Published online Jan 7, 2023. doi: 10.3748/wjg.v29.i1.43
Peer-review started: September 21, 2022
First decision: October 18, 2022
Revised: October 27, 2022
Accepted: December 13, 2022
Article in press: December 13, 2022
Published online: January 7, 2023
Processing time: 104 Days and 10.7 Hours
Given the frequent co-existence of an aggressive tumor and underlying chronic liver disease, the management of hepatocellular carcinoma (HCC) patients requires experienced multidisciplinary team discussion. Moreover, imaging plays a key role in the diagnosis, staging, restaging, and surveillance of HCC. Currently, imaging assessment of HCC entails the assessment of qualitative characteristics which are prone to inter-reader variability. Radiomics is an emerging field that extracts high-dimensional mineable quantitative features that cannot be assessed visually with the naked eye from medical imaging. The main potential applications of radiomic models in HCC are to predict histology, response to treatment, genetic signature, recurrence, and survival. Despite the encouraging results to date, there are challenges and limitations that need to be overcome before radiomics implementation in clinical practice. The purpose of this article is to review the main concepts and challenges pertaining to radiomics, and to review recent studies and potential applications of radiomics in HCC.
Core Tip: Radiomics is an emerging field that extracts high-dimensional mineable quantitative features that cannot be assessed visually with the naked eye from medical imaging. The main potential applications of radiomic models in hepatocellular carcinoma (HCC) are to predict histology, predict response to treatment, predict genetic signature, predict recurrence, and predict survival. The purpose of this article is to review the main concepts and challenges pertaining to radiomics, and to review recent studies and potential applications of radiomics in HCC.
- Citation: Miranda J, Horvat N, Fonseca GM, Araujo-Filho JAB, Fernandes MC, Charbel C, Chakraborty J, Coelho FF, Nomura CH, Herman P. Current status and future perspectives of radiomics in hepatocellular carcinoma. World J Gastroenterol 2023; 29(1): 43-60
- URL: https://www.wjgnet.com/1007-9327/full/v29/i1/43.htm
- DOI: https://dx.doi.org/10.3748/wjg.v29.i1.43
Hepatocellular carcinoma (HCC) is the third most common cause of cancer-related deaths worldwide[1]. Liver cancer is especially common in Asia, where 72.5% of all new liver cancer cases worldwide are diagnosed[2]. HCC accounts for over 90% of all primary liver cancer cases[3]. The main risk factors for HCC in the West is viral hepatitis (hepatitis C virus in the West and hepatitis B virus in Asia and in developing countries) and alcohol intake. In addition, non-alcoholic steatohepatitis is becoming a common risk factor, particularly in the West[3,4]. HCC patient prognosis depends on the stage of HCC at the time of diagnosis[5]; and advanced-staged patients at the time of diagnosis have a poor prognosis[5-7].
The treatment of HCC is based on tumor burden, clinical performance of the patient, and liver function[8]. Given the frequent co-existence of an aggressive tumor and underlying chronic liver disease, the management of HCC requires experienced multidisciplinary team discussion[9]. Moreover, radiology plays a key role in the screening, diagnosis, staging, restaging, and surveillance of HCC. Currently, imaging assessment is based on qualitative characteristics, such as size and enhancement pattern, which are prone to inter-reader variability. Reliable tools that can potentially address this variability as well as deal with the vast amount of imaging data are warranted[10]. Over the last decade, radiomics has become a popular quantitative tool that can potentially address these challenges and provide information not previously available for precision decision-making[11].
Radiomics is an emerging field that extracts high-dimensional mineable quantitative features that cannot be assessed visually with the naked eye from medical imaging[12]. The main potential applications of radiomic models in HCC are to predict histology, response to treatment, genetic signature, recurrence, and survival[13]. Despite the encouraging results to date, there are several challenges and limitations that need to be overcome before the implementation of radiomics in clinical practice. The purpose of this study is to review the main concepts, challenges pertaining to radiomics and recent studies and potential applications of radiomics in HCC.
In the new era of precision medicine, artificial intelligence (AI) and in its various branches, such as machine learning (ML) and deep learning (DL), have provided new imaging biomarkers that can potentially provide new data that are useful for clinical decision-making. ML is related to a set of computational systems that improve with experience. DL is a subset of ML based on series of layers (trainable nonlinear operations), each of which transforms input data into a representation that facilitates pattern recognition[14].
Radiomics has recently emerged as a translational research field that proposes to discover new associations between clinical data and quantitative data extracted from medical images using conventional biostatistics or AI methods[12] and become popular, particularly in oncologic imaging. Radiomics involves mineable high-dimensional data extraction, characterizing intensity, shape, size, and/or texture from images to create big-data datasets that are then used to identify distinct sub-visual imaging patterns[15]. Radiomics models usually use magnetic resonance imaging (MRI), computed tomography (CT) and positron emission tomography (PET) images data. Fundamentally, radiomics is motivated by the observation that these imaging characteristics reflect phenotype and genotype of underlying tissue and thus can help in clinical decision making[16].
Radiomic can be subdivided into texture, size and shape, and transformed based features. The most common radiomic features is texture. It can be subdivided into first-order, second-order and higher-order statistical features. First-order features reflect the distribution of values of individual voxels without concern for spatial relationships; they are generally histogram-based, such as mean (average intensity), entropy (quantify randomness of intensity), kurtosis (flatness) and skewness (asymmetry). Second-order features reflect the statistical interrelationships between voxels with similar (or dissimilar) contrast values[12] and some of the commonly used 2nd order features are: Grey level co-occurrence matrix, grey level run length matrix, and grey level size zone matrix features. Taking into account the repetitive patterns in radiological images, higher-order statistical methods use sophisticated filter grids on the images - such as Minkowski functionals (to evaluate voxels whose intensity is above a determined threshold), Wavelet and Laplacian transforms (to identify coarse texture patterns) and fractal analysis (to assess the irregularity of a surface)[12]. In practice, standard libraries with predefined feature configurations and validated reference values (such as PyRadiomics) are frequently used to increase the reproducibility of radiomic models.
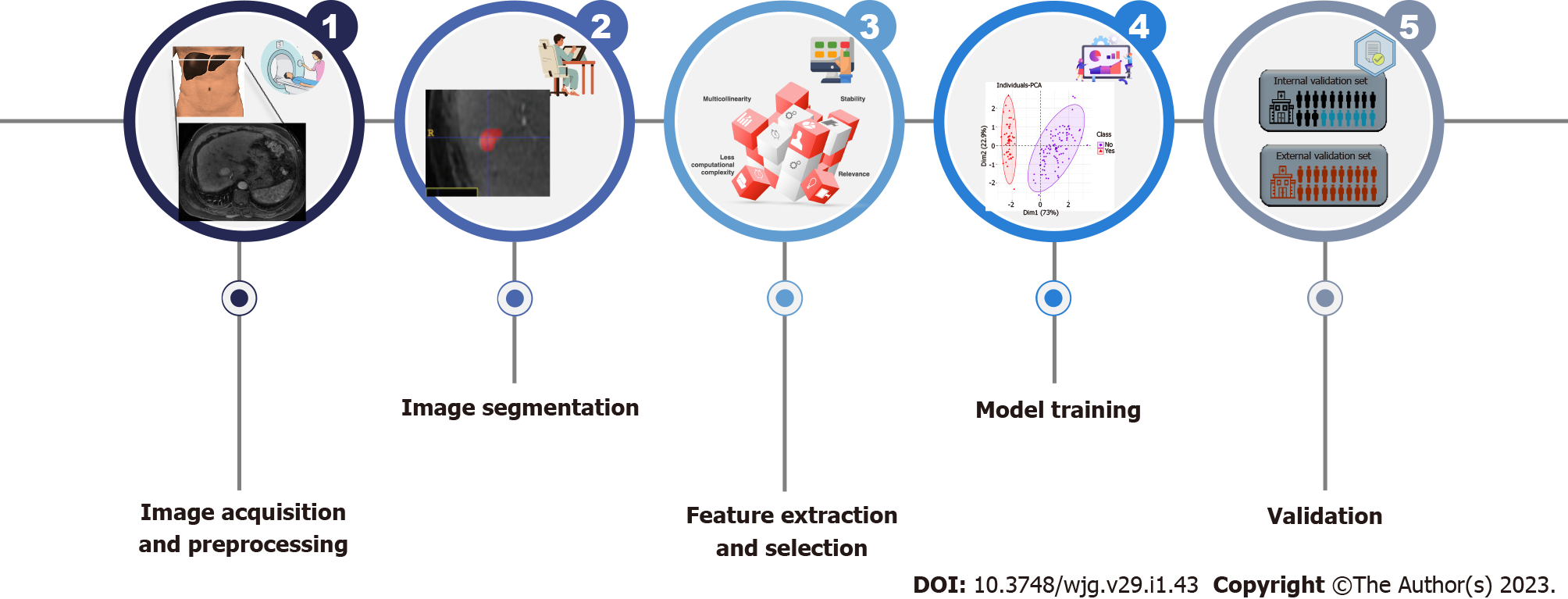
Radiomic analysis is a multistep process involving the processing of medical images to generate different features from segmented images. The typical radiomics workflow can be summarized in the following steps (Figure 1):
Image acquisition and preprocessing: Standardized imaging protocols should be used to avoid reproducibility issues related to noise and confounding. However, standardized imaging protocols also decrease the generalizability of the results. Once a patient dataset has been identified, images should be anonymized as well as exported as Digital Imaging and Communication in Medicine files[17]. De-noising and motion correction steps may be needed.
Segmentation: Segmentation involves the delineation of region of interests (ROIs) on the tumor or peritumoral zones. ROIs can be delineated manually, semiautomatically, or automatically (using ML tools) in either two-dimensional (2D) or three-dimensional (3D) views (Figure 2). Whenever possible, segmentations should be checked by a radiologist to ensure accuracy.
Feature extraction, feature selection and model building: A wide range of statistical models are commonly used to choose a subset of optimal features that correlate with the predenid outcome[15]. Many of the extracted features are in fact redundant and supervised or unsupervised approaches can be applied to achieve dimension reduction. ML and DL techniques are emerging as useful tools to achieve more accurate feature selection[18,19]. The features should be selected only based on the training data to avoid bias.
