Published online Jan 7, 2023. doi: 10.3748/wjg.v29.i1.19
Peer-review started: September 21, 2022
First decision: October 18, 2022
Revised: November 5, 2022
Accepted: December 16, 2022
Article in press: December 16, 2022
Published online: January 7, 2023
Processing time: 105 Days and 3 Hours
The gut microbiota is currently considered an external organ of the human body that provides important mechanisms of metabolic regulation and protection. The gut microbiota encodes over 3 million genes, which is approximately 150 times more than the total number of genes present in the human genome. Changes in the qualitative and quantitative composition of the microbiome lead to disruption in the synthesis of key bacterial metabolites, changes in intestinal barrier function, and inflammation and can cause the development of a wide variety of diseases, such as diabetes, obesity, gastrointestinal disorders, cardiovascular issues, neurological disorders and oncological concerns. In this review, I consider issues related to the role of the microbiome in the regulation of intestinal barrier function, its influence on physiological and pathological processes occurring in the body, and potential new therapeutic strategies aimed at restoring the gut microbiome. Herewith, it is important to understand that the gut microbiota and human body should be considered as a single biological system, where change of one element will inevitably affect its other components. Thus, the study of the impact of the intestinal microbiota on health should be considered only taking into account numerous factors, the role of which has not yet been fully elucidated.
Core Tip: The gut microbiota affects the development and functioning of all body systems, providing metabolic, physiological, regulatory and protective functions. Violations in the qualitative and quantitative composition of the microbiome lead to the development of a wide variety of diseases, such as diabetes, obesity, cardiovascular issues, neurological disorders and oncological concerns. Considering that intestinal dysbiosis plays a key role in the development of a number of diseases, aim to normalize the microbiome seems to be a greatly perspective direction in their prevention and treatment.
- Citation: Senchukova MA. Microbiota of the gastrointestinal tract: Friend or foe? World J Gastroenterol 2023; 29(1): 19-42
- URL: https://www.wjgnet.com/1007-9327/full/v29/i1/19.htm
- DOI: https://dx.doi.org/10.3748/wjg.v29.i1.19
Trillions of microorganisms, known as microbiota, colonize the human body. The human gastrointestinal tract harbours more than 1000 species of bacteria belonging to a relatively few known bacterial phyla[1]. Features of their mutual coexistence determine the nature of various physiological and pathological processes occurring in the human body. To discuss these issues, a search was made in the PubMed database (https://pubmed.ncbi.nlm.nih.gov/) and Reference Citation Analysis (https://www.referencecitationanalysis.com/) for studies published up to July 1, 2022 using a combination of text keywords "gut microbiota", "bacterial metabolites", "intestinal barrier", "dysbiosis", "fecal microbiota transplantation", and "probiotics". A total of 846 unique results were identified, which were screened individually by title and abstract and were included based on the role of the microbiome in the regulation of intestinal barrier function, its influence on physiological and pathological processes occurring in the body, and new therapeutic strategies aimed at restoring the gut microbiome.
It is believed that bacteria begin to colonize the human intestine immediately after birth and, possibly, even in utero[2,3]. Breast milk plays a crucial role in the composition of the microbial community within newborns through transfer of the milk microbiota to the infant's gut[4]. In the first 6 mo of a child's life, there is a steady increase in the number of Enterobacteriaceae, Bifidobacteriaceae and Clostridiaceae. However, the microbiome of children differs depending on diet, gender, race and ethnicity[5,6]. In addition, the mode of delivery may affect the composition of the gut microbiota of early infants[3,7]. In particular, in children born by cesarean section, there is a high abundance of Bifidobacterium, and Clostridium genera and the family Enterobacteriaceae, along with a low abundance of Streptococcus and Ruminococcus genera[8]. Moreover, in children born by cesarean section, the Bacteroides genus is not detected in the feces until 6-18 mo after birth[9].
