Published online May 21, 2023. doi: 10.3748/wjg.v29.i19.2961
Peer-review started: February 28, 2023
First decision: March 14, 2023
Revised: March 24, 2023
Accepted: April 23, 2023
Article in press: April 23, 2023
Published online: May 21, 2023
Processing time: 76 Days and 24 Hours
Intrahepatic cholangiocarcinoma (ICC) is a malignant tumor of the hepatobiliary system with concealed onset, strong invasiveness and poor prognosis.
To explore the disease characteristic genes that may be helpful in the diagnosis of ICC and affect immune cell infiltration.
We downloaded two ICC-related human gene expression profiles from GEO database as the training group (GSE26566 and GSE32958 datasets) for difference analysis, and performed enrichment analysis on differential genes. The least absolute shrinkage and selection operator (LASSO), support vector machine-recursive feature elimination (SVM-RFE) and random forest (RF), three machine learning algorithms, were used to screen the characteristic genes. Double veri
A total of 1091 differential genes were obtained in the training group. Enrichment analysis showed that the above genes were mainly enriched in small molecular catabolism, complement and coagulation cascade, bile secretion and other functions and pathways. Twenty-five characteristic genes were screened by LASSO regression, 19 by SVM-RFE algorithm, and 30 by RF algorithm. Three algorithms were used in combination to determine the characteristic gene of ICC: MMP14. The verification group confirmed that the genes had a high diagnostic accuracy (AUC values of the training group and the verification group were 0.960, 0.999, and 0.977, respectively). Comprehensive analysis of immune infiltration showed that MMP14 could affect the infiltration of monocytes, activated memory CD4 T cells, resting memory CD4 T cells, and other immune cells, and was closely related to the expression of CD200, cytotoxic T-lymphocyte-associated antigen 4, CD14, CD44, and other immune checkpoints. The results of immunohistochemistry in HPA database showed was indeed overexpressed in ICC.
MMP14 can be used as a disease characteristic gene of ICC, and may regulate the distribution of immune-infiltrating cells in the ICC tumor microenvironment, which provides a new method for the determination of ICC diagnostic markers and screening of therapeutic targets.
Core Tip: This study demonstrates that a new candidate molecular marker: MMP14 that is important for the diagnosis of intrahepatic cholangiocarcinoma, and MMP14 was related to a variety of immune cell components and participated in the occurrence and development in intrahepatic cholangiocarcinoma.
- Citation: Wu J, Guo Y, Zuo ZF, Zhu ZW, Han L. MMP14 is a diagnostic gene of intrahepatic cholangiocarcinoma associated with immune cell infiltration. World J Gastroenterol 2023; 29(19): 2961-2978
- URL: https://www.wjgnet.com/1007-9327/full/v29/i19/2961.htm
- DOI: https://dx.doi.org/10.3748/wjg.v29.i19.2961
Intrahepatic cholangiocarcinoma (ICC) is the second most common primary tumor of the liver, which originates from intrahepatic bile duct epithelial cells and accounts for about 20% of primary liver malignant tumors[1]. Its incidence is on the rise worldwide, with the highest incidence in Southeast Asia, and the second highest incidence in China after Thailand[2,3]. Pathological changes such as chronic cholangitis, cholestasis, intrahepatic bile duct stones, and liver cirrhosis all increase the incidence of ICC[4]. At present, due to the poor efficacy of radiotherapy and chemotherapy for ICC, surgical treatment is currently the main treatment method for ICC. However, patients with ICC may be asymptomatic in the early stage, and most patients have intrahepatic metastasis, lymph node metastasis, or even distant metastasis at the first time of diagnosis. Only 9% of patients with advanced ICC can survive for 5 years[5-8]. Despite the continuous improvement in imaging and laboratory tests, the early diagnosis of ICC is still not ideal. Currently, the biomarkers of ICC mainly include carcinoembryonic antigen, carbohydrate antigen 19-9 and a-fetoprotein, but the sensitivity and specificity are not satisfactory[9]. Hence, it is crucial to find new biomarkers for early screening and diagnosis of ICC.
Many studies on the tumor microenvironment have confirmed that a variety of immune regulatory mechanisms play an important role in the malignant biological behavior of tumors. Therefore, immunotherapy has gradually been paid attention in various treatments for solid tumors, and as an emerging and effective anticancer treatment, it has also been widely used for ICC. However, the improvement of the efficacy of immunotherapy and the development of effective drugs targeting specific immune targets are still problems to be solved[10].
Machine learning is a part of the field of artificial intelligence, which can find rules through the analysis of data, and can be conducive to the prediction of similar unknown data. And the existing knowledge structure can be constantly updated to improve its own performance[11]. With the continuous progress of machine learning, the development of more accurate machine learning algorithms makes it possible for us to find new disease-related genes[12]. At the same time, second-generation sequencing technology has made convenient screening of ICC diagnostic targets, and provided a solid foundation for precise treatment of patients and selection of personalized drugs[13].
In this study, using the ICC-related data in the GEO and The Cancer Genome Atlas (TCGA) databases, bioinformatics and machine learning algorithms were used to comprehensively analyze and obtain reliable disease-characteristic genes, and in-depth exploration found that this gene may regulate the progression of ICC by mediating the composition of immune-infiltrating cells. The above preliminary research results can provide important references for the identification of early diagnostic markers for ICC and the mining of potential intervention targets for immunotherapy.
The gene expression data of ICC cancer tissues and paracancerous tissues were obtained by using the three datasets GSE26566, GSE32958, and GSE107943 in GEO database. The transcriptome gene expression data of cancer and paracancerous samples of ICC patients were obtained from TCGA database. The relevant datasets were sorted, and samples other than cancer and paracancerous tissues of ICC patients were deleted. The two datasets of GSE26566 and GSE32958 were combined as the training group. The GSE107943 dataset was used as the verification group, while the TCCG dataset was used as the external database verification group.
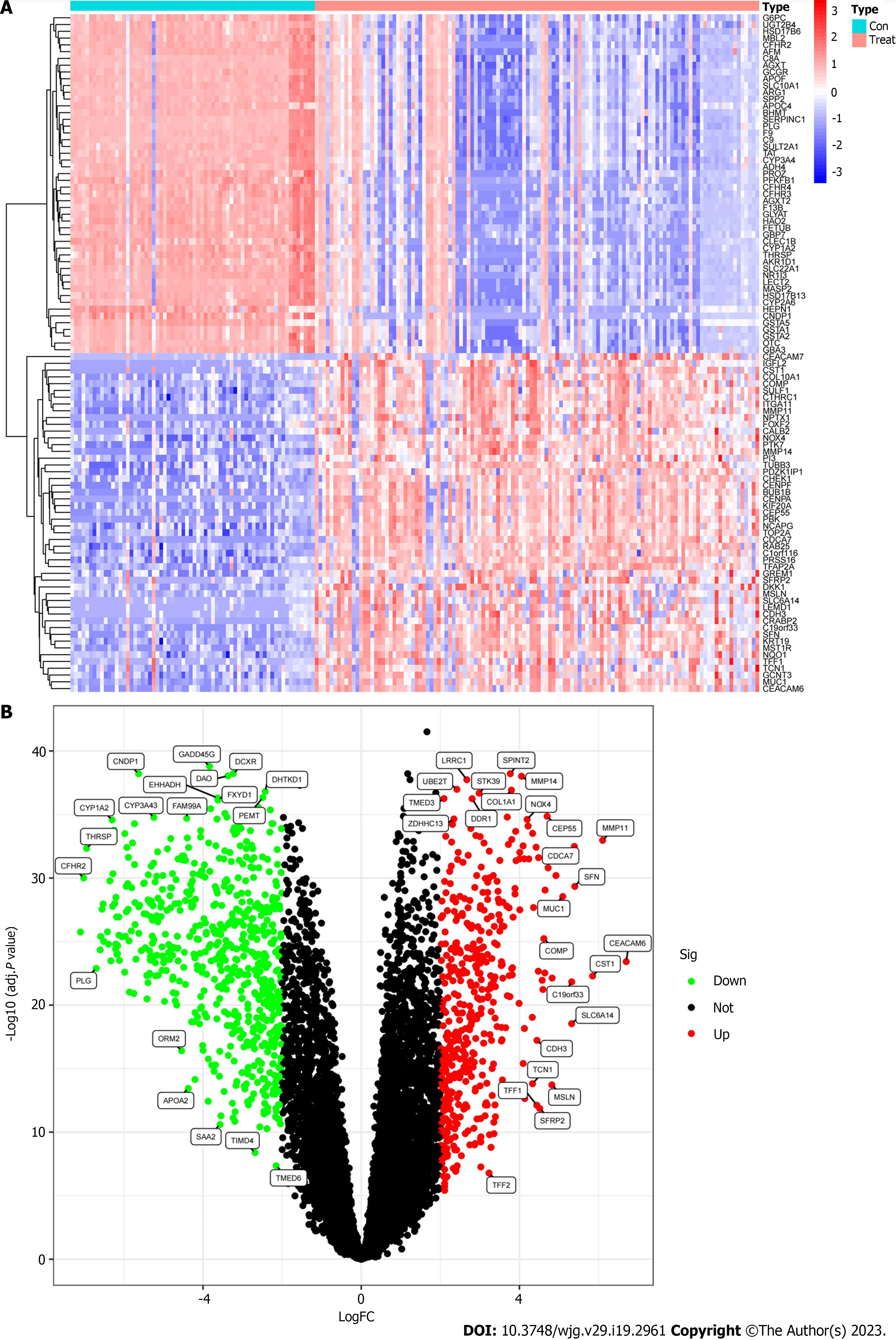
The Limma package was used to analyze the differences between the GSE26566 and GSE32958 datasets after merging the training groups. Genes that met the conditions |log2FC| > 2 and adj. P < 0.05 were considered to be differential genes. The first 50 genes with the most significant difference between upregulated and downregulated genes were selected for heat mapping using the pheatmap package. The volcanic map was plotted using ggplot2 and ggrepel packages to visualize significantly different genes.
