Published online Aug 7, 2023. doi: 10.3748/wjg.v29.i29.4499
Peer-review started: March 28, 2023
First decision: May 12, 2023
Revised: May 23, 2023
Accepted: July 3, 2023
Article in press: July 3, 2023
Published online: August 7, 2023
Processing time: 126 Days and 9.6 Hours
Cancer cells exhibit metabolic reprogramming and bioenergetic alteration, utilizing glucose fermentation for energy production, known as the Warburg effect. However, there are a lack of comprehensive reviews summarizing the me
Core Tip: This review discusses the bioenergetic alteration and metabolic reprogramming in gastrointestinal (GI) cancers, including the interplay between aerobic glycolysis, tricarboxylic acid cycle, and oxidative phosphorylation. The review also highlights potential strategies for targeting bioenergetic regulators for anti-cancer therapy in GI cancers, summarizing the efficacy and challenges of several drugs.
- Citation: Chu YD, Chen CW, Lai MW, Lim SN, Lin WR. Bioenergetic alteration in gastrointestinal cancers: The good, the bad and the ugly. World J Gastroenterol 2023; 29(29): 4499-4527
- URL: https://www.wjgnet.com/1007-9327/full/v29/i29/4499.htm
- DOI: https://dx.doi.org/10.3748/wjg.v29.i29.4499
Cells require energy to carry out their functions, and the most common form of cellular energy is adenosine triphosphate (ATP). This energy is typically produced by oxidative phosphorylation (OXPHOS) in the mitochondria of normal cells[1]. However, in cancer cells, there is a shift in the way energy is generated. Instead of using OXPHOS, cancer cells use glycolysis, a process that results in increased uptake of glucose and secretion of lactate[2]. This phenomenon is known as the Warburg effect and is observed in many types of cancer[3,4]. By understanding the altered energy metabolism in cancer cells, researchers can gain new insights into cancer cell biology and identify potential targets for cancer therapy.
Glycolysis is the process by which glucose is broken down to produce ATP, and it does not require oxygen (Figure 1). Glucose enters cells through glucose transporters and is converted to glucose-6-phosphate (G6P) by hexokinase (HK). Glucose-6-phosphate isomerase (G6PI) converts G6P to fructose-6-phosphate (F6P), which is used in both the glycolytic pathway to generate pyruvate or lactate and the pentose phosphate pathway (PPP) to produce nucleotides and nicotinamide adenine dinucleotide phosphate (NADPH). Phosphofructokinase-1 (PFK1) converts F6P and fructose-2,6-bisphosphate (F2,6BP), a metabolite from a branch driven by fructose-2,6-biphosphatase 3 (PFKBP3), to fructose-1,6-bisphosphate (F1,6BP), which is further processed by aldolase to generate glyceraldehyde-3-phosphate (G3P) and dihydroxyacetone phosphate (DHAP). G3P is converted by glyceraldehyde 3-phosphate dehydrogenase (GAPDH) to 1,3-bisphosphoglycerate (1,3BPG), which is further converted to 3-phosphoglycerate (3PG) by phosphoglycerate kinase (PGK1). The 3PG is subsequently converted by phosphoglycerate mutase (PGAM) to 2-phosphoglycerate (2PG). The 2PG then serves as a substrate for enolase (ENO) to convert to phosphoenolpyruvate (PEP). Pyruvate kinase isozyme M1/M2 (PKM1/2) catalyzes the conversion of PEP to pyruvate, which can be converted to acetyl-CoA or lactate. This process generates NAD+ from NADH, which is important for the continuation of the glycolysis process. Although glycolysis itself does not require oxygen, the fate of the pyruvate produced by glycolysis depends on the availability of oxygen, and the overall efficiency of ATP production is much higher when oxygen is present[5].
Pyruvate, a product of glycolysis, enters the mitochondria where it is converted to acetyl-CoA. The resulting acetyl-CoA can then enter the tricarboxylic acid (TCA) cycle, also known as the Krebs cycle, which plays a pivotal role in generating ATP through the electron transport chain (ETC). The TCA cycle completes the breakdown of glucose by breaking down acetyl-CoA into carbon dioxide (CO2) and water, releasing energy in the form of NADH and flavin adenine dinucleotide (FADH2). NADH and FADH2 donate their electrons to the ETC at Complex I and II, respectively. The ETC, specifically Complexes I-IV, transfers electrons from NADH and FADH2 to generate a proton gradient across the inner mitochondrial membrane. This gradient is then used by ATP synthase to produce ATP. Complex I, also known as NADH dehydrogenase or NADH ubiquinone oxidoreductase, is the largest of the five mitochondrial complexes and marks the initiation of the ETC[6]. Electrons are transferred from Complex I to coenzyme Q (CoQ) across the inner mitochondrial membrane and then from CoQ to Complex III, although an alternative pathway exists via Complex II, succinate dehydrogenase (SDH)[7,8]. Following reduction of succinate by Complex II, electrons are transported to CoQ and then transferred to Complex III. Complex III and cytochrome c transfer electrons to Complex IV, cytochrome c oxidase (COX). The ETC complexes act as proton pumps, creating an electrochemical gradient across the inner mito
In cancer cells, certain enzymes and molecules involved in the conversion of glucose to energy are upregulated, which provides an attractive target for anti-cancer therapies[14]. Disrupting this process could prevent cancer cells from producing energy and lead to their death. In addition to the upregulation of these enzymes, alterations in certain mitochondrial enzymes and oncometabolites have been identified in cancer cells. Oncometabolites are small molecules that are produced in cancer cells and contribute to their growth and proliferation[15]. These alterations can be caused by genetic and epigenetic changes in the genes involved in energy production[13,16]. Recent research has focused on understanding these bioenergetic alterations in gastrointestinal (GI) cancers, such as esophageal cancer (ESCA), gastric cancer (GC), hepatocellular carcinoma (HCC), cholangiocarcinoma (CCA), pancreatic cancer (PAC), and colorectal cancer (CRC). Understanding these specific metabolic changes in cancer cells can provide insight into developing more effective targeted therapies for GI cancers. In addition to the potential for targeted therapy, these metabolic changes could also serve as biomarkers for cancer diagnosis and prognosis. By identifying alterations in the genes and molecules involved in energy production, clinicians may be able to more accurately diagnose and predict the course of the disease. Overall, understanding the bioenergetic alterations in cancer cells is a promising avenue for developing new therapies and improving cancer diagnosis and treatment. In this review, we summarize the latest findings on bioenergetic alterations in various GI cancers, and discuss the potential therapeutic strategies that target these alterations. Such strategies may include inhibitors of specific enzymes or molecules involved in energy production, as well as interventions aimed at modulating the metabolic environment of cancer cells. Further research in this area could lead to new and more effective treatments for GI cancers.
The process of bioenergetic alteration in cancer involves changes in the way cancer cells generate energy. One well-known component of bioenergetic alteration is the Warburg effect. This phenomenon describes how cancer cells prefer to use glucose fermentation to produce energy even in the presence of oxygen[2]. This process, called aerobic glycolysis, is less efficient than mitochondrial OXPHOS in terms of ATP production[17,18]. However, it has been noted that respiration alone can maintain tumor viability, suggesting that glucose and oxygen must be eliminated to kill cancer cells by depriving them of energy[2]. The underlying mechanisms of the Warburg effect have been investigated for decades. Otto Warburg originally proposed that mitochondrial dysfunction could be responsible for aerobic glycolysis[19]. This theory was later confirmed and explored by another group that demonstrated the Warburg effect could be caused by an imbalance of intracellular pH and mitochondrial ATPase dysfunction[20]. Moreover, it was observed that aerobic glycolysis could be controlled by cascade signaling mediated by growth factors and oncogenes, questioning whether the Warburg effect was a mere bystander in the pathogenesis of cancer[21-24]. It was not until later that the Warburg effect was discovered to be crucial for tumor growth in genetic and pharmacological studies[25,26].
Scientists have been trying to understand why cancer cells prefer aerobic glycolysis to mitochondrial OXPHOS for decades, given that the ATP generated by aerobic glycolysis is much lower than that produced by mitochondrial OXPHOS[27-29]. Recent studies have shed light on this phenomenon. For example, when changes in the cellular environment increase ATP demand through alteration of ATP-dependent membrane activity, aerobic glycolysis increases rapidly and OXPHOS remains unchanged[30]. Another study showed high aerobic glycolysis as a metabolic strategy which cancer cells use to optimally respond to fluctuating energy availability[31]. Together, this literature suggests that the Warburg effect is a metabolic strategy that allows flexibility among cancer cells under an unpredictable tumor microenvironment.
Not all pyruvate produced during glycolysis is converted to lactate. Indeed, a significant amount of pyruvate can enter the TCA cycle for oxidation and further metabolism. The intermediates generated during the TCA cycle, such as NAD+/NADH and NADP+/NADPH, can continue to enter the OXPHOS pathway, which can further generate bioenergy[32,33]. Although the role of the Warburg effect in cancers remains controversial, interfering with tumor metabolism and targeting both aerobic glycolysis and mitochondrial OXPHOS pathways have been shown to be necessary[34-37]. It is evident from current literature that there exists crosstalk between aerobic glycolysis, the TCA cycle, and coupled OXPHOS, suggesting cooperative and competitive roles in cancer. Interestingly, some studies suggest that targeting mitochondrial metabolism alone may not be sufficient to inhibit tumor growth, as cancer cells can redirect their metabolism to rely on other energy sources. In such cases, blocking both the glycolytic and mitochondrial pathways may be necessary to prevent cancer cell growth[34-37]. Therefore, a better understanding of the metabolic pathways in cancer cells and their interactions is required to develop effective cancer therapies.
Although the exact molecular mechanism that triggers the Warburg effect in cancer remains unclear, multiple hypotheses have been proposed, including the involvement of tumor suppressors (e.g., p53) and oncogenes (e.g., PI3K, AKT, mTOR), all of which appear to converge on the role of hypoxia-inducible transcription factors (HIFs), particularly HIF-1. HIF-1 is a transcription factor that regulates cellular responses to oxygen deprivation, and it was initially identified as a protein that is present only under hypoxic conditions[38-41]. However, it was later discovered that HIF-1 can also be stabilized under normoxia in a microenvironment with high lactate concentration[42,43]. Under normal conditions, HIF-1α, a subunit of HIF-1, is targeted for degradation by prolyl hydroxylases (PHDs), which utilize molecular oxygen to hydroxylate HIF-1α, leading to its recognition by the von Hippel-Lindau tumor suppressor (VHL), and degradation via proteasome-mediated pathways[44-47].
HIF-1 regulates the expression of several key glycolytic enzymes, such as glucose transporter-1 (GLUT1), GLUT3, HK, aldolase A (ALDOA), PGK1, PKM1/2, ENO1, pyruvate dehydrogenase kinase (PDKs), and lactate dehydrogenase subunit A (LDHA), by directly promoting their expression[48-54]. This leads to an increased level of pyruvate, the final product of glycolysis. However, it is important to note that cancer cells with high glycolytic activity are not guaranteed to catabolize all pyruvate to lactate, as significant amounts of pyruvate can enter the TCA cycle for oxidation and metabolism. In cancer cells, it is suggested that the HIF-1 induced increased expression of PDKs can inhibit the function of pyruvate dehydrogenase (PDH), which blocks pyruvate entry into the TCA cycle and promotes lactate production. Since HIF-1 also promotes the expression of LDHA, an important subunit of LDH necessary for lactate biosynthesis from pyruvate, it is thought to be crucial in cancers affecting terminal lactate levels[55] (Figure 2). Therefore, HIF-1 plays a significant role in the Warburg effect, which may have implications for cancer diagnosis and treatment. While the precise molecular mechanism behind the Warburg effect remains to be elucidated, the involvement of HIF-1 is clear. Understanding the interplay between HIF-1, glycolysis, and OXPHOS in cancer cells may lead to the development of novel cancer therapies that target both pathways.
The concept of lactate as a metabolic waste product has been revised with the latest findings in lactate metabolism and transport. It is now known that lactate can serve as an alternative fuel for certain types of cells, including cancer cells[56,57]. In cancer, the excess lactate is transported between the intracellular and extracellular matrix by the monocarboxylate transporter family (MCT1-4), which depends on the gradients of the protons and monocarboxylate ions[58,59]. Imported extracellular lactate can be converted to pyruvate via LDH primarily composed by the LDHB subunit[60,61]. In oxidative cancer cells with a functional TCA cycle and OXPHOS, pyruvate can be further converted to acetyl-CoA through PDH, thus linking aerobic glycolysis and OXPHOS[62,63]. It has been demonstrated that HIF-1 and downstream oncometabolite lactate play causal roles in these regulatory events. Therefore, current findings provide a possible explanation for the Warburg effect and crosstalk of bioenergetic homeostatic transition between aerobic glycolysis and OXPHOS observed in cancer. The importance of lactate in cancer metabolism and its potential as a therapeutic target have been recognized by others in the field. Thus, a better understanding of the metabolic pathways and their interactions could lead to the development of new strategies for cancer treatment.
Cancer cells often undergo a metabolic shift characterized by increased glycolysis and decreased mitochondrial respiration, a phenomenon known as the Warburg effect. This metabolic reprogramming has been linked to the activity of HIF-1 under low-oxygen conditions[64,65]. Genetic and epigenetic alterations in HIF-1 regulatory genes contribute to the development of the Warburg effect in cancer. Methylation-induced epigenetic changes can drive transcriptional changes, leading to impaired expression of key enzymes involved in bioenergetic homeostasis. Additionally, mutations in nuclear and mitochondrial genomes may cause a loss of function or decreased expression of glycolytic/OXPHOS enzymes. Therefore, mutations, transcriptional changes, or epigenetic alterations that enhance HIF-1 stability or activity can lead to increased aerobic glycolysis, resembling the Warburg effect (Table 1).
| Cancer type | Gene | Type of change | Consequence | Model | Ref. |
| HCC and CCA | PHD2 | Haplo-deficiency | Stabilized HIF-1 and promoted carcinogenesis and progression of HCC/CCA | Mice | [66,67] |
| HCC | PHD3 | Reduced tumor level | Correlated with elevated levels of HIF-1, aggressive tumor behavior, and a poor prognosis in HCC patients | HCC patient | [68] |
| GC | PHD3 | Reduced tumor level | Correlated negatively with tumor size and stage, as well as HIF-1 and VEGF expression | GC patient | [69,70] |
| GC | PHD2 | Reduced tumor level | Correlated with shortened overall survival | GC patient | [71] |
| CRC | PHD1-3 | Reduced tumor level | Although not correlated with HIF-1 expression, PHD2 was the only factor found to be associated with unfavorable overall survival | CRC patient | [72] |
| PAC | PHD1-3 | Increased tumor level | PHD1-3 expression was elevated, and specifically PHD3 expression was found to be associated with unfavorable overall disease-specific survival | PAC patient | [73] |
| PAC | VHL | Promoter methylation or deletion of VHL | Correlated with decreased VHL expression and poor prognosis | PAC patient | [74] |
| CRC | VHL | VHL mutation | Elevated cytoplasmic expression of HIF-1 in tumors | CRC patient | [75] |
| HCC | VHL | Reduced tumor level | Negative VHL expression was correlated with an unfavorable prognosis | HCC patient | [76] |
Studies have found that alterations in PHD enzymes, which target HIF-1 for degradation, contribute to cancer development and progression. Reduced expression or loss-of-function due to PHD2 mutations lead to constitutive activation of HIF-1 and have been found to stimulate HCC and CC development and progression in mouse models[66,67]. In contrast, decreased PHD1-3 expression correlates with increased HIF-1 and vascular endothelial growth factor (VEGF) levels, invasive tumor behavior, and poor prognosis in certain GI cancers such as HCC[68], GC[69-71], and CRC[72]. Interestingly, the opposite effect has been observed in patients with PAC[73]. Another protein involved in HIF-1 stabilization, VHL, also plays a role in GI cancers. Mutations or promoter methylation within the VHL gene lead to increased cytoplasmic HIF-1 levels and an unfavorable prognosis in patients with PAC and CRC[74,75]. However, the general status of VHL protein expression in GI cancers remains unclear, with the exception of HCC, whose levels have been shown to decrease, and low levels correlate with poor prognosis[76]. Further investigation is needed to determine the impact of mutations, genetic, or epigenetic alterations in these hypoxia-associated enzymes on bioenergetic alterations in GI cancers, since understanding the mechanisms behind the Warburg effect and the role of HIF-1 regulatory genes could potentially provide new therapeutic targets for treating GI cancers.
Cancer development and progression are often accompanied by changes in cellular metabolism that contribute to tumor growth and survival. In addition to genetic and epigenetic alterations in hypoxia-associated regulatory enzymes that promote aerobic glycolysis, emerging evidence suggests that changes in nuclear-encoded genes for enzymes and subunits involved in OXPHOS and the TCA cycle may also play a role in driving the switch to glycolysis and altering bioenergetic homeostasis in cancer. Studies have shown that changes in the expression of key enzymes involved in OXPHOS, such as cytochrome c oxidase (COX) and ATP synthase, as well as the TCA cycle enzymes isocitrate dehydrogenase (IDH), fumarate hydratase (FH), and succinate dehydrogenase (SDH), may contribute to glycolysis transition and cancer progression[77-80]. Furthermore, mutations and copy number alterations in mtDNA have also been identified as important factors in the development and progression of GI cancer by altering bioenergetic homeostasis[81]. These emerging factors and their potential contribution to the complex mechanisms underlying the progression of GI cancer are discussed in more detail in the following sections.
The COX complex, also known as respiratory chain complex IV, is a multi-subunit enzyme complex, consisting of 14 subunits, and a vital component of the final step in the mitochondrial ETC responsible for catalyzing the transfer of electrons from cytochrome c to oxygen, a crucial step in the process of OXPHOS[82]. Recent studies have shown that alterations in the expression of both mtDNA-encoded and nuclear-encoded COX subunits are associated with tumorigenesis, cancer progression, and bioenergetic homeostasis in cancer. In GI cancers, alterations in the expression of the mitochondrial-nuclear encoded subunits of the COX complex have been implicated in driving disease progression. Studies have shown that the overall levels of the COX complex are increased in GI cancers, and higher levels have been associated with poor clinical outcomes[83,84]. Of the three mtDNA-encoded core subunits essential for the basic functions of the COX complex, including MTCO1, MTCO2, and MTCO3[85], MTCO1 is the most frequently investigated in GI cancers (Table 2). In ESCA, MTCO1 expression was found to be elevated but did not correlate with clinicopathological variables or survival[86]. On the other hand, elevated levels of MTCO1 were associated with diffuse GC types, suggesting a link between MTCO1 expression and GC carcinogenesis, de-differentiation, and distant metastasis[87,88]. In contrast, defective MTCO1 expression was observed in patients with HCC and CCA, while MTCO1 levels have been shown to predict postoperative survival in patients with HCC[89,90]. Elevated MTCO3 levels have been observed only in HCC, especially among patients with hepatitis B virus (HBV)-related HCC. This is likely due to the ability of the HBV X protein (HBx) to interact and increase MTCO3 expression[91,92]. Additionally, genetic variants identified within MTCO1 and MTCO3 are associated with increased carcinogenic risk in CRC[93,94], GC[95], and HCC[96], possibly due to reduced COX activity leading to intrinsic proton leak and a reduction in overall bioenergetic production efficiency[93,94]. However, studies on the expression or genetic variation of MTCO2 in GI cancers are relatively few and need further investigation.
| Type | Gene | Type of defect | Consequence | Model | Ref. |
| GC | Full COX complex | Increased expression | Correlated with poor prognosis | GC patient | [83] |
| CRC | Full COX complex | Increased expression | May be involved in the initiation of carcinogenesis, but not in cancer progression | CRC patient | [84] |
| ESCA | MTCO1 | Increased expression | There is no correlation with clinical variables or survival | ESCA patient | [86] |
| GC | MTCO1 | Increased expression | Correlated with gastric tumorigenesis, de-differentiation, and distant metastasis, but showed no significant correlation with prognosis | GC patient | [87,88] |
| HCC | MTCO1 | Reduced expression | Correlated with postoperative prognosis | HCC patient | [89] |
| CCA | MTCO1 | Reduced expression | Reduced MTCO1 correlates with increased VDAC1 expression but not with other clinicopathological factors | CCA patient | [90] |
| HCC | MTCO3 | Increased expression | HBx interacted with MTCO3, leading to an increase in MTCO3 expression levels and an enhancement in OXPHOS activity | Cell line | [91,92] |
| CRC | MTCO1 | Genetic variation | The Gly125Asp substitution in MTCO1 correlated with an increased risk of CRC and caused proton leak in COX | CRC patient | [93,94] |
| GC | MTCO3 | Genetic variation | Polymorphisms at mtDNA positions 9540 and 9548 correlated with an increased risk of GC | GC patient | [95] |
| HCC | MTCO3 | Genetic variation | Polymorphisms at mtDNA position 9545 correlated with an increased risk of HCC | HCC patient | [96] |
| ESCA | COX4I1 | Expression silenced | Promotes alterations in cellular bioenergetics and increases cancer cell aggressiveness | ESCA Cell line | [99] |
| ESCA | COX5B | Expression silenced | Promotes alterations in cellular bioenergetics and increases cancer cell aggressiveness | ESCA Cell line | [99] |
| HCC | COX5B | Increased in tumor | Correlated with prognosis, regulated bioenergetic alterations, and influenced cell proliferation, tumor growth, and migration | HCC patient, cell line, mouse model | [100] |
| CRC | COX5B | Reduced in tumor | Correlated with prognosis, modulated COX activity, and controlled cell proliferation, apoptosis, and response to chemotherapy | CRC patient and cell line | [101,102] |
| CRC | COX4I2 | Increased in tumor | Promoted cell proliferation, migration, tumorigenesis, and angiogenesis | CRC patient and cell line | [103] |
| PAC | COX6C | Increased expression | Modulated COX activity and cell proliferation | PAC cell line | [104] |
| PAC | COX6B2 | Increased in tumor | Correlated with prognosis, and modulated cancer cell metastatic potential, and altered bioenergetic homeostasis | PCA patient and cell line | [105] |
While the three core mtDNA-encoded COX subunits have been extensively studied, 11 nuclear-encoded protein subunits are also required for the full functionality of the COX complex[97]. Of these 11 subunits, six can be replaced by isoforms, leading to heterogeneity in the composition and activity of this large complex[98]. In GI cancers, altered expression of nuclear-encoded COX subunits has been shown to play a crucial role in the switch to glycolysis and the promotion of tumor growth and progression (Table 2). For example, in ESCA, the silencing of COX4I1 and COX5B has been shown to promote bioenergetic changes and increased aggressiveness of ESCA cells in vitro[99]. In HCC and CRC, COX5B levels were found to correlate with prognosis, and changes in COX5B expression were associated with alterations in bioenergetics, cell proliferation, tumor growth, migration, and chemosensitivity. HCC and CRC, however, showed different COX5B expression patterns[100-102]. Similarly in CRC, increased COX4I2 has been shown to promote cell proliferation, migration, tumorigenesis, and angiogenesis[103]. COX6C and COX6B2 were also found to be increased in PAC, with changes in expression levels of COX6C affecting COX activity and cell growth in vitro. Meanwhile, COX6B2 levels were associated with prognosis, metastatic potential in PAC cells, and altered bioenergetic homeostasis[104,105].