Microorganisms living in the gut of adults include bacteria, fungi, protozoa, archaea, and some viruses[10]. The total number of bacteria in a 70 kg "reference man" is estimated at 3.8 × 1013 cells, which is comparable to the number of human host cells (3.0 × 1013)[11]. The specific microbiota at the genus and species levels varies depending on geography, environment, diet, age, genotype, presence of diseases and lifestyle[12,13]. For instance, if the prevalence of proteins and animal fats in the diet exists, Bacteroides will predominate in the microbiota, and if there is a high level of carbohydrates, then Prevotella will ascendant. The gut microbiota encodes over 3 million genes, which is around 150 times more than the number of genes in the host genome[14]. Approximately 90% of the composition of the gut microbiome is represented by Firmicutes (79.4%), Bacteroidetes (16.9%), Actinobacteria (2.5%) and Proteobacteria (1%)[5,15]. The number of microorganisms increases from the proximal to the distal gastrointestinal tract and from the epithelial layer to the lumen. This difference can be explained by the presence of a more aggressive environment in the upper intestines due to the incoming gastric acid, action of digestive enzymes, rapid movement of chyme and the decrease in partial pressure of oxygen in the distal gastrointestinal tract. For this reason, aerobic bacteria predominate in the small intestine, while facultative and obligate anaerobes predominate in the lower gastrointestinal tract. Furthermore, the distribution and organization of the gut microbiota is determined by intestinal mucins, which protect intestinal epithelial cells (IECs) from bacterial colonization. At the same time, the presence of the gut microbiota is a necessary condition for normal functioning of the mucosal barrier[16]. For example, mice treated with antibiotics had a thinner layer of mucus[17,18].
Partition of the body’s internal environment from the intestinal microbiota is carried out by three types of barriers: physical, chemical and immunological. The physical barrier consists of epithelial cells, glycocalyx and a layer of mucus covering the surface of the gastrointestinal wall[19].
The intestinal epithelium is the most rapidly self-renewing tissue in adult mammals. To maintain epithelial layer integrity, IECs are continuously replaced by proliferating progenitor cells derived from multipotent intestinal stem cells (ISCs) localized in the base of the crypts of Lieberkühn and the colon[20]. IECs differ in their proliferation ability, renewal rate and age. Aged IECs undergo apoptosis and are later ejected into the intestinal lumen, whereas Paneth cells leave the crypt bottom by cellular fragmentation and phagocytosis by macrophages infiltrating the lamina propria mucosae[21]. Under homeostatic conditions, the entire ileal crypt is replaced every 4-5 d[22].
It is customary to distinguish the following populations of the intestinal epithelium[22-24].
Columnar cells: Columnar cells (colonocytes) are the most numerous population of enterocytes. They participate in digestion due to the secretion of digestive enzymes, the absorption of digested products and transcellular transfer of dissociated monomers into the blood and lymph and take part in the exchange of bile acids and humoral immune response. Absorbent enterocytes produce polymeric immunoglobulin (Ig) receptor, which mediates transcytosis of dimeric IgA and polymeric IgM from the lamina propria through the epithelial barrier to the mucosal surface, ensuring the binding of bacteria and viruses on their surface and thereby preventing the penetration of pathogens into the internal environment of the body. In addition, during transcytosis of IgA through the epithelium, it can neutralize viruses that have entered cells, as well as bind and excrete proteins and other immune complexes on the surface of mucous[20,25].
Goblet cells: Goblet cells are a source of mucus. In addition, they can deliver small soluble antigens to dendritic cells (DCs) localized in the lamina propria and thus participate in the formation of immune tolerance to food antigens and the gut microbiome[20,26].
Enteroendocrine cells: Enteroendocrine cells secrete peptides and hormones (cholecystokinin, serotonin) to stimulate intestinal motility.
Tuft cells: Tuft cells participate in the clearance of parasites from the intestinal lumen due to the synthesis of interleukin (IL)-25, which is the key activator of type 2 innate lymphoid cells.
Paneth cells: Paneth cells in the small intestine or deep crypt secretory cells in the large intestine are the main regulators of microbial density in the intestines. When interacting with gram-negative bacteria, gram-positive bacteria or their products (lipopolysaccharides, lipoteichoic acids, lipid A, muramyl peptide), they secrete antimicrobial peptides (AMPs)[20,27,28]. Moreover, Paneth cells secrete important factors, such as epidermal growth factor, transforming growth factor-α, and Wnt ligands involved in stem cell maintenance[29].
Microfold cells: Microfold cells (M) are located in the follicle-associated epithelium overlying Peyer's patches and stimulate an immune response by binding luminal antigens for their further transport to subepithelial regions, where they are captured by DCs migrating to the mesenteric lymph nodes and stimulating the immune response[20]. The interaction of DCs with T cells stimulates an antigen-specific immune response directed against the pathogen or, conversely, leads to the induction of tolerance. Activation of B cells leads to the secretion of IgA, which plays an important role in the regulation of the gut microbiome. Immunoglobulin A, on the one hand, is involved in the binding and elimination of pathogenic bacteria, and on the other hand, it facilitates the translocation of commensals into Peyer's patches, activating the mechanisms of immunological tolerance and thereby stimulating the growth of intestinal symbionts[30,31].