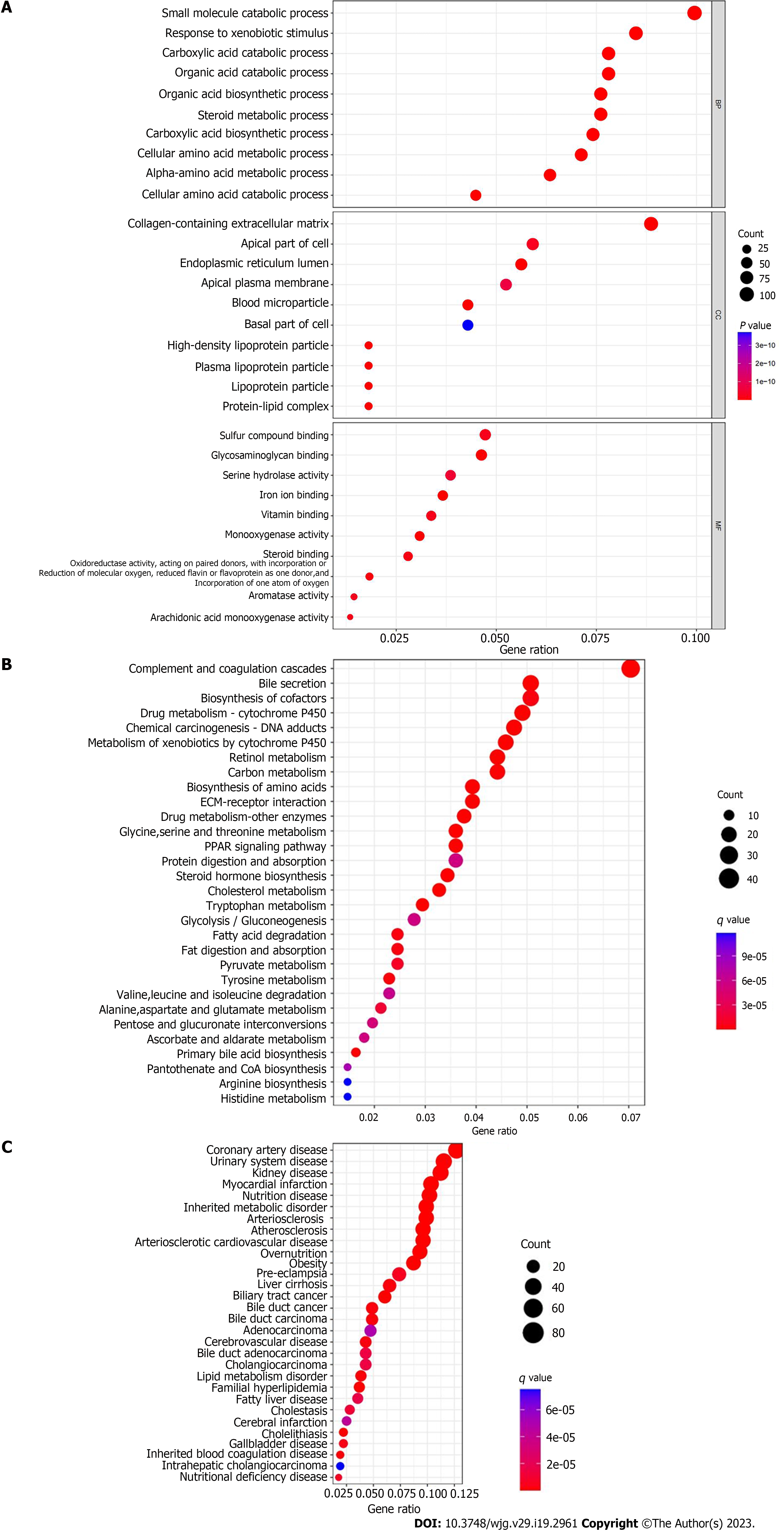
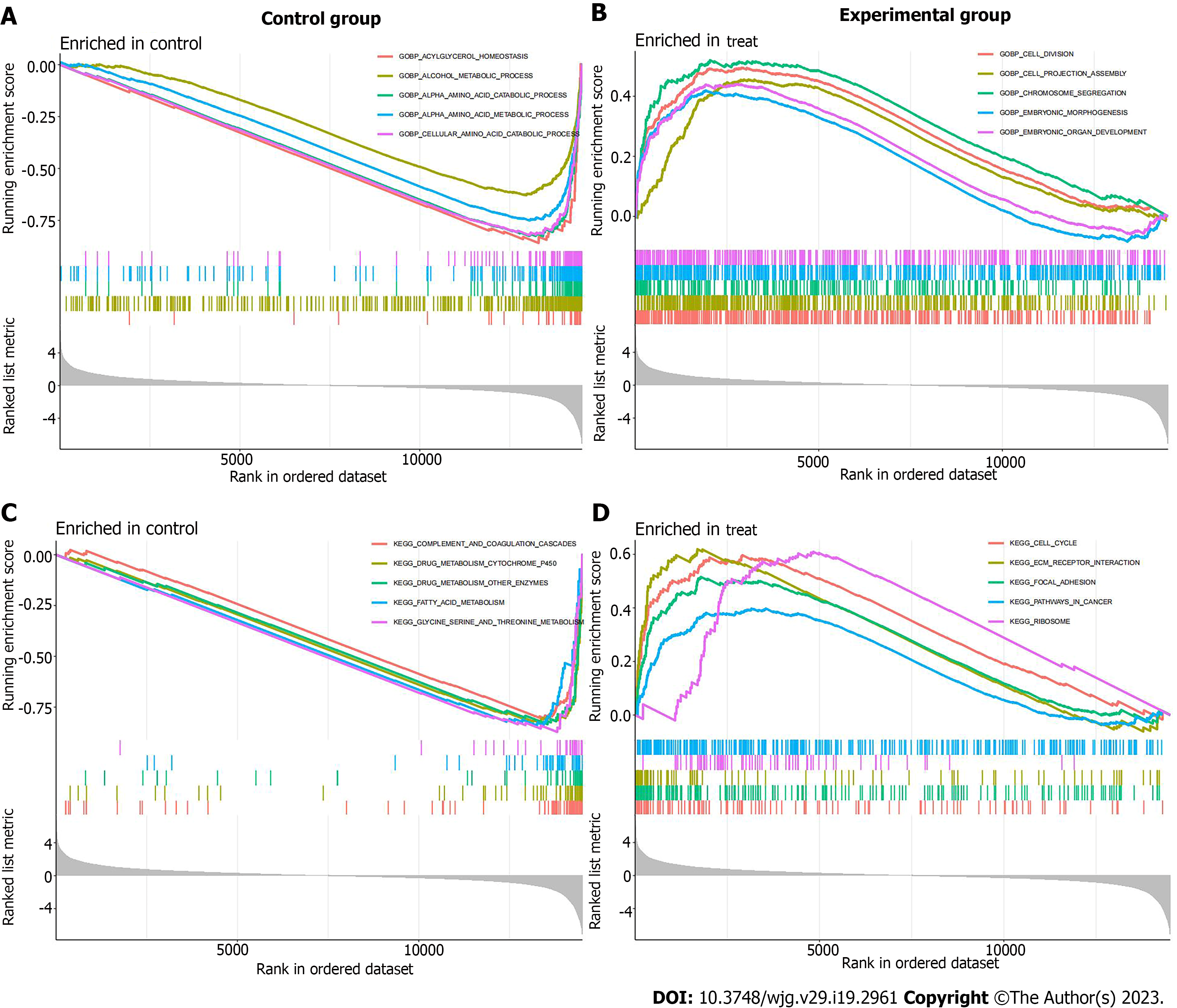
According to the results of differential expression analysis, the org.Hs.eg.db, enrichplot, clusterProfiler, DOSE and GSEABase package were used for GO functional enrichment analysis, KEGG pathway enrichment analysis, DO disease enrichment analysis and GSEA enrichment analysis, respectively. The top 10 most significantly enriched functions in biological processes (BP), CC and molecular functions (MF) were visualized by drawing bubble plots. The top 30 pathways or diseases with the most significant enrichment were selected for KEGG enrichment analysis and DO enrichment analysis bubble diagram drawing. GSEA enrichment analysis was carried out on the experimental group (cancer tissue samples) and the control group (paracancerous tissue samples), and the first five functions and pathways that were most significantly enriched in the experimental group and the control group were visualized respectively.
