Published online Dec 14, 2023. doi: 10.3748/wjg.v29.i46.6060
Peer-review started: September 29, 2023
First decision: October 16, 2023
Revised: October 24, 2023
Accepted: November 17, 2023
Article in press: November 17, 2023
Published online: December 14, 2023
Processing time: 74 Days and 18.5 Hours
Mesenchymal stem cells (MSCs) exert anti-oncogenic effects via exosomes containing non-coding RNA (ncRNA), which play important roles in tumor biology. Our preliminary study identified the interaction of the ncRNA hsa_
To identify the clinical significance, functional implications, and mechanisms of circ563 in HCC.
The expression levels of miR-148a-3p and MTF-1 in exosomes derived from MSC and HCC cells were compared, and their effects on HCC cells were assessed. Using a dual-luciferase reporter assay, miR-148a-3p was identified as an asso
The silencing of circ563 blocked the HCC cell proliferation and invasion and induced apoptosis. Co-culturing of HCC cells with MSC-derived exosomes following circ563 overexpression promoted cell proliferation and metastasis and elicited changes in miR-148a-3p and MTF-1 expression. The tumor-promoting effects of circ563 were partially suppressed by miR-148a-3p overexpression or MTF-1 depletion. Xenograft experiments performed in nude mice confirmed that circ563-enriched exosomes facilitated tumor growth by upregulating the ex
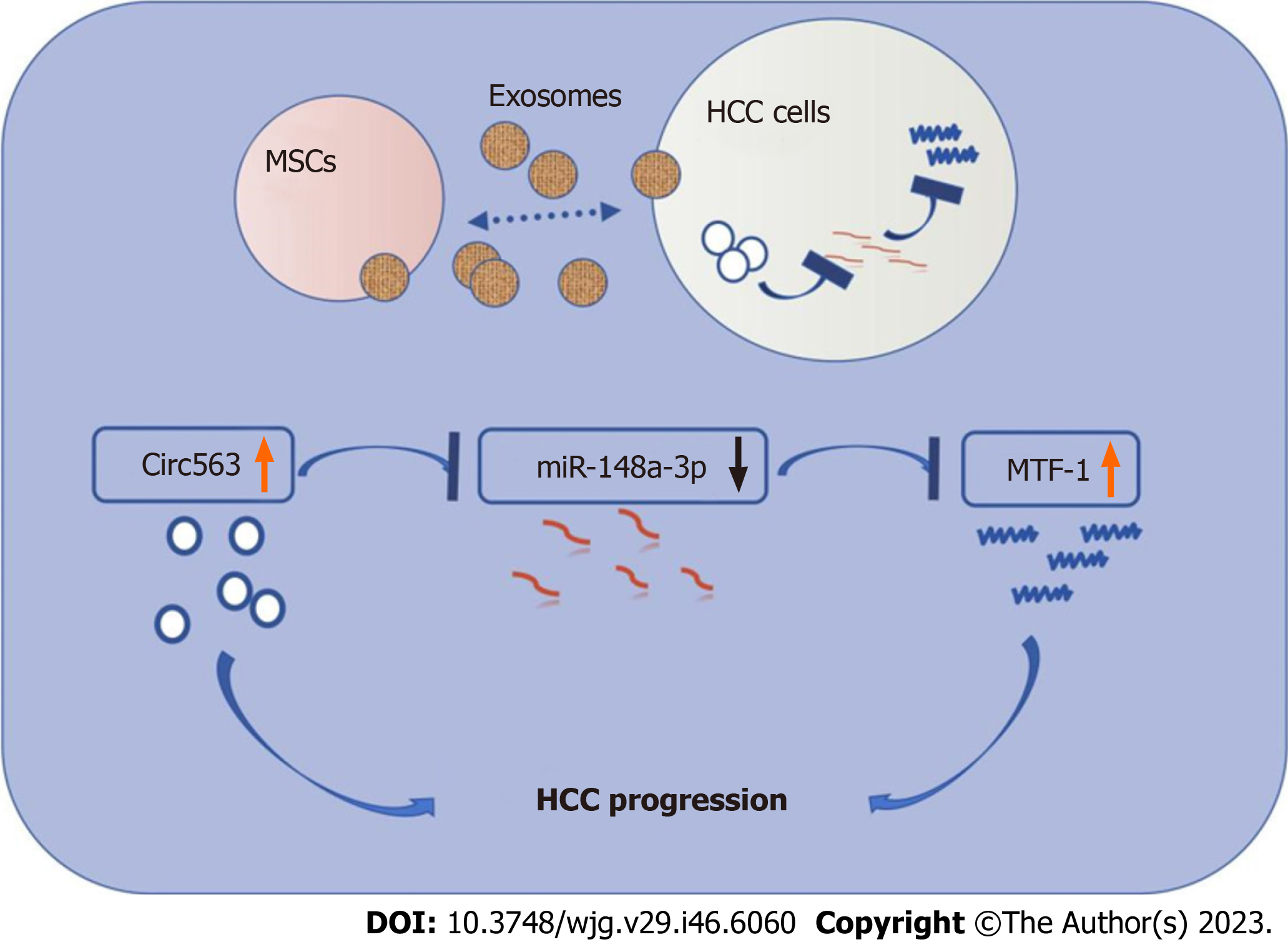
MSCs may exhibit anti-HCC activity through the exosomal circ563/miR-148a-3p/MTF-1 pathway, while exosomes can transmit circ563 to promote oncogenic behavior by competitively binding to miR-148a-3p to activate MTF-1.
Core Tip: We identified the functional implications and mechanisms of exosomal hsa_circ_0000563 (circ563) in hepatocellular carcinoma (HCC). Mesenchymal stem cells suppressed HCC proliferation and invasion via their exosomes. The circ563 interacts with miR-148a-3p, a molecule involved in HCC progression, and both are identified at different levels in exosomes. The tumor-promoting effects of circ563 were partially suppressed by miR-148a-3p overexpression or depletion of metal-regulatory transcription factor-1 (MTF-1). Xenograft experiments performed in nude mice confirmed that circ563-enriched exosomes facilitated tumor growth via MTF-1 upregulation. Our findings provide new insights into circ563/miR-148a-3p/MTF-1 signaling as a potential therapeutic target for HCC.
- Citation: Lyu ZZ, Li M, Yang MY, Han MH, Yang Z. Exosome-mediated transfer of circRNA563 promoting hepatocellular carcinoma by targeting the microRNA148a-3p/metal-regulatory transcription factor-1 pathway. World J Gastroenterol 2023; 29(46): 6060-6075
- URL: https://www.wjgnet.com/1007-9327/full/v29/i46/6060.htm
- DOI: https://dx.doi.org/10.3748/wjg.v29.i46.6060
Hepatocellular carcinoma (HCC) is a fatal disease and the most common form of primary liver cancer. Although novel chemotherapeutic agents and treatment modalities have been clinically implemented, the survival rate of patients with HCC has not yet improved[1]. Therefore, a deeper understanding of the mechanisms facilitating HCC progression is paramount for the development of more effective therapeutic strategies.
The tumor microenvironment (TME) has been an important aspect of tumor research. Recent studies indicate that mesenchymal stem cells (MSCs) are essential stromal cells in the TME and exert their effects via exosomes, as demonstrated by multiple studies[2,3]. Furthermore, the efficacy of therapeutic MSC transplantation has been reported in both experimental and clinical research[4-6]. As a result, MSC-derived exosomes have attracted significant attention due to their implications in intercellular communication.
