Published online Aug 21, 2023. doi: 10.3748/wjg.v29.i31.4783
Peer-review started: April 4, 2023
First decision: April 12, 2023
Revised: April 29, 2023
Accepted: July 27, 2023
Article in press: July 27, 2023
Published online: August 21, 2023
Processing time: 136 Days and 5.5 Hours
Bioinformatics analysis showed that the expression of the poly(A)-specific ribonuclease (PARN) gene in gastric cancer, head and neck squamous cell carcinoma, melanoma, cervical cancer and lung squamous cell carcinoma tissues was significantly higher than that in normal tissues and was associated with high stage and poor prognosis. The expression of the PARN gene in esophageal cancer (EC) tissue is also significantly higher than that in normal tissues, but the effect of PARN on the proliferation, migration and invasion of EC cells remains unclear.
To investigate the relationship between PARN and the proliferation, migration and invasion of EC cells.
The EC tissues of 91 patients after EC surgery and 63 paired precancerous healthy tissues were collected. PARN mRNA levels were measured using a tissue microarray, and the PARN expression level was evaluated using immunohistochemistry to analyze the relationship between PARN expression and clinicopathologic features as well as the survival and prognosis of patients. In addition, the effects of PARN gene knockout on tumor cell proliferation, invasion and migration were studied by using shRNA during the in vitro culture of EC cell lines Eca-109 and TE-1, and the effects of the PARN gene on tumor growth in vivo were verified by a xenotransplantation nude mice model.
The expression of PARN in EC tissues was higher than that in adjacent normal tissues, and the level of PARN expression was significantly positively correlated with lymphatic metastasis. Patients with high PARN levels had poor overall survival. BIM, IGFBP-5 and p21 levels were significantly increased in the PARN knockout group, while the expression levels of the antiapoptotic proteins Survivin and sTNF-R1 were significantly decreased in the apoptotic antibody array data. In addition, the expression levels of Akt, p-Akt, PIK3CA and CCND1 in the downstream signaling pathway regulating EC progression were significantly decreased. The culture of EC cell lines confirmed that the apoptosis rate of EC cells was significantly increased, the growth and proliferation of tumor cells were significantly inhibited, and the invasion and migration ability of tumor cells were significantly decreased after PARN gene knockout. In vivo experiments of BALB/c nude mice transfected with Eca-109 cells expressing control shRNA (sh-NC) and PARN shRNA (sh-PARN) showed that the tumor volume and weight of nude mice treated with sh-PARN were significantly decreased compared with those of nude mice treated with sh-NC, indicating that PARN knockdown significantly inhibited tumor growth in vivo.
PARN has antiapoptotic effects on EC cells and promotes their proliferation, invasion and migration, which is associated with the development of EC and poor patient prognosis. PARN may become a potential target for the diagnosis, prognosis prediction and treatment of EC.
Core Tip: Bioinformatics analysis showed that the expression of the poly(A)-specific ribonuclease (PARN) gene in gastric cancer, head and neck squamous cell carcinoma, melanoma, cervical cancer and lung squamous cell carcinoma tissues was significantly higher than that in normal tissues and was associated with high stage and poor prognosis. The expression of the PARN gene in esophageal cancer (EC) tissue is also significantly higher than that in normal tissues, but the effect of PARN on the proliferation, migration and invasion of EC cells remains unclear. This study investigated the relationship between PARN and the proliferation, migration and invasion of EC cells.
- Citation: Zhang FW, Xie XW, Chen MH, Tong J, Chen QQ, Feng J, Chen FT, Liu WQ. Poly(A)-specific ribonuclease protein promotes the proliferation, invasion and migration of esophageal cancer cells. World J Gastroenterol 2023; 29(31): 4783-4796
- URL: https://www.wjgnet.com/1007-9327/full/v29/i31/4783.htm
- DOI: https://dx.doi.org/10.3748/wjg.v29.i31.4783
Esophageal cancer (EC) is one of the most common malignancies in the world and the sixth leading cause of cancer-related mortality worldwide[1]. The main pathological types of esophageal carcinoma include esophageal squamous cell carcinoma (ESCC) and esophageal adenocarcinoma. In contrast to Europe and America, Asia has the most ESCC cases, which account for the vast majority of EC cases[2,3]. Currently, the main treatments for EC include surgery, radiotherapy and chemotherapy[4]. Since the clinical symptoms of early EC are not obvious, more than half of patients are in the advanced stage at the time of detection, and only 20% of patients with EC can be treated with surgery. Inoperable EC patients can only use radiotherapy, chemotherapy and other comprehensive treatments. Since it is highly invasive and has a high recurrence and metastasis rate, the prognosis for ESCC patients remains poor despite the use of multidisciplinary therapies[5]. The overall 5-year survival rate for ESCC patients is only 30%-40%. Patients with advanced or metastatic EC have a worse prognosis, with a 5-year overall survival of less than 15 years[2,4,5].
Increasing evidence shows that the resistance of some ECs to radiotherapy and chemotherapy is the cause of relapse and metastasis. Obviously, it is necessary to study the molecular mechanism of EC progression through genomics and proteomics and explore effective cancer biomarkers to predict treatment efficacy and EC patient prognosis.
Bioinformatics analysis from The Cancer Genome Atlas (TCGA) database showed that the expression of the PARN gene in breast cancer, head and neck squamous cell carcinoma, skin cancer, testicular cancer, thymic carcinoma and lung cancer tissues was significantly higher than that in normal tissues and was positively correlated with high stage and poor prognosis. In addition, the expression of the PARN gene in EC tissues in the TCGA database was also significantly higher than that in adjacent tissues. However, the effect of PARN on the proliferation, migration and invasion of EC cells is not clear and is worth further discussion.