Of note, the number of extracted features is commonly larger than the study sample, which can contribute to overfitting of the model and to overoptimistic results. Some strategies can be done, for example, select the features in such a way to maintain the ratio or regularization methods are used to minimize the complexity of the respective models[20]. Once the optimal features are identified, a statistical model can be proposed to predict a specific clinical question using different classifiers such as generalized linear models, random forests, support vector machines, or neural networks[20,21].
Validation: Validation is essential to estimate model performance and can be done using subsets of the original training dataset (i.e., cross-validation) or using a separate hold-out dataset containing either internal or external data[17].
To date, radiomic models reproducibility is often poor, due to insufficient reporting or limited open-source code and data, which undermines external validation and increases the subsequent risk of false-positive results[22]. Further, researchers often face great difficulty in acquiring unbiased and homogeneous datasets across multiple institutions, thus hampering multi-institutional collaborations involving large multi-institutional datasets for the training and validation of radiomic models[14]. For successful multi-institutional cooperation for building large multi-institutional datasets for radiomic models training and validation, radiomics workflow standardization, clear reporting of study methodology, and data sharing across different institutions are needed[17]. Additionally, an effective means to interpret the vast and varied data derived from radiomics analysis is another key obstacle to the clinical implementation of radiomic models. Therefore, a balanced interpretation of results and an increased focus on interpretable models are essential to their successful integration into clinical practice[23]. Finally, manual segmentation is a time-consuming process and one of the most common limitations that should be managed with automatic or semiautomatic strategies before widespread use of radiomics tools.
Table 1 summarizes the studies in the literature to date that have evaluated the use of radiomics to preoperatively predict HCC histology.
| Ref. | Country | n | Imaging modality | Endpoint | Segmentation | ROI/VOI | No. of readers | Main results | Validation |
| Wang et al[92], 2022 | China | 196 | MRI | cHCC-CC vs HCC | Manual, intratumoral | ROI | 1 | AUC (delayed phase MRI): 0.91 | None |
| Liu et al[24], 2021 | Canada | 85 | MRI and CT | cHCC-CC vs HCC vs CC | Manual, intratumoral | ROI | 2 | AUC (MRI): 0.77-0.81. AUC (CT): 0.71-0.81 | Cross-validation |
| Lewis et al[25], 2019 | United States | 63 | MRI | cHCC-CC vs HCC vs CC | Manual, intratumoral | VOI | 2 | AUC (LI-RADS and male gender): 0.90 | None |
| Nie et al[27], 2020 | China | 156 | CT | HCC vs FNH | Manual, intratumoral | ROI | 2 | AUC (radiomics): 0.96 training, 0.87 validation. AUC (radiomics + clinical factors): 0.98 training, 0.92 validation | None |
| Wu et al[28], 2019 | China | 369 | MRI | HCC vs hemangioma | Manual, intratumoral | ROI | 2 | AUC: 0.86 training, 0.89 testing | None |
| Mokrane et al[29], 2020 | United States | 178 | CT | HCC diagnosis | Manual, intratumoral | VOI | 2 | AUC: 0.70 training, 0.66 validation | External |
| Brancato et al[34], 2022 | Italy | 38 | MRI | Tumor grade | Manual, intratumoral | VOI | 1 | AUC: 0.89 | None |
| Gao et al[93], 2018 | China | Training: 125. Validation: 45 | MRI | Tumor grade | Manual, intratumoral | N/A | N/A | AUC: 0.83 training, 0.74 validation | None |
| Wu et al[30], 2019 | China | Training: 125. Validation: 45 | MRI | Tumor grade | Manual, intratumoral | ROI | 1 | AUC: 0.83 training, 0.74 validation | Internal |
| Zhou et al[94], 2017 | China | 46 | MRI | Tumor grade | Manual, intratumoral | ROI | 1 | AUC: 0.83-0.92 | None |
| Mao et al[31], 2022 | China | Training: 85. Validation: 37 | MRI | Tumor grade | Manual, intratumoral | ROI | 2 | AUC: 0.97 training, 0.94 validation | Internal |
| Chen et al[33], 2021 | China | Training: 112. Validation: 49 | CT | Tumor grade | Manual, intratumoral | VOI | 2 | AUC: 0.90 training, 0.94 validation | Internal |
| Yang et al[95], 2019 | China | Training: 146. Validation: 62 | Gadoxetic acid-enhanced MRI | MVI | Manual, intratumoral | ROI | 2 (consensus) | AUC: 0.94 training, 0.86 validation | Internal |
| Xu et al[39], 2019 | China | 495 | CT | MVI | Semi-automatic, intratumoral and peritumoral | VOI | 3 | AUC: 0.91 training, 0.89 validation | Internal |
| Feng et al[40], 2019 | China | 160 | Gadoxetic acid-enhanced MRI | MVI | Manual, intratumoral and peritumoral | VOI | 3 | AUC: 0.85 training, 0.83 validation | Internal |
| Zheng et al[41], 2017 | United States | 120 | CT | MVI | Semi-automatic | ROI | 1 | AUC: 0.80 | None |
| Bakr et al[96], 2017 | United States | 28 | CT | MVI | Manual, intratumoral | ROI | 4 | AUC: 0.76 | None |
| Ma et al[97], 2019 | China | 157 | CT | MVI | Manual, intratumoral | VOI | 1 | AUC (portal venous phase CT): 0.79 | Cross-validation |
Distinguishing between HCC and other malignant or benign lesions: The distinction between HCC and other primary hepatobiliary malignancies can be challenging on imaging, because of the overlap of some features, especially for combined tumors[24]. In light of this, many studies have investigated radiomics performance in differentiating HCC from other malignant and benign hepatic lesions. For instance, Liu et al[24] studied the use of MRI- and CT-based radiomics to differentiate between HCC, cholangiocarcinoma, and combined HCC-cholangiocarcinoma. Using MRI, radiomic features derived from contrast-enhanced phases demonstrated excellent performance to differentiate HCC from non-HCC [area under the curve (AUC) ≥ 0.79], with the highest AUC obtained from the arterial phase (AUC of 0.81); meanwhile, using CT, radiomic features derived from the pre-contrast and portal venous phase yielded AUC values of 0.81 and 0.71, respectively. In another study, Lewis et al[25] found that the combination of the apparent diffusion coefficient 5th percentile radiomic feature with Liver Imaging Reporting and Data System classification and male gender achieved an accuracy of 80%-81.5% in distinguishing HCC from intrahepatic cholangiocarcinoma (ICC) and combined HCC-ICC, and outperformed either measure alone. Other studies showed that radiomics is helpful to distinguish between HCC and benign tumors in non-cirrhotic livers, e.g., from hepatocellular adenoma (AUC of 0.96 in the training set and 0.94 in the test set)[26], from focal nodular hyperplasia (AUC of 0.979 in the training set and 0.917 in the test set)[27], and from hemangioma (AUC: 0.86 in the training set and 0.89 in the test set)[28]. Mokrane et al[29] validated a radiomics signature to diagnose HCC in patients with cirrhosis and increased radiologists’ confidence.
Prediction of histologic grade: Histologic grade is an important prognostic factor in patients with HCC and is only available preoperatively in patients who undergo biopsy. Therefore, studies have aimed to identify non-invasive imaging features such as radiomic features that could potentially predict the tumor grade. Wu et al[30] found that MRI-based radiomics can successfully categorize low-grade and high-grade HCC, with the radiomic model outperforming the clinical model (AUC 0.742 for the combined T1-weighted and T2-weighted MRI-based radiomic model vs AUC 0.6 for the clinical one) and the combined radiomic and clinical model (AUC 0.8) outperforming both models alone. Mao et al[31] also investigated MRI-based radiomic features, with Gd-EOB-DTPA contrast administered for the MRI exams, finding that the artificial neural network combining radiomic features from the contrast-enhanced arterial phase and hepatobiliary phase yielded the highest AUC of 0.944. Moreover, they found that the artificial neural network models were superior to the logistic regression models. In other studies, CT-based radiomics has been found to have high performance in distinguishing between low- and high-grade tumors[32-34]; for instance, Chen et al[33] found an AUC of 0.937 for a ML-based radiomics model based on the CT portal phase.
Prediction of microvascular invasion: Microvascular invasion (MVI) is found in about 15%-57% of patients with HCC who undergo surgery[35,36] and is associated with higher rates of recurrence and shorter survival after surgery[37]. Although imaging can be used to diagnose macrovascular invasion (or tumor in vein), preoperative imaging identification of MVI is difficult. Studies have evaluated the performance of radiomics as a tool to predict MVI, with most predictive models combining radiomics and clinical biomarkers[38]. For instance, Xu et al[39] proposed a model combining CT-based radiomic features with radiologic and clinical parameters; the model was not only an independent predictor of histologic MVI (AUC of 0.909 in the training/validation set and 0.889 in the test set) but was also an independent predictor of worse prognosis (disease-specific recurrence and disease-specific mortality). Of note, the radiomics-only model did not add significant value to radiologist scores alone. Since MVI occurs primarily at the tumor periphery (approximately 85% of MVI is located within one centimeter from the tumor margin), studies have investigated radiomic features derived from the peritumoral tissue. For instance, Feng et al[40] demonstrated that a model combining intratumoral and peritumoral radiomic features was superior in predicting MVI using Gd-EOB-DTPA-enhanced MRI compared to the model containing only intratumoral radiomics features. Additionally, Zheng et al[41] demonstrated that peritumoral textural features had an AUC of 0.80 and a multivariate model combining alfa-fetoprotein, tumor size, hepatitis status and quantitative features achieved an AUC of 0.88.
