Published online May 14, 2023. doi: 10.3748/wjg.v29.i18.2864
Peer-review started: December 31, 2022
First decision: February 28, 2023
Revised: March 14, 2023
Accepted: April 18, 2023
Article in press: April 18, 2023
Published online: May 14, 2023
Processing time: 130 Days and 10.9 Hours
Genetic tests are increasingly performed for the management of unresectable pancreatic cancer. For genotyping aimed samples current guidelines recommend using core specimens, although based on moderate quality evidence. However, in clinical practice among the endoscopic ultrasound (EUS) guided tissue acquisition methods, fine needle aspiration (FNA) is the most widely performed.
To assess the adequacy for next generation sequencing (NGS) of the DNA yielded from EUS-FNA pancreatic adenocarcinoma (PDAC) samples.
Between November 2018 and December 2021, 105 patients with PDAC confirmed by EUS-FNA were included in the study at our tertiary gastroenterology center. Either 22 gauge (G) or 19G FNA needles were used. One pass was dedicated to DNA extraction. DNA concentration and purity (A260/280, A260/230) were assessed by spectrophotometry. We assessed the differences in DNA parameters according to needle size and tumor characteristics (size, location) and the adequacy of the extracted DNA for NGS (defined as A260/280 ≥ 1.7, and DNA yield: ≥ 10 ng for amplicon based NGS, ≥ 50 ng for whole exome sequencing [WES], ≥ 100 ng for whole genome sequencing [WGS]) by analysis of variance and t-test respectively. Moreover, we compared DNA purity parameters across the different DNA yield categories.
Our cohort included 49% male patients, aged 67.02 ± 8.38 years. The 22G needle was used in 71% of the cases. The DNA parameters across our samples varied as follows: DNA yield: 1289 ng (inter quartile range: 534.75-3101), A260/280 = 1.85 (1.79-1.86), A260/230 = 2.2 (1.72-2.36). DNA yield was > 10 ng in all samples and > 100 ng in 93% of them (one sample < 50 ng). There were no significant differences in the concentration and A260/280 between samples by needle size. Needle size was the only independent predictor of A260/230 which was higher in the 22G samples (P = 0.038). NGS adequacy rate was 90% for 19G samples regardless of NGS type, and for 22G samples it reached 89% for WGS adequacy and 91% for WES and amplicon based NGS. Samples with DNA yield > 100 ng had significantly higher A260/280 (1.89 ± 0.32 vs 1.34 ± 0.42, P = 0.013). Tumor characteristics were not corelated with the DNA parameters.
EUS-FNA PDAC samples yield DNA adequate for subsequent NGS. DNA amount was similar between 22G and 19G FNA needles. DNA purity parameters may vary indirectly with needle size.
Core Tip: Genetic testing is increasingly undertaken in pancreatic adenocarcinoma (PDAC). The main diagnostic method is endoscopic ultrasound (EUS)-guided tissue acquisition and fine needle aspiration (FNA) is most frequently performed in clinical practice. However, current guidelines recommend using core specimens for genotyping aimed samples. In our cohort analysis, we show that EUS-FNA PDAC samples yield DNA of adequate amount and purity for subsequent next-generation sequencing (NGS). DNA amount was similar between 22G and 19G FNA needles. DNA purity parameters may vary indirectly with needle size. EUS FNA PDAC samples may be used for NGS.
- Citation: Bunduc S, Varzaru B, Iacob RA, Sorop A, Manea I, Spiridon A, Chelaru R, Croitoru AE, Becheanu G, Dumbrava M, Dima S, Popescu I, Gheorghe C. Endoscopic ultrasound-guided fine-needle aspiration pancreatic adenocarcinoma samples yield adequate DNA for next-generation sequencing: A cohort analysis. World J Gastroenterol 2023; 29(18): 2864-2874
- URL: https://www.wjgnet.com/1007-9327/full/v29/i18/2864.htm
- DOI: https://dx.doi.org/10.3748/wjg.v29.i18.2864
Pancreatic cancer (PC) is among the most the most aggressive malignancies with a mortality rate comparable to the incidence[1,2]. In more than 80% of cases it is detected in advanced stages that require systemic treatment[3]. Moreover, there is growing evidence about the benefits of neoadjuvant treatment over upfront surgery for the management of resectable pancreatic cancer[4]. Regardless of stage, anatomopathological confirmation of the disease is mandatory prior to chemo or radiotherapy initiation[5].
Endoscopic ultrasound (EUS) guided tissue acquisition (TA) is the main sampling method for the diagnosis of pancreatic solid lesions with high sensitivity (85%-89%) and specificity (96%-99%)[6]. Among the EUS-TA methods, fine needle aspiration (FNA) is the most widely performed[5].
Despite the high diagnostic accuracy, EUS-TA samples are still associated with an inconclusive diagnosis in 10% to 15% of cases[7]. Detection for KRAS mutations in inconclusive samples could decrease with up to 50% the rates of false negative results[6].
Precision medicine has changed the paradigm in oncologic treatment and although the targeted therapies are not yet routinely used in PC, it is known that up to 25% of the tumors harbor actionable alterations[8]. Molecular testing on tumor tissue for guiding personalized treatment is recommended in the current guidelines for the management of unresectable pancreatic adenocarcinoma (PDAC) amenable for anti-cancer therapy[9].