The roles of remaining subunits in GI cancer are currently unknown, and studies focusing on the level of nuclear-encoded COX subunit in GI cancer largely suggest that altered expression leads to decreased OXPHOS activity in a Warburg effect-like phenotype. Increased GI cancer growth and/or progression is also suggested. Together, these findings highlight the crucial role COX subunits play in GI cancer progression and underscore the need for continued research. The identification of altered COX subunit expression and function may lead to the development of novel therapeutic targets for the treatment of GI cancers. Therefore, further research on the COX complex and its subunits is needed to fully elucidate their role in GI cancer.
ATP synthase, also known as Complex V, is a crucial mitochondrial protein complex that plays a vital role in cellular ATP synthesis. The F1 beta-catalytic subunit (ATP5F1B) is a critical component that has been extensively studied to find a significant reduction in various cancer types, including GI cancers[106] (Table 3). However, the expression patterns of ATP5F1B in patients with GC remain controversial. While one study reported increased ATP5F1B expression in tumors, correlating with poor prognosis[107], consistent findings from other GI cancer studies indicate that decreased ATP5F1B expression results in reduced ATP production efficiency from OXPHOS and a subsequent shift towards the glycolysis-dependent Warburg effect phenotype[108-111]. These findings highlight the critical role of ATP synthase in GI cancer progression, suggesting that mitochondrial defects in ATP synthesis may contribute to the bioenergetic alterations observed in these cancers.
| Type | Gene | Type of defect | Consequence | Model | Ref. |
| GC | ATP5F1B | Increased in tumor | Higher ATP5B expression correlated with poor prognosis. Over-expression of ATP5F1B increased intracellular and extracellular ATP levels, cell proliferation, migration, and invasion | GC patient, cell line, and xeno-transplantation mouse model | [107] |
| GC | ATP5F1B | Reduced in tumor | Reduced ATP5F1B expression correlated with elevated glycolytic enzyme levels | GC patient | [108] |
| HCC | ATP5F1B | Reduced in tumor | Reduced ATP5F1B expression correlated with impaired OXPHOS | HCC patient | [109,110] |
| ESCA | ATP5F1B | Reduced in tumor | Reduced ATP5F1B expression correlated with elevated glycolytic enzyme levels | ESCA patient | [108] |
| CRC | ATP5F1B | Reduced in tumor | Reduced ATP5F1B expression correlated with poor prognosis in CRC patients | CRC patient | [109] |
| PAC | ATP5F1B | Reduced in tumor | Unknown | PAC patient and cell line | [111] |
| CRC | ATP5F1A | Increased in liver metastasized tumor | Silencing of ATP5F1A inhibited cell invasion and reduced cell proliferation in CRC cancer cells | CRC patient and cell line | [112] |
| CRC | ATP5F1E | Increased in tumor | Higher ATP5E levels correlated with poor prognosis. Silencing of ATP5F1E inhibited cancer cell migration and invasion in vitro, and distal metastasis in vivo | CRC patient, cell line, and tail vein injected mouse model | [113] |
| CRC | ATP5F1D | Increased in liver metastasized tumor | Higher ATP5F1D expression correlated with poor prognosis, and silencing of ATP5F1D inhibited cell invasion | CRC patient and cell line | [112] |
Apart from the F1 beta-subunit, other subunits of the ATP synthase F1 region have been implicated as crucial to CRC carcinogenesis/progression. Interestingly, in contrast to the finding that ATP5F1B generally decreases in tumors, ATP5F1A, ATP5F1E, and ATP5F1D were found to be increased in patients with CRC. Moreover, higher levels correlated with poorer prognosis as well as increased risk of CRC liver metastasis[112,113]. Currently, there are no reports on the expression patterns or role of ATP synthase subunits in CCA. The mechanisms underlying opposing expression patterns in ATP synthase subunits are thus unknown pending further investigation.
To provide more insight into the development of novel therapeutic targets for the treatment of GI cancers, further research on ATP synthase expression and function is necessary. In this regard, potential avenues of research may focus on clarifying the controversial findings regarding ATP5F1B expression patterns in GC and elucidating the mechanisms underlying these opposing expression patterns seen in differing ATP synthase subunits in CRC. Such research may uncover novel therapeutic targets, leading to improved treatment outcomes.
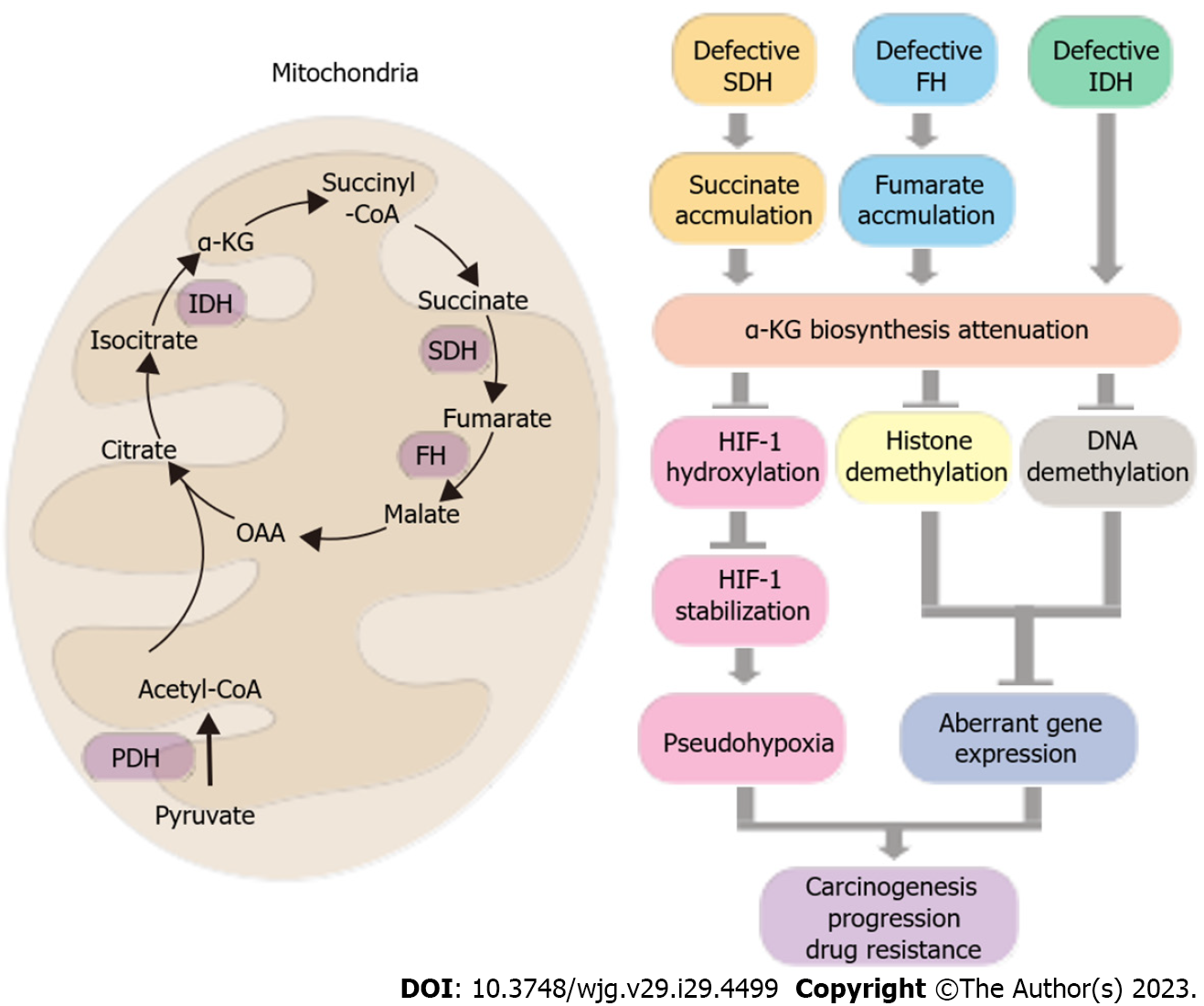
Fumarate and succinate are critical metabolites that are produced during the TCA cycle, which is an essential process for energy production in cells. While these metabolites are important for normal cellular function, they have been shown to act as oncometabolites in various types of cancer by inducing pseudohypoxia[114]. Specifically, aberrant fumarate and succinate accumulation resulting from mutations or abnormal expression in FH and SDH, respectively, can impede the production of α-ketoglutarate in the TCA cycle, which is a key substrate in tumor suppression pathways. Similarly, mutations in IDH enzymes, which are responsible for α-ketoglutarate synthesis, can directly reduce the levels of α-ketoglutarate. This reduction in α-ketoglutarate can limit the availability of substrate for the hydroxylation of HIF-1 by PHDs for subsequent degradation by the proteasome. Consequently, stabilized HIFs activate the transcription of genes involved in cancer-related processes such as angiogenesis, glucose metabolism, and cell proliferation, thereby promoting cancer development and progression[114].
In addition to their effects on HIFs, high levels of fumarate and succinate have been shown to cause abnormal methylation of DNA and histones, leading to dysregulation of gene expression and cell function. This is due to attenuation of enzymes responsible for DNA and histone demethylation such as tet-eleven translocation methyl-cytosine dioxygenase (TET) and lysine demethylase (KDM, also known as the Jumonji C domain-containing histone demethylase, JHDM)). Dysregulation of gene expression, increased carcinogenicity, and cancer progression can result from decreased α-ketoglutarate under high fumarate and succinate levels[115,116] (Figure 3).
The FH and SDH enzymes responsible for the catabolism of fumarate and succinate have been implicated as tumor suppressors[117]. Genetic variants in FH or SDH complex subunits, including SDHA, SDHB, SDHC, and SDHD, have been associated with increased risk of certain cancers such as hereditary leiomyomatosis and renal cell cancer (HLRCC)[118,119] as well as paraganglioma and pheochromocytoma[120-123]. Although there is limited evidence involving genetic mutants of FH or SDH complex subunit genes in GI cancer, an unusual mutation of the FH gene was found to be associated with the development of gastric leiomyoma following cutaneous and uterine leiomyomatosis[124]. Except for loss-of-function mutations, some researchers have revealed FH and SDH complex subunit gene single nucleotide polymorphisms (SNP) in patients with HCC and CRC[125,126]. Interestingly, FH was found to be downregulated in HCC patients with portal vein thrombosis due to currently unknown underlying mechanisms[127]. However, the role of FH and SDH in GI cancer remains largely unknown. Further investigation is thus necessary.
Understanding the role of oncometabolites in GI cancer could provide valuable insights into the development of novel therapeutic targets for the treatment of these cancers. Further research should be conducted to investigate the potential roles of FH and SDH in the development and progression of GI cancer and explore the possible therapeutic targets associated with the regulation of these enzymes. By gaining a better understanding of oncometabolites in GI cancer, we may be able to develop more effective therapies and improve patient outcome.
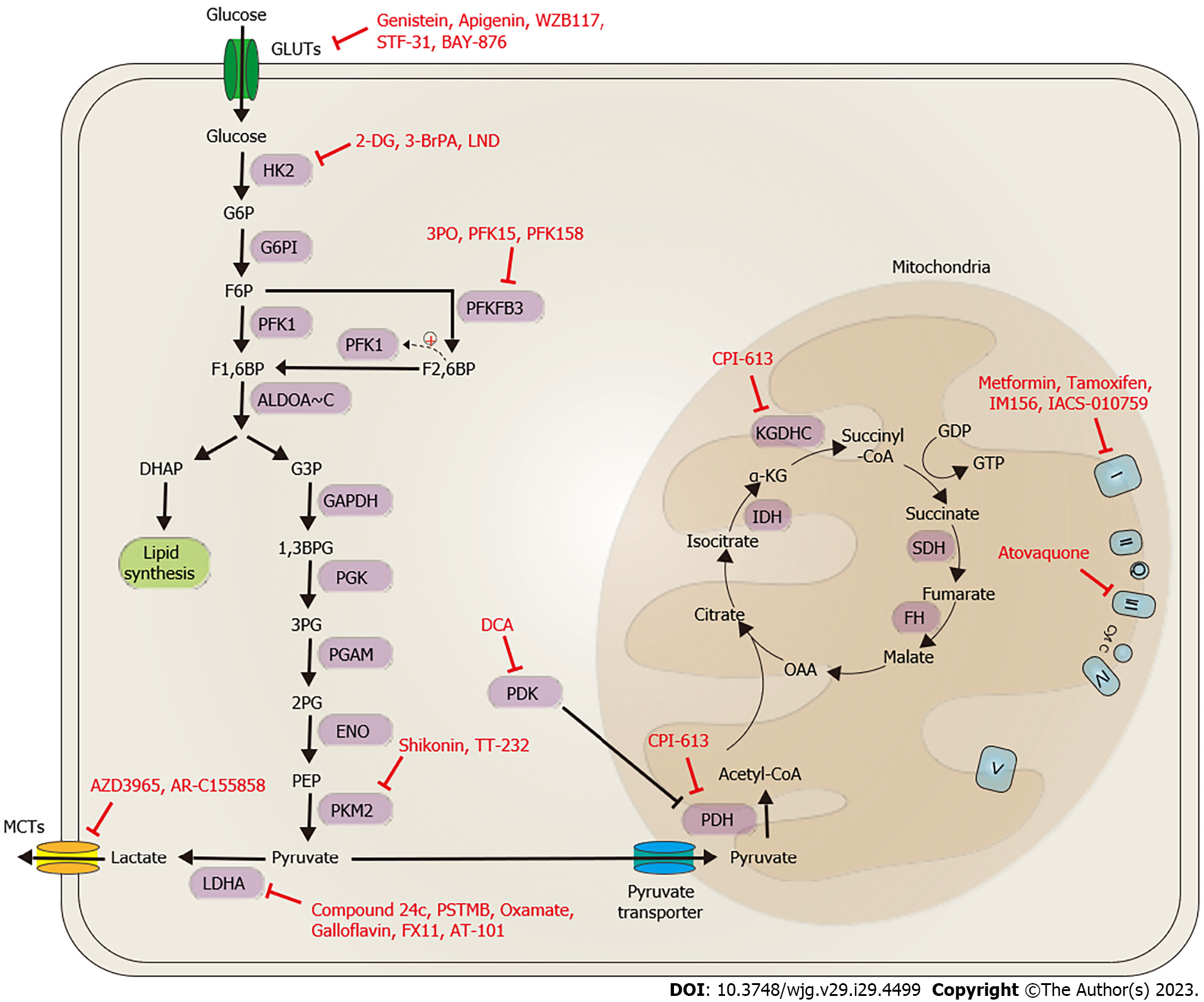
Our current understanding of metabolic reprogramming and bioenergetic alterations in cancer has led to the emergence of several potential drugs that target the bioenergetics of cancer cells, offering a promising avenue for anti-cancer therapy. These drugs can be classified into two main categories based on their mode of action: targeting aerobic glycolysis/lactate biosynthesis and transportation, or targeting the TCA cycle and coupled OXPHOS (Figure 4).
To target aerobic glycolysis, several strategies have been developed including blocking glucose import by targeting GLUT1, reducing glycolysis activity by targeting hexokinase 2 (HK2), PKMFB3, and PKM2, inhibiting lactate biosynthesis by targeting LDHA and PDK, and blocking lactate transportation through targeting MCT1/2. Targeting the TCA cycle and OXPHOS involves PDH and mitochondrial complex inhibitors. Several bioenergetic-targeted drugs have provided pre-clinical or clinical evidence in treating GI cancers. Table 4 provides a summary of these drugs. In the following sections, we will discuss the details of such strategies and the drugs used to target bioenergetic regulators during GI cancer therapy.