Compared to the prediction of histology, fewer researches in the literature have evaluated the use of radiomics to predict genetic expression in patients with HCC (Table 2). Overall, studies on the use of radiomics to predict genetic expression have focused on using radiomics to predict Ki67 expression as well as cytokeratin 19 (CK19), P53, and phosphatidylinositol-3 kinase (PI3K) status. Of note, in 2007, Segal et al[42] investigated for the first time the correlation between HCC genetic expression and CT imaging traits, finding 32 CT imaging traits that were correlated with the expression levels of 116 genetic markers.
| Ref. | Country | n | Imaging modality | Endpoint | Segmentation | ROI/VOI | No. of readers | Main results | Validation |
| Xia et al[98], 2018 | China | 38 | CT | Association with gene expression profile | Manual, intratumoral | ROI | 1 | Individual textural features predicted gene modules | No |
| Wu et al[44], 2022 | China | Training: 120. Validation: 52 | CT | Ki-67 expression | Manual, intratumoral | VOI | 2 | AUC: 0.85 (training), 0.74 (validation) | Internal |
| Li et al[45], 2019 | China | 83 | MRI | Ki-67 expression | Manual, intratumoral | ROI | 2 | Some features were associated, no model | No |
| Ye et al[47], 2019 | China | 89 | MRI | Ki-67 expression | Manual, intratumoral | VOI | 2 | C-index: 0.878 | No |
| Fan et al[46], 2021 | China | Training: 103. Validation: 48 | MRI | Ki-67 expression | Manual, intratumoral | VOI | 2 | AUC: 0.88 (training), 0.80 (validation) | Internal |
| Hu et al[48], 2022 | China | Training: 87. Validation: 21 | MRI | Ki-67 expression | Manual, intratumoral | ROI | 1 | AUC: 0.90 (training), 0.83 (validation) | Internal |
| Wang et al[50], 2019 | China | 78 | MRI | CK19 positivity | Manual, intra- and peritumoral | ROI | 1 | AUC: 0.76 | No |
| Chen et al[51], 2021 | China | Training: 102. Validation: 19 | MRI | CK19 positivity | Manual, intratumoral | ROI | 2 | AUC: 0.82 (training), 0.78 (external validation) | Internal and external |
| Yang et al[52], 2021 | China (multi-center) | Training: 143. Validation: 75 | MRI | CK19 positivity | Manual, intratumoral | ROI | 2 | AUC: 0.85 (training), 0.79 (external validation) | Internal and external |
| Wu et al[55], 2019 | China | 63 | CT | P53 mutation status | Manual, intratumoral | ROI | 2 | AUC: 0.62-0.79 | No |
| Li et al[99], 2022 | China | 92 | MRI | Gene signatures associated with disease recurrence | Manual, intratumoral | ROI | 2 | MRI radiomics features could help quantify GOLM1, SETD7, and RND1 expression levels | Internal |
| Liao et al[56], 2022 | China | Training: 86. Validation: 46 | CT | Somatic mutations of the PI3K signaling pathway | Manual, intratumoral and peritumoral | VOI | 2 | AUC: 0.74 (training), 0.73 (external validation) | Internal and external |
| Che et al[60], 2022 | China | Training: 69. Validation: 30 | CT | β-arrestin1 phosphorylation | Manual, intratumoral | ROI | 1 | AUC: 0.89 (training), 0.74 (validation) | Internal |
Ki67 expression: High Ki-67 expression in HCC patients is associated with fast progression and poor prognosis[43]. To determine if radiomics can be useful to predict Ki67 expression, Wu et al[44] developed and validated a radiomic nomogram based on the combination of CT-based radiomic features and clinical factors. Using Gd-EOB-DTPA-enhanced MRI, Li et al[45] found that texture analysis of the hepatobiliary phase, arterial phase, and portal vein phase were helpful for predicting Ki67 expression. In their study, a single slice with the largest proportion of the lesion was delineated, and the predictive performance of models were compared by misclassification rate. In another study by Fan et al[46] using Gd-EOB-DTPA-enhanced MRI, the authors delineated the whole lesion, and the predictive performance of different models were compared using the receiver operating curve, calibration curve, and decision cure analysis. The optimal model combining the arterial phase radiomic score and serum alpha-fetoprotein (AFP) levels showed high AUCs (AUC of 0.922 and 0.863 in the training and validation cohorts, respectively) for the preoperative Ki-67 expression prediction. In yet another study using Gd-EOB-DTPA-enhanced MRI, Ye et al[47] showed that the nomogram combining the texture signature (using the segmentation of the whole lesion) and clinical factors demonstrated a high discrimination ability (C-index of 0.936) for predicting Ki-67 group (high vs low). Finally, Hu et al[48] explored the added value of viscoelasticity measured by magnetic resonance elastography to predict Ki-67 expression, showing that shear wave speed and phase angle significantly improved the performance of the radiomic model.
CK19 expression: CK19 expression is associated with aggressive tumor behavior, resistance to therapy, and poor outcomes including worse overall survival and recurrence[49]. To date, three studies have focused on developing radiomic models to predict CK19 expression[50-52], all using MRI. Wang et al[50] showed that their texture model independently predicted CK19-positive HCC cases and improved the diagnostic performance of AFP level ≥ 400 ng/mL and arterial rim enhancement. The two remaining studies developed a radiomics model based on Gd-EOB-DTPA-enhanced MRI, with external validation AUC varying from 0.78-0.79; of note, one of the studies was a multicenter study with over 250 patients[51,52].
P53, PI3K, and other genetic expression: P53 can be used as a tumor biomarker, since it plays an important role in the pathogenesis of HCC[53]. P53 mutation has also been suggested as a feasible target for antitumor therapy[54]. Wu et al[55] demonstrated a direct relationship between P53 mutations in patients with HCC and the gray-level co-occurrence matrix on CT. PI3K signaling is a key pathway regulating HCC aggressiveness and is associated with response to sorafenib. Liao et al[56] developed a CT-based radiomics model that yielded an AUC of 0.73 in the external validation set for prediction of PI3K status.
The phosphorylation status of β-arrestin1 is associated with sorafenib resistance[57-59]. Che et al[60] developed a model combining a CT-based radiomics score with clinico-radiological risk factors which yielded an AUC of 0.898 in predicting β-arrestin1 phosphorylation, and the predicted β-arrestin1 phosphorylation was in turn significantly associated with overall survival in both the training and validation cohorts (P < 0.05).
Tumor recurrence, liver failure and treatment response rates are major concerns during HCC treatment. Radiomics has emerged as a promising tool to predict recurrence and treatment response beyond the current predictive criteria[61,62]. Table 3 summarizes the studies to date that have evaluated the use of radiomic models to predict recurrence and treatment response. Most of these studies were single-center studies performed in China, with only a few studies incorporating external validation[63,64]. Segmentation strategies were predominantly manual strategies, including manual segmentation of the tumor region or area of interest, with only a few studies involving the segmentation of the peritumoral liver parenchyma[63,65-67]. Overall, the radiomic models yielded an AUC between 0.59 and 0.94 (see Table 3).
| Ref. | Country | n | Imaging modality | Endpoint | Treatment type | Segmentation | ROI/VOI | No. of readers | Main results | Validation |
| Hui et al[100], 2018 | Singapore | 50 | MRI | Recurrence | Hepatic resection | Manual, intratumoral | ROI | 3 | AUC: 0.78-0.84 | None |
| Kim et al[65], 2019 | South Korea | Training: 128. Validation: 39 | MRI | Recurrence | Hepatic resection | Semiautomatic, intra- and peritumoral | VOI | 2 | C-index: 0.716 | Internal |
| Zhao et al[101], 2021 | China | Training: 78. Validation: 35 | MRI | Recurrence | Hepatic resection | Manual, intratumoral | VOI | 2 | AUC: 0.83 (training), 0.77 (validation) | Internal |
| Zhou et al[68], 2017 | China | 215 | CT | Recurrence | Hepatic resection | Manual, intratumoral | ROI | 2 | AUC: 0.84 (combined model) | None |
| Ji et al[64], 2020 | China | Internal: 177. External: 118 | CT | Recurrence | Hepatic resection | Manual, intratumoral | VOI | 1 | AUC: 0.77 (internal), 0.78 (external) | External |
| Guo et al[69], 2019 | China | Training: 93. Validation: 40 | CT | Recurrence | Liver transplant | Semiautomatic, intratumoral | ROI | 1 | AUC: 0.79 (training), 0.79 (validation) | Internal |
| Shan et al[66], 2019 | China | Training: 109. Validation: 47 | CT | Recurrence | Hepatic resection or ablation | Manual, intra- and peritumoral | ROI | 2 | AUC: 0.80 (training), 0.79 (validation) | Internal |
| Zheng et al[79], 2018 | China | Training: 212. Validation: 107 | CT | Recurrence and survival | Hepatic resection | Manual, intratumoral | ROI | 2 | AUC: 0.64 (training), 0.59 (validation) | Internal |
| Song et al[67], 2020 | China | Training: 110. Validation: 74 | MRI | Recurrence | TACE | Semiautomatic, intra- and peritumoral | VOI | 2 | C-index: 0.82 | Internal |
| Lv et al[71], 2021 | China | Training: 40. Validation: 18 | MRI | Recurrence | RFA | Semiautomatic, intratumoral | VOI | 2 | AUC: 0.94 (training), 0.82 (validation) | Internal |
| Sun et al[70], 2020 | China | Training: 67. Validation: 17 | MRI | Recurrence | TACE | Manual (intratumoral) | VOI | 2 | AUC: 0.71-0.79 | Internal |
| Cai et al[75], 2019 | China | Training: 80. Validation: 32 | CT | Liver failure | Hepatic resection | Semiautomatic, intratumoral | ROI | 2 | AUC: 0.82 (training), 0.76 (validation) | Internal |
| Zhu et al[76], 2020 | China | 101 | MRI | Liver failure | Hepatic resection | Manual, entire liver | ROI | 2 | AUC: 0.81-0.89 | None |
| Ivanics et al[73], 2021 | Canada | 88 | CT | Treatment response | TACE | Manual, intratumoral | VOI | 1 | AUC: 0.70-0.87 | None |
| Kong et al[72], 2021 | China | Training: 69. Validation: 30 | MRI | Treatment response | TACE | Manual, intratumoral | VOI | 2 | AUC: 0.81 (training), 0.87 (validation) | Internal |
| Chen et al[63], 2021 | China | Training: 355. Internal: 118. External: 122 | CT | Treatment response | TACE | Semiautomatic, intra- and peritumoral | ROI | 2 | AUC: 0.94 (internal), 0.90 (external) | Internal and external |
| Horvat et al[74], 2021 | Brazil | 34 | MRI | Treatment response | RFA | Manual, intratumoral | VOI | 1 | AUC: 0.76 | None |
Of the studies evaluating the use of radiomics to predict recurrence, most involved the prediction of recurrence after surgical resection on CT or MRI, demonstrating a validation AUC between 0.59 and 0.84 (Table 3). Zhou et al[68] demonstrated that combining the radiomic signature with conventional preoperative variables significantly improved clinical model accuracy in early recurrence prediction (AUC of 0.84). Ji et al[64] developed and externally validated a radiomic model with better prognostic ability (C index ≥ 0.77, AUC of 0.78), lower prediction error (Brier score ≤ 0.14), and better clinical use compared with other staging systems and models. A few other studies evaluating the use of radiomics to predict recurrence involved the prediction of recurrence after liver transplant[69], transarterial chemoembolization (TACE)[67,70], and radiofrequency ablation (RFA)[71], demonstrating a validation AUC between 0.71 and 0.82.