It is worth noting that at the level of IECs, a structure composed of three junctions is formed: tight junctions (TJs), adherens junctions and desmosomes[20,32-34]. They provide mechanical strength between cells, intercellular adhesion and polarization and are also involved in cell signaling pathways[19]. The strength of the mechanical barrier also depends on the regeneration rate of the intestinal epithelium. It was established that the presence of intestinal microorganisms affects the number of Paneth cells and hence the integrity of the epithelial barrier, as Paneth cells regulate stem cell homeostasis[35]. Bacteria can directly damage TJ proteins or interfere with their synthesis via type 3 or type 4 secretion systems. The disruption of cell contacts can also be mediated by some bacterial enzymes and toxins, such as hemagglutinin/protease and ZO toxin, as well as bacterial metabolites, such as ethanol and acetaldehyde[13,36]. Apart from that, the gut microbiota can alter the mitochondrial metabolism of epithelial and immune cells, thereby activating inflammation and disrupting epithelial barrier function[37]. Integrity violation of the intestinal epithelium, the mucus barrier and cell contacts leads to the translocation of bacteria[38].
Recognition of the bacterial microbiota is carried out by TLR and NOD receptors, which are expressed by most IECs, including stem cells, enterocytes, goblet cells, enteroendocrine cells, Paneth cells, and M cells[19]. Most TLRs (TLR2, TLR3, TLR4, TLR5, and TLR9) are present on the basolateral membrane, while TLR2, TLR3, and TLR9 are also expressed on the apical surface. TLR5 expression is limited to Paneth cells in the epithelium of the small intestine and proximal colon[39,40]. Activation of innate immune system receptors, including TLR and NOD receptors, as well as inflammasomes, leads to a signaling cascade that triggers the secretion of cytokines and chemokines, including IL-1β, IL-6, IL-12, IL-18, tumor necrosis factor alpha (TNF-α), CXCL8 and CCL20, which activate immunocompetent cells localized in the lamina propria[41-43]. Mitochondrial reactive oxygen species (mtROS), produced by immune cells, play a key role in the eradication of invading pathogens through direct bactericidal action or indirect impact on the activation of the NLRP3 inflammasome and the production of proinflammatory cytokines. Invading bacteria, as well as gut microbiome fermentation products such as short-chain fatty acids (SCFAs), induce mtROS production in immune cells through increased mitochondrial respiration and increased oxidative phosphorylation[44]. Hypoxia inducible factor-1α is believed to be the main regulator of mitochondrial responses during bacterial infection[37]. At the same time, damage to the intestinal epithelium mitochondria by toxins of pathogenic bacteria leads to the accumulation of mtROS and disruption of the barrier function of the epithelium[45,46].
Intestinal mucus is the first barrier for microorganisms in the gastrointestinal tract. It regulates nutrient and drug delivery, regulates symbiosis with the gut microbiota and protects the epithelium from dietary antigens and food toxins[20,47,48]. The thickness of the mucus layer in the small and large intestine is not the same. The main function of the small intestine is food digestion and absorption of nutrients, so the small intestine has a loose, discontinuous layer of mucus that can be easily removed. In the large intestine, where the density of microorganisms is much higher than in the small intestine, the number of mucus-producing goblet cells is significantly larger[20]. In the large intestine, the mucus layer is organized into two layers: an inner, dense, microbiota-free mucus layer and an outer layer, which is friable and permeable for microorganisms[13,17,31,49,50].
More than 20 mucin subtypes have been identified in humans. The best known and most studied one, found in the small and large intestines, is MUC2[17,49]. MUC2 is a highly O-glycosylated gelling mucin that forms polymeric networks via C-terminal dimerization and N-terminal trimerization. MUC2 monomers are glycosylated in proline, threonine, and serine-rich domains[51,52].
Mucus is secreted by goblet cells, grows rapidly and forms a stratified, dense layer that adheres to the epithelium[53]. On the one hand, mucus is necessary for the normal functioning of the gut microbiome, but on the other hand, the presence of the microbiome is a necessary condition for the normal functioning of the mucus barrier[54]. As noted above, mice treated with antibiotics have a thinner layer of mucus[17,18].
The barrier function of the mucus is confirmed by the fact that mice genetically deficient in Muc2 (Muc2 -/-) have bacteria invading the normally sterile distal colon crypts, which results in the development of spontaneous colitis[55], adenomas arising in the small intestine and an invasive cancer[56]. However, intestinal bacteria can directly influence the production and quality of intestinal mucus and hence the intestinal barrier permeability[57]. Bacteria and their metabolites that enter crypts are endocytosed by specialized goblet cells known as ‘sentinel’ goblet cells. This leads to the activation of TLR2/1, TLR4, and TLR5 Ligands with activated ROS synthesis, triggering the formation of the NLRP6 inflammasome and Ca2+-dependent compound exocytosis of MUC2-containing granules[58,59]. Importantly, increased regulatory secretion leads to the secretion of large amounts of MUC2 and the physical removal of bacteria from the crypt opening, thereby protecting the inferior crypt and multipotent ISCs, located at the bottom of the crypts, from bacterial invasion[20,60].