| Inhibitor | Target | GI model | Consequence | Clinical trial | Ref. |
| Targeting glucose transportation | |||||
| Genistein | HIF1A, GLUT1 and HK2 | GC, ESCA, HCC, CCA, PCA, and CRC cell lines | Inhibited cancer cell proliferation, cell cycle progression, migration, invasion, angiogenesis, stemness, spheroid formation, EMT, and promoted apoptosis | CRC patient, phase I/II (NCT10985763), and PAC patient, phase I/II (NCT02336087, NCT00376948 and NCT00882765) | [131-140] |
| Apigenin | HIF1A, GLUT1 and HK2 | GC, ESCA, HCC, CCA, PCA, and CRC cell lines | Inhibited cancer cell proliferation, colony-forming, cell cycle progression, migration, invasion, angiogenesis, and induced apoptosis | CRC patient, phase II (NCT00609310) | [141-146] |
| WZB117 | GLUT1 | HCC, CCA, PAC, and CRC cell lines, and xenograft models | Reduced glucose uptake, inhibits cell proliferation, and invasion, and enhanced chemosensitivity | None in GI cancers | [148-151] |
| STF-31 | GLUT1 | PAC and CRC cell lines, and xenograft model | Reduced cancer stem cell properties, such as stemness, and inhibits cell proliferation, viability, and tumor growth | None in GI cancers | [152,153] |
| BAY-876 | GLUT1 | ESCA, PCA, and CRC cell lines, and xenograft mouse models | Reduced cancer cell proliferation, tumor growth, and glucose uptake, while also increased chemosensitivity | None in GI cancers | [154-156] |
| Targeting glucose metabolism | |||||
| 2-Deoxy-D-glucose (2-DG) | HK2 | GC, ESCA, HCC, PAC and CRC cell lines, xenograft models, and rat HCC and hamster PAC models | Inhibited cell proliferation, tumor growth, and promoted chemosensitivity | PAC patient, phase I (NCT00096707) | [159-165] |
| 3-Bromopyruvate (3-BrPA) | HK2 | GC, HCC, PCA, and CRC cell lines, and rabbit, transgenic mouse and xenograft mouse models | Inhibited cellular ATP generation, cell proliferation, and tumor growth. Also induced mitochondrial depolarization, reduced animal serum VEGF levels, and promoted cell death and chemosensitivity | HCC patient, case report[170] | [167-170] |
| Lonidamine (LND) | HK2 | HCC, CCA, and CRC cell lines, hamster CCA model, and GC and CRC patients | Inhibited cell proliferation, migration, invasion, and cell cycle progression. Increased chemosensitivity, patient overall response rate, and duration of disease progression in GC patients. However, was ineffective and toxic in advanced CRC patients | GC patient, phase II[172], CRC patients, phase II[176,177] | [174-179] |
| 3-(3-pyridinyl)-1-(4-pyridinyl)-2-propen-1-one (3PO) | PFKFB3 | HCC, PAC, and CRC cell lines, and transgenic and xenograft mouse models | Inhibited glucose uptake, cell proliferation, tumor growth, angiogenesis, fibrogenesis, and promoted cell death | None in GI cancers | [182-184] |
| 1-(4-pyridinyl)-3-(2-quinolinyl)-2-propen-1-one (PFK15) | PFKFB3 | GC, HCC, PAC, and CRC cell lines, xenograft models, and HCC rat model | Inhibited cell proliferation, migration, invasion, cell cycle progression, tumor growth, and enhanced cell death | None in GI cancers | [185-189] |
| 1-pyridin-4-yl-3-[7-(trifluoromethyl)-quinolin-2-yl]-prop-2-en-1-one (PFK158) | PFKFB3 | None in GI cancers | None in GI cancers | Solid tumor patients, phase I (NCT02044861) | [190] |
| Shikonin | PKM2 | GC, ESCA, HCC, CCA, PCA, and CRC cell lines, and xenograft mouse models | Inhibited cell proliferation, migration, invasion, cell cycle progression, tumor growth, and enhanced cell death | None in GI cancers | [192-197] |
| TT-232 | PKM2 | HCC, PAC, and CRC cell lines, and xenograft mouse models | Inhibited cell proliferation, tumor growth, and enhanced cell death | None in GI cancers | [198-200] |
| Targeting lactate biosynthesis | |||||
| Dichloroacetate (DCA) | PDK | GC, ESCA, HCC, PAC, and CRC cell lines, xenograft models, and B6C3F1 mice | Reduced lactate production, cell proliferation, migration, and increased chemosensitivity. Showed synergistic anti-cancer effects in HCC. However, promoted hepatocarcinogenesis in B6C3F1 mice | CRC patient, phase I (NCT00566410) | [203-207] |
| Compound 24c | LDHA | PAC cell lines, and xenograft model | Suppressed cell proliferation, colony formation, enhanced cell apoptosis, arrested cell at G2 phase, repressed xenograft growth, and re-programmed cancer metabolism, with minimal impact on mouse weight | None in GI cancers | [210] |
| 1-(Phenylseleno)-4-(Trifluoromethyl) Benzene (PSTMB) | LDHA | HCC and CRC cell lines | Inhibited cell proliferation, reduced cell viability, attenuated LDHA activity, lowered lactate levels, and induced mitochondria-mediated apoptosis | None in GI cancers | [211] |
| Oxamate | LDHA | GC, ESCA, HCC, PCA, and CRC cell lines | Suppressed LDHA activity, lactate production, cell proliferation, migration, MMP9 expression, pro-inflammatory cytokines, EMT transition, and AKT/ERK/mTOR signaling pathways, while enhanced apoptosis, senescence, protective autophagy, and metabolic rewiring | None in GI cancers | [212-218] |
| Galloflavin | LDHA | HCC, PCA, and CRC cell lines | Reduced ATPase activity and expression levels of heat shock proteins, inhibited cell proliferation, lactate production, pro-inflammatory cytokines, and EMT transition, while promoting apoptosis and senescence | None in GI cancers | [215,218-220] |
| FX11 | LDHA | HCC, PCA, and CRC cell lines, and xenograft mouse models | FX11 reduced lactate production and ATP levels, suppressed cell proliferation, migration, invasion, and xenograft tumor growth, while enhancing apoptosis. However, in a PCA patient-derived mouse xenograft model, FX11 was only effective in attenuating tumor growth in the presence of mutant TP53 | None in GI cancers | [221-225] |
| Gossypol (AT-101) or its derivatives | LDHA | GC, ESCA, HCC, PAC and CRC cell lines, GC and xenograft mouse models, and ESCA patient | Reduced cell viability, suppressed cell proliferation, migration, and tumor growth, down-regulated cancer stem cell markers CD133, Nanog, LC3, and YAP-1, enhanced apoptosis, protective autophagy. and complete response rate/prognosis | ESCA patient, phase I/II (NCT00561197) | [226-240] |
| Targeting lactate transportation | |||||
| AZD3965 | MCT1/2 | GC, ESCA, HCC, CRC cell lines | Inhibited cell proliferation and tumor growth, while increasing intracellular lactate concentration, TCA-related metabolites, mitochondrial metabolism, and chemosensitivity. Also decreased intracellular pH | None in GI cancers | [242-246] |
| AR-C155858 | MCT1/2 | GC, PAC, and CRC cell lines, and xenograft mouse models | Inhibited cell proliferation, spheroid forming ability, and tumor growth, while decreased glycolysis and increased intracellular lactate concentration, TCA-related metabolites, mitochondrial metabolism, and chemosensitivity | None in GI cancers | [247-249] |
| Targeting mitochondrial OXPHOS | |||||
| Metformin | Mitochondrial complex I | GC, ESCA, HCC, CCA, PAC, and CRC cell lines, xenograft models, and ESCA, HCC, CCA, PCA and CRC patients | Suppressed cell proliferation, migration, cell cycle progression, and tumor growth while increasing chemosensitivity and cell death. Also re-programmed the tumor immune microenvironment in ESCA patients | ESCA patient, phase II (ChiCTR-ICR-15005940), HCC patient, phase I (CTRI/2018/07/014865), CCA patient, phase Ib (NCT0249674), PCA patient, phase II (NCT01210911 and NCT01167738), and CRC patient, phase II (NCT01312467, NCT03047837, and NCT01941953) | [252-265] |
| Tamoxifen | Mitochondrial complex I | GC, ESCA, HCC, CCA, PAC and CRC cell lines, CRC murine model, and ESCA, HCC and PAC patients | Inhibited cell proliferation, tumor growth, metastasis, and increased chemosensitivity. However, no prolonged survival benefits have been observed in HCC patients, and in some cases, there may even be a higher risk of death | ESCA patient, phase I (NCT02513849), PAC patient, phase II[272-274], and HCC patient, phase III (NCT00003424) | [267-273,277] |
| IM156 | Mitochondrial complex I | GC and CRC patients | Considered tolerable in human subjects, with stable disease being the most common response. Combinatorial therapy may be necessary for improved efficacy | GC and CRC patients, phase I (NCT03272256), and PAC patient, phase Ib (NCT05497778) | [278] |
| IACS-010759 | Mitochondrial complex I | PAC cell lines, and CCA, PAC, and CRC patients | Reduced cell viability and generally well tolerated, but may induce neurotoxicity, peripheral neuropathy, and behavioral/physiological changes in mice. Increased blood lactate levels | CCA, PAC, and CRC patient, phase I (NCT03291938) | [279,280] |
| Atovaquone | Mitochondrial complex III | GC, HCC, PAC and CRC cell lines, and xenograft models | Reduced OXPHOS, oxygen consumption rate, cell viability, cell proliferation, and cell cycle progression. Inhibited tumor growth and enhanced cell death | None in GI cancers | [283-285] |
| Targeting TCA cycle | |||||
| CPI-613 | PDH and KGDHC | GC, ESCA, PAC and CRC cell lines, xenograft mouse models, and GC mouse model | Inhibited cell proliferation, cell viability, tumor growth, and metastasis, while increased cell death and chemosensitivity. In PAC patients, also increased the overall response rate | PAC patient, phase I (NCT01835041) and III (NCT03504423), HCC and CCA patients, phase I/II (NCT01766219), and CRC patients, phase I (NCT05070104 and NCT02232152) | [287-291] |
Cancer cells typically rely on increased glucose uptake, a phenomenon known as the Warburg effect, to meet energy requirements, making glucose uptake a promising target for anti-cancer therapy. As a result, GLUT1 has been identified as a potential drug target for blocking glucose uptake. Several GLUT1 inhibitors, including genistein, apigenin, fasentin, WZB117, WZB27, WZB115, STF-31, and BAY-876 have shown an ability to block glucose uptake[14]. Genistein and apigenin are natural compounds belonging to the flavonoid group, and they have been shown to inhibit hypoxia-inducible factor 1A (HIF1A) mRNA and protein expression, which leads to inactivation of downstream genes such as GLUT1 and HK2, thereby attenuating glycolysis activity[128-130]. In GI cancers, these compounds have demonstrated the ability to inhibit cancer cell proliferation, cell cycle progression, colony formation, migration, invasion, angiogenesis, stemness, spheroid formation, EMT, and to enhance cell death[131-146]. Although the majority of evidence pertaining to efficacy comes from in vitro cell-based assays, genistein and apigenin have entered clinical trials as a combination anti-cancer therapy for patients with CRC (NCT10985763 and NCT00609310) and PAC (NCT02336087, NCT00376948 and NCT00882765). Moreover, dietary supplementation with apigenin has been shown to significantly prevent CRC recurrence in a prospective study[147]. Fasentin, WZB117, WZB27, WZB115, STF-31, and BAY-876 are synthetic chemicals with selective activity on GLUT1 inhibition. Fasentin, WZB27, and WZB115 have shown anti-cancer potential in other pre-clinical cancer models, although there is currently little to no research on GI cancers. WZB117 has been shown to reduce glucose uptake, inhibit cell proliferation/invasion, and enhance chemosensitivity in GI cancer cell lines, as well as in xenograft models[148-151]. STF-31 has been implicated in reducing cancer stem cell stemness, cell proliferation, viability, and tumor growth in PAC and CRC cell lines, as well as in xenograft models[152,153]. BAY-876 has been found to inhibit cell proliferation, tumor growth, glucose uptake, and promote chemosensitivity in ESCA, PCA, and CRC cell lines, and in xenograft mouse models[154-156]. Although these findings are promising, WZB117, STF-31, and BAY-876 are not currently in clinical trials for GI cancer. Thus, their safety, dosage, and therapeutic response in GI cancer patients remain to be determined in future studies.
Another strategy to block glycolysis is by targeting glycolytic enzymes or attenuating glycolytic activity. A well-studied example of this strategy is the use of 2-deoxy-D-glucose (2-DG), a glucose molecule with a 2-hydroxyl group replaced by hydrogen. 2-DG is taken up by cells with high glucose uptake ability, such as cancer cells, where it serves as a competitive inhibitor of glucose[157]. Once inside the cell, 2-DG enters the glycolytic pathway and is phosphorylated by HK2 to become 2-DG-6-phosphate (2-DG-6-P), which cannot be further processed by G6P isomerase and therefore accumulates. Accumulated 2-DG-6-P reversely negatively inhibits HK2 activity, leading to a reduction in glycolytic activity. A derivative of 2-DG, fluorodeoxyglucose (18F-FDG), has been extensively employed in positron emission tomography (PET) to visualize the location and status of certain types of cancers[158]. In pre-clinical studies using GI cancer cell lines, as well as xenograft models and rat HCC and hamster PAC models, 2-DG has been shown to inhibit cell proliferation, tumor growth, and promote chemosensitivity[159-165]. Although 2-DG has entered clinical trials for other cancer types, only a phase I trial (NCT00096707) was conducted for patients with PAC, and the safety, dose, and efficacy of 2-DG in treating patients with other GI cancers are unknown.
Several other chemical drugs have been claimed to inhibit HK2 function, but their roles in GI cancers are unclear, with the exception of 3-bromopyruvate (3-BrPA) and lonidamine (LND). 3-BrPA is an analog of both lactate and pyruvate and shows an inhibitory effect on HK2, possibly due to its ability to induce protein alkylation[166,167]. In pre-clinical studies of GI cancers, 3-BrPA has shown its ability to inhibit cellular ATP generation, cell proliferation, tumor growth, induce mitochondrial depolarization, reduce animal serum VEGF levels, and promote cell death and chemosensitivity in GC, HCC, PAC, and CRC cell lines, as well as rabbit, transgenic mice, and xenograft mouse models[167-171]. Therapeutic efficacy and safety were only evaluated in a case report study, providing a safe and tolerable dose of 3-BrPA in patients with fibrolamellar HCC[172].
LND is an indazole derivative that was previously utilized as an anti-spermatogenic agent. In drug re-purposing studies, LND was found to have anti-cancer activity by affecting bioenergetic homeostasis, including the glycolytic pathway, through targeting HK2 via currently unclear mechanisms[173]. LND showed promising therapeutic efficacy by inhibiting cell proliferation, migration, invasion, cell cycle progression, and increasing chemosensitivity in HCC, CCA, and CRC cell lines, as well as in a hamster CCA model[174-179]. Encouraging results were observed in a clinical trial recruiting patients with GC, showing improved overall response rate and duration of disease progression[174]. Reversely, it was reported that administration of LND was ineffective and toxic in clinical trials recruiting patients with CRC[178,179].
Targeting PFKFB3 is another approach to block cancer glycolysis, as it is considered an oncogene in cancers due to its high expression and role in glycolysis[180]. PFKFB3 is activated by multiple cancer-associated stimuli, including cytokines, chemokines, growth factors, and hypoxia, and then participates in glycolysis through catalyzing fructose-6-P to become F2,6BP, which can further positively enhance PFK1 activity and thus accelerate glycolysis[180]. Accordingly, PFKFB3 drugs have been identified and tested in pre-clinical and clinical studies. Among the list of candidate drugs that target PFKFB3, 3-(3-pyridinyl)-1-(4-pyridinyl)-2-propen-1-one (3PO), 1-(4-pyridinyl)-3-(2-quinolinyl)-2-propen-1-one (PFK15), and 1-pyridin-4-yl-3-[7-(trifluoromethyl)-quinolin-2-yl]-prop-2-en-1-one (PFK158) have drawn more attention than others[181]. It was found that 3PO and PFK15 inhibit cell proliferation, reduce tumor growth, attenuate angio
One strategy proposed to inhibit glycolysis activity is to target the last enzyme in the glycolytic pathway –PKM2. PKM2 targeting is based on its glycolysis role as well as aberrant expression in cancer-associated events[191]. While many drugs have shown the ability to inhibit PKM activity, only two, TT-232 and Shikonin, have been confirmed efficacious in pre-clinical studies. Both TT-232 and Shikonin have been found to inhibit GI cancer cell proliferation, migration, invasion, cell cycle progression, and tumor growth, as well as enhance cell death[192-200]. However, the efficacy of these drugs in treating GI cancers is still unclear and requires further investigation. Both drugs have entered clinical trials for specific cancers, showing promise as cancer therapy targets.
As mentioned above, the Warburg effect is a common phenomenon in many cancers for which glycolysis is upregulated even in the presence of oxygen. This results in the accumulation of lactate, which is the last product of glycolysis. The PDK class of enzymes play a key role in deciding whether pyruvate is converted to lactate or enters the TCA cycle. Under hypoxia, PDKs are transcriptionally upregulated by HIF1A in cancers, promoting the inactivation of PDH through PDK-mediated phosphorylation. This leads to elevated lactate biosynthesis, resulting in excessive lactate levels that can promote carcinogenesis or progression[201]. Therefore, targeting PDKs is a potential strategy to inhibit lactate synthesis. Although several candidate drugs that target PDKs have been proposed, dichloroacetate (DCA) has been the most convincing inactivator of PDKs[202]. DCA has been shown in numerous pre-clinical studies on GI cancer to reduce lactate production, cell proliferation, migration, and increase chemosensitivity[203-207]. It has also shown synergistic anti-cancer activity in HCC despite concerns that it may promote hepatic carcinogenesis in B6C3F1 mice[205,208]. Despite promising pre-clinical results, clinical studies are still necessary to determine the efficacy and safety of DCA during cancer therapy. A clinical trial recruiting patients with CRC has been conducted to evaluate DCA as a potential anti-cancer drug (NCT00566410).
In previous studies on lactic acid inhibitors for anti-cancer therapy, the focus has been on inhibiting the enzymes responsible for lactate biosynthesis, namely LDH. TLDH complex composition has been investigated as a crucial factor in determining the fate of lactate biosynthesis or catabolism, and LDHA homo-tetramer (LDH5 or A4) has been considered the most effective complex for lactate biosynthesis. Accordingly, the currently established strategy is to identify LDH inhibitors with high selectivity against LDHA[209]. Although many candidates exist, including small peptides, small interfering RNAs (siRNAs), small chemical molecules, and natural compounds, only a few have progressed towards clinical use in anti-cancer therapy. Compound 24c and 1-(Phenylseleno)-4-(Trifluoromethyl) Benzene (PSTMB) are small compounds that have recently been identified as capable of selectively inhibiting LDHA, suppressing cancer cell aggressiveness, and enhancing cell death in both PCA cells and xenograft mouse models[210] as well as HCC and CRC cells[211]. Notably, Compound 24c has little effect on mouse weight, perhaps due to its relatively strong activity to reprogram metabolic profiling[210]. In contrast, oxamate, galloflavin, and FX11 have a longer history than Compound 24c and PSTMB in targeting LDHA. Pre-clinical evidence shows promise in suppressing GI cancer cell aggressiveness by targeting LDHA and other cancer-associated signaling pathways, suggesting possible treatment of GI cancers[212-225]. Despite this evidence, there is still a lack of clinical results to support the safety and efficacy of these LDHA-targeting drugs in GI cancer patients. An early natural compound, gossypol (AT-101), derived from the cotton plant, is one exception. Gossypol and its derivatives have proven potent inhibitors of LDHA[226]. Gossypol not only reduces the aggressiveness of GI and other cancers, but also has a strong cytotoxic effect on cancer cells[226-240]. Most importantly, gossypol has entered a phase I/II clinical trial (NCT00561197) to evaluate its safety and efficacy in treating patients with esophageal cancer, showing significant improvement in complete response and survival rates[231]. Therefore, gossypol may be the most promising clinical drug targeting LDHA to date for use in GI cancers.
Excessive intracellular accumulation of lactate is a hallmark of many cancer types, which necessitates MCTs in transporting lactate from highly glycolytic cancer cells. Secretory lactate can acidify the extracellular microenvironment, which can impact the tumor microenvironment[241]. While secretory lactate was initially considered a waste product of cancer cells, recent evidence has suggested that it serves as an alternative fuel for oxidative cancer cells, leading to enhanced aggressiveness[56]. Therefore, MCT targets have emerged as an alternative strategy for anti-cancer therapy[241]. Among the various compounds proposed to target MCTs in cancer, AZD3965 and AR-C155858 have received more attention from researchers. Both drugs have demonstrated potential in targeting MCTs, inhibiting GI cancer cell aggressiveness, and stunting tumor growth both in vitro and in vivo[242-249]. While AZD3965 has entered the clinical trial phase, further investigation is needed to determine the safety and therapeutic efficacy of these drugs in patients with GI cancer. Notably, the development of MCT inhibitors has faced several challenges, including the presence of MCT isoforms and the need for inhibitors that selectively target cancer cells without affecting normal tissues[58,250]. In this regard, approaches and strategies to develop selective MCT inhibitors are being actively pursued. While MCT inhibitors hold promise as a potential anti-cancer therapy, further research is needed to fully understand their mechanisms of action and optimize their clinical applications.
Excessive OXPHOS activity has been observed in certain cancers and has been associated with more aggressive phenotypes/unfavorable clinical outcomes, making it a novel target for anti-cancer therapy[251]. Attenuating OXPHOS activity has been proposed as the best strategy to target OXPHOS, leading to the identification of a large number of candidate compounds that target mitochondrial complex I. Metformin, a compound that has long been used to treat diabetes, has been reported to exhibit mitochondrial complex I inhibition activity and can impact cancer cell aggressiveness/tumor growth in both GI cancer cell lines and xenograft models[252-265]. Metformin has advanced to clinical trials in combination with other anti-cancer regimens for patients with GI cancers, such as ESCA patients in Phase II (ChiCTR-ICR-15005940), HCC patients in Phase I (CTRI/2018/07/014865), CCA patients in Phase Ib (NCT0249674), PCA patients in Phase II (NCT01210911 and NCT01167738), and CRC patients in Phase II (NCT01312467, NCT03047837, and NCT01941953). It was found that metformin combination therapy can provide benefit to patients, perhaps through reprogramming the tumor immune microenvironment[258].
Recent studies have proposed several candidates as mitochondrial complex I-targeting compounds in addition to metformin. Among them, tamoxifen, IM156, and IACS-010759 have gained attention as potential anti-cancer agents. Tamoxifen is an anti-estrogen agent that has been clinically used to treat breast cancer patients with positive estrogen-receptor (ER) expression[266]. Interestingly, tamoxifen has also been shown to inhibit cancer cell aggressiveness, tumor growth, metastasis, and increase chemosensitivity in GI cancers[267-273]. This effect is thought to be through an ER-independent anti-cancer pathway[269]. Tamoxifen has been used as a monotherapy or combined therapy in several clinical trials, including an early phase trial in ESCA patients, Phase II trials in PAC patients[274-276], and a Phase III trial in HCC patients (NCT00003424). Tamoxifen has been found to be tolerable, safe, and with manageable adverse effects, while a Phase III trial in HCC patients found that tamoxifen monotherapy either offered no effect or decreased survival in patients with unresectable HCC[277]. This result has slowed the advancement of tamoxifen in GI cancers and requires further investigation.
