis and Monitoring Diseases: Challenges and Possibilities. Metabolites. 2021;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 48] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 30. | Ayata CK, Ganal SC, Hockenjos B, Willim K, Vieira RP, Grimm M, Robaye B, Boeynaems JM, Di Virgilio F, Pellegatti P, Diefenbach A, Idzko M, Hasselblatt P. Purinergic P2Y₂ receptors promote neutrophil infiltration and hepatocyte death in mice with acute liver injury. Gastroenterology. 2012;143:1620-1629.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 79] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 31. | Oliveira AG, Marques PE, Amaral SS, Quintão JL, Cogliati B, Dagli ML, Rogiers V, Vanhaecke T, Vinken M, Menezes GB. Purinergic signalling during sterile liver injury. Liver Int. 2013;33:353-361. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 32. | Wang P, Jia J, Zhang D. Purinergic signalling in liver diseases: Pathological functions and therapeutic opportunities. JHEP Rep. 2020;2:100165. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 27] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 33. | Hoque R, Sohail MA, Salhanick S, Malik AF, Ghani A, Robson SC, Mehal WZ. P2X7 receptor-mediated purinergic signaling promotes liver injury in acetaminophen hepatotoxicity in mice. Am J Physiol Gastrointest Liver Physiol. 2012;302:G1171-G1179. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 116] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 34. | Amaral SS, Oliveira AG, Marques PE, Quintão JL, Pires DA, Resende RR, Sousa BR, Melgaço JG, Pinto MA, Russo RC, Gomes AK, Andrade LM, Zanin RF, Pereira RV, Bonorino C, Soriani FM, Lima CX, Cara DC, Teixeira MM, Leite MF, Menezes GB. Altered responsiveness to extracellular ATP enhances acetaminophen hepatotoxicity. Cell Commun Signal. 2013;11:10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 44] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 35. | Xie Y, Williams CD, McGill MR, Lebofsky M, Ramachandran A, Jaeschke H. Purinergic receptor antagonist A438079 protects against acetaminophen-induced liver injury by inhibiting p450 isoenzymes, not by inflammasome activation. Toxicol Sci. 2013;131:325-335. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 57] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 36. | Cao J, Mi Y, Shi C, Bian Y, Huang C, Ye Z, Liu L, Miao L. First-line anti-tuberculosis drugs induce hepatotoxicity: A novel mechanism based on a urinary metabolomics platform. Biochem Biophys Res Commun. 2018;497:485-491. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 33] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 37. | Schomaker S, Warner R, Bock J, Johnson K, Potter D, Van Winkle J, Aubrecht J. Assessment of emerging biomarkers of liver injury in human subjects. Toxicol Sci. 2013;132:276-283. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 96] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 38. | Nagao H, Nishizawa H, Tanaka Y, Fukata T, Mizushima T, Furuno M, Bamba T, Tsushima Y, Fujishima Y, Kita S, Funahashi T, Maeda N, Mori M, Fukusaki E, Shimomura I. Hypoxanthine Secretion from Human Adipose Tissue and its Increase in Hypoxia. Obesity (Silver Spring). 2018;26:1168-1178. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 58] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 39. | Farthing DE, Farthing CA, Xi L. Inosine and hypoxanthine as novel biomarkers for cardiac ischemia: from bench to point-of-care. Exp Biol Med (Maywood). 2015;240:821-831. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 75] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 40. | Fisher O, Benson RA, Imray CH. The clinical application of purine nucleosides as biomarkers of tissue Ischemia and hypoxia in humans in vivo. Biomark Med. 2019;13:953-965. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 21] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 41. | Kim YJ, Ryu HM, Choi JY, Cho JH, Kim CD, Park SH, Kim YL. Hypoxanthine causes endothelial dysfunction through oxidative stress-induced apoptosis. Biochem Biophys Res Commun. 2017;482:821-827. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 56] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 42. | Furuhashi M. New insights into purine metabolism in metabolic diseases: role of xanthine oxidoreductase activity. Am J Physiol Endocrinol Metab. 2020;319:E827-E834. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 187] [Article Influence: 37.4] [Reference Citation Analysis (0)] |
| 43. | Nguyen P, Leray V, Diez M, Serisier S, Le Bloc'h J, Siliart B, Dumon H. Liver lipid metabolism. J Anim Physiol Anim Nutr (Berl). 2008;92:272-283. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 471] [Cited by in RCA: 653] [Article Influence: 38.4] [Reference Citation Analysis (1)] |
| 44. | Houten SM, Violante S, Ventura FV, Wanders RJ. The Biochemistry and Physiology of Mitochondrial Fatty Acid β-Oxidation and Its Genetic Disorders. Annu Rev Physiol. 2016;78:23-44. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 301] [Cited by in RCA: 591] [Article Influence: 59.1] [Reference Citation Analysis (0)] |
| 45. | Li L, Wang B, Yu P, Wen X, Gong D, Zeng Z. Medium and Long Chain Fatty Acids Differentially Modulate Apoptosis and Release of Inflammatory Cytokines in Human Liver Cells. J Food Sci. 2016;81:H1546-H1552. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 17] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 46. | Rui L. Energy metabolism in the liver. Compr Physiol. 2014;4:177-197. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 951] [Cited by in RCA: 1427] [Article Influence: 129.7] [Reference Citation Analysis (0)] |
| 47. | Kawaguchi M, Nukaga T, Sekine S, Takemura A, Susukida T, Oeda S, Kodama A, Hirota M, Kouzuki H, Ito K. Mechanism-based integrated assay systems for the prediction of drug-induced liver injury. Toxicol Appl Pharmacol. 2020;394:114958. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |