Published online Jul 28, 2023. doi: 10.3748/wjg.v29.i28.4397
Peer-review started: May 25, 2023
First decision: June 14, 2023
Revised: June 27, 2023
Accepted: July 7, 2023
Article in press: July 7, 2023
Published online: July 28, 2023
Processing time: 61 Days and 16.4 Hours
Over the past decade, the advent of single cell RNA-sequencing has revolution
Core Tip: Single-cell techniques and omics have taken off in the last few years and the ability to detect individual cellular transcript details has revolutionized the world of research. In the field of gastroenterology in just the last five years, several single-cell techniques have been applied to inflammatory bowel disease research with the identification of novel cellular immune players in the pathogenesis of both ulcerative colitis and Crohn’s disease.
- Citation: Zheng HB. Application of single-cell omics in inflammatory bowel disease. World J Gastroenterol 2023; 29(28): 4397-4404
- URL: https://www.wjgnet.com/1007-9327/full/v29/i28/4397.htm
- DOI: https://dx.doi.org/10.3748/wjg.v29.i28.4397
The gastrointestinal (GI) tract houses a complex network of immune cells, cellular signaling, and intestinal flora. By balancing host tolerance and microbial defense response, the mucosal immune system of the GI tract performs constant surveillance and sampling of antigens and microbes to induce either tolerance or mounts a vigorous immune response to pathogens[1]. Intestinal inflammation and disease occur when this delicate balance breaks down leading to over activation of mucosal immunity and aberrant response to host antigen and commensal organisms[2]. Microscopic disruption of the epithelial barrier, over activation of proinflammatory cells, and the lack of regulatory mechanisms are some pathways that present in clinical chronic GI disorders and distinguishes mucosal immunity from systemic immu
The ability to detect pathogenic pathophysiology is rooted in the organ, the GI tract, and may be missed through studying the systemic immune system. In order to understand the target organ and mucosal immune system, technical advances have been made in the last few years giving us the ability to interrogate down to the microscopic single cell level, thus revolutionizing GI research. Since its inception in 2009, single cell RNA-sequencing (scRNA-seq) has enhanced our ability to comprehensively map and resolve cell types, cellular subsets, and cells states present in both healthy GI tissue and diseased states[3]. The novelty of single-cell technologies vs previous technologies such as bulk-sequencing is the ability to detect rare subsets of cells that may be the aberrant drivers of disease[4]. The homogeneity of bulk-sequen