Exosomes are small extracellular vesicles approximately 30-150 nm in diameter that are formed by cells through a series of regulatory processes. These vesicles are enriched in bioactive molecules like non-coding RNA (ncRNA) with crucial biological functions, whose delivery to recipient cells is involved in various physiological and pathological processes, including carcinogenesis and metastasis[7,8]. Circular RNA (circRNA) is a new class of endogenous ncRNA, which has selectively conserved microRNA (miRNA) target sites and exerts regulatory effects in cancer biology by regulating miRNA gene expression[9,10]. Such interactions between circRNA and miRNA are known as the competitive endogenous RNA theory[11]. miRNAs represent the ncRNA of 20-25 nucleotides, which mediate post-transcriptional gene silencing by binding to the 3’-untranslated regions of target mRNAs. Exosomal circRNA and miRNA are involved in tumor initiation and progression[7,8,12,13]; however, their expression profiles, functions, and dysregulation in HCC require further clarification.
This study found that MSCs suppressed HCC proliferation and invasion via their exosomes. The ncRNA circ563 interacts with miR-148a-3p, and both are identified at different levels in exosomes. Metal-regulatory transcription factor-1 (MTF-1) as the direct target of miR-148a-3p is implicated in HCC progression. Therefore, further studies should be performed to determine the possible role and significance of circ563/miR-148a-3p/MTF-1 axis in the development of HCC.
The human HCC cell lines Hep3B (RRID: CVCL_0326, iCell Bioscience, Shanghai, China) and SNU387 (RRID: CVCL_0250, Procell, Wuhan, China), MSC (Procell, Wuhan, China), and the human embryonic kidney cell line HEK 293T (RRID: CVCL_0045, Cell Research, Shanghai, China) were cultured according to the manufacturer’s instructions. The cell lines used in this study were authenticated, and no cellular cross-contamination or mycoplasma infection was detected.
The exosomes were isolated as previously described[14], and their morphology was observed under a transmission electron microscope. The size distribution of exosomes was analyzed using the NanoSight LM10 system (NanoSight, Amesbury, United Kingdom) equipped with fast video capture and particle-tracking software.
MSCs were modified to express fused CD63-eGFP via lentiviral transduction following the manufacturer’s instructions. The HCC cells were seeded on glass coverslips coated with poly-L-lysine (Sigma, Shanghai, China). MSC-derived exosomes were co-cultured with HCC cells for 24 h at 37 °C before washing and fixation at room temperature. The uptake of labeled exosomes by recipient cells was observed under a fluorescence microscope (Leica TCS SP8 X, Leica, Shanghai, China).
Exosomes were harvested for protein extraction. Equal amounts of protein were subjected to sodium dodecyl-sulfate polyacrylamide gel electrophoresis and then electroblotted onto a polyvinylidene difluoride membrane (Merck, Nantong, Jiangsu, China) followed by western immunoblotting. Membranes were incubated with primary antibodies overnight at 4 °C, followed by incubation with horseradish peroxidase-conjugated secondary antibody (ZSGB-BIO, Beijing, China). The bands were visualized using a Tanon 5200 Chemiluminescent imaging system (Molecular Devices, Beijing, China) and analyzed using ImageJ software (version 1.52, National Institutes of Health, Bethesda, United States). The following primary antibodies were used: Anti-TSG101, anti-CD9, and anti-CD81 (Abcam, Shanghai, China).
Total RNA was extracted using the Ultrapure RNA Kit (CoWin Biotech, Beijing, China) according to the manufacturer’s protocol. Equal amounts of the samples were amplified, and real-time polymerase chain reaction (RT-PCR) was performed as previously described[14]. The following primer pairs were used: MiR-148a-3p, forward: 5’-CTCAGTGCACTACAGAACTTTGT-3’, reverse: 5’-ATCCAGTGCAGGGTCCGAGG-3’; MTF-1, forward: 5’-GAAAAGCCATTTCGGTGCGA-3’, reverse: 5’-CAGCCATTACTGGGGCAGAA-3’ (DingGuo Biotechnology, Beijing, China); hsa_
Overexpression plasmids of miR-148a-3p, knockdown plasmids of MTF-1, and control empty vectors were obtained from GenePharma (Shanghai, China). Small interfering RNA targeting circ562 (sense: 5’-GGCUCAGCAAAAUGGAUGAAAdTdT-3’; antisense: 5’-UUUCAUCCAUUUUGCUGAGCCdTdT-3’), circ563 (sense: 5’-GCUU
Cell viability was determined using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. The cells were seeded into 96-well plates (approximately 1500 cells/well) before treatment. At the end of each treatment, the MTT reagent was added to the medium. The cells were incubated for an additional 4 h, and the absorbance of the samples was measured at 490 nm using a plate reader (Thermo Fisher Scientific, MA, United States). Experiments were performed in triplicate, and data were expressed as the mean optical density ± SD.
The cells (approximately 1000 cells/well) were seeded into six-well plates with different treatments, either simultaneously or subsequently. After 14 d, the cells were fixed with 4% paraformaldehyde and stained with 0.5% crystal violet, and the individual clones (> 50 cells/clone) were counted. Each treatment was performed in triplicate. Data were expressed as the mean ± SD.
Transwell assays were performed to assess cell invasion and migration abilities in 24-well plates with BD chambers (8-mm pores; BD Biosciences, Shanghai, China). Approximately 1 × 105 cells/well were seeded into the upper chambers and cultured in a serum-free medium. The medium containing 30% serum was placed in the lower chambers. After migration through the Transwell membrane, the cells were fixed with 4% paraformaldehyde and stained with crystal violet (Solarbio, Beijing, China). Compared with the invasion assay, the Transwell chambers for the migration assay were not coated with Matrigel. All Transwell treatments were conducted in triplicate.
Following different treatments, cell apoptosis was detected using the phycoerythrin (PE) Annexin V Apoptosis Detection Kit I (BD Pharmingen, China) according to the manufacturer’s instructions. The cells were harvested after trypsin digestion, centrifuged, and washed with annexin V binding buffer three times. The cells were then stained with PE annexin V and 7-amino-actinomycin D at room temperature for 15 min and analyzed using flow cytometry (BD FACSAria II, Shanghai, China), and the results were analyzed using FlowJo software (version 10; Ashland, OR, United States).
A wild-type (wt) or mutant circ563 fragment was constructed and cloned into plasmids containing luciferase. The recombinant circ563-wt/mt plasmids were co-transfected into HEK 293T cells with either a negative control or a miR-148a-3p mimic using Lipofectamine 2000 (Invitrogen Inc., CA, United States). After 48 h, the luciferase activity was measured using the Dual-Luciferase Reporter 1000 Assay System (Promega, Madison, WI, United States). The firefly luciferase enzyme activity was normalized to the Renilla luciferase activity, and the ratio of firefly to Renilla luciferase activity was evaluated. The experiments were independently performed in triplicate.
Human HCC tissues and paired tumor-adjacent noncancerous liver tissues were obtained from 14 patients with HCC who underwent tumor resection at the Shandong Provincial Hospital, China. None of the patients had undergone radio- or chemotherapy before surgery. Informed consent was obtained from each participant and approved by the Institutional Review Board. The study was approved by the Institutional Medical Ethics Committee of Shandong Provincial Hospital. Patients aged ≥ 18 years and diagnosed with HCC without any local or systemic treatment were included in the study. By contrast, patients with other types of cancers, liver failure, and taking medications that affect liver function were excluded.