The roles of EUS-TA in PC have therefore broadened from tumor diagnosis to obtaining adequate samples for further molecular testing that enable the use of targeted therapies and may also increase diagnostic accuracy[6,9,10]. Although according to the current guidelines in pancreatic solid masses core tissue specimens should be used in downstream applications, there is increasing evidence on the feasibility of performing comprehensive genomic profiling based on FNA samples[5,11].
In our study we evaluated the adequacy for NGS [targeted, amplicon based, whole exome sequencing (WES) and whole genome sequencing (WGS)] of the DNA extracted from fresh EUS FNA PDAC samples, in terms of quantity and purity as measured by spectrophotometry. Moreover, we assessed the influence of needle the size and tumor characteristics on the DNA parameters.
The research was conducted in Fundeni Clinical Institute-a tertiary gastroenterology referral center in Bucharest, Romania, between November 2018 and December 2021. This prospective observational study was approved by the Internal Review Board of Fundeni Clinical Institute. Patients provided informed consent for EUS-FNA and study enrolment before the procedure.
For enrolment, we assessed the cases with pancreatic lesions referred to our department for EUS-TA. The eligibility criterion was-diagnosis of PDAC established by EUS-FNA. We only considered for inclusion in the study patients with a high suspicion of PDAC (as evaluated by clinical, biological, and imaging criteria). Patients with subsequent diagnoses other than PDAC were excluded from our analysis.
Highly experienced endosonographers performed all the procedures and all patients were under propofol sedation. A linear echoendoscope was used (EG-3870UTK, Pentax, equipped with a Hitachi Arietta v70 processor, Tokyo, Japan) and either 19 gauge (G) or 22G FNA needles (EchoTip Ultra Endoscopic Ultrasound Needle; Cook Medical, Bloomington, IN, United States). The endosonographer decided on the needle type. After obtaining the samples for the pathological diagnosis, one pass was dedicated to sample acquisition for DNA extraction. The needle stylet or air flushing were used to facilitate the specimen extrusion. The samples dedicated to diagnosis were smeared onto slides and fixed with ethanol and/or placed into 10% formalin for further paraffin fixation and cell block analysis, FNA yielded small tissue fragments (about 50% of cases). Dedicated pathologists with extensive experience in cytology techniques assessed the samples on site. The diagnosis of PDAC was confirmed if the pathology findings were positive or suspicious for malignancy[12]. The samples purposed for DNA extraction were placed directly in 1.5 mL of RNA later solution, in 2 mL Eppendorf tubes and kept at room temperature until DNA extraction that was performed within 4 h after sampling. For DNA extraction we used a commercially available silica membrane-based column DNA extraction kit (PureLink™ Genomic DNA, Invitrogen™, Waltham, MA, United States) according to the manufac
The purity and quantity of the extracted DNA were assessed immediately after extraction by spectrophotometry (Nanodrop, Thermo Fisher™, Waltham, MA, United States). The DNA concentration was measured in ng/μL and the purity was assessed by the absorbance ratios-A260/280 and A260/230 (the ratios between absorbances of the samples at 260 nm-which is characteristic for nucleic acids, and 280 nm and 230 nm respectively)[13]. The optimal DNA parameters for second generation high throughput sequencing were considered as follows: For DNA yield ≥ 100 ng will be sufficient for NGS of any type (whole genome-WGS, whole exome-WES or amplicon based targeted NGS), for WES at least ≥ 50 ng are required, whereas for amplicon based targeted NGS a minimum of 10 ng is necessary[14-16]. The main purity parameter assessed in terms of NGS adequacy is A260/280 and values ≥ 1.7 are considered optimal[17]. A260/230 is a secondary purity parameter with optimal values ranging between 2.0-2.2[17]. Abnormal values of the purity parameters reflect mainly a contaminated sample with proteins, phenols, carbohydrates, salts among others[17]. To calculate the DNA yield we multiplied the measured concentration with the elution volume which was 25 μL in all cases. The DNA samples were stored at -80 °C until further analysis. We have successfully performed targeted amplicon based NGS in 20 of the collected samples, using the Illumina NextSeq500 platform.
Two investigators, trainees in gastroenterology, collected the data using a customized form. Among the gathered information, we used in our study: Patients demographics (age, gender), tumor characteristics-size (mm)-the maximum diameter measured during EUS) and location (coded as head/neck or body/tail respectively), if cross-sectional imaging was performed we also collected-TNM (tumor, node, metastasis) stage according to the American Joint Committee on 8th Cancer edition[18], and the vascular invasion status; information about the procedure: total number of passes, needle size (recorded either during the procedure or retrieved from the EUS registry of the department), the parameters for the extracted DNA-concentration and absorbance ratios. We included in our analysis only samples from patients with confirmed PDAC on the FNA specimens.
Our end-points were as follows: To evaluate the adequacy for NGS of the DNA extracted from EUS-FNA PDAC samples we generated several categorical variables based on the previously mentioned criteria: Three of them defining optimal DNA yield for the main types of NGS with the cut-offs: ≥ 100 ng for WGS, ≥ 50 ng for WES and ≥ 10 ng for amplicon based targeted NGS, and one for optimal DNA purity: A260/280 ≥ 1.7. NGS adequacy was defined as optimal parameters for both DNA yield and purity for each type of NGS (WGS, WES and amplicon based NGS adequacy respectively). The definitions of the categorical variables are detailed in Supplementary Table 1. The variables were coded as 1 if the corresponding criteria were accomplished and 0 if not. We also compared the purity parameters between the predefined categories of optimal DNA yield. Furthermore, we assessed the association between the FNA needle size and tumor characteristics (diameter and location) and the DNA parameters (DNA concentration [ng/μL], A260/280, A260/230).