IBD which includes both ulcerative colitis (UC) and Crohn’s disease (CD) is a chronic complex autoimmune condition characterized by inflammation of the GI tract. The pathogenesis of IBD is thought to develop from an inappropriate immune response towards self-antigens and commensal microbiota in a genetically susceptible host. The advent of scRNA-seq has led to a boom in the number of publications with the application of single cell techniques in IBD research[6,7]. As a disclaimer, this is by no means a comprehensive review of all single-cell studies or single-cell techniques avai
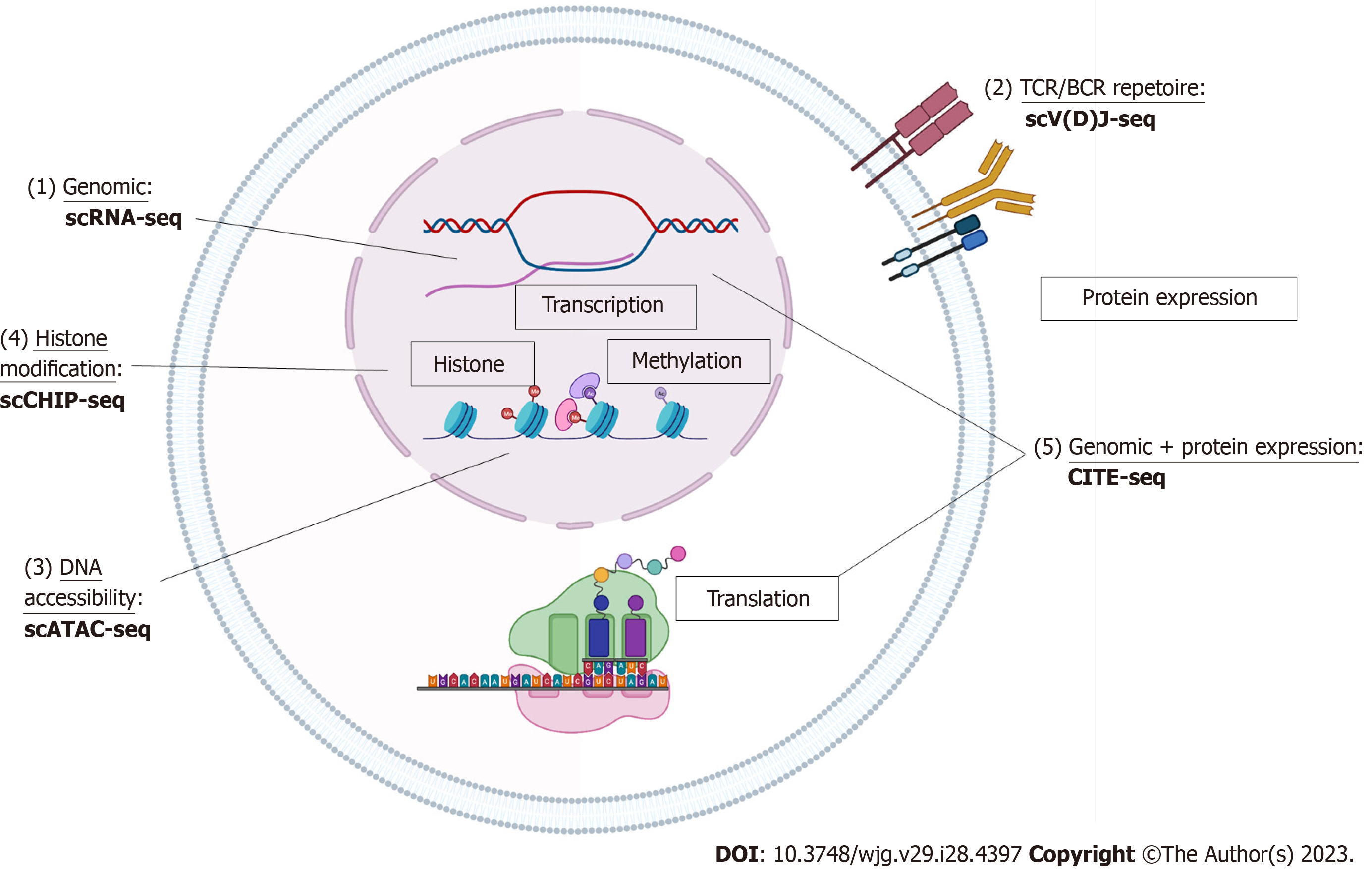
With the commercialization of products and increasing user compatibility, scRNA-seq can be done using a variety of platforms and through multiple approaches (10 × Genomics, Fluidigm, BD Rhapsody, SmartSeq, etc.) whether with multiple well plates, microfluidics, or drop-seq[8,9]. Single-cell data sets can also be created from the 3’ end or 5’ end of mRNA. The basis behind the technology is partitioning individual cells with one uniquely barcoded mRNA-capture medium which has unique molecular code tag for each cell and its contained transcripts. The cell is then lysed and the mRNA is captured by these coded tags and each cell’s mRNA is reverse transcribed into uniquely barcoded cDNA. cDNA libraries are amplified, indexed and sequenced using next-generation sequencing platforms (NextGen). Using the unique molecular code tags, the cDNA can be linked to the cell of origin and abundance of transcripts can be deciphered using the number of copies of the certain cDNA (Figure 1).
With large sets of data, bioinformatics analysis can be challenging. To perform this analysis, FASTQ files are generated from binary base call output from sequencing using software programs that then align the reads from the FASTQ files to a human reference genome/transcriptome using STAR[10]. Gene-cell matrices with principle component analysis can then be used to quantify transcripts of interest using standard workflows to give traditional visualization of single-cell datasets in the form of uniform manifold approximation and projection algorithms or T-distributed stochastic neighbor embedding plots (Figure 1).
In some cases, studies are interested not only in the current state of the cell but what the possible future of how the cell may develop (“pseudotime”). Software programs including Monocle[11] and Census[12] offer toolkits of computational bioinformatic programs specifically designed for the analysis of scRNA-Seq data to look at the temporal resolution of transcriptome dynamics using unsupervised algorithms on scRNA-Seq data collected at multiple time points. Thes programs can be used to recover single-cell gene expression kinetics from a wide array of cellular processes, including differentiation, proliferation and oncogenic transformation to identify branching patterns in the cells. The algorithm finds the longest path through the minimum spanning tree, one that corresponds to the longest sequence of transcriptionally similar cells and produces a ‘trajectory’ of an individual cell’s progress through differentiation that is expressed in units of pseudotime (Figure 1).
T cell receptor (TCR) and B cell receptor (BCR) drive a range of antigen specific adaptive immune responses to pathogens with large highly diverse repertoires to allow for recognition of antigens[9]. The advent of 5’ single-cell sequencing allows researchers to better understand lymphocyte diversity and antigen specificity or essentially the clonality of T and B cells present in disease states. The technique of TCR/BCR is similar to scRNA-seq but utilizes a specialized switch oligo nucleotides and poly dT tail primers[9]. Untemplated C (cytosine) nucleotides to the 3’ end will pair with the end of switch oligo and reverse transcription occurs to capture the TCR/BCR sequences[9]. cDNA libraries of TCR/BCR are created and sequenced per usual protocol (Figure 1).
The modification in gene expression through epigenetics can also be studied down to a single-cell level. Through epigenetics, although the DNA sequence is unaltered, expression patterns can be affected by DNA methylation or chromatin structure. DNA chromatic accessibility is maintained by regulatory elements such as transcription factors, DNA methylation, and histone modification where DNA is wound around into nucleosomes. Single-cell assay for transposase-accessible chromatin-seq (scATAC-seq) takes advantage of the Tn5 transposase which is a bacterial enzyme primed to find and cut open DNA positions[13]. Through scATAC-seq, cells are lysed and nuclei are harvested and then undergo transposition where open DNA fragments are “cut and tagged” with adaptors[13]. Single nuclei run through a similar process as the scRNA-seq mechanism to create barcoded cDNA and sequenced and mapped to reference genome and accessible chromatic regions. These peak calling reads can then be linked to areas such as promoters and enhancers. The distribution of reads across the whole genome, functional analysis the genes associated with the peaks, and peak distribution on functional gene elements can also be done with further analysis[13] (Figure 1).
Single-cell epigenetics in the measurement of transcription factor binding and histone modification can also be studied through single-cell chromatin immunoprecipitation-seq. Generally, cells are encapsulated and lysed within the droplets and chromatin is fragmented. DNA barcodes and chromatin fragments are merged via the microfluidics device[14]. DNA barcodes are ligated to chromatin fragments and droplets are then immunoprecipitated with antibody with a carrier chromatin and library construction[14] (Figure 1).
Cell cellular indexing of transcriptomes and epitopes-seq (CITE)-seq combines traditional protein marker detection (such as flow cytometry) with scRNA-seq to provide phenotypic information such as cell-surface protein expression with transcript information[15]. Antibodies are conjugated to oligonucleotides with antibody specific barcodes using streptavidin-biotin interaction and cells are processed per scRNAseq technique[15]. The cells are then lysed and the Oligo-dT primers capture the oligonucleotides and mRNA to create cDNA[15]. cDNA is then processed into libraries and sequenced (Figure 1).
Kinchen et al[16] published a study describing the activation of intestinal mesenchymal cells subpopulations in adult human UC and dextran-sodium sulfate colitis murine models. Through scRNA-seq, the authors identified SOX6, CD142, and WNT expressing colonic crypt mesenchymal cells consisting of fibroblasts subsets that when dysregulated can lead to impaired epithelial function and inflammation driving the UC state[16]. The human colonic epithelial layer in the UC state is again studied by Parikh et al[17] with further identification of various progenitor cells, colonocytes, and goblets cell also implicated in states of inflammation. The authors go on to describe a new subset of absorptive BEST4+ colono
Boland et al[20] integrated scRNA-seq with scTCR-seq and scBCR-seq to describe cellular states and clonal relation