Of the studies evaluating the use of radiomics to predict treatment response, a few involved the prediction of treatment response post-TACE[63,72,73]. In Canada, Ivanics et al[73] developed a CT-based radiomic model and achieved an AUC of 0.87 on the internal validation set. A large multi-center Chinese study by Chen et al[63] evaluating treatment response after TACE performed semi-automatic segmentation of the tumor and of the peritumoral region on contrast-enhanced CT in 585 patients, and the validation AUC was 0.90. One small study by Horvat et al[74] assessed treatment response after RFA using tumor 3D volumes of interest on MRI, yielding an AUC of 0.76 for the radiomics model, although the model lacked validation. Finally, two studies from China evaluated the use of radiomics to predict liver failure after surgical resection[75,76].
Table 4 summarizes the studies to date that have evaluated the use of radiomics to predict survival in patients with HCC. Four studies evaluated the use of CT-based radiomics to predict survival after hepatic resection, demonstrating an AUC between 0.71 and 0.81, with two of the four studies performing internal validation[39,77-79]. A few other studies evaluated the use of radiomics to predict survival after TACE[80], TARE[81], and RFA[82], all without validation.
| Ref. | Country | n | Imaging modality | Endpoint | Treatment type | Segmentation | ROI/VOI | No. of readers | Main results | Validation |
| Kiryu et al[77], 2017 | Japan | 122 | CT | Survival | Hepatic resection | Manual, intra- and peritumoral | ROI | 1 | OS and DFS were significantly different between 2 rad score groups | None |
| Xu et al[39], 2019 | China | Training: 350. Validation: 145 | CT | Survival | Hepatic resection | Semiautomatic, intratumoral | VOI | 3 | AUC: 0.91 (training), 0.81 (validation) | Internal |
| Akai et al[78], 2018 | Japan | 127 | CT | Survival | Hepatic resection | Manual, intratumoral | ROI | 1 | OS and DFS were significantly different between 2 rad score groups | None |
| Kim et al[80], 2018 | South Korea | 88 | CT | Survival | TACE | Manual, intratumoral | ROI | 1 | Combined clinical and radiomics score was a better predictor of survival | None |
| Blanc-Durand et al[81], 2018 | Switzerland | 47 | 18F-FDG PET-CT | Survival | TARE | Semiautomatic, whole liver | VOI | N/A | PFS-Rad Score and OS-Rad Score were independent negative predictors | None |
| Petukhova-Greenstein et al[82], 2022 | United States | 65 | MRI | Survival | RFA | Semiautomatic, intra- and peritumoral | VOI | 2 | OS was significantly different between 2 rad score groups | None |
| Zheng et al[79], 2018 | China | Training: 212. Validation: 107 | CT | Survival | Hepatic resection | Manual, intratumoral | ROI | 2 | AUC: 0.71 (training and validation) | Internal |
Of the studies that involved the prediction of survival after hepatic resection, Xu et al[39] had the largest sample size. In their study, a risk model integrating clinico-radiological factors and a high CT-based radiomic score was independently associated with long-term mortality and disease-specific recurrence. Kim et al[80] evaluated the use of CT-based radiomics in survival prediction in patients after TACE. They demonstrated a combined model integrating radiomic features and clinical data (HCC size, Child-Pugh score and AFP) outperformed the clinical sore model or the radiomic score model. Petukhova-Greenstein et al[82] found that a higher MRI-based radiomic signature based on nodular and perinodular radiomic features predicted poorer survival after RFA. A study evaluated the survival prediction after TARE, using 18-fuoro-deoxyglucose PET-based radiomics[81]. They observed that whole-liver radiomics textural features were an independent negative predictor of survival. Furthermore, radiomic scoring system did not differ after stratification by tumor size and Barcelona Clinic Liver Cancer staging.
Immunotherapy represents a paradigm shift in the management of patients with advanced HCC. Preoperatively assessing the immune status can assist the multidisciplinary team to identify which patients are suitable for immunotherapy, potentially improving treatment efficiency and overall survival rate. A few studies have evaluated the use of radiomics to predict programmed cell death ligand 1 (PD-L1) expression[83], CD8+ T cell infiltration[84], immunoscore[85,86], and anti-PD-1 treatment efficacy[87] in patients with HCC, with none of them performing external validation. Tian et al[83] were the first group to explore the efficacy of MRI-based radiomics to predict PD-L1 status. They proposed a model integrating radiomic and DL features for the quick and accurate assessment of PD-L1 expression levels in HCC patients before immune checkpoint inhibitor therapy which yielded an AUC of 0.897. Chen et al[85] demonstrated in 207 patients that radiomic features including those from the peritumoural region were associated with a validated “immunoscore”. This score characterizes the tumor infiltrating lymphocyte population and theoretically reflects the immune phenotype of the tumor microenvironment.
Reproducibility refers to variations of the same patient across different imaging scenarios (e.g., scanner or imaging parameters), while repeatability refers to variations of the same patient using the same imaging protocol. Table 5 summarizes the 13 studies to date that have studied the reproducibility of radiomics in HCC patients. Most of these studies were conducted in China (8/13; 62%). Seven were performed using CT (54%), 5 using MRI (38%), and 1 using both CT scan and MRI (8%). Different software programs were used for segmentation and feature extraction. Most studies adopted manual segmentation (11/13; 85%), and most evaluated first- and second-order features, with a few including shape and higher-order features. In addition to intra and inter-reader reproducibility, some also assessed the repeatability of radiomic features obtained through two separate exams from the same scanner, different scanners from different vendors and centers, 3D vs 2D segmentation, different contrast imaging phases, injection rates and pixel resolutions on contrast-enhanced CT, and different b-values on diffusion-weighted imaging on MRI.
| Ref. | Country | n | Imaging modality | Segmentation | Segmentation software | ROI/VOI | No. of readers | Intra-reader reproducibility | Inter-reader reproducibility | Other reproducibility |
| Duan et al[88], 2022 | China | 19 | CT, MRI | Manual, intra- and peritumoral | 3D-Slicer | ROI | 2 (1 radiologist and 1 radiation oncologist) | Features with ICC ≥ 0.75 in both tumoral and peritumoral tissue greatest in MR | Features with ICC ≥ 0.75 in both tumoral and peritumoral tissue greatest in MR | N/A |
| Zhang et al[102], 2022 | China | 90 (31 HCC) | MRI | Manual, intratumoral | ITK-SNAP | ROI and VOI | 2 radiologists | N/A | ICC > 0.8 used | N/A |
| Carbonell et al[89], 2022 | United States | 55 (16 HCC) | MRI | Manual, intratumoral and liver parenchyma | Olea sphere 3.0, Olea Medical | ROI for normal liver, VOI for HCC | 2 radiologists | N/A | CCC: 0.80-0.99 | For test-retest (same MRI system, 2 different MRI exams): ICC: 0.53-0.99; and in liver parenchyma: ICC: 0.53-0.73. For inter-platform reproducibility (MRI systems from 2 different vendors): CCC: 0.58-0.99 |
| Park et al[103], 2022 | South Korea | 249 | CT | Manual followed by automatic segmentation, intratumoral | MEDIP PRO | ROI and VOI | 1 radiologist | For VOI: Manual: ICC 0.594-0.998 for FO, 0.764-0.997 for shape, and 0.190-0.926 for SO; DL-AS: ICC > 0.75 for all. For ROI: Manual: 0.698-0.997 for FO, 0.556-0.997 for shape, and 0.341-0.935 for SO; DL-AS ICC > 0.75 for all | N/A | |
| Haniff et al[104], 2021 | Malaysia | 30 | MRI | Manual and semi-automatic, intratumoral | 3D-Slicer | VOI | Manual: 4 readers. Semi-automatic: 2 readers | N/A | Manual segmentation: ICC 0.897. Semi-automatic segmentation: ICC 0.952 | NA |
| Ibrahim et al[90], 2021 | Germany | 61 patients, 104 lesions | CT | Manual, intratumoral | MIM software | ROI | 1 nonradiologist revised by radiologist | N/A | N/A | Across different contrast imaging phases: 25% of extracted features had CCC > 0.9 across arterial and portal venous phases |
| Hu et al[105], 2021 | China | 30 | CT | Manual, intratumoral | MaZda software | ROI | 2 radiologists | ICC > 0.7 | ICC > 0.7 | N/A |
| Mao et al[32], 2020 | China | 30 | CT | Manual, intratumoral | ITK-SNAP | ROI | 2 radiologists | N/A | ICC ≥ 0.8 | N/A |
| Hu et al[106], 2020 | China | 50 | CT | Semi-automatic, peritumoral | Not mentioned | ROI | 2 radiologists | N/A | ICC > 0.6 | N/A |
| Qiu et al[107], 2019 | China | 26 | CT | Manual and semi-automatic, intratumoral | GrowCut and GraphCut | ROI | Manual: 5 radiation oncologists. Semi-automatic: 2 radiation oncologists | N/A | ICC ≥ 0.75 in 69% of features extracted from manual segmentation, 73% from GraphCut, and 79% from GrowCut | Across different centers: Poor reproducibility of CT-based peritumoral-radiomics model |
| Zhang et al[108], 2019 | China | 46 (34 HCC) | MRI | Manual, intratumoral | MIM software | VOI | 1 radiologist | N/A | N/A | Across different b-values: radiomic features extracted from b = 0, 20, 50, 100, 200 s/mm2 and b = 1000 s/mm2 and nearby b-values DWIs showed a high reproducibility (ICC ≥ 0.8) |
| Feng et al[40], 2019 | China | 160 (110) | MRI | Manual, intra- and peritumoral | ITK-SNAP | VOI | 3 radiologists | 85% ICC ≥ 0.8 | 82% ICC ≥ 0.8 | N/A |
| Perrin et al[91], 2018 | United States | 38 (6 HCC) | CT | Semi-automatic, intratumoral and liver parenchyma | Scout Liver | VOI | 1 research fellow under supervision of radiologist | N/A | N/A | Across different contrast injection rates, pixel resolutions, and scanner models: Number of reproducible radiomic features (CCC > 0.9) decreased with variations in contrast injection rate, pixel resolution, and scanner model |
Of note, one study showed that intra-reader tumoral and peritumoral reproducibility were greatest in MRI[88]. Another study showed that for test-retest (same MRI system, 2 different MRI exams), the intraclass correlation coefficient varied from 0.53-0.99 and the inter-platform reproducibility (MRI systems from 2 different vendors) varied from 0.58-0.99[89]. Regarding different contrast phases, Ibrahim et al[90] showed that 25% of extracted features had a concordance correlation coefficient (CCC) > 0.9 across arterial and portal venous phases. Perrin et al[91] demonstrated that the number of reproducible features decreased with variations in contrast injection rate, pixel resolution, and scanner model.