Continuous variables are computed as either mean and standard deviation (mean ± SD) or median and inter quartile range. We analyzed the impact of needle size on DNA parameters by one way analysis of variance. We assessed the association between needle size and NGS adequacy using chi-square test. The impact of both needle size and tumor characteristics on the DNA parameters was evaluated by linear regression, while their impact on DNA NGS adequacy was evaluated by logistic regression respectively. The differences in purity parameters across the predefined DNA yield categories were assessed with independent sample t-test. Data was normalized using a two-step approach-transforming the variable into a percentile rank and subsequent inverse-normal transformation[19]. Two-tailed alpha of 0.05 defined statistical significance. Data analysis was performed with SPSS (IBM Corp. Released 2020. IBM SPSS Statistics for Windows, Version 27.0. IBM Corp, NY, Armonk, United States). The statistical review of the study was reviewed by Daniel Veres, MD, PhD who is a biomedical statistician at Center for Translational Medicine of Semmelweis University.
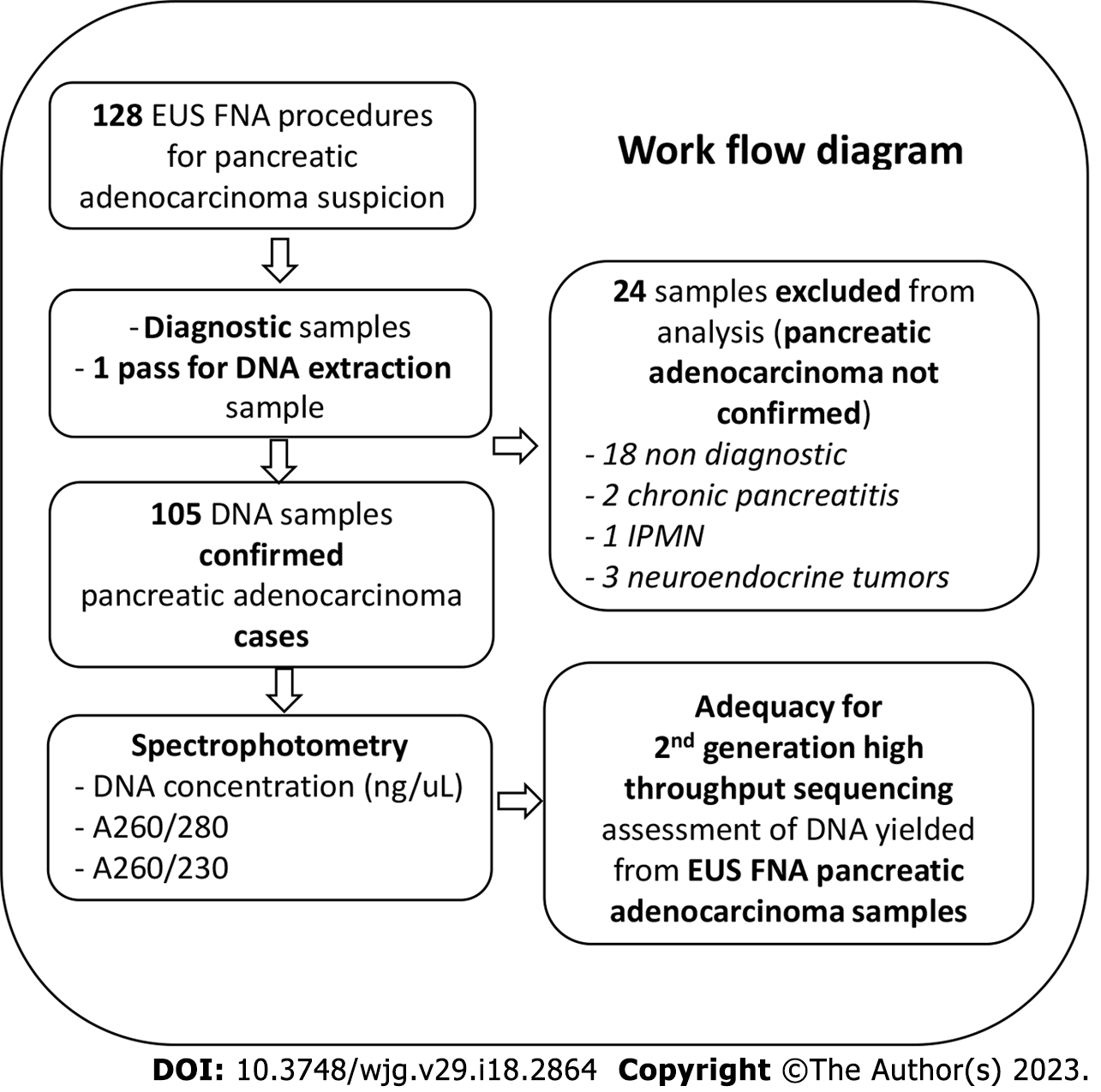
The characteristics of patients included in our analysis are summarized in Table 1. Out of the 128 patients who accepted to participate in our study, 105 were confirmed with PDAC subsequent to EUS-FNA (Figure 1). Sex distribution was almost equal and the mean age at diagnosis was 67.02 ± 8.38 years. Tumors were more frequently located in the head or neck of the pancreas than in the body or tail, and the tumor diameter was in average 43.66 ± 14.83 mm. More than half of the cases were metastatic at diagnosis. Most of the samples were acquired with 22G FNA needles-75 (71%).