Martin et al[22] published one of the first CD scRNA-seq data sets linking anti-TNF therapy resistance with a cellular module they entitled GIMATS which stands for IgG plasma cells, inflammatory mononuclear phagocytes, activated T cells, and stromal cells. Through utilizing resected inflamed terminal ileum from adult human CD patients who did not respond to anti-TNF therapy along with uninflamed samples, the authors identified this unique cellular signature to potentially eventually develop biomarkers in prediction of therapy response[22]. Jaeger et al[23] further looked at the T cell composition of the terminal ileum from adult human patients with CD and resected terminal ileums, distinguishing between lamina propria and epithelial layers. Within the epithelial layer, intraepithelial lymphocytes from inflamed tissue included specific NKp30+ gamma delta T cells that expressed ROR gamma which produced IL-26 with an increased in active T helper 17 (Th17) cells[23]. The lamina propria layer also found a Th17 signature with increased CD8+ cells implicating the Th17 pathway in CD[23]. Yokoi et al[24] further classified T cell subsets in CD to identify CD4+ tissue-resident memory T cells that were increased in CD that expressed CD161, CCR5, and CD103 using scRNA-seq and CyTOF. Rosati et al[25] used both bulk TCR repertoire and scRNA-seq on peripheral blood in CD samples to identify a subpopulation of unconventional Crohn-associated invariant T (CAIT) cells with a distinctly unique TCR. These peripheral CAIT cells seem to show a gene expression similar to cells of the innate immune system and mucosal associ
Huang et al[27] describes a pediatric cohort of patients with UC, Crohn’s colitis, and undefined pediatric IBD using scRNA-seq, scTCR-seq, and scBCR-seq. The authors describe a common pathway of impaired cyclic AMP-response signaling in all three pediatric cohorts along with infiltration of PDE4B-expresing and TNF-expressing macrophages within the mucosal samples. The authors also describe a decreased abundance of CD39 expressing intraepithelial T cells along with platelet aggregation and release of 5-hydroxytryptamine at the mucosal level[27]. Futhermore, they demons
Mitsialis et al[28] also reports a study with scRNA-seq to confirm their mass cytometry (CyTOF) findings in patients with UC and CD. Within both the CD and UC cohorts, the authors found an expansion of HLA-DR+CD38+ T cells, CXCR+ plasmablasts, and IL1B+ macrophages and monocytes[28]. Expansion of IL17A+CD161+ effector memory T cells and IL17A+ T regulatory cells along with HLA-DR+CD56+ granulocytes was found within the UC cohort. Within CD, IL1B+HLA-DR+CD38+ T cells, ILB+TNF+IFNG+ naïve B cells, and IL1B+ dendritic cells, and IL1B+ plasmacytoid dendritic cells were expanded in the mucosal samples. Expanded IL1B+ T regulatory cells, IL-B+ dendritic cells, IL1B+ plasmacytoid dendritic cells, and IL1B+ monocytes were found in the peripheral blood of patients with CD but not UC[28].
The future of single-cell technologies within gastroenterology research is already here in the ability to perform single-cell multi-omics and spatial transcriptomic on the target organ (GI tract) and peripheral blood. The ability to multiplex single-cell technologies is in development and the ability to use them on clinical samples will lead to even further growth in the field of IBD. The ability to combine transcriptomics (scRNA-seq), epitopes, (protein expression), and chromatin accessibility (scATAC-seq) from single cells was described by Swanson et al[29] on peripheral blood using transcriptomics, epitopes, accessibility-seq. These multimodal single-cell assays may provide a novel way to uncover and link gene expression, gene regulation, and phenotypic gene expression within a single cell and begin to fully cover genotype with phenotype in specific diseases such as IBD.
Another exciting technique that is more readily available now is spatial transcriptomics where architectural infor
Given the limited scope of this minireview, we have described these technologies only in the field of IBD and did not elaborate into its use in other GI disorders such as GI cancers and other GI diseases such as allergies of the GI tract. Spatial transcriptomics and various single-cell techniques have been successfully applied to colorectal cancers using the GI cancer tissue and resections allowing researchers to better understand the tumor microenvironment to derive better chemotherapeutic targets[32-36]. Allergic disease of the GI tract such as eosinophilic esophagitis is also under interro
The limitation to single-cell technologies is most importantly cost of reagents and cost of sequencing[38]. The ability to integrate multiple single-cell data across multiple platforms and techniques is also a challenge as there are many commercial products out in the market[38].