IM156 and IACS-010759 are two novel mitochondria-targeting drugs that specifically inhibit mitochondrial complex I. While both compounds have shown promising results in pre-clinical studies against certain cancer cell lines, their potential in treating GI cancers involves limited evidence. Interestingly, IM156 has entered Phase I clinical trials in patients with GC, CRC and PCA (NCT03272256 and Janku
On the other hand, IACS-010759 has shown significant cell viability reduction in PCA cell lines[279], leading to the initiation of a Phase I clinical trial (NCT03291938) to evaluate clinical efficacy and safety in patients with solid tumors due to CCA, PAC, and CRC. However, a recent publication reported that although IACS-010759 was tolerable and safe, it increased blood lactate levels and neurotoxicity while offering only limited anti-cancer efficacy. A reverse translational study using mice also found IACS-010759 to induce behavioral and physiological changes indicative of peripheral neuropathy, minimizing the possibility of combined therapy with specific anti-cancer compounds (e.g., histone deacetylase 6 inhibitor). The development of mitochondrial complex I inhibitors is ongoing[280].
While the mitochondrial complex I inhibitors metformin, tamoxifen, IM156, and IACS-010759 hold promise as potential treatments for GI cancer, further studies are needed to evaluate their efficacy and safety, particularly in combination with other anti-cancer compounds. The development of more selective and potent mitochondrial complex I inhibitors may help overcome side effects and improve efficacy in cancer treatment.
The targeting of mitochondrial complexes other than complex I has also been proposed as a strategy for anti-cancer therapy[281]. One such compound of note is atovaquone, which was identified as a mitochondrial complex III inhibitor during a drug re-purposing study[282]. Pre-clinical studies have evaluated the potential of atovaquone as an anti-cancer agent in GI cancer cell lines and xenograft models, and have shown its ability to reduce OXPHOS, OCR, cell viability, cell proliferation, cell cycle progression, and tumor growth, while enhancing cell death[283-285]. Despite promising results, atovaquone is currently in clinical trials for patients with non-small cell lung cancer (NCT04648033) and acute myeloid leukemia (NCT03568994) but not for patients with GI cancer. Further studies are needed to determine drug tolerability, safety, and therapeutic efficacy in patients with GI cancer. Nonetheless, the potential benefits of targeting OXPHOS make for a promising strategy in GI cancer therapy. However, the potential toxicity of these inhibitors in normal cells must be carefully evaluated before being considered as viable anti-cancer agents. In addition, the development of resistance to mitochondrial inhibitors, similar to the resistance seen with other anti-cancer agents, highlights the need for combination therapy.
The TCA cycle is a critical metabolic pathway that fuels bioenergetic processes in cells. Targeting the TCA cycle has emerged as a potential strategy for anti-cancer therapy[286]. Various agents have been tested for their anti-cancer efficacy, including AGI-5195, AG-221, AG-881, and CPI-613[286]. Among these compounds, CPI-613 is the only PDH and alpha-ketoglutarate dehydrogenase complex (KGDHC) dual targeting agent that has shown promising anti-cancer properties in GI cancer models both in vitro and in vivo[287-291]. The tolerability and safety of CPI-613, alone or in combination with other agents, has been evaluated or is currently being studied in patients with HCC, CCA, and CRC (NCT01766219, NCT05070104 and NCT02232152). However, a recent Phase III trial (NCT03504423) evaluating the anti-cancer efficacy of CPI-613 in patients with advanced PAC failed to improve survival rate but improved overall response rate[292]. This outcome is disappointing, combining CPI-613 with other drugs such as gemcitabine or nab-paclitaxel may provide better results.
The TCA cycle is a complex pathway, and there are multiple enzymes and metabolites that could be targeted for anti-cancer therapy. For example, the isocitrate dehydrogenase 1 and 2 (IDH1/2) enzymes play a crucial role in the TCA cycle, and mutations in these enzymes have been observed in several types of cancer, including gliomas and acute myeloid leukemia (AML)[293]. Enasidenib and ivosidenib are two IDH1/2 inhibitors that have been approved for the treatment of relapsed or refractory AML[294,295]. In GI cancers, however, the efficacy of IDH1/2 inhibitors is still under investigation[296]. In addition to IDH1/2 inhibitors, other TCA cycle inhibitors are being explored for anti-cancer therapy. For example, IDH1/2 mutant tumors are sensitive to glutaminase inhibitor CB-839, which targets glutamine metabolism[297]. Another TCA cycle inhibitor, BPTES, has shown anti-cancer efficacy in pre-clinical studies by blocking the activity of the glutaminase enzyme[298]. However, our understanding of these inhibitors in GI cancer treatment is still limited.
Targeting the TCA cycle and associated bioenergetic processes is a promising approach for anti-cancer therapy. While CPI-613 has shown some success in GI cancer models, the failure in Phase III trial underscores the need for continued research and combination therapy. Other TCA cycle inhibitors, such as IDH1/2 and glutaminase inhibitors, are being evaluated for their anti-cancer efficacy in GI cancers, offering hope for future treatments.
Cancer cells undergo significant metabolic changes which involve alteration to the nuclear and mitochondrial genomes as well as cell microenvironment. Understanding the molecular mechanisms behind these alterations is critical for the development of effective cancer therapies. Next-generation technologies such as metabolic profiling, single-cell sequencing, and metabolic tracing can provide insights into the regulation of mitochondrial metabolism in different cancer types. However, developing therapies based on altered metabolism is challenging due to the diverse metabolic patterns observed across different cancer cells.
Simply targeting a single bioenergetic enzyme or pathway may not be enough to effectively inhibit cancer cell growth, as metabolic symbiosis enables cancer cells to adapt to harsh tumor environments. One potential strategy is to treat the metabolic patterns of different cellular subpopulations in the tumor microenvironment to create a homogeneous metabolic population for targeting.
Bioenergetic enzymes have been explored as a way to inhibit cancer cell growth, with some small-molecule inhibitors of glucose metabolism showing significant inhibition in various cancers. However, clinical translation of these inhibitors has been limited by side effects. Other small-molecule inhibitors and natural products that regulate key bioenergy enzymes have also shown promise, but their specific mechanisms and targets require further investigation. Developing anticancer drugs targeting bioenergetic enzymes remains a significant challenge due to the unique metabolic features of cancer cells. Targeted drugs have shown anticancer effects in various tumor models, and combining them with conventional anticancer drugs is a promising strategy.
High-throughput multi-omics and spatial omics can help elucidate the heterogeneity of cancer cells and provide opportunities for therapeutic drugs targeting the bioenergetics of malignant tumors. Unbiased CRISPR-Cas9 synthetic lethality screening of metabolic genes that favor anti-cancer responses, particularly in vivo, could provide an avenue towards the identification of bioenergetic targets of interest. The ultimate goal is to develop drugs that simultaneously disable cancer cells while synergizing with targeted therapies.
However, while targeting bioenergetic pathways in cancer cells shows promise, it also has the potential to affect normal cells and tissues that rely on these pathways. Therefore, careful consideration and further research are needed to ensure that therapies targeting bioenergetics in cancer cells are specific and effective while minimizing potential side effects on normal cells and tissues. Additionally, combination therapies that target multiple pathways may be necessary to achieve optimal therapeutic effects.
The metabolic reprogramming and bioenergetic alteration of cancer cells, particularly their utilization of glucose fermentation (the Warburg effect) for energy production, are well-known phenomena. However, comprehensive summaries of these alterations and their oncogenetic links in GI cancers are lacking. This review provides a summary of the interplay between aerobic glycolysis, the TCA cycle, and OXPHOS in cancer cells, including the molecular mechanisms that trigger these alterations. It also explores the role of HIFs, tumor suppressors, and the oncogenetic link between hypoxia-related enzymes, bioenergetic changes, and GI cancer. Additionally, this review details various anti-cancer drugs and strategies for treating GI cancers, along with the challenges associated with them. Understanding dysregulated cancer cell bioenergetics is critical for effective treatments, although the diverse metabolic patterns present challenges for targeted therapies. Further research is needed to comprehensively understand the specific mechanisms of inhibiting bioenergetic enzymes, address side effects, and utilize high-throughput multi-omics and spatial omics for insights into the heterogeneity of GI cancer cells in targeted bioenergetic therapies.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Corresponding Author's Membership in Professional Societies: The Gastroenterological Society of Taiwan, No. 001565; American Gastroenterological Association, No. 337214; American Society for Gastrointestinal Endoscopy, No. 161580.
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Taiwan
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Masaru T, Hungary; Sun G, China S-Editor: Yan JP L-Editor: Webster JR P-Editor: Yu HG
| 1. | Herrera AS, Del C A Esparza M, Md Ashraf G, Zamyatnin AA, Aliev G. Beyond mitochondria, what would be the energy source of the cell? Cent Nerv Syst Agents Med Chem. 2015;15:32-41. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 22] [Article Influence: 2.2] [Reference Citation Analysis (1)] |
| 2. | Warburg O, Wind F, Negelein E. The metabolism of tumors in the body. J Gen Physiol. 1927;8:519-530. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2361] [Cited by in RCA: 3021] [Article Influence: 201.4] [Reference Citation Analysis (0)] |
| 3. | Liberti MV, Locasale JW. The Warburg Effect: How Does it Benefit Cancer Cells? Trends Biochem Sci. 2016;41:211-218. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1910] [Cited by in RCA: 3225] [Article Influence: 358.3] [Reference Citation Analysis (1)] |
| 4. | Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646-674. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51728] [Cited by in RCA: 46917] [Article Influence: 3351.2] [Reference Citation Analysis (5)] |
| 5. | El Hassouni B, Granchi C, Vallés-Martí A, Supadmanaba IGP, Bononi G, Tuccinardi T, Funel N, Jimenez CR, Peters GJ, Giovannetti E, Minutolo F. The dichotomous role of the glycolytic metabolism pathway in cancer metastasis: Interplay with the complex tumor microenvironment and novel therapeutic strategies. Semin Cancer Biol. 2020;60:238-248. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 66] [Article Influence: 13.2] [Reference Citation Analysis (1)] |
| 6. | Mimaki M, Wang X, McKenzie M, Thorburn DR, Ryan MT. Understanding mitochondrial complex I assembly in health and disease. Biochim Biophys Acta. 2012;1817:851-862. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 284] [Cited by in RCA: 334] [Article Influence: 23.9] [Reference Citation Analysis (0)] |
| 7. | Banerjee R, Purhonen J, Kallijärvi J. The mitochondrial coenzyme Q junction and complex III: biochemistry and pathophysiology. FEBS J. 2022;289:6936-6958. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 66] [Article Influence: 22.0] [Reference Citation Analysis (0)] |
| 8. | Bénit P, Goncalves J, El Khoury R, Rak M, Favier J, Gimenez-Roqueplo AP, Rustin P. Succinate Dehydrogenase, Succinate, and Superoxides: A Genetic, Epigenetic, Metabolic, Environmental Explosive Crossroad. Biomedicines. 2022;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 14] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 9. | Lin YH, Lim SN, Chen CY, Chi HC, Yeh CT, Lin WR. Functional Role of Mitochondrial DNA in Cancer Progression. Int J Mol Sci. 2022;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 35] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 10. | Zhao RZ, Jiang S, Zhang L, Yu ZB. Mitochondrial electron transport chain, ROS generation and uncoupling (Review). Int J Mol Med. 2019;44:3-15. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 236] [Cited by in RCA: 543] [Article Influence: 90.5] [Reference Citation Analysis (0)] |
| 11. | Moloney JN, Cotter TG. ROS signalling in the biology of cancer. Semin Cell Dev Biol. 2018;80:50-64. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 754] [Cited by in RCA: 1302] [Article Influence: 162.8] [Reference Citation Analysis (0)] |
| 12. | Kowalczyk P, Sulejczak D, Kleczkowska P, Bukowska-Ośko I, Kucia M, Popiel M, Wietrak E, Kramkowski K, Wrzosek K, Kaczyńska K. Mitochondrial Oxidative Stress-A Causative Factor and Therapeutic Target in Many Diseases. Int J Mol Sci. 2021;22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 224] [Article Influence: 56.0] [Reference Citation Analysis (0)] |
| 13. | Kim M, Mahmood M, Reznik E, Gammage PA. Mitochondrial DNA is a major source of driver mutations in cancer. Trends Cancer. 2022;8:1046-1059. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 47] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 14. | Zhang Y, Li Q, Huang Z, Li B, Nice EC, Huang C, Wei L, Zou B. Targeting Glucose Metabolism Enzymes in Cancer Treatment: Current and Emerging Strategies. Cancers (Basel). 2022;14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 66] [Reference Citation Analysis (0)] |
| 15. | Godel M, Ortone G, Anobile DP, Pasino M, Randazzo G, Riganti C, Kopecka J. Targeting Mitochondrial Oncometabolites: A New Approach to Overcome Drug Resistance in Cancer. Pharmaceutics. 2021;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 17] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 16. | Wagner A, Kosnacova H, Chovanec M, Jurkovicova D. Mitochondrial Genetic and Epigenetic Regulations in Cancer: Therapeutic Potential. Int J Mol Sci. 2022;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 19] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 17. | Schiliro C, Firestein BL. Mechanisms of Metabolic Reprogramming in Cancer Cells Supporting Enhanced Growth and Proliferation. Cells. 2021;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 313] [Article Influence: 78.3] [Reference Citation Analysis (0)] |
| 18. | Schmidt CA, Fisher-Wellman KH, Neufer PD. From OCR and ECAR to energy: Perspectives on the design and interpretation of bioenergetics studies. J Biol Chem. 2021;297:101140. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 105] [Article Influence: 26.3] [Reference Citation Analysis (0)] |
| 19. | WARBURG O. On the origin of cancer cells. Science. 1956;123:309-314. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9117] [Cited by in RCA: 9883] [Article Influence: 143.2] [Reference Citation Analysis (0)] |
| 21. | Boerner P, Resnick RJ, Racker E. Stimulation of glycolysis and amino acid uptake in NRK-49F cells by transforming growth factor beta and epidermal growth factor. Proc Natl Acad Sci U S A. 1985;82:1350-1353. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 66] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 22. | Flier JS, Mueckler MM, Usher P, Lodish HF. Elevated levels of glucose transport and transporter messenger RNA are induced by ras or src oncogenes. Science. 1987;235:1492-1495. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 578] [Cited by in RCA: 582] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 23. | Birnbaum MJ, Haspel HC, Rosen OM. Transformation of rat fibroblasts by FSV rapidly increases glucose transporter gene transcription. Science. 1987;235:1495-1498. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 186] [Cited by in RCA: 214] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 24. | Hiraki Y, Rosen OM, Birnbaum MJ. Growth factors rapidly induce expression of the glucose transporter gene. J Biol Chem. 1988;263:13655-13662. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 183] [Cited by in RCA: 165] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 25. | Fantin VR, St-Pierre J, Leder P. Attenuation of LDH-A expression uncovers a link between glycolysis, mitochondrial physiology, and tumor maintenance. Cancer Cell. 2006;9:425-434. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1141] [Cited by in RCA: 1226] [Article Influence: 64.5] [Reference Citation Analysis (0)] |
| 26. | Shim H, Chun YS, Lewis BC, Dang CV. A unique glucose-dependent apoptotic pathway induced by c-Myc. Proc Natl Acad Sci U S A. 1998;95:1511-1516. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 213] [Cited by in RCA: 220] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 27. | Locasale JW, Cantley LC. Metabolic flux and the regulation of mammalian cell growth. Cell Metab. 2011;14:443-451. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 308] [Cited by in RCA: 352] [Article Influence: 25.1] [Reference Citation Analysis (0)] |
| 28. | Lunt SY, Vander Heiden MG. Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annu Rev Cell Dev Biol. 2011;27:441-464. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1757] [Cited by in RCA: 2244] [Article Influence: 172.6] [Reference Citation Analysis (0)] |
| 29. | Slavov N, Budnik BA, Schwab D, Airoldi EM, van Oudenaarden A. Constant growth rate can be supported by decreasing energy flux and increasing aerobic glycolysis. Cell Rep. 2014;7:705-714. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 62] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 30. | Epstein T, Xu L, Gillies RJ, Gatenby RA. Separation of metabolic supply and demand: aerobic glycolysis as a normal physiological response to fluctuating energetic demands in the membrane. Cancer Metab. 2014;2:7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 82] [Cited by in RCA: 113] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 31. | Epstein T, Gatenby RA, Brown JS. The Warburg effect as an adaptation of cancer cells to rapid fluctuations in energy demand. PLoS One. 2017;12:e0185085. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 91] [Cited by in RCA: 119] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 32. | DeBerardinis RJ, Mancuso A, Daikhin E, Nissim I, Yudkoff M, Wehrli S, Thompson CB. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci U S A. 2007;104:19345-19350. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1731] [Cited by in RCA: 1994] [Article Influence: 110.8] [Reference Citation Analysis (0)] |
| 33. | Grashei M, Biechl P, Schilling F, Otto AM. Conversion of Hyperpolarized [1-(13)C]Pyruvate in Breast Cancer Cells Depends on Their Malignancy, Metabolic Program and Nutrient Microenvironment. Cancers (Basel). 2022;14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 8] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 34. | Birsoy K, Wang T, Chen WW, Freinkman E, Abu-Remaileh M, Sabatini DM. An Essential Role of the Mitochondrial Electron Transport Chain in Cell Proliferation Is to Enable Aspartate Synthesis. Cell. 2015;162:540-551. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 786] [Cited by in RCA: 1014] [Article Influence: 101.4] [Reference Citation Analysis (0)] |
| 35. | Flaveny CA, Griffett K, El-Gendy Bel-D, Kazantzis M, Sengupta M, Amelio AL, Chatterjee A, Walker J, Solt LA, Kamenecka TM, Burris TP. Broad Anti-tumor Activity of a Small Molecule that Selectively Targets the Warburg Effect and Lipogenesis. Cancer Cell. 2015;28:42-56. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 133] [Cited by in RCA: 158] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 36. | Sullivan LB, Gui DY, Hosios AM, Bush LN, Freinkman E, Vander Heiden MG. Supporting Aspartate Biosynthesis Is an Essential Function of Respiration in Proliferating Cells. Cell. 2015;162:552-563. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 684] [Cited by in RCA: 861] [Article Influence: 86.1] [Reference Citation Analysis (0)] |
| 37. | Viale A, Pettazzoni P, Lyssiotis CA, Ying H, Sánchez N, Marchesini M, Carugo A, Green T, Seth S, Giuliani V, Kost-Alimova M, Muller F, Colla S, Nezi L, Genovese G, Deem AK, Kapoor A, Yao W, Brunetto E, Kang Y, Yuan M, Asara JM, Wang YA, Heffernan TP, Kimmelman AC, Wang H, Fleming JB, Cantley LC, DePinho RA, Draetta GF. Oncogene ablation-resistant pancreatic cancer cells depend on mitochondrial function. Nature. 2014;514:628-632. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 887] [Cited by in RCA: 976] [Article Influence: 88.7] [Reference Citation Analysis (0)] |
| 38. | Semenza GL, Wang GL. A nuclear factor induced by hypoxia via de novo protein synthesis binds to the human erythropoietin gene enhancer at a site required for transcriptional activation. Mol Cell Biol. 1992;12:5447-5454. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 742] [Article Influence: 22.5] [Reference Citation Analysis (0)] |
| 39. | Wang GL, Semenza GL. Characterization of hypoxia-inducible factor 1 and regulation of DNA binding activity by hypoxia. J Biol Chem. 1993;268:21513-21518. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 629] [Cited by in RCA: 612] [Article Influence: 19.1] [Reference Citation Analysis (0)] |