The animal study was approved by the Animal Ethics Committee of Shandong Provincial Hospital. The animal protocol was designed to minimize pain or discomfort during the experiments. The animals were acclimatized to laboratory conditions (23 °C, 12 h/12 h light/dark, 50% humidity, ad libitum access to food and water) for 2 wk before experimentation. Five-week-old BALB/c nude mice (Provincial Laboratory Animal Center, Jinan, China) were used for this study. Following different treatments, the Hep3B cells were prepared at 3 × 106 cells/100 μL. Initially, the HCC-bearing nude mice were prepared to develop subcutaneous tumors. After four weeks, circ563-enriched exosomes (approximately 106) were injected intratumorally into the tumor mass twice a week for two weeks. The tumor size was measured weekly, and the tumor volume was calculated using the following formula: Volume (mm3) = tumor length × width2/2. After six weeks, all mice were sacrificed to compare the tumor volumes and weights. The tumors were fixed in 10% formalin and embedded in paraffin for subsequent immunohistochemical examination.
The sections were stained with an anti-MTF-1 sary antibody (Zhongshan Biotechnology Co., Beijing, China). The slides were imaged and analyzed using Aipathwell software (Servicebio, Wuhan, China). The staining intensity was characterized as not present (0), weak but detectable above control (1), moderate (2), and very strong (3). The H-scores were calculated as follows: H-score = ∑(pi × i) = (percentage of weak intensity cells × 1) + (percentage of moderate intensity cells × 2) + (percentage of strong intensity cells × 3). Data were presented as the mean ± SD.
Statistical analyses were performed using SPSS 21.0 software. Data were presented in bar plots as the mean ± SD of at least three independent experiments. A P value of < 0.05 (two-tailed) was considered significant. Student’s t-test was performed to determine whether the two groups were significantly different. Pearson’s correlation analysis was used to assess the correlations between circ563, miR-148a-3p, and MTF-1.
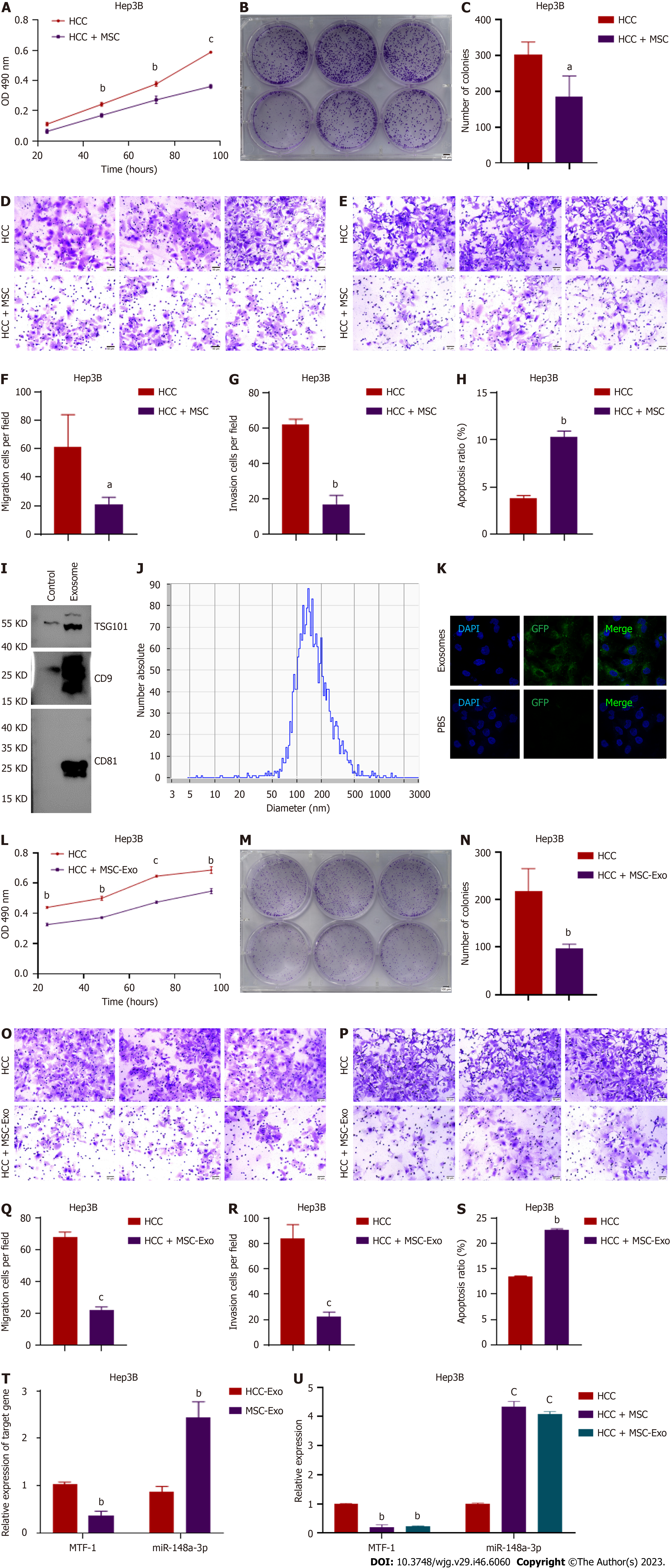
HCC cell proliferation was inhibited by co-culturing with MSCs, which was consistent with the reduced cell viability, migration, and invasion observed under these conditions (Figures 1A-G and Supplementary Figures 1A-G). However, as shown in Figure 1H and Supplementary Figure 1H, apoptosis was induced in HCC cells. To further verify the regulatory role of MSC-derived exosomes in HCC, the exosomes were isolated and characterized by specific marker expression (Figure 1I) and particle size analysis (Figure 1J). In addition, we observed and verified the uptake of labeled exosomes in HCC cells (Figure 1K and Supplementary Figure 1I). MSC-derived exosomes also exerted substantial anti-oncogenic effects in HCC cells by blocking cell proliferation, migration, and invasion while inducing apoptosis (Figures 1L-S and Supplementary Figures 1J-Q). Therefore, MSCs inhibit HCC proliferation and metastasis via their exosomes.
A previous study from our laboratory demonstrated that exosomal miR-148a-3p functions as a tumor suppressor in HCC progression, with MTF-1 being its direct target[14]. In the present study, MSC-derived exosomes displayed increased miR-148a-3p and decreased MTF-1 expression levels compared with HCC-derived exosomes (Figure 1T and Supplementary Figure 1R). Co-culturing HCC cells with either MSCs or MSC-derived exosomes led to the corresponding changes in miR-148a-3p and MTF-1 expression levels (Figure 1U and Supplementary Figure 1S). This finding indicates that exosomes contain bioactive materials involved in the regulation of miR-148a-3p and MTF-1 expression in HCC cells.