| Variable | Amount |
| Number of PDAC cases | 105 |
| Age (yr) | 67.02 ± 8.38 |
| Male (number of cases) | 51 |
| Tumor diameter (mm) | 43.66 ± 14.83 |
| Tumor location (number of cases) | |
| Head/neck | 60 |
| Body/tail | 44 |
| Tumor stage (number of cases)1 | |
| IA | 1 |
| IB | 3 |
| IIA | 3 |
| IIB | 12 |
| III | 31 |
| IV | 55 |
| Needle size (number of samples) | |
| 22G | 75 |
| 19G | 30 |
In our cohort the DNA yield was in average 1289 ng (inter-quartile range: 534.75-2995). All samples resulted in more than 10 ng of DNA while 98 (93%) of them yielded more than 100 ng of DNA. All 7 samples from which less than 100 ng of DNA were extracted, were acquired with 22G needles, and only one yielded below 50 ng of DNA. In our analysis, needle size was not correlated with DNA NGS adequacy rate regardless of NGS type (Supplementary Table 2). For 19G needles, NGS adequacy rate was 90% for all types of NGS while for the 22 G needles the adequacy rate was 89% for WGS and 91% for both WES and amplicon based NGS. Tumor location and diameter did not significantly influence DNA NGS adequacy regardless of NGS type. When comparing the purity parameters across the predefined DNA yield categories (Table 2), the only significant difference we found was for the A260/280 parameter which was significantly higher in the samples with DNA yield ≥ 100 ng (1.89 ± 0.32 vs 1.34 ± 0.42, P = 0.013).
| DNA yield | Purity parameters | |
| A260/280 | A260/230 | |
| ≥ 100 ng | 1.86 (1.80-1.89) | 2.23 (1.79-2.35) |
| ≥ 50 ng | 1.85 (1.80-1.89) | 2.21 (1.7-2.36) |
| ≥ 10 ng | 1.85 (1.79-1.86) | 2.2 (1.72-2.36) |
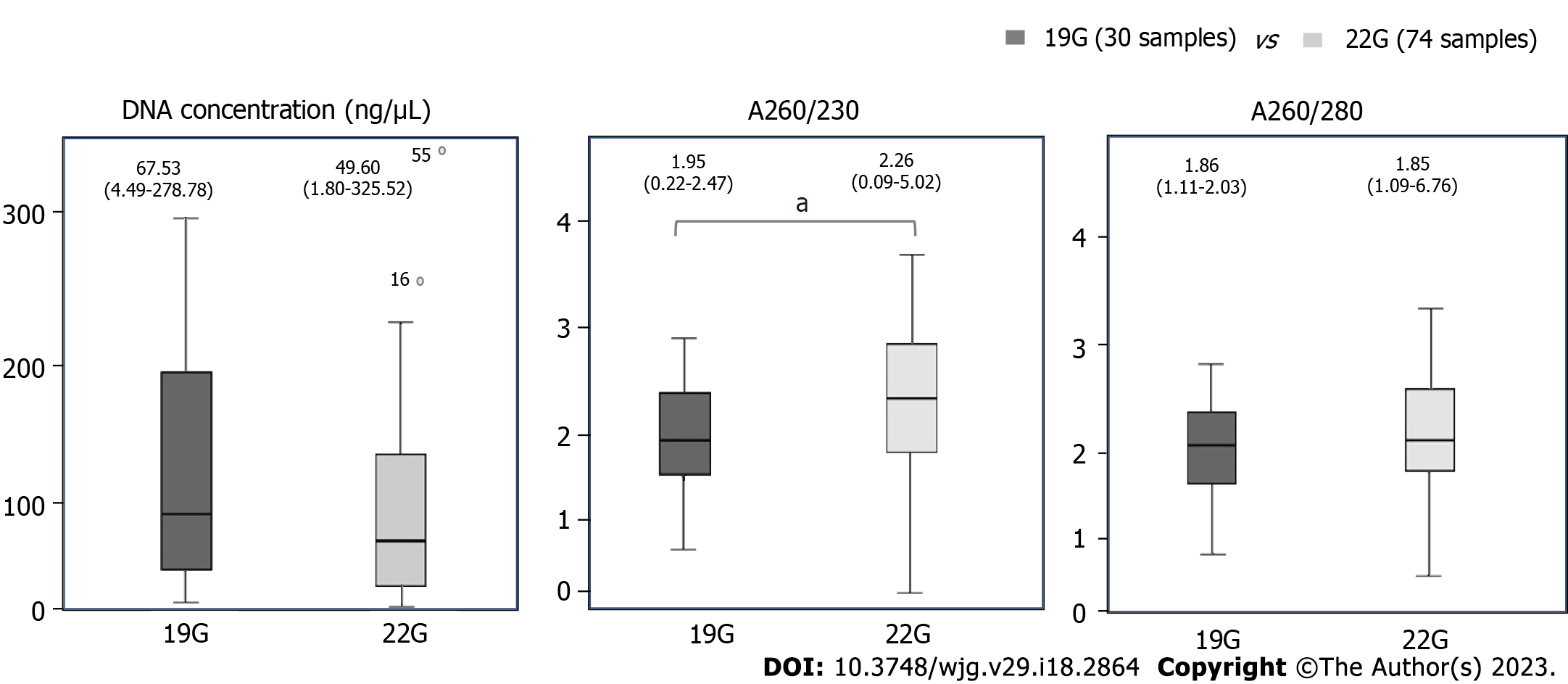
The DNA parameters across our samples were as follows: median concentration 51.56 ng/μL (21.39-124.04), A260/280 = 1.85 (1.79-1.86), A260/230 = 2.2 (1.72-2.36). The association between FNA needle size and the extracted DNA parameters are summarized in Figure 2. Needle size did not influence the concentration of the extracted DNA nor the A260/280 ratio. The median A260/230 was significantly higher in the 22G samples than in 19G samples (P = 0.038). In multivariate analysis (on needle size, tumor location and tumor diameter) the only independent predictor of A260/230 was needle size (β = 0.36, t(104) = 2.1, P = 0.038). None of the DNA parameters were significantly influenced by the tumor size or location.