An important function of the chemical barrier is to maintain the abundance and composition of the gut microbiome. The chemical barrier includes AMPs, gastric acid, digestive enzymes, mucopolysaccharides, glycoproteins, glycolipids, and other compounds[19,61,62]. In addition, the composition of the microbiota can be influenced by various factors, such as hygiene, diet (especially the "Western diet" low in fiber and high in sugar and fat), oxygen concentration, microbial adhesion, host stress and other factors[17,49,63,64]. It is believed that microbiome regulation in the small intestine is mainly carried out by antibacterial peptides, while in the large intestine, this regulation is carried out through pattern recognition receptors[5]. The microbiome population is maintained, either by preventing colonization or through direct killing mechanisms.
The production of antimicrobials that lyse target cells is one of the main mechanisms for regulating the homeostasis of the gut microbiome. The contact of bacteria with Toll (TLR2, TLR4, TLR7 and TLR9), NOD1 and NOD2 receptors activates adapter proteins (for example, MyD88) and genes responsible for the synthesis of cytokines and chemokines in IECs[65-68]. In turn, the synthesis of cytokines by immune cells activates the genes responsible for the synthesis of AMPs[69]. Thus, mice deficient in MyD88 exhibit a 100-fold higher bacterial load in the gut than wild-type mice, and this increase is correlated with a decrease in the antibacterial peptide RegIIIgγ[70]. Paneth cells expressing TLR5 are major producers of antimicrobials, many of which are cationic AMPs that interact with negatively charged bacterial membranes and destroy them[12]. Interestingly, TLR5 expression occurs predominantly in intestinal crypts and is genetically determined, as TLR5 expression does not require bacterial or immune signals. It is believed that their main function may be related to the protection of Paneth cells and stem cells[31,71].
Other AMPs are also involved in the regulation of the gut microbiome population. For example, protein 8 (Lypd8) is highly expressed by colonocytes and facilitates segregation of microorganisms in the colon via flagella binding[61]. The lectin-like protein ZG16 specifically binds the peptidoglycan of gram-positive bacteria and thereby inhibits their penetration into the inner layer of the colon[62]. Lectins RegIIIγ from IECs and beta-defensins from neutrophils have a bactericidal effect against a number of bacteria. However, Firmicutes and Bacteroidetes living in the small intestine are resistant to these antibacterial agents[72]. It is also worth noting that some bacteria can synthesize bacteriocins (for example, colicin and microcins), which inhibit the growth of competitors[73,74].
The gut immune barrier is represented by single lymphoid follicles and Peyer's patches — peripheral accumulations of lymphoid cells located in the lamina propria of the small intestine mucosa[20]. Within the follicles, there are various immune elements, including B and T lymphocytes, DCs, and neutrophils, that secrete cytokines and antibodies in response to antigen entry. Goblet cells are involved in the presentation of luminal antigens to the CD103+ DC complex of the intestinal mucosa lamina propria, forming antigenic complexes (goblet cell-associated antigen passages)[75]. Secretory IgA (SIgA), another component of the intestinal barrier, is produced by plasma cells (50 mg/kg SIgA daily in an adult) and localized predominantly in the lamina propria of the intestinal mucosa[31]. It is believed that SIgA is able to interact with commensal intestinal bacteria, mediating the formation of a bacterial biofilm. SIgA is resistant to the action of intestinal proteases, which provide protection for bacteria. SIgA can penetrate through the epithelial lining into the intestinal lumen, bind antigens and deliver them to the immune cells of the lymphoid tissue[76].
Elements of immune protection can also include an increase in "tolerance" to a microbe (or toxin of microbial origin)[52] and the death of infected cells. In particular, flagellins of pathogenic bacteria that have overcome the epithelial barrier are able to activate NAIP/NLRC4 in macrophages, which causes the death of infected epithelial cells and their expulsion into the intestinal lumen[77].
It is important to note that the intestine is the most important immune organ, which not only protects against external pathogens but also participates in the formation of immune tolerance to food substrates and the normal gut microbiome. The main cytokines involved in the formation of immunological tolerance are IL-10 and TGF-beta, which are produced by CD4+ T cells, some populations of macrophages and other cells and have an anti-inflammatory effect, limiting the expansion of effector cells and inducing the proliferation of regulatory T cells[78].
