Published online Oct 21, 2023. doi: 10.3748/wjg.v29.i39.5435
Peer-review started: May 26, 2023
First decision: July 23, 2023
Revised: August 13, 2023
Accepted: October 16, 2023
Article in press: October 16, 2023
Published online: October 21, 2023
Processing time: 146 Days and 0.9 Hours
Small extracellular vesicles (exosomes) are important components of the tumor microenvironment. They are small membrane-bound vesicles derived from almost all cell types and play an important role in intercellular communication. Exosomes transmit biological molecules obtained from parent cells, such as proteins, lipids, and nucleic acids, and are involved in cancer development. MicroRNAs (miRNAs), the most abundant contents in exosomes, are selectively packaged into exosomes to carry out their biological functions. Recent studies have revealed that exosome-delivered miRNAs play crucial roles in the tumorigenesis, progression, and drug resistance of hepatocellular carcinoma (HCC). In addition, exosomes have great industrial prospects in the diagnosis, treatment, and prognosis of patients with HCC. This review summarized the composition and function of exosomal miRNAs of different cell origins in HCC and highlighted the association between exosomal miRNAs from stromal cells and immune cells in the tumor microenvironment and the progression of HCC. Finally, we described the potential applicability of exosomal miRNAs derived from mesenchymal stem cells in the treatment of HCC.
Core Tip: Hepatocellular carcinoma (HCC) is one of the most serious cancers in adults, and microRNAs (miRNAs) in small extracellular vesicles (exosomes) play a vital role in the pathological processes of HCC. Recent studies on exosomal miRNAs in HCC mainly focus on miRNA profiling but place little emphasis on where miRNAs come from and what target cells they act on. This review focused on the origin of exosomal miRNAs according to their parent cells in the tumor microenvironment and their role in HCC pathogenesis, contributing to a better understanding of exosomal miRNAs in the tumor microenvironment.
- Citation: Wang HC, Yin WX, Jiang M, Han JY, Kuai XW, Sun R, Sun YF, Ji JL. Function and biomedical implications of exosomal microRNAs delivered by parenchymal and nonparenchymal cells in hepatocellular carcinoma. World J Gastroenterol 2023; 29(39): 5435-5451
- URL: https://www.wjgnet.com/1007-9327/full/v29/i39/5435.htm
- DOI: https://dx.doi.org/10.3748/wjg.v29.i39.5435
In 2020, liver cancer was ranked the sixth most frequent malignant solid cancer globally. It was also the third-leading cause of cancer-related deaths in the world[1]. Hepatocellular carcinoma (HCC) is the primary histological type of liver cancer, comprising 80% of primary liver cancer cases[2]. It is characterized by the high degree of malignancy and poor prognosis. It is a threat to the health of humans. The symptoms of incipient-stage HCC are strong concealment, and it is challenging to diagnose HCC early. In addition, approximately 70% of patients undergo recurrence and experience metastasis within 5 years after surgical resection[3].
The tumor microenvironment (TME) is important in the development of HCC[3] and primarily comprises host cells, both resident and recruited, along with the secreted molecules and extracellular matrix (ECM) proteins[4]. Nonparenchymal cells in the liver, such as sinusoidal endothelial cells, hepatic stellate cells (HSCs), and macrophages, have a critical role in establishing the TME and mediating tumorigenesis by paracrine communication via cytokines and/or angiocrine factors[5]. Accumulating investigations on the TME have revealed novel perceptions of tumor growth as well as metastasis therein exosomes play a crucial function[6-8].
Small extracellular vesicles, also known as exosomes, refer to a specific type of extracellular vesicles with a size of 40-160 nm that originate from multivesicular bodies (MVBs), which act as carriers for biological information exchange to shape the cellular microenvironment[9]. To maintain consistency in nomenclature across studies published at different stages, we will use the name exosome for the rest of this review. Studies have shown that exosomes contain various cargoes including DNA, lipids, proteins, and RNA such as microRNAs (miRNAs), circular RNAs (circRNAs), long noncoding RNAs, and messenger RNAs, which are involved in intercellular communication[10,11].
More and more molecules of different classes carried by exosomes have been reported. Based on data retrieved from the ExoCarta database (http://www.exocarta.org), the identified components within exosomes consist of 9769 unique proteins, 3408 distinct messenger RNAs, 2838 different miRNAs, and 1116 lipids. Initially, exosomes were considered carriers of cellular waste, and their functions were underestimated[12]. Over the past few decades, the crucial functions of exosomes in facilitating intercellular communication in both physiological and pathological processes have been extensively studied and validated[13].
In 1996, exosomes derived from murine and human B lymphocytes were proven to execute a crucial function in transporting MHC molecules and eliciting MHC-II restricted T-cell responses[14]. Later, cancer cells and non-tumor cells in the TME were also found to be able to deliver exosomes and thereby participate in the malignant progression of tumors through molecular exchanges mediated by them[15,16]. Exosomes, hence, are recognized as important contributors to cancer initiation and progression[17-19].
MiRNAs represent an extensive collection of post-transcriptional gene expression regulators in eukaryotes. These regulatory molecules typically consist of 20-24 nucleotides and exert their function over various developmental and cellular processes[20]. Due to their essential role in gene expression, exosomal miRNAs have also been widely studied. In 2007, Valadi et al[21] reported that exosomes contained miRNAs, which could be delivered to other cells and exert their functions. Studies have demonstrated that exosomes are loaded with a high abundance of miRNAs, which play a crucial role in immune modulation, resistance to chemotherapy, and metastasis in diverse malignancies[22]. These miRNAs can promote tumor development in a paracrine manner in the surrounding microenvironment[23-25]. Furthering the comprehension of cancer mechanisms will require the identification of exosomal miRNAs, which are abnormally expressed in pathological states.
Numerous scientific studies have demonstrated that exosomes play a critical role in the genesis and malignant progression of tumors by transmitting signals between cells and regulating the TME[26]. This paper summarizes the studies of exosomal miRNAs released from nonparenchymal cells in the TME of HCC and discusses the association between these exosomal miRNAs and HCC. This study will help researchers in the field to better understand the role of exosomal miRNAs from stromal cells and immune cells in HCC and develop innovative strategies for HCC prevention and treatment.
Unlike other types of vesicles, exosomes have a different formation mechanism. First, the plasma membrane germinates inwards to form early endosomes (membrane-bound vacuoles)[27,28]. By further inwards budding of early endosomes encompassing miRNAs, proteins, and other selected substances, late endosomes called MVBs are formed[29]. Following this, the MVBs undergo fusion with the cell membrane, and the intraluminal endosomal vesicles are released into the extracellular area. These vesicles subsequently form exosomes[30] or fuse with the lysosome to decompose the biological information[31].
Studies revealed that the essential system involved in the biogenesis of exosomes is the endosomal sorting complex required for transport[32]. The endosomal sorting complex required for transport- identifies the ubiquitin-labeled “cargo” protein, guides it to MVBs, and subsequently separates the MVB from the peripheral membrane in a highly conserved process similar to the process of cytokinesis and virus budding[33].
Exosomes can be produced by any cell under normal or pathological conditions and might be taken up by other cells, hereby executing their designated tasks[34,35]. Exosomes transport multiple biologically active substances, such as proteins, RNA, DNA, and cholesterol[36-38]. The sucrose gradient density range in which exosomes float is 1.13-1.19 g/mL[39]. Of note, the composition of exosomes varies depending on their cellular origin[40], and different cell-derived exosomes or even the same cell-derived exosomes contain different components in different physiological or pathological states[41]. The amount of exosomal miRNAs secreted by hepatoma cells could also vary under different stimuli[42]. Research has shown that 55 miRNAs in Heb3B cell-derived exosomes were expressed at levels that were four times higher than those in donor cells, while 30 miRNAs were expressed at lower levels, and 11 miRNAs were expressed only in exosomes[43]. These changes may be a potential mechanism for disease progression.
In the past few years, exosomes have been shown to be crucial mediators of intercellular material and information exchange that can modulate the TME by transmitting nucleic acids and proteins between cells; hence, they are involved in tumor cell proliferation and migration, immune regulation, and drug resistance[44,45]. As an essential component of exosomes, exosomal miRNAs exert crucial functions in HCC tumorigenesis and progression.
First, we will review the function of exosomal miRNAs derived from HCC cells. MiR-122, which proved to be the most enriched miRNA in the human liver, is found to be decreased in the liver of HCC patients[46-48]. It is expressed and delivered by Huh7 cells (human HCC cell line) and can be transferred into HepG2 cells (human HCC cell line, of which the basal expression of miR-122 is low) in the form of exosomes, reducing the growth and proliferation of recipient HepG2 cells. The restoration of miR-122 inhibits HCC growth and enhances HCC sensitivity to chemotherapeutic drugs[49]. In addition, exosomes delivered by liver cancer cells can affect nonparenchymal cells in the microenvironment, promoting the malignant progression of tumors, which will be discussed in subsequent sections.
On the other hand, exosomal miRNAs secreted by tumor cells other than liver cancer cells can also promote the formation of premetastatic niches in the liver. Colon cancer cell-derived exosomes are able to deliver miR-21, miR-192, and miR-221 to hepatoma cells[50]. Exosomal miR-25-3p delivered by colon cancer cells promotes premetastatic niche formation in the liver by improving vascular permeability and angiogenesis[51]. Exosomes from colorectal cancer highly expressed miR-135a-5p, which could be transmitted to hepatic Kupffer cells to regulate the LATS2-YAP1/TEAD1-matrix metalloproteinase (MMP) 7 pathway and promote cell adhesion, forming premetastatic niches[52]. These results showed that exosomes could communicate between different types of cancers, even remodeling the microenvironment to boost liver metastasis[53].
Exosomal miRNAs might also be linked to different etiology of liver disease related to HCC. The connection between miRNAs and different liver diseases covering hepatitis B virus (HBV) infection, hepatitis C virus (HCV) infection, alcohol-associated liver disease (ALD), nonalcoholic steatohepatitis (NASH), nonalcoholic fatty liver disease, autoimmune hepatitis, and drug-induced liver injury has been discussed in-depth in previous high-quality reviews[54-56]. In the liver of ALD, NASH, and HCC patients, the level of hepatocyte-specific miR-122 exhibits a remarkable decrease. This specific miRNA directly targets distinct regions at the 5′-UTR of the HCV RNA genome, thereby facilitating the replication of HCV RNA[57]. When it comes to HBV replication, miR-122 functions oppositely. It acts as an inhibitor by downregulating the cyclin G1-p53 complex and preventing the specific interaction between p53 and HBV enhancers[58].
In simple steatosis, the liver shows an increase in the expression of miR-192, which is enriched in hepatocytes. However, this elevation is not observed in NASH[59]. On the other hand, the expression of miR-192 is decreased in HCC[60]. It is the most significantly downregulated miRNA in hepatic cancer stem cells and plays a role in the activation of cancer stem cells. Due to the anti-tumor property of miR-192, administering miR-192 to individuals with HCC can be a potential strategy for HCC therapy[60].
The expression of miR-155, a highly abundant miRNA in immune cells, including macrophages, is elevated in the liver tissues of patients with ALD, autoimmune hepatitis, and HCC. It is an oncogenic miRNA that links inflammation with tumorigenesis[61,62]. The activation of NF-κB signaling was reported to induce an upregulation in miR-155 levels in hepatocytes and liver cancer when mice were fed a choline-deficient and amino acid-defined diet[61] or in HCV infection in patients[62]. However, few studies have focused on the etiology of HCC and miRNAs delivered by exosomes.
According to a recent investigation, extracellular vesicles derived from neutrophils have the capability to transfer miR-223 to macrophages, stimulating the resolution of liver fibrosis[63]. Neutrophil/myeloid-specific miR-223 has been extensively studied for its anti-inflammatory properties. Its function involves the suppression of IL-6 expression, effectively reducing the activation of the IL-6-p47phox-ROS pathway within neutrophils[64]. The upregulation of miR-223 is observed in the serum and/or liver of patients or mouse models experiencing ALD or NASH, both diseases characterized by significant hepatic neutrophil infiltration. Consequently, the compensatory increase in miR-223 expression is a protective mechanism against ALD[64] and NASH[65]. At the same time, the reduction of miR-223 in HCC might be a causal factor in promoting HCC progression[66]. Therefore, the administration of miR-223 is thought to be a potent treatment in murine models of acute hepatitis and NASH[67]. Future studies of the above-reported miRNAs associated with different etiologies of liver diseases underlying HCC could be extended to the area of exosomes.