The quantity and quality of the genetic material extracted from tumor samples are essential for their adequacy for downstream genetic analyses performed to further guide the individualized cancer therapy[21]. The requirements for NGS dedicated specimens vary with extraction protocols and testing method, but also with acquisition method, sample manipulation, disease type[22]. Although prior research has demonstrated the suitability of several pancreatic EUS-TA sample types for a range of comprehensive genetic testing techniques, the best sampling protocol for this use is still up for debate[23]. The current guidelines recommend the use of core samples for molecular testing in pancreatic solid lesions based on a moderate level of evidence[6,11].
Our cohort analysis revealed that FNA needle size does not influence the concentration and the primary purity parameter (A260/280) of the DNA extracted from 19G or 22G EUS-FNA PDAC samples. However, needle size was the only independent predictor of A260/230. Tumor diameter and location did not influence the DNA purity and concentration parameters. Based on the purity and DNA yield thresholds there were no significant differences in NGS adequacy rates between the two FNA needle sizes. NGS adequacy rate was 90% for 19G needle samples regardless of NGS type, while for the 22G needle samples it was 89% for WGS and 91% for both WES and amplicon based NGS. Regarding purity parameters, A260/280 was significantly better in samples with DNA yield above 100 ng. We further successfully performed targeted amplicon based NGS on a subgroup of 20 samples.
Park et al[24] evaluated which of the EUS-TA related factors and tumor characteristics have a significant impact on the NGS success rate on a cohort of 190 PDAC confirmed cases. They used for DNA analysis the remaining material after acquiring cytologic or histologic specimens for PDAC diagnosis[24]. As needed, 19G, 22G or 25G FNA or Fine needle biopsy (FNB) needles were used[24]. While tumor location in the body or tail of the pancreas was associated with a higher NGS success rate in their cohort than head or uncinate process tumor location, in multivariate analysis the only independent predictor of NGS success was a greater needle size (19G and 22G vs 25G)[24]. Although we were able to evaluate only a surrogate marker of NGS success, respectively-the adequacy of the DNA parameters for high throughput analysis, did we did not find a significant impact of tumor characteristics and neither of needle size on this outcome. Notably, we only used higher gauge needles (19G and 22G and not 25 G).
A group from Johns Hopkins evaluated the concordance of the mutational profiles assessed by targeted NGS between EUS-FNA snap frozen PDAC samples and the corresponding biopsy specimens from the primary tumors that were subsequently resected [25]. Although on a pool of 16 cases only, they detected a 100% concordance between the two sample types for KRAS mutations. They concluded that it is feasible to perform NGS on FNA samples and the results are reliable in complementing pathology results[25].
Besides the parameters of the extracted DNA, the total cellularity and the tumor cell content of the samples are also essential for the assessment of adequacy for NGS[22,26]. We did not evaluate these metrics in our cohort, since we used dedicated FNA samples, processed directly after collection, for DNA extraction. However, we must emphasize that the same needle types were used for the samples dedicated to diagnosis and we only report on confirmed PDAC cases. Moreover, more than 50% of specimens obtained by FNA yielded small tissue fragments and were subjected to cell block techniques to increase the diagnostic accuracy. The most widely used NGS platforms to date require a sample cellularity between 1000 and 5000 cells and a minimum 10% “lesional to-non-lesional cell ratio”[26-28]. A group from MD Anderson compared these parameters between concurrently acquired, percutaneous, image guided FNA and core needle biopsy samples in 24 various malignancy cases. They obtained higher cellularity and better tumor fraction in the cytologic specimens, arguing that by comparison with core biopsies, during FNA the proportion of acquired stromal cells is lower[27].
In their study Berry et al[21] performed molecular profiling of 66 snap frozen EUS-FNA pancreatic cancer samples. They revealed a high epithelial and tumor cell content in their specimens and a low contamination with inflammatory, gastric or duodenal cells[21]. Moreover, they compared the DNA yield between the FNA samples dedicated to molecular testing and formalin fixed paraffin embedded cell-block preparations used for PDAC diagnosis, obtaining 10 times higher amounts of DNA from the FNA samples which was in average 4.8 ± 3.7 μg[21]. This could be explained by the degrading effect of formaldehyde on nucleic acids and the cross-links formation between protein and DNA that it determines[22].
Cytologic specimens yield optimal DNA and have adequate cellularity for further NGS. The quality of nucleic acids is altered in samples previously fixed with formalin. More standardized procedures that are focused on specimen sparing and keeping costs and processing times within accessible ranges are needed for the optimization of the sample assessment in the preanalytical phase of NGS.