| 40. | Beck I, Weinmann R, Caro J. Characterization of hypoxia-responsive enhancer in the human erythropoietin gene shows presence of hypoxia-inducible 120-Kd nuclear DNA-binding protein in erythropoietin-producing and nonproducing cells. Blood. 1993;82:704-711. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 89] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 41. | Soni S, Padwad YS. HIF-1 in cancer therapy: two decade long story of a transcription factor. Acta Oncol. 2017;56:503-515. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 137] [Cited by in RCA: 198] [Article Influence: 24.8] [Reference Citation Analysis (0)] |
| 42. | De Saedeleer CJ, Copetti T, Porporato PE, Verrax J, Feron O, Sonveaux P. Lactate activates HIF-1 in oxidative but not in Warburg-phenotype human tumor cells. PLoS One. 2012;7:e46571. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 171] [Cited by in RCA: 200] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 43. | Sonveaux P, Copetti T, De Saedeleer CJ, Végran F, Verrax J, Kennedy KM, Moon EJ, Dhup S, Danhier P, Frérart F, Gallez B, Ribeiro A, Michiels C, Dewhirst MW, Feron O. Targeting the lactate transporter MCT1 in endothelial cells inhibits lactate-induced HIF-1 activation and tumor angiogenesis. PLoS One. 2012;7:e33418. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 319] [Cited by in RCA: 405] [Article Influence: 31.2] [Reference Citation Analysis (0)] |
| 44. | To KK, Huang LE. Suppression of hypoxia-inducible factor 1alpha (HIF-1alpha) transcriptional activity by the HIF prolyl hydroxylase EGLN1. J Biol Chem. 2005;280:38102-38107. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 80] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 45. | Berra E, Benizri E, Ginouvès A, Volmat V, Roux D, Pouysségur J. HIF prolyl-hydroxylase 2 is the key oxygen sensor setting low steady-state levels of HIF-1alpha in normoxia. EMBO J. 2003;22:4082-4090. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 998] [Cited by in RCA: 1074] [Article Influence: 48.8] [Reference Citation Analysis (0)] |
| 46. | Maxwell PH, Wiesener MS, Chang GW, Clifford SC, Vaux EC, Cockman ME, Wykoff CC, Pugh CW, Maher ER, Ratcliffe PJ. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399:271-275. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3728] [Cited by in RCA: 3831] [Article Influence: 147.3] [Reference Citation Analysis (0)] |
| 47. | Ohh M, Park CW, Ivan M, Hoffman MA, Kim TY, Huang LE, Pavletich N, Chau V, Kaelin WG. Ubiquitination of hypoxia-inducible factor requires direct binding to the beta-domain of the von Hippel-Lindau protein. Nat Cell Biol. 2000;2:423-427. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1151] [Cited by in RCA: 1187] [Article Influence: 47.5] [Reference Citation Analysis (0)] |
| 48. | Semenza GL, Roth PH, Fang HM, Wang GL. Transcriptional regulation of genes encoding glycolytic enzymes by hypoxia-inducible factor 1. J Biol Chem. 1994;269:23757-23763. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1282] [Cited by in RCA: 1325] [Article Influence: 42.7] [Reference Citation Analysis (0)] |
| 49. | Jiang BH, Agani F, Passaniti A, Semenza GL. V-SRC induces expression of hypoxia-inducible factor 1 (HIF-1) and transcription of genes encoding vascular endothelial growth factor and enolase 1: involvement of HIF-1 in tumor progression. Cancer Res. 1997;57:5328-5335. [PubMed] |
| 50. | Semenza GL, Jiang BH, Leung SW, Passantino R, Concordet JP, Maire P, Giallongo A. Hypoxia response elements in the aldolase A, enolase 1, and lactate dehydrogenase A gene promoters contain essential binding sites for hypoxia-inducible factor 1. J Biol Chem. 1996;271:32529-32537. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1226] [Cited by in RCA: 1338] [Article Influence: 46.1] [Reference Citation Analysis (0)] |
| 51. | O'Rourke JF, Pugh CW, Bartlett SM, Ratcliffe PJ. Identification of hypoxically inducible mRNAs in HeLa cells using differential-display PCR. Role of hypoxia-inducible factor-1. Eur J Biochem. 1996;241:403-410. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 83] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 52. | Ebert BL, Firth JD, Ratcliffe PJ. Hypoxia and mitochondrial inhibitors regulate expression of glucose transporter-1 via distinct Cis-acting sequences. J Biol Chem. 1995;270:29083-29089. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 366] [Cited by in RCA: 391] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 53. | Riddle SR, Ahmad A, Ahmad S, Deeb SS, Malkki M, Schneider BK, Allen CB, White CW. Hypoxia induces hexokinase II gene expression in human lung cell line A549. Am J Physiol Lung Cell Mol Physiol. 2000;278:L407-L416. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 115] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 54. | Kim JW, Tchernyshyov I, Semenza GL, Dang CV. HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 2006;3:177-185. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2552] [Cited by in RCA: 2878] [Article Influence: 151.5] [Reference Citation Analysis (0)] |
| 55. | Nagao A, Kobayashi M, Koyasu S, Chow CCT, Harada H. HIF-1-Dependent Reprogramming of Glucose Metabolic Pathway of Cancer Cells and Its Therapeutic Significance. Int J Mol Sci. 2019;20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 294] [Cited by in RCA: 315] [Article Influence: 52.5] [Reference Citation Analysis (0)] |
| 56. | de la Cruz-López KG, Castro-Muñoz LJ, Reyes-Hernández DO, García-Carrancá A, Manzo-Merino J. Lactate in the Regulation of Tumor Microenvironment and Therapeutic Approaches. Front Oncol. 2019;9:1143. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 273] [Cited by in RCA: 602] [Article Influence: 100.3] [Reference Citation Analysis (0)] |
| 57. | Muñoz-Pinedo C, El Mjiyad N, Ricci JE. Cancer metabolism: current perspectives and future directions. Cell Death Dis. 2012;3:e248. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 276] [Cited by in RCA: 296] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 58. | Halestrap AP. The SLC16 gene family - structure, role and regulation in health and disease. Mol Aspects Med. 2013;34:337-349. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 388] [Cited by in RCA: 548] [Article Influence: 45.7] [Reference Citation Analysis (0)] |
| 59. | Payen VL, Mina E, Van Hée VF, Porporato PE, Sonveaux P. Monocarboxylate transporters in cancer. Mol Metab. 2020;33:48-66. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 184] [Cited by in RCA: 388] [Article Influence: 64.7] [Reference Citation Analysis (0)] |
| 60. | Urbańska K, Orzechowski A. Unappreciated Role of LDHA and LDHB to Control Apoptosis and Autophagy in Tumor Cells. Int J Mol Sci. 2019;20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 74] [Cited by in RCA: 182] [Article Influence: 30.3] [Reference Citation Analysis (0)] |
| 61. | Mishra D, Banerjee D. Lactate Dehydrogenases as Metabolic Links between Tumor and Stroma in the Tumor Microenvironment. Cancers (Basel). 2019;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 106] [Cited by in RCA: 195] [Article Influence: 32.5] [Reference Citation Analysis (0)] |
| 62. | Mendes C, Serpa J. Revisiting lactate dynamics in cancer-a metabolic expertise or an alternative attempt to survive? J Mol Med (Berl). 2020;98:1397-1414. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 63. | Moldogazieva NT, Mokhosoev IM, Terentiev AA. Metabolic Heterogeneity of Cancer Cells: An Interplay between HIF-1, GLUTs, and AMPK. Cancers (Basel). 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 92] [Cited by in RCA: 110] [Article Influence: 22.0] [Reference Citation Analysis (0)] |
| 64. | Liu Y, Chen C, Wang X, Sun Y, Zhang J, Chen J, Shi Y. An Epigenetic Role of Mitochondria in Cancer. Cells. 2022;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 71] [Article Influence: 23.7] [Reference Citation Analysis (0)] |
| 65. | Zhu X, Xuan Z, Chen J, Li Z, Zheng S, Song P. How DNA methylation affects the Warburg effect. Int J Biol Sci. 2020;16:2029-2041. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 24] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 66. | Bogaerts E, Paridaens A, Verhelst X, Carmeliet P, Geerts A, Van Vlierberghe H, Devisscher L. Effect of prolyl hydroxylase domain 2 haplodeficiency on liver progenitor cell characteristics in early mouse hepatocarcinogenesis. EXCLI J. 2016;15:687-698. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 67. | Heindryckx F, Kuchnio A, Casteleyn C, Coulon S, Olievier K, Colle I, Geerts A, Libbrecht L, Carmeliet P, Van Vlierberghe H. Effect of prolyl hydroxylase domain-2 haplodeficiency on the hepatocarcinogenesis in mice. J Hepatol. 2012;57:61-68. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 18] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 68. | Ma M, Hua S, Li G, Wang S, Cheng X, He S, Wu P, Chen X. Prolyl hydroxylase domain protein 3 and asparaginyl hydroxylase factor inhibiting HIF-1 levels are predictive of tumoral behavior and prognosis in hepatocellular carcinoma. Oncotarget. 2017;8:12983-13002. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 69. | Cui L, Qu J, Dang S, Mao Z, Wang X, Fan X, Sun K, Zhang J. Prolyl hydroxylase 3 inhibited the tumorigenecity of gastric cancer cells. Mol Carcinog. 2014;53:736-743. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 70. | Xia YJ, Jiang XT, Jiang SB, He XJ, Luo JG, Liu ZC, Wang L, Tao HQ, Chen JZ. PHD3 affects gastric cancer progression by negatively regulating HIF1A. Mol Med Rep. 2017;16:6882-6889. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 10] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 71. | Kamphues C, Wittschieber D, Klauschen F, Kasajima A, Dietel M, Schmidt SC, Glanemann M, Bahra M, Neuhaus P, Weichert W, Stenzinger A. Prolyl hydroxylase domain 2 protein is a strong prognostic marker in human gastric cancer. Pathobiology. 2012;79:11-17. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 9] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 72. | Xie G, Zheng L, Ou J, Huang H, He J, Li J, Pan F, Liang H. Low expression of prolyl hydroxylase 2 is associated with tumor grade and poor prognosis in patients with colorectal cancer. Exp Biol Med (Maywood). 2012;237:860-866. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 15] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 73. | Gossage L, Zaitoun A, Fareed KR, Turley H, Aloysius M, Lobo DN, Harris AL, Madhusudan S. Expression of key hypoxia sensing prolyl-hydroxylases PHD1, -2 and -3 in pancreaticobiliary cancer. Histopathology. 2010;56:908-920. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 28] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 74. | Schmitt AM, Schmid S, Rudolph T, Anlauf M, Prinz C, Klöppel G, Moch H, Heitz PU, Komminoth P, Perren A. VHL inactivation is an important pathway for the development of malignant sporadic pancreatic endocrine tumors. Endocr Relat Cancer. 2009;16:1219-1227. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 73] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 75. | Kuwai T, Kitadai Y, Tanaka S, Hiyama T, Tanimoto K, Chayama K. Mutation of the von Hippel-Lindau (VHL) gene in human colorectal carcinoma: association with cytoplasmic accumulation of hypoxia-inducible factor (HIF)-1alpha. Cancer Sci. 2004;95:149-153. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 23] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 76. | Li G, Shen Y, Wang F, Hong S, Cai M. Correlation Between von Hippel-Lindau Gene Expression and Tumor SUVmax and Survival Prognosis in Hepatocellular Carcinoma. Med Sci Monit. 2020;26:e920473. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 77. | Linehan WM, Rouault TA. Molecular pathways: Fumarate hydratase-deficient kidney cancer--targeting the Warburg effect in cancer. Clin Cancer Res. 2013;19:3345-3352. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 138] [Cited by in RCA: 165] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 78. | Rasheed MRHA, Tarjan G. Succinate Dehydrogenase Complex: An Updated Review. Arch Pathol Lab Med. 2018;142:1564-1570. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 35] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 79. | Herrmann PC, Herrmann EC. Oxygen metabolism and a potential role for cytochrome c oxidase in the Warburg effect. J Bioenerg Biomembr. 2007;39:247-250. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 13] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 80. | Sánchez-Aragó M, Formentini L, Cuezva JM. Mitochondria-mediated energy adaption in cancer: the H(+)-ATP synthase-geared switch of metabolism in human tumors. Antioxid Redox Signal. 2013;19:285-298. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 55] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 81. | Lee HC, Wei YH. Mitochondrial DNA instability and metabolic shift in human cancers. Int J Mol Sci. 2009;10:674-701. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 96] [Cited by in RCA: 96] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 82. | Balsa E, Marco R, Perales-Clemente E, Szklarczyk R, Calvo E, Landázuri MO, Enríquez JA. NDUFA4 is a subunit of complex IV of the mammalian electron transport chain. Cell Metab. 2012;16:378-386. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 281] [Cited by in RCA: 298] [Article Influence: 22.9] [Reference Citation Analysis (0)] |
| 83. | Kim SH, Choi SI, Won KY, Lim SJ. Distinctive interrelation of p53 with SCO2, COX, and TIGAR in human gastric cancer. Pathol Res Pract. 2016;212:904-910. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 19] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 84. | Zhang K, Chen Y, Huang X, Qu P, Pan Q, Lü L, Jiang S, Ren T, Su H. Expression and clinical significance of cytochrome c oxidase subunit IV in colorectal cancer patients. Arch Med Sci. 2016;12:68-77. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 15] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 85. | Ramzan R, Kadenbach B, Vogt S. Multiple Mechanisms Regulate Eukaryotic Cytochrome C Oxidase. Cells. 2021;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 20] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 86. | Huhta H, Helminen O, Palomäki S, Kauppila JH, Saarnio J, Lehenkari PP, Karttunen TJ. Intratumoral lactate metabolism in Barrett's esophagus and adenocarcinoma. Oncotarget. 2017;8:22894-22902. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 25] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 87. | Ma JT, Han CB, Zhou Y, Zhao JZ, Jing W, Zou HW. Altered expression of mitochondrial cytochrome c oxidase I and NADH dehydrogenase 4 transcripts associated with gastric tumorigenesis and tumor dedifferentiation. Mol Med Rep. 2012;5:1526-1530. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 4] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 88. | Eskuri M, Kemi N, Kauppila JH. Monocarboxylate Transporters 1 and 4 and MTCO1 in Gastric Cancer. Cancers (Basel). 2021;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 8] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 89. | LE PH, Huang SC, Lim SN, Chou CH, Yeh TS, Chen TC, Yen TH, Su MY, Chiu CT, Yeh CT, Lin WR. Complex IV subunit 1 defect predicts postoperative survival in hepatocellular carcinoma. Oncol Lett. 2014;7:1430-1438. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 7] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 90. | Feichtinger RG, Neureiter D, Kemmerling R, Mayr JA, Kiesslich T, Kofler B. Low VDAC1 Expression Is Associated with an Aggressive Phenotype and Reduced Overall Patient Survival in Cholangiocellular Carcinoma. Cells. 2019;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 8] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 91. | Li D, Wang XZ, Yu JP, Chen ZX, Huang YH, Tao QM. Cytochrome C oxidase III interacts with hepatitis B virus X protein in vivo by yeast two-hybrid system. World J Gastroenterol. 2004;10:2805-2808. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 10] [Cited by in RCA: 12] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 92. | Zou LY, Zheng BY, Fang XF, Li D, Huang YH, Chen ZX, Zhou LY, Wang XZ. HBx co-localizes with COXIII in HL-7702 cells to upregulate mitochondrial function and ROS generation. Oncol Rep. 2015;33:2461-2467. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 27] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 93. | Greaves LC, Preston SL, Tadrous PJ, Taylor RW, Barron MJ, Oukrif D, Leedham SJ, Deheragoda M, Sasieni P, Novelli MR, Jankowski JA, Turnbull DM, Wright NA, McDonald SA. Mitochondrial DNA mutations are established in human colonic stem cells, and mutated clones expand by crypt fission. Proc Natl Acad Sci U S A. 2006;103:714-719. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 242] [Cited by in RCA: 228] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 94. | Namslauer I, Brzezinski P. A mitochondrial DNA mutation linked to colon cancer results in proton leaks in cytochrome c oxidase. Proc Natl Acad Sci U S A. 2009;106:3402-3407. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 35] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 95. | Wang Y, Wang H, Yin S, Zhang J, Zhang R, Guo Z. Identification of polymorphisms in mitochondrial cytochrome c oxidase genes as risk factors for gastric cancer. Transl Cancer Res. 2020;9:3854-3859. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 96. | Wang H, Xu J, Li D, Zhang S, Guo Z. Identification of sequence polymorphisms in the mitochondrial cytochrome c oxidase genes as risk factors for hepatocellular carcinoma. J Clin Lab Anal. 2018;32. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 6] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 97. | Zong S, Wu M, Gu J, Liu T, Guo R, Yang M. Structure of the intact 14-subunit human cytochrome c oxidase. Cell Res. 2018;28:1026-1034. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 167] [Article Influence: 23.9] [Reference Citation Analysis (0)] |
| 98. | Kadenbach B. Regulation of Mammalian 13-Subunit Cytochrome c Oxidase and Binding of other Proteins: Role of NDUFA4. Trends Endocrinol Metab. 2017;28:761-770. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 52] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 99. | Srinivasan S, Guha M, Dong DW, Whelan KA, Ruthel G, Uchikado Y, Natsugoe S, Nakagawa H, Avadhani NG. Disruption of cytochrome c oxidase function induces the Warburg effect and metabolic reprogramming. Oncogene. 2016;35:1585-1595. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 76] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 100. | Chu YD, Lin WR, Lin YH, Kuo WH, Tseng CJ, Lim SN, Huang YL, Huang SC, Wu TJ, Lin KH, Yeh CT. COX5B-Mediated Bioenergetic Alteration Regulates Tumor Growth and Migration by Modulating AMPK-UHMK1-ERK Cascade in Hepatoma. Cancers (Basel). 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 22] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 101. | Chen ZX, Pervaiz S. Involvement of cytochrome c oxidase subunits Va and Vb in the regulation of cancer cell metabolism by Bcl-2. Cell Death Differ. 2010;17:408-420. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 115] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 102. | Chu YD, Lim SN, Yeh CT, Lin WR. COX5B-Mediated Bioenergetic Alterations Modulate Cell Growth and Anticancer Drug Susceptibility by Orchestrating Claudin-2 Expression in Colorectal Cancers. Biomedicines. 2021;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 5] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 103. | Li JP, Liu YJ, Zeng SH, Gao HJ, Chen YG, Zou X. Identification of COX4I2 as a hypoxia-associated gene acting through FGF1 to promote EMT and angiogenesis in CRC. Cell Mol Biol Lett. 2022;27:76. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 13] [Reference Citation Analysis (0)] |
| 104. | Yang J, Liu J, Zhang S, Yang Y, Gong J. The overexpression of cytochrome c oxidase subunit 6C activated by Kras mutation is related to energy metabolism in pancreatic cancer. Transl Cancer Res. 2018;7:290-300. [RCA] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 105. | Nie K, Li J, He X, Wang Y, Zhao Q, Du M, Sun H, Wang J, Lyu J, Fang H, Jin L. COX6B2 drives metabolic reprogramming toward oxidative phosphorylation to promote metastasis in pancreatic ductal cancer cells. Oncogenesis. 2020;9:51. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 38] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 106. | Esparza-Moltó PB, Cuezva JM. The Role of Mitochondrial H(+)-ATP Synthase in Cancer. Front Oncol. 2018;8:53. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 59] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 107. | Wang X, Chang X, He C, Fan Z, Yu Z, Yu B, Wu X, Hou J, Li J, Su L, Liu B, Zhu Z. ATP5B promotes the metastasis and growth of gastric cancer by activating the FAK/AKT/MMP2 pathway. FASEB J. 2021;35:e20649. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 11] [Reference Citation Analysis (0)] |
| 108. | Isidoro A, Martínez M, Fernández PL, Ortega AD, Santamaría G, Chamorro M, Reed JC, Cuezva JM. Alteration of the bioenergetic phenotype of mitochondria is a hallmark of breast, gastric, lung and oesophageal cancer. Biochem J. 2004;378:17-20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 143] [Cited by in RCA: 146] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 109. | Cuezva JM, Krajewska M, de Heredia ML, Krajewski S, Santamaría G, Kim H, Zapata JM, Marusawa H, Chamorro M, Reed JC. The bioenergetic signature of cancer: a marker of tumor progression. Cancer Res. 2002;62:6674-6681. [PubMed] |
| 110. | Capuano F, Varone D, D'Eri N, Russo E, Tommasi S, Montemurro S, Prete F, Papa S. Oxidative phosphorylation and F(O)F(1) ATP synthase activity of human hepatocellular carcinoma. Biochem Mol Biol Int. 1996;38:1013-1022. [PubMed] |