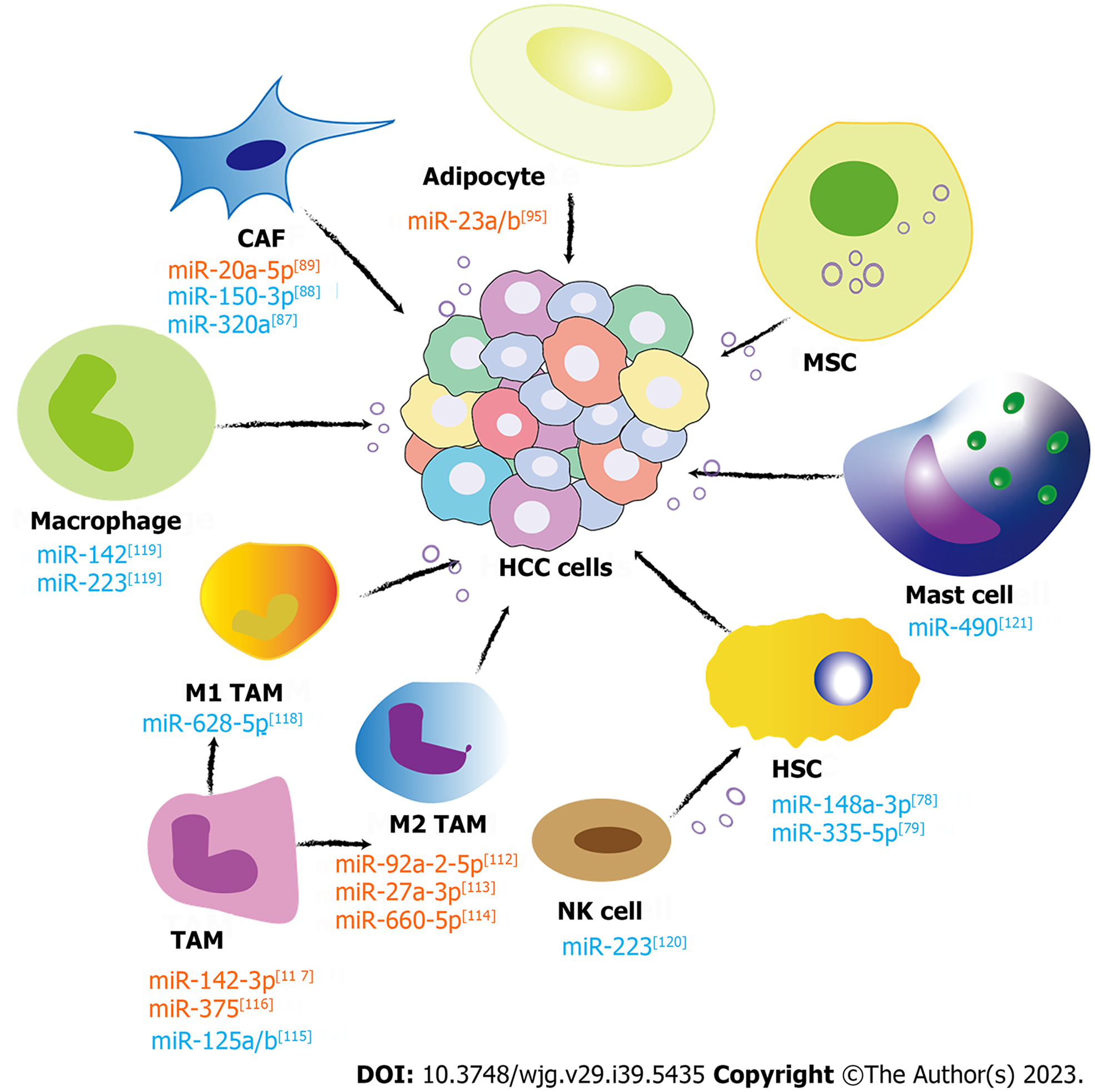
Since Stephen Paget proposed the “seed-soil” theory of tumor metastasis in 1889 to explain the organ specificity of tumor metastasis, there has been increasing evidence that tumor metastasis requires coordination between tumor cells and the TME, which has been identified as an evolutionary and ecological process characterized by constant, dynamic, and reciprocal action upon each other. Nonparenchymal cells in the liver cancer TME, such as HSCs, cancer-associated fibroblasts (CAFs), immune cells [T lymphocytes, B lymphocytes, natural killer (NK) cells, NK T cells, and tumor-associated macrophages (TAMs)], and endothelial cells, are pivotal in mediating tumor-stromal communications, thus regulating the biological processes of HCC[68]. Noncellular components are composed of growth factors like transforming growth factor-β (TGF-β), insulin-like growth factor, fibroblast growth factor, hepatocyte growth factor, vascular endothelial growth factor, proteolytic enzymes, ECM, and inflammatory cytokines. These components create a beneficial environment for the formation and proliferation of HCCs. Exosomal miRNAs, a crucial element of the TME, play a significant role in transmitting signals between cells and contribute to the development and advancement of tumors. In the next section, the role of the exosomal miRNAs from different nonparenchymal cells in HCC formation and metastasis is thoroughly discussed. The related investigations are paving the way for novel strategies in clinical diagnosis and treatments aimed at HCC (Figure 1).
HSCs can be observed in the space of Disse, located between liver sinusoidal endothelial cells and hepatocytes. These cells are responsible for storing lipid droplets containing vitamin A[69,70]. When there is damage to the liver, quiescent HSCs transform to activated HSCs, which resemble myofibroblasts and produce excessive fibrotic ECM[70]. The migration and accumulation of myofibroblasts are thought to be the key events that initiate liver fibrosis. Although many cell types, such as HSCs[71-73], portal fibroblasts[71,72], mesenchymal stem cell (MSC)-like cells[74], mesothelial cells[75] and bone marrow-derived cells[76], have been reported to contribute to the myofibroblast pool, recently researchers have evidence that 82%-96% of myofibroblasts in models with toxic, cholestatic, and fatty liver diseases are generated from activated HSCs[73].
The initiation and promotion of liver cancer are significantly correlated to the existence of liver fibrosis[70]. Activated HSC is a major factor mediating liver fibrosis and promotes liver cancer progression. Activated HSCs cocultured with HCC cells promoted tumor growth and invasiveness in nude mice[77]. In 2022, Zhang et al[78] reported that reducing activated HSC-delivered exosomal miR-148a-3p inhibited HCC initiation through the ITGA5/PI3K/Akt pathway. Another group found that HSC-HCC cell coculture reduced intracellular miR-335-5p expression in both types of cells. Additionally, in vitro and in vivo experiments showed that miR-335-5p-loaded HSC exosomes inhibited cancer growth and invasion[79]. In summary, activated HSCs can promote the development of HCC via various miRNAs delivered by exosomes, and targeting activated HSC-exosome miRNAs represents an innovative therapeutic strategy in HCC. At the same time, exosomes derived from HCC cells also promote the activation of HSCs. The HCC cell-derived exosome-miRNA-21, which targets the PTEN gene in HSCs, activates the PDK1/AKT pathway and converts HSCs to CAFs[80]. The progression of cancer was further accelerated by the activation of CAFs, which release angiogenic cytokines such as vascular endothelial growth factor, basic fibroblast growth factor, TGF-β, MMP2, and MMP9[80]. Another study suggested that a high level of serum exosomal miRNA-21 is associated with increased activation of CAFs and a higher vessel density in patients with HCC[80].
CAFs are an important component of the TME[81]. However, the concepts of HSCs and CAFs in early literature sometimes need to be clarified. Researchers used to believe that in the HCC microenvironment, HSCs frequently differentiated into CAFs, which have been extensively reported to influence HCC progression[81-84]. In the latest study, Zhu et al[85] successfully identified five CAF subtypes within HCC tumors through single-cell RNA sequencing data obtained from both mouse and human HCC tumors. The subtypes include vascular CAFs, matrix CAFs, lipid processing-matrix CAFs (also known as CD36+ CAFs), lipid-processing CAFs, and antigen-presenting CAFs. In these cells, CD36+ CAFs are derived from HSCs[85]. Another group also showed that Tcf21 was explicitly expressed in HSCs in mouse and human livers. Tcf21-positive HSCs, representing approximately 10% of all HSCs, can transdifferentiate into the majority of myofibroblasts in fibrotic liver and CAFs in HCC[86].
As crucial contributors to the alterations of the ECM that contribute to the development of HCC, CAFs have the potential to stimulate the progression of HCC through communication mediated by exosomes. A recent study found that the miR-320a level was remarkably decreased in CAF-derived exosomes compared with corresponding para-neoplastic fibroblast-derived exosomes in HCC patients. In vitro and in vivo experiments showed the anti-tumor effects of miR-320a when it was delivered to malignant cells through exosomes. The anti-tumor effect of miR-320a might be achieved by effectively targeting PBX3, thereby impeding the activation of the MAPK pathway[87]. Another study confirmed that miR-150-3p was lost in exosomes released by CAFs. CAF-delivered exosomes potently accelerate the malignant progression of HCC due to the absence of anti-tumoral miR-150-3p. Restoring the expression level of miR-150-3p by delivering miR-150-3p-loaded exosomes to HCC cells can effectively suppress their migration and invasiveness. Therefore, exosomal miR-150-3p can serve as a prognostic biomarker for HCC, and a supplement with exosomal miR-150-3p might be a potential treatment option[88].
Apart from those underexpressed anti-tumor miRNAs found in CAF-derived exosomes, the oncogenic miR-20a-5p was enriched in CAFs compared to HCC cells. MiR-20a-5p can be transferred from CAFs to HCC cells through exosomes and thereby suppress the expression of the tumor suppressor LIM domain and actin binding 1, which in turn inhibits the Wnt/β-catenin signaling pathway in HCC[89]. Thus, the distinct expression of exosomal miRNAs in CAFs plays a crucial part in the malignant progression of HCC. Therefore, potential therapeutic implications can be expected from anti-CAF medications that aim at certain exosomal miRNAs.
However, exosomal noncoding RNAs other than miRNAs also participate in the CAF-tumor cell communication. Chemoresistance in HCC can be influenced by CAF-exosomal circRNAs. Circular RNA ZFR is highly expressed in CAFs and CAF exosomes. CAF-exosomes transfer circular RNA ZFR to tumor cells, suppress the STAT3/NF-κB signaling pathway, and consequently enhance the growth of HCC cells as well as stimulate chemoresistance to cisplatin[90]. In addition, the migration, invasion, and glycolytic abilities of HCC cells were enhanced by long noncoding RNA TUG1 loaded in CAF-exosomes by targeting the miR-524-5p/SIX1 axis[91].
The involvement of adipose tissue in tumor progression has long been recognized[92]. Adipocytes play a crucial role in the hepatic microenvironment of nonalcoholic fatty liver disease, which is also a proven risk factor for HCC[44]. There is a close association between the adipocyte-HCC cell interaction and the risk of HCC development and progression[93]. Adipocyte-derived exosomes can affect the gene expression of liver cancer cells. In 2014, Koeck et al[94] found that exosomes from obese donors’ visceral adipose tissues caused dysregulation of genes involved in the TGF-β pathway in HepG2 cells. Recently, Liu et al[95] found that the levels of miR-23a/b in serum exosomes and tumor tissues were significantly elevated in high-body fat ratio (BFR) HCC patients compared to their low-BFR counterparts. In tumor tissues, it is highly probable that miR-23a/b can be transported from adipocytes into cancer cells via exosomes, thus promoting the malignant progression of HCC[95]. Moreover, exosomal miR-23a/b affects the von Hippel-Lindau/hypoxia-inducible factor pathway, thus promoting chemoresistance[95]. Exosomal circRNAs also play a role. Adipocyte exosomal circDB can suppress miR-34a expression in HCC cells and subsequently activate the deubiquitination-related USP7/cyclin A2 signaling pathway and promote tumor growth of HCC[96]. These studies provided evidence that high BFR-related exosomal miRNA could be valuable therapeutic targets for HCC.
On the other hand, HCC cell-derived exosomes can educate adjacent adipocytes and generate a microenvironment that promotes tumor formation and progression. HepG2-exosomes induced an inflammatory phenotype in adipocytes by activating several phosphorylated kinases (p-AKT, p-Erk1/2, p-GSKb, p-stat5a, and p-p38) and the NF-kB signaling pathway[44]. Adipocytes treated by tumor-derived exosomes enhance tumor development, angiogenesis, and macrophage recruitment in a mouse xenograft model[44]. The specific exosomal miRNAs that play a role in the process remain to be revealed.
In addition, it was observed in experimental models and human studies that the exposure to the adipocyte exosome increased the expression of various profibrotic molecules in HSCs, including tissue inhibitor of metal protease 1 and 4, Smad-3, integrins ανβ-5 and ανβ-8, and MMP-9[94].
It is widely acknowledged that angiogenic factors from tumor cells activate vascular endothelial cells, promote their proliferation and migration, and contribute to aberrant tumor angiogenesis[97]. HCC is a typical hypervascular tumor, and understanding the mechanisms of angiogenesis in HCC is very important[98]. In an early study, Shih et al[99] discovered that the decrease of miR-214 in HCC cells contributed to the upregulation of hepatoma-derived growth factor, stimulating vascular endothelial cells to promote angiogenesis and tumor growth. Therefore, miR-214 is a potent suppressor of angiogenesis. It was also shown that exosomes derived from HCC cells are able to induce the formation of lumens of human umbilical vein endothelial cells[98].