Poly(A)-specific ribonuclease (PARN) is a deadenylase enzyme that is present in mammalian cells[6,7]. As a deadenylase, PARN interacts with the cap and the poly(A) tail of mRNA to control the length of the poly(A) tail and regulate gene expression. Therefore, it plays a role in mRNA degradation in the nucleus and cytoplasm[8,9]. Interference in RNA stability is closely related to tumorigenesis and tumor development, and factors that affect RNA stability may become new targets for the treatment of malignant tumors[10]. Recent studies have shown that RNA-degrading enzymes, called RNases, are involved in the development of malignant tumors, so the regulation of mRNA turnover is a promising mechanism[11].
However, the biological function of PARN and its basic molecular mechanism in the carcinogenesis of the esophagus are still unclear and are worthy of further investigation. In the present study, EC tissues and adjacent normal tissues were immunohistochemically stained and analyzed to detect PARN expression. Moreover, we also investigated the clinicopathological characteristics of PARN and explored the value of PARN expression in predicting prognosis. In addition, we investigated the role of PARN knockdown in the biological characteristics of EC cells in vitro and in vivo.
Data on PARN expression in EC cell lines were obtained from the Cancer Cell Line Encyclopedia (CCLE) (www.broadinstitute.org/ccle). Mutation data were obtained from cBioPortal (https://www.cbioportal.org/). The mRNA expression level data of EC patients were downloaded from TCGA Portal (https://tcga-data.nci.nih.gov/tcga/).
Ninety-one EC tissues and 63 healthy mucosa tissues were taken from EC patients who underwent resection at the Second Affiliated Hospital of Guangxi Medical University and the Affiliated Zhujiang Hospital of Southern Medical University from 2017 to 2018. The study ethics were approved by the Research Ethics Committee of the Second Affiliated Hospital of Guangxi Medical University and Zhujiang Hospital Affiliated to Southern Medical University, and written patient consent was obtained from all patients.
Immunohistochemistry (IHC) was conducted as previously described[12]. Briefly, the unstained tissue sections were deparaffinized by xylene and then rehydrated with a graded alcohol series. The sections were placed in EDTA buffer (pH = 8.0) buffer at 95-100 °C for 20 min to retrieve antigens. Then, the sections were incubated with rabbit anti-human PARN primary antibodies at 4 °C (1:100; Abcam, ab188333) overnight and then incubated with secondary antibodies for 1 h at room temperature.
The mRNA transcription levels of PARN in tumor tissues and adjacent normal mucosal tissues were measured using a tissue microarray (TMA).
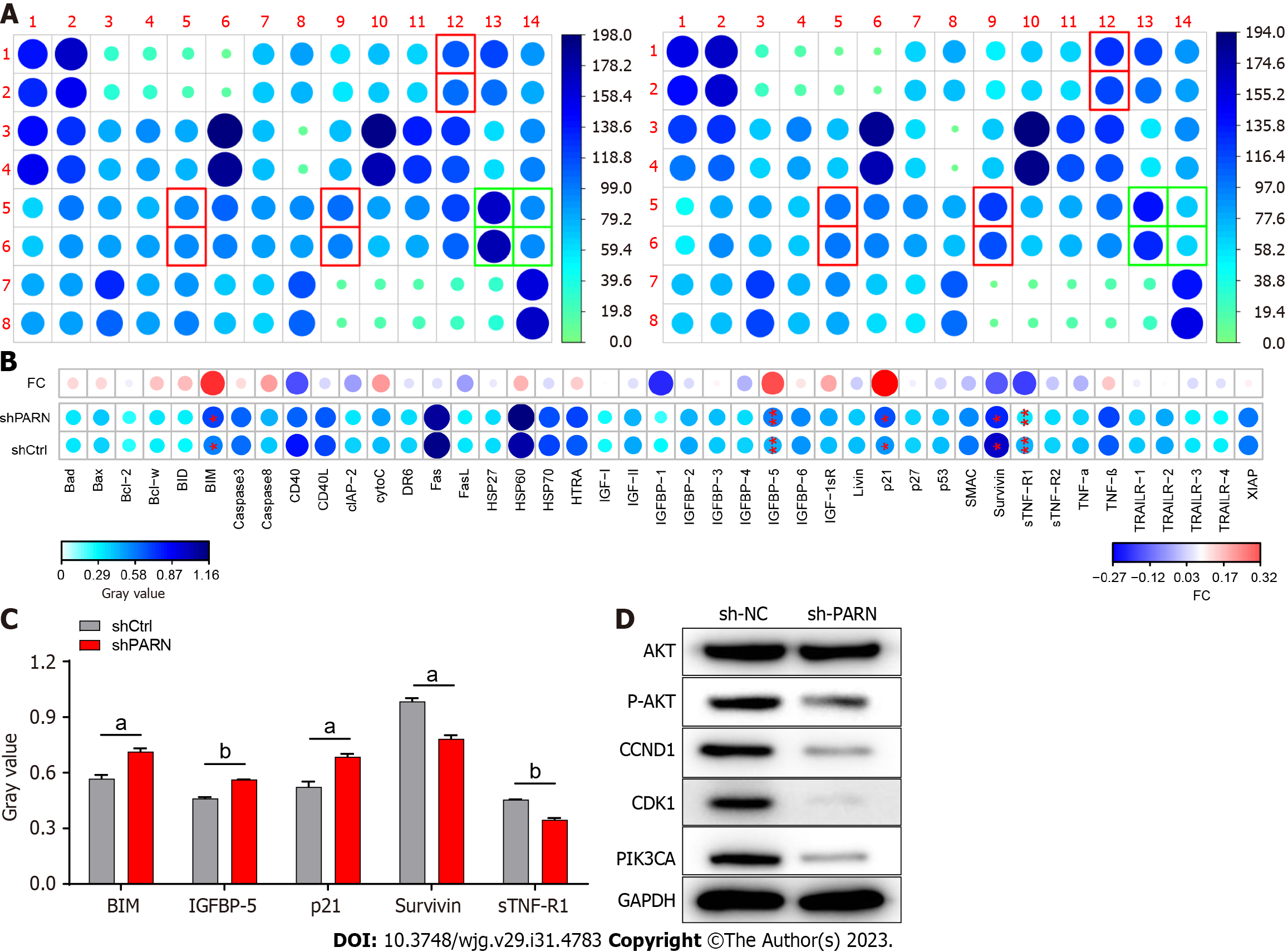
In this research, we obtained the protein-protein interaction by using an online tool, the String Database (STRING, https://string-db.org/). Then, the protein-protein interaction (PPI) we obtained was analyzed by using the software CytoScape (https://cytoscape.org/) and its plug-ins ClueGO and CluePedia. Then, Gene Ontology and Kyoto Encyclopedia of Genes and Genomes analyses were performed. The results of the PPI network and functional and pathway enrichment analyses are shown in Figures 1A and B.