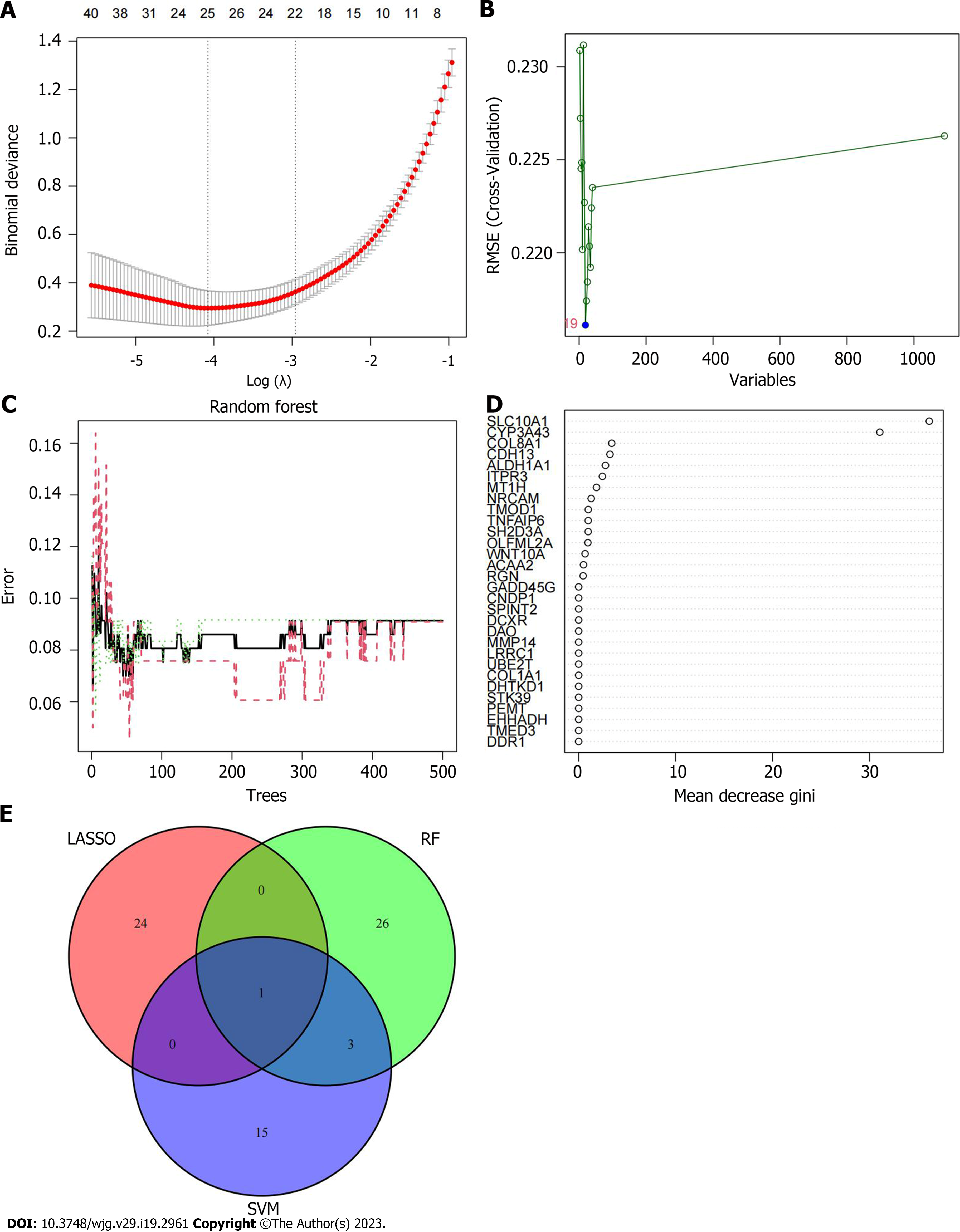
Based on the differential gene expression, least absolute shrinkage and selection operator (LASSO) regression, support vector machine-recursive feature elimination (SVM-RFE) algorithm and random forest (RF) algorithm were used to screen disease characteristic genes. The genes screened by the three algorithms were intersected to obtain the final disease characteristic gene, which was the new characteristic gene for ICC diagnosis. The diagnostic genes obtained by the comprehensive analysis of the above three machine learning algorithms had stronger persuasion and higher accuracy. The LASSO regression algorithm was based on the glmnet package. When the parameters were set to a = 1, type.measure = deviance and nfolds = 10, the number of genes corresponding to the point with the smallest cross-validation error was the target gene. The SVM-RFE algorithm was based on the e1071, kernlab and caret packages. The method adopted was svmRadial, and the number of genes corresponding to the point with the smallest cross-validation error was selected as the disease characteristic gene. The RF algorithm was based on the randomForest package. The differentially expressed genes were used to train the RF model. The number of forest trees was set to 500, and the number of corresponding trees with the smallest cross-verification error was found to reconstruct the RF model for training. The importance score of the gene was obtained. We sorted from high to low according to the scores, and selected the 30 genes with the highest importance scores as the feature gene set.
The limma package was used to analyze the differences between the two verification groups of GSE107943 and TCGA to determine whether the differential expression of disease characteristic genes in the verification group was consistent with that of the training group. We used the ggpubr package to visualize and draw a boxplot. The receiver operating characteristic (ROC) curve was drawn based on the pROC package to evaluate the efficiency of characteristic genes in the diagnosis of ICC. Area under the curve (AUC) value was used as an index to evaluate the diagnostic value of characteristic genes in the training and test groups. The 95% confidence interval (CI) of AUC value was extracted and calculated by bootstrap method.
According to the gene expression level of each sample, CIBERSORT algorithm was used to quantify the relative content of immune cells in each sample with the help of e1071 and preprocessCore package, so as to obtain the proportion of different immune cells in each sample, and a bar chart was drawn. The corrplot package was used to analyze the correlation of immune cells and draw the correlation heat map. The vioplot package was used to analyze the difference in immune cell infiltration between cancer tissue and paracancerous tissue of ICC patients, and the violin picture was drawn. The CIBERSORT and ssGSEA algorithms were used to evaluate the relationship between ICC characteristic genes and tumor immune-infiltrating cells, in order to explore the effect of ICC characteristic genes on tumor immune microenvironment. Finally, 46 immune checkpoint related genes (including CD274, PDCD1, CTLA4, CD14, LAG3, and TNFRSF9) were found through the literature. We used the limma and corrplot packages, etc. to analyze whether the disease signature genes had a regulatory effect on the expression of immune checkpoints.
The Human Protein Atlas (HPA) database provides the expression and distribution of all available proteins in human tissues and organs. Therefore, protein expression data of disease-characteristic genes in ICC and paracancerous tissues were obtained from this database for comparative analysis.
All statistical analyses in this study were conducted in R version 4.2.0. ICC characteristic genes were screened by LASSO regression, SVM-RFE and RF machine learning algorithms. The diagnostic efficacy of diagnostic biomarkers was evaluated by ROC curve. The relationship between the expression of characteristic genes and immune cell infiltration and immune checkpoint was analyzed by Spearman and Pearson correlation. Statistical significance was identified based on P < 0.05.
Table 1 shows the three datasets (GSE26566, GSE32958, and GSE107943) in the GEO database and the ICC-related information obtained from the TCGA database. Two hundred and eighty-four samples were collected, including 120 cancer tissues and 66 paracancerous tissues in the training group. In the two verification groups, cancer tissue samples were from 30 and 33 cases, respectively, and paracancerous tissue samples from 27 and eight cases, respectively.
| Dataset | Number of cancer samples | Number of paracancerous samples | Total |
| GSE26566 | 104 | 59 | 163 |
| GSE32958 | 16 | 7 | 23 |
| GSE107943 | 30 | 27 | 57 |
| TCGA | 33 | 8 | 41 |
| Total | 183 | 101 | 284 |
By the difference analysis of the training group, 1091 differential mRNAs were obtained, including 463 upregulated and 628 downregulated mRNAs (Figure 1A and B). There were many genes with multiple differences in ICC cancer and paracancerous tissues; therefore, it was important to screen characteristic genes that mediated the occurrence and development of ICC, which laid a foundation for the following mechanistic exploration and target gene identification.
Through GO functional enrichment analysis, we found that in terms of BP, processes such as small molecule catabolic process, response to xenobiotic stimulus, and carboxylic acid catabolic process had the largest number of enrichment genes. In terms of CC, collagen-containing extracellular matrix (ECM), apical part of cell and other CC had the largest number of enriched genes. In terms of MF, sulfur compound binding, glycosaminoglycan binding, serine hydrolase activity and iron ion binding had the largest number of enriched genes (Figure 2A). KEGG pathway enrichment analysis showed that the differential genes were mainly enriched in 56 related regulatory mechanisms, which were mainly involved in complement and coagulation cascades, bile secretion, biosynthesis of cofactors, drug metabolism cytochrome P450, chemical carcinogenesis DNA adducts, retinol metabolism and other pathways (Figure 2B). DO disease enrichment analysis showed that differential genes were closely related to cardiovascular disease, urinary system disease, fatty liver and cholangiocarcinoma (Figure 2C). GSEA enrichment analysis was carried out on the experimental and control groups. GO enrichment analysis showed that acylglycerol homeostasis and alcohol metabolic process were mainly enriched in paracancerous tissues (Figure 3A). In ICC cancer tissues, differential genes were mainly enriched in cell division, chromosome segregation, embryonic organ development and other functions (Figure 3B). KEGG enrichment analysis showed that complement, coagulation cascades and drug metabolism cytochrome P450 were actively expressed in para
To identify the specific genes of value in the diagnosis of ICC, we used machine learning to filter the above 1091 differential genes. A total of 25 genes were screened by LASSO regression algorithm, namely: MMP14, CDCA8, ZDHHC13, NOX4, HEPN1, LRRC49, PDZK1IP1, OR1L4, FAP, NDST3, FXN, PYCR1, PRR11, SPAM1, MAD2L1, APOBEC3G, PI3, CYP7A1, RNF165, MT1M, GREM1, EME1, ENAH, CDKN2B and FOSB (Figure 4A). After screening by SVM-RFE algorithm, 19 genes were retained, namely: CFHR2, PROC, UBE2T, HGD, AGMAT, ACSL1, GADD45G, DCXR, HGFAC, DTL, MMP14, ADHFE1, SLC22A1, CBS, PIPOX, SHMT1, RDH5, F12, and F9 (Figure 4B). After screening by RF algorithm, the first 30 genes obtained good diagnostic effect, namely: SLC10A1, CYP3A43, COL8A1, CDH13, ALDH1A1, ITPR3, MT1H, NRCAM, TMOD1, TNFAIP6, SH2D3A, OLFML2A, WNT10A, ACAA2, RGN, GADD45G, CNDP1, SPINT2, DCXR, DAO, MMP14, LRRC1, UBE2T, COL1A1, DHTKD1, STK39, PEMT, EHHADH, TMED3, and DDR1 (Figure 4C and D). After the intersection of the genes determined by the above three results, the best disease characteristic gene was obtained: MMP14 (Figure 4E).