Recently, several HCC cell-derived exosomal miRNAs were found to be vital to angiogenesis. Fang et al[100] reported that hepatoma cell-derived exosomal miR-103 can be internalized by endothelial cells and damage the integrity of endothelial junctions and a subsequent elevation in vascular permeability that facilitates tumor metastasis. The underlying mechanism involves the specific targeting of crucial endothelial junction proteins, such as vascular endothelial-cadherin and p120-catenin, by exosomal miR-103[100]. Exosomal miR-210, derived from HCC cells, can be delivered to endothelial cells and lead to the promotion of tumor angiogenesis. This effect is mediated by the specific targeting of SMAD4 and STAT6, key regulators involved in modulating angiogenic processes[101]. Exosomal miRNAs (miR-638, miR-663a, miR-3648, and miR-4258) from HuH-7M (which is established from luciferase-expressing human hepatoma Huh-7 and deemed as a new, highly intrahepatic metastatic cell line) are able to attenuate the integrity of endothelial junctions, thus enhancing permeability by reducing vascular endothelial cadherin and zonula occludens-1 expression[102]. These findings revealed that HCC-exosomal miRNAs could be delivered to endothelial cells to promote HCC progression.
On the other hand, the exosomes released by endothelial cells might also affect tumor cells. A recent study showed that engineered human cerebral endothelial cell-derived exosomes containing increased miR-214 (hCEC-Exo-214) could enhance the sensitivity of HCC cells to anticancer drugs, such as oxaliplatin and sorafenib[103]. However, how endothelial cell-derived exosomes and exosomal miRNAs act on HCC cells is poorly studied. It is worth paying attention to in the follow-up studies.
The tumor immune microenvironment (TIME) is an important part of the TME[104]. The complicated interactions between cancer cells and host immune cells significantly influence TIME[105]. In HCC, the poor overall survival outcome arises as a result of immune surveillance disruption, which is strongly associated with the suppression of host immune reactions[105-107]. The growing evidence shows that the intricate interplay of exosome exchange-based cancer immunity shapes the tumor microenvironment, causing immune suppression and immune tolerance.
TAM presents the major leukocyte component infiltrating the HCC TIME[107]. Hepatic macrophages, also known as Kupffer cells, are the most abundant immune cells in the liver[108]. During the early stages of carcinogenesis, proinflammatory activation of Kupffer cells is important in tumor development. Once the primary tumor is established, the liver-infiltrating macrophages play a more critical role than Kupffer cells in HCC progression[109]. M2-polarized TAMs promote HCC progression by preventing T cells from recognizing and killing cancer cells, promoting tumor growth, angiogenesis, invasion, metastasis, and evasion of immune attack[110,111].
The role of TAM-derived exosomes is now attracting more and more attention. Liu et al[112] found a role of exosomal miR-92a-2-5p derived from M2 macrophages in promoting HCC cell invasion. This process is mediated through the regulation of the AR/PHLPP/p-AKT/β-catenin signaling pathway by miR-92a-2-5p. Increased expression of miR-27a-3p and miR-660-5p in M2 macrophage-derived exosomes facilitates HCC development by downregulating thioredoxin-interacting protein and KLF Transcription Factor 3 (KLF3)[113,114]. Exosomes derived from TAMs exhibit a reduction of miR-125a and miR-125b expression, which have been proven to promote HCC cell proliferation, sphere cell formation, and metastasis. MiR-125a/b exerts inhibitory effects on the HCC proliferation and attenuates their stem cell-like characteristics by specifically targeting CD90, a recognized stem cell marker in HCC[115].
Modulating TAM exosomal miRNAs provides a new way to suppress HCC. A tumor suppressor miRNA, miR-375, which is enriched in exosomes from IL-2 modulated TAMs, can ameliorate HCC development[116]. Moreover, propofol can stimulate TAMs to secrete exosomes overexpressing miR-142-3p. When miR-142-3p exosomes are transferred to HCC cells, they can inhibit HCC cell invasion[117]. Conversely, M1 macrophages contribute to proinflammatory and anti-tumor effects. M1 macrophage-derived exosomal miR-628-5p suppresses HCC development by restraining the m6A modification of circFUT8[118]. Peripheral blood monocyte-derived exosomal miR-142 and miR-223 can directly inhibit the proliferation of HCC[119].
The exosomes from other immune cells are also involved in HCC. In mice, NK-exosomes rich in miR-223 inhibited carbon tetrachloride-induced liver fibrosis by inhibiting TGF-β1 induced HSC activation by directly targeting ATG7. Therefore, the overexpression of ATG7 in HSCs abolished the HSC activation-suppressive effect of NK cell exosomes[120]. Hepatitis C virus E2 envelope glycoprotein can stimulate mast cells, which in turn secrete a considerable amount of miR-490 enriched exosomes. When these exosomes are transferred into HCC cells, they inhibit tumor cell metastasis through the ERK1/2 pathway[121]. In addition, miR-150-5p and miR-142-3p can be transferred from regulatory T cells (Tregs) to dendritic cells via exosomes, resulting in the induction of a tolerant phenotype in these cells, characterized by elevated IL-10 production and decreased IL-6 production upon lipopolysaccharide stimulation[122].
On the other hand, tumor-derived exosomal miRNAs also affect the distribution and function of immune cells. Tregs constitute the most prominent subset of suppressor cells in the TME and release immunosuppressive factors, including IL-10 and TGF-β, contributing to tumor progression. Tregs also present various chemokine receptors and surface molecules like CTLA4 and PD-1, which make them susceptible to immune checkpoint inhibitor immunotherapy. The development of immune-related adverse events may partly be attributed to Treg destabilization[123]. Tumor cell-delivered miR-214 has the potential to augment the population of CD4+CD25highFoxp3+ Treg by reducing the expression of PTEN in CD4+ T cells, which results in the suppression of the host immune response and accelerates tumor development[124]. Indeed, the expansion of Treg populations through tumor-secreted miR-214 is believed to be a shared mechanism employed by various cancer cells to establish an immune-tolerant environment. This miRNA is crucial in modulating immune responses and promoting immune tolerance within the tumor microenvironment. Consequently, the inhibition of tumor-secreted miR-214 transportation to immune cells shows potential as an innovative approach to counteract tumor-induced immune tolerance[124].
In summary, exosome-delivered miRNAs from immune cells were intensely involved in the biological processes of HCC, and HCC-derived exosomal miRNAs also affect the distribution and function of immune cells.
Radical resection and transarterial chemoembolization remain the most efficacious therapeutic approaches for patients with early-stage liver cancer. Still, the treatment efficacy remains unsatisfactory due to the compensatory effect of vascular proliferation after hypoxia[125,126]. For patients with advanced liver cancer, targeted therapy and traditional chemotherapy can only prolong the survival of these patients to a certain extent. Innovative and alternative therapies are continuously needed to improve the prognosis of HCC patients.
Studies have recently confirmed that specific miRNAs can be transported through exosomes, thereby controlling tumor growth and achieving therapeutic effects[127]. Since exosomes exhibit distinct characteristics as a vehicle for drug transport, encompassing diminished immunogenicity, enhanced biocompatibility, reduced toxicity, and the capacity to traverse the blood-brain barrier, exosomes have garnered considerable attention as an innate delivery vector for conveying miRNA molecules[128]. Among the various cell types recognized for their ability to produce exosomes, MSCs are a promising choice for the large-scale production of exosomes for drug delivery. It has been shown in regenerative medicine and tumor treatment studies that MSC-derived exosomes can serve as effective vehicles for drug delivery[129,130]. Based on the above findings, engineered MSC-derived exosomes loaded with specific miRNAs present a novel therapeutic approach for HCC treatment.
Exosomal miRNAs have been utilized to enhance the chemosensitivity of tumor cells[131,132]. Recent research demonstrated that miR-122 overexpression could sensitize the response of HCC cells to chemotherapy drugs by suppressing multidrug resistance-associated genes, the anti-apoptotic gene Bcl-w, and the cell cycle-related gene cyclin B1[47]. The miR-122 overexpression amniotic membrane MSCs (AMSCs) can effectively encapsulate miR-122 to secreted exosomes, which are in turn delivered to HCC cells and further increase the sensitivity of HCC cells to sorafenib[133]. The miR-199a loaded AMSC exosomes produced through miR-199a overexpression lentivirus infection and subsequent puromycin selection are able to potently transport miR-199a and enhance the sensitivity of HCC cells towards doxorubicin by specifically targeting the mTOR pathway. Furthermore, tumor tissue can be effectively targeted by AMSC exosomal miRNA-199a through intravenous injection, thereby enhancing the therapeutic effect of Dox on HCC in vivo[134].
Liver fibrosis is the precursor stage of cirrhosis and liver cancer. MSC-derived exosomes alleviated carbon tetrachloride-induced liver fibrosis in mice through the expression of miR-148a. MiR-148a directly targets KLF transcription factor 6 and successfully converts the M1 macrophages to M2 macrophages in vitro and liver fibrosis models[135]. In vitro studies have shown that transplanted human chorionic plate-derived MSCs reduce lung and liver fibrosis in murine models[136,137]. One study supported that chorionic plate-derived MSCs released exosomes containing miRNA-125b into hedgehog-responsive HSCs and restrained the activation of hedgehog signaling by blocking the expression of smoothened receptors, consequently reducing the severity of hepatic fibrosis[138]. As a new candidate therapeutic strategy, MSC exosomes have excellent application prospects for HCC.
In addition, human liver stem cell-derived exosomes are loaded with multiple antitumor miRNAs (miR451, miR223, miR24, miR31, miR214, and miR122), which can downregulate multi-drug resistance 1, migration inhibitory factor, ras-associated protein 14, and E2F transcription factor 1. These exosomes have been proven to be able to inhibit the growth of hepatoma cells both in vitro and in vivo[139].
Despite significant advances in diagnosis and therapeutics, HCC remains exceedingly fatal. In most cases, HCC develops from chronic liver inflammation, which provides a tumor-promoting microenvironment composed of immune and stromal cells. As a novel cellular communicator in TME, exosomes mediate the intricate interaction of nonparenchymal cells (including immune and stromal cells) with cancer cells. They are involved in the etiology of HCC and multiple processes related to tumor initiation, development, metastasis, and drug resistance. Exosome cargoes, especially miRNAs, are key communication factors in the complicated cross-talk, indicating that they are promising prognostic markers and therapeutic targets for HCC. In this review, we emphasized the function and mechanism of exosomal miRNAs from nonparenchymal cells for the initiation and malignant progression of HCC. Also, we introduced the influences of exosomal miRNAs delivered by tumor cells on nonparenchymal cells. The functions of the exosomal miRNAs in HCC were also summarized (Table 1). Finally, the therapeutic potential of exosomes for HCC was discussed. With the development of nanoengineering technology, exosomes can be modified to carry specific miRNAs and target specific cells, thus enabling precision and individualized treatment of HCC.