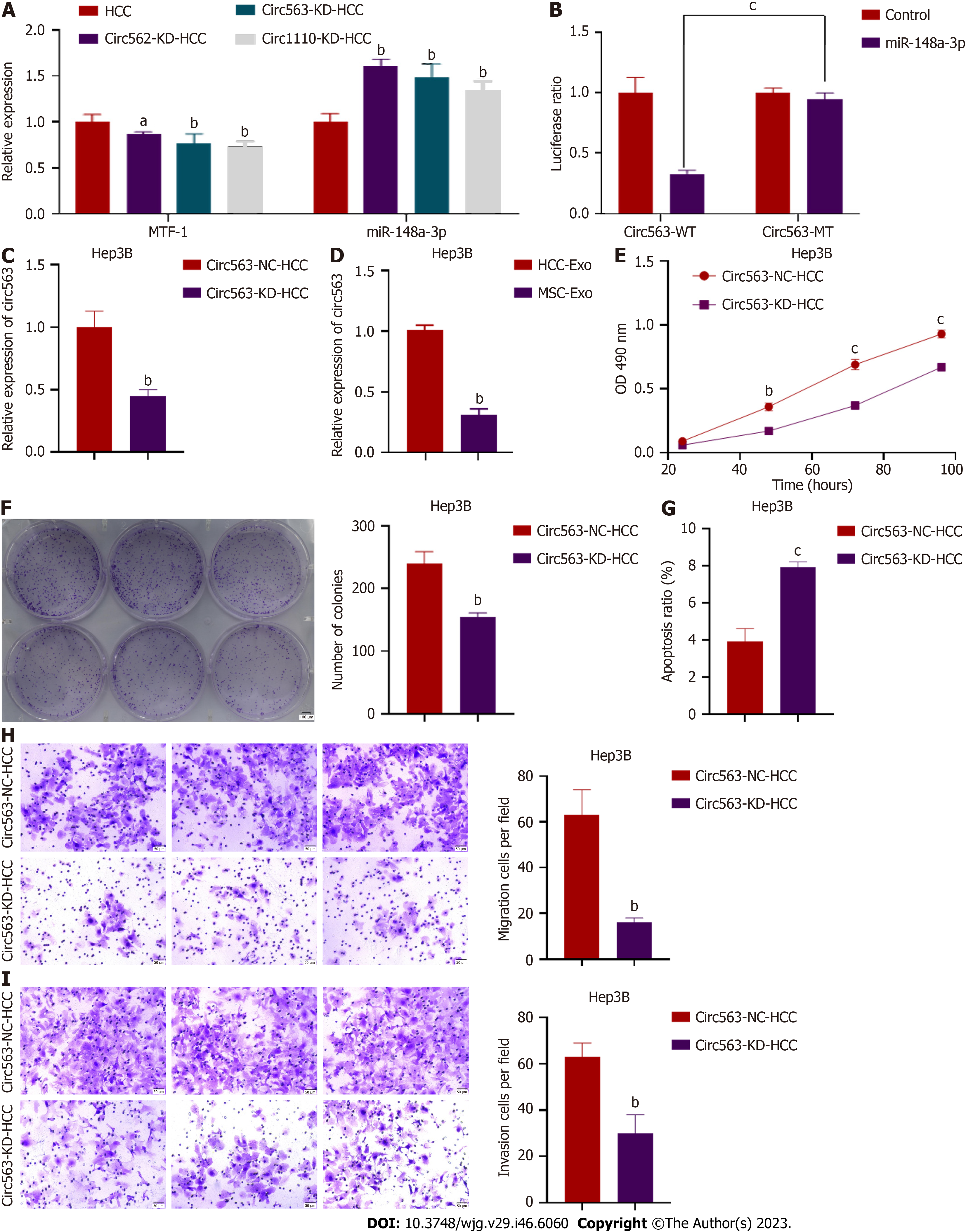
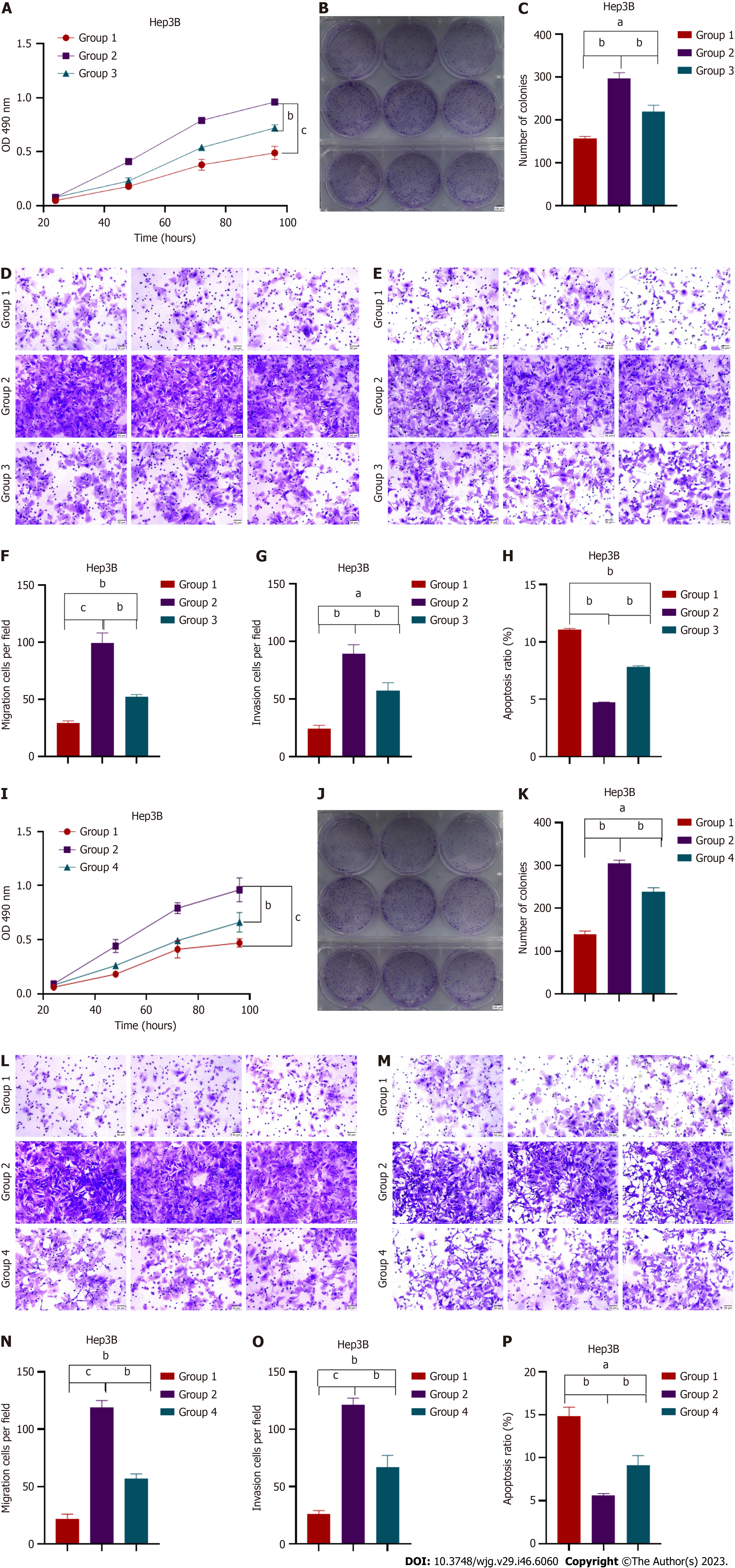
A search of the CircInteractome databases (https://circinteractome.nia.nih.gov/) yielded three potential circRNAs [circ563, hsa_circRNA_0000562 (circ562), and hsa_circRNA_00001110 (circ1110)] that are most likely to bind to miR-148a-3p based on potential complementary sequences. Further analysis was performed using dual-luciferase reporter assays and RT-PCR. The RT-PCR analyses showed that circRNA was positively correlated with MTF-1 but negatively correlated with miR-148a-3p (Figure 2A). The overexpression of miR-148a-3p considerably inhibited the luciferase activity of circ563-wt. However, the luciferase activity of circ563-mutant-type (mt) cells transfected with miR-148a-3p mimics remained unaffected, thus indicating that miR-148a-3p is a target miRNA of circ563 (Figure 2B). Subsequently, knockdown experiments were performed to investigate whether circ563 affects the biological processes of HCC cells. si-circ563 was transfected into Hep3B and SNU387 cells, both strongly expressing circ563, and the knockdown efficiency was determined by RT-PCR (Figure 2C and Supplementary Figure 2A). Exosomal circ563 (exo-circ563) expression