Despite the increasing and encouraging results in the literature concerning radiomics in patients with HCC, there are challenges and limitations to be overcome before its clinical implementation, particularly related to reproducibility and repeatability, lesion segmentation, model overfitting, multidisciplinary acceptance, and multi-modal data integration[23].
Patient selection, imaging data, segmentation strategy, image processing, feature selection, and computational processing are some factors that may affect the reproducibility and repeatability. Transparent patient accrual, data normalization, standard image manipulation, and feature extraction data are some strategies that may improve these challenges. Additionally, multi-center studies are recommended to increase reproducibility of the results.
Overfitting occurs when the model performs better in the training set with limited generalization of the results. The main factors contributing to overfitting are the number of included features being higher than the number of events and overoptimistic feature selection. Multiple strategies can be implemented to decrease overfitting, such as increasing the number of patients and events, using regularization methods, and including external validation cohorts. Multidisciplinary acceptance may improve with clear methods and a close relationship between radiologists, surgeons, oncologists, statistician, and data scientists to improve the interpretability of the results and to make way for clinical translation.
Multi-omics data integration is an additional step to improve the clinical acceptance of radiomics. Radiomics requires a multistep workflow process using different software and expertise; technological investments to create integrated and user-friendly tools are necessary to facilitate its widespread use in clinical practice. Finally, segmentation is a time-consuming process, susceptible to intra and inter-observer variability. Automatic and semi-automatic segmentations are required, particularly using DL strategies to facilitate this crucial step.
Additionally, some heterogeneity related to patients with HCC should be take into consideration. Since pathological confirmation is not always performed, the definition of clear and reproducible endpoints, like the LI-RADS criteria, are relevant strategies. Combined data integrating imaging and clinical variables are important to address the issue that patients with HCC are also dealing with systemic consequences related to cirrhosis.
Radiomics is an evolving computer-assisted tool with the potential to improve the multidisciplinary management of patients with HCC and to provide personalized treatment optimizing the available resources. Multiple studies have evaluated the use of radiomics in HCC with promising applications, including the prediction of pre-surgical histology, genetic signature, recurrence, and treatment response, as well as survival rates. Although promising, several challenges need to be overcome before radiomics can achieve clinical translation, including workflow optimization, model validation in multi-center studies, and the development of integrated models to facilitate clinical use and acceptance.
The authors would like to express their deepest gratitude to Joanne Chin, MFA, ELS, for her editorial support on this manuscript.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Brazil
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: D’Alterio C, Italy; Wang Y, China S-Editor: Wang JJ L-Editor: A P-Editor: Wang JJ
| 1. | Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53206] [Cited by in RCA: 55842] [Article Influence: 7977.4] [Reference Citation Analysis (132)] |
| 2. | World Health Organization. Cancer fact sheets. [cited 10 August 2022]. Available from: https://gco.iarc.fr/today/fact-sheets-cancers. |
| 3. | Llovet JM, Kelley RK, Villanueva A, Singal AG, Pikarsky E, Roayaie S, Lencioni R, Koike K, Zucman-Rossi J, Finn RS. Hepatocellular carcinoma. Nat Rev Dis Primers. 2021;7:6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4432] [Cited by in RCA: 3887] [Article Influence: 971.8] [Reference Citation Analysis (3)] |
| 4. | Lopes Fde L, Coelho FF, Kruger JA, Fonseca GM, Araujo RL, Jeismann VB, Herman P. Influence of hepatocellular carcinoma etiology in the survival after resection. Arq Bras Cir Dig. 2016;29:105-108. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 13] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 5. | Llovet JM, Bustamante J, Castells A, Vilana R, Ayuso Mdel C, Sala M, Brú C, Rodés J, Bruix J. Natural history of untreated nonsurgical hepatocellular carcinoma: rationale for the design and evaluation of therapeutic trials. Hepatology. 1999;29:62-67. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 839] [Cited by in RCA: 907] [Article Influence: 34.9] [Reference Citation Analysis (1)] |
| 6. | Mittal S, El-Serag HB. Epidemiology of hepatocellular carcinoma: consider the population. J Clin Gastroenterol. 2013;47 Suppl:S2-S6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 770] [Cited by in RCA: 880] [Article Influence: 73.3] [Reference Citation Analysis (0)] |
| 7. | Venook AP, Papandreou C, Furuse J, de Guevara LL. The incidence and epidemiology of hepatocellular carcinoma: a global and regional perspective. Oncologist. 2010;15 Suppl 4:5-13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 626] [Cited by in RCA: 737] [Article Influence: 49.1] [Reference Citation Analysis (0)] |
| 8. | Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma. Lancet. 2012;379:1245-1255. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3249] [Cited by in RCA: 3597] [Article Influence: 276.7] [Reference Citation Analysis (4)] |
| 9. | Herman P, Fonseca GM, Coelho FF, Kruger JAP, Makdissi FF, Jeismann VB, Carrilho FJ, D'Albuquerque LAC, Nahas SC. Two decades of liver resection with a multidisciplinary approach in a single institution: What has changed? Clinics (Sao Paulo). 2022;77:100088. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 10. | Jiménez Pérez M, Grande RG. Application of artificial intelligence in the diagnosis and treatment of hepatocellular carcinoma: A review. World J Gastroenterol. 2020;26:5617-5628. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 52] [Cited by in RCA: 37] [Article Influence: 7.4] [Reference Citation Analysis (2)] |
| 11. | Liu X, Elbanan MG, Luna A, Haider MA, Smith AD, Sabottke CF, Spieler BM, Turkbey B, Fuentes D, Moawad A, Kamel S, Horvat N, Elsayes KM. Radiomics in Abdominopelvic Solid-Organ Oncologic Imaging: Current Status. AJR Am J Roentgenol. 2022;219:985-995. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 10] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 12. | Gillies RJ, Kinahan PE, Hricak H. Radiomics: Images Are More than Pictures, They Are Data. Radiology. 2016;278:563-577. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4541] [Cited by in RCA: 5556] [Article Influence: 617.3] [Reference Citation Analysis (3)] |
| 13. | Miranda Magalhaes Santos JM, Clemente Oliveira B, Araujo-Filho JAB, Assuncao-Jr AN, de M Machado FA, Carlos Tavares Rocha C, Horvat JV, Menezes MR, Horvat N. State-of-the-art in radiomics of hepatocellular carcinoma: a review of basic principles, applications, and limitations. Abdom Radiol (NY). 2020;45:342-353. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 48] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 14. | Bera K, Braman N, Gupta A, Velcheti V, Madabhushi A. Predicting cancer outcomes with radiomics and artificial intelligence in radiology. Nat Rev Clin Oncol. 2022;19:132-146. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 501] [Cited by in RCA: 421] [Article Influence: 140.3] [Reference Citation Analysis (1)] |
| 15. | Rizzo S, Botta F, Raimondi S, Origgi D, Fanciullo C, Morganti AG, Bellomi M. Radiomics: the facts and the challenges of image analysis. Eur Radiol Exp. 2018;2:36. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 413] [Cited by in RCA: 684] [Article Influence: 97.7] [Reference Citation Analysis (0)] |
| 16. | Wagner MW, Namdar K, Biswas A, Monah S, Khalvati F, Ertl-Wagner BB. Radiomics, machine learning, and artificial intelligence-what the neuroradiologist needs to know. Neuroradiology. 2021;63:1957-1967. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 89] [Article Influence: 22.3] [Reference Citation Analysis (0)] |
| 17. | Shur JD, Doran SJ, Kumar S, Ap Dafydd D, Downey K, O'Connor JPB, Papanikolaou N, Messiou C, Koh DM, Orton MR. Radiomics in Oncology: A Practical Guide. Radiographics. 2021;41:1717-1732. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 213] [Article Influence: 53.3] [Reference Citation Analysis (0)] |
| 18. | Giger ML. Machine Learning in Medical Imaging. J Am Coll Radiol. 2018;15:512-520. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 260] [Cited by in RCA: 282] [Article Influence: 40.3] [Reference Citation Analysis (0)] |
| 19. | Suzuki K. Overview of deep learning in medical imaging. Radiol Phys Technol. 2017;10:257-273. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 380] [Cited by in RCA: 421] [Article Influence: 52.6] [Reference Citation Analysis (0)] |
| 20. | Larue RT, Defraene G, De Ruysscher D, Lambin P, van Elmpt W. Quantitative radiomics studies for tissue characterization: a review of technology and methodological procedures. Br J Radiol. 2017;90:20160665. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 209] [Cited by in RCA: 249] [Article Influence: 27.7] [Reference Citation Analysis (0)] |
| 21. | Thawani R, McLane M, Beig N, Ghose S, Prasanna P, Velcheti V, Madabhushi A. Radiomics and radiogenomics in lung cancer: A review for the clinician. Lung Cancer. 2018;115:34-41. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 235] [Cited by in RCA: 319] [Article Influence: 45.6] [Reference Citation Analysis (0)] |
| 22. | van Timmeren JE, Cester D, Tanadini-Lang S, Alkadhi H, Baessler B. Radiomics in medical imaging-"how-to" guide and critical reflection. Insights Imaging. 2020;11:91. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 588] [Cited by in RCA: 771] [Article Influence: 154.2] [Reference Citation Analysis (0)] |