| miRNA species in exosomes | Exosome secreting cells | Exosome isolation methods | Target cells | miRNA expression of exosome | Downstream targets | Functions of miRNA | Additional information | Ref. | Year |
| miR-148a-3p | Primary fibroblasts (the HSC cell line LX-2) | The ExoQuick-TC kit | Human HCC cell lines PLC, HCCLM3, and SMMC-7721 | Reduced in the exosomes of HSCs after cocultivation with primary liver cancer-associated fibroblasts | ITGA5/PI3K/Akt axis | Inhibited HCC cell malignancy | Primary fibroblasts were isolated from primary HCC tumor and paired peritumor tissues in 17 primary HCC patient samples | [78] | 2022 |
| miR-335-5p | The HSC cell line LX-2 | Ultracentrifugation | Human HCC cell lines MHCC97H, MHCC97L, HepG2, and Huh7 | Reduced in the exosomes of fibroblasts as well as in HCC cells after cocultivation | CDC42? CDK2? | Inhibited neighboring cancer cell proliferation, invasion, and motility | - | [79] | 2019 |
| miR-320a | CAFs | Life Technology exosome precipitation solution | Human HCC cell lines MHCC97-H, SMMC-7721, Huh7, and the human normal liver cell line 7702 | Reduced in the exosomes of CAFs derived from human HCC patients | PBX3 | Inhibited HCC cell proliferation and metastasis ability | PAFs and CAFs derived from 6 pairs of matched primary hepatocarcinoma and adjacent tumor-free tissues (5 cm from the cut edge of the tumor edge) | [87] | 2017 |
| miR-150-3p | CAFs | 0.22-µm PVDF filter and Total Exosome Isolation Reagent | Human HCC cell lines Huh7 and Hep3B | Decreased in CAF-derived exosomes | - | Inhibited HCC proliferation and metastasis | Stromal fibroblasts isolated from tumor tissue and adjacent (> 5 cm from the tumor edge) tissues from 6 HCC patients | [88] | 2021 |
| miR-20a-5p | CAFs | Centrifuged and filtered through a 0.22-µm PVDF membrane | Human HCC cell lines SMMC7721, Huh7, YY8103, Hep3B, Focus, HepG2, and HCCLM3 and a normal liver cell line MIHA, WRL68 | Higher in exosomes from cancer tissues than in matched adjacent para-tumoral tissues | LIMA1 | Contributed to HCC cell proliferation, metastasis, and EMT | CAFs were from the HCC tissues and NFs in paired adjacent normal tissues from 92 HCC patients | [89] | 2022 |
| miR-214 | hCECs | Centrifuged and filtered through a 0.22-µm PVDF membrane and ultracentrifugation | Human HCC cell lines HepG2, Hep3B, the human liver epithelial cell line THLE-2 | Lower levels in HCC cells than in normal human liver epithelial cells | P-gp/SF3B3 | Reduced cancer cell viability and invasion compared with monotherapy with oxaliplatin or sorafenib | - | 2021 | |
| miR-23a/b | Adipose cell mouse preadipocyte 3T3-L1 cells | Differential centrifugation | The human HCC cell lines BEL-7402 and BEL-7402/5-Fu murine hepatoma cell line Hepa1-6 | High in exosomes from HCC patients with high BFR | VHL/HIF-1α | Promoted HCC cell growth and migration | Adipose cells were isolated from human tumor tissues from obese and nonobese patients | [95] | 2019 |
| miR-142, miR-223 | Monocyte-derived macrophages; human acute monocytic leukemia THP-1, B-lymphoblastoid 721.221, and murine lymphoblast-like mastocytoma P815 cell lines | Microfiltration and ultracentrifugation | The human HCC cell lines Huh7 and HepG2 | High when cocultured with HCC cells | STMN-1 | Inhibited HCC proliferation | PBMCs were isolated from lymphocyte cones or fresh blood by density gradient centrifugation and were incubated for 2 h in plastic plates before the flask was washed intensively to remove any nonadherent cells. After 4 d of incubation in serum-free medium supplemented with 1% autologous serum, adherent cells were washed with PBS and cultured in standard DMEM-based medium for 3-6 extra days to generate monocyte-derived macrophages phenotyped to be CD14+, CD11a+, CD3−, CD56−, and CD19− | [119] | 2013 |
| miR-490 | Human MC line HMC-1 (treated with HCV-E2) | Total exosome separation reagent from Invitrogen | The human HCC cell lines HepG2 and HepG3b | High when HCV-E2-stimulated MC-derived exosomes were incubated with the two types of HCC cells for 24 h compared with the incubation with normal MC-derived exosomes | ERK1/2 | Inhibited HCC proliferation | [121] | 2017 | |
| miR-223 | Human NK cell line NK92-MI | Differential centrifugation | The human HSC line LX-2 | Higher in exosomes derived from NK cells than in parental NK-92MI cells | AGT7 | Attenuated TGF-β1-induced HSC activation and inhibited liver fibrosis | LX-2 cells were treated with TGF-β1 (5 ng/mL) for 24 h to stimulate HSC activation. LX-2 cells in the exosomes derived from NK cells-treated groups were pretreated with exosomes derived from NK cells (10 μg/mL) before TGF-β1 treatment. LX-2 cells in the rapamycin-treated groups were pretreated with the autophagy activator rapamycin (2 mM) in DMSO for 12 h before TGF-β1 treatment | [120] | 2020 |
| miR-125a/b | TAMs | ExoQuick exosome precipitation solution | The human HCC cell lines Huh7, HepG2, and BEL-7404 | Downregulated in exosomes from HCC-associated macrophages | CD90 | Suppressed HCC cell growth and sphere formation | TAMs and nontumor macrophages were isolated from primary human HCC, adjacent nontumor liver tissues from 6 patients with HCC | [115] | 2019 |
| miR-628-5p | M1 macrophage | - | The human HCC cell lines Huh7, HCCLM3, Hep3B, and MHCC97H, immortalized human liver epithelial THLE-3 cell line | High in M1-exosomes | METTL14/circFUT4/CHMP14B | Inhibited HCC cell progression | THP-1 cells were differentiated into M0 macrophages by a 24 h incubation with 150 nM phorbol 12-myristate 13-acetate followed by a 24 h incubation in RPMI medium. M0 macrophages were polarized to M1 macrophages by incubation with 20 ng/mL IFN-γ and 10 pg/mL lipopolysaccharide | [118] | 2022 |
| miR-92a-2-5p | M2 macrophage (monocytic leukemia cell line THP-1) | Centrifuged and filtered through a 0.22-µm PVDF membrane and ultracentrifugation | Human liver cancer SK-HEP-1 and HepG2 cell lines, HA22T cell line, and murine HCC Hepa 1-6 cell line | Increased after coculture with liver cancer cells | AR/PHLPP/p-AKT/β-catenin signaling | Promoted HCC growth and invasiveness | To induce differentiation into macrophages, THP-1 cells were cultured with 100 ng/mL PMA (Sigma) for 48 h, and the macrophage was cultured with the addition of DMSO to promote M2 polarization | [112] | 2020 |
| miR-660-5p | M2 macrophage (monocytic leukemia cell line THP-1) | Differential centrifugation | Human HCC cell lines HepG2 and Bel-7402 | High | KLF3 | Augmented the tumorigenic ability of HCC cells | THP-1 monocytes were stimulated by 100 ng of phorbol 12-myristate 13-acetate (Sigma-Aldrich, MO, United States) for 48 h, thus differentiating into M0 macrophages. Then, M0 macrophages were treated with 20 ng/mL interleukin 4 (AF-200–04-5, PeproTech, NJ, United States) for 72 h to polarize into M2 macrophages | [114] | 2021 |
| miR-27a-3p | M2 macrophage (monocytic leukemia cell line THP-1) | SBI ExoQuick-TC Kit | Human HCC cell lines Huh7, 97H, HepG2, LM3, and SMMC-7721 | - | TXNIP | Induced the cancer stemness of HCC | Differentiation of THP-1 cells to macrophages was performed using 200 ng/mL phorbol myristic acetate, and the cells were then cultured with 20 ng/mL interleukin-4 for 72 h to induce M2-type polarization | [113] | 2021 |
| miR-142-3p | TAMs treated by propofol (the murine macrophage cell line Raw 264.7 cells) | Differential centrifugation | The murine HCC cell line Hepa1-6 | Dose-dependent increase when treated with propofol | RAC1 | Enhanced the antitumor activity of propofol | Raw 264.7 cells were cultured in complete RPMI 1640 with 10% FBS and treated with propofol (dissolved in RPMI 1640) in complete medium. TAMs were isolated from tumor-bearing mice treated with 0 mg/kg, 20 mg/kg, and 50 mg/kg propofol by i.p. injection | [117] | 2014 |
| miR-375 | TAMs (IL-2 induced) | Total exosome isolation reagent | The human HCC cell lines HepG2 and QJY–7703 | High | - | Ameliorated HCC development and progression | Primary human HCC specimens were collected from patients who suffered from hepatectomy. The macrophages were isolated and cultured by Percoll (GE Healthcare) density gradient centrifugation. TAMs were treated with IL-2 for 24 h before the supernatants were collected. The treatment concentration was 20 ng/mL | [116] | 2022 |
Although significant progress has been achieved in elucidating the functions of exosomes and their miRNA cargoes in HCC, some challenges remain. Sometimes, different investigators reported different experimental observations for the same exosomal miRNAs. The inconsistency of experimental subjects and study designs might cause these discrepancies. Therefore, factors such as the environment, age and sex of the subjects, cause of HCC occurrence, and data collection from multiple centers should be considered to produce more accurate results. Moreover, different techniques can lead to the isolation of varied subtypes of extracellular vesicles, each exhibiting unique miRNA profiles, protein compositions, and biological functions[140-142]. In clinical applications, problems include low targeting efficiency and easily phagocytosed by the immune system. The exosome separation and purification method also have limitations and could be time-consuming and laborious. Therefore, further research must be done to address these problems and determine more feasible and effective clinical translational applications of exosomes. The integration of nanoengineering and molecular biology allows for the utilization of exosome-mediated miRNAs in precision nanomedicine, presenting novel approaches for the diagnosis and treatment of HCC.
The authors would like to thank the anonymous reviewers whose feedback substantially improved the quality of this article.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): B, B, B, B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Ghazy A, Egypt; Granito A, Italy; Haque N, Bangladesh; Tsai HW, Taiwan; Wang YG, China S-Editor: Yan JP L-Editor: Filipodia P-Editor: Cai YX
| 1. | Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75126] [Cited by in RCA: 64711] [Article Influence: 16177.8] [Reference Citation Analysis (177)] |
| 2. | Rumgay H, Ferlay J, de Martel C, Georges D, Ibrahim AS, Zheng R, Wei W, Lemmens VEPP, Soerjomataram I. Global, regional and national burden of primary liver cancer by subtype. Eur J Cancer. 2022;161:108-118. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 125] [Cited by in RCA: 258] [Article Influence: 86.0] [Reference Citation Analysis (0)] |
| 3. | Villanueva A. Hepatocellular Carcinoma. N Engl J Med. 2019;380:1450-1462. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2066] [Cited by in RCA: 3177] [Article Influence: 529.5] [Reference Citation Analysis (37)] |
| 4. | Anderson NM, Simon MC. The tumor microenvironment. Curr Biol. 2020;30:R921-R925. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 232] [Cited by in RCA: 1536] [Article Influence: 384.0] [Reference Citation Analysis (0)] |
| 5. | Lu C, Rong D, Zhang B, Zheng W, Wang X, Chen Z, Tang W. Current perspectives on the immunosuppressive tumor microenvironment in hepatocellular carcinoma: challenges and opportunities. Mol Cancer. 2019;18:130. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 142] [Cited by in RCA: 303] [Article Influence: 50.5] [Reference Citation Analysis (0)] |
| 6. | Wu Q, Zhou L, Lv D, Zhu X, Tang H. Exosome-mediated communication in the tumor microenvironment contributes to hepatocellular carcinoma development and progression. J Hematol Oncol. 2019;12:53. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 113] [Cited by in RCA: 212] [Article Influence: 35.3] [Reference Citation Analysis (0)] |
| 7. | Luo C, Xin H, Zhou Z, Hu Z, Sun R, Yao N, Sun Q, Borjigin U, Wu X, Fan J, Huang X, Zhou S, Zhou J. Tumor-derived exosomes induce immunosuppressive macrophages to foster intrahepatic cholangiocarcinoma progression. Hepatology. 2022;76:982-999. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 69] [Article Influence: 23.0] [Reference Citation Analysis (0)] |
| 8. | Martínez-Reyes I, Chandel NS. Cancer metabolism: looking forward. Nat Rev Cancer. 2021;21:669-680. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 290] [Cited by in RCA: 1019] [Article Influence: 254.8] [Reference Citation Analysis (0)] |