Single-cell techniques and omics have taken off in the last few years and the ability to detect individual cellular transcript details has revolutionized the world of research. In the field of gastroenterology in just the last five years, several single-cell techniques have been applied to IBD research with the identification of novel cellular immune players in the pathogenesis of both UC and CD. As we continue to develop further immunological techniques, we may begin to detect signals of treatment response in IBD and tailor therapies to immune signatures present in disease state.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: United States
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Miyoshi E, Japan; Xiao Y, China; Yang JS, China S-Editor: Wang JJ L-Editor: A P-Editor: Wang JJ
| 1. | Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336:1268-1273. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3003] [Cited by in RCA: 2984] [Article Influence: 229.5] [Reference Citation Analysis (0)] |
| 2. | Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, Lee JC, Schumm LP, Sharma Y, Anderson CA, Essers J, Mitrovic M, Ning K, Cleynen I, Theatre E, Spain SL, Raychaudhuri S, Goyette P, Wei Z, Abraham C, Achkar JP, Ahmad T, Amininejad L, Ananthakrishnan AN, Andersen V, Andrews JM, Baidoo L, Balschun T, Bampton PA, Bitton A, Boucher G, Brand S, Büning C, Cohain A, Cichon S, D'Amato M, De Jong D, Devaney KL, Dubinsky M, Edwards C, Ellinghaus D, Ferguson LR, Franchimont D, Fransen K, Gearry R, Georges M, Gieger C, Glas J, Haritunians T, Hart A, Hawkey C, Hedl M, Hu X, Karlsen TH, Kupcinskas L, Kugathasan S, Latiano A, Laukens D, Lawrance IC, Lees CW, Louis E, Mahy G, Mansfield J, Morgan AR, Mowat C, Newman W, Palmieri O, Ponsioen CY, Potocnik U, Prescott NJ, Regueiro M, Rotter JI, Russell RK, Sanderson JD, Sans M, Satsangi J, Schreiber S, Simms LA, Sventoraityte J, Targan SR, Taylor KD, Tremelling M, Verspaget HW, De Vos M, Wijmenga C, Wilson DC, Winkelmann J, Xavier RJ, Zeissig S, Zhang B, Zhang CK, Zhao H; International IBD Genetics Consortium (IIBDGC), Silverberg MS, Annese V, Hakonarson H, Brant SR, Radford-Smith G, Mathew CG, Rioux JD, Schadt EE, Daly MJ, Franke A, Parkes M, Vermeire S, Barrett JC, Cho JH. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119-124. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3979] [Cited by in RCA: 3588] [Article Influence: 276.0] [Reference Citation Analysis (0)] |
| 3. | Tang F, Barbacioru C, Wang Y, Nordman E, Lee C, Xu N, Wang X, Bodeau J, Tuch BB, Siddiqui A, Lao K, Surani MA. mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods. 2009;6:377-382. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3043] [Cited by in RCA: 2432] [Article Influence: 152.0] [Reference Citation Analysis (0)] |
| 4. | Yu X, Abbas-Aghababazadeh F, Chen YA, Fridley BL. Statistical and Bioinformatics Analysis of Data from Bulk and Single-Cell RNA Sequencing Experiments. Methods Mol Biol. 2021;2194:143-175. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 20] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 5. | Nath A, Bild AH. Leveraging Single-Cell Approaches in Cancer Precision Medicine. Trends Cancer. 2021;7:359-372. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 25] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 6. | Corridoni D, Chapman T, Antanaviciute A, Satsangi J, Simmons A. Inflammatory Bowel Disease Through the Lens of Single-cell RNA-seq Technologies. Inflamm Bowel Dis. 2020;26:1658-1668. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 32] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 7. | Corridoni D, Pizarro TT. Single-cell Transcriptomics Reveal the Importance of Distinct Epithelial Cell Populations in Ileal-specific, Treatment-naïve, and Treated Crohn's Disease Patients. Inflamm Bowel Dis. 2023;29:334-336. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 8. | Valihrach L, Androvic P, Kubista M. Platforms for Single-Cell Collection and Analysis. Int J Mol Sci. 2018;19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 93] [Cited by in RCA: 117] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 9. | Pai JA, Satpathy AT. High-throughput and single-cell T cell receptor sequencing technologies. Nat Methods. 2021;18:881-892. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 158] [Cited by in RCA: 168] [Article Influence: 42.0] [Reference Citation Analysis (0)] |