levels were also compared with those of MSCs (Figure 2D and Supplementary Figure 2B). Silencing of circ563 expression blocked the proliferation and invasion of HCC cells and increased their apoptosis rate (Figures 2E-I and Supple
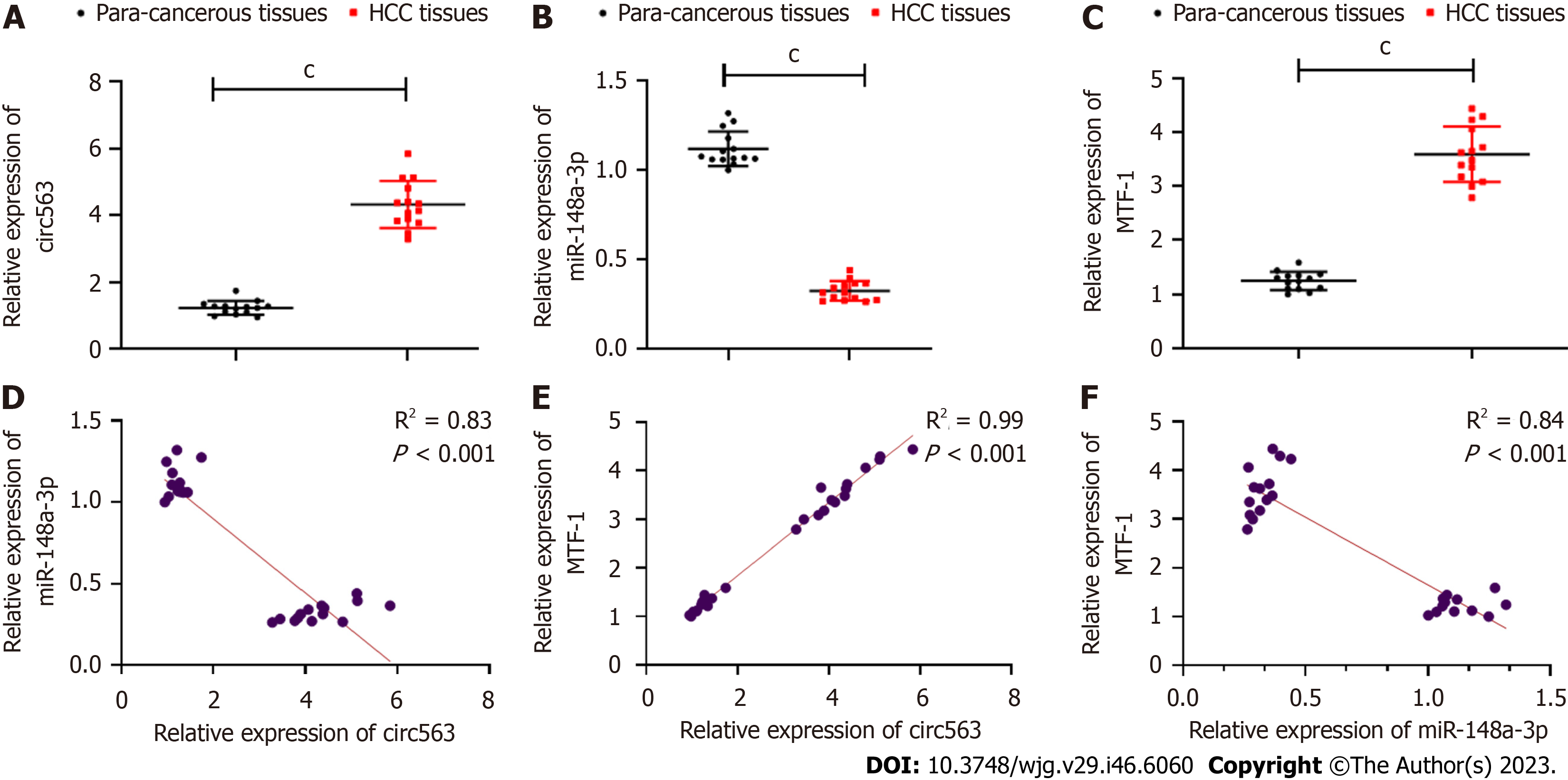
Next, circ563 expression was analyzed in human HCC tissues, which showed higher circ563 levels with downregulated miR-148a-3p and upregulated MTF-1 expression levels compared with tumor-adjacent tissues (Figures 3A-C). Correlation analyses further revealed the relationships between circ563, miR-148a-3p, and MTF-1 levels in HCC (Figures 3D-F). These findings suggest that circ563 may be involved in potentiating HCC cell growth and metastasis, considering the observed correlation with miR-148a-3p and MTF-1.
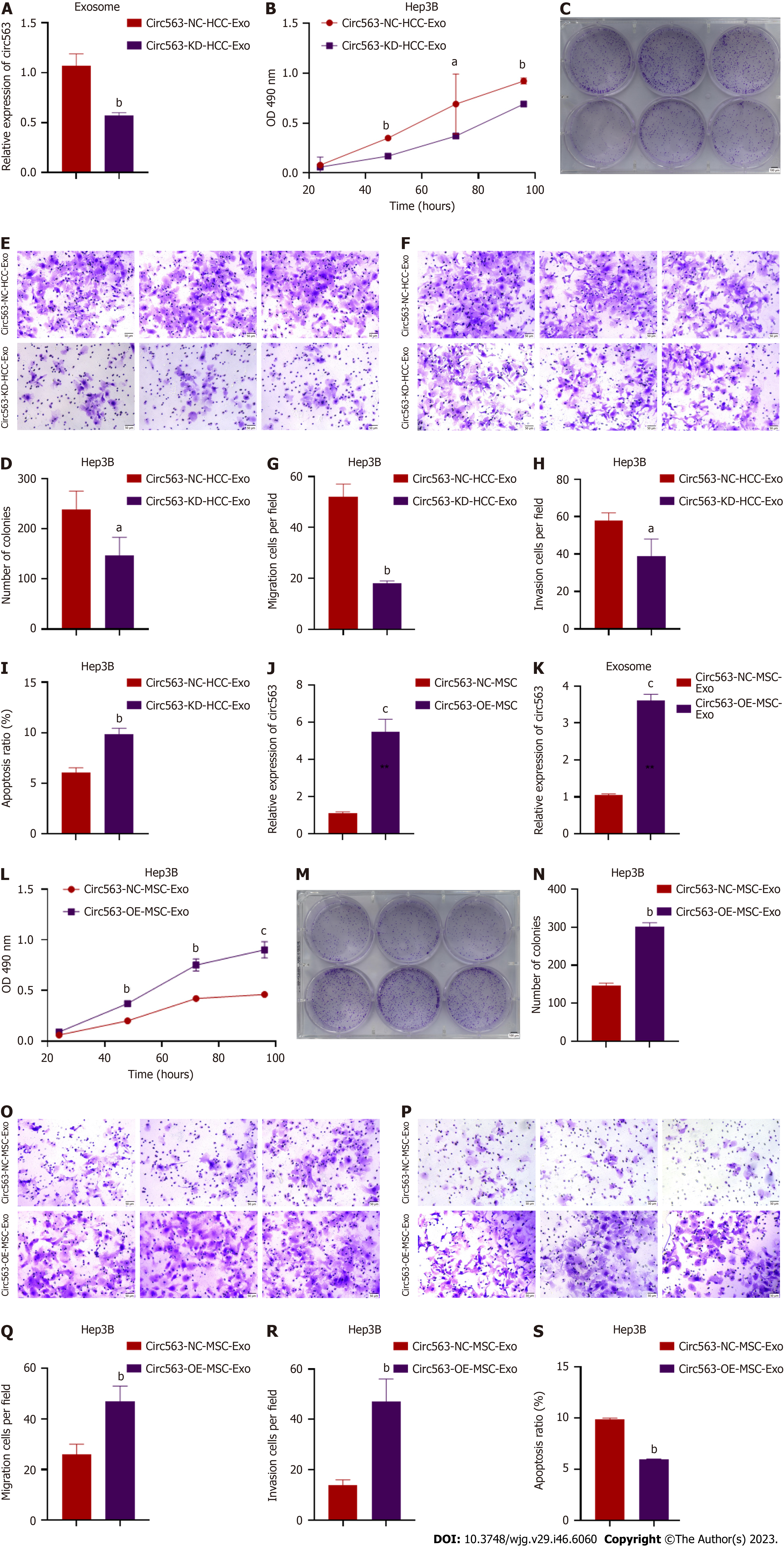
To further explore whether exo-circ563 exerts an oncogenic role, we suppressed the expression of circ563 in HCC cells and detected a reduction in exo-circ563 levels using RT-PCR (Figure 4A). The HCC cells co-treated with exosomes containing downregulated circ563 levels led to a marked reduction in cell proliferation and invasion and enhanced apoptosis (Figures 4B-I and Supplementary Figures 3A-H).