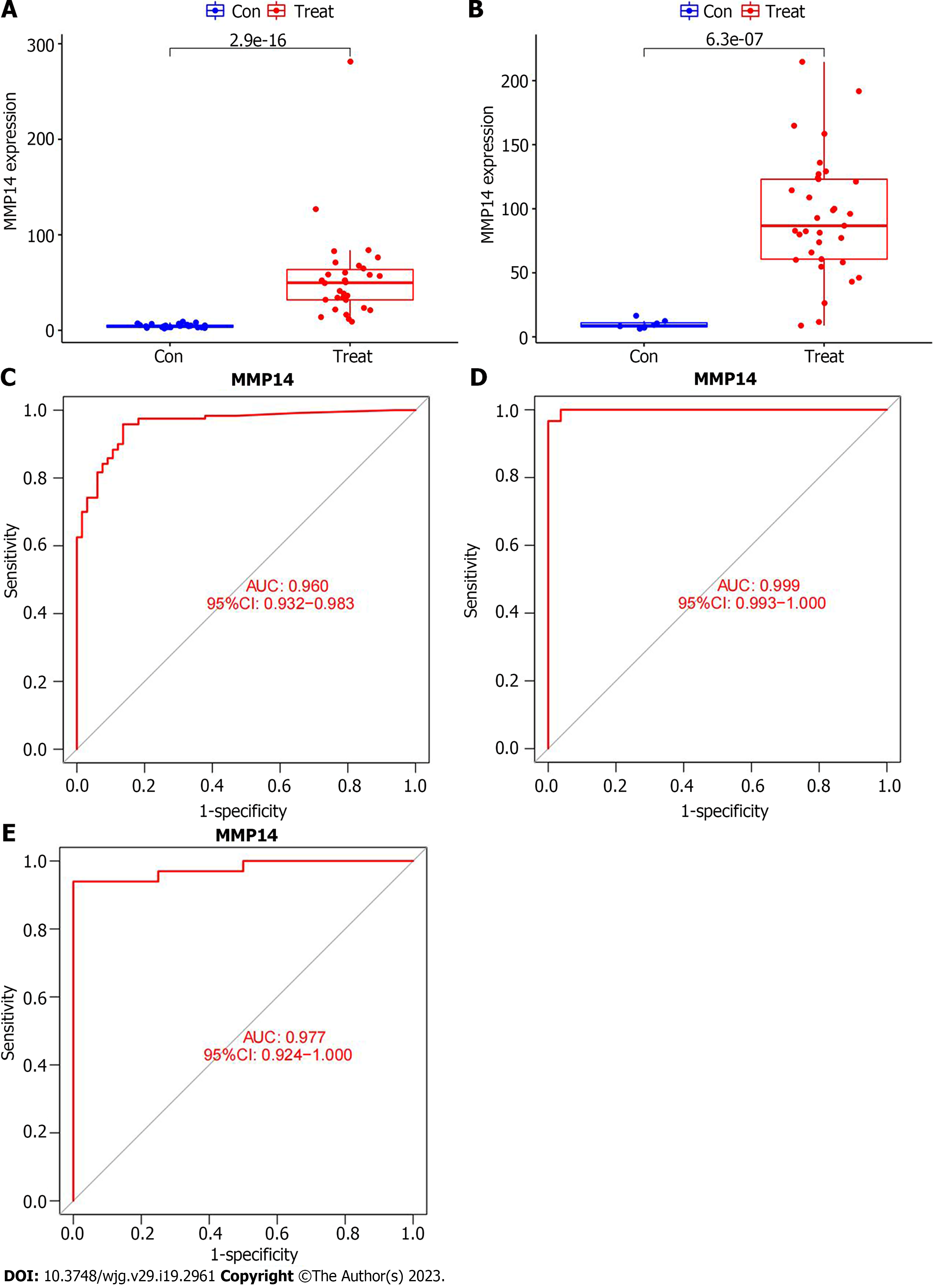
Previous studies had shown that MMP14 was highly expressed in cancer tissues of ICC patients in the training group, so it was important to clarify its expression in two independent verification groups before evaluating the accuracy of this gene as a disease signature gene. In the GSE107943 dataset, the expression of MMP14 in ICC cancer tissues was apparently higher than that in paracancer tissue (Figure 5A). The same result was obtained by external database verification of MMP14 in the TCGA database (Figure 5B). ROC curves were drawn for the training and validation groups, which showed that in the training group, AUC was 0.960 (95%CI: 0.932-0.983) (Figure 5C). In the GSE107943 verification group, AUC was 0.999 (95%CI: 0.993-1.000) (Figure 5D), and in the TCGA verification group, AUC was 0.977 (95%CI: 0.924-1.000) (Figure 5E). AUC in the verification group was higher than in the training group, and AUC values were all > 0.9. This showed that MMP14 had good diagnostic efficiency and could be used as a characteristic gene of ICC, which provided a new reference marker for the selection of gene targets for diagnosis of ICC.
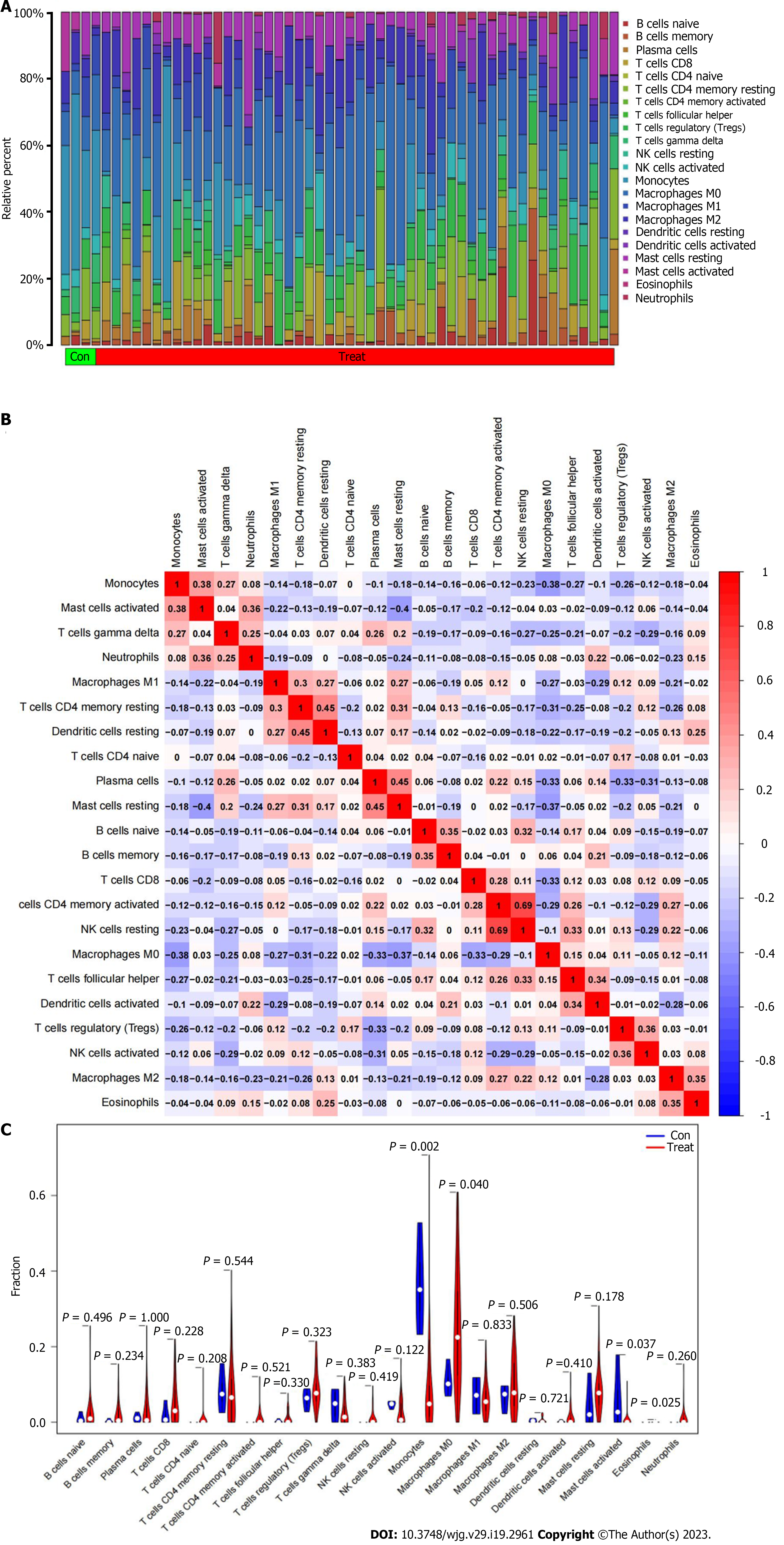
To explore the infiltration of immune cells in the immune microenvironment of ICC, the proportion of 22 types of immune cells in each sample of ICC cancer and paracancerousl tissues was clarified (Figure 6A). The positive correlation between resting dendritic cells resting and resting memory CD4 T cells resting was the strongest (r = 0.45), and the negative correlation between mast cells resting and mast cells activated was the strongest (r = 0.40) (Figure 6B). We then analyzed the infiltration of immune cells in cancer and paracancerous tissues, and was significant for monocytes, M0 macrophages, activated mast cells, and eosinophils. Compared with paracancerous tissues, expression of M0 macrophages in tumor tissues was greatly higher (P = 0.040), while expression of monocytes (P = 0.002) and activated mast cells (P = 0.037) was significantly lower in tumor tissues (Figure 6C).