Regarding the implications for practice our study has shown that both 19G and 22G fresh EUS-FNA PDAC samples yield DNA of adequate amount and purity for next generation high throughput sequencing, regardless of tumor size or location within the pancreas. Overall, the NGS adequacy rates corresponded to the diagnostic accuracy of EUS-FNA samples in PDAC in our cohort[6]. A randomized controlled trial could reliably evaluate the cause-effect relationship between EUS needle G and DNA output from pancreatic cancer samples. Since the technical requirements in various clinical scenarios may limit the feasibility of randomization, prospective cohort studies of greater sizes that allow propensity score matching could decrease the influence of confounders in analyzing this association. Besides targeted NGS, the EUS-FNA samples yielding below 100 ng of DNA could be also used in other downstream applications for genetic testing like droplet digital PCR or TaqMan assays, which, although addressing fewer targets, have the advantage of good sensitivity for samples with low DNA amount.
Even though relatively similar analyses have been previously reported, our study included a high number of samples[21,27]. Besides measuring the yielded DNA concentration and purity ratios, we were able to successfully perform NGS on subgroup of samples, functionality in the downstream application being a reliable method of sample adequacy evaluation[26]. Nevertheless, several limitations should be pointed out: (1) Study design-one of the main limitations of our work-since lack of patients randomization precludes the evaluation of causality between needle size and samples’ NGS adequacy; (2) our study was performed in a tertiary gastroenterology center and all involved personnel were experts in their fields (endosonographers, pathologists, biologists); (3) the analysis was not based on a prior sample size calculation therefore our results must be interpreted with caution especially since 71% of the procedures were performed with one EUS-FNA needle type; (4) we did not use 25G needles for our samples therefore our conclusions cannot be extrapolated to all FNA needle sizes; and (5) lack of a comparison group comprising samples obtained by EUS-FNB-another main limitation of our study; to this end however we cite the study of Razzano et al[29] that compared the performance for NGS between FNA, FNB and resection PDAC specimens. They obtained similar success rates for mutation and amplification analysis between FNA and FNB samples and proposed FNA material as a source for comprehensive molecular testing[29]. Moreover, the spectrophotometric methods may overestimate the quantity of amplifiable DNA by measuring not only the double stranded fragments, but also single stranded DNA, free and oligonucleotides[22].
Acquiring EUS-FNA specimens dedicated for genetic testing is associated with a high adequacy rate for NGS of the extracted DNA. Nineteen and 22G EUS-FNA PDAC samples generate similar amount of DNA. DNA purity may vary indirectly with needle size in EUS-FNA samples. Further standardization in sample handling and cellularity assessment in the pre-analytical phase of NGS will increase reliability of the results for EUS-FNA PDAC samples and hence their usability in current practice.
Due to the opportunities for personalized treatment in pancreatic adenocarcinoma (PDAC), genetic testing is increasingly performed. Fine needle biopsy is the method recommended for endoscopic ultrasound (EUS) guided tissue acquisition to obtain samples dedicated to downstream comprehensive molecular analyses. In current practice however, fine needle aspiration (FNA) is more widely accessible.
We evaluated the EUS-FNA PDAC samples in terms of adequacy for next generation sequencing (NGS) of the yielded DNA to assess the possibility of using this type of samples for genetic testing.
To investigate the association between DNA parameters (amount and purity) measured by spectrophotometry and FNA needle size (19 gauge [G] or 22G), and also tumor characteristics.
We performed an observational prospective study on PDAC cases diagnosed through EUS-FNA at a tertiary center of Gastroenterology in Romania. During EUS one pass acquired samples dedicated to genetic testing. NGS adequacy was a dichotomus variable defined based on DNA parameters (purity: A260/280 ≥ 1.7 and DNA amount: ≥ 100 ng for whole genome sequencing, ≥ 50 ng for whole exome sequencing or ≥ 10 ng for amplicon based targeted NGS).
Our cohort analysis comprised 105 confirmed PDAC cases. The majority of samples were acquired with 22G FNA needles-75 (71%). The DNA amount was in average 1289 ng (inter-quartile range: 534.75-2995). All samples yielded more than 10 ng of DNA while 98 (93%) of them yielded more than 100 ng of DNA. Needle size was not correlated with DNA NGS adequacy rate regardless of NGS type. Needle size did not influence the concentration or A260/280 ratio of the extracted DNA. The median A260/230 was significantly higher in the 22G samples than in 19G samples (P = 0.038). In multivariate analysis (on needle size, tumor location and tumor diameter) the only independent predictor of A260/230 was needle size (β = 0.36, t(104) = 2.1, P = 0.038).
Both 22G an 19G EUS-FNA PDAC samples are adequate for downstream NGS. FNA needle size and tumor characteristics did not significantly influence sample NGS adequacy rate. Greater FNA needle size might be associated with decreased sample purity.