Consistent with previous results, exo-circ563 overexpression facilitated HCC cell progression. The exo-circ563 overexpression efficiency and upregulation were ascertained through RT-PCR (Figures 4J and K). We co-cultured HCC cells with MSC-derived exosomes following the circ563 overexpression and observed an increase in HCC cell proliferation and metastasis and a suppression of apoptosis (Figures 4L-S and Supplementary Figures 3I-P). These effects were partially prevented by the overexpression of miR-148a-3p or depletion of MTF-1 (Figure 5 and Supplementary Figure 4). These data indicate the involvement of a circ563-dependent mechanism in regulating miR-148a-3p expression as described by the competitive endogenous RNA theory.
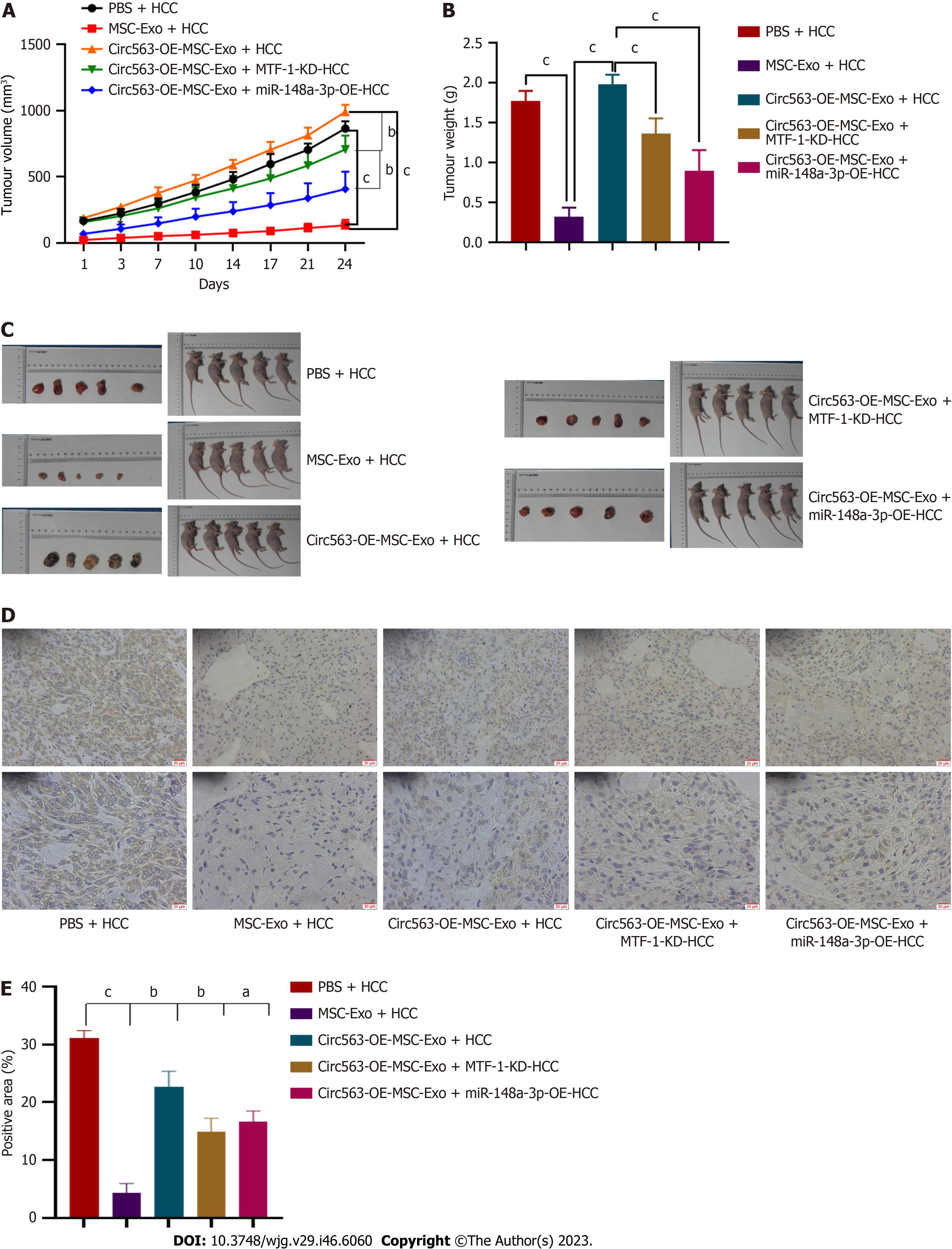
To further confirm the significance of exo-circ563 in HCC tumorigenesis and progression, in vivo experiments were performed. The upregulation of circ563 dramatically accelerated tumor growth in nude mice as evidenced by the detection of larger tumors. This effect could be reversed by miR-148a-3p overexpression or MTF-1 depletion (Figures 6A-C). Therefore, the effects of circ563, by facilitating HCC progression via miR-148a-3p and MTF-1 regulation, were also present in vivo. Correspondingly, the expression of MTF-1 in murine tumor tissues had changed similarly to tumor growth (Figures 6D and E). All these results show that circ563 accelerates HCC progression by sponging miR-148a-3p to elicit MTF-1-dependent oncogenic effects via the circ563/miR-148a-3p/MTF-1 axis (Figure 7).
Liver cancer remains a global health challenge, and its incidence is expected to reach over 1 million cases by 2025[15]. HCC is the most common primary liver cancer. It is characterized by aggressive progression, broad metastasis, frequent recurrence, and high mortality. Therefore, a better understanding of the processes involved in HCC tumorigenesis and metastasis is urgently needed to discover new treatment strategies and improve patient prognosis.
Recent studies have established a comprehensive role of TME in disease development, making it one of the most promising areas of oncological research[16,17]. Moreover, HCC cells can alter their surrounding microenvironment to promote their growth and metastasis[18,19]. The crosstalk between tumor cells and their microenvironment is important for MSC recruitment to HCC tissues, which has also been observed in HCC animal models[19,20]. Although MSCs were initially considered as oncological delivery vehicles due to their migratory and homing capacity toward tumors[21], increasing evidence suggests that they can modulate tumorigenesis via the exosomes[2,3]. In the present study, co-incubation with MSCs inhibited the proliferation and invasion of HCC cells but enhanced their apoptotic capabilities. Further investigation on the mediating role of MSC-derived exosomes in the regulation of HCC cells also led to the discovery of their anti-oncogenic potential by blocking the development of HCC cells. Based on our findings, an exosome-based adjuvant intervention could offer a novel therapeutic approach to HCC by delivering the protected “cargo” directly to the tumor site.