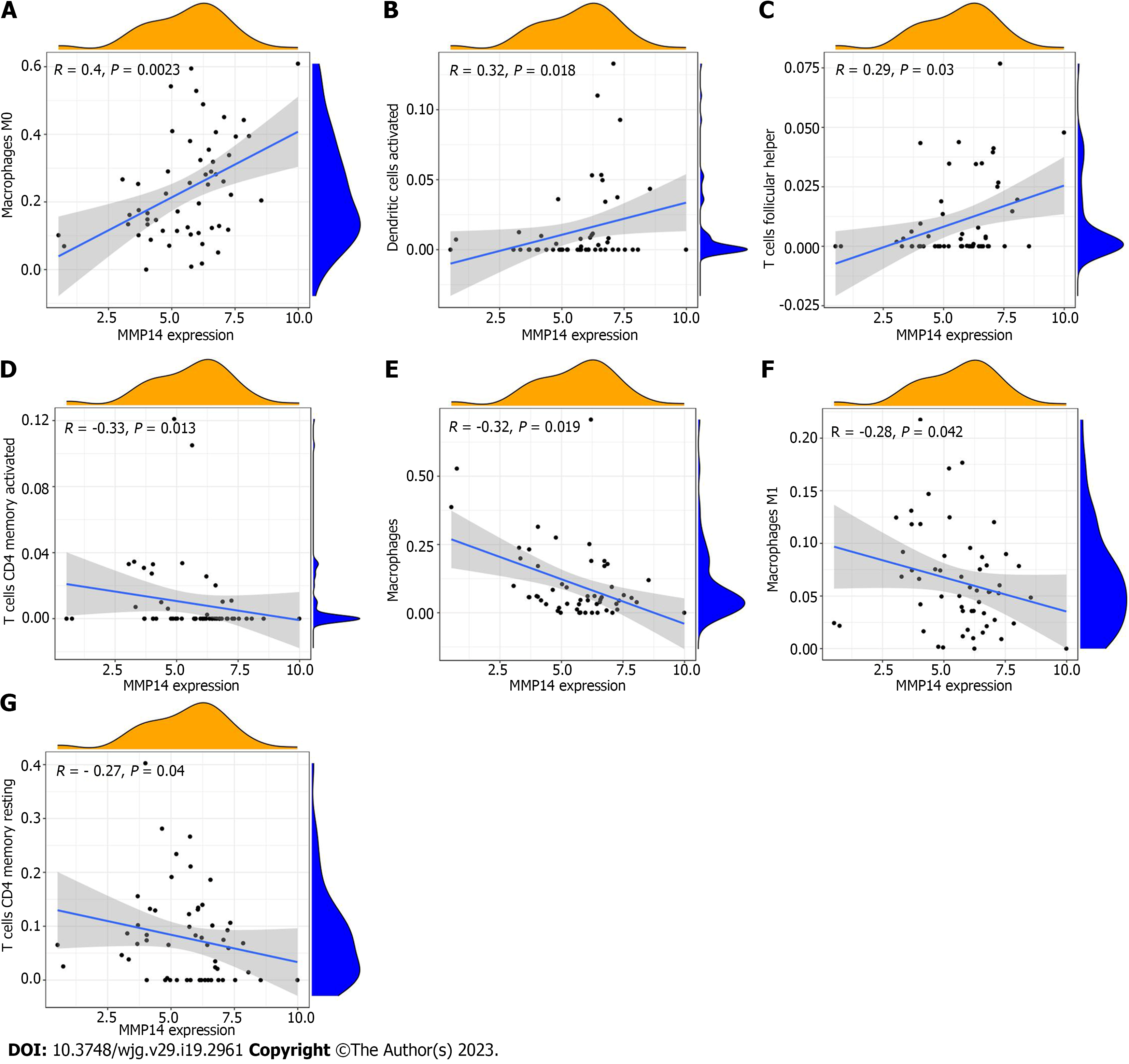
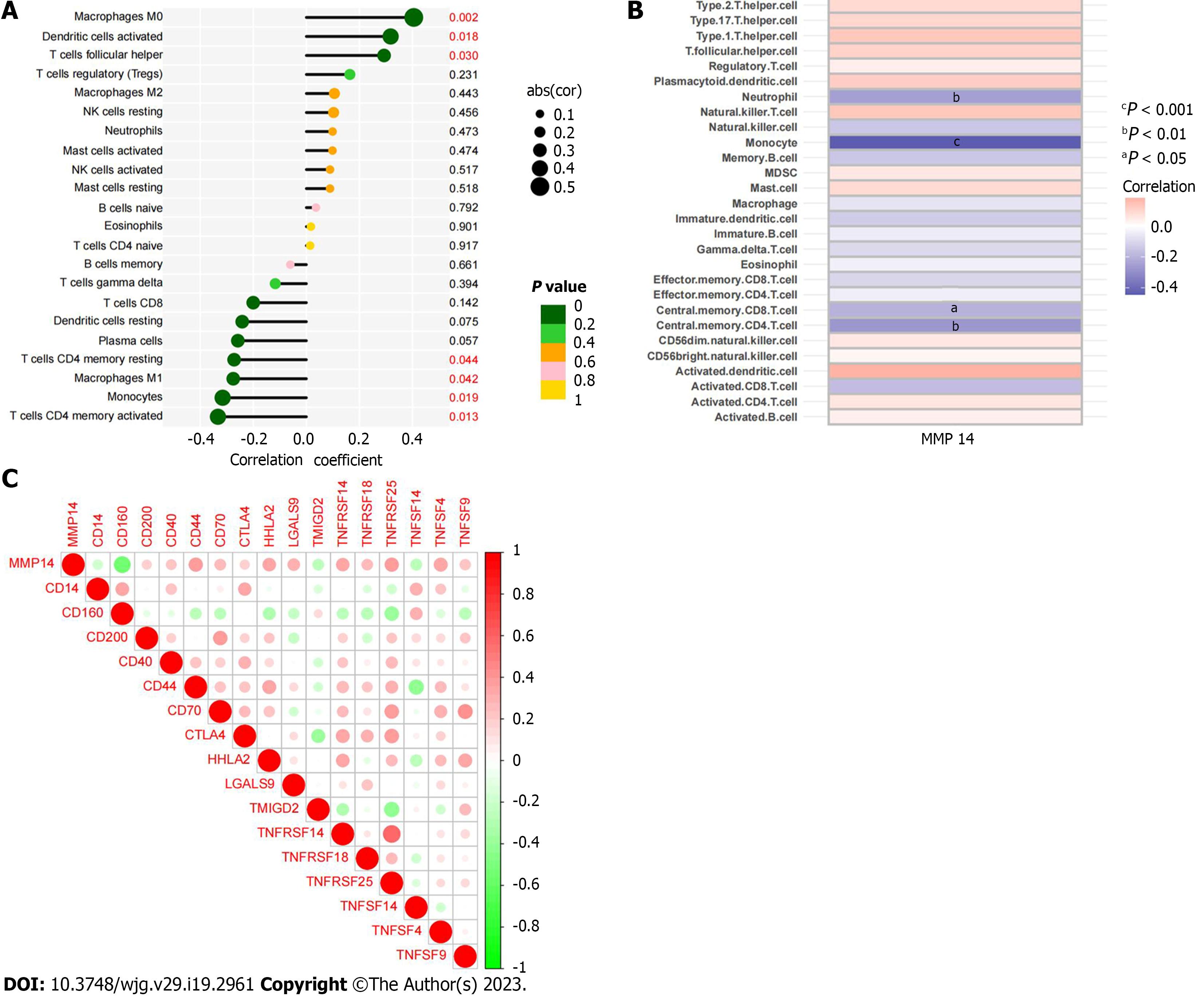
There were differences in the distribution of immune infiltrating cells between ICC cancer tissues and paracancerous tissues. To analyze its potential regulatory factors, we explored the correlation between ICC disease signature genes and immune microenvironment. CIBERSORT algorithm analysis showed that among the immune cells, MMP14 was significantly correlated with the number of seven types of immune cells. Expression of MMP14 was positively related with M0 macrophages M0 (r = 0.40, P = 0.0023), activated dendritic cells (r = 0.32, P = 0.0180) and follicular helper T cells (r = 0.29, P = 0.0300) (Figure 7A-C), and negatively related with activated memory CD4 T cells (r = -0.33, P = 0.0130), monocytes (r = -0.32, P = 0.0190), M1 macrophages (r = -0.28, P = 0.0420) and resting memory CD4 T cells (r = -0.27, P = 0.0440) (Figure 7D-G). The final results of MMP14 and immune cell infiltration based on the above CIBERSORT algorithm are summarized (Figure 8A). ssGSEA algorithm analysis showed that there was a significant correlation between MMP14 and the number of four types of immune cells and 28 types of immune cells. It was significantly negatively correlated with the number of neutrophils, monocytes, central memory CD8 T cells and central memory CD4 T cells (Figure 8B). MMP14 was positively correlated with the expression of CD200, CD40, CD44, CD70, CTLA4, HHLA2, LGALS9, TNFRSF14, TNFRSF18, TNFRSF25, TNFSF4 and TNFSF9. However, it was negatively correlated with the expression of CD14, CD160, TMIGD2, and TNFSF14 (Figure 8C). In summary, MMP14 regulates the composition of immune infiltrating cells in the tumor microenvironment of ICC and may affect the efficacy of immunotherapy by mediating the expression of immune cell surface markers.