| 10. | Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15-21. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22949] [Cited by in RCA: 32861] [Article Influence: 2527.8] [Reference Citation Analysis (0)] |
| 11. | Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, Lennon NJ, Livak KJ, Mikkelsen TS, Rinn JL. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol. 2014;32:381-386. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3105] [Cited by in RCA: 4332] [Article Influence: 393.8] [Reference Citation Analysis (0)] |
| 12. | Qiu X, Hill A, Packer J, Lin D, Ma YA, Trapnell C. Single-cell mRNA quantification and differential analysis with Census. Nat Methods. 2017;14:309-315. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 762] [Cited by in RCA: 1135] [Article Influence: 141.9] [Reference Citation Analysis (0)] |
| 13. | Satpathy AT, Granja JM, Yost KE, Qi Y, Meschi F, McDermott GP, Olsen BN, Mumbach MR, Pierce SE, Corces MR, Shah P, Bell JC, Jhutty D, Nemec CM, Wang J, Wang L, Yin Y, Giresi PG, Chang ALS, Zheng GXY, Greenleaf WJ, Chang HY. Massively parallel single-cell chromatin landscapes of human immune cell development and intratumoral T cell exhaustion. Nat Biotechnol. 2019;37:925-936. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 517] [Cited by in RCA: 611] [Article Influence: 101.8] [Reference Citation Analysis (0)] |
| 14. | Rotem A, Ram O, Shoresh N, Sperling RA, Goren A, Weitz DA, Bernstein BE. Single-cell ChIP-seq reveals cell subpopulations defined by chromatin state. Nat Biotechnol. 2015;33:1165-1172. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 620] [Cited by in RCA: 642] [Article Influence: 64.2] [Reference Citation Analysis (0)] |
| 15. | Stoeckius M, Hafemeister C, Stephenson W, Houck-Loomis B, Chattopadhyay PK, Swerdlow H, Satija R, Smibert P. Simultaneous epitope and transcriptome measurement in single cells. Nat Methods. 2017;14:865-868. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2072] [Cited by in RCA: 1939] [Article Influence: 242.4] [Reference Citation Analysis (0)] |
| 16. | Kinchen J, Chen HH, Parikh K, Antanaviciute A, Jagielowicz M, Fawkner-Corbett D, Ashley N, Cubitt L, Mellado-Gomez E, Attar M, Sharma E, Wills Q, Bowden R, Richter FC, Ahern D, Puri KD, Henault J, Gervais F, Koohy H, Simmons A. Structural Remodeling of the Human Colonic Mesenchyme in Inflammatory Bowel Disease. Cell. 2018;175:372-386.e17. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 282] [Cited by in RCA: 505] [Article Influence: 72.1] [Reference Citation Analysis (0)] |
| 17. | Parikh K, Antanaviciute A, Fawkner-Corbett D, Jagielowicz M, Aulicino A, Lagerholm C, Davis S, Kinchen J, Chen HH, Alham NK, Ashley N, Johnson E, Hublitz P, Bao L, Lukomska J, Andev RS, Björklund E, Kessler BM, Fischer R, Goldin R, Koohy H, Simmons A. Colonic epithelial cell diversity in health and inflammatory bowel disease. Nature. 2019;567:49-55. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 289] [Cited by in RCA: 553] [Article Influence: 92.2] [Reference Citation Analysis (0)] |
| 18. | Smillie CS, Biton M, Ordovas-Montanes J, Sullivan KM, Burgin G, Graham DB, Herbst RH, Rogel N, Slyper M, Waldman J, Sud M, Andrews E, Velonias G, Haber AL, Jagadeesh K, Vickovic S, Yao J, Stevens C, Dionne D, Nguyen LT, Villani AC, Hofree M, Creasey EA, Huang H, Rozenblatt-Rosen O, Garber JJ, Khalili H, Desch AN, Daly MJ, Ananthakrishnan AN, Shalek AK, Xavier RJ, Regev A. Intra- and Inter-cellular Rewiring of the Human Colon during Ulcerative Colitis. Cell. 2019;178:714-730.e22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1042] [Cited by in RCA: 855] [Article Influence: 142.5] [Reference Citation Analysis (0)] |
| 19. | Uzzan M, Martin JC, Mesin L, Livanos AE, Castro-Dopico T, Huang R, Petralia F, Magri G, Kumar S, Zhao Q, Rosenstein AK, Tokuyama M, Sharma K, Ungaro R, Kosoy R, Jha D, Fischer J, Singh H, Keir ME, Ramamoorthi N, O'Gorman WE, Cohen BL, Rahman A, Cossarini F, Seki A, Leyre L, Vaquero ST, Gurunathan S, Grasset EK, Losic B, Dubinsky M, Greenstein AJ, Gottlieb Z, Legnani P, George J, Irizar H, Stojmirovic A, Brodmerkel C, Kasarkis A, Sands BE, Furtado G, Lira SA, Tuong ZK, Ko HM, Cerutti A, Elson CO, Clatworthy MR, Merad M, Suárez-Fariñas M, Argmann C, Hackney JA, Victora GD, Randolph GJ, Kenigsberg E, Colombel JF, Mehandru S. Ulcerative colitis is characterized by a plasmablast-skewed humoral response associated with disease activity. Nat Med. 2022;28:766-779. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 133] [Article Influence: 44.3] [Reference Citation Analysis (0)] |