| 9. | Théry C, Witwer KW, Aikawa E, Alcaraz MJ, Anderson JD, Andriantsitohaina R, Antoniou A, Arab T, Archer F, Atkin-Smith GK, Ayre DC, Bach JM, Bachurski D, Baharvand H, Balaj L, Baldacchino S, Bauer NN, Baxter AA, Bebawy M, Beckham C, Bedina Zavec A, Benmoussa A, Berardi AC, Bergese P, Bielska E, Blenkiron C, Bobis-Wozowicz S, Boilard E, Boireau W, Bongiovanni A, Borràs FE, Bosch S, Boulanger CM, Breakefield X, Breglio AM, Brennan MÁ, Brigstock DR, Brisson A, Broekman ML, Bromberg JF, Bryl-Górecka P, Buch S, Buck AH, Burger D, Busatto S, Buschmann D, Bussolati B, Buzás EI, Byrd JB, Camussi G, Carter DR, Caruso S, Chamley LW, Chang YT, Chen C, Chen S, Cheng L, Chin AR, Clayton A, Clerici SP, Cocks A, Cocucci E, Coffey RJ, Cordeiro-da-Silva A, Couch Y, Coumans FA, Coyle B, Crescitelli R, Criado MF, D'Souza-Schorey C, Das S, Datta Chaudhuri A, de Candia P, De Santana EF, De Wever O, Del Portillo HA, Demaret T, Deville S, Devitt A, Dhondt B, Di Vizio D, Dieterich LC, Dolo V, Dominguez Rubio AP, Dominici M, Dourado MR, Driedonks TA, Duarte FV, Duncan HM, Eichenberger RM, Ekström K, El Andaloussi S, Elie-Caille C, Erdbrügger U, Falcón-Pérez JM, Fatima F, Fish JE, Flores-Bellver M, Försönits A, Frelet-Barrand A, Fricke F, Fuhrmann G, Gabrielsson S, Gámez-Valero A, Gardiner C, Gärtner K, Gaudin R, Gho YS, Giebel B, Gilbert C, Gimona M, Giusti I, Goberdhan DC, Görgens A, Gorski SM, Greening DW, Gross JC, Gualerzi A, Gupta GN, Gustafson D, Handberg A, Haraszti RA, Harrison P, Hegyesi H, Hendrix A, Hill AF, Hochberg FH, Hoffmann KF, Holder B, Holthofer H, Hosseinkhani B, Hu G, Huang Y, Huber V, Hunt S, Ibrahim AG, Ikezu T, Inal JM, Isin M, Ivanova A, Jackson HK, Jacobsen S, Jay SM, Jayachandran M, Jenster G, Jiang L, Johnson SM, Jones JC, Jong A, Jovanovic-Talisman T, Jung S, Kalluri R, Kano SI, Kaur S, Kawamura Y, Keller ET, Khamari D, Khomyakova E, Khvorova A, Kierulf P, Kim KP, Kislinger T, Klingeborn M, Klinke DJ 2nd, Kornek M, Kosanović MM, Kovács ÁF, Krämer-Albers EM, Krasemann S, Krause M, Kurochkin IV, Kusuma GD, Kuypers S, Laitinen S, Langevin SM, Languino LR, Lannigan J, Lässer C, Laurent LC, Lavieu G, Lázaro-Ibáñez E, Le Lay S, Lee MS, Lee YXF, Lemos DS, Lenassi M, Leszczynska A, Li IT, Liao K, Libregts SF, Ligeti E, Lim R, Lim SK, Linē A, Linnemannstöns K, Llorente A, Lombard CA, Lorenowicz MJ, Lörincz ÁM, Lötvall J, Lovett J, Lowry MC, Loyer X, Lu Q, Lukomska B, Lunavat TR, Maas SL, Malhi H, Marcilla A, Mariani J, Mariscal J, Martens-Uzunova ES, Martin-Jaular L, Martinez MC, Martins VR, Mathieu M, Mathivanan S, Maugeri M, McGinnis LK, McVey MJ, Meckes DG Jr, Meehan KL, Mertens I, Minciacchi VR, Möller A, Møller Jørgensen M, Morales-Kastresana A, Morhayim J, Mullier F, Muraca M, Musante L, Mussack V, Muth DC, Myburgh KH, Najrana T, Nawaz M, Nazarenko I, Nejsum P, Neri C, Neri T, Nieuwland R, Nimrichter L, Nolan JP, Nolte-'t Hoen EN, Noren Hooten N, O'Driscoll L, O'Grady T, O'Loghlen A, Ochiya T, Olivier M, Ortiz A, Ortiz LA, Osteikoetxea X, Østergaard O, Ostrowski M, Park J, Pegtel DM, Peinado H, Perut F, Pfaffl MW, Phinney DG, Pieters BC, Pink RC, Pisetsky DS, Pogge von Strandmann E, Polakovicova I, Poon IK, Powell BH, Prada I, Pulliam L, Quesenberry P, Radeghieri A, Raffai RL, Raimondo S, Rak J, Ramirez MI, Raposo G, Rayyan MS, Regev-Rudzki N, Ricklefs FL, Robbins PD, Roberts DD, Rodrigues SC, Rohde E, Rome S, Rouschop KM, Rughetti A, Russell AE, Saá P, Sahoo S, Salas-Huenuleo E, Sánchez C, Saugstad JA, Saul MJ, Schiffelers RM, Schneider R, Schøyen TH, Scott A, Shahaj E, Sharma S, Shatnyeva O, Shekari F, Shelke GV, Shetty AK, Shiba K, Siljander PR, Silva AM, Skowronek A, Snyder OL 2nd, Soares RP, Sódar BW, Soekmadji C, Sotillo J, Stahl PD, Stoorvogel W, Stott SL, Strasser EF, Swift S, Tahara H, Tewari M, Timms K, Tiwari S, Tixeira R, Tkach M, Toh WS, Tomasini R, Torrecilhas AC, Tosar JP, Toxavidis V, Urbanelli L, Vader P, van Balkom BW, van der Grein SG, Van Deun J, van Herwijnen MJ, Van Keuren-Jensen K, van Niel G, van Royen ME, van Wijnen AJ, Vasconcelos MH, Vechetti IJ Jr, Veit TD, Vella LJ, Velot É, Verweij FJ, Vestad B, Viñas JL, Visnovitz T, Vukman KV, Wahlgren J, Watson DC, Wauben MH, Weaver A, Webber JP, Weber V, Wehman AM, Weiss DJ, Welsh JA, Wendt S, Wheelock AM, Wiener Z, Witte L, Wolfram J, Xagorari A, Xander P, Xu J, Yan X, Yáñez-Mó M, Yin H, Yuana Y, Zappulli V, Zarubova J, Žėkas V, Zhang JY, Zhao Z, Zheng L, Zheutlin AR, Zickler AM, Zimmermann P, Zivkovic AM, Zocco D, Zuba-Surma EK. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles. 2018;7:1535750. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6453] [Cited by in RCA: 7703] [Article Influence: 1100.4] [Reference Citation Analysis (1)] |
| 10. | Isaac R, Reis FCG, Ying W, Olefsky JM. Exosomes as mediators of intercellular crosstalk in metabolism. Cell Metab. 2021;33:1744-1762. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 464] [Article Influence: 116.0] [Reference Citation Analysis (0)] |
| 11. | Krylova SV, Feng D. The Machinery of Exosomes: Biogenesis, Release, and Uptake. Int J Mol Sci. 2023;24. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 299] [Reference Citation Analysis (0)] |
| 12. | Johnstone RM, Adam M, Hammond JR, Orr L, Turbide C. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes). J Biol Chem. 1987;262:9412-9420. [PubMed] |
| 13. | Kalluri R, LeBleu VS. The biology, function, and biomedical applications of exosomes. Science. 2020;367. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6920] [Cited by in RCA: 6574] [Article Influence: 1314.8] [Reference Citation Analysis (0)] |
| 14. | Raposo G, Nijman HW, Stoorvogel W, Liejendekker R, Harding CV, Melief CJ, Geuze HJ. B lymphocytes secrete antigen-presenting vesicles. J Exp Med. 1996;183:1161-1172. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2293] [Cited by in RCA: 2691] [Article Influence: 92.8] [Reference Citation Analysis (0)] |
| 15. | Rak J. Microparticles in cancer. Semin Thromb Hemost. 2010;36:888-906. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 200] [Cited by in RCA: 217] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 16. | Hood JL, San RS, Wickline SA. Exosomes released by melanoma cells prepare sentinel lymph nodes for tumor metastasis. Cancer Res. 2011;71:3792-3801. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 719] [Cited by in RCA: 785] [Article Influence: 56.1] [Reference Citation Analysis (0)] |
| 17. | Zhang L, Yu D. Exosomes in cancer development, metastasis, and immunity. Biochim Biophys Acta Rev Cancer. 2019;1871:455-468. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 215] [Cited by in RCA: 676] [Article Influence: 112.7] [Reference Citation Analysis (0)] |
| 18. | Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16:203-222. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2492] [Cited by in RCA: 3513] [Article Influence: 439.1] [Reference Citation Analysis (0)] |
| 19. | Kim SB. Function and therapeutic development of exosomes for cancer therapy. Arch Pharm Res. 2022;45:295-308. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 30] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 20. | Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597-610. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3165] [Cited by in RCA: 3676] [Article Influence: 245.1] [Reference Citation Analysis (0)] |
| 21. | Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654-659. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8246] [Cited by in RCA: 9801] [Article Influence: 544.5] [Reference Citation Analysis (0)] |
| 22. | Qiu S, Xie L, Lu C, Gu C, Xia Y, Lv J, Xuan Z, Fang L, Yang J, Zhang L, Li Z, Wang W, Xu H, Li B, Xu Z. Gastric cancer-derived exosomal miR-519a-3p promotes liver metastasis by inducing intrahepatic M2-like macrophage-mediated angiogenesis. J Exp Clin Cancer Res. 2022;41:296. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 142] [Reference Citation Analysis (0)] |
| 23. | Nallasamy P, Nimmakayala RK, Parte S, Are AC, Batra SK, Ponnusamy MP. Tumor microenvironment enriches the stemness features: the architectural event of therapy resistance and metastasis. Mol Cancer. 2022;21:225. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 123] [Article Influence: 41.0] [Reference Citation Analysis (0)] |
| 24. | Yuan X, Qian N, Ling S, Li Y, Sun W, Li J, Du R, Zhong G, Liu C, Yu G, Cao D, Liu Z, Wang Y, Qi Z, Yao Y, Wang F, Liu J, Hao S, Jin X, Zhao Y, Xue J, Zhao D, Gao X, Liang S, Song J, Yu S. Breast cancer exosomes contribute to pre-metastatic niche formation and promote bone metastasis of tumor cells. Theranostics. 2021;11:1429-1445. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 69] [Cited by in RCA: 217] [Article Influence: 54.3] [Reference Citation Analysis (0)] |
| 25. | Chen B, Sang Y, Song X, Zhang D, Wang L, Zhao W, Liang Y, Zhang N, Yang Q. Exosomal miR-500a-5p derived from cancer-associated fibroblasts promotes breast cancer cell proliferation and metastasis through targeting USP28. Theranostics. 2021;11:3932-3947. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 163] [Article Influence: 40.8] [Reference Citation Analysis (0)] |
| 26. | Han QF, Li WJ, Hu KS, Gao J, Zhai WL, Yang JH, Zhang SJ. Exosome biogenesis: machinery, regulation, and therapeutic implications in cancer. Mol Cancer. 2022;21:207. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 341] [Article Influence: 113.7] [Reference Citation Analysis (0)] |
| 27. | Tan S, Xia L, Yi P, Han Y, Tang L, Pan Q, Tian Y, Rao S, Oyang L, Liang J, Lin J, Su M, Shi Y, Cao D, Zhou Y, Liao Q. Exosomal miRNAs in tumor microenvironment. J Exp Clin Cancer Res. 2020;39:67. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 126] [Article Influence: 25.2] [Reference Citation Analysis (0)] |
| 28. | Shao H, Im H, Castro CM, Breakefield X, Weissleder R, Lee H. New Technologies for Analysis of Extracellular Vesicles. Chem Rev. 2018;118:1917-1950. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1114] [Cited by in RCA: 1157] [Article Influence: 165.3] [Reference Citation Analysis (0)] |
| 29. | Han J, Zhang Y, Ge P, Dakal TC, Wen H, Tang S, Luo Y, Yang Q, Hua B, Zhang G, Chen H, Xu C. Exosome-derived CIRP: An amplifier of inflammatory diseases. Front Immunol. 2023;14:1066721. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 12] [Reference Citation Analysis (0)] |
| 30. | Sun Z, Shi K, Yang S, Liu J, Zhou Q, Wang G, Song J, Li Z, Zhang Z, Yuan W. Effect of exosomal miRNA on cancer biology and clinical applications. Mol Cancer. 2018;17:147. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 311] [Cited by in RCA: 589] [Article Influence: 84.1] [Reference Citation Analysis (0)] |
| 31. | Fei X, Li Z, Yang D, Kong X, Lu X, Shen Y, Li X, Xie S, Wang J, Zhao Y, Sun Y, Zhang J, Ye Z, Cai Z. Neddylation of Coro1a determines the fate of multivesicular bodies and biogenesis of extracellular vesicles. J Extracell Vesicles. 2021;10:e12153. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 34] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 32. | Lee YJ, Shin KJ, Jang HJ, Ryu JS, Lee CY, Yoon JH, Seo JK, Park S, Lee S, Je AR, Huh YH, Kong SY, Kwon T, Suh PG, Chae YC. GPR143 controls ESCRT-dependent exosome biogenesis and promotes cancer metastasis. Dev Cell. 2023;58:320-334. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 84] [Article Influence: 42.0] [Reference Citation Analysis (0)] |
| 33. | Shinde SR, Mick DU, Aoki E, Rodrigues RB, Gygi SP, Nachury MV. The ancestral ESCRT protein TOM1L2 selects ubiquitinated cargoes for retrieval from cilia. Dev Cell. 2023;58:677-693.e9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 13] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 34. | Hirsova P, Ibrahim SH, Verma VK, Morton LA, Shah VH, LaRusso NF, Gores GJ, Malhi H. Extracellular vesicles in liver pathobiology: Small particles with big impact. Hepatology. 2016;64:2219-2233. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 185] [Cited by in RCA: 187] [Article Influence: 20.8] [Reference Citation Analysis (0)] |
| 35. | Yokoi A, Yoshioka Y, Yamamoto Y, Ishikawa M, Ikeda SI, Kato T, Kiyono T, Takeshita F, Kajiyama H, Kikkawa F, Ochiya T. Malignant extracellular vesicles carrying MMP1 mRNA facilitate peritoneal dissemination in ovarian cancer. Nat Commun. 2017;8:14470. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 176] [Cited by in RCA: 249] [Article Influence: 31.1] [Reference Citation Analysis (0)] |
| 36. | Wang Y, Balaji V, Kaniyappan S, Krüger L, Irsen S, Tepper K, Chandupatla R, Maetzler W, Schneider A, Mandelkow E, Mandelkow EM. The release and trans-synaptic transmission of Tau via exosomes. Mol Neurodegener. 2017;12:5. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 349] [Cited by in RCA: 497] [Article Influence: 62.1] [Reference Citation Analysis (0)] |