| 111. | Tanton H, Voronina S, Evans A, Armstrong J, Sutton R, Criddle DN, Haynes L, Schmid MC, Campbell F, Costello E, Tepikin AV. F(1)F(0)-ATP Synthase Inhibitory Factor 1 in the Normal Pancreas and in Pancreatic Ductal Adenocarcinoma: Effects on Bioenergetics, Invasion and Proliferation. Front Physiol. 2018;9:833. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 7] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 112. | Chang HJ, Lee MR, Hong SH, Yoo BC, Shin YK, Jeong JY, Lim SB, Choi HS, Jeong SY, Park JG. Identification of mitochondrial FoF1-ATP synthase involved in liver metastasis of colorectal cancer. Cancer Sci. 2007;98:1184-1191. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 28] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 113. | Huang YJ, Jan YH, Chang YC, Tsai HF, Wu AT, Chen CL, Hsiao M. ATP Synthase Subunit Epsilon Overexpression Promotes Metastasis by Modulating AMPK Signaling to Induce Epithelial-to-Mesenchymal Transition and Is a Poor Prognostic Marker in Colorectal Cancer Patients. J Clin Med. 2019;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 17] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 114. | Martínez-Reyes I, Chandel NS. Mitochondrial TCA cycle metabolites control physiology and disease. Nat Commun. 2020;11:102. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 605] [Cited by in RCA: 1585] [Article Influence: 317.0] [Reference Citation Analysis (0)] |
| 115. | Beyoğlu D, Idle JR. Metabolic Rewiring and the Characterization of Oncometabolites. Cancers (Basel). 2021;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 37] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 116. | Missiroli S, Perrone M, Genovese I, Pinton P, Giorgi C. Cancer metabolism and mitochondria: Finding novel mechanisms to fight tumours. EBioMedicine. 2020;59:102943. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 167] [Article Influence: 33.4] [Reference Citation Analysis (0)] |
| 117. | Kaelin WG Jr. Cancer and altered metabolism: potential importance of hypoxia-inducible factor and 2-oxoglutarate-dependent dioxygenases. Cold Spring Harb Symp Quant Biol. 2011;76:335-345. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 135] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 118. | Carvajal-Carmona LG, Alam NA, Pollard PJ, Jones AM, Barclay E, Wortham N, Pignatelli M, Freeman A, Pomplun S, Ellis I, Poulsom R, El-Bahrawy MA, Berney DM, Tomlinson IP. Adult leydig cell tumors of the testis caused by germline fumarate hydratase mutations. J Clin Endocrinol Metab. 2006;91:3071-3075. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 87] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 119. | Tomlinson IP, Alam NA, Rowan AJ, Barclay E, Jaeger EE, Kelsell D, Leigh I, Gorman P, Lamlum H, Rahman S, Roylance RR, Olpin S, Bevan S, Barker K, Hearle N, Houlston RS, Kiuru M, Lehtonen R, Karhu A, Vilkki S, Laiho P, Eklund C, Vierimaa O, Aittomäki K, Hietala M, Sistonen P, Paetau A, Salovaara R, Herva R, Launonen V, Aaltonen LA; Multiple Leiomyoma Consortium. Germline mutations in FH predispose to dominantly inherited uterine fibroids, skin leiomyomata and papillary renal cell cancer. Nat Genet. 2002;30:406-410. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1139] [Cited by in RCA: 1117] [Article Influence: 48.6] [Reference Citation Analysis (0)] |
| 120. | Baysal BE, Ferrell RE, Willett-Brozick JE, Lawrence EC, Myssiorek D, Bosch A, van der Mey A, Taschner PE, Rubinstein WS, Myers EN, Richard CW 3rd, Cornelisse CJ, Devilee P, Devlin B. Mutations in SDHD, a mitochondrial complex II gene, in hereditary paraganglioma. Science. 2000;287:848-851. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1235] [Cited by in RCA: 1210] [Article Influence: 48.4] [Reference Citation Analysis (0)] |
| 121. | Niemann S, Müller U. Mutations in SDHC cause autosomal dominant paraganglioma, type 3. Nat Genet. 2000;26:268-270. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 654] [Cited by in RCA: 648] [Article Influence: 25.9] [Reference Citation Analysis (0)] |
| 122. | Burnichon N, Brière JJ, Libé R, Vescovo L, Rivière J, Tissier F, Jouanno E, Jeunemaitre X, Bénit P, Tzagoloff A, Rustin P, Bertherat J, Favier J, Gimenez-Roqueplo AP. SDHA is a tumor suppressor gene causing paraganglioma. Hum Mol Genet. 2010;19:3011-3020. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 489] [Cited by in RCA: 505] [Article Influence: 33.7] [Reference Citation Analysis (0)] |
| 123. | Astuti D, Latif F, Dallol A, Dahia PL, Douglas F, George E, Sköldberg F, Husebye ES, Eng C, Maher ER. Gene mutations in the succinate dehydrogenase subunit SDHB cause susceptibility to familial pheochromocytoma and to familial paraganglioma. Am J Hum Genet. 2001;69:49-54. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 794] [Cited by in RCA: 797] [Article Influence: 33.2] [Reference Citation Analysis (0)] |
| 124. | Serra D, Amaro P, Gonçalo M, Silva M, Ferrando B, Pasini B, Figueiredo A. Gastric leiomyoma and hyperplastic polyposis coli in a patient with multiple cutaneous and uterine leiomyomatosis. J Cutan Med Surg. 2012;16:208-211. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 125. | Du X, Wan S, Chen Y, Qu P, Huang X, Yu X, Yang H, Zhang Y, Xing J. Genetic variants in genes of tricarboxylic acid cycle key enzymes predict postsurgical overall survival of patients with hepatocellular carcinoma. Ann Surg Oncol. 2014;21:4300-4307. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 126. | Dong G, He X, Chen Y, Cao H, Wang J, Liu X, Wang S, Wan S, Xing J. Genetic variations in genes of metabolic enzymes predict postoperational prognosis of patients with colorectal cancer. Mol Cancer. 2015;14:171. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 11] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 127. | Lee WC, Chou HS, Wu TJ, Lee CF, Hsu PY, Hsu HY, Wu TH, Chan KM. Down-regulation of metabolic proteins in hepatocellular carcinoma with portal vein thrombosis. Clin Proteomics. 2017;14:29. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 6] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 128. | Tuli HS, Tuorkey MJ, Thakral F, Sak K, Kumar M, Sharma AK, Sharma U, Jain A, Aggarwal V, Bishayee A. Molecular Mechanisms of Action of Genistein in Cancer: Recent Advances. Front Pharmacol. 2019;10:1336. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 293] [Cited by in RCA: 223] [Article Influence: 37.2] [Reference Citation Analysis (0)] |
| 129. | Samec M, Liskova A, Koklesova L, Mersakova S, Strnadel J, Kajo K, Pec M, Zhai K, Smejkal K, Mirzaei S, Hushmandi K, Ashrafizadeh M, Saso L, Brockmueller A, Shakibaei M, Büsselberg D, Kubatka P. Flavonoids Targeting HIF-1: Implications on Cancer Metabolism. Cancers (Basel). 2021;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 65] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 130. | Ashrafizadeh M, Bakhoda MR, Bahmanpour Z, Ilkhani K, Zarrabi A, Makvandi P, Khan H, Mazaheri S, Darvish M, Mirzaei H. Apigenin as Tumor Suppressor in Cancers: Biotherapeutic Activity, Nanodelivery, and Mechanisms With Emphasis on Pancreatic Cancer. Front Chem. 2020;8:829. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 65] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 131. | Cao X, Ren K, Song Z, Li D, Quan M, Zheng Y, Cao J, Zeng W, Zou H. 7-Difluoromethoxyl-5,4'-di-n-octyl genistein inhibits the stem-like characteristics of gastric cancer stem-like cells and reverses the phenotype of epithelial-mesenchymal transition in gastric cancer cells. Oncol Rep. 2016;36:1157-1165. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 25] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 132. | Yanagihara K, Ito A, Toge T, Numoto M. Antiproliferative effects of isoflavones on human cancer cell lines established from the gastrointestinal tract. Cancer Res. 1993;53:5815-5821. [PubMed] |
| 133. | Gao J, Xia R, Chen J, Gao J, Luo X, Ke C, Ren C, Li J, Mi Y. Inhibition of esophageal-carcinoma cell proliferation by genistein via suppression of JAK1/2-STAT3 and AKT/MDM2/p53 signaling pathways. Aging (Albany NY). 2020;12:6240-6259. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 37] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 134. | Mousavi Y, Adlercreutz H. Genistein is an effective stimulator of sex hormone-binding globulin production in hepatocarcinoma human liver cancer cells and suppresses proliferation of these cells in culture. Steroids. 1993;58:301-304. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 113] [Cited by in RCA: 97] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 135. | Dai W, Wang F, He L, Lin C, Wu S, Chen P, Zhang Y, Shen M, Wu D, Wang C, Lu J, Zhou Y, Xu X, Xu L, Guo C. Genistein inhibits hepatocellular carcinoma cell migration by reversing the epithelial-mesenchymal transition: partial mediation by the transcription factor NFAT1. Mol Carcinog. 2015;54:301-311. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 69] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 136. | Li S, Li J, Dai W, Zhang Q, Feng J, Wu L, Liu T, Yu Q, Xu S, Wang W, Lu X, Chen K, Xia Y, Lu J, Zhou Y, Fan X, Mo W, Xu L, Guo C. Genistein suppresses aerobic glycolysis and induces hepatocellular carcinoma cell death. Br J Cancer. 2017;117:1518-1528. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 128] [Cited by in RCA: 116] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 137. | Tanjak P, Thiantanawat A, Watcharasit P, Satayavivad J. Genistein reduces the activation of AKT and EGFR, and the production of IL6 in cholangiocarcinoma cells involving estrogen and estrogen receptors. Int J Oncol. 2018;53:177-188. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 20] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 138. | Mouria M, Gukovskaya AS, Jung Y, Buechler P, Hines OJ, Reber HA, Pandol SJ. Food-derived polyphenols inhibit pancreatic cancer growth through mitochondrial cytochrome C release and apoptosis. Int J Cancer. 2002;98:761-769. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 206] [Cited by in RCA: 202] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 139. | Büchler P, Reber HA, Büchler MW, Friess H, Lavey RS, Hines OJ. Antiangiogenic activity of genistein in pancreatic carcinoma cells is mediated by the inhibition of hypoxia-inducible factor-1 and the down-regulation of VEGF gene expression. Cancer. 2004;100:201-210. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 84] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 140. | Yu Z, Li W, Liu F. Inhibition of proliferation and induction of apoptosis by genistein in colon cancer HT-29 cells. Cancer Lett. 2004;215:159-166. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 91] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 141. | Wu K, Yuan LH, Xia W. Inhibitory effects of apigenin on the growth of gastric carcinoma SGC-7901 cells. World J Gastroenterol. 2005;11:4461-4464. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 22] [Cited by in RCA: 27] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 142. | Zhang Q, Zhao XH, Wang ZJ. Flavones and flavonols exert cytotoxic effects on a human oesophageal adenocarcinoma cell line (OE33) by causing G2/M arrest and inducing apoptosis. Food Chem Toxicol. 2008;46:2042-2053. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 135] [Cited by in RCA: 141] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 143. | Chiang LC, Ng LT, Lin IC, Kuo PL, Lin CC. Anti-proliferative effect of apigenin and its apoptotic induction in human Hep G2 cells. Cancer Lett. 2006;237:207-214. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 103] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 144. | Subhasitanont P, Chokchaichamnankit D, Chiablaem K, Keeratichamroen S, Ngiwsara L, Paricharttanakul NM, Lirdprapamongkol K, Weeraphan C, Svasti J, Srisomsap C. Apigenin inhibits growth and induces apoptosis in human cholangiocarcinoma cells. Oncol Lett. 2017;14:4361-4371. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 14] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 145. | Ujiki MB, Ding XZ, Salabat MR, Bentrem DJ, Golkar L, Milam B, Talamonti MS, Bell RH Jr, Iwamura T, Adrian TE. Apigenin inhibits pancreatic cancer cell proliferation through G2/M cell cycle arrest. Mol Cancer. 2006;5:76. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 110] [Cited by in RCA: 132] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 146. | Engelmann C, Blot E, Panis Y, Bauer S, Trochon V, Nagy HJ, Lu H, Soria C. Apigenin--strong cytostatic and anti-angiogenic action in vitro contrasted by lack of efficacy in vivo. Phytomedicine. 2002;9:489-495. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 21] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 147. | Hoensch H, Groh B, Edler L, Kirch W. Prospective cohort comparison of flavonoid treatment in patients with resected colorectal cancer to prevent recurrence. World J Gastroenterol. 2008;14:2187-2193. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 107] [Cited by in RCA: 82] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 148. | Hung MH, Chen YL, Chen LJ, Chu PY, Hsieh FS, Tsai MH, Shih CT, Chao TI, Huang CY, Chen KF. Canagliflozin inhibits growth of hepatocellular carcinoma via blocking glucose-influx-induced β-catenin activation. Cell Death Dis. 2019;10:420. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 66] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 149. | Tiemin P, Peng X, Qingfu L, Yan W, Junlin X, Zhefeng H, Ming Z, Desen L, Qinghui M. Dysregulation of the miR-148a-GLUT1 axis promotes the progression and chemoresistance of human intrahepatic cholangiocarcinoma. Oncogenesis. 2020;9:19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 21] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 150. | Shibuya K, Okada M, Suzuki S, Seino M, Seino S, Takeda H, Kitanaka C. Targeting the facilitative glucose transporter GLUT1 inhibits the self-renewal and tumor-initiating capacity of cancer stem cells. Oncotarget. 2015;6:651-661. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 124] [Cited by in RCA: 143] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 151. | Liu W, Fang Y, Wang XT, Liu J, Dan X, Sun LL. Overcoming 5-Fu resistance of colon cells through inhibition of Glut1 by the specific inhibitor WZB117. Asian Pac J Cancer Prev. 2014;15:7037-7041. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 43] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 152. | Cao X, Cao Y, Zhao H, Wang P, Zhu Z. Prolyl 4-hydroxylase P4HA1 Mediates the Interplay Between Glucose Metabolism and Stemness in Pancreatic Cancer Cells. Curr Stem Cell Res Ther. 2023;18:712-719. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 153. | Kraus D, Reckenbeil J, Veit N, Kuerpig S, Meisenheimer M, Beier I, Stark H, Winter J, Probstmeier R. Targeting glucose transport and the NAD pathway in tumor cells with STF-31: a re-evaluation. Cell Oncol (Dordr). 2018;41:485-494. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 61] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 154. | Sawayama H, Ogata Y, Ishimoto T, Mima K, Hiyoshi Y, Iwatsuki M, Baba Y, Miyamoto Y, Yoshida N, Baba H. Glucose transporter 1 regulates the proliferation and cisplatin sensitivity of esophageal cancer. Cancer Sci. 2019;110:1705-1714. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 57] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 155. | Gauthier-Coles G, Bröer A, McLeod MD, George AJ, Hannan RD, Bröer S. Identification and characterization of a novel SNAT2 (SLC38A2) inhibitor reveals synergy with glucose transport inhibition in cancer cells. Front Pharmacol. 2022;13:963066. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 12] [Reference Citation Analysis (0)] |
| 156. | Guo L, Zhang W, Xie Y, Chen X, Olmstead EE, Lian M, Zhang B, Zaytseva YY, Evers BM, Spielmann HP, Liu X, Watt DS, Liu C. Diaminobutoxy-substituted Isoflavonoid (DBI-1) Enhances the Therapeutic Efficacy of GLUT1 Inhibitor BAY-876 by Modulating Metabolic Pathways in Colon Cancer Cells. Mol Cancer Ther. 2022;21:740-750. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 15] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 157. | Laussel C, Léon S. Cellular toxicity of the metabolic inhibitor 2-deoxyglucose and associated resistance mechanisms. Biochem Pharmacol. 2020;182:114213. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 58] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 158. | Shen K, Liu B, Zhou X, Ji Y, Chen L, Wang Q, Xue W. The Evolving Role of (18)F-FDG PET/CT in Diagnosis and Prognosis Prediction in Progressive Prostate Cancer. Front Oncol. 2021;11:683793. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 27] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 159. | Xu Y, Wang Q, Zhang L, Zheng M. 2-Deoxy-D-glucose enhances TRAIL-induced apoptosis in human gastric cancer cells through downregulating JNK-mediated cytoprotective autophagy. Cancer Chemother Pharmacol. 2018;81:555-564. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 14] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 160. | Mack P, Ahrén B, Jeppsson B, Kan Z, Bengmark S. Influence of 2-deoxy-D-glucose and arterial ischaemia on glucose oxidation and growth of liver cancer in the rat. Eur J Cancer Clin Oncol. 1988;24:1433-1437. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 161. | Saydjari R, Alexander RW, Barranco SC, Townsend CM Jr, Thompson JC. The effects of 2-deoxy-D-glucose and alpha-difluoromethylornithine on the growth of pancreatic cancer in vivo. Pancreas. 1989;4:38-43. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 162. | Takemura A, Che XF, Tabuchi T, Moriya S, Miyazawa K, Tomoda A. Enhancement of cytotoxic and pro-apoptotic effects of 2-aminophenoxazine-3-one on the rat hepatocellular carcinoma cell line dRLh-84, the human hepatocellular carcinoma cell line HepG2, and the rat normal hepatocellular cell line RLN-10 in combination with 2-deoxy-D-glucose. Oncol Rep. 2012;27:347-355. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 6] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 163. | Maher JC, Savaraj N, Priebe W, Liu H, Lampidis TJ. Differential sensitivity to 2-deoxy-D-glucose between two pancreatic cell lines correlates with GLUT-1 expression. Pancreas. 2005;30:e34-e39. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 35] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 164. | Zhang XD, Deslandes E, Villedieu M, Poulain L, Duval M, Gauduchon P, Schwartz L, Icard P. Effect of 2-deoxy-D-glucose on various malignant cell lines in vitro. Anticancer Res. 2006;26:3561-3566. [PubMed] |
| 165. | Cui Y, Yang D, Wang W, Zhang L, Liu H, Ma S, Guo W, Yao M, Zhang K, Li W, Zhang Y, Guan F. Nicotinamide N-methyltransferase decreases 5-fluorouracil sensitivity in human esophageal squamous cell carcinoma through metabolic reprogramming and promoting the Warburg effect. Mol Carcinog. 2020;59:940-954. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 29] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 166. | Fan T, Sun G, Sun X, Zhao L, Zhong R, Peng Y. Tumor Energy Metabolism and Potential of 3-Bromopyruvate as an Inhibitor of Aerobic Glycolysis: Implications in Tumor Treatment. Cancers (Basel). 2019;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 131] [Cited by in RCA: 124] [Article Influence: 20.7] [Reference Citation Analysis (0)] |
| 167. | Ko YH, Pedersen PL, Geschwind JF. Glucose catabolism in the rabbit VX2 tumor model for liver cancer: characterization and targeting hexokinase. Cancer Lett. 2001;173:83-91. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 260] [Cited by in RCA: 274] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 168. | Xian SL, Cao W, Zhang XD, Lu YF. Inhibitory effects of 3-bromopyruvate on human gastric cancer implant tumors in nude mice. Asian Pac J Cancer Prev. 2014;15:3175-3178. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 169. | Cao X, Jia G, Zhang T, Yang M, Wang B, Wassenaar PA, Cheng H, Knopp MV, Sun D. Non-invasive MRI tumor imaging and synergistic anticancer effect of HSP90 inhibitor and glycolysis inhibitor in RIP1-Tag2 transgenic pancreatic tumor model. Cancer Chemother Pharmacol. 2008;62:985-994. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 29] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 170. | Ihrlund LS, Hernlund E, Khan O, Shoshan MC. 3-Bromopyruvate as inhibitor of tumour cell energy metabolism and chemopotentiator of platinum drugs. Mol Oncol. 2008;2:94-101. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 95] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 171. | Sun X, Sun G, Huang Y, Hao Y, Tang X, Zhang N, Zhao L, Zhong R, Peng Y. 3-Bromopyruvate regulates the status of glycolysis and BCNU sensitivity in human hepatocellular carcinoma cells. Biochem Pharmacol. 2020;177:113988. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 22] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 172. | Ko YH, Verhoeven HA, Lee MJ, Corbin DJ, Vogl TJ, Pedersen PL. A translational study "case report" on the small molecule "energy blocker" 3-bromopyruvate (3BP) as a potent anticancer agent: from bench side to bedside. J Bioenerg Biomembr. 2012;44:163-170. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 120] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 173. | Huang Y, Sun G, Sun X, Li F, Zhao L, Zhong R, Peng Y. The Potential of Lonidamine in Combination with Chemotherapy and Physical Therapy in Cancer Treatment. Cancers (Basel). 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 81] [Cited by in RCA: 75] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 174. | Calabresi F, Di Lauro L, Marolla P, Curcio CG, Paoletti G, Calabró A, Giannarelli D, Ballatore P, Foggi CM, Di Palma M. Fluorouracil, doxorubicin, and cyclophosphamide versus fluorouracil, doxorubicin, and cyclophosphamide plus lonidamine for the treatment of advanced breast cancer: a multicentric randomized clinical study. Semin Oncol. 1991;18:66-72. [PubMed] |