The microbiota are currently considered an external organ of the human body, which provides important mechanisms of metabolic regulation and protection, alongside the development and functioning of all organ systems[79]. It performs the following functions, the list of which is incomplete.
Digestion of plant polysaccharides. Approximately 17 carbohydrate-active enzymes are formed in the human body, while the microbiota provides around 89[80]. The gut microbiota actively digests dietary fiber, which the human body is unable to digest. These processes take place in the large intestine via the most actively involved enzymatic anaerobes, which decompose polysaccharides, particularly representatives of the Bacteroidaceae and Clostridiaceae families[12,81]. As a result of their digestion, compounds are produced that have a positive effect on the intestinal mucosa. In addition, the mucus layer is an alternative source of glycans for bacteria[12,51,81,82].
Participation in the metabolism of proteins, lipids and fatty acids[83-87]. In particular, gram-negative (Bacteroides thetaiotamicron) and gram-positive (Lactobacillus rhamnosus gg) bacteria are involved in the regulation of lipid absorption by activating cholecystokinin and secretin receptors expressed by epithelial endocrine cells of the proximal small intestine[88].
Energy supply of IECs[15,81,89] and regulation of their proliferative activity[90,91].
Modulation of goblet cell functions and mucin secretion[16].
The presence of intestinal microorganisms affects the number of Paneth cells and hence the integrity of the epithelial barrier, as Paneth cells regulate ISC homeostasis[35].
Stimulation of local and systemic immunity due to activation of the synthesis of IgA, interferons, and activation of immune cells (macrophages, lymphocytes, and DCs), influence on the development of the intestinal lymphoid apparatus in newborns[92-94].
Synthesis of group B, K vitamins, a number of coenzymes, for example, tocopherols[95-97].
Participation in the regulation of intestinal peristalsis[98-100].
Influence on bone metabolism and pathogenesis of osteoporosis[14,101,102]. The bacterial synthesis of SCFA leads to a decrease in pH in the intestinal lumen and an increase in calcium solubility, an increase in its absorption, and a decrease in bone resorption. Bifidobacterium and Lactobacillus are mainly involved in these processes. In addition, Fusobacterium nucleatum can enhance osteoclast differentiation through increased expression of IL-17A, TNF-alpha, and trimethylamine N-oxide (TMAO), while Bacteroides, Lactobacillus, and Bifidobacterium can promote the development of Treg cells and thereby increase osteoblast activity[14,101,103,104].
Influence on the processes associated with the synthesis of neurotransmitters, myelination of neurons in the prefrontal cortex, with the development of the amygdala and hippocampus[105,106]. In dysbacteriosis, the response to antidepressant therapy may be impaired[107]. Germ-free mice show hyperactivity, memory and learning deficits and impaired expression of the serotonin 5-HT1A and NMDA receptors in the hippocampus[108,109].
Inhibition of the growth of pathogenic microorganisms is due to the activation of phagocytosis, the synthesis of antibacterial peptides or the synthesis of bacteriocins that inhibit the growth of competitors[69,73,74,81,100,110].
Influence the effectiveness of several drugs, in particular antibiotics, proton pump inhibitors, metformin, vitamin D and laxatives. It has been shown that the use of these drugs disrupts both the composition of the microbiota and its functional activity[89,111,112].
As already noted, health and disease conditions are largely dependent on the functioning microbiome. Products of bacterial metabolism can be crucial for maintaining both the health of an organism and the development of various diseases[113].
The most important products of bacterial fermentation are SCFAs: butyrate, acetate and propionate. The main producers of SCFAs are Firmicutes and Actinobacteria.