The Homo sapiens apoptosis antibody array kit was purchased from Abcam (ab134001). Total protein samples were prepared as described above and incubated with the array according to the kit instructions. After the addition of chemiluminescence detection reagent, a signal proportional to the protein binding amount was detected. After RNA interference with the PARN gene in Eca-109 cells, the expression levels of apoptosis signaling pathway-related genes and proteins BIM, IGFBP-5 and p21 were evaluated.
We purchased human EC cell lines (Eca-109 and TE-1) from the Chinese Academy of Sciences Cell Bank (Shanghai, China). We cultured all cells in RPMI 1640 (Gibco, Grand Island, NY, United States) supplemented with 10% fetal bovine serum and maintained all cells in a humidified chamber at 37 °C with 5% CO2. Control shRNA, shPARN-1, shPARN-2, and shPARN-3 were constructed by our group (the list of shRNA sequences is shown in Table 1). Viral transduction and selection of stable transfectants were carried out as mentioned above.
| Target number | Target sequence |
| Human-PARN-1 | TATGACACAGCCTCTGAACA |
| Human-PARN-2 | TGGATACTAAATTGATGGCCA |
| Human-PARN-3 | CAACACATCCCTTGCGGAATT |
We collected cells and lysed them in RIPA lysis buffer. Aliquots of protein were then loaded and separated on SDS-PAGE gels and transferred to a polyvinylidene difluoride membrane. After blocking, the membrane was incubated with the appropriate primary antibody, followed by incubation with the corresponding secondary antibody. The primary antibodies and dilution factors were as follows: Rabbit anti-PARN (ab1883331, 1:1000 dilution; Abcam, United States), rabbit anti-CDK1 (ab133327, 1:3000 dilution, Abcam, United States), rabbit anti-PIK3CA (ab40776, 1:1000 dilution, Abcam, United States), rabbit anti-AKT [4685, 1:1000 dilution, Cell Signaling Technology (CST), United States], rabbit anti-CCND1 (2978, 1:1000 dilution, CST, United States), rabbit anti-P-AKT (AF887-sp, 1:500 dilution, R&D Systems, United States), and rabbit anti-GAPDH (AP0063, 1:3000 dilution, Bioworld, United States).
Total RNA was isolated using TRIzol (Sigma-Aldrich, T9424-100 m), and cDNA was obtained with HiScript Q RT SuperMix for quantitative real-time polymerase chain reaction qRT-PCR (+gDNA wiper) (Vazyme, R123-01). PCRs were performed on an ABI 7500 qRT-PCR machine. GAPDH was used as an endogenous control. We assessed the qualified expression by employing the 2-ΔΔCt formula, while statistical analysis was conducted by using the fold change. All primer sequences used in this research are available in Table 2.
| Gene | Sequence (5’-3’) | Tm (°C) | |
| GAPDH | Forward primer | GAAAGCCTGCCGGTGACTAA | 60.32 |
| Reverse primer | GCCCAATACGACCAAATCAGAG | 59.39 | |
| PARN | Forward primer | GCCGCGGAATTCGATTTTAAG | 58.63 |
| Reverse primer | ATCGATGGCGAAGAAGTCGG | 60.25 |
The cell viability of the TE-1 and Eca-109 cell lines was calculated using the Celigo cell counting assay after transfection. Logarithmic-phase transfected cells were collected, and cell suspensions were obtained by trypsin digestion. Then, we seeded the cell suspensions (2000 cells/well) into 96-well plates and cultured them for 5 d. The cell number was recorded, and the cell growth curve was plotted.
Apoptosis and cell cycle analyses were performed by flow cytometry. As mentioned above, transfected TE-1 and Eca-109 cells were cultured in 6-well plates and then collected, washed with D-Hanks buffer and incubated with 1 × binding buffer. The cells were centrifuged and resuspended in 200 μL 1 × binding buffer, and an additional 10 μL Annexin V from the Annexin V Apoptosis Detection kit APC (cat. no. 88-8007; eBioscience; Thermo Fisher Scientific, Inc.) was added. Then, the cells were incubated in the dark for 15 min. The cell cycle distribution phases were detected based on propidium iodide (PI; Sigma, P4170) staining. The samples were analyzed on an easy Cyte HT flow cytometer (Merck Millipore).
In this study, we detected cell migratory abilities by using a wound healing assay. Cells were seeded into 96-well plates (5.0 × 104) and cultured to 90% confluence. Confluent monolayer cells were scratched gently with a 96 Wounding Replicator (VP scientific, VP408FH), and images were captured using a Cellomics ArrayScan VTI (Thermo Scientific) at 0 h, 4 h, 8 h, 24 h and 72 h. The wound area was quantified using a Cellomics ArrayScan HCS Reader (Thermo Scientific).
Transwell plates (Corning, 3422) were used for migration assays, which were carried out according to the manufacturer’s protocol. Cells were seeded in transwell inserts (100 μL, 6.0 × 104/inserts), and RPMI 1640 medium with 30% foetal bovine serum (600 μL) was added to the bottom chamber. Following incubation for 24 h, the inserts were removed and stained with crystal violet. Photos were obtained using an inverted microscope (Olympus I × 73) and analyzed by ImageJ software.