| 37. | Chivet M, Javalet C, Hemming F, Pernet-Gallay K, Laulagnier K, Fraboulet S, Sadoul R. Exosomes as a novel way of interneuronal communication. Biochem Soc Trans. 2013;41:241-244. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 93] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 38. | Kowal J, Arras G, Colombo M, Jouve M, Morath JP, Primdal-Bengtson B, Dingli F, Loew D, Tkach M, Théry C. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc Natl Acad Sci U S A. 2016;113:E968-E977. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2360] [Cited by in RCA: 2577] [Article Influence: 286.3] [Reference Citation Analysis (0)] |
| 39. | Pužar Dominkuš P, Stenovec M, Sitar S, Lasič E, Zorec R, Plemenitaš A, Žagar E, Kreft M, Lenassi M. PKH26 labeling of extracellular vesicles: Characterization and cellular internalization of contaminating PKH26 nanoparticles. Biochim Biophys Acta Biomembr. 2018;1860:1350-1361. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 135] [Cited by in RCA: 230] [Article Influence: 32.9] [Reference Citation Analysis (0)] |
| 40. | Mathivanan S, Lim JW, Tauro BJ, Ji H, Moritz RL, Simpson RJ. Proteomics analysis of A33 immunoaffinity-purified exosomes released from the human colon tumor cell line LIM1215 reveals a tissue-specific protein signature. Mol Cell Proteomics. 2010;9:197-208. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 415] [Cited by in RCA: 457] [Article Influence: 28.6] [Reference Citation Analysis (0)] |
| 41. | Ruan Z, Liang Y, Chen Z, Yin J, Li C, Pan P, Zhang Q, Wu J, Luo Z. Enterovirus 71 non-structural protein 3A hijacks vacuolar protein sorting 25 to boost exosome biogenesis to facilitate viral replication. Front Microbiol. 2022;13:1024899. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 15] [Reference Citation Analysis (0)] |
| 42. | Lin H, Zhang R, Wu W, Lei L. miR-4454 Promotes Hepatic Carcinoma Progression by Targeting Vps4A and Rab27A. Oxid Med Cell Longev. 2021;2021:9230435. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 15] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 43. | Kogure T, Lin WL, Yan IK, Braconi C, Patel T. Intercellular nanovesicle-mediated microRNA transfer: a mechanism of environmental modulation of hepatocellular cancer cell growth. Hepatology. 2011;54:1237-1248. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 433] [Cited by in RCA: 441] [Article Influence: 31.5] [Reference Citation Analysis (0)] |
| 44. | Wang S, Xu M, Li X, Su X, Xiao X, Keating A, Zhao RC. Exosomes released by hepatocarcinoma cells endow adipocytes with tumor-promoting properties. J Hematol Oncol. 2018;11:82. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 78] [Cited by in RCA: 127] [Article Influence: 18.1] [Reference Citation Analysis (0)] |
| 45. | Zhang H, Deng T, Liu R, Ning T, Yang H, Liu D, Zhang Q, Lin D, Ge S, Bai M, Wang X, Zhang L, Li H, Yang Y, Ji Z, Wang H, Ying G, Ba Y. CAF secreted miR-522 suppresses ferroptosis and promotes acquired chemo-resistance in gastric cancer. Mol Cancer. 2020;19:43. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 445] [Cited by in RCA: 730] [Article Influence: 146.0] [Reference Citation Analysis (0)] |
| 46. | Bandiera S, Pfeffer S, Baumert TF, Zeisel MB. miR-122--a key factor and therapeutic target in liver disease. J Hepatol. 2015;62:448-457. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 399] [Cited by in RCA: 490] [Article Influence: 49.0] [Reference Citation Analysis (0)] |
| 47. | Xu Y, Xia F, Ma L, Shan J, Shen J, Yang Z, Liu J, Cui Y, Bian X, Bie P, Qian C. MicroRNA-122 sensitizes HCC cancer cells to adriamycin and vincristine through modulating expression of MDR and inducing cell cycle arrest. Cancer Lett. 2011;310:160-169. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 104] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 48. | Girard M, Jacquemin E, Munnich A, Lyonnet S, Henrion-Caude A. miR-122, a paradigm for the role of microRNAs in the liver. J Hepatol. 2008;48:648-656. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 281] [Cited by in RCA: 295] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 49. | Basu S, Bhattacharyya SN. Insulin-like growth factor-1 prevents miR-122 production in neighbouring cells to curtail its intercellular transfer to ensure proliferation of human hepatoma cells. Nucleic Acids Res. 2014;42:7170-7185. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 76] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 50. | Chiba M, Kimura M, Asari S. Exosomes secreted from human colorectal cancer cell lines contain mRNAs, microRNAs and natural antisense RNAs, that can transfer into the human hepatoma HepG2 and lung cancer A549 cell lines. Oncol Rep. 2012;28:1551-1558. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 172] [Cited by in RCA: 200] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 51. | Zeng Z, Li Y, Pan Y, Lan X, Song F, Sun J, Zhou K, Liu X, Ren X, Wang F, Hu J, Zhu X, Yang W, Liao W, Li G, Ding Y, Liang L. Cancer-derived exosomal miR-25-3p promotes pre-metastatic niche formation by inducing vascular permeability and angiogenesis. Nat Commun. 2018;9:5395. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 383] [Cited by in RCA: 708] [Article Influence: 101.1] [Reference Citation Analysis (0)] |
| 52. | Sun H, Meng Q, Shi C, Yang H, Li X, Wu S, Familiari G, Relucenti M, Aschner M, Wang X, Chen R. Hypoxia-Inducible Exosomes Facilitate Liver-Tropic Premetastatic Niche in Colorectal Cancer. Hepatology. 2021;74:2633-2651. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 94] [Article Influence: 23.5] [Reference Citation Analysis (0)] |
| 53. | Xie Z, Gao Y, Ho C, Li L, Jin C, Wang X, Zou C, Mao Y, Li Q, Fu D, Zhang YF. Exosome-delivered CD44v6/C1QBP complex drives pancreatic cancer liver metastasis by promoting fibrotic liver microenvironment. Gut. 2022;71:568-579. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 130] [Cited by in RCA: 127] [Article Influence: 42.3] [Reference Citation Analysis (0)] |
| 54. | Xie KL, Zhang YG, Liu J, Zeng Y, Wu H. MicroRNAs associated with HBV infection and HBV-related HCC. Theranostics. 2014;4:1176-1192. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 63] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 55. | Wang X, He Y, Mackowiak B, Gao B. MicroRNAs as regulators, biomarkers and therapeutic targets in liver diseases. Gut. 2021;70:784-795. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 139] [Cited by in RCA: 301] [Article Influence: 75.3] [Reference Citation Analysis (0)] |
| 56. | Hochreuter MY, Dall M, Treebak JT, Barrès R. MicroRNAs in non-alcoholic fatty liver disease: Progress and perspectives. Mol Metab. 2022;65:101581. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 92] [Reference Citation Analysis (0)] |
| 57. | Sarnow P, Sagan SM. Unraveling the Mysterious Interactions Between Hepatitis C Virus RNA and Liver-Specific MicroRNA-122. Annu Rev Virol. 2016;3:309-332. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 45] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 58. | Wang S, Qiu L, Yan X, Jin W, Wang Y, Chen L, Wu E, Ye X, Gao GF, Wang F, Chen Y, Duan Z, Meng S. Loss of microRNA 122 expression in patients with hepatitis B enhances hepatitis B virus replication through cyclin G(1) -modulated P53 activity. Hepatology. 2012;55:730-741. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 213] [Cited by in RCA: 209] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 59. | Pirola CJ, Fernández Gianotti T, Castaño GO, Mallardi P, San Martino J, Mora Gonzalez Lopez Ledesma M, Flichman D, Mirshahi F, Sanyal AJ, Sookoian S. Circulating microRNA signature in non-alcoholic fatty liver disease: from serum non-coding RNAs to liver histology and disease pathogenesis. Gut. 2015;64:800-812. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 373] [Cited by in RCA: 445] [Article Influence: 44.5] [Reference Citation Analysis (0)] |
| 60. | Gu Y, Wei X, Sun Y, Gao H, Zheng X, Wong LL, Jin L, Liu N, Hernandez B, Peplowska K, Zhao X, Zhan QM, Feng XH, Tang ZY, Ji J. miR-192-5p Silencing by Genetic Aberrations Is a Key Event in Hepatocellular Carcinomas with Cancer Stem Cell Features. Cancer Res. 2019;79:941-953. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 67] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 61. | Wang B, Majumder S, Nuovo G, Kutay H, Volinia S, Patel T, Schmittgen TD, Croce C, Ghoshal K, Jacob ST. Role of microRNA-155 at early stages of hepatocarcinogenesis induced by choline-deficient and amino acid-defined diet in C57BL/6 mice. Hepatology. 2009;50:1152-1161. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 255] [Cited by in RCA: 249] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 62. | Zhang Y, Wei W, Cheng N, Wang K, Li B, Jiang X, Sun S. Hepatitis C virus-induced up-regulation of microRNA-155 promotes hepatocarcinogenesis by activating Wnt signaling. Hepatology. 2012;56:1631-1640. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 235] [Cited by in RCA: 257] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 63. | Calvente CJ, Tameda M, Johnson CD, Del Pilar H, Lin YC, Adronikou N, De Mollerat Du Jeu X, Llorente C, Boyer J, Feldstein AE. Neutrophils contribute to spontaneous resolution of liver inflammation and fibrosis via microRNA-223. J Clin Invest. 2019;129:4091-4109. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 153] [Cited by in RCA: 195] [Article Influence: 32.5] [Reference Citation Analysis (0)] |
| 64. | Li M, He Y, Zhou Z, Ramirez T, Gao Y, Ross RA, Cao H, Cai Y, Xu M, Feng D, Zhang P, Liangpunsakul S, Gao B. MicroRNA-223 ameliorates alcoholic liver injury by inhibiting the IL-6-p47(phox)-oxidative stress pathway in neutrophils. Gut. 2017;66:705-715. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 137] [Cited by in RCA: 186] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 65. | He Y, Hwang S, Cai Y, Kim SJ, Xu M, Yang D, Guillot A, Feng D, Seo W, Hou X, Gao B. MicroRNA-223 Ameliorates Nonalcoholic Steatohepatitis and Cancer by Targeting Multiple Inflammatory and Oncogenic Genes in Hepatocytes. Hepatology. 2019;70:1150-1167. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 120] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 66. | Wong QW, Lung RW, Law PT, Lai PB, Chan KY, To KF, Wong N. MicroRNA-223 is commonly repressed in hepatocellular carcinoma and potentiates expression of Stathmin1. Gastroenterology. 2008;135:257-269. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 337] [Cited by in RCA: 365] [Article Influence: 21.5] [Reference Citation Analysis (0)] |
| 67. | Jimenez Calvente C, Del Pilar H, Tameda M, Johnson CD, Feldstein AE. MicroRNA 223 3p Negatively Regulates the NLRP3 Inflammasome in Acute and Chronic Liver Injury. Mol Ther. 2020;28:653-663. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 82] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 68. | Ogunwobi OO, Harricharran T, Huaman J, Galuza A, Odumuwagun O, Tan Y, Ma GX, Nguyen MT. Mechanisms of hepatocellular carcinoma progression. World J Gastroenterol. 2019;25:2279-2293. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 116] [Cited by in RCA: 170] [Article Influence: 28.3] [Reference Citation Analysis (0)] |
| 69. | Jenne CN, Kubes P. Immune surveillance by the liver. Nat Immunol. 2013;14:996-1006. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 619] [Cited by in RCA: 781] [Article Influence: 65.1] [Reference Citation Analysis (0)] |
| 70. | Tsuchida T, Friedman SL. Mechanisms of hepatic stellate cell activation. Nat Rev Gastroenterol Hepatol. 2017;14:397-411. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1221] [Cited by in RCA: 2002] [Article Influence: 250.3] [Reference Citation Analysis (0)] |
| 71. | Lua I, Li Y, Zagory JA, Wang KS, French SW, Sévigny J, Asahina K. Characterization of hepatic stellate cells, portal fibroblasts, and mesothelial cells in normal and fibrotic livers. J Hepatol. 2016;64:1137-1146. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 122] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 72. | Iwaisako K, Jiang C, Zhang M, Cong M, Moore-Morris TJ, Park TJ, Liu X, Xu J, Wang P, Paik YH, Meng F, Asagiri M, Murray LA, Hofmann AF, Iida T, Glass CK, Brenner DA, Kisseleva T. Origin of myofibroblasts in the fibrotic liver in mice. Proc Natl Acad Sci U S A. 2014;111:E3297-E3305. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 309] [Cited by in RCA: 408] [Article Influence: 37.1] [Reference Citation Analysis (0)] |
| 73. | Mederacke I, Hsu CC, Troeger JS, Huebener P, Mu X, Dapito DH, Pradere JP, Schwabe RF. Fate tracing reveals hepatic stellate cells as dominant contributors to liver fibrosis independent of its aetiology. Nat Commun. 2013;4:2823. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 750] [Cited by in RCA: 1055] [Article Influence: 95.9] [Reference Citation Analysis (0)] |