Butyrate: Butyrate is the primary metabolite of Firmicutes. It can be synthesized through condensation of 2 molecules of acetyl-CoA, which are reduced to butyryl-CoA and then converted to butyric acid by phosphotransferase and butyrate kinase[114]. It can also be synthesized from butyryl-CoA, lactate and acetate using the acetyl-CoA transferase pathway[115] and from proteins using lysine[116]. It has anti-inflammatory, antitumour, antiproliferative and immunomodulatory properties and is involved in genetic/epigenetic regulation[117,118]. In particular: (1) Regulates antigen-specific adaptive immunity mediated by T- and B-cells: induce T-cells to produce IL-10; regulate the transcription of some cytokine genes, such as IFN-γ and TNF-α, and the activity of the nuclear factor kappa B (NF-κB) signaling pathway; reduce the production of proinflammatory mediators (TNF-α, IL-6, IFN-γ and NO); increase the production of antibodies by B-cells and promote the differentiation of B-cells into plasma B-cells[119-121]; (2) Participates in fat metabolism, reduces insulin resistance, hyperglycaemia, hyperinsulinaemia, and lipid concentrations in the liver and pancreas, thereby reducing the risk of obesity[122]; (3) Affects fatty acid receptors in epithelial, enteroendocrine, neuronal and glial cells, which leads to the production of serotonin by enterochromaffin cells. This may affect the peripheral and central nervous systems of experimental model organisms[50,123]; (4) Helps improve memory[124]; (5) Involved in maintaining the mechanical integrity of the intestinal barrier by inducing the expression of occludin, ZO-1 mRNA and claudin-1 mRNA, thereby reducing intestinal permeability and increasing intestinal villus growth[13,122,125,126]; and (6) Inhibits the rate of cancer cell migration and invasion by increasing the expression of antimetastatic genes (e.g., metalloproteinases) and inhibiting the activation of prometastatic genes (e.g., matrix metalloproteinases)[85,127].
Acetate: Acetate is a fermentation product of various bacteria and is produced from pyruvate using acetyl coenzyme A[128]. It performs the following functions: (1) Participates in the regulation of cholesterol synthesis and activation of local immunity[129]; (2) Helps increasing physical endurance[130]; (3) Influences cognitive functions by activating synaptophysin synthesis[131]; (4) Promotes appetite reduction, fat oxidation, increased levels of proinflammatory cytokines through activated secretion of intestinal hormones such as glucagon-like peptide-1 and peptide YY and increased insulin sensitivity[86]. Nonobese patients show higher production of acetate by gut microbiota than obese patients[132]; (5) Regulates the gut microbiome by increasing the production of IgA and its selective binding to certain microorganisms[86].
Propionate: Propionate is the primary metabolite of Bacteroidetes fermentation and is formed from the conversion of succinate to methylmalonyl-CoA by the succinate pathway or from acrylate by the acrylate pathway using lactate as a precursor[133]. Additionally, fucose and rhamnose can be used as substrates for the synthesis of propionic acid via the propanediol pathway[134]. Propionate performs the following functions: (1) Participates in maintaining the mechanical integrity of the intestinal barrier by increasing the expression of gut TJ proteins ZO-1, occludin and cadherin[135,136], as well as the synthesis of the antimicrobial protein Regenerating islet-derived protein type 3 (Reg3)[137]; (2) Reduces the risk of developing atherosclerosis and the development of cardiovascular disease by increasing insulin sensitivity and reducing the levels of proinflammatory IL-8[138] and IL-17[139], as well as by reducing the absorption of cholesterol in the intestine by an immune-mediated mechanism through an increase in the number of regulatory T cells and the level of IL-10, which suppress the expression of C1-like 1 Niemann-Pick (Npc1 L1), the main cholesterol transporter in the intestine[140]; (3) Influences physical endurance[141] and motor functions[136]; (4) Promotes regeneration and functional recovery of sensory axons through an immune-mediated mechanism[142]; and (5) Involved in the regulation of the gut microbiome, possibly through direct suppression of the growth of pathogenic microorganisms[143].
A number of other intestinal metabolites can also affect the health of individuals. Among these metabolites, a special place is occupied by tryptophan derivatives: serotonin, tryptamine, kynurenine, and indoles. They perform the following functions.
Act on the central nervous system through the brain-gut axis[144]. The influence is implemented due to the impact on the glutamatergic receptor N-methyl-d-aspartate (NMDA). It has been established that kynurenine, a breakdown product of tryptophan, easily penetrates the blood-brain barrier, where it is metabolized to form neuroactive glutamatergic compounds, kynurenic acid or quinoline acid, acting in the opposite way. Kynurenic acid acts as an NMDA receptor antagonist and has a neuroprotective effect, while quinolinic acid acts as an NMDA receptor agonist and exhibits a neurotoxic effect[145]. In major depressive disorder and bipolar disorder, a decrease in tryptophan and kynurenine was noted. In these mental disorders, there is a shift in tryptophan metabolism from the serotonin pathway to the kynurenine pathway[146]. Tryptophan metabolites are involved in the pathogenesis of various neurodegenerative disorders (Alzheimer's disease, amyotrophic lateral sclerosis, Huntington's disease, Parkinson's disease) as well as other diseases such as AIDS, cancer, cardiovascular disease, inflammation and irritable bowel syndrome[147,148].
Participate in the regulation of activation, proliferation and migration of immune surveillance cells (T- and B-lymphocytes, macrophages and natural killer cells) and the production of inflammatory signaling molecules, cytokines, nitric oxides and superoxides[149,150].
