Published online Sep 15, 1997. doi: 10.3748/wjg.v3.i3.143
Revised: January 24, 1997
Accepted: February 15, 1997
Published online: September 15, 1997
AIM: To elucidate the role of hepatitis G virus (HGV) infection in chronic non-A–E hepatitis and sequence the partial NS5 genome of HGV isolated from the serum of a Chinese patient with chronic non-A–E hepatitis
METHODS: Serum samples of patients with chronic non-A–E hepatitis were collected and total nucleic acids were extracted and subjected to reverse transcriptase-nested-polymerase chain reaction (RT-nested-PCR) using primers from the putative NS5 region of HGV genome. Then, 994bp cDNA was prepared from the positive serum, purified with electrophoresis of polyacrylamide gels, and directly sequenced using the dideoxy-mediated chain-termination method.
RESULTS: HGV-RNA was detected in 1 of the 35 patients with chronic non-A–E hepatitis. Compared with the 2 HGV isolates (PNF2161 and R10291) obtained from American patients, the HGV NS5 gene of this Beijing isolate (HG-G) showed homology of 88.0% and 89.2% respectively. On the other hand, in comparison with the West African isolate (GBV-C), the Beijing isolate showed homology of 93.5%. The patient showed persistent increase of alanine transaminase, but normal levels were achieved after interferon therapy with persistent positive HGV RNA.
CONCLUSION: HGV is one of the causes of chronic non-A–E hepatitis, but it may not be a very important cause. The nucleotide sequence of partial NS5 gene of HG-G was found to be highly homologous to the West Africa isolate.
- Citation: Chang JH, Wei L, Du SC, Wang H, Sun Y, Tao QM. Hepatitis G virus infection in patients with chronic non-A–E hepatitis. World J Gastroenterol 1997; 3(3): 143-146
- URL: https://www.wjgnet.com/1007-9327/full/v3/i3/143.htm
- DOI: https://dx.doi.org/10.3748/wjg.v3.i3.143
Since reliable detection of hepatitis C virus (HCV) and E virus (HEV) infections has now become possible, most of the cases of non-A, non-B hepatitis have been found to be caused by HCV and HEV. However, the etiology of some non-A, non-B hepatitis still remains unknown[1], which highlights the possibility of additional causes of human non-A, non-B, non-C, non-D, non-E (non-A–E) hepatitis. Recently, a new RNA virus genome associated with human hepatitis, termed HGV (or GBV-C), was identified simultaneously by two American laboratories[2,3]; this virus is considered to be one of the causes of human cryptogenic (non-A–E) hepatitis.
Thus far, the role of HGV infection in chronic non-A–E hepatitis and the primary structure of HGV in Chinese patients have been seldom reported. We detected HGV-RNA in the sera of 35 patients with chronic non-A–E hepatitis in Beijing by using the RT-nested PCR method and sequenced the partial NS5 gene from one isolate positive for HGV RNA.
Of 35 patients with chronic non-A–E hepatitis, 20 were male and 15 were female and aged at an average of 41.2 years. Six patients had history of transfusion, and the remaining 29 were sporadic cases. All were patients treated at the Institute of Hepatology, People′s Hospital, BMU. The serum samples were stored at -20 °C. They all tested negative for anti-HAV IgM, HBsAg, anti-HBc, HCV-RNA, anti-HCV, and anti-HEV.
AMV-RT,dNTP, Taq DNA polymerase and DNA sequence kit used in this study are products of Promega.