PDAC FNA samples (22 and 19G) yield samples of adequate purity and amount for NGS and can be used both in current practice and for research purposes.
we would like to give special thanks to Daniel Veres, MD, PhD, for his guidance in performing the formal analysis for the project and Mihaela Topala, MD for her help with patients’ enrolment.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Romania
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Li JJ, China; Martino A, Italy; Wang D, China S-Editor: Chang KL L-Editor: A P-Editor: Zhao S
| 1. | Lu Y, Gentiluomo M, Macauda A, Gioffreda D, Gazouli M, Petrone MC, Kelemen D, Ginocchi L, Morelli L, Papiris K, Greenhalf W, Izbicki JR, Kiudelis V, Mohelníková-Duchoňová B, Bueno-de-Mesquita B, Vodicka P, Brenner H, Diener MK, Pezzilli R, Ivanauskas A, Salvia R, Szentesi A, Aoki MN, Németh BC, Sperti C, Jamroziak K, Chammas R, Oliverius M, Archibugi L, Ermini S, Novák J, Kupcinskas J, Strouhal O, Souček P, Cavestro GM, Milanetto AC, Vanella G, Neoptolemos JP, Theodoropoulos GE, van Laarhoven HWM, Mambrini A, Moz S, Kala Z, Loveček M, Basso D, Uzunoglu FG, Hackert T, Testoni SGG, Hlaváč V, Andriulli A, Lucchesi M, Tavano F, Carrara S, Hegyi P, Arcidiacono PG, Busch OR, Lawlor RT, Puzzono M, Boggi U, Guo F, Małecka-Panas E, Capurso G, Landi S, Talar-Wojnarowska R, Strobel O, Gao X, Vashist Y, Campa D, Canzian F. Identification of Recessively Inherited Genetic Variants Potentially Linked to Pancreatic Cancer Risk. Front Oncol. 2021;11:771312. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 5] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 2. | GLOBOCAN. Estimated number of deaths in 2020, both sexes, all ages. [cited 31 December 2022]. Available from: https://gco.iarc.fr/today/fact-sheets-cancers. |
| 3. | Winter K, Talar-Wojnarowska R, Dąbrowski A, Degowska M, Durlik M, Gąsiorowska A, Głuszek S, Jurkowska G, Kaczka A, Lampe P, Marek T, Nasierowska-Guttmejer A, Nowakowska-Duława E, Rydzewska G, Strzelczyk J, Śledziński Z, Małecka-Panas E. Diagnostic and therapeutic recommendations in pancreatic ductal adenocarcinoma. Recommendations of the Working Group of the Polish Pancreatic Club. Prz Gastroenterol. 2019;14:1-18. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 17] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 4. | Ghanem I, Lora D, Herradón N, de Velasco G, Carretero-González A, Jiménez-Varas MÁ, Vázquez de Parga P, Feliu J. Neoadjuvant chemotherapy with or without radiotherapy versus upfront surgery for resectable pancreatic adenocarcinoma: a meta-analysis of randomized clinical trials. ESMO Open. 2022;7:100485. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 23] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 5. | Elhanafi S, Mahmud N, Vergara N, Kochman ML, Das KK, Ginsberg GG, Rajala M, Chandrasekhara V. Comparison of endoscopic ultrasound tissue acquisition methods for genomic analysis of pancreatic cancer. J Gastroenterol Hepatol. 2019;34:907-913. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 67] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 6. | Dumonceau JM, Deprez PH, Jenssen C, Iglesias-Garcia J, Larghi A, Vanbiervliet G, Aithal GP, Arcidiacono PG, Bastos P, Carrara S, Czakó L, Fernández-Esparrach G, Fockens P, Ginès À, Havre RF, Hassan C, Vilmann P, van Hooft JE, Polkowski M. Indications, results, and clinical impact of endoscopic ultrasound (EUS)-guided sampling in gastroenterology: European Society of Gastrointestinal Endoscopy (ESGE) Clinical Guideline - Updated January 2017. Endoscopy. 2017;49:695-714. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 264] [Cited by in RCA: 238] [Article Influence: 29.8] [Reference Citation Analysis (0)] |
| 7. | De Palo GM, De Lena M, Bonadonna G. Adriamycin vs adriamycin plus melphalan in advanced ovarian carcinoma. Cancer Treat Rep. 1977;61:355-357. [PubMed] |
| 8. | Reed JK, Raftery MA. Properties of the tetrodotoxin binding component in plasma membranes isolated from Electrophorus electricus. Biochemistry. 1976;15:944-953. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 97] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 9. | Tempero MA, Malafa MP, Al-Hawary M, Behrman SW, Benson AB, Cardin DB, Chiorean EG, Chung V, Czito B, Del Chiaro M, Dillhoff M, Donahue TR, Dotan E, Ferrone CR, Fountzilas C, Hardacre J, Hawkins WG, Klute K, Ko AH, Kunstman JW, LoConte N, Lowy AM, Moravek C, Nakakura EK, Narang AK, Obando J, Polanco PM, Reddy S, Reyngold M, Scaife C, Shen J, Vollmer C, Wolff RA, Wolpin BM, Lynn B, George GV. Pancreatic Adenocarcinoma, Version 2.2021, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw. 2021;19:439-457. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 160] [Cited by in RCA: 695] [Article Influence: 173.8] [Reference Citation Analysis (0)] |
| 10. | Triplett RG, Gillmore JD, Armstrong GC. An experimental model for osteomyelitis in the mandible. J Dent Res. 1977;56:1520. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 11. | Pouw RE, Barret M, Biermann K, Bisschops R, Czakó L, Gecse KB, de Hertogh G, Hucl T, Iacucci M, Jansen M, Rutter M, Savarino E, Spaander MCW, Schmidt PT, Vieth M, Dinis-Ribeiro M, van Hooft JE. Endoscopic tissue sampling - Part 1: Upper gastrointestinal and hepatopancreatobiliary tracts. European Society of Gastrointestinal Endoscopy (ESGE) Guideline. Endoscopy. 2021;53:1174-1188. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 93] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 12. | Saik RP, Brown D. Lithium: not a sensitive indicator of hydrogen ion diffusion. J Surg Res. 1978;25:163-165. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.0] [Reference Citation Analysis (0)] |