| 74. | Kramann R, Schneider RK, DiRocco DP, Machado F, Fleig S, Bondzie PA, Henderson JM, Ebert BL, Humphreys BD. Perivascular Gli1+ progenitors are key contributors to injury-induced organ fibrosis. Cell Stem Cell. 2015;16:51-66. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 602] [Cited by in RCA: 713] [Article Influence: 64.8] [Reference Citation Analysis (0)] |
| 75. | Li Y, Wang J, Asahina K. Mesothelial cells give rise to hepatic stellate cells and myofibroblasts via mesothelial-mesenchymal transition in liver injury. Proc Natl Acad Sci U S A. 2013;110:2324-2329. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 144] [Cited by in RCA: 166] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 76. | Kisseleva T, Uchinami H, Feirt N, Quintana-Bustamante O, Segovia JC, Schwabe RF, Brenner DA. Bone marrow-derived fibrocytes participate in pathogenesis of liver fibrosis. J Hepatol. 2006;45:429-438. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 360] [Cited by in RCA: 366] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 77. | Amann T, Bataille F, Spruss T, Mühlbauer M, Gäbele E, Schölmerich J, Kiefer P, Bosserhoff AK, Hellerbrand C. Activated hepatic stellate cells promote tumorigenicity of hepatocellular carcinoma. Cancer Sci. 2009;100:646-653. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 211] [Cited by in RCA: 219] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 78. | Zhang X, Chen F, Huang P, Wang X, Zhou K, Zhou C, Yu L, Peng Y, Fan J, Zhou J, Lu Z, Hu J, Wang Z. Exosome-depleted MiR-148a-3p derived from Hepatic Stellate Cells Promotes Tumor Progression via ITGA5/PI3K/Akt Axis in Hepatocellular Carcinoma. Int J Biol Sci. 2022;18:2249-2260. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 44] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 79. | Wang F, Li L, Piontek K, Sakaguchi M, Selaru FM. Exosome miR-335 as a novel therapeutic strategy in hepatocellular carcinoma. Hepatology. 2018;67:940-954. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 193] [Cited by in RCA: 214] [Article Influence: 30.6] [Reference Citation Analysis (0)] |
| 80. | Zhou Y, Ren H, Dai B, Li J, Shang L, Huang J, Shi X. Hepatocellular carcinoma-derived exosomal miRNA-21 contributes to tumor progression by converting hepatocyte stellate cells to cancer-associated fibroblasts. J Exp Clin Cancer Res. 2018;37:324. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 329] [Cited by in RCA: 356] [Article Influence: 50.9] [Reference Citation Analysis (0)] |
| 81. | Song M, He J, Pan QZ, Yang J, Zhao J, Zhang YJ, Huang Y, Tang Y, Wang Q, Gu J, Li Y, Chen S, Zeng J, Zhou ZQ, Yang C, Han Y, Chen H, Xiang T, Weng DS, Xia JC. Cancer-Associated Fibroblast-Mediated Cellular Crosstalk Supports Hepatocellular Carcinoma Progression. Hepatology. 2021;73:1717-1735. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 217] [Article Influence: 54.3] [Reference Citation Analysis (0)] |
| 82. | Song T, Dou C, Jia Y, Tu K, Zheng X. TIMP-1 activated carcinoma-associated fibroblasts inhibit tumor apoptosis by activating SDF1/CXCR4 signaling in hepatocellular carcinoma. Oncotarget. 2015;6:12061-12079. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 72] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 83. | Fang T, Lv H, Lv G, Li T, Wang C, Han Q, Yu L, Su B, Guo L, Huang S, Cao D, Tang L, Tang S, Wu M, Yang W, Wang H. Tumor-derived exosomal miR-1247-3p induces cancer-associated fibroblast activation to foster lung metastasis of liver cancer. Nat Commun. 2018;9:191. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 407] [Cited by in RCA: 750] [Article Influence: 107.1] [Reference Citation Analysis (0)] |
| 84. | Zheng X, Xu M, Yao B, Wang C, Jia Y, Liu Q. IL-6/STAT3 axis initiated CAFs via up-regulating TIMP-1 which was attenuated by acetylation of STAT3 induced by PCAF in HCC microenvironment. Cell Signal. 2016;28:1314-1324. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 58] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 85. | Zhu GQ, Tang Z, Huang R, Qu WF, Fang Y, Yang R, Tao CY, Gao J, Wu XL, Sun HX, Zhou YF, Song SS, Ding ZB, Dai Z, Zhou J, Ye D, Wu DJ, Liu WR, Fan J, Shi YH. CD36(+) cancer-associated fibroblasts provide immunosuppressive microenvironment for hepatocellular carcinoma via secretion of macrophage migration inhibitory factor. Cell Discov. 2023;9:25. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 189] [Reference Citation Analysis (0)] |
| 86. | Wang SS, Tang XT, Lin M, Yuan J, Peng YJ, Yin X, Shang G, Ge G, Ren Z, Zhou BO. Perivenous Stellate Cells Are the Main Source of Myofibroblasts and Cancer-Associated Fibroblasts Formed After Chronic Liver Injuries. Hepatology. 2021;74:1578-1594. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 47] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 87. | Zhang Z, Li X, Sun W, Yue S, Yang J, Li J, Ma B, Wang J, Yang X, Pu M, Ruan B, Zhao G, Huang Q, Wang L, Tao K, Dou K. Loss of exosomal miR-320a from cancer-associated fibroblasts contributes to HCC proliferation and metastasis. Cancer Lett. 2017;397:33-42. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 166] [Cited by in RCA: 240] [Article Influence: 30.0] [Reference Citation Analysis (0)] |
| 88. | Yugawa K, Yoshizumi T, Mano Y, Itoh S, Harada N, Ikegami T, Kohashi K, Oda Y, Mori M. Cancer-associated fibroblasts promote hepatocellular carcinoma progression through downregulation of exosomal miR-150-3p. Eur J Surg Oncol. 2021;47:384-393. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 77] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 89. | Qi Y, Wang H, Zhang Q, Liu Z, Wang T, Wu Z, Wu W. CAF-Released Exosomal miR-20a-5p Facilitates HCC Progression via the LIMA1-Mediated β-Catenin Pathway. Cells. 2022;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 31] [Reference Citation Analysis (0)] |
| 90. | Zhou Y, Tang W, Zhuo H, Zhu D, Rong D, Sun J, Song J. Cancer-associated fibroblast exosomes promote chemoresistance to cisplatin in hepatocellular carcinoma through circZFR targeting signal transducers and activators of transcription (STAT3)/ nuclear factor -kappa B (NF-κB) pathway. Bioengineered. 2022;13:4786-4797. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 58] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 91. | Lu L, Huang J, Mo J, Da X, Li Q, Fan M, Lu H. Exosomal lncRNA TUG1 from cancer-associated fibroblasts promotes liver cancer cell migration, invasion, and glycolysis by regulating the miR-524-5p/SIX1 axis. Cell Mol Biol Lett. 2022;27:17. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 94] [Article Influence: 31.3] [Reference Citation Analysis (0)] |
| 92. | Chang KS, Ng PN, Lee MM, Chan SJ. Sexual maturation of chinese boys in Hong Kong. Pediatrics. 1966;37:804-811. [PubMed] |
| 93. | Fujiwara N, Nakagawa H, Kudo Y, Tateishi R, Taguri M, Watadani T, Nakagomi R, Kondo M, Nakatsuka T, Minami T, Sato M, Uchino K, Enooku K, Kondo Y, Asaoka Y, Tanaka Y, Ohtomo K, Shiina S, Koike K. Sarcopenia, intramuscular fat deposition, and visceral adiposity independently predict the outcomes of hepatocellular carcinoma. J Hepatol. 2015;63:131-140. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 571] [Cited by in RCA: 566] [Article Influence: 56.6] [Reference Citation Analysis (0)] |
| 94. | Koeck ES, Iordanskaia T, Sevilla S, Ferrante SC, Hubal MJ, Freishtat RJ, Nadler EP. Adipocyte exosomes induce transforming growth factor beta pathway dysregulation in hepatocytes: a novel paradigm for obesity-related liver disease. J Surg Res. 2014;192:268-275. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 140] [Cited by in RCA: 159] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 95. | Liu Y, Tan J, Ou S, Chen J, Chen L. Adipose-derived exosomes deliver miR-23a/b to regulate tumor growth in hepatocellular cancer by targeting the VHL/HIF axis. J Physiol Biochem. 2019;75:391-401. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 79] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 96. | Zhang H, Deng T, Ge S, Liu Y, Bai M, Zhu K, Fan Q, Li J, Ning T, Tian F, Li H, Sun W, Ying G, Ba Y. Exosome circRNA secreted from adipocytes promotes the growth of hepatocellular carcinoma by targeting deubiquitination-related USP7. Oncogene. 2019;38:2844-2859. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 227] [Cited by in RCA: 272] [Article Influence: 45.3] [Reference Citation Analysis (0)] |
| 97. | Viallard C, Larrivée B. Tumor angiogenesis and vascular normalization: alternative therapeutic targets. Angiogenesis. 2017;20:409-426. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 561] [Cited by in RCA: 1016] [Article Influence: 127.0] [Reference Citation Analysis (0)] |
| 98. | Yukawa H, Suzuki K, Aoki K, Arimoto T, Yasui T, Kaji N, Ishikawa T, Ochiya T, Baba Y. Imaging of angiogenesis of human umbilical vein endothelial cells by uptake of exosomes secreted from hepatocellular carcinoma cells. Sci Rep. 2018;8:6765. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 57] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 99. | Shih TC, Tien YJ, Wen CJ, Yeh TS, Yu MC, Huang CH, Lee YS, Yen TC, Hsieh SY. MicroRNA-214 downregulation contributes to tumor angiogenesis by inducing secretion of the hepatoma-derived growth factor in human hepatoma. J Hepatol. 2012;57:584-591. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 147] [Cited by in RCA: 159] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 100. | Fang JH, Zhang ZJ, Shang LR, Luo YW, Lin YF, Yuan Y, Zhuang SM. Hepatoma cell-secreted exosomal microRNA-103 increases vascular permeability and promotes metastasis by targeting junction proteins. Hepatology. 2018;68:1459-1475. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 197] [Cited by in RCA: 264] [Article Influence: 37.7] [Reference Citation Analysis (0)] |
| 101. | Lin XJ, Fang JH, Yang XJ, Zhang C, Yuan Y, Zheng L, Zhuang SM. Hepatocellular Carcinoma Cell-Secreted Exosomal MicroRNA-210 Promotes Angiogenesis In Vitro and In Vivo. Mol Ther Nucleic Acids. 2018;11:243-252. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 118] [Cited by in RCA: 186] [Article Influence: 26.6] [Reference Citation Analysis (0)] |
| 102. | Yokota Y, Noda T, Okumura Y, Kobayashi S, Iwagami Y, Yamada D, Tomimaru Y, Akita H, Gotoh K, Takeda Y, Tanemura M, Murakami T, Umeshita K, Doki Y, Eguchi H. Serum exosomal miR-638 is a prognostic marker of HCC via downregulation of VE-cadherin and ZO-1 of endothelial cells. Cancer Sci. 2021;112:1275-1288. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 92] [Article Influence: 23.0] [Reference Citation Analysis (0)] |
| 103. | Semaan L, Zeng Q, Lu Y, Zhang Y, Zreik MM, Chamseddine MB, Chopp M, Zhang ZG, Moonka D. MicroRNA-214 enriched exosomes from human cerebral endothelial cells (hCEC) sensitize hepatocellular carcinoma to anti-cancer drugs. Oncotarget. 2021;12:185-198. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 4] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 104. | Binnewies M, Roberts EW, Kersten K, Chan V, Fearon DF, Merad M, Coussens LM, Gabrilovich DI, Ostrand-Rosenberg S, Hedrick CC, Vonderheide RH, Pittet MJ, Jain RK, Zou W, Howcroft TK, Woodhouse EC, Weinberg RA, Krummel MF. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat Med. 2018;24:541-550. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2019] [Cited by in RCA: 3834] [Article Influence: 547.7] [Reference Citation Analysis (0)] |
| 105. | Ma L, Hernandez MO, Zhao Y, Mehta M, Tran B, Kelly M, Rae Z, Hernandez JM, Davis JL, Martin SP, Kleiner DE, Hewitt SM, Ylaya K, Wood BJ, Greten TF, Wang XW. Tumor Cell Biodiversity Drives Microenvironmental Reprogramming in Liver Cancer. Cancer Cell. 2019;36:418-430.e6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 390] [Cited by in RCA: 547] [Article Influence: 91.2] [Reference Citation Analysis (0)] |
| 106. | Zhang Q, He Y, Luo N, Patel SJ, Han Y, Gao R, Modak M, Carotta S, Haslinger C, Kind D, Peet GW, Zhong G, Lu S, Zhu W, Mao Y, Xiao M, Bergmann M, Hu X, Kerkar SP, Vogt AB, Pflanz S, Liu K, Peng J, Ren X, Zhang Z. Landscape and Dynamics of Single Immune Cells in Hepatocellular Carcinoma. Cell. 2019;179:829-845.e20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 763] [Cited by in RCA: 1007] [Article Influence: 167.8] [Reference Citation Analysis (0)] |
| 107. | Lu Y, Yang A, Quan C, Pan Y, Zhang H, Li Y, Gao C, Lu H, Wang X, Cao P, Chen H, Lu S, Zhou G. A single-cell atlas of the multicellular ecosystem of primary and metastatic hepatocellular carcinoma. Nat Commun. 2022;13:4594. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 242] [Reference Citation Analysis (0)] |