This study was approved by the Guangxi Medical University Ethics Committee. All BALB/c nude mice (male; 4 wk old) were purchased from Charles River (Beijing, China). All BALB/c nude mice (male; 4 wk old) were maintained in a specific-pathogen-free environment. Cells were collected and suspended in D-Hanks solution (1 × 107 cells/mL) and then subcutaneously injected into the right front limb of 20 nude mice (n = 10 for each group). After injection, the size of the tumor was measured with calipers every 4 d for 23 d. On day 23 after injection, the mice were deeply anesthetized with isoflurane gas, and tumor growth and metastases were visualized and analyzed using a whole-body fluorescence imaging system (Berthold Technologies, LB983). After the live imaging experiment, mice were sacrificed by cervical dislocation under anesthesia. Additionally, the tumors were isolated, weighed and photographed. Tumor tissues were saved for further experimentation.
In this study, data are reported as the mean ± SD, and statistical analysis was carried out using GraphPad Prism 8.3 and SPSS 23.0. The Kaplan-Meier log rank test was used for survival curve analysis. Unpaired student’s t test was used to assess statistically significant differences between two groups, and one-way ANOVA with Dunnett’s posttest was used to compare the differences among three or more groups. P values < 0.05 represented a statistically significant difference.
Analysis of EC datasets from TCGA demonstrated that PARN mRNA levels were significantly increased in EC tissues compared with adjacent nontumor tissues (Figure 2B). Then, we extended the detailed annotation process of the preclinical human cancer model by compiling CCLE data and proved that PARN was abnormally upregulated in EC cell lines (Figures 2D and E). We found that PARN expression was upregulated in various EC cell lines, including Eca-109, KYSE450 and TE-1 cells (Figure 2F). In this study, we observed PARN mutations in < 2% of EC patients based on the cBioportal datasets, suggesting that gene mutation is not a major mechanism contributing to the frequent upregulation of PARN in EC patients (Supplementary Figure 1).
The IHC staining and the TMA assay results indicated that PARN protein expression is exceedingly upregulated in EC tissues compared with nontumor tissues (Figure 2A). Additionally, to assess the clinical significance of PARN expression in EC, we investigated the relationship between PARN expression and clinicopathologic data in 91 EC patients (Table 3). After statistical analysis, we found that PARN expression was correlated with lymphatic metastasis (P = 0.028). The expression of PARN was positively correlated with tumor lymph node metastasis (N value). With increasing tumor malignancy, the expression levels of PARN increased (Table 4). However, PARN expression was not correlated with tumor size (P = 0.110), T cell infiltration (P = 0.680) or stage (P = 0.336). To determine the relationship between PARN expression and EC patient clinical prognosis, we performed survival analysis using the survival data of 91 EC patients. The Kaplan-Meier analysis results indicated that high PARN levels were linked with poor survival (Figure 2C and Table 5). In summary, the above results demonstrated that high PARN expression may be implicated in the progression and metastasis of EC and that high PARN expression may predict a worse prognosis.
| Features | No. of patients | PARN expression | P value | |
| Low | High | |||
| All patients | 91 | 48 | 43 | |
| Age (yr) | 0.758 | |||
| < 65 | 45 | 23 | 22 | |
| ≥ 65 | 46 | 25 | 21 | |
| Gender | 0.433 | |||
| Male | 73 | 40 | 33 | |
| Female | 18 | 8 | 10 | |
| Tumor size | 0.110 | |||
| ≤ 5 cm | 46 | 28 | 18 | |
| > 5 cm | 35 | 15 | 20 | |
| T Infiltrate | 0.680 | |||
| T0 | 1 | 1 | 0 | |
| T1 | 3 | 1 | 2 | |
| T2 | 15 | 8 | 7 | |
| T3 | 39 | 22 | 17 | |
| T4 | 10 | 4 | 6 | |
| Lymphatic metastasis (n) | 0.028a | |||
| N0 | 31 | 20 | 11 | |
| N1 | 18 | 10 | 8 | |
| N2 | 11 | 4 | 7 | |
| N3 | 8 | 2 | 6 | |
| Stage | 0.336 | |||
| I | 3 | 2 | 1 | |
| II | 30 | 18 | 12 | |
| III | 33 | 14 | 19 | |
| IV | 2 | 2 | 0 | |
| Lymphoid positive number | 0.164 | |||
| < 1 | 43 | 26 | 17 | |
| ≥ 1 | 46 | 21 | 25 | |
| Grade | 0.516 | |||
| I | 7 | 4 | 3 | |
| II | 49 | 25 | 24 | |
| III | 26 | 16 | 10 | |
| PARN | ||
| Lymphatic metastasis (N) | Spearman correlation analysis | 0.269 |
| Significance (two-tailed) | 0.027a | |
| N | 68 |
| Score type | Score point | Score |
| Positive cell score | No positive signal | 0 (negative) |
| 0% < the proportion of positive cells < 25% | 1 | |
| 25% ≤ the proportion of positive cells < 50% | 2 | |
| 50% ≤ the proportion of positive cells < 75% | 3 | |
| 75% ≤ the proportion of positive cells | 4 | |
| Staining intensity score (the staining intensity of cytoplasm, membrane or nucleus) | No signal color | 0 (negative) |
| Pale yellow | 1 | |
| Brown yellow | 2 | |
| Dark brown | 3 |
Functional and pathway enrichment and differential gene analysis showed that PARN plays a role in tumor apoptosis. To further identify the potential mechanism by which PARN induces apoptosis, a Human Apoptosis Antibody Array kit (ab134001), including 43 human apoptosis-related proteins, was used to investigate the mechanisms of PARN knockdown treatment-induced apoptosis. Among all detected proteins, BIM, IGFBP-5 and p21 were found to be significantly upregulated in the PARN knockdown group, while knockdown of PARN significantly downregulated the expression of the antiapoptotic proteins Surviving and sTNF-R1 (Figures 1A-C). Changes in apoptosis-associated proteins strongly demonstrated that PARN is involved in preventing apoptosis of EC cells by regulating these apoptotic proteins. The original figures of the Human Apoptosis Antibody Array in this study are provided (Supplementary Figure 2). Furthermore, the expression of Akt, p-Akt, CCND1, CDK1 and PIK3CA was downregulated in the sh-PARN group compared with the sh-NC group (Figure 1D).