Influence the motility of the gastrointestinal tract. In particular, as a result of tryptamine action on the serotonin receptor 5-HT4R[151].
Indoles may contribute to the development of cardiovascular, metabolic and psychiatric diseases[152].
It is worth noting that secondary bile acids play a special role in the development of inflammation and colorectal cancer (CRC). It is known that between 5% and 10% of nonreabsorbed bile acids can undergo biotransformation into secondary bile acids as a result of bacterial metabolism involving bacterial bile acid hydrolases (BSHs). Most BSH bacteria are gram-positive enteric bacteria, including Clostridium, Enterococcus, Bifidobacterium, and Lactobacilli. The only gram-negative bacteria with BSH activity are members of the genus Bacteroides[85,153]. Interestingly, secondary bile acids at high concentrations associated with Western diets can induce oxidative/nitrosative stress, ROS production, DNA damage, apoptosis and mutations[85,154,155] and induce proinflammatory macrophage M1 polarization[154] by binding secondary bile acids with Takeda G protein-coupled receptor 5 (TGR5)[156], thereby initiating inflammation[157]. Several studies have noted that through the activation of the epidermal growth factor receptor (EGFR), secondary bile acids can induce COX-2 expression, stimulate EGFR-MARK signaling[155,158], activate cellular β-catenin signaling and the NF-κB pathway[155,159], and provide transfer of extracellular signal-regulated kinase 1 and 2 via activator protein 1 and c-Myc, thereby stimulating the proliferation and invasiveness of colon cancer cells[85,160,161]. At the same time, at low and physiological concentrations, secondary bile acids can have an anti-inflammatory and antitumour effect through a reduction in proinflammatory cytokine levels[162,163]. In particular, an antitumour effect has been noted for lithocholic and ursodeoxycholic acid (UDCA), which are secondary bile acids produced by Clostridium species. Lithocholic acid (LCA) at concentrations corresponding to its tissue reference concentrations (< 1 μmol/L) has an antitumour effect on breast cancer cells by inhibiting the epithelial-mesenchymal transition, reducing the production of vascular endothelial growth factor, and activating antitumour immunity[164]. UDCA may prevent the development of CRC by regulating oxidative stress in colon cancer cells[165]. However, the preventive effect of UDCA on CRC is not universally accepted[166].
TMAO is a molecule resulting from the oxidation in the liver of a microbial metabolism product, trimethylamine (TMA). TMA is formed in the colon from choline, betaine, and carnitine. The main food precursors of TMA are red meat, fish, poultry, and eggs. TMA from the colon is absorbed into the bloodstream, where it is oxidized by the hepatic enzyme flavin-containing monooxygenase-3 to TMAO, most of which is then excreted unchanged in the urine. Plasma TMAO levels are determined by several factors, including diet, age, gut microbiota, drug intake, and liver flavin monooxygenase activity. The main TMA producers are Clostridia, Shigella, Proteus, Aerobacter, and Eubacterium sp.[167,168]. Some bacterial enzymes are able to oxidize TMA to TMAO in the colon[169]. At the same time, TMAO can be metabolized into dimethylamine, formaldehyde, ammonia and methane by some methanogenic bacteria, which leads to its depletion in the colon[170]. It cannot be excluded that the production of formaldehyde under oxidative stress conditions caused by TMAO metabolism may be one of the factors contributing to the induction of CRC. In the experiment, intragastric administration of a suspension of CaCO3 in a mixture with formaldehyde and hydrogen peroxide induced tumors of the stomach and cecum in rats[171].
It has now been established that elevated plasma levels of TMAO correlate with the risk of developing atherosclerosis[172-175], obesity[175,176], cardiovascular diseases[174,177,178], type 2 diabetes[175], chronic kidney disease[179,180] and CRC[181,182]. Elevated TMAO levels have been associated with endothelial dysfunction and inflammatory damage to the vascular endothelium[183,184], an increase in the level of proinflammatory cytokines, and a decrease in the level of anti-inflammatory cytokines[185,186] with the activation of the MAPK and NF-κB transcriptional pathways[185], oxidative stress[177], cell proliferation and angiogenesis[187]. Xu et al[188] provided evidence that these risks may be genetically determined. Moreover, it is worth noting that the effects of TMAO may differ between healthy and diseased conditions. In healthy individuals, TMAO can demonstrate protective, antioxidant or anti-inflammatory effects, while in patients, especially under conditions of oxidative stress, it can have a negative impact on human health[189]. Further research is needed to elucidate the effects of TMAO on human health.