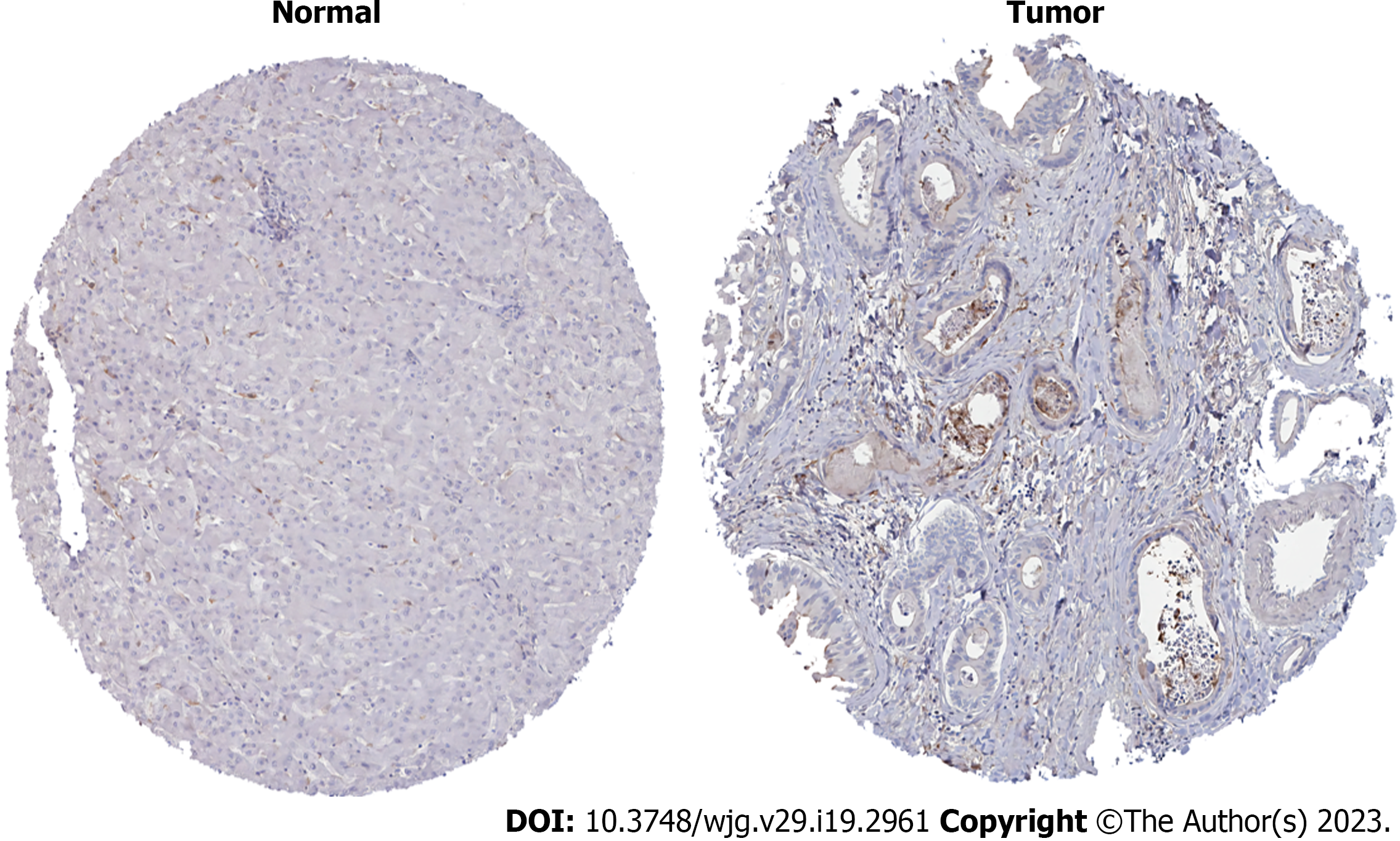
HPA database analysis showed that expression of MMP14 protein in ICC cancer tissue was higher than that in paracancerous tissue (Figure 9). It was consistent with the trend in mRNA expression levels in the training and verification groups, which confirmed that MMP14 could be used as a disease characteristic diagnostic gene of ICC.
Although there has been some improvement in the treatment of ICC in recent years, the disease control rate is still low[14]. There is no definite marker for early diagnosis of ICC[15]. Individualized treatment has not yet achieved satisfactory results in ICC patients[16]. At present, immunotherapy is a research hot spot in the field of cancer treatment worldwide, and it has widespread prospects for treatment of cholangiocarcinoma[17]. Therefore, the present study focused on discovering new diagnostic markers and improving immunotherapy and determining synergistic therapeutic targets for ICC. Recently, similar ideas have been developed in the research of a variety of tumors and other diseases. More researchers have begun to identify new disease signature genes and explore their potential links with immune cell infiltration. For example, in chronic obstructive pulmonary disease (COPD), STAU1 and SLC27A3 are considered to be important diagnostic biomarkers. The pathogenesis of COPD is largely affected by the pattern of immune cell infiltration. It has been confirmed that STAU1 and SLC27A3 are important factors in regulating the content of plasma cells, resting NK cells, CD8 T cells, and other immune cells[18]. Studies have pointed out that LTBP2 is a highly effective biomarker for prostate cancer, which may inhibit the proliferation and metastasis of prostate cancer through the PI3K/AKT signaling pathway, and is related to CD4 T-cell infiltration and the response to anti-PD-1/PD-L1 immunotherapy[19]. However, for ICC with poor prognosis, there have been few studies on potential diagnostic markers and their correlation with immune cell infiltration. Therefore, we hope to find diagnostic biomarkers for ICC and investigate their relationship with immune cell infiltration.
The present study was a series of retrospective studies based on the gene expression matrix of ICC patients in the GEO and TCGA databases, looking for new diagnostic genes for ICC and analyzing the significance of immune cell infiltration. The results obtained are consistent with previous studies that complex metabolic changes occurred in the entire process of cholangiocarcinoma development, and metabolic reprogramming mechanisms such as glucose metabolism, amino acid metabolism, and lipid metabolism all mediate tumor proliferation and invasion are involved[20]. Mechanisms such as ECM-receptor interaction, cell cycle disorder, and stem cell self-renewal have also been confirmed to affect the development of ICC, which is consistent with our enrichment analysis[21-23]. To accurately screen for disease characteristic genes, we used LASSO regression, SVM-RFE, and RF learning algorithms for joint analysis. After the joint verification of another data set in the GEO and TCGA databases, we identified MMP14 as having high diagnostic accuracy for ICC.
The MMP family are proteolytic enzymes closely related to angiogenesis and tumor progression. MMP14 was the first transmembrane protein found in the family[24]. Previous studies have indicated that MMP14 plays a key role in the malignant transformation of liver cancer, pancreatic cancer, colorectal cancer and other tumors[25-28]. In primary liver cancer, the expression of MMP14 in cancer tissues is significantly higher than that in paracancerous tissues. The expression level of MMP14 is positively correlated with tumor size, Edmondson-Steiner grade and α-fetoprotein level, and mediates the poor prognosis of patients with primary liver cancer[29]. In renal cell carcinoma, MMP14, as a member of the circPTCH1/miR-485-5p/MMP14 endogenous competitive network, participates in promoting metastasis and activating epithelial-mesenchymal transformation of renal cell carcinoma[30]. Direct degradation of ECM or indirect activation of MMP2 is also the main way for MMP14 to promote the invasion and metastasis of many types of tumors[31,32]. Ragusa and colleagues found that, in a chemotherapy-resistant, aggressive, matrix-rich colorectal cancer subtype, fluctuations in tumor signals triggered a decrease in PROX1 Levels and upregulation of MMP14 through the WNT and Notch pathways, which promoted a series of adverse tumor microenvironmental changes, including activation of fibroblasts, vascular dysfunction, and inability of cytotoxic T cells to enter the tumor[33]. It can be seen that, as a clear tumor-promoting factor, MMP14 is a key regulator in the tumor microenvironment, tumor immune infiltration and tumorigenesis, but its effect on disease diagnosis and tumor immune microenvironment in ICC has not been reported, so our research is important.