In addition, the exosomes from MSCs have shown tumor-homing capabilities in xenograft mouse models[22]. The uptake of labeled exosomes from MSCs into HCC cells was also reported in the present study. Interestingly, the exosome function most likely depends on the biological materials they encapsulate[23,24]. Mounting evidence suggests that exosomal ncRNAs (miRNA, lncRNA, and circRNA) are actively involved in the initiation and progression of various diseases by regulating cellular proliferation, differentiation, angiogenesis, and immune response[25-27].
circRNAs, a subset of ncRNA that is enriched and more stable in exosomes[28], were differentially expressed in tumor tissues than in normal tissues, suggesting their involvement in tumorigenesis and disease development[29]. In vitro and in vivo experiments revealed that circRNA commonly sponges miRNA, a major class of small ncRNA closely associated with HCC[30,31], and mediates post-transcriptional gene silencing by binding to the 3’-untranslated region or open reading frames of target mRNA. Previously, our data revealed that exosomal miR-148a-3p functions as a tumor suppressor in HCC with MTF-1 as its direct target. MTF-1 maintains metal homeostasis to protect cells against injury by excess metals and exerts oncogenic potential by manipulating metal or redox homeostasis, enhancing angiogenesis, and inducing tumor development in HCC. The current findings confirmed an increase in miR-148a-3p and a decrease in MTF-1 expression levels in MSC-derived exosomes compared with HCC-derived exosomes. Following HCC cell co-culture with MSC or MSC-derived exosomes, consistent changes were observed in the miR-148a-3p and MTF-1 expression levels. This finding indicates that exosomes may contain specific bioactive materials involved in regulating the expression of miR-148a-3p and MTF-1.
Considering this, we searched the CircInteractome databases and identified hsa_circ_0000563 (circ563) as the specific target that competitively binds to or sponges miR-148a-3p. Circ563 has been implicated in the changes in the patho
Furthermore, rescue experiments demonstrated that the potentiating effects of exo-circ563 could be partially blocked by miR-148a-3p upregulation or MTF-1 knockdown. The oncogenic role of circ563 was verified in vivo, as exosomes enriched with circ563 facilitated HCC cell growth in nude mice. Additionally, we assessed the expression patterns of circ563, miR-148a-3p, and MTF-1 in HCC tissues and compared them with those in tumor-adjacent tissues; the trends were consistent with those in HCC cells and exosomes. Collectively, the relative stability, high abundance, and conserved nature of exosomal circRNA across species make them promising candidates as oncogenic biomarkers; a better understanding of the exo-circ563-mediated intercellular network could facilitate the development of therapeutic strategies for patients with HCC.
We clarified the regulatory mechanisms of circ563, which can sponge miR-148a-3p to elicit MTF-1-dependent oncogenic effects, eventually acting as a tumor-potentiating factor in HCC. Thus, the ncRNA circ563 could be a promising the
Mesenchymal stem cells (MSCs) exert anti-oncogenic effects via exosomes containing non-coding RNA (ncRNA), and the efficacy of MSC-derived exosome therapies has been demonstrated. Our preliminary study identified the interaction of the ncRNA hsa_circ_0000563 (circ563) and the circ563-associated miR-148a-3p which are both enclosed in exosomes, as miR-148a-3p and its target metal-regulatory transcription factor-1 (MTF-1) are implicated in hepatocellular carcinoma (HCC) progression.
This study is to identify the clinical significance, functional implications, and mechanisms of circ563 in HCC.
This study aims to investigate the role of circ563 in modulating HCC functions.
The expression levels of miR-148a-3p and MTF-1 in exosomes derived from MSC and HCC cells were compared, and their effects on HCC cells were assessed. Using a dual-luciferase reporter assay, miR-148a-3p was identified as an associated miRNA of circ563, whose role in HCC regulation was assessed in vitro and in vivo.
Circ563 silencing blocked the HCC cell proliferation and invasion and induced apoptosis. Co-culturing of HCC cells with MSC-derived exosomes following circ563 overexpression contributed to cell proliferation and metastasis and elicited changes in miR-148a-3p and MTF-1 expression. The tumor-promoting effects of circ563 were partially suppressed by miR-148a-3p overexpression or MTF-1 depletion. Xenograft experiments confirmed that circ563-enriched exosomes facilitated tumor growth by upregulating the expression of MTF-1. In HCC tissues, circ563 expression was negatively correlated with miR-148a-3p expression but positively correlated with MTF-1 levels.
Our findings suggested MSCs may exhibit anti-HCC activity through the exosomal circ563/miR-148a-3p/MTF-1 pathway.
The study presents a new dataset related to HCC. We clarified the regulatory mechanisms of circ563, which can sponge miR-148a-3p to elicit MTF-1-dependent oncogenic effects in HCC. Thus, the ncRNA circ563 could be a potential the
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Jeyaraman M, India; Shalaby MN, Egypt S-Editor: Wang JJ L-Editor: A P-Editor: Xu ZH
| 1. | Ingle PV, Samsudin SZ, Chan PQ, Ng MK, Heng LX, Yap SC, Chai AS, Wong AS. Development and novel therapeutics in hepatocellular carcinoma: a review. Ther Clin Risk Manag. 2016;12:445-455. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 30] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 2. | Atiya H, Frisbie L, Pressimone C, Coffman L. Mesenchymal Stem Cells in the Tumor Microenvironment. Adv Exp Med Biol. 2020;1234:31-42. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 89] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 3. | Li H, Li F. Exosomes from BM-MSCs increase the population of CSCs via transfer of miR-142-3p. Br J Cancer. 2018;119:744-755. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 72] [Cited by in RCA: 82] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 4. | Lou G, Chen Z, Zheng M, Liu Y. Mesenchymal stem cell-derived exosomes as a new therapeutic strategy for liver diseases. Exp Mol Med. 2017;49:e346. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 264] [Cited by in RCA: 449] [Article Influence: 56.1] [Reference Citation Analysis (0)] |
| 5. | Dabrowska S, Andrzejewska A, Lukomska B, Janowski M. Neuroinflammation as a target for treatment of stroke using mesenchymal stem cells and extracellular vesicles. J Neuroinflammation. 2019;16:178. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 123] [Cited by in RCA: 221] [Article Influence: 36.8] [Reference Citation Analysis (0)] |
| 6. | Li A, Guo F, Pan Q, Chen S, Chen J, Liu HF. Mesenchymal Stem Cell Therapy: Hope for Patients With Systemic Lupus Erythematosus. Front Immunol. 2021;12:728190. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 59] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 7. | Xu Z, Chen Y, Ma L, Liu J, Guo Y, Yu T, Zhang L, Zhu L, Shu Y. Role of exosomal non-coding RNAs from tumor cells and tumor-associated macrophages in the tumor microenvironment. Mol Ther. 2022;30:3133-3154. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 199] [Article Influence: 66.3] [Reference Citation Analysis (0)] |
| 8. | Lin H, Yu J, Gu X, Ge S, Fan X. Novel insights into exosomal circular RNAs: Redefining intercellular communication in cancer biology. Clin Transl Med. 2021;11:e636. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 17] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 9. | Fan HN, Chen ZY, Chen XY, Chen M, Yi YC, Zhu JS, Zhang J. METTL14-mediated m(6)A modification of circORC5 suppresses gastric cancer progression by regulating miR-30c-2-3p/AKT1S1 axis. Mol Cancer. 2022;21:51. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 138] [Article Influence: 46.0] [Reference Citation Analysis (0)] |