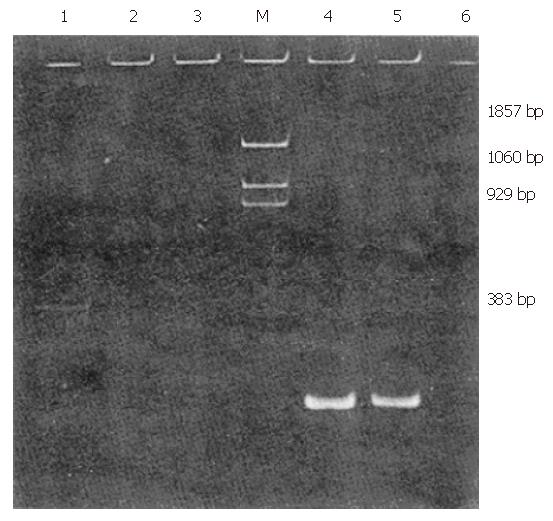
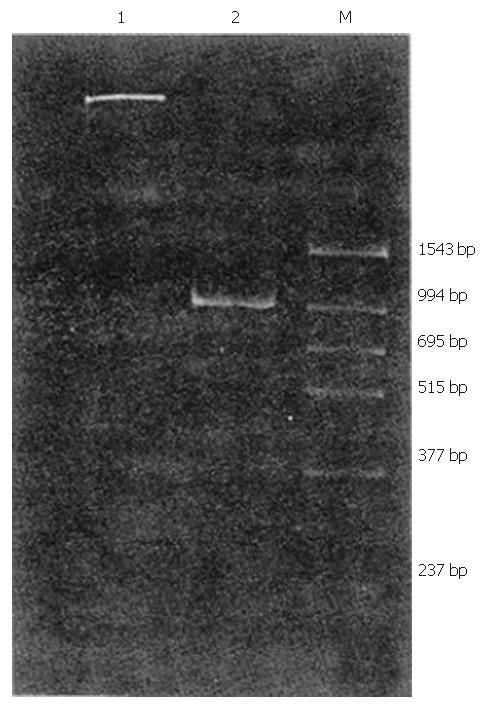
Oligonucleotide primers were designed as described previously[3] and were derived from the putative NS5 region of the HGV genome (Table 1). 51# and 52# (outer primers) as well as 53#, 54#(inner primers) were used for clinical detection, with the first PCR product being 400-bp in length and the second PCR product being 210-bp in length. Further, 52# and 55#(outer primers) as well as 56# and 57#(inner primers) were used for the amplification of a longer fragment as a sequence template (994 bp).
| Primer | GBV-C position | Sequence (5'-3') |
| 51# | 7614~7632 | GTTACACTTATGAGGARGC |
| 52# | 7994~8013 | GCRTCCACACAGATGGCGCA |
| 53# | 7739~7758 | GAGATACTTGAAGGGACTCC |
| 54# | 7930~7948 | CTGGTTGGGGGTGTACTGG |
| 55# | 6885~6909 | CTCTTTGTGGTAGTAGCCGAGAGAT |
| 56# | 7925~7952 | CYCGCTCRTTTGGGGTGTACTGGAAGGC |
| 57# | 6959~6981 | TCGGTTACTGAGTGCAGCTCAGATGAG |
Total nucleic acid was extracted as described before[4]. The extracted H GV RNA was resuspended in diethylpyrocarbonate-treated water and subjected to denaturation at 70 °C for 1 min, followed by a quickly chilling it on ice. Thereafter, the reverse reaction mixture (10 μL reaction-containing AMV-RT buffer, 50 pmol of primer 52#, dNTPs 200 μmol/L each, 2 units of AMV-RT) was added to synthesize cDNA at 43 °C for one hour. The first PCR was performed in a volume of 50 μL, containing 50 pmol primer 51# or 55#, 1.5 units of Taq P, dNTPs 200 μmol/L each. For the second PCR reaction, a volume of 5 μL was removed from the first reaction and added to a tube containing the inner primers 53# and 54# or 56# and 57#, along with dNTPs, Taq polymerase and Taq buffer, as was the case in the first reaction. The first and second PCRs were carried out for 30 cycles, consisting of 94 °C for 60 s, 55 °C for 90 s and 72 °C for 120 s, with a 600-s extension at 72 °C at the end. The second amplified product was analyzed by electrophoresis in 6% polyacrylamide gels. The positive products were purified and directly sequenced using primers 56# and 57#, as per our previously described method[5].
Of 35 patients with chronic non-A–E hepatitis, 1 tested positive for HGV RNA (Figures 1 and 2).
Clinical data of the HGV RNA positive patient with chronic non-A–E hepatitis
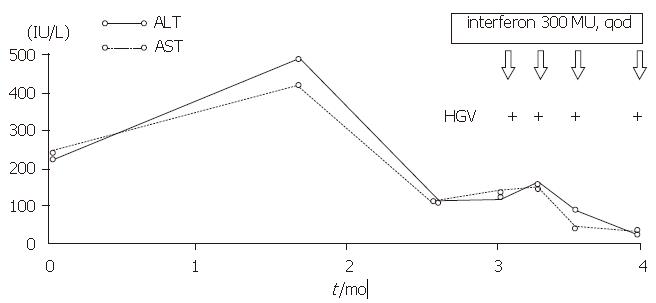
The patient who tested positive for HGV RNA was a 64-year-old woman who had received transfusion of 400 mL blood in 1979. She had not developed any symptoms until February 1996. She was found to have elevated serum ALT level at a physical check-up but tested negative for anti-HAV IgM, HBsAg, anti-HBc, HCV RNA, anti-HCV, and anti-HEV (Figure 3).