| 23. | Horvat N, Miranda J, El Homsi M, Peoples JJ, Long NM, Simpson AL, Do RKG. A primer on texture analysis in abdominal radiology. Abdom Radiol (NY). 2022;47:2972-2985. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 12] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 24. | Liu X, Khalvati F, Namdar K, Fischer S, Lewis S, Taouli B, Haider MA, Jhaveri KS. Can machine learning radiomics provide pre-operative differentiation of combined hepatocellular cholangiocarcinoma from hepatocellular carcinoma and cholangiocarcinoma to inform optimal treatment planning? Eur Radiol. 2021;31:244-255. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 71] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 25. | Lewis S, Peti S, Hectors SJ, King M, Rosen A, Kamath A, Putra J, Thung S, Taouli B. Volumetric quantitative histogram analysis using diffusion-weighted magnetic resonance imaging to differentiate HCC from other primary liver cancers. Abdom Radiol (NY). 2019;44:912-922. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 42] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 26. | Nie P, Wang N, Pang J, Yang G, Duan S, Chen J, Xu W. CT-Based Radiomics Nomogram: A Potential Tool for Differentiating Hepatocellular Adenoma From Hepatocellular Carcinoma in the Noncirrhotic Liver. Acad Radiol. 2021;28:799-807. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 30] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 27. | Nie P, Yang G, Guo J, Chen J, Li X, Ji Q, Wu J, Cui J, Xu W. A CT-based radiomics nomogram for differentiation of focal nodular hyperplasia from hepatocellular carcinoma in the non-cirrhotic liver. Cancer Imaging. 2020;20:20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 50] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 28. | Wu J, Liu A, Cui J, Chen A, Song Q, Xie L. Radiomics-based classification of hepatocellular carcinoma and hepatic haemangioma on precontrast magnetic resonance images. BMC Med Imaging. 2019;19:23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 70] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 29. | Mokrane FZ, Lu L, Vavasseur A, Otal P, Peron JM, Luk L, Yang H, Ammari S, Saenger Y, Rousseau H, Zhao B, Schwartz LH, Dercle L. Radiomics machine-learning signature for diagnosis of hepatocellular carcinoma in cirrhotic patients with indeterminate liver nodules. Eur Radiol. 2020;30:558-570. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 125] [Article Influence: 25.0] [Reference Citation Analysis (0)] |
| 30. | Wu M, Tan H, Gao F, Hai J, Ning P, Chen J, Zhu S, Wang M, Dou S, Shi D. Predicting the grade of hepatocellular carcinoma based on non-contrast-enhanced MRI radiomics signature. Eur Radiol. 2019;29:2802-2811. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 134] [Article Influence: 19.1] [Reference Citation Analysis (0)] |
| 31. | Mao Y, Wang J, Zhu Y, Chen J, Mao L, Kong W, Qiu Y, Wu X, Guan Y, He J. Gd-EOB-DTPA-enhanced MRI radiomic features for predicting histological grade of hepatocellular carcinoma. Hepatobiliary Surg Nutr. 2022;11:13-24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 30] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 32. | Mao B, Zhang L, Ning P, Ding F, Wu F, Lu G, Geng Y, Ma J. Preoperative prediction for pathological grade of hepatocellular carcinoma via machine learning-based radiomics. Eur Radiol. 2020;30:6924-6932. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 81] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 33. | Chen W, Zhang T, Xu L, Zhao L, Liu H, Gu LR, Wang DZ, Zhang M. Radiomics Analysis of Contrast-Enhanced CT for Hepatocellular Carcinoma Grading. Front Oncol. 2021;11:660509. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 16] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 34. | Brancato V, Garbino N, Salvatore M, Cavaliere C. MRI-Based Radiomic Features Help Identify Lesions and Predict Histopathological Grade of Hepatocellular Carcinoma. Diagnostics (Basel). 2022;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 14] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 35. | Du M, Chen L, Zhao J, Tian F, Zeng H, Tan Y, Sun H, Zhou J, Ji Y. Microvascular invasion (MVI) is a poorer prognostic predictor for small hepatocellular carcinoma. BMC Cancer. 2014;14:38. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 69] [Cited by in RCA: 104] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 36. | Ueda K, Tokugawa K. Rubella vaccination and congenital rubella syndrome in Japan. Acta Paediatr Jpn. 1988;30:163-166. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 37. | Lauwers GY, Terris B, Balis UJ, Batts KP, Regimbeau JM, Chang Y, Graeme-Cook F, Yamabe H, Ikai I, Cleary KR, Fujita S, Flejou JF, Zukerberg LR, Nagorney DM, Belghiti J, Yamaoka Y, Vauthey JN; International Cooperative Study Group on Hepatocellular Carcinoma. Prognostic histologic indicators of curatively resected hepatocellular carcinomas: a multi-institutional analysis of 425 patients with definition of a histologic prognostic index. Am J Surg Pathol. 2002;26:25-34. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 152] [Cited by in RCA: 149] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 38. | Zhong X, Long H, Su L, Zheng R, Wang W, Duan Y, Hu H, Lin M, Xie X. Radiomics models for preoperative prediction of microvascular invasion in hepatocellular carcinoma: a systematic review and meta-analysis. Abdom Radiol (NY). 2022;47:2071-2088. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 23] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 39. | Xu X, Zhang HL, Liu QP, Sun SW, Zhang J, Zhu FP, Yang G, Yan X, Zhang YD, Liu XS. Radiomic analysis of contrast-enhanced CT predicts microvascular invasion and outcome in hepatocellular carcinoma. J Hepatol. 2019;70:1133-1144. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 278] [Cited by in RCA: 495] [Article Influence: 82.5] [Reference Citation Analysis (0)] |
| 40. | Feng ST, Jia Y, Liao B, Huang B, Zhou Q, Li X, Wei K, Chen L, Li B, Wang W, Chen S, He X, Wang H, Peng S, Chen ZB, Tang M, Chen Z, Hou Y, Peng Z, Kuang M. Preoperative prediction of microvascular invasion in hepatocellular cancer: a radiomics model using Gd-EOB-DTPA-enhanced MRI. Eur Radiol. 2019;29:4648-4659. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149] [Cited by in RCA: 147] [Article Influence: 24.5] [Reference Citation Analysis (0)] |
| 41. | Zheng J, Chakraborty J, Chapman WC, Gerst S, Gonen M, Pak LM, Jarnagin WR, DeMatteo RP, Do RKG, Simpson AL; Hepatopancreatobiliary Service in the Department of Surgery of the Memorial Sloan Kettering Cancer Center; Research Staff in the Department of Surgery at Washington University School of Medicine. Preoperative Prediction of Microvascular Invasion in Hepatocellular Carcinoma Using Quantitative Image Analysis. J Am Coll Surg. 2017;225:778-788.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 66] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 42. | Segal E, Sirlin CB, Ooi C, Adler AS, Gollub J, Chen X, Chan BK, Matcuk GR, Barry CT, Chang HY, Kuo MD. Decoding global gene expression programs in liver cancer by noninvasive imaging. Nat Biotechnol. 2007;25:675-680. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 400] [Cited by in RCA: 443] [Article Influence: 24.6] [Reference Citation Analysis (0)] |
| 43. | Luo Y, Ren F, Liu Y, Shi Z, Tan Z, Xiong H, Dang Y, Chen G. Clinicopathological and prognostic significance of high Ki-67 labeling index in hepatocellular carcinoma patients: a meta-analysis. Int J Clin Exp Med. 2015;8:10235-10247. [PubMed] |
| 44. | Wu C, Chen J, Fan Y, Zhao M, He X, Wei Y, Ge W, Liu Y. Nomogram Based on CT Radiomics Features Combined With Clinical Factors to Predict Ki-67 Expression in Hepatocellular Carcinoma. Front Oncol. 2022;12:943942. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 18] [Reference Citation Analysis (0)] |
| 45. | Li Y, Yan C, Weng S, Shi Z, Sun H, Chen J, Xu X, Ye R, Hong J. Texture analysis of multi-phase MRI images to detect expression of Ki67 in hepatocellular carcinoma. Clin Radiol. 2019;74:813.e19-813.e27. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 31] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 46. | Fan Y, Yu Y, Wang X, Hu M, Hu C. Radiomic analysis of Gd-EOB-DTPA-enhanced MRI predicts Ki-67 expression in hepatocellular carcinoma. BMC Med Imaging. 2021;21:100. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 33] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 47. | Ye Z, Jiang H, Chen J, Liu X, Wei Y, Xia C, Duan T, Cao L, Zhang Z, Song B. Texture analysis on gadoxetic acid enhanced-MRI for predicting Ki-67 status in hepatocellular carcinoma: A prospective study. Chin J Cancer Res. 2019;31:806-817. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 35] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 48. | Hu X, Zhou J, Li Y, Wang Y, Guo J, Sack I, Chen W, Yan F, Li R, Wang C. Added Value of Viscoelasticity for MRI-Based Prediction of Ki-67 Expression of Hepatocellular Carcinoma Using a Deep Learning Combined Radiomics (DLCR) Model. Cancers (Basel). 2022;14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 33] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 49. | Zhuo JY, Lu D, Tan WY, Zheng SS, Shen YQ, Xu X. CK19-positive Hepatocellular Carcinoma is a Characteristic Subtype. J Cancer. 2020;11:5069-5077. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 71] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 50. | Wang HQ, Yang C, Zeng MS, Rao SX, Ji Y, Weng X, Wang JY, Sheng RF. Magnetic resonance texture analysis for the identification of cytokeratin 19-positive hepatocellular carcinoma. Eur J Radiol. 2019;117:164-170. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 24] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 51. | Chen Y, Chen J, Zhang Y, Lin Z, Wang M, Huang L, Huang M, Tang M, Zhou X, Peng Z, Huang B, Feng ST. Preoperative Prediction of Cytokeratin 19 Expression for Hepatocellular Carcinoma with Deep Learning Radiomics Based on Gadoxetic Acid-Enhanced Magnetic Resonance Imaging. J Hepatocell Carcinoma. 2021;8:795-808. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 22] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 52. | Yang F, Wan Y, Xu L, Wu Y, Shen X, Wang J, Lu D, Shao C, Zheng S, Niu T, Xu X. MRI-Radiomics Prediction for Cytokeratin 19-Positive Hepatocellular Carcinoma: A Multicenter Study. Front Oncol. 