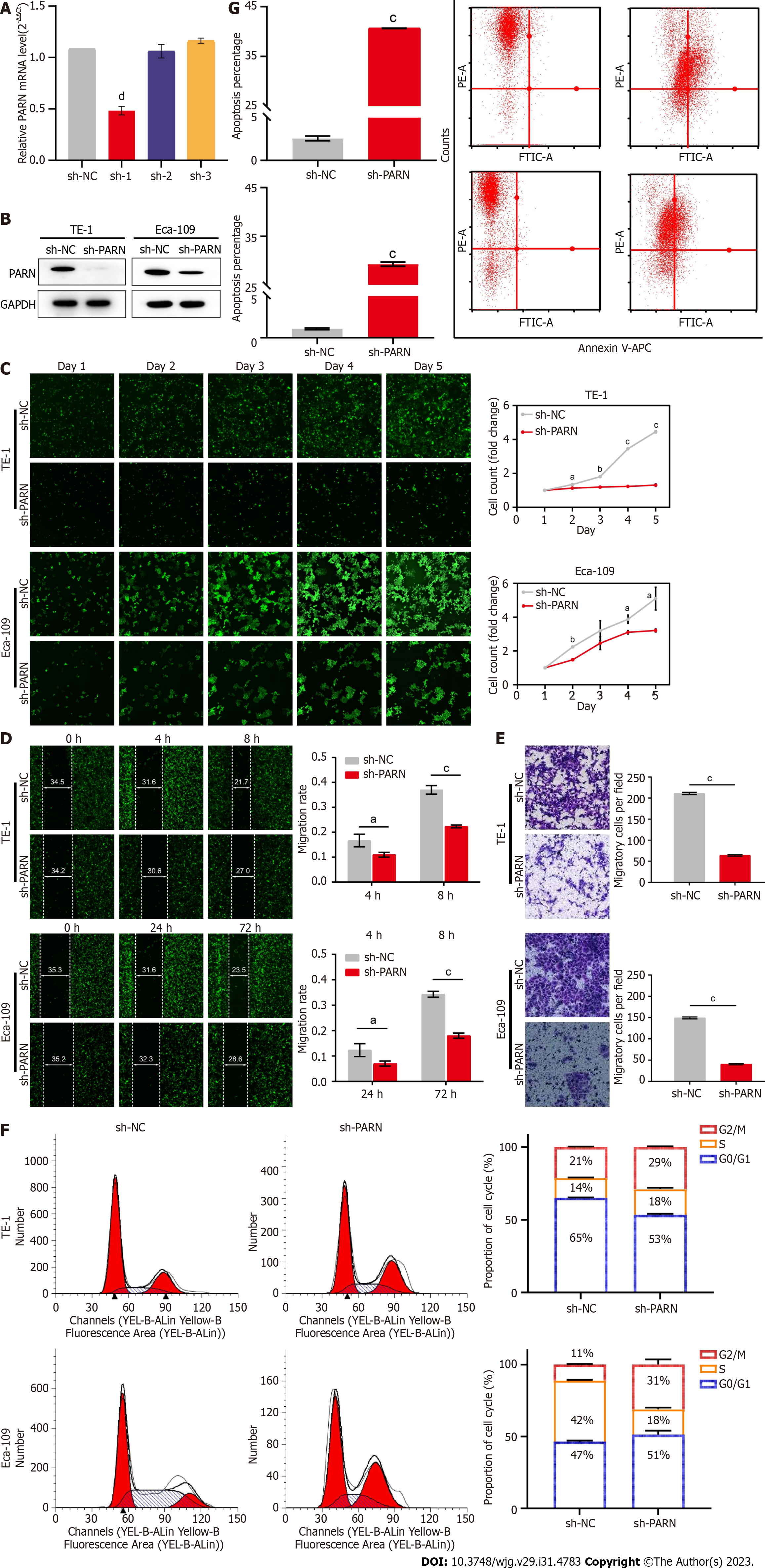
To investigate the effect of PARN on the biological characteristics of esophageal tumor cells, TE-1 and Eca-109 cells were transfected with sh-NC, shPARN-1, shPARN-2, and shPARN-3. The knockdown efficiency of PARN was validated by RT-qPCR. The results showed that shPARN-1 achieved the most efficient knockdown of PARN (Figure 3A), and thereafter, it was used to represent PARN knockdown in all follow-up experiments. The knockdown efficiency was verified at the protein level by western blotting (Figure 3B). The potential effect of PARN on the proliferation of Eca-109 and TE-1 cells was evaluated using the Celigo cell counting method. The results showed that the proliferation rate of Eca-109 and TE-1 cells infected with sh-PARN was significantly reduced compared with that of control cells (Figure 3C). It was confirmed that PARN knockout could significantly inhibit the proliferation of tumor cells. Apoptosis was analyzed by flow cytometry with Annexin V staining and showed a significantly increased percentage of apoptotic cells in sh-PARN-infected TE-1 and Eca-109 cells compared to sh-NC cells (Figure 3G). These results suggest that PARN knockout inhibits cell proliferation by inducing apoptosis. It was also found that compared with the sh-NC group, the sh-PARN group exhibited a significantly increased percentage of cells in the G2/M phase (Figure 3F), suggesting that PARN affects the proliferation of TE-1 and Eca-109 cells by regulating the cell cycle. Together, these findings confirm that PARN promotes EC cell proliferation by blocking cell cycle arrest and apoptosis.