Tumor microenvironment plays an important role in the occurrence, development and prognosis of tumor. As an important part of tumor microenvironment, immune cells are involved in the prognosis of the disease and guidance of clinical treatment[34-36]. We calculated the profiles of immune cell infiltration in ICC cancer and paracancerous tissues, and elucidated ICC-associated immune cell subtypes and their potential relationships. Compared with paracancerous tissues, the infiltration of M0 macrophages in ICC tissues was significantly increased, while the proportion of monocytes and mast cells activated was significantly decreased. The correlation analysis between MMP14 and immune cells showed that it was positively correlated with M0 macrophages, activated dendritic cells and follicular helper T cells, and negatively correlated with activated memory CD4 T cells, monocytes, M1 macrophages and resting memory CD4 T cells. The ssGSEA algorithm showed that MMP14 was closely related to the degree of immune cell infiltration, and the number of central memory CD8 T cells, neutrophils, monocytes and central memory CD4 T cells decreased significantly in ICC patients with high expression of MMP14. We found that MMP14 was positively correlated with expression of 12 molecules, including CTLA4 and CD40, and negatively correlated with expressions of four molecules, including CD14 and CD160. Differential regulatory mechanisms of the tumor microenvironment in ICC patients may help explain individual differences in immunotherapy. The expression of MMP14 was negatively regulated by the distribution of monocytes and CD4 T cells in the analysis of CIBERSORT and ssGSEA algorithms. Coincidentally, in the correlation analysis with immune checkpoints, we found that MMP14 was related to the expression of many important molecules, such as monocyte surface marker CD14 and transmembrane receptor CTLA4 on T cells. As an important part of effective antitumor immunity, CD4 T cells can directly eliminate tumor cells through cytolytic mechanisms, or indirectly target tumor cells by regulating the tumor microenvironment to kill tumor cells[37]. Different subpopulations of monocytes perform exclusive functions for tumor promotion or antitumor immunity[38]. It has been reported that CD14 and CD16 monocyte subsets can induce cancer cell death through antibody-dependent cytotoxicity[24,39]. Therefore, we have the reason to speculate that MMP14 may accelerate the progression of ICC by interfering with the abundance of monocytes and CD4 T cells.
In our study, we found new ICC characteristic gene, MMP14, and clarified that it might influence the infiltration pattern of immune cells in complex tumor microenvironments. It could be helpful for the clinical diagnosis and immunotherapy of ICC, and provides new ideas for the future study of the occurrence and molecular mechanism of ICC.
However, our research still had some limitations. Firstly, all data came from public databases, which could have led to inevitable errors or biases. Secondly, the expression and specific function of disease characteristic genes need to be verified by further experiments. Although bioinformatics and machine learning have been used to evaluate the diagnostic efficacy of ICC feature genes and their relationship with immune infiltration, a larger prospective study is needed to confirm our conclusions.
Here we investigated new candidate molecular markers that are important for the diagnosis of ICC, which was related to a variety of immune cell components and participated in the occurrence and development of ICC. The findings may have a beneficial impact on the diagnosis of ICC and the provision of precise immunotherapy in future clinical work.
Intrahepatic cholangiocarcinoma (ICC) has a high mortality rate. Its early diagnosis is very important.
At present, there are no diagnostic markers of ICC, and most patients are in the middle and late stage when the tumors are found, and the treatment options for these patients are limited, resulting in a poor prognosis.
It is urgent to find new biomarkers for early screening and diagnosis of ICC.
Bioinformatics analysis and machine learning algorithm were used to screen for ICC disease characteristic genes, and the diagnostic performance of characteristic gene MMP14 was verified by different databases. The relationship between different immune cell infiltration in MMP14 and ICC tumor microenvironment was analyzed.
We discovered for the first time a new gene MMP14 for the diagnosis of ICC. We further analyzed the expression of MMP14 and its significance in immune cell infiltration in ICC tumor microenvironment, indicating that MMP14 is closely related to a variety of immune cell infiltration, which may provide a new direction for immune-related research.
In this study, we found that MMP14 can be used as a disease characteristic gene for ICC diagnosis using a variety of machine learning algorithms. There is a relationship between different immune cell infiltration in MMP14 and ICC tumor microenvironment, which plays an important role in the occurrence and development of tumor. MMP14 may be a potential target for ICC immunotherapy.
Based on bioinformatics analysis and machine learning algorithm, MMP14 was identified as the characteristic gene of ICC and was associated with ICC immune cell infiltration. In the future research, it will be of great significance to explore the signal pathway mediated by MMP14 in the occurrence and development of ICC and the mechanism of immune cell infiltration.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Bleszynski MS, Canada; Ishii T, Japan; Quaresima S, Italy S-Editor: Chen YL L-Editor: A P-Editor: Cai YX
| 1. | Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022;72:7-33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4235] [Cited by in RCA: 11313] [Article Influence: 3771.0] [Reference Citation Analysis (4)] |
| 2. | Bridgewater J, Galle PR, Khan SA, Llovet JM, Park JW, Patel T, Pawlik TM, Gores GJ. Guidelines for the diagnosis and management of intrahepatic cholangiocarcinoma. J Hepatol. 2014;60:1268-1289. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 862] [Cited by in RCA: 1067] [Article Influence: 97.0] [Reference Citation Analysis (0)] |
| 3. | Khan SA, Tavolari S, Brandi G. Cholangiocarcinoma: Epidemiology and risk factors. Liver Int. 2019;39 Suppl 1:19-31. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 273] [Cited by in RCA: 491] [Article Influence: 81.8] [Reference Citation Analysis (0)] |
| 4. | Clements O, Eliahoo J, Kim JU, Taylor-Robinson SD, Khan SA. Risk factors for intrahepatic and extrahepatic cholangiocarcinoma: A systematic review and meta-analysis. J Hepatol. 2020;72:95-103. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 173] [Cited by in RCA: 336] [Article Influence: 67.2] [Reference Citation Analysis (1)] |
| 5. | Mazzaferro V, Gorgen A, Roayaie S, Droz Dit Busset M, Sapisochin G. Liver resection and transplantation for intrahepatic cholangiocarcinoma. J Hepatol. 2020;72:364-377. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 237] [Article Influence: 47.4] [Reference Citation Analysis (0)] |
| 6. | Chen Q, Wang H, Li Z, Li F, Liang L, Zou Y, Shen H, Li J, Xia Y, Cheng Z, Yang T, Wang K, Shen F. Circular RNA ACTN4 promotes intrahepatic cholangiocarcinoma progression by recruiting YBX1 to initiate FZD7 transcription. J Hepatol. 2022;76:135-147. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 157] [Cited by in RCA: 143] [Article Influence: 47.7] [Reference Citation Analysis (0)] |
| 7. | Chen C, Wu YH, Zhang JW, Qiu YH, Wu H, Li Q, Song TQ, He Y, Mao XH, Zhai WL, Cheng ZJ, Li JD, Si SB, Cai ZQ, Geng ZM, Tang ZH. [A prognostic model of intrahepatic cholangiocarcinoma after curative intent resection based on Bayesian network]. Zhonghua Wai Ke Za Zhi. 2021;59:265-271. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 8. | Moris D, Palta M, Kim C, Allen PJ, Morse MA, Lidsky ME. Advances in the treatment of intrahepatic cholangiocarcinoma: An overview of the current and future therapeutic landscape for clinicians. CA Cancer J Clin. 2023;73:198-222. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 262] [Cited by in RCA: 249] [Article Influence: 124.5] [Reference Citation Analysis (0)] |
| 9. | Izquierdo-Sanchez L, Lamarca A, La Casta A, Buettner S, Utpatel K, Klümpen HJ, Adeva J, Vogel A, Lleo A, Fabris L, Ponz-Sarvise M, Brustia R, Cardinale V, Braconi C, Vidili G, Jamieson NB, Macias RI, Jonas JP, Marzioni M, Hołówko W, Folseraas T, Kupčinskas J, Sparchez Z, Krawczyk M, Krupa Ł, Scripcariu V, Grazi GL, Landa-Magdalena A, Ijzermans JN, Evert K, Erdmann JI, López-López F, Saborowski A, Scheiter A, Santos-Laso A, Carpino G, Andersen JB, Marin JJ, Alvaro D, Bujanda L, Forner A, Valle JW, Koerkamp BG, Banales JM. Cholangiocarcinoma landscape in Europe: Diagnostic, prognostic and therapeutic insights from the ENSCCA Registry. J Hepatol. 2022;76:1109-1121. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 206] [Cited by in RCA: 192] [Article Influence: 64.0] [Reference Citation Analysis (0)] |
| 10. | Kam AE, Masood A, Shroff RT. Current and emerging therapies for advanced biliary tract cancers. Lancet Gastroenterol Hepatol. 2021;6:956-969. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 107] [Article Influence: 26.8] [Reference Citation Analysis (0)] |