2021;11:672126. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 24] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 53. | He X, Liu F, Yan J, Zhang Y, Shang H, Dou Q, Zhao Q, Song Y. Trans-splicing repair of mutant p53 suppresses the growth of hepatocellular carcinoma cells in vitro and in vivo. Sci Rep. 2015;5:8705. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 23] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 54. | Mantovani F, Walerych D, Sal GD. Targeting mutant p53 in cancer: a long road to precision therapy. FEBS J. 2017;284:837-850. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 50] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 55. | Wu H, Chen X, Chen J, Luo Y, Jiang X, Wei X, Tang W, Liu Y, Liang Y, Liu W, Guo Y. Correlations between P53 Mutation Status and Texture Features of CT Images for Hepatocellular Carcinoma. Methods Inf Med. 2019;58:42-49. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 1] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 56. | Liao H, Jiang H, Chen Y, Duan T, Yang T, Han M, Xue Z, Shi F, Yuan K, Bashir MR, Shen D, Song B, Zeng Y. Predicting Genomic Alterations of Phosphatidylinositol-3 Kinase Signaling in Hepatocellular Carcinoma: A Radiogenomics Study Based on Next-Generation Sequencing and Contrast-Enhanced CT. Ann Surg Oncol. 2022;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 57. | Ezzoukhry Z, Louandre C, Trécherel E, Godin C, Chauffert B, Dupont S, Diouf M, Barbare JC, Mazière JC, Galmiche A. EGFR activation is a potential determinant of primary resistance of hepatocellular carcinoma cells to sorafenib. Int J Cancer. 2012;131:2961-2969. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 140] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 58. | Ma Y, Xu R, Liu X, Zhang Y, Song L, Cai S, Zhou S, Xie Y, Li A, Cao W, Tang X. LY3214996 relieves acquired resistance to sorafenib in hepatocellular carcinoma cells. Int J Med Sci. 2021;18:1456-1464. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 28] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 59. | Negri FV, Dal Bello B, Porta C, Campanini N, Rossi S, Tinelli C, Poggi G, Missale G, Fanello S, Salvagni S, Ardizzoni A, Maria SE. Expression of pERK and VEGFR-2 in advanced hepatocellular carcinoma and resistance to sorafenib treatment. Liver Int. 2015;35:2001-2008. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 49] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 60. | Che F, Xu Q, Li Q, Huang ZX, Yang CW, Wang LY, Wei Y, Shi YJ, Song B. Radiomics signature: A potential biomarker for β-arrestin1 phosphorylation prediction in hepatocellular carcinoma. World J Gastroenterol. 2022;28:1479-1493. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 8] [Cited by in RCA: 6] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 61. | Villanueva A. Hepatocellular Carcinoma. N Engl J Med. 2019;380:1450-1462. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2066] [Cited by in RCA: 3176] [Article Influence: 529.3] [Reference Citation Analysis (37)] |
| 62. | Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet. 2018;391:1301-1314. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2800] [Cited by in RCA: 4110] [Article Influence: 587.1] [Reference Citation Analysis (6)] |
| 63. | Chen M, Cao J, Hu J, Topatana W, Li S, Juengpanich S, Lin J, Tong C, Shen J, Zhang B, Wu J, Pocha C, Kudo M, Amedei A, Trevisani F, Sung PS, Zaydfudim VM, Kanda T, Cai X. Clinical-Radiomic Analysis for Pretreatment Prediction of Objective Response to First Transarterial Chemoembolization in Hepatocellular Carcinoma. Liver Cancer. 2021;10:38-51. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 82] [Article Influence: 20.5] [Reference Citation Analysis (0)] |
| 64. | Ji GW, Zhu FP, Xu Q, Wang K, Wu MY, Tang WW, Li XC, Wang XH. Radiomic Features at Contrast-enhanced CT Predict Recurrence in Early Stage Hepatocellular Carcinoma: A Multi-Institutional Study. Radiology. 2020;294:568-579. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 193] [Cited by in RCA: 191] [Article Influence: 38.2] [Reference Citation Analysis (0)] |
| 65. | Kim S, Shin J, Kim DY, Choi GH, Kim MJ, Choi JY. Radiomics on Gadoxetic Acid-Enhanced Magnetic Resonance Imaging for Prediction of Postoperative Early and Late Recurrence of Single Hepatocellular Carcinoma. Clin Cancer Res. 2019;25:3847-3855. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 151] [Article Influence: 25.2] [Reference Citation Analysis (0)] |
| 66. | Shan QY, Hu HT, Feng ST, Peng ZP, Chen SL, Zhou Q, Li X, Xie XY, Lu MD, Wang W, Kuang M. CT-based peritumoral radiomics signatures to predict early recurrence in hepatocellular carcinoma after curative tumor resection or ablation. Cancer Imaging. 2019;19:11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 133] [Article Influence: 22.2] [Reference Citation Analysis (1)] |
| 67. | Song W, Yu X, Guo D, Liu H, Tang Z, Liu X, Zhou J, Zhang H, Liu Y. MRI-Based Radiomics: Associations With the Recurrence-Free Survival of Patients With Hepatocellular Carcinoma Treated With Conventional Transcatheter Arterial Chemoembolization. J Magn Reson Imaging. 2020;52:461-473. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 65] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 68. | Zhou Y, He L, Huang Y, Chen S, Wu P, Ye W, Liu Z, Liang C. CT-based radiomics signature: a potential biomarker for preoperative prediction of early recurrence in hepatocellular carcinoma. Abdom Radiol (NY). 2017;42:1695-1704. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 128] [Cited by in RCA: 164] [Article Influence: 20.5] [Reference Citation Analysis (0)] |
| 69. | Guo D, Gu D, Wang H, Wei J, Wang Z, Hao X, Ji Q, Cao S, Song Z, Jiang J, Shen Z, Tian J, Zheng H. Radiomics analysis enables recurrence prediction for hepatocellular carcinoma after liver transplantation. Eur J Radiol. 2019;117:33-40. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 59] [Article Influence: 9.8] [Reference Citation Analysis (1)] |
| 70. | Sun Y, Bai H, Xia W, Wang D, Zhou B, Zhao X, Yang G, Xu L, Zhang W, Liu P, Xu J, Meng S, Liu R, Gao X. Predicting the Outcome of Transcatheter Arterial Embolization Therapy for Unresectable Hepatocellular Carcinoma Based on Radiomics of Preoperative Multiparameter MRI. J Magn Reson Imaging. 2020;52:1083-1090. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 44] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 71. | Lv X, Chen M, Kong C, Shu G, Meng M, Ye W, Cheng S, Zheng L, Fang S, Chen C, Wu F, Weng Q, Tu J, Zhao Z, Ji J. Construction of a novel radiomics nomogram for the prediction of aggressive intrasegmental recurrence of HCC after radiofrequency ablation. Eur J Radiol. 2021;144:109955. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 13] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 72. | Kong C, Zhao Z, Chen W, Lv X, Shu G, Ye M, Song J, Ying X, Weng Q, Weng W, Fang S, Chen M, Tu J, Ji J. Prediction of tumor response via a pretreatment MRI radiomics-based nomogram in HCC treated with TACE. Eur Radiol. 2021;31:7500-7511. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 86] [Article Influence: 21.5] [Reference Citation Analysis (0)] |
| 73. | Ivanics T, Salinas-Miranda E, Abreu P, Khalvati F, Namdar K, Dong X, Deniffel D, Gorgen A, Erdman L, Jhaveri K, Haider M, Veit-Haibach P, Sapisochin G. A Pre-TACE Radiomics Model to Predict HCC Progression and Recurrence in Liver Transplantation: A Pilot Study on a Novel Biomarker. Transplantation. 2021;105:2435-2444. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 24] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 74. | Horvat N, Araujo-Filho JAB, Assuncao-Jr AN, Machado FAM, Sims JA, Rocha CCT, Oliveira BC, Horvat JV, Maccali C, Puga ALBL, Chagas AL, Menezes MR, Cerri GG. Radiomic analysis of MRI to Predict Sustained Complete Response after Radiofrequency Ablation in Patients with Hepatocellular Carcinoma - A Pilot Study. Clinics (Sao Paulo). 2021;76:e2888. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 10] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 75. | Cai W, He B, Hu M, Zhang W, Xiao D, Yu H, Song Q, Xiang N, Yang J, He S, Huang Y, Huang W, Jia F, Fang C. A radiomics-based nomogram for the preoperative prediction of posthepatectomy liver failure in patients with hepatocellular carcinoma. Surg Oncol. 2019;28:78-85. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 46] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 76. | Zhu WS, Shi SY, Yang ZH, Song C, Shen J. Radiomics model based on preoperative gadoxetic acid-enhanced MRI for predicting liver failure. World J Gastroenterol. 2020;26:1208-1220. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 25] [Cited by in RCA: 21] [Article Influence: 4.2] [Reference Citation Analysis (2)] |
| 77. | Kiryu S, Akai H, Nojima M, Hasegawa K, Shinkawa H, Kokudo N, Yasaka K, Ohtomo K. Impact of hepatocellular carcinoma heterogeneity on computed tomography as a prognostic indicator. Sci Rep. 2017;7:12689. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 40] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 78. | Akai H, Yasaka K, Kunimatsu A, Nojima M, Kokudo T, Kokudo N, Hasegawa K, Abe O, Ohtomo K, Kiryu S. Predicting prognosis of resected hepatocellular carcinoma by radiomics analysis with random survival forest. Diagn Interv Imaging. 2018;99:643-651. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 75] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 79. | Zheng BH, Liu LZ, Zhang ZZ, Shi JY, Dong LQ, Tian LY, Ding ZB, Ji Y, Rao SX, Zhou J, Fan J, Wang XY, Gao Q. Radiomics score: a potential prognostic imaging feature for postoperative survival of solitary HCC patients. BMC Cancer. 2018;18:1148. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 112] [Cited by in RCA: 107] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 80. | Kim J, Choi SJ, Lee SH, Lee HY, Park H. Predicting Survival Using Pretreatment CT for Patients With Hepatocellular Carcinoma Treated With Transarterial Chemoembolization: Comparison of Models Using Radiomics. AJR Am J Roentgenol. 2018;211:1026-1034. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 94] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 81. | Blanc-Durand P, Van Der Gucht A, Jreige M, Nicod-Lalonde M, Silva-Monteiro M, Prior JO, Denys A, Depeursinge A, Schaefer N. Signature of survival: a (18)F-FDG PET based whole-liver radiomic analysis predicts survival after (90)Y-TARE for hepatocellular carcinoma. Oncotarget. 2018;9:4549-4558. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 43] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 82. | Petukhova-Greenstein A, Zeevi T, Yang J, Chai N, DiDomenico P, Deng Y, Ciarleglio M, Haider SP, Onyiuke I, Malpani R, Lin M, Kucukkaya AS, Gottwald LA, Gebauer B, Revzin M, Onofrey J, Staib L, Gunabushanam G, Taddei T, Chapiro J. MR Imaging Biomarkers for the Prediction of Outcome after Radiofrequency Ablation of Hepatocellular Carcinoma: Qualitative and Quantitative Assessments of the Liver Imaging Reporting and Data System and Radiomic Features. J Vasc Interv Radiol. 2022;33:814-824.e3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 12] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 83. | Tian Y, Komolafe TE, Zheng J, Zhou G, Chen T, Zhou B, Yang X. Assessing PD-L1 Expression Level via Preoperative MRI in HCC Based on Integrating Deep Learning and Radiomics Features. Diagnostics (Basel). 2021;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 27] [Article Influence: 6.8] [Reference Citation Analysis (1)] |