| 20. | Boland BS, He Z, Tsai MS, Olvera JG, Omilusik KD, Duong HG, Kim ES, Limary AE, Jin W, Milner JJ, Yu B, Patel SA, Louis TL, Tysl T, Kurd NS, Bortnick A, Quezada LK, Kanbar JN, Miralles A, Huylebroeck D, Valasek MA, Dulai PS, Singh S, Lu LF, Bui JD, Murre C, Sandborn WJ, Goldrath AW, Yeo GW, Chang JT. Heterogeneity and clonal relationships of adaptive immune cells in ulcerative colitis revealed by single-cell analyses. Sci Immunol. 2020;5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 176] [Article Influence: 35.2] [Reference Citation Analysis (0)] |
| 21. | Corridoni D, Antanaviciute A, Gupta T, Fawkner-Corbett D, Aulicino A, Jagielowicz M, Parikh K, Repapi E, Taylor S, Ishikawa D, Hatano R, Yamada T, Xin W, Slawinski H, Bowden R, Napolitani G, Brain O, Morimoto C, Koohy H, Simmons A. Single-cell atlas of colonic CD8(+) T cells in ulcerative colitis. Nat Med. 2020;26:1480-1490. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 144] [Article Influence: 28.8] [Reference Citation Analysis (0)] |
| 22. | Martin JC, Chang C, Boschetti G, Ungaro R, Giri M, Grout JA, Gettler K, Chuang LS, Nayar S, Greenstein AJ, Dubinsky M, Walker L, Leader A, Fine JS, Whitehurst CE, Mbow ML, Kugathasan S, Denson LA, Hyams JS, Friedman JR, Desai PT, Ko HM, Laface I, Akturk G, Schadt EE, Salmon H, Gnjatic S, Rahman AH, Merad M, Cho JH, Kenigsberg E. Single-Cell Analysis of Crohn's Disease Lesions Identifies a Pathogenic Cellular Module Associated with Resistance to Anti-TNF Therapy. Cell. 2019;178:1493-1508.e20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 544] [Cited by in RCA: 579] [Article Influence: 96.5] [Reference Citation Analysis (0)] |
| 23. | Jaeger N, Gamini R, Cella M, Schettini JL, Bugatti M, Zhao S, Rosadini CV, Esaulova E, Di Luccia B, Kinnett B, Vermi W, Artyomov MN, Wynn TA, Xavier RJ, Jelinsky SA, Colonna M. Single-cell analyses of Crohn's disease tissues reveal intestinal intraepithelial T cells heterogeneity and altered subset distributions. Nat Commun. 2021;12:1921. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 140] [Article Influence: 35.0] [Reference Citation Analysis (0)] |
| 24. | Yokoi T, Murakami M, Kihara T, Seno S, Arase M, Wing JB, Søndergaard JN, Kuwahara R, Minagawa T, Oguro-Igashira E, Motooka D, Okuzaki D, Mori R, Ikeda A, Sekido Y, Amano T, Iijima H, Ozono K, Mizushima T, Hirota S, Ikeuchi H, Takeda K. Identification of a unique subset of tissue-resident memory CD4(+) T cells in Crohn's disease. Proc Natl Acad Sci U S A. 2023;120:e2204269120. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 34] [Reference Citation Analysis (0)] |
| 25. | Rosati E, Rios Martini G, Pogorelyy MV, Minervina AA, Degenhardt F, Wendorff M, Sari S, Mayr G, Fazio A, Dowds CM, Hauser C, Tran F, von Schönfels W, Pochhammer J, Salnikova MA, Jaeckel C, Gigla JB, Sabet SS, Hübenthal M, Schiminsky E, Schreiber S, Rosenstiel PC, Scheffold A, Thomas PG, Lieb W, Bokemeyer B, Witte M, Aden K, Hendricks A, Schafmayer C, Egberts JH, Mamedov IZ, Bacher P, Franke A. A novel unconventional T cell population enriched in Crohn's disease. Gut. 2022;71:2194-2204. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 35] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 26. | Maddipatla SC, Kolachala VL, Venkateswaran S, Dodd AF, Pelia RS, Geem D, Yin H, Sun Y, Xu C, Mo A, Kosters A, Yang J, Matthews JD, Ghosn E, Kugathasan S, Qiu P. Assessing Cellular and Transcriptional Diversity of Ileal Mucosa Among Treatment-Naïve and Treated Crohn's Disease. Inflamm Bowel Dis. 2023;29:274-285. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 15] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 27. | Huang B, Chen Z, Geng L, Wang J, Liang H, Cao Y, Chen H, Huang W, Su M, Wang H, Xu Y, Liu Y, Lu B, Xian H, Li H, Ren L, Xie J, Ye L, Zhao J, Chen P, Zhang L, Zhao S, Zhang T, Xu B, Che D, Si W, Gu X, Zeng L, Wang Y, Li D, Zhan Y, Delfouneso D, Lew AM, Cui J, Tang WH, Zhang Y, Gong S, Bai F, Yang M. Mucosal Profiling of Pediatric-Onset Colitis and IBD Reveals Common Pathogenics and Therapeutic Pathways. Cell. 2019;179:1160-1176.e24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 199] [Article Influence: 39.8] [Reference Citation Analysis (0)] |