| 11. | Greener JG, Kandathil SM, Moffat L, Jones DT. A guide to machine learning for biologists. Nat Rev Mol Cell Biol. 2022;23:40-55. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 185] [Cited by in RCA: 813] [Article Influence: 271.0] [Reference Citation Analysis (0)] |
| 12. | Handelman GS, Kok HK, Chandra RV, Razavi AH, Lee MJ, Asadi H. eDoctor: machine learning and the future of medicine. J Intern Med. 2018;284:603-619. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 215] [Cited by in RCA: 483] [Article Influence: 69.0] [Reference Citation Analysis (0)] |
| 13. | Mosele F, Remon J, Mateo J, Westphalen CB, Barlesi F, Lolkema MP, Normanno N, Scarpa A, Robson M, Meric-Bernstam F, Wagle N, Stenzinger A, Bonastre J, Bayle A, Michiels S, Bièche I, Rouleau E, Jezdic S, Douillard JY, Reis-Filho JS, Dienstmann R, André F. Recommendations for the use of next-generation sequencing (NGS) for patients with metastatic cancers: a report from the ESMO Precision Medicine Working Group. Ann Oncol. 2020;31:1491-1505. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 569] [Cited by in RCA: 758] [Article Influence: 151.6] [Reference Citation Analysis (0)] |
| 14. | Nault JC, Villanueva A. Biomarkers for Hepatobiliary Cancers. Hepatology. 2021;73 Suppl 1:115-127. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 106] [Article Influence: 26.5] [Reference Citation Analysis (0)] |
| 15. | Banales JM, Cardinale V, Carpino G, Marzioni M, Andersen JB, Invernizzi P, Lind GE, Folseraas T, Forbes SJ, Fouassier L, Geier A, Calvisi DF, Mertens JC, Trauner M, Benedetti A, Maroni L, Vaquero J, Macias RI, Raggi C, Perugorria MJ, Gaudio E, Boberg KM, Marin JJ, Alvaro D. Expert consensus document: Cholangiocarcinoma: current knowledge and future perspectives consensus statement from the European Network for the Study of Cholangiocarcinoma (ENS-CCA). Nat Rev Gastroenterol Hepatol. 2016;13:261-280. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 731] [Cited by in RCA: 961] [Article Influence: 106.8] [Reference Citation Analysis (0)] |
| 16. | Rizvi S, Khan SA, Hallemeier CL, Kelley RK, Gores GJ. Cholangiocarcinoma - evolving concepts and therapeutic strategies. Nat Rev Clin Oncol. 2018;15:95-111. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1052] [Cited by in RCA: 1134] [Article Influence: 162.0] [Reference Citation Analysis (0)] |
| 17. | Charalampakis N, Papageorgiou G, Tsakatikas S, Fioretzaki R, Kole C, Kykalos S, Tolia M, Schizas D. Immunotherapy for cholangiocarcinoma: a 2021 update. Immunotherapy. 2021;13:1113-1134. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 12] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 18. | Zhang Y, Xia R, Lv M, Li Z, Jin L, Chen X, Han Y, Shi C, Jiang Y, Jin S. Machine-Learning Algorithm-Based Prediction of Diagnostic Gene Biomarkers Related to Immune Infiltration in Patients With Chronic Obstructive Pulmonary Disease. Front Immunol. 2022;13:740513. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 29] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 19. | Zhang X, Tian C, Cheng J, Mao W, Li M, Chen M. LTBP2 inhibits prostate cancer progression and metastasis via the PI3K/AKT signaling pathway. Exp Ther Med. 2022;24:563. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 20. | Raggi C, Taddei ML, Rae C, Braconi C, Marra F. Metabolic reprogramming in cholangiocarcinoma. J Hepatol. 2022;77:849-864. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 84] [Article Influence: 28.0] [Reference Citation Analysis (0)] |
| 21. | Li H, Long J, Xie F, Kang K, Shi Y, Xu W, Wu X, Lin J, Xu H, Du S, Xu Y, Zhao H, Zheng Y, Gu J. Transcriptomic analysis and identification of prognostic biomarkers in cholangiocarcinoma. Oncol Rep. 2019;42:1833-1842. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 22. | Fan S, Ge Y, Liu J, Liu H, Yan R, Gao T, Fan X, Xiao Z, An G. Combination of anlotinib and gemcitabine promotes the G0/G1 cell cycle arrest and apoptosis of intrahepatic cholangiocarcinoma in vitro. J Clin Lab Anal. 2021;35:e23986. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 19] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 23. | Lin Y, Cai Q, Chen Y, Shi T, Liu W, Mao L, Deng B, Ying Z, Gao Y, Luo H, Yang X, Huang X, Shi Y, He R. CAFs shape myeloid-derived suppressor cells to promote stemness of intrahepatic cholangiocarcinoma through 5-lipoxygenase. Hepatology. 2022;75:28-42. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 107] [Article Influence: 35.7] [Reference Citation Analysis (0)] |
| 24. | Claesson-Welsh L. How the matrix metalloproteinase MMP14 contributes to the progression of colorectal cancer. J Clin Invest. 2020;130:1093-1095. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 16] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 25. | Määttä M, Soini Y, Liakka A, Autio-Harmainen H. Differential expression of matrix metalloproteinase (MMP)-2, MMP-9, and membrane type 1-MMP in hepatocellular and pancreatic adenocarcinoma: implications for tumor progression and clinical prognosis. Clin Cancer Res. 2000;6:2726-2734. [PubMed] |
| 26. | Qiang L, Cao H, Chen J, Weller SG, Krueger EW, Zhang L, Razidlo GL, McNiven MA. Pancreatic tumor cell metastasis is restricted by MT1-MMP binding protein MTCBP-1. J Cell Biol. 2019;218:317-332. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 31] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 27. | Yu J, He Z, He X, Luo Z, Lian L, Wu B, Lan P, Chen H. Comprehensive Analysis of the Expression and Prognosis for MMPs in Human Colorectal Cancer. Front Oncol. 2021;11:771099. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 20] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 28. | Decotret LR, Wadsworth BJ, Li LV, Lim CJ, Bennewith KL, Pallen CJ. Receptor-type protein tyrosine phosphatase alpha (PTPα) mediates MMP14 Localization and facilitates triple-negative breast cancer cell invasion. Mol Biol Cell. 2021;32:567-578. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 6] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 29. | Jin Y, Liang ZY, Zhou WX, Zhou L. High MMP14 expression is predictive of poor prognosis in resectable hepatocellular carcinoma. Pathology. 2020;52:359-365. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 16] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 30. | Liu H, Hu G, Wang Z, Liu Q, Zhang J, Chen Y, Huang Y, Xue W, Xu Y, Zhai W. circPTCH1 promotes invasion and metastasis in renal cell carcinoma via regulating miR-485-5p/MMP14 axis. Theranostics. 2020;10:10791-10807. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 65] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 31. | Poincloux R, Lizárraga F, Chavrier P. Matrix invasion by tumour cells: a focus on MT1-MMP trafficking to invadopodia. J Cell Sci. 2009;122:3015-3024. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 366] [Cited by in RCA: 371] [Article Influence: 23.2] [Reference Citation Analysis (0)] |
| 32. | Gonzalez-Molina J, Gramolelli S, Liao Z, Carlson JW, Ojala PM, Lehti K. MMP14 in Sarcoma: A Regulator of Tumor Microenvironment Communication in Connective Tissues. Cells. 2019;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 61] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 33. | DeBerardinis RJ. Tumor Microenvironment, Metabolism, and Immunotherapy. N Engl J Med. 2020;382:869-871. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 125] [Cited by in RCA: 186] [Article Influence: 37.2] [Reference Citation Analysis (0)] |
| 34. | Vitale I, Manic G, Coussens LM, Kroemer G, Galluzzi L. Macrophages and Metabolism in the Tumor Microenvironment. Cell Metab. 2019;30:36-50. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 613] [Cited by in RCA: 1145] [Article Influence: 190.8] [Reference Citation Analysis (0)] |
| 35. | Bader JE, Voss K, Rathmell JC. Targeting Metabolism to Improve the Tumor Microenvironment for Cancer Immunotherapy. Mol Cell. 2020;78:1019-1033. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 566] [Cited by in RCA: 618] [Article Influence: 123.6] [Reference Citation Analysis (2)] |
| 36. | Borst J, Ahrends T, Bąbała N, Melief CJM, Kastenmüller W. CD4(+) T cell help in cancer immunology and immunotherapy. Nat Rev Immunol. 2018;18:635-647. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 582] [Cited by in RCA: 1085] [Article Influence: 180.8] [Reference Citation Analysis (0)] |
| 37. | Olingy CE, Dinh HQ, Hedrick CC. Monocyte heterogeneity and functions in cancer. J Leukoc Biol. 2019;106:309-322. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 338] [Cited by in RCA: 378] [Article Influence: 63.0] [Reference Citation Analysis (0)] |
| 38. | Gordon IO, Freedman RS. Defective antitumor function of monocyte-derived macrophages from epithelial ovarian cancer patients. Clin Cancer Res. 2006;12:1515-1524. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 69] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 39. | Yeap WH, Wong KL, Shimasaki N, Teo EC, Quek JK, Yong HX, Diong CP, Bertoletti A, Linn YC, Wong SC. CD16 is indispensable for antibody-dependent cellular cytotoxicity by human monocytes. Sci Rep. 2016;6:34310. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 115] [Cited by in RCA: 164] [Article Influence: 18.2] [Reference Citation Analysis (0)] |