In the wound healing assay, the migration rates were significantly decreased in the sh-PARN group compared to the sh-NC group after lentivirus transfection. The migration abilities of Eca-109 cells in the sh-PARN group (72 h) were 47% lower than those in the sh-NC group (P < 0.001). In TE-1 cells, migration abilities in the sh-PARN group (8 h) were reduced by 40% (P < 0.001). (Figures 3D and E). Transwell invasion assays showed that after lentivirus transfection, the invasion ability of Eca-109 cells in the sh-PARN group was reduced by 73% compared with that in the sh-NC group (P < 0.001). In TE-1 cells, cell invasion was 70% lower in the sh-PARN group than in the sh-NC group (P < 0.001).
The above results showed that knockdown of PARN inhibits cell proliferation and promotes cell apoptosis in vitro. We next wanted to examine the tumor suppression effects of PARN in vivo. To establish the xenograft tumor model, Eca-109 cells that were stably transfected with sh-NC and sh-PARN were subcutaneously implanted into BALB/c nude mice. Tumor weight and volume in the shPARN group were significantly reduced compared with those in the sh-NC-treated group (Figures 4A-C). Additionally, bioluminescence imaging suggested that tumor growth in the sh-PARN group was substantially suppressed (Figures 4D and E), which was similar to the results presented above. The sh-PARN group exhibited significantly repressed tumor development in vivo, suggesting that the tumor-forming capacity of Eca-109 cells in nude mice was significantly accelerated by PARN downregulation. Additionally, Ki67 staining was used to examine the proliferative cells in tumors. IHC data of tumor tissue showed that the number of Ki-67-positive tumor cells in the sh-PARN group markedly declined in comparison to that in the sh-NC group (Figure 4F). These in vivo results confirmed the in vitro results and showed that PARN significantly accelerated EC formation in nude mice.
Tumor progression is a complicated process, and increasing the degradation rate of homologous mRNAs affects the expression of dominant oncogenes, dysfunctional trans-acting factors and/or destruction of specific tumor suppressor genes. Therefore, precise control of mRNA levels is important for the regulation of gene expression[13,14].
As an RNA-processing enzymes, PARN may have an important role in tumor development and progression[15-17]. Currently, there is little evidence indicating a correlation between PARN expression and EC. Our study found that PARN expression levels in EC tissues are clearly higher than those in adjacent normal tissues and are significantly correlated with lymph node metastasis and poor patient survival. Since PARN expression has potential clinical implications in EC, the investigation of its regulatory mechanisms attracted our attention.
The main function of PARN is to cut the mRNA poly(A) tail and produce AMP in the process. The shortening of the eukaryotic poly(A) mRNA tail inhibits mRNA translation and induces transcript renewal; the deregulation of this process is common in cancer. A shortened poly(A) tail destabilizes mRNA and induces degradation. Thus, PARN is considered to be one of the important posttranscriptional regulators in cells. Previous studies have reported that the poly(A) tail of mRNA transcripts is removed by 3’ to 5’ exonucleases (deadenylases), and this process is referred to as the rate-limiting step of mRNA degradation[9,18-21]. After mRNA transcript degradation, the protein expression levels also change accordingly. The levels of these mRNAs are low under normal conditions due to deadenylase activity[22-24].
In our study, compared with normal tissues, EC tissues exhibited significantly increased mRNA levels of PARN, and we also observed this trend in EC cell lines. Interestingly, we found that high PARN expression predicted a poor prognosis in EC patients.
Furthermore, compared with the those of the respective control cell lines, the growth and proliferation of EC cells were significantly inhibited after PARN knockdown. In contrast, it was confirmed that high PARN levels can promote the growth and proliferation of EC cells.
In addition, it was also found that compared with the sh-NC group, the sh-PARN group exhibited a significantly increased percentage of cells in the G2/M phase, suggesting that PARN affects the proliferation of carcinoma cells by regulating the cell cycle.
Escape from apoptosis is beneficial for malignant cell survival and thus could be one of the important mechanisms in cancer pathogenesis[25,26]. Apoptosis involves many biochemical processes that are induced by multiple signaling pathways[27,28]. In our study, PARN knockdown significantly downregulated multiple apoptosis-related proteins (for example, the antiapoptotic proteins Surviving and sTNF-R1) and promoted apoptosis. Therefore, knockdown of the PARN gene promotes apoptosis by regulating apoptotic proteins. In addition, PARN knockdown not only affects the apoptosis rate but also regulates a wide range of downstream signaling factors, including Akt, p-Akt, PIK3CA and CCND1. There has been much evidence suggesting that the PI3K/Akt pathway is one of the most important signaling pathways for cell proliferation, survival, apoptosis and malignant transformation[29,30]. These results suggest that PARN may inhibit tumor cell apoptosis and promote tumor proliferation through the PI3K/Akt pathway in EC. Subsequent wound healing tests and transwell invasion tests confirmed the promoting effect of PARN on the migration and invasion of EC cells. In a BALB/c nude mouse xenograft model, the apoptosis ratio of Eca-109 and TE-1 cells was significantly increased after PARN knockout, and the cell proliferation rate was significantly decreased, as well as the percentage of cells in G2/M phase arrest. It has been confirmed that PARN promotes tumor cell proliferation by blocking cell cycle arrest and apoptosis in EC.
Our study preliminarily concluded that PARN may inhibit in EC cell apoptosis and cell cycle arrest through the PI3K/Akt pathway, thus promoting tumor proliferation, invasion and migration and further accelerating the progression of EC. Therefore, PARN can be used as a prognostic marker and therapeutic target for the diagnosis and treatment of EC.
Esophageal cancer (EC) is a common malignant cancer type and the sixth leading cause of cancer-related mortality worldwide. The main treatment options for esophageal squamous cell carcinoma (ESCC) include surgery, radiotherapy, and chemotherapy. Due to its high invasiveness and high recurrence and metastasis rates, the prognosis of ESCC patients remains poor despite the use of multidisciplinary treatment. The 5-year overall survival rate of ESCC patients is only 30%-40%. However, the prognosis of patients with advanced or metastatic EC is even worse, with a 5-year overall survival rate of less than 15%.