| 13. | Thermo Fisher Scientfic. DNA purification and concentration. 2019. [cited 31 December 2022]. Available from https://assets.thermofisher.com/TFS-Assets/BID/brochures/dna-purification-analysis-brochure.pdf. |
| 14. | Asokkumar R, Yung Ka C, Loh T, Kah Ling L, Gek San T, Ying H, Tan D, Khor C, Lim T, Soetikno R. Comparison of tissue and molecular yield between fine-needle biopsy (FNB) and fine-needle aspiration (FNA): a randomized study. Endosc Int Open. 2019;7:E955-E963. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 74] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 15. | Marg-Haufe B. TECAN. Evaluating the quality of DNA for Next Generation Sequencing, genotyping, and other downstream applications. [cited 31 December 2022]. Available from: https://www.tecan.com/blog/evaluating-the-quality-of-dna-for-next-generation-sequencing. |
| 16. | How do I determine the concentration, yield and purity of a DNA sample? [Internet]. 2022 Promega Corporation. [cited 20 May 2022]. Available from: https://www.promega.ro/resources/pubhub/enotes/how-do-i-determine-the-concentration-yield-and-purity-of-a-dna-sample/. |
| 17. | Assessment of Nucleic Acid Purity. Thermo Fisher Scientific – NanoDrop Products. [cited 31 December 2022]. https://tools.thermofisher.com/content/sfs/brochures/TN52646-E-0215M-NucleicAcid.pdf. |
| 18. | James D. Brierley, Mary K. Gospodarowicz, Christian Wittekind BO, Mason, H. Asamura, A. Lee EVE, L. Denny, M.B. Amin SG. TNM Classification of Malignant Tumors. 8th ed. John Wiley & Sons: WILEY Balckwell, 2017: 102-107. |
| 19. | Templeton GF. A two-step approach for transforming continuous variables to normal: Implications and recommendations for IS research. Commun Assoc Inf Syst. 2011;28:41-58. [RCA] [DOI] [Full Text] [Cited by in Crossref: 174] [Cited by in RCA: 177] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 20. | Gharaibeh A, Koppikar S, J. Bonilla-Escobar. F. Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) in the International Journal of Medical Students. Int J Med Students. 2014;2:36-37. [RCA] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 17] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 21. | Berry W, Algar E, Kumar B, Desmond C, Swan M, Jenkins BJ, Croagh D. Endoscopic ultrasound-guided fine-needle aspirate-derived preclinical pancreatic cancer models reveal panitumumab sensitivity in KRAS wild-type tumors. Int J Cancer. 2017;140:2331-2343. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 33] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 22. | Mäkinen KK, Virtanen KK. Effect of 4.5--year use of xylitol and sorbitol on plaque. J Dent Res. 1978;57:441-446. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 12] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 23. | Archibugi L, Testoni SGG, Redegalli M, Petrone MC, Reni M, Falconi M, Doglioni C, Capurso G, Arcidiacono PG. New era for pancreatic endoscopic ultrasound: From imaging to molecular pathology of pancreatic cancer. World J Gastrointest Oncol. 2019;11:933-945. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 7] [Cited by in RCA: 8] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 24. | Park JK, Lee JH, Noh DH, Park JK, Lee KT, Lee JK, Lee KH, Jang KT, Cho J. Factors of Endoscopic Ultrasound-Guided Tissue Acquisition for Successful Next-Generation Sequencing in Pancreatic Ductal Adenocarcinoma. Gut Liver. 2020;14:387-394. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 41] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 25. | Bratfos O. [Drug therapy of anxiety]. Tidsskr Nor Laegeforen. 1979;99:333-334. [PubMed] |
| 26. | Fabbri C, Gibiino G, Fornelli A, Cennamo V, Grifoni D, Visani M, Acquaviva G, Fassan M, Fiorino S, Giovanelli S, Bassi M, Ghersi S, Tallini G, Jovine E, Gasbarrini A, de Biase D. Team work and cytopathology molecular diagnosis of solid pancreatic lesions. Dig Endosc. 2017;29:657-666. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 9] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 27. | Higa HH, Manzi A, Varki A. O-acetylation and de-O-acetylation of sialic acids. Purification, characterization, and properties of a glycosylated rat liver esterase specific for 9-O-acetylated sialic acids. J Biol Chem. 1989;264:19435-19442. [PubMed] |
| 28. | Ronai SE, Hyman PE. Gianetti v Norwalk Hospital: Connecticut Supreme Court rules on hospital and medical staff member contractual relationship and judicial review of medical staff termination decisions. Conn Med. 1989;53:425-426. [PubMed] |
| 29. | Razzano D, Bouza SJ, Hernandez PV, Wang M, Robert ME, Walther Z, Cai G. Comprehensive molecular profiling of pancreatic ductal adenocarcinoma in FNA, biopsy, and resection specimens. Cancer Cytopathol. 2022;130:726-734. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 8] [Article Influence: 2.7] [Reference Citation Analysis (0)] |