| 108. | Ju C, Tacke F. Hepatic macrophages in homeostasis and liver diseases: from pathogenesis to novel therapeutic strategies. Cell Mol Immunol. 2016;13:316-327. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 400] [Cited by in RCA: 410] [Article Influence: 45.6] [Reference Citation Analysis (0)] |
| 109. | Wu J, Li J, Salcedo R, Mivechi NF, Trinchieri G, Horuzsko A. The proinflammatory myeloid cell receptor TREM-1 controls Kupffer cell activation and development of hepatocellular carcinoma. Cancer Res. 2012;72:3977-3986. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 163] [Cited by in RCA: 198] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 110. | Chanmee T, Ontong P, Konno K, Itano N. Tumor-associated macrophages as major players in the tumor microenvironment. Cancers (Basel). 2014;6:1670-1690. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 899] [Cited by in RCA: 1196] [Article Influence: 108.7] [Reference Citation Analysis (0)] |
| 111. | Mantovani A, Marchesi F, Malesci A, Laghi L, Allavena P. Tumour-associated macrophages as treatment targets in oncology. Nat Rev Clin Oncol. 2017;14:399-416. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1778] [Cited by in RCA: 2856] [Article Influence: 357.0] [Reference Citation Analysis (0)] |
| 112. | Liu G, Ouyang X, Sun Y, Xiao Y, You B, Gao Y, Yeh S, Li Y, Chang C. The miR-92a-2-5p in exosomes from macrophages increases liver cancer cells invasion via altering the AR/PHLPP/p-AKT/β-catenin signaling. Cell Death Differ. 2020;27:3258-3272. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 67] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 113. | Li W, Xin X, Li X, Geng J, Sun Y. Exosomes secreted by M2 macrophages promote cancer stemness of hepatocellular carcinoma via the miR-27a-3p/TXNIP pathways. Int Immunopharmacol. 2021;101:107585. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 43] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 114. | Tian B, Zhou L, Wang J, Yang P. miR-660-5p-loaded M2 macrophages-derived exosomes augment hepatocellular carcinoma development through regulating KLF3. Int Immunopharmacol. 2021;101:108157. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 28] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 115. | Wang Y, Wang B, Xiao S, Li Y, Chen Q. miR-125a/b inhibits tumor-associated macrophages mediated in cancer stem cells of hepatocellular carcinoma by targeting CD90. J Cell Biochem. 2019;120:3046-3055. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 90] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 116. | Chen H, Tang C, Tan C, Wu F, Li Z, Ji W, Lu L, Xu C, Shen Z, Huang Y. IL-2 Modulates TAMs Derived Exosomal MiRNAs to Ameliorate Hepatocellular Carcinoma Development and Progression. J Oncol. 2022;2022:3445350. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 117. | Zhang J, Shan WF, Jin TT, Wu GQ, Xiong XX, Jin HY, Zhu SM. Propofol exerts anti-hepatocellular carcinoma by microvesicle-mediated transfer of miR-142-3p from macrophage to cancer cells. J Transl Med. 2014;12:279. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 63] [Cited by in RCA: 84] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 118. | Wang L, Yi X, Xiao X, Zheng Q, Ma L, Li B. Exosomal miR-628-5p from M1 polarized macrophages hinders m6A modification of circFUT8 to suppress hepatocellular carcinoma progression. Cell Mol Biol Lett. 2022;27:106. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 57] [Reference Citation Analysis (0)] |
| 119. | Aucher A, Rudnicka D, Davis DM. MicroRNAs transfer from human macrophages to hepato-carcinoma cells and inhibit proliferation. J Immunol. 2013;191:6250-6260. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 171] [Cited by in RCA: 201] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 120. | Wang L, Wang Y, Quan J. Exosomal miR-223 derived from natural killer cells inhibits hepatic stellate cell activation by suppressing autophagy. Mol Med. 2020;26:81. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 47] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 121. | Xiong L, Zhen S, Yu Q, Gong Z. HCV-E2 inhibits hepatocellular carcinoma metastasis by stimulating mast cells to secrete exosomal shuttle microRNAs. Oncol Lett. 2017;14:2141-2146. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 60] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 122. | Tung SL; Boardman DA; Sen M, Letizia M, Peng Q, Cianci N, Dioni L, Carlin LM, Lechler R, Bollati V, Lombardi G, Smyth LA. Regulatory T cell-derived extracellular vesicles modify dendritic cell function. Sci Rep. 2018;8:6065. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 90] [Cited by in RCA: 162] [Article Influence: 23.1] [Reference Citation Analysis (0)] |
| 123. | Granito A, Muratori L, Lalanne C, Quarneti C, Ferri S, Guidi M, Lenzi M, Muratori P. Hepatocellular carcinoma in viral and autoimmune liver diseases: Role of CD4+ CD25+ Foxp3+ regulatory T cells in the immune microenvironment. World J Gastroenterol. 2021;27:2994-3009. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 122] [Cited by in RCA: 131] [Article Influence: 32.8] [Reference Citation Analysis (1)] |
| 124. | Yin Y, Cai X, Chen X, Liang H, Zhang Y, Li J, Wang Z, Zhang W, Yokoyama S, Wang C, Li L, Hou D, Dong L, Xu T, Hiroi T, Yang F, Ji H, Zhang J, Zen K, Zhang CY. Tumor-secreted miR-214 induces regulatory T cells: a major link between immune evasion and tumor growth. Cell Res. 2014;24:1164-1180. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 183] [Cited by in RCA: 232] [Article Influence: 21.1] [Reference Citation Analysis (0)] |
| 125. | Kudo M, Ueshima K, Ikeda M, Torimura T, Tanabe N, Aikata H, Izumi N, Yamasaki T, Nojiri S, Hino K, Tsumura H, Kuzuya T, Isoda N, Yasui K, Aino H, Ido A, Kawabe N, Nakao K, Wada Y, Yokosuka O, Yoshimura K, Okusaka T, Furuse J, Kokudo N, Okita K, Johnson PJ, Arai Y; TACTICS study group. Randomised, multicentre prospective trial of transarterial chemoembolisation (TACE) plus sorafenib as compared with TACE alone in patients with hepatocellular carcinoma: TACTICS trial. Gut. 2020;69:1492-1501. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 221] [Cited by in RCA: 504] [Article Influence: 100.8] [Reference Citation Analysis (0)] |
| 126. | Chan SL, Yeo W, Mo F, Chan AWH, Koh J, Li L, Hui EP, Chong CCN, Lai PBS, Mok TSK, Yu SCH. A phase 2 study of the efficacy and biomarker on the combination of transarterial chemoembolization and axitinib in the treatment of inoperable hepatocellular carcinoma. Cancer. 2017;123:3977-3985. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 23] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 127. | Inoue J, Inazawa J. Cancer-associated miRNAs and their therapeutic potential. J Hum Genet. 2021;66:937-945. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 71] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 128. | Liao W, Du Y, Zhang C, Pan F, Yao Y, Zhang T, Peng Q. Exosomes: The next generation of endogenous nanomaterials for advanced drug delivery and therapy. Acta Biomater. 2019;86:1-14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 161] [Cited by in RCA: 287] [Article Influence: 47.8] [Reference Citation Analysis (0)] |
| 129. | Shi Y, Du L, Lin L, Wang Y. Tumour-associated mesenchymal stem/stromal cells: emerging therapeutic targets. Nat Rev Drug Discov. 2017;16:35-52. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 246] [Cited by in RCA: 349] [Article Influence: 38.8] [Reference Citation Analysis (0)] |
| 130. | Zhang Z, Mi T, Jin L, Li M, Zhanghuang C, Wang J, Tan X, Lu H, Shen L, Long C, Wei G, He D. Comprehensive proteomic analysis of exosome mimetic vesicles and exosomes derived from human umbilical cord mesenchymal stem cells. Stem Cell Res Ther. 2022;13:312. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 46] [Reference Citation Analysis (0)] |
| 131. | Jayaraj R, Raymond G, Krishnan S, Tzou KS, Baxi S, Ram MR, Govind SK, Chandramoorthy HC, Abu-Khzam FN, Shaw P. Clinical Theragnostic Potential of Diverse miRNA Expressions in Prostate Cancer: A Systematic Review and Meta-Analysis. Cancers (Basel). 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 16] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 132. | Zuo Y, Zheng W, Liu J, Tang Q, Wang SS, Yang XS. MiR-34a-5p/PD-L1 axis regulates cisplatin chemoresistance of ovarian cancer cells. Neoplasma. 2020;67:93-101. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 55] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 133. | Lou G, Song X, Yang F, Wu S, Wang J, Chen Z, Liu Y. Exosomes derived from miR-122-modified adipose tissue-derived MSCs increase chemosensitivity of hepatocellular carcinoma. J Hematol Oncol. 2015;8:122. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 387] [Cited by in RCA: 556] [Article Influence: 55.6] [Reference Citation Analysis (0)] |
| 134. | Lou G, Chen L, Xia C, Wang W, Qi J, Li A, Zhao L, Chen Z, Zheng M, Liu Y. MiR-199a-modified exosomes from adipose tissue-derived mesenchymal stem cells improve hepatocellular carcinoma chemosensitivity through mTOR pathway. J Exp Clin Cancer Res. 2020;39:4. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 96] [Cited by in RCA: 216] [Article Influence: 43.2] [Reference Citation Analysis (0)] |
| 135. | Tian S, Zhou X, Zhang M, Cui L, Li B, Liu Y, Su R, Sun K, Hu Y, Yang F, Xuan G, Ma S, Zheng X, Guo C, Shang Y, Wang J, Han Y. Mesenchymal stem cell-derived exosomes protect against liver fibrosis via delivering miR-148a to target KLF6/STAT3 pathway in macrophages. Stem Cell Res Ther. 2022;13:330. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 74] [Reference Citation Analysis (0)] |
| 136. | Lee MJ, Jung J, Na KH, Moon JS, Lee HJ, Kim JH, Kim GI, Kwon SW, Hwang SG, Kim GJ. Anti-fibrotic effect of chorionic plate-derived mesenchymal stem cells isolated from human placenta in a rat model of CCl(4)-injured liver: potential application to the treatment of hepatic diseases. J Cell Biochem. 2010;111:1453-1463. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 104] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 137. | Cargnoni A, Gibelli L, Tosini A, Signoroni PB, Nassuato C, Arienti D, Lombardi G, Albertini A, Wengler GS, Parolini O. Transplantation of allogeneic and xenogeneic placenta-derived cells reduces bleomycin-induced lung fibrosis. Cell Transplant. 2009;18:405-422. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 193] [Cited by in RCA: 186] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 138. | Hyun J, Wang S, Kim J, Kim GJ, Jung Y. MicroRNA125b-mediated Hedgehog signaling influences liver regeneration by chorionic plate-derived mesenchymal stem cells. Sci Rep. 2015;5:14135. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 111] [Cited by in RCA: 110] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 139. | Fonsato V, Collino F, Herrera MB, Cavallari C, Deregibus MC, Cisterna B, Bruno S, Romagnoli R, Salizzoni M, Tetta C, Camussi G. Human liver stem cell-derived microvesicles inhibit hepatoma growth in SCID mice by delivering antitumor microRNAs. Stem Cells. 2012;30:1985-1998. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 141] [Cited by in RCA: 160] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 140. | Nordin JZ, Lee Y, Vader P, Mäger I, Johansson HJ, Heusermann W, Wiklander OP, Hällbrink M, Seow Y, Bultema JJ, Gilthorpe J, Davies T, Fairchild PJ, Gabrielsson S, Meisner-Kober NC, Lehtiö J, Smith CI, Wood MJ, El Andaloussi S. Ultrafiltration with size-exclusion liquid chromatography for high yield isolation of extracellular vesicles preserving intact biophysical and functional properties. Nanomedicine. 2015;11:879-883. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 354] [Cited by in RCA: 485] [Article Influence: 48.5] [Reference Citation Analysis (0)] |
| 141. | Van Deun J, Mestdagh P, Sormunen R, Cocquyt V, Vermaelen K, Vandesompele J, Bracke M, De Wever O, Hendrix A. The impact of disparate isolation methods for extracellular vesicles on downstream RNA profiling. J Extracell Vesicles. 2014;3. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 535] [Cited by in RCA: 702] [Article Influence: 63.8] [Reference Citation Analysis (0)] |
| 142. | Nawaz M, Camussi G, Valadi H, Nazarenko I, Ekström K, Wang X, Principe S, Shah N, Ashraf NM, Fatima F, Neder L, Kislinger T. The emerging role of extracellular vesicles as biomarkers for urogenital cancers. Nat Rev Urol. 2014;11:688-701. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 206] [Cited by in RCA: 236] [Article Influence: 21.5] [Reference Citation Analysis (0)] |