Poly(A)-specific ribonuclease (PARN) is a multifunctional enzyme that plays a crucial role in the occurrence and development of a variety of cancer types. The aim of this study was to explore the relationship between PARN and the proliferation, metastasis and invasion of EC cells to evaluate whether PARN could be a potential biomarker and drug target for the treatment of EC.
The objects of this study are as follows: (1) EC tissues and paired adjacent normal tissues were obtained from 91 patients with EC after surgery; (2) EC lines Eca-109 and TE-1; and (3) Nude mice.
The expression of PARN mRNA was measured using a tissue microarray, and the expression of PARN was also detected using immunohistochemistry. The relationship between PARN expression and clinicopathological features and the survival prognosis of patients was analyzed. The effect of PARN on the proliferation, invasion and migration of Eca-109 and TE-1 EC cells was investigated in vitro by knocking down PARN using shRNA. The effect of PARN on tumor growth in vivo was verified by a nude mouse xenograft model.
Our study found that PARN expression in EC tissues is clearly higher than that in adjacent healthy tissues and is significantly correlated with lymph node metastasis and poor survival. It was confirmed that PARN can promote the growth and proliferation of EC cells. Compared with the control shRNA group, the PARN shRNA group exhibited a significantly increased percentage of cells in the G2/M phase, suggesting that PARN affects the proliferation of carcinoma cells by regulating their cell cycle. Knockdown of the PARN gene promoted apoptosis by regulating apoptotic proteins. Wound healing tests and transwell invasion tests confirmed the promoting effect of PARN on the migration and invasion of EC cells. In the BALB/c nude mouse xenograft model, the apoptosis ratio of Eca-109 and TE-1 cells was significantly increased after PARN knockout, and the cell proliferation rate was significantly decreased, as well as the percentage of cells in G2/M phase arrest.
Our study preliminarily concluded that PARN may inhibit EC cell apoptosis and cell cycle arrest through the PI3K/Akt pathway, thus promoting tumor cell proliferation, invasion and migration and further accelerating the progression of EC.
PARN can be used as a prognostic marker and therapeutic target for the diagnosis and treatment of EC.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Arigami T, Japan; Luyer MDP, Netherlands S-Editor: Wang JJ L-Editor: A P-Editor: Zhao S
| 1. | Kashyap MK, Abdel-Rahman O. Expression, regulation and targeting of receptor tyrosine kinases in esophageal squamous cell carcinoma. Mol Cancer. 2018;17:54. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 69] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 2. | van den Ende T, Ter Veer E, Mali RMA, van Berge Henegouwen MI, Hulshof MCCM, van Oijen MGH, van Laarhoven HWM. Prognostic and Predictive Factors for the Curative Treatment of Esophageal and Gastric Cancer in Randomized Controlled Trials: A Systematic Review and Meta-Analysis. Cancers (Basel). 2019;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 3. | Tepper J, Krasna MJ, Niedzwiecki D, Hollis D, Reed CE, Goldberg R, Kiel K, Willett C, Sugarbaker D, Mayer R. Phase III trial of trimodality therapy with cisplatin, fluorouracil, radiotherapy, and surgery compared with surgery alone for esophageal cancer: CALGB 9781. J Clin Oncol. 2008;26:1086-1092. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 941] [Cited by in RCA: 1050] [Article Influence: 61.8] [Reference Citation Analysis (0)] |
| 4. | Sjoquist KM, Burmeister BH, Smithers BM, Zalcberg JR, Simes RJ, Barbour A, Gebski V; Australasian Gastro-Intestinal Trials Group. Survival after neoadjuvant chemotherapy or chemoradiotherapy for resectable oesophageal carcinoma: an updated meta-analysis. Lancet Oncol. 2011;12:681-692. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1141] [Cited by in RCA: 1261] [Article Influence: 90.1] [Reference Citation Analysis (0)] |
| 5. | van Hagen P, Hulshof MC, van Lanschot JJ, Steyerberg EW, van Berge Henegouwen MI, Wijnhoven BP, Richel DJ, Nieuwenhuijzen GA, Hospers GA, Bonenkamp JJ, Cuesta MA, Blaisse RJ, Busch OR, ten Kate FJ, Creemers GJ, Punt CJ, Plukker JT, Verheul HM, Spillenaar Bilgen EJ, van Dekken H, van der Sangen MJ, Rozema T, Biermann K, Beukema JC, Piet AH, van Rij CM, Reinders JG, Tilanus HW, van der Gaast A; CROSS Group. Preoperative chemoradiotherapy for esophageal or junctional cancer. N Engl J Med. 2012;366:2074-2084. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3288] [Cited by in RCA: 4051] [Article Influence: 311.6] [Reference Citation Analysis (0)] |
| 6. | Balatsos NA, Maragozidis P, Anastasakis D, Stathopoulos C. Modulation of poly(A)-specific ribonuclease (PARN): current knowledge and perspectives. Curr Med Chem. 2012;19:4838-4849. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 23] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 7. | Dejene EA, Li Y, Showkatian Z, Ling H, Seto E. Regulation of poly(a)-specific ribonuclease activity by reversible lysine acetylation. J Biol Chem. 2020;295:10255-10270. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 8] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 8. | Delis C, Krokida A, Tomatsidou A, Tsikou D, Beta RA, Tsioumpekou M, Moustaka J, Stravodimos G, Leonidas DD, Balatsos NA, Papadopoulou KK. AtHESPERIN: a novel regulator of circadian rhythms with poly(A)-degrading activity in plants. RNA Biol. 2016;13:68-82. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 9. | Virtanen A, Henriksson N, Nilsson P, Nissbeck M. Poly(A)-specific ribonuclease (PARN): an allosterically regulated, processive and mRNA cap-interacting deadenylase. Crit Rev Biochem Mol Biol. 2013;48:192-209. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 55] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 10. | Dehan E, Ben-Dor A, Liao W, Lipson D, Frimer H, Rienstein S, Simansky D, Krupsky M, Yaron P, Friedman E, Rechavi G, Perlman M, Aviram-Goldring A, Izraeli S, Bittner M, Yakhini Z, Kaminski N. Chromosomal aberrations and gene expression profiles in non-small cell lung cancer. Lung Cancer. 2007;56:175-184. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 109] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 11. | Valles I, Pajares MJ, Segura V, Guruceaga E, Gomez-Roman J, Blanco D, Tamura A, Montuenga LM, Pio R. Identification of novel deregulated RNA metabolism-related genes in non-small cell lung cancer. PLoS One. 2012;7:e42086. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 47] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 12. | Yu K, Ahrens S, Zhang X, Schiff H, Ramakrishnan C, Fenno L, Deisseroth K, Zhao F, Luo MH, Gong L, He M, Zhou P, Paninski L, Li B. The central amygdala controls learning in the lateral amygdala. Nat Neurosci. 2017;20:1680-1685. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 143] [Cited by in RCA: 129] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 13. | Audic Y, Hartley RS. Post-transcriptional regulation in cancer. Biol Cell. 2004;96:479-498. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 161] [Cited by in RCA: 175] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 14. | Tang Y, Shu G, Yuan X, Jing N, Song J. FOXA2 functions as a suppressor of tumor metastasis by inhibition of epithelial-to-mesenchymal transition in human lung cancers. Cell Res. 2011;21:316-326. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 126] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 15. | Maragozidis P, Karangeli M, Labrou M, Dimoulou G, Papaspyrou K, Salataj E, Pournaras S, Matsouka P, Gourgoulianis KI, Balatsos NA. Alterations of deadenylase expression in acute leukemias: evidence for poly(a)-specific ribonuclease as a potential biomarker. Acta Haematol. 2012;128:39-46. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 32] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 16. | Maragozidis P, Papanastasi E, Scutelnic D, Totomi A, Kokkori I, Zarogiannis SG, Kerenidi T, Gourgoulianis KI, Balatsos NA. Poly(A)-specific ribonuclease and Nocturnin in squamous cell lung cancer: prognostic value and impact on gene expression. Mol Cancer. 2015;14:187. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 29] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 17. | Zhang LN, Yan YB. Depletion of poly(A)-specific ribonuclease (PARN) inhibits proliferation of human gastric cancer cells by blocking cell cycle progression. Biochim Biophys Acta. 2015;1853:522-534. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 26] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 18. | Funakoshi Y, Doi Y, Hosoda N, Uchida N, Osawa M, Shimada I, Tsujimoto M, Suzuki T, Katada T, Hoshino S. Mechanism of mRNA deadenylation: evidence for a molecular interplay between translation termination factor eRF3 and mRNA deadenylases. Genes Dev. 2007;21:3135-3148. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 135] [Cited by in RCA: 145] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 19. | Nousch M, Techritz N, Hampel D, Millonigg S, Eckmann CR. The Ccr4-Not deadenylase complex constitutes the main poly(A) removal activity in C. elegans. J Cell Sci. 2013;126:4274-4285. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 55] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 20. | Wilson T, Treisman R. Removal of poly(A) and consequent degradation of c-fos mRNA facilitated by 3' AU-rich sequences. Nature. 1988;336:396-399. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 482] [Cited by in RCA: 556] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 21. | Wolf J, Passmore LA. mRNA deadenylation by Pan2-Pan3. Biochem Soc Trans. 2014;42:184-187. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 47] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 22. | Cevher MA, Zhang X, Fernandez S, Kim S, Baquero J, Nilsson P, Lee S, Virtanen A, Kleiman FE. Nuclear deadenylation/polyadenylation factors regulate 3' processing in response to DNA damage. EMBO J. 2010;29:1674-1687. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 67] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 23. | Devany E, Zhang X, Park JY, Tian B, Kleiman FE. Positive and negative feedback loops in the p53 and mRNA 3' processing pathways. Proc Natl Acad Sci U S A. 2013;110:3351-3356. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 46] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 24. | Moraes KC, Wilusz CJ, Wilusz J. CUG-BP binds to RNA substrates and recruits PARN deadenylase. RNA. 2006;12:1084-1091. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 134] [Cited by in RCA: 148] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 25. | Chen T, Dai X, Dai J, Ding C, Zhang Z, Lin Z, Hu J, Lu M, Wang Z, Qi Y, Zhang L, Pan R, Zhao Z, Lu L, Liao W, Lu X. AFP promotes HCC progression by suppressing the HuR-mediated Fas/FADD apoptotic pathway. Cell Death Dis. 2020;11:822. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 105] [Article Influence: 21.0] [Reference Citation Analysis (0)] |
| 26. | Zhivotovsky B, Kroemer G. Apoptosis and genomic instability. Nat Rev Mol Cell Biol. 2004;5:752-762. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 209] [Cited by in RCA: 216] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 27. | Fuchs Y, Steller H. Live to die another way: modes of programmed cell death and the signals emanating from dying cells. Nat Rev Mol Cell Biol. 2015;16:329-344. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 384] [Cited by in RCA: 481] [Article Influence: 48.1] [Reference Citation Analysis (0)] |
| 28. | Jeong SY, Seol DW. The role of mitochondria in apoptosis. BMB Rep. 2008;41:11-22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 350] [Cited by in RCA: 504] [Article Influence: 29.6] [Reference Citation Analysis (0)] |
| 29. | Downward J. Mechanisms and consequences of activation of protein kinase B/Akt. Curr Opin Cell Biol. 1998;10:262-267. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1011] [Cited by in RCA: 1016] [Article Influence: 37.6] [Reference Citation Analysis (0)] |
| 30. | Duan C, Bauchat JR, Hsieh T. Phosphatidylinositol 3-kinase is required for insulin-like growth factor-I-induced vascular smooth muscle cell proliferation and migration. Circ Res. 2000;86:15-23. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 110] [Article Influence: 4.4] [Reference Citation Analysis (0)] |