| 10. | Wang Y, Liu J, Ma J, Sun T, Zhou Q, Wang W, Wang G, Wu P, Wang H, Jiang L, Yuan W, Sun Z, Ming L. Exosomal circRNAs: biogenesis, effect and application in human diseases. Mol Cancer. 2019;18:116. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 252] [Cited by in RCA: 491] [Article Influence: 81.8] [Reference Citation Analysis (0)] |
| 11. | Li LJ, Zhao W, Tao SS, Leng RX, Fan YG, Pan HF, Ye DQ. Competitive endogenous RNA network: potential implication for systemic lupus erythematosus. Expert Opin Ther Targets. 2017;21:639-648. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 63] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 12. | Xie M, Yu T, Jing X, Ma L, Fan Y, Yang F, Ma P, Jiang H, Wu X, Shu Y, Xu T. Exosomal circSHKBP1 promotes gastric cancer progression via regulating the miR-582-3p/HUR/VEGF axis and suppressing HSP90 degradation. Mol Cancer. 2020;19:112. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 136] [Cited by in RCA: 291] [Article Influence: 58.2] [Reference Citation Analysis (0)] |
| 13. | Shi L, Zhu W, Huang Y, Zhuo L, Wang S, Chen S, Zhang B, Ke B. Cancer-associated fibroblast-derived exosomal microRNA-20a suppresses the PTEN/PI3K-AKT pathway to promote the progression and chemoresistance of non-small cell lung cancer. Clin Transl Med. 2022;12:e989. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 109] [Article Influence: 36.3] [Reference Citation Analysis (0)] |
| 14. | Lyu Z, Yang M, Yang T, Ma M, Yang Z. Metal-Regulatory Transcription Factor-1 Targeted by miR-148a-3p Is Implicated in Human Hepatocellular Carcinoma Progression. Front Oncol. 2021;11:700649. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 13] [Reference Citation Analysis (0)] |
| 15. | Hepatocellular carcinoma. Nat Rev Dis Primers. 2021;7:7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 145] [Article Influence: 36.3] [Reference Citation Analysis (0)] |
| 16. | Romano V, Belviso I, Venuta A, Ruocco MR, Masone S, Aliotta F, Fiume G, Montagnani S, Avagliano A, Arcucci A. Influence of Tumor Microenvironment and Fibroblast Population Plasticity on Melanoma Growth, Therapy Resistance and Immunoescape. Int J Mol Sci. 2021;22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 37] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 17. | Ruocco MR, Lamberti A, Serrano MJ, Fiume G, Arcucci A. Editorial: Tumor Microenvironment and Cancer Cell Interactions in Solid Tumor Growth and Therapy Resistance. Front Cell Dev Biol. 2022;10:896194. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 7] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 18. | Urtasun R, Latasa MU, Demartis MI, Balzani S, Goñi S, Garcia-Irigoyen O, Elizalde M, Azcona M, Pascale RM, Feo F, Bioulac-Sage P, Balabaud C, Muntané J, Prieto J, Berasain C, Avila MA. Connective tissue growth factor autocriny in human hepatocellular carcinoma: oncogenic role and regulation by epidermal growth factor receptor/yes-associated protein-mediated activation. Hepatology. 2011;54:2149-2158. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 99] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 19. | Bayo J, Marrodán M, Aquino JB, Silva M, García MG, Mazzolini G. The therapeutic potential of bone marrow-derived mesenchymal stromal cells on hepatocellular carcinoma. Liver Int. 2014;34:330-342. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 16] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 20. | Niess H, Bao Q, Conrad C, Zischek C, Notohamiprodjo M, Schwab F, Schwarz B, Huss R, Jauch KW, Nelson PJ, Bruns CJ. Selective targeting of genetically engineered mesenchymal stem cells to tumor stroma microenvironments using tissue-specific suicide gene expression suppresses growth of hepatocellular carcinoma. Ann Surg. 2011;254:767-74; discussion 774. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 93] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 21. | Studeny M, Marini FC, Dembinski JL, Zompetta C, Cabreira-Hansen M, Bekele BN, Champlin RE, Andreeff M. Mesenchymal stem cells: potential precursors for tumor stroma and targeted-delivery vehicles for anticancer agents. J Natl Cancer Inst. 2004;96:1593-1603. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 621] [Cited by in RCA: 620] [Article Influence: 29.5] [Reference Citation Analysis (0)] |
| 22. | Zhou Y, Zhou W, Chen X, Wang Q, Li C, Chen Q, Zhang Y, Lu Y, Ding X, Jiang C. Bone marrow mesenchymal stem cells-derived exosomes for penetrating and targeted chemotherapy of pancreatic cancer. Acta Pharm Sin B. 2020;10:1563-1575. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 123] [Article Influence: 24.6] [Reference Citation Analysis (1)] |
| 23. | Jeppesen DK, Fenix AM, Franklin JL, Higginbotham JN, Zhang Q, Zimmerman LJ, Liebler DC, Ping J, Liu Q, Evans R, Fissell WH, Patton JG, Rome LH, Burnette DT, Coffey RJ. Reassessment of Exosome Composition. Cell. 2019;177:428-445.e18. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1476] [Cited by in RCA: 2067] [Article Influence: 344.5] [Reference Citation Analysis (1)] |
| 24. | Liu Y, Gu Y, Cao X. The exosomes in tumor immunity. Oncoimmunology. 2015;4:e1027472. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 134] [Cited by in RCA: 182] [Article Influence: 18.2] [Reference Citation Analysis (0)] |
| 25. | Couzin J. Cell biology: The ins and outs of exosomes. Science. 2005;308:1862-1863. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 108] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 26. | Ni YQ, Lin X, Zhan JK, Liu YS. Roles and Functions of Exosomal Non-coding RNAs in Vascular Aging. Aging Dis. 2020;11:164-178. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 41] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 27. | Dolati S, Shakouri SK, Dolatkhah N, Yousefi M, Jadidi-Niaragh F, Sanaie S. The role of exosomal non-coding RNAs in aging-related diseases. Biofactors. 2021;47:292-310. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 15] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 28. | Bao C, Lyu D, Huang S. Circular RNA expands its territory. Mol Cell Oncol. 2016;3:e1084443. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 39] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 29. | Li C, Ni YQ, Xu H, Xiang QY, Zhao Y, Zhan JK, He JY, Li S, Liu YS. Roles and mechanisms of exosomal non-coding RNAs in human health and diseases. Signal Transduct Target Ther. 2021;6:383. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 245] [Cited by in RCA: 243] [Article Influence: 60.8] [Reference Citation Analysis (1)] |
| 30. | Fang T, Lv H, Lv G, Li T, Wang C, Han Q, Yu L, Su B, Guo L, Huang S, Cao D, Tang L, Tang S, Wu M, Yang W, Wang H. Tumor-derived exosomal miR-1247-3p induces cancer-associated fibroblast activation to foster lung metastasis of liver cancer. Nat Commun. 2018;9:191. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 407] [Cited by in RCA: 749] [Article Influence: 107.0] [Reference Citation Analysis (0)] |
| 31. | Lin XJ, Fang JH, Yang XJ, Zhang C, Yuan Y, Zheng L, Zhuang SM. Hepatocellular Carcinoma Cell-Secreted Exosomal MicroRNA-210 Promotes Angiogenesis In Vitro and In Vivo. Mol Ther Nucleic Acids. 2018;11:243-252. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 118] [Cited by in RCA: 186] [Article Influence: 26.6] [Reference Citation Analysis (0)] |
| 32. | Zhou H, Gan X, He S, Wang Y, Zhang S, Chen J, Zhou Y, Hou C, Hua L, Zhang Q, Jia E. Identification of circular RNA BTBD7_hsa_circ_0000563 as a novel biomarker for coronary artery disease and the functional discovery of BTBD7_hsa_circ_0000563 based on peripheral blood mononuclear cells: a case control study. Clin Proteomics. 2022;19:37. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 12] [Reference Citation Analysis (0)] |