| 175. | Ricotti L, Tesei A, De Paola F, Milandri C, Amadori D, Frassineti GL, Ulivi P, Zoli W. Potentiation of antiproliferative drug activity by lonidamine in hepatocellular carcinoma cells. J Chemother. 2003;15:480-487. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 4] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 176. | Thamrongwaranggoon U, Seubwai W, Phoomak C, Sangkhamanon S, Cha'on U, Boonmars T, Wongkham S. Targeting hexokinase II as a possible therapy for cholangiocarcinoma. Biochem Biophys Res Commun. 2017;484:409-415. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 29] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 177. | Villa R, Zaffaroni N, Orlandi L, Bearzatto A, Costa A, Silvestrini R. In vitro effect of lonidamine on the cytotoxicity of mitomycin C and BCNU in human colon adenocarcinoma cells. Eur J Cancer. 1994;30A:1534-1540. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 9] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 178. | Zaniboni A, Meriggi F, Alghisi A, Mutti S, Distefano L, Rizzi A, Bettini L, Simoncini E, Marpicati P, Montini E. Mitomycin-C and lonidamine as second-line therapy for colorectal cancer: a phase II study. Tumori. 1995;81:435-437. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 179. | Gebbia V, Testa A, Cannata G, Tirrito M, Longo A, Sciume C, Valdesi M, Salamone G, Gebbia N, Leo P. Second line chemotherapy for metastatic colorectal carcinoma. Oncol Rep. 1996;3:867-869. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 180. | Shi L, Pan H, Liu Z, Xie J, Han W. Roles of PFKFB3 in cancer. Signal Transduct Target Ther. 2017;2:17044. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 107] [Cited by in RCA: 200] [Article Influence: 25.0] [Reference Citation Analysis (0)] |
| 181. | Varghese E, Samuel SM, Líšková A, Samec M, Kubatka P, Büsselberg D. Targeting Glucose Metabolism to Overcome Resistance to Anticancer Chemotherapy in Breast Cancer. Cancers (Basel). 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 107] [Cited by in RCA: 140] [Article Influence: 28.0] [Reference Citation Analysis (0)] |
| 182. | Mejias M, Gallego J, Naranjo-Suarez S, Ramirez M, Pell N, Manzano A, Suñer C, Bartrons R, Mendez R, Fernandez M. CPEB4 Increases Expression of PFKFB3 to Induce Glycolysis and Activate Mouse and Human Hepatic Stellate Cells, Promoting Liver Fibrosis. Gastroenterology. 2020;159:273-288. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 88] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 183. | Conradi LC, Brajic A, Cantelmo AR, Bouché A, Kalucka J, Pircher A, Brüning U, Teuwen LA, Vinckier S, Ghesquière B, Dewerchin M, Carmeliet P. Tumor vessel disintegration by maximum tolerable PFKFB3 blockade. Angiogenesis. 2017;20:599-613. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 72] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 184. | Klarer AC, O'Neal J, Imbert-Fernandez Y, Clem A, Ellis SR, Clark J, Clem B, Chesney J, Telang S. Inhibition of 6-phosphofructo-2-kinase (PFKFB3) induces autophagy as a survival mechanism. Cancer Metab. 2014;2:2. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 68] [Cited by in RCA: 80] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 185. | Zhu W, Ye L, Zhang J, Yu P, Wang H, Ye Z, Tian J. PFK15, a Small Molecule Inhibitor of PFKFB3, Induces Cell Cycle Arrest, Apoptosis and Inhibits Invasion in Gastric Cancer. PLoS One. 2016;11:e0163768. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 83] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 186. | Shi WK, Zhu XD, Wang CH, Zhang YY, Cai H, Li XL, Cao MQ, Zhang SZ, Li KS, Sun HC. PFKFB3 blockade inhibits hepatocellular carcinoma growth by impairing DNA repair through AKT. Cell Death Dis. 2018;9:428. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 40] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 187. | Dou Q, Grant AK, Callahan C, Coutinho de Souza P, Mwin D, Booth AL, Nasser I, Moussa M, Ahmed M, Tsai LL. PFKFB3-mediated Pro-glycolytic Shift in Hepatocellular Carcinoma Proliferation. Cell Mol Gastroenterol Hepatol. 2023;15:61-75. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 14] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 188. | Richardson DA, Sritangos P, James AD, Sultan A, Bruce JIE. Metabolic regulation of calcium pumps in pancreatic cancer: role of phosphofructokinase-fructose-bisphosphatase-3 (PFKFB3). Cancer Metab. 2020;8:2. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 22] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 189. | Horváthová J, Moravčík R, Matúšková M, Šišovský V, Boháč A, Zeman M. Inhibition of Glycolysis Suppresses Cell Proliferation and Tumor Progression In Vivo: Perspectives for Chronotherapy. Int J Mol Sci. 2021;22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 12] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 190. | Redman RA, Pohlmann PR, Kurman MR, Tapolsky G, Chesney JA. A phase I, dose-escalation, multi-center study of PFK-158 in patients with advanced solid malignancies explores a first-in-man inhbibitor of glycolysis. J Clin Oncol. 2015;33:tps2606. [RCA] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 10] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 191. | Chhipa AS, Patel S. Targeting pyruvate kinase muscle isoform 2 (PKM2) in cancer: What do we know so far? Life Sci. 2021;280:119694. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 22] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 192. | Kim SJ, Kim JM, Shim SH, Chang HI. Shikonin induces cell cycle arrest in human gastric cancer (AGS) by early growth response 1 (Egr1)-mediated p21 gene expression. J Ethnopharmacol. 2014;151:1064-1071. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 29] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 193. | Tang JC, Zhao J, Long F, Chen JY, Mu B, Jiang Z, Ren Y, Yang J. Efficacy of Shikonin against Esophageal Cancer Cells and its possible mechanisms in vitro and in vivo. J Cancer. 2018;9:32-40. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 39] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 194. | Yingkun N, Lvsong Z, Huimin Y. Shikonin inhibits the proliferation and induces the apoptosis of human HepG2 cells. Can J Physiol Pharmacol. 2010;88:1138-1146. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 39] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 195. | Zhou G, Yang Z, Wang X, Tao R, Zhou Y. TRAIL Enhances Shikonin Induced Apoptosis through ROS/JNK Signaling in Cholangiocarcinoma Cells. Cell Physiol Biochem. 2017;42:1073-1086. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 35] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 196. | Wang Y, Zhou Y, Jia G, Han B, Liu J, Teng Y, Lv J, Song Z, Li Y, Ji L, Pan S, Jiang H, Sun B. Shikonin suppresses tumor growth and synergizes with gemcitabine in a pancreatic cancer xenograft model: Involvement of NF-κB signaling pathway. Biochem Pharmacol. 2014;88:322-333. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 60] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 197. | Li MY, Mi C, Wang KS, Wang Z, Zuo HX, Piao LX, Xu GH, Li X, Ma J, Jin X. Shikonin suppresses proliferation and induces cell cycle arrest through the inhibition of hypoxia-inducible factor-1α signaling. Chem Biol Interact. 2017;274:58-67. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 26] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 198. | Diaconu CC, Szathmári M, Kéri G, Venetianer A. Apoptosis is induced in both drug-sensitive and multidrug-resistant hepatoma cells by somatostatin analogue TT-232. Br J Cancer. 1999;80:1197-1203. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 22] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 199. | Lee JU, Hosotani R, Wada M, Doi R, Koshiba T, Fujimoto K, Miyamoto Y, Tsuji S, Nakajima S, Hirohashi M, Uehara T, Arano Y, Fujii N, Imamura M. Antiproliferative activity induced by the somatostatin analogue, TT-232, in human pancreatic cancer cells. Eur J Cancer. 2002;38:1526-1534. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 15] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 200. | Tejeda M, Gaal D, Barna K, Csuka O, Kéri G. The antitumor activity of the somatostatin structural derivative (TT-232) on different human tumor xenografts. Anticancer Res. 2003;23:4061-4066. [PubMed] |
| 201. | Zhao Y, Butler EB, Tan M. Targeting cellular metabolism to improve cancer therapeutics. Cell Death Dis. 2013;4:e532. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 668] [Cited by in RCA: 793] [Article Influence: 66.1] [Reference Citation Analysis (0)] |
| 202. | Michelakis ED, Webster L, Mackey JR. Dichloroacetate (DCA) as a potential metabolic-targeting therapy for cancer. Br J Cancer. 2008;99:989-994. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 469] [Cited by in RCA: 512] [Article Influence: 30.1] [Reference Citation Analysis (0)] |
| 203. | Hur H, Xuan Y, Kim YB, Lee G, Shim W, Yun J, Ham IH, Han SU. Expression of pyruvate dehydrogenase kinase-1 in gastric cancer as a potential therapeutic target. Int J Oncol. 2013;42:44-54. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 108] [Cited by in RCA: 111] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 204. | Twarock S, Reichert C, Bach K, Reiners O, Kretschmer I, Gorski DJ, Gorges K, Grandoch M, Fischer JW. Inhibition of the hyaluronan matrix enhances metabolic anticancer therapy by dichloroacetate in vitro and in vivo. Br J Pharmacol. 2019;176:4474-4490. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 15] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 205. | Sun L, Jiang Y, Yan X, Dai X, Huang C, Chen L, Li T, Zhang Y, Xiao H, Yang M, Xiang L, Chen S, Li S, Chen A, He F, Lian J. Dichloroacetate enhances the anti-tumor effect of sorafenib via modulating the ROS-JNK-Mcl-1 pathway in liver cancer cells. Exp Cell Res. 2021;406:112755. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 2] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 206. | Tataranni T, Agriesti F, Pacelli C, Ruggieri V, Laurenzana I, Mazzoccoli C, Sala GD, Panebianco C, Pazienza V, Capitanio N, Piccoli C. Dichloroacetate Affects Mitochondrial Function and Stemness-Associated Properties in Pancreatic Cancer Cell Lines. Cells. 2019;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 42] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 207. | Babu E, Ramachandran S, CoothanKandaswamy V, Elangovan S, Prasad PD, Ganapathy V, Thangaraju M. Role of SLC5A8, a plasma membrane transporter and a tumor suppressor, in the antitumor activity of dichloroacetate. Oncogene. 2011;30:4026-4037. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 70] [Cited by in RCA: 68] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 208. | Bull RJ, Sanchez IM, Nelson MA, Larson JL, Lansing AJ. Liver tumor induction in B6C3F1 mice by dichloroacetate and trichloroacetate. Toxicology. 1990;63:341-359. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 191] [Cited by in RCA: 178] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 209. | Claps G, Faouzi S, Quidville V, Chehade F, Shen S, Vagner S, Robert C. The multiple roles of LDH in cancer. Nat Rev Clin Oncol. 2022;19:749-762. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 136] [Reference Citation Analysis (0)] |
| 210. | Zhou Y, Tao P, Wang M, Xu P, Lu W, Lei P, You Q. Development of novel human lactate dehydrogenase A inhibitors: High-throughput screening, synthesis, and biological evaluations. Eur J Med Chem. 2019;177:105-115. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 28] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 211. | Kim EY, Chung TW, Han CW, Park SY, Park KH, Jang SB, Ha KT. A Novel Lactate Dehydrogenase Inhibitor, 1-(Phenylseleno)-4-(Trifluoromethyl) Benzene, Suppresses Tumor Growth through Apoptotic Cell Death. Sci Rep. 2019;9:3969. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 70] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 212. | Liu X, Yang Z, Chen Z, Chen R, Zhao D, Zhou Y, Qiao L. Effects of the suppression of lactate dehydrogenase A on the growth and invasion of human gastric cancer cells. Oncol Rep. 2015;33:157-162. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 47] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 213. | Zhao Z, Han F, Yang S, Wu J, Zhan W. Oxamate-mediated inhibition of lactate dehydrogenase induces protective autophagy in gastric cancer cells: involvement of the Akt-mTOR signaling pathway. Cancer Lett. 2015;358:17-26. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 124] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 214. | Forkasiewicz A, Stach W, Wierzbicki J, Stach K, Tabola R, Hryniewicz-Jankowska A, Augoff K. Effect of LDHA Inhibition on TNF-α-Induced Cell Migration in Esophageal Cancers. Int J Mol Sci. 2022;23. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 215. | Manerba M, Di Ianni L, Govoni M, Roberti M, Recanatini M, Di Stefano G. LDH inhibition impacts on heat shock response and induces senescence of hepatocellular carcinoma cells. Eur J Pharm Sci. 2017;105:91-98. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 27] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 216. | Lu QY, Zhang L, Yee JK, Go VW, Lee WN. Metabolic Consequences of LDHA inhibition by Epigallocatechin Gallate and Oxamate in MIA PaCa-2 Pancreatic Cancer Cells. Metabolomics. 2015;11:71-80. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 59] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 217. | Miskimins WK, Ahn HJ, Kim JY, Ryu S, Jung YS, Choi JY. Synergistic anti-cancer effect of phenformin and oxamate. PLoS One. 2014;9:e85576. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 97] [Cited by in RCA: 104] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 218. | Manerba M, Di Ianni L, Govoni M, Roberti M, Recanatini M, Di Stefano G. Lactate dehydrogenase inhibitors can reverse inflammation induced changes in colon cancer cells. Eur J Pharm Sci. 2017;96:37-44. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 44] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 219. | Wendt EHU, Schoenrogge M, Vollmar B, Zechner D. Galloflavin Plus Metformin Treatment Impairs Pancreatic Cancer Cells. Anticancer Res. 2020;40:153-160. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 220. | Moir JAG, Long A, Haugk B, French JJ, Charnley RM, Manas DM, Wedge SR, Mann J, Robinson SM, White SA. Therapeutic Strategies Toward Lactate Dehydrogenase Within the Tumor Microenvironment of Pancreatic Cancer. Pancreas. 2020;49:1364-1371. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 8] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 221. | Yang J, Wang C, Zhao F, Luo X, Qin M, Arunachalam E, Ge Z, Wang N, Deng X, Jin G, Cong W, Qin W. Loss of FBP1 facilitates aggressive features of hepatocellular carcinoma cells through the Warburg effect. Carcinogenesis. 2017;38:134-143. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 30] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 222. | Le A, Cooper CR, Gouw AM, Dinavahi R, Maitra A, Deck LM, Royer RE, Vander Jagt DL, Semenza GL, Dang CV. Inhibition of lactate dehydrogenase A induces oxidative stress and inhibits tumor progression. Proc Natl Acad Sci U S A. 2010;107:2037-2042. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 893] [Cited by in RCA: 1131] [Article Influence: 75.4] [Reference Citation Analysis (0)] |
| 223. | Rajeshkumar NV, Dutta P, Yabuuchi S, de Wilde RF, Martinez GV, Le A, Kamphorst JJ, Rabinowitz JD, Jain SK, Hidalgo M, Dang CV, Gillies RJ, Maitra A. Therapeutic Targeting of the Warburg Effect in Pancreatic Cancer Relies on an Absence of p53 Function. Cancer Res. 2015;75:3355-3364. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 127] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 224. | Mohammad GH, Vassileva V, Acedo P, Olde Damink SWM, Malago M, Dhar DK, Pereira SP. Targeting Pyruvate Kinase M2 and Lactate Dehydrogenase A Is an Effective Combination Strategy for the Treatment of Pancreatic Cancer. Cancers (Basel). 2019;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 41] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 225. | Kim D, Koh B, Kim KR, Kim KY, Jung WH, Kim HY, Kim S, Dal Rhee S. Anticancer effect of XAV939 is observed by inhibiting lactose dehydrogenase A in a 3-dimensional culture of colorectal cancer cells. Oncol Lett. 2019;18:4858-4864. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 226. | Yu Y, Deck JA, Hunsaker LA, Deck LM, Royer RE, Goldberg E, Vander Jagt DL. Selective active site inhibitors of human lactate dehydrogenases A4, B4, and C4. Biochem Pharmacol. 2001;62:81-89. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 103] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 227. | Gunassekaran GR, Priya DK, Gayathri R, Sakthisekaran D. In vitro and in vivo studies on antitumor effects of gossypol on human stomach adenocarcinoma (AGS) cell line and MNNG induced experimental gastric cancer. Biochem Biophys Res Commun. 2011;411:661-666. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 13] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 228. | Xin J, Zhan YH, Xia LM, Zhu HW, Nie YZ, Liang JM, Tian J. ApoG2 as the most potent gossypol derivatives inhibits cell growth and induces apoptosis on gastric cancer cells. Biomed Pharmacother. 2013;67:88-95. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 229. | Xin J, Zhan Y, Liu M, Hu H, Xia L, Nie Y, Wu K, Liang J, Tian J. ApoG2 induces ER stress-dependent apoptosis in gastric cancer cells in vitro and its real-time evaluation by bioluminescence imaging in vivo. Cancer Lett. 2013;336:260-269. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 11] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 230. | Wei X, Duan W, Li Y, Zhang S, Xin X, Sun L, Gao M, Li Q, Wang D. AT101 exerts a synergetic efficacy in gastric cancer patients with 5-FU based treatment through promoting apoptosis and autophagy. Oncotarget. 2016;7:34430-34441. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 23] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 231. | Song S, Chen Q, Li Y, Lei G, Scott A, Huo L, Li CY, Estrella JS, Correa A, Pizzi MP, Ma L, Jin J, Liu B, Wang Y, Xiao L, Hofstetter WL, Lee JH, Weston B, Bhutani M, Shanbhag N, Johnson RL, Gan B, Wei S, Ajani JA. Targeting cancer stem cells with a pan-BCL-2 inhibitor in preclinical and clinical settings in patients with gastroesophageal carcinoma. Gut. 2021;70:2238-2248. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 42] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 232. | Wang Y, Lonard DM, Yu Y, Chow DC, Palzkill TG, O'Malley BW. Small molecule inhibition of the steroid receptor coactivators, SRC-3 and SRC-1. Mol Endocrinol. 2011;25:2041-2053. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 97] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 233. | Cheng P, Ni Z, Dai X, Wang B, Ding W, Rae Smith A, Xu L, Wu D, He F, Lian J. The novel BH-3 mimetic apogossypolone induces Beclin-1- and ROS-mediated autophagy in human hepatocellular carcinoma [corrected] cells. Cell Death Dis. 2013;4:e489. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 62] [Cited by in RCA: 73] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 234. | Mohammad RM, Wang S, Banerjee S, Wu X, Chen J, Sarkar FH. Nonpeptidic small-molecule inhibitor of Bcl-2 and Bcl-XL, (-)-Gossypol, enhances biological effect of genistein against BxPC-3 human pancreatic cancer cell line. Pancreas. 2005;31:317-324. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 53] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 235. | Thakur A, Lum LG, Schalk D, Azmi A, Banerjee S, Sarkar FH, Mohommad R. Pan-Bcl-2 inhibitor AT-101 enhances tumor cell killing by EGFR targeted T cells. PLoS One. 2012;7:e47520. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 9] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 236. | Yuan Y, Tang AJ, Castoreno AB, Kuo SY, Wang Q, Kuballa P, Xavier R, Shamji AF, Schreiber SL, Wagner BK. Gossypol and an HMT G9a inhibitor act in synergy to induce cell death in pancreatic cancer cells. Cell Death Dis. 2013;4:e690. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 68] [Cited by in RCA: 89] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 237. | Lee S, Hong E, Jo E, Kim ZH, Yim KJ, Woo SH, Choi YS, Jang HJ. Gossypol Induces Apoptosis of Human Pancreatic Cancer Cells via CHOP/Endoplasmic Reticulum Stress Signaling Pathway. J Microbiol Biotechnol. 2022;32:645-656. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 5] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 238. | Wang X, Wang J, Wong SC, Chow LS, Nicholls JM, Wong YC, Liu Y, Kwong DL, Sham JS, Tsa SW. Cytotoxic effect of gossypol on colon carcinoma cells. Life Sci. 2000;67:2663-2671. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 59] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 239. | Zhang M, Liu H, Guo R, Ling Y, Wu X, Li B, Roller PP, Wang S, Yang D. Molecular mechanism of gossypol-induced cell growth inhibition and cell death of HT-29 human colon carcinoma cells. Biochem Pharmacol. 2003;66:93-103. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 121] [Cited by in RCA: 131] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 240. | Wang X, Zhang C, Yan X, Lan B, Wang J, Wei C, Cao X, Wang R, Yao J, Zhou T, Zhou M, Liu Q, Jiang B, Jiang P, Kesari S, Lin X, Guo F. A Novel Bioavailable BH3 Mimetic Efficiently Inhibits Colon Cancer via Cascade Effects of Mitochondria. Clin Cancer Res. 2016;22:1445-1458. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 18] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 241. | Sun X, Wang M, Yao L, Li X, Dong H, Li M, Sun T, Liu X, Liu Y, Xu Y. Role of Proton-Coupled Monocarboxylate Transporters in Cancer: From Metabolic Crosstalk to Therapeutic Potential. Front Cell Dev Biol. 2020;8:651. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 46] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 242. | Tao Z, Huang C, Wang D, Wang Q, Gao Q, Zhang H, Zhao Y, Wang M, Xu J, Shen B, Zhou C, Zhu W. Lactate induced mesenchymal stem cells activation promotes gastric cancer cells migration and proliferation. Exp Cell Res. 2023;424:113492. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 14] [Reference Citation Analysis (0)] |