| 28. | Mitsialis V, Wall S, Liu P, Ordovas-Montanes J, Parmet T, Vukovic M, Spencer D, Field M, McCourt C, Toothaker J, Bousvaros A; Boston Children’s Hospital Inflammatory Bowel Disease Center; Brigham and Women’s Hospital Crohn’s and Colitis Center, Shalek AK, Kean L, Horwitz B, Goldsmith J, Tseng G, Snapper SB, Konnikova L. Single-Cell Analyses of Colon and Blood Reveal Distinct Immune Cell Signatures of Ulcerative Colitis and Crohn's Disease. Gastroenterology. 2020;159:591-608.e10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 233] [Article Influence: 46.6] [Reference Citation Analysis (0)] |
| 29. | Swanson E, Lord C, Reading J, Heubeck AT, Genge PC, Thomson Z, Weiss MD, Li XJ, Savage AK, Green RR, Torgerson TR, Bumol TF, Graybuck LT, Skene PJ. Simultaneous trimodal single-cell measurement of transcripts, epitopes, and chromatin accessibility using TEA-seq. Elife. 2021;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 119] [Cited by in RCA: 150] [Article Influence: 37.5] [Reference Citation Analysis (0)] |
| 30. | Williams CG, Lee HJ, Asatsuma T, Vento-Tormo R, Haque A. An introduction to spatial transcriptomics for biomedical research. Genome Med. 2022;14:68. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 301] [Cited by in RCA: 369] [Article Influence: 123.0] [Reference Citation Analysis (0)] |
| 31. | Marx V. Method of the Year: spatially resolved transcriptomics. Nat Methods. 2021;18:9-14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 172] [Cited by in RCA: 506] [Article Influence: 126.5] [Reference Citation Analysis (0)] |
| 32. | Guo W, Zhang C, Wang X, Dou D, Chen D, Li J. Resolving the difference between left-sided and right-sided colorectal cancer by single-cell sequencing. JCI Insight. 2022;7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 50] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 33. | Mei Y, Xiao W, Hu H, Lu G, Chen L, Sun Z, Lü M, Ma W, Jiang T, Gao Y, Li L, Chen G, Wang Z, Li H, Wu D, Zhou P, Leng Q, Jia G. Single-cell analyses reveal suppressive tumor microenvironment of human colorectal cancer. Clin Transl Med. 2021;11:e422. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 79] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 34. | Qi J, Sun H, Zhang Y, Wang Z, Xun Z, Li Z, Ding X, Bao R, Hong L, Jia W, Fang F, Liu H, Chen L, Zhong J, Zou D, Liu L, Han L, Ginhoux F, Liu Y, Ye Y, Su B. Single-cell and spatial analysis reveal interaction of FAP(+) fibroblasts and SPP1(+) macrophages in colorectal cancer. Nat Commun. 2022;13:1742. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 476] [Article Influence: 158.7] [Reference Citation Analysis (0)] |
| 35. | Roerink SF, Sasaki N, Lee-Six H, Young MD, Alexandrov LB, Behjati S, Mitchell TJ, Grossmann S, Lightfoot H, Egan DA, Pronk A, Smakman N, van Gorp J, Anderson E, Gamble SJ, Alder C, van de Wetering M, Campbell PJ, Stratton MR, Clevers H. Intra-tumour diversification in colorectal cancer at the single-cell level. Nature. 2018;556:457-462. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 329] [Cited by in RCA: 389] [Article Influence: 55.6] [Reference Citation Analysis (0)] |
| 36. | Wu Y, Yang S, Ma J, Chen Z, Song G, Rao D, Cheng Y, Huang S, Liu Y, Jiang S, Liu J, Huang X, Wang X, Qiu S, Xu J, Xi R, Bai F, Zhou J, Fan J, Zhang X, Gao Q. Spatiotemporal Immune Landscape of Colorectal Cancer Liver Metastasis at Single-Cell Level. Cancer Discov. 2022;12:134-153. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 548] [Article Influence: 137.0] [Reference Citation Analysis (0)] |
| 37. | Ben-Baruch Morgenstern N, Ballaban AY, Wen T, Shoda T, Caldwell JM, Kliewer K, Felton JM, Abonia JP, Mukkada VA, Putnam PE, Bolton SM, Dwyer DF, Barrett NA, Rothenberg ME. Single-cell RNA sequencing of mast cells in eosinophilic esophagitis reveals heterogeneity, local proliferation, and activation that persists in remission. J Allergy Clin Immunol. 2022;149:2062-2077. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 59] [Article Influence: 19.7] [Reference Citation Analysis (0)] |
| 38. | Ziegenhain C, Vieth B, Parekh S, Reinius B, Guillaumet-Adkins A, Smets M, Leonhardt H, Heyn H, Hellmann I, Enard W. Comparative Analysis of Single-Cell RNA Sequencing Methods. Mol Cell. 2017;65:631-643.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 855] [Cited by in RCA: 958] [Article Influence: 119.8] [Reference Citation Analysis (0)] |