The nucleotide sequences of HG-G in HGV NS5 as well as those of West African isolate (GBV-C) and American isolates (PNF 2161, R10291) for comparison are shown in Figure 4. The amino acid sequences of the 4 isolates are shown in Figure 5. At a nucleotide level, HG-G homology was 87.95% and 89.23% when compared with two American isolates, and all isolates showed homology of 96.49% at the amino acid level. When compared with the West African isolate, HG-G showed homology of 93.50% and 97.77% at the nucleotide level and amino acid level, respectively. In the region sequenced, 16 proline, 8 cysteine and 3 glycosylated residues were detected.
As reported before, about 20% patients with chronic hepatitis tested negative for serum markers of HBV and HCV, thereby indicating the existence of an unknown etiology in human hepatitis[1]. In 1995, HGV was found to be responsible for 9.49% of the cases of non-B non-C hepatitis and 8.33% of the cases of chronic non-A–E hepatitis[3], which indicates that HGV is one of the causes of non-A–E hepatitis. In our research, 1 (2.9%) of 35 patients with chronic non-A–E hepatitis tested positively for HGV RNA. This relatively low detection rate may be related to the insufficient exclusion of other non-virus elements and racial/ethnic and geographic differences. Consistent with other reports[3], many patients with chronic non-A–E hepatitis in our series tested negative for serum HGV RNA; this may be attributed to the lack of sufficient knowledge about the newly discovered virus in its affective factors of molecular biologic detection. In addition, it must be recognized that there may be other causes of human hepatitis besides the known factors, that is to say, HGV may not be a very important cause of chronic non-A–E hepatitis.
The patient, who tested positive for HGV RNA in our research had a history of receiving blood transfusion 17 years before, and she did not show any symptoms and an elevated serum ALT level until February 1996. She did not respond very well to that treatment, but her serum ALT level was normalized after interferon treatment although HGV RNA continued to persist in her serum. Since we cannot find evidence of other hepatitis virus infection, we believe that HGV must be responsible for the chronic hepatitis in this case, although the clinical features are similar to those of hepatitis C.
The primary structure in HGV NS5 region in the Beijing isolate (HG-G) was also analyzed. In the region sequenced, 16 conserved proline residues and 8 conserved cysteine residues were detected; these residues may play an important role in the formation of the spatial configuration, indicating that the product encoded by this region may be important in maintaining HGV function. With respect to the nucleic acid sequence and amino acid sequence, HG-G showed marked homology to the West African isolate. Since only 3 isolates of HGV sequences have been reported thus far, it is difficult to decide whether HG-G was of the same genotype as the West African isolate. Future studies on sequence analyses are needed for further research.
Original title:
S- Editor: Filipodia L- Editor: Jennifer E- Editor: Hu S
| 1. | Tao QM, Wang Y, Du SC, Guo JP. Epidemiology of hepatitis B and C in China. Tokyo: Springs 1994; 412-415. [DOI] [Full Text] |
| 2. | Leary TP, Muerhoff AS, Simons JN, Pilot-Matias TJ, Erker JC, Chalmers ML, Schlauder GG, Dawson GJ, Desai SM, Mushahwar IK. Sequence and genomic organization of GBV-C: a novel member of the flaviviridae associated with human non-A-E hepatitis. J Med Virol. 1996;48:60-67. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 11] [Reference Citation Analysis (0)] |
| 3. | Linnen J, Wages J, Zhang-Keck ZY, Fry KE, Krawczynski KZ, Alter H, Koonin E, Gallagher M, Alter M, Hadziyannis S. Molecular cloning and disease association of hepatitis G virus: a transfusion-transmissible agent. Science. 1996;271:505-508. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 957] [Cited by in RCA: 893] [Article Influence: 30.8] [Reference Citation Analysis (0)] |
| 4. | Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156-159. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40518] [Cited by in RCA: 39094] [Article Influence: 1028.8] [Reference Citation Analysis (0)] |
| 5. | Wei L, Wang Y, Chen HS, Tao QM. Sequencing of hepatitis C virus cDNA with polymerase chain reaction directed sequencing. Chinese Journal of Hepatology. 1996;4:103-105. |