| 84. | Liao H, Zhang Z, Chen J, Liao M, Xu L, Wu Z, Yuan K, Song B, Zeng Y. Preoperative Radiomic Approach to Evaluate Tumor-Infiltrating CD8(+) T Cells in Hepatocellular Carcinoma Patients Using Contrast-Enhanced Computed Tomography. Ann Surg Oncol. 2019;26:4537-4547. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 63] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 85. | Chen S, Feng S, Wei J, Liu F, Li B, Li X, Hou Y, Gu D, Tang M, Xiao H, Jia Y, Peng S, Tian J, Kuang M. Pretreatment prediction of immunoscore in hepatocellular cancer: a radiomics-based clinical model based on Gd-EOB-DTPA-enhanced MRI imaging. Eur Radiol. 2019;29:4177-4187. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 124] [Article Influence: 20.7] [Reference Citation Analysis (0)] |
| 86. | Hectors SJ, Lewis S, Besa C, King MJ, Said D, Putra J, Ward S, Higashi T, Thung S, Yao S, Laface I, Schwartz M, Gnjatic S, Merad M, Hoshida Y, Taouli B. MRI radiomics features predict immuno-oncological characteristics of hepatocellular carcinoma. Eur Radiol. 2020;30:3759-3769. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 113] [Article Influence: 22.6] [Reference Citation Analysis (0)] |
| 87. | Yuan G, Song Y, Li Q, Hu X, Zang M, Dai W, Cheng X, Huang W, Yu W, Chen M, Guo Y, Zhang Q, Chen J. Development and Validation of a Contrast-Enhanced CT-Based Radiomics Nomogram for Prediction of Therapeutic Efficacy of Anti-PD-1 Antibodies in Advanced HCC Patients. Front Immunol. 2020;11:613946. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 53] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 88. | Duan J, Qiu Q, Zhu J, Shang D, Dou X, Sun T, Yin Y, Meng X. Reproducibility for Hepatocellular Carcinoma CT Radiomic Features: Influence of Delineation Variability Based on 3D-CT, 4D-CT and Multiple-Parameter MR Images. Front Oncol. 2022;12:881931. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Reference Citation Analysis (0)] |
| 89. | Carbonell G, Kennedy P, Bane O, Kirmani A, El Homsi M, Stocker D, Said D, Mukherjee P, Gevaert O, Lewis S, Hectors S, Taouli B. Precision of MRI radiomics features in the liver and hepatocellular carcinoma. Eur Radiol. 2022;32:2030-2040. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 27] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 90. | Ibrahim A, Widaatalla Y, Refaee T, Primakov S, Miclea RL, Öcal O, Fabritius MP, Ingrisch M, Ricke J, Hustinx R, Mottaghy FM, Woodruff HC, Seidensticker M, Lambin P. Reproducibility of CT-Based Hepatocellular Carcinoma Radiomic Features across Different Contrast Imaging Phases: A Proof of Concept on SORAMIC Trial Data. Cancers (Basel). 2021;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 14] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 91. | Perrin T, Midya A, Yamashita R, Chakraborty J, Saidon T, Jarnagin WR, Gonen M, Simpson AL, Do RKG. Short-term reproducibility of radiomic features in liver parenchyma and liver malignancies on contrast-enhanced CT imaging. Abdom Radiol (NY). 2018;43:3271-3278. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 41] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 92. | Wang X, Wang S, Yin X, Zheng Y. MRI-based radiomics distinguish different pathological types of hepatocellular carcinoma. Comput Biol Med. 2022;141:105058. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 17] [Article Influence: 4.3] [Reference Citation Analysis (61)] |
| 93. | Gao F, Yan B, Chen J, Wu M, Shi D. Pathological grading of Hepatocellular Carcinomas in MRI using a LASSO algorithm. J Physic Confer Series. 2018;1053:012095. [RCA] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 94. | Zhou W, Zhang L, Wang K, Chen S, Wang G, Liu Z, Liang C. Malignancy characterization of hepatocellular carcinomas based on texture analysis of contrast-enhanced MR images. J Magn Reson Imaging. 2017;45:1476-1484. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 88] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 95. | Yang L, Gu D, Wei J, Yang C, Rao S, Wang W, Chen C, Ding Y, Tian J, Zeng M. A Radiomics Nomogram for Preoperative Prediction of Microvascular Invasion in Hepatocellular Carcinoma. Liver Cancer. 2019;8:373-386. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 238] [Article Influence: 39.7] [Reference Citation Analysis (0)] |
| 96. | Bakr S, Echegaray S, Shah R, Kamaya A, Louie J, Napel S, Kothary N, Gevaert O. Noninvasive radiomics signature based on quantitative analysis of computed tomography images as a surrogate for microvascular invasion in hepatocellular carcinoma: a pilot study. J Med Imaging (Bellingham). 2017;4:041303. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 45] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 97. | Ma X, Wei J, Gu D, Zhu Y, Feng B, Liang M, Wang S, Zhao X, Tian J. Preoperative radiomics nomogram for microvascular invasion prediction in hepatocellular carcinoma using contrast-enhanced CT. Eur Radiol. 2019;29:3595-3605. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 163] [Article Influence: 27.2] [Reference Citation Analysis (1)] |
| 98. | Xia W, Chen Y, Zhang R, Yan Z, Zhou X, Zhang B, Gao X. Radiogenomics of hepatocellular carcinoma: multiregion analysis-based identification of prognostic imaging biomarkers by integrating gene data-a preliminary study. Phys Med Biol. 2018;63:035044. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 51] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 99. | Li X, Cheng L, Li C, Hu X, Tan L, Li Q, Liu C, Wang J. Associating Preoperative MRI Features and Gene Expression Signatures of Early-stage Hepatocellular Carcinoma Patients using Machine Learning. J Clin Transl Hepatol. 2022;10:63-71. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 6] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 100. | Hui TCH, Chuah TK, Low HM, Tan CH. Predicting early recurrence of hepatocellular carcinoma with texture analysis of preoperative MRI: a radiomics study. Clin Radiol. 2018;73:1056.e11-1056.e16. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 53] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 101. | Zhao Y, Wu J, Zhang Q, Hua Z, Qi W, Wang N, Lin T, Sheng L, Cui D, Liu J, Song Q, Li X, Wu T, Guo Y, Cui J, Liu A. Radiomics Analysis Based on Multiparametric MRI for Predicting Early Recurrence in Hepatocellular Carcinoma After Partial Hepatectomy. J Magn Reson Imaging. 2021;53:1066-1079. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 55] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 102. | Zhang H, Guo D, Liu H, He X, Qiao X, Liu X, Liu Y, Zhou J, Zhou Z, Fang Z. MRI-Based Radiomics Models to Discriminate Hepatocellular Carcinoma and Non-Hepatocellular Carcinoma in LR-M According to LI-RADS Version 2018. Diagnostics (Basel). 2022;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 8] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 103. | Park S, Kim JH, Kim J, Joseph W, Lee D, Park SJ. Development of a deep learning-based auto-segmentation algorithm for hepatocellular carcinoma (HCC) and application to predict microvascular invasion of HCC using CT texture analysis: preliminary results. Acta Radiol. 2022;2841851221100318. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 2] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 104. | Haniff NSM, Abdul Karim MK, Osman NH, Saripan MI, Che Isa IN, Ibahim MJ. Stability and Reproducibility of Radiomic Features Based Various Segmentation Technique on MR Images of Hepatocellular Carcinoma (HCC). Diagnostics (Basel). 2021;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 19] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 105. | Hu MJ, Yu YX, Fan YF, Hu CH. CT-based radiomics model to distinguish necrotic hepatocellular carcinoma from pyogenic liver abscess. Clin Radiol. 2021;76:161.e11-161.e17. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 8] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 106. | Hu HT, Shan QY, Chen SL, Li B, Feng ST, Xu EJ, Li X, Long JY, Xie XY, Lu MD, Kuang M, Shen JX, Wang W. CT-based radiomics for preoperative prediction of early recurrent hepatocellular carcinoma: technical reproducibility of acquisition and scanners. Radiol Med. 2020;125:697-705. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 58] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 107. | Qiu Q, Duan J, Duan Z, Meng X, Ma C, Zhu J, Lu J, Liu T, Yin Y. Reproducibility and non-redundancy of radiomic features extracted from arterial phase CT scans in hepatocellular carcinoma patients: impact of tumor segmentation variability. Quant Imaging Med Surg. 2019;9:453-464. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 52] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 108. | Zhang J, Qiu Q, Duan J, Gong G, Jiang Q, Sun G, Yin Y. Variability of radiomic features extracted from multi-b-value diffusion-weighted images in hepatocellular carcinoma. Transl Cancer Res. 2019;8:130-140. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 11] [Article Influence: 1.8] [Reference Citation Analysis (0)] |