| 243. | Wang C, Wen Z, Xie J, Zhao Y, Zhao L, Zhang S, Liu Y, Xue Y, Shi M. MACC1 mediates chemotherapy sensitivity of 5-FU and cisplatin via regulating MCT1 expression in gastric cancer. Biochem Biophys Res Commun. 2017;485:665-671. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 27] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 244. | Grasa L, Chueca E, Arechavaleta S, García-González MA, Sáenz MÁ, Valero A, Hördnler C, Lanas Á, Piazuelo E. Antitumor effects of lactate transport inhibition on esophageal adenocarcinoma cells. J Physiol Biochem. 2023;79:147-161. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 245. | Behrends V, Giskeødegård GF, Bravo-Santano N, Letek M, Keun HC. Acetaminophen cytotoxicity in HepG2 cells is associated with a decoupling of glycolysis from the TCA cycle, loss of NADPH production, and suppression of anabolism. Arch Toxicol. 2019;93:341-353. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 26] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 246. | Beloueche-Babari M, Wantuch S, Casals Galobart T, Koniordou M, Parkes HG, Arunan V, Chung YL, Eykyn TR, Smith PD, Leach MO. MCT1 Inhibitor AZD3965 Increases Mitochondrial Metabolism, Facilitating Combination Therapy and Noninvasive Magnetic Resonance Spectroscopy. Cancer Res. 2017;77:5913-5924. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 99] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 247. | Lee JY, Lee I, Chang WJ, Ahn SM, Lim SH, Kim HS, Yoo KH, Jung KS, Song HN, Cho JH, Kim SY, Kim KM, Lee S, Kim ST, Park SH, Lee J, Park JO, Park YS, Lim HY, Kang WK. MCT4 as a potential therapeutic target for metastatic gastric cancer with peritoneal carcinomatosis. Oncotarget. 2016;7:43492-43503. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 46] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 248. | Kong SC, Nøhr-Nielsen A, Zeeberg K, Reshkin SJ, Hoffmann EK, Novak I, Pedersen SF. Monocarboxylate Transporters MCT1 and MCT4 Regulate Migration and Invasion of Pancreatic Ductal Adenocarcinoma Cells. Pancreas. 2016;45:1036-1047. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 71] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 249. | Le Floch R, Chiche J, Marchiq I, Naiken T, Ilc K, Murray CM, Critchlow SE, Roux D, Simon MP, Pouysségur J. CD147 subunit of lactate/H+ symporters MCT1 and hypoxia-inducible MCT4 is critical for energetics and growth of glycolytic tumors. Proc Natl Acad Sci U S A. 2011;108:16663-16668. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 294] [Cited by in RCA: 342] [Article Influence: 24.4] [Reference Citation Analysis (0)] |
| 250. | Fisel P, Schaeffeler E, Schwab M. Clinical and Functional Relevance of the Monocarboxylate Transporter Family in Disease Pathophysiology and Drug Therapy. Clin Transl Sci. 2018;11:352-364. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 63] [Cited by in RCA: 90] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 251. | Frattaruolo L, Brindisi M, Curcio R, Marra F, Dolce V, Cappello AR. Targeting the Mitochondrial Metabolic Network: A Promising Strategy in Cancer Treatment. Int J Mol Sci. 2020;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 48] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 252. | Kato K, Gong J, Iwama H, Kitanaka A, Tani J, Miyoshi H, Nomura K, Mimura S, Kobayashi M, Aritomo Y, Kobara H, Mori H, Himoto T, Okano K, Suzuki Y, Murao K, Masaki T. The antidiabetic drug metformin inhibits gastric cancer cell proliferation in vitro and in vivo. Mol Cancer Ther. 2012;11:549-560. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 148] [Cited by in RCA: 186] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 253. | Feng Y, Ke C, Tang Q, Dong H, Zheng X, Lin W, Ke J, Huang J, Yeung SC, Zhang H. Metformin promotes autophagy and apoptosis in esophageal squamous cell carcinoma by downregulating Stat3 signaling. Cell Death Dis. 2014;5:e1088. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 196] [Cited by in RCA: 230] [Article Influence: 20.9] [Reference Citation Analysis (0)] |
| 254. | Cai X, Hu X, Cai B, Wang Q, Li Y, Tan X, Hu H, Chen X, Huang J, Cheng J, Jing X. Metformin suppresses hepatocellular carcinoma cell growth through induction of cell cycle G1/G0 phase arrest and p21CIP and p27KIP expression and downregulation of cyclin D1 in vitro and in vivo. Oncol Rep. 2013;30:2449-2457. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 63] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 255. | Fujimori T, Kato K, Fujihara S, Iwama H, Yamashita T, Kobayashi K, Kamada H, Morishita A, Kobara H, Mori H, Okano K, Suzuki Y, Masaki T. Antitumor effect of metformin on cholangiocarcinoma: In vitro and in vivo studies. Oncol Rep. 2015;34:2987-2996. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 31] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 256. | Wang LW, Li ZS, Zou DW, Jin ZD, Gao J, Xu GM. Metformin induces apoptosis of pancreatic cancer cells. World J Gastroenterol. 2008;14:7192-7198. [RCA] [PubMed] [DOI] [Full Text] [Cited by in CrossRef: 120] [Cited by in RCA: 121] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 257. | Tsai CC, Chuang TW, Chen LJ, Niu HS, Chung KM, Cheng JT, Lin KC. Increase in apoptosis by combination of metformin with silibinin in human colorectal cancer cells. World J Gastroenterol. 2015;21:4169-4177. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 35] [Cited by in RCA: 33] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 258. | Wang S, Lin Y, Xiong X, Wang L, Guo Y, Chen Y, Chen S, Wang G, Lin P, Chen H, Yeung SJ, Bremer E, Zhang H. Low-Dose Metformin Reprograms the Tumor Immune Microenvironment in Human Esophageal Cancer: Results of a Phase II Clinical Trial. Clin Cancer Res. 2020;26:4921-4932. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 109] [Article Influence: 21.8] [Reference Citation Analysis (0)] |
| 259. | Ostwal V, Ramaswamy A, Gota V, Bhargava PG, Srinivas S, Shriyan B, Jadhav S, Goel M, Patkar S, Mandavkar S, Naughane D, Daddi A, Nashikkar C, Shetty N, Ankathi SK, Banavali SD. Phase I Study Evaluating Dose De-escalation of Sorafenib with Metformin and Atorvastatin in Hepatocellular Carcinoma (SMASH). Oncologist. 2022;27:165-e222. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 12] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 260. | Khurshed M, Molenaar RJ, van Linde ME, Mathôt RA, Struys EA, van Wezel T, van Noorden CJF, Klümpen HJ, Bovée JVMG, Wilmink JW. A Phase Ib Clinical Trial of Metformin and Chloroquine in Patients with IDH1-Mutated Solid Tumors. Cancers (Basel). 2021;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 19] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 261. | Kordes S, Pollak MN, Zwinderman AH, Mathôt RA, Weterman MJ, Beeker A, Punt CJ, Richel DJ, Wilmink JW. Metformin in patients with advanced pancreatic cancer: a double-blind, randomised, placebo-controlled phase 2 trial. Lancet Oncol. 2015;16:839-847. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 247] [Cited by in RCA: 312] [Article Influence: 31.2] [Reference Citation Analysis (0)] |
| 262. | Reni M, Dugnani E, Cereda S, Belli C, Balzano G, Nicoletti R, Liberati D, Pasquale V, Scavini M, Maggiora P, Sordi V, Lampasona V, Ceraulo D, Di Terlizzi G, Doglioni C, Falconi M, Piemonti L. (Ir)relevance of Metformin Treatment in Patients with Metastatic Pancreatic Cancer: An Open-Label, Randomized Phase II Trial. Clin Cancer Res. 2016;22:1076-1085. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 145] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 263. | Zell JA, McLaren CE, Morgan TR, Lawson MJ, Rezk S, Albers CG, Chen WP, Carmichael JC, Chung J, Richmond E, Rodriguez LM, Szabo E, Ford LG, Pollak MN, Meyskens FL. A Phase IIa Trial of Metformin for Colorectal Cancer Risk Reduction among Individuals with History of Colorectal Adenomas and Elevated Body Mass Index. Cancer Prev Res (Phila). 2020;13:203-212. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 19] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 264. | Petrera M, Paleari L, Clavarezza M, Puntoni M, Caviglia S, Briata IM, Oppezzi M, Mislej EM, Stabuc B, Gnant M, Bachleitner-Hofmann T, Roth W, Scherer D, Haefeli WE, Ulrich CM, DeCensi A. The ASAMET trial: a randomized, phase II, double-blind, placebo-controlled, multicenter, 2 × 2 factorial biomarker study of tertiary prevention with low-dose aspirin and metformin in stage I-III colorectal cancer patients. BMC Cancer. 2018;18:1210. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 27] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 265. | Miranda VC, Braghiroli MI, Faria LD, Bariani G, Alex A, Bezerra Neto JE, Capareli FC, Sabbaga J, Lobo Dos Santos JF, Hoff PM, Riechelmann RP. Phase 2 Trial of Metformin Combined With 5-Fluorouracil in Patients With Refractory Metastatic Colorectal Cancer. Clin Colorectal Cancer. 2016;15:321-328.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 53] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 266. | Hoskins JM, Carey LA, McLeod HL. CYP2D6 and tamoxifen: DNA matters in breast cancer. Nat Rev Cancer. 2009;9:576-586. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 223] [Cited by in RCA: 225] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 267. | Hosoya Y, Kitoh Y, Kobayashi E, Okabe R, Fujimura A, Kanazawa K. Combination effects of tamoxifen plus 5-fluorouracil on gastric cancer cell lines in vitro. Cancer Lett. 1999;140:139-143. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 7] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 268. | Due SL, Watson DI, Bastian I, Ding GQ, Sukocheva OA, Astill DS, Vat L, Hussey DJ. Tamoxifen enhances the cytotoxicity of conventional chemotherapy in esophageal adenocarcinoma cells. Surg Oncol. 2016;25:269-277. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 13] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 269. | Jiang SY, Shyu RY, Yeh MY, Jordan VC. Tamoxifen inhibits hepatoma cell growth through an estrogen receptor independent mechanism. J Hepatol. 1995;23:712-719. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 32] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 270. | Sampson LK, Vickers SM, Ying W, Phillips JO. Tamoxifen-mediated growth inhibition of human cholangiocarcinoma. Cancer Res. 1997;57:1743-1749. [PubMed] |
| 271. | Xie X, Wu MY, Shou LM, Chen LP, Gong FR, Chen K, Li DM, Duan WM, Xie YF, Mao YX, Li W, Tao M. Tamoxifen enhances the anticancer effect of cantharidin and norcantharidin in pancreatic cancer cell lines through inhibition of the protein kinase C signaling pathway. Oncol Lett. 2015;9:837-844. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 24] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 272. | Lointier P, Wildrick DM, Boman BM. Growth effects of tamoxifen on Lovo colon carcinoma cells and cultured cells from normal colonic mucosa. Anticancer Res. 1992;12:1523-1525. [PubMed] |
| 273. | Kuruppu D, Christophi C, Bertram JF, O'Brien PE. Tamoxifen inhibits colorectal cancer metastases in the liver: a study in a murine model. J Gastroenterol Hepatol. 1998;13:521-527. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 14] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 274. | Tomao S, Romiti A, Massidda B, Ionta MT, Farris A, Zullo A, Brescia A, Santuari L, Frati L. A phase II study of gemcitabine and tamoxifen in advanced pancreatic cancer. Anticancer Res. 2002;22:2361-2364. [PubMed] |
| 275. | Eckel F, Lersch C, Lippl F, Assmann G, Schulte-Frohlinde E. Phase II trial of cyclophosphamide, leucovorin, 5-fluorouracil 24-hour infusion and tamoxifen in pancreatic cancer. J Exp Clin Cancer Res. 2000;19:295-300. [PubMed] |
| 276. | Lissoni P, Paolorossi F, Tancini G, Ardizzoia A, Barni S, Brivio F, Maestroni GJ, Chilelli M. A phase II study of tamoxifen plus melatonin in metastatic solid tumour patients. Br J Cancer. 1996;74:1466-1468. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 40] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 277. | Chow PK, Tai BC, Tan CK, Machin D, Win KM, Johnson PJ, Soo KC; Asian-Pacific Hepatocellular Carcinoma Trials Group. High-dose tamoxifen in the treatment of inoperable hepatocellular carcinoma: A multicenter randomized controlled trial. Hepatology. 2002;36:1221-1226. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 155] [Cited by in RCA: 156] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 278. | Janku F, Beom SH, Moon YW, Kim TW, Shin YG, Yim DS, Kim GM, Kim HS, Kim SY, Cheong JH, Lee YW, Geiger B, Yoo S, Thurston A, Welsch D, Rudoltz MS, Rha SY. First-in-human study of IM156, a novel potent biguanide oxidative phosphorylation (OXPHOS) inhibitor, in patients with advanced solid tumors. Invest New Drugs. 2022;40:1001-1010. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 31] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 279. | Aguilar-Valdés A, Noriega LG, Tovar AR, Ibarra-Sánchez MJ, Sosa-Hernández VA, Maravillas-Montero JL, Martínez-Aguilar J. SWATH-MS proteomics of PANC-1 and MIA PaCa-2 pancreatic cancer cells allows identification of drug targets alternative to MEK and PI3K inhibition. Biochem Biophys Res Commun. 2021;552:23-29. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 280. | Yap TA, Daver N, Mahendra M, Zhang J, Kamiya-Matsuoka C, Meric-Bernstam F, Kantarjian HM, Ravandi F, Collins ME, Francesco MED, Dumbrava EE, Fu S, Gao S, Gay JP, Gera S, Han J, Hong DS, Jabbour EJ, Ju Z, Karp DD, Lodi A, Molina JR, Baran N, Naing A, Ohanian M, Pant S, Pemmaraju N, Bose P, Piha-Paul SA, Rodon J, Salguero C, Sasaki K, Singh AK, Subbiah V, Tsimberidou AM, Xu QA, Yilmaz M, Zhang Q, Li Y, Bristow CA, Bhattacharjee MB, Tiziani S, Heffernan TP, Vellano CP, Jones P, Heijnen CJ, Kavelaars A, Marszalek JR, Konopleva M. Complex I inhibitor of oxidative phosphorylation in advanced solid tumors and acute myeloid leukemia: phase I trials. Nat Med. 2023;29:115-126. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 195] [Cited by in RCA: 186] [Article Influence: 93.0] [Reference Citation Analysis (0)] |
| 281. | Roth KG, Mambetsariev I, Kulkarni P, Salgia R. The Mitochondrion as an Emerging Therapeutic Target in Cancer. Trends Mol Med. 2020;26:119-134. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 137] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 282. | Aminzadeh-Gohari S, Weber DD, Vidali S, Catalano L, Kofler B, Feichtinger RG. From old to new - Repurposing drugs to target mitochondrial energy metabolism in cancer. Semin Cell Dev Biol. 2020;98:211-223. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 24] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 283. | Li X, Wang H, Li Z, Li D, Lu X, Ai S, Dong Y, Liu S, Wu J, Guan W. Oxygen tank for synergistic hypoxia relief to enhance mitochondria-targeted photodynamic therapy. Biomater Res. 2022;26:47. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 19] [Reference Citation Analysis (0)] |
| 284. | Gao X, Liu X, Shan W, Liu Q, Wang C, Zheng J, Yao H, Tang R. Anti-malarial atovaquone exhibits anti-tumor effects by inducing DNA damage in hepatocellular carcinoma. Am J Cancer Res. 2018;8:1697-1711. [PubMed] |
| 285. | Cheng G, Hardy M, You M, Kalyanaraman B. Combining PEGylated mito-atovaquone with MCT and Krebs cycle redox inhibitors as a potential strategy to abrogate tumor cell proliferation. Sci Rep. 2022;12:5143. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 11] [Reference Citation Analysis (0)] |
| 286. | Sainero-Alcolado L, Liaño-Pons J, Ruiz-Pérez MV, Arsenian-Henriksson M. Targeting mitochondrial metabolism for precision medicine in cancer. Cell Death Differ. 2022;29:1304-1317. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 177] [Article Influence: 59.0] [Reference Citation Analysis (0)] |
| 287. | Rabben HL, Andersen GT, Olsen MK, Øverby A, Ianevski A, Kainov D, Wang TC, Lundgren S, Grønbech JE, Chen D, Zhao CM. Neural signaling modulates metabolism of gastric cancer. iScience. 2021;24:102091. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 6] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 288. | Inoue J, Kishikawa M, Tsuda H, Nakajima Y, Asakage T, Inazawa J. Identification of PDHX as a metabolic target for esophageal squamous cell carcinoma. Cancer Sci. 2021;112:2792-2802. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 12] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 289. | Zachar Z, Marecek J, Maturo C, Gupta S, Stuart SD, Howell K, Schauble A, Lem J, Piramzadian A, Karnik S, Lee K, Rodriguez R, Shorr R, Bingham PM. Non-redox-active lipoate derivates disrupt cancer cell mitochondrial metabolism and are potent anticancer agents in vivo. J Mol Med (Berl). 2011;89:1137-1148. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 169] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 290. | Alistar A, Morris BB, Desnoyer R, Klepin HD, Hosseinzadeh K, Clark C, Cameron A, Leyendecker J, D'Agostino R Jr, Topaloglu U, Boteju LW, Boteju AR, Shorr R, Zachar Z, Bingham PM, Ahmed T, Crane S, Shah R, Migliano JJ, Pardee TS, Miller L, Hawkins G, Jin G, Zhang W, Pasche B. Safety and tolerability of the first-in-class agent CPI-613 in combination with modified FOLFIRINOX in patients with metastatic pancreatic cancer: a single-centre, open-label, dose-escalation, phase 1 trial. Lancet Oncol. 2017;18:770-778. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 136] [Cited by in RCA: 175] [Article Influence: 21.9] [Reference Citation Analysis (0)] |
| 291. | Arnold C, Demuth P, Seiwert N, Wittmann S, Boengler K, Rasenberger B, Christmann M, Huber M, Brunner T, Linnebacher M, Fahrer J. The Mitochondrial Disruptor Devimistat (CPI-613) Synergizes with Genotoxic Anticancer Drugs in Colorectal Cancer Therapy in a Bim-Dependent Manner. Mol Cancer Ther. 2022;21:100-112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 7] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 292. | Philip PA, Buyse ME, Alistar AT, Rocha Lima CM, Luther S, Pardee TS, Van Cutsem E. A Phase III open-label trial to evaluate efficacy and safety of CPI-613 plus modified FOLFIRINOX (mFFX) versus FOLFIRINOX (FFX) in patients with metastatic adenocarcinoma of the pancreas. Future Oncol. 2019;15:3189-3196. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 70] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 293. | Dang L, Su SM. Isocitrate Dehydrogenase Mutation and (R)-2-Hydroxyglutarate: From Basic Discovery to Therapeutics Development. Annu Rev Biochem. 2017;86:305-331. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 131] [Cited by in RCA: 134] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 294. | DiNardo CD, Stein EM, de Botton S, Roboz GJ, Altman JK, Mims AS, Swords R, Collins RH, Mannis GN, Pollyea DA, Donnellan W, Fathi AT, Pigneux A, Erba HP, Prince GT, Stein AS, Uy GL, Foran JM, Traer E, Stuart RK, Arellano ML, Slack JL, Sekeres MA, Willekens C, Choe S, Wang H, Zhang V, Yen KE, Kapsalis SM, Yang H, Dai D, Fan B, Goldwasser M, Liu H, Agresta S, Wu B, Attar EC, Tallman MS, Stone RM, Kantarjian HM. Durable Remissions with Ivosidenib in IDH1-Mutated Relapsed or Refractory AML. N Engl J Med. 2018;378:2386-2398. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 888] [Cited by in RCA: 1067] [Article Influence: 152.4] [Reference Citation Analysis (0)] |
| 295. | Stein EM, DiNardo CD, Fathi AT, Pollyea DA, Stone RM, Altman JK, Roboz GJ, Patel MR, Collins R, Flinn IW, Sekeres MA, Stein AS, Kantarjian HM, Levine RL, Vyas P, MacBeth KJ, Tosolini A, VanOostendorp J, Xu Q, Gupta I, Lila T, Risueno A, Yen KE, Wu B, Attar EC, Tallman MS, de Botton S. Molecular remission and response patterns in patients with mutant-IDH2 acute myeloid leukemia treated with enasidenib. Blood. 2019;133:676-687. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 264] [Cited by in RCA: 255] [Article Influence: 42.5] [Reference Citation Analysis (0)] |
| 296. | Khurshed M, Aarnoudse N, Hulsbos R, Hira VVV, van Laarhoven HWM, Wilmink JW, Molenaar RJ, van Noorden CJF. IDH1-mutant cancer cells are sensitive to cisplatin and an IDH1-mutant inhibitor counteracts this sensitivity. FASEB J. 2018;32:fj201800547R. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 29] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 297. | Gross MI, Demo SD, Dennison JB, Chen L, Chernov-Rogan T, Goyal B, Janes JR, Laidig GJ, Lewis ER, Li J, Mackinnon AL, Parlati F, Rodriguez ML, Shwonek PJ, Sjogren EB, Stanton TF, Wang T, Yang J, Zhao F, Bennett MK. Antitumor activity of the glutaminase inhibitor CB-839 in triple-negative breast cancer. Mol Cancer Ther. 2014;13:890-901. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 587] [Cited by in RCA: 788] [Article Influence: 71.6] [Reference Citation Analysis (0)] |
| 298. | Le A, Lane AN, Hamaker M, Bose S, Gouw A, Barbi J, Tsukamoto T, Rojas CJ, Slusher BS, Zhang H, Zimmerman LJ, Liebler DC, Slebos RJ, Lorkiewicz PK, Higashi RM, Fan TW, Dang CV. Glucose-independent glutamine metabolism via TCA cycling for proliferation and survival in B cells. Cell Metab. 2012;15:110-121. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 770] [Cited by in RCA: 877] [Article Influence: 67.5] [Reference Citation Analysis (0)] |