Hydrogen sulfide is a metabolite of sulfate-reducing bacteria that metabolize dietary sulfates and other sulfur-containing compounds, including taurine[155]. It is produced by a wide range of Enterobacteria, primarily of the genus γ-Proteobacteria. The hydrogen sulfide concentration in the large intestine is more than that 5 times higher than in the small intestine[190]. Hydrogen sulfide, similarly to secondary bile acids and TMAO, can multidirectional affect inflammation, oxidative stress, and carcinogenesis[155,191]. Various authors have demonstrated both its inflammatory[192] and anti-inflammatory effects[193,194], as well as its carcinogenic[195,196] and anticancer properties[197,198]. For example, some researchers have demonstrated that hydrogen sulfide may be associated with the breakdown of disulfide bonds in the mucus double layer in the colon wall, leading to inflammation and translocation of bacteria and toxins[199]. At the same time, other researchers suggest that hydrogen sulfide can protect the mucus layer and repair this already destroyed, thereby preventing inflammation[190]. It is believed that the physiological and pathological effects of hydrogen sulfide are associated with its concentration[200]. Given that intracellular hydrogen sulfide has a significant impact on many cellular functions, such as TJs, autophagy, apoptosis, vesicle trafficking, cell signaling, epigenetics and inflammasomes[201], and can be used as a therapeutic agent, its further study may open new opportunities in understanding the mechanisms of the development of pathological conditions and their treatment.
Polyamines are versatile polyfunctional molecules involved in cell proliferation and differentiation, apoptosis, angiogenesis, immune response, signaling, and gene expression[202,203]. They can be supplied with food and can form as a result of endogenous synthesis, as well as a result of bacterial metabolism, such as putrescine, spermidine and cadaverine[204,205]. In particular, cadaverine is formed from lysine by decarboxylation with the participation of lysine decarboxylase (LDC). Putrescine is formed by decarboxylation of ornithine catalyzed by ornithine decarboxylase. Spermidine synthase is involved in the formation of spermidine from putrescine. These enzymes are produced by most gram-negative bacteria[205,206]. Polyamines synthesized by the intestinal microbiota are then transported into the bloodstream through the colonic mucosa[207].
It has been established that polyamines produced by intestinal bacteria suppress chronic inflammation and strengthen the intestinal barrier in the colon, contribute to a significant improvement in the host's cognitive functions and increase life expectancy in experimental animals and have a cardioprotective effect[203,208,209]. In a number of studies, the antitumour effect of cadaverine was noted. In an experiment, cadaverine caused a decrease in the proliferative activity and invasiveness of breast cancer cells and contributed to the induction of mesenchymal-epithelial transition, a decrease in the stemness of cancer cells and their ability to metastasize[210]. In breast cancer, a decrease in the intestinal biosynthesis of cadaverine was noted, especially in patients with carcinoma in situ and stage I of the disease. With a high expression of bacterial LDC in the gut contents, a significantly longer survival was noted than with a low expression[210].
At the same time, a number of studies have noted that high levels of polyamines may be associated with tumor transformation and cancer progression[211]. Thus, in CRC, an increase in the levels of bacterial cadaverine and putrescine in the feces was noted[212,213]. Huang et al[214] reported that increased spermine intake is associated with an increased risk of CRC, while a higher intake of total polyamine, putrescine and spermidine is significantly associated with a reduced risk of CRC. It is believed that the procarcinogenic effect of polyamines is associated with the activation of the PTEN-PI3K-mTOR (TORC1), WNT, and RAS pathways[211].
The gut microbiome can influence the biotransformation of a number of xenobiotics with known carcinogenic properties, such as heterocyclic amines (HCAs). HCAs are formed during thermal processing (frying, baking, grilling, etc.) of various food products, including oils, grains and vegetables, but especially processed meat[215]. HCAs have pronounced genotoxic and mutagenic properties, contributing to the development of malignant neoplasms of the intestine, liver, lungs, breast and other tumors. The carcinogenicity of HCAs is associated with mutations in proto-oncogenes and tumor suppressor genes, including K-ras, Haras, Apc, β-catenin and TP53[216]. The intestinal microbiota can metabolize HCAs into molecules with increased mutagenic activity[217]. At the same time, the intestinal microbiota can bind or metabolize food-derived HCAs, facilitating their excretion with feces or conversion into less toxic compounds[215,218]. These processes involve bacterial beta-glucuronidase (B-GUS) and glycerol/diol dehydratase (GDH) produced by some lactic acid bacteria and probiotics. A decrease in the number of taxa with B-GUS and GDH activity was noted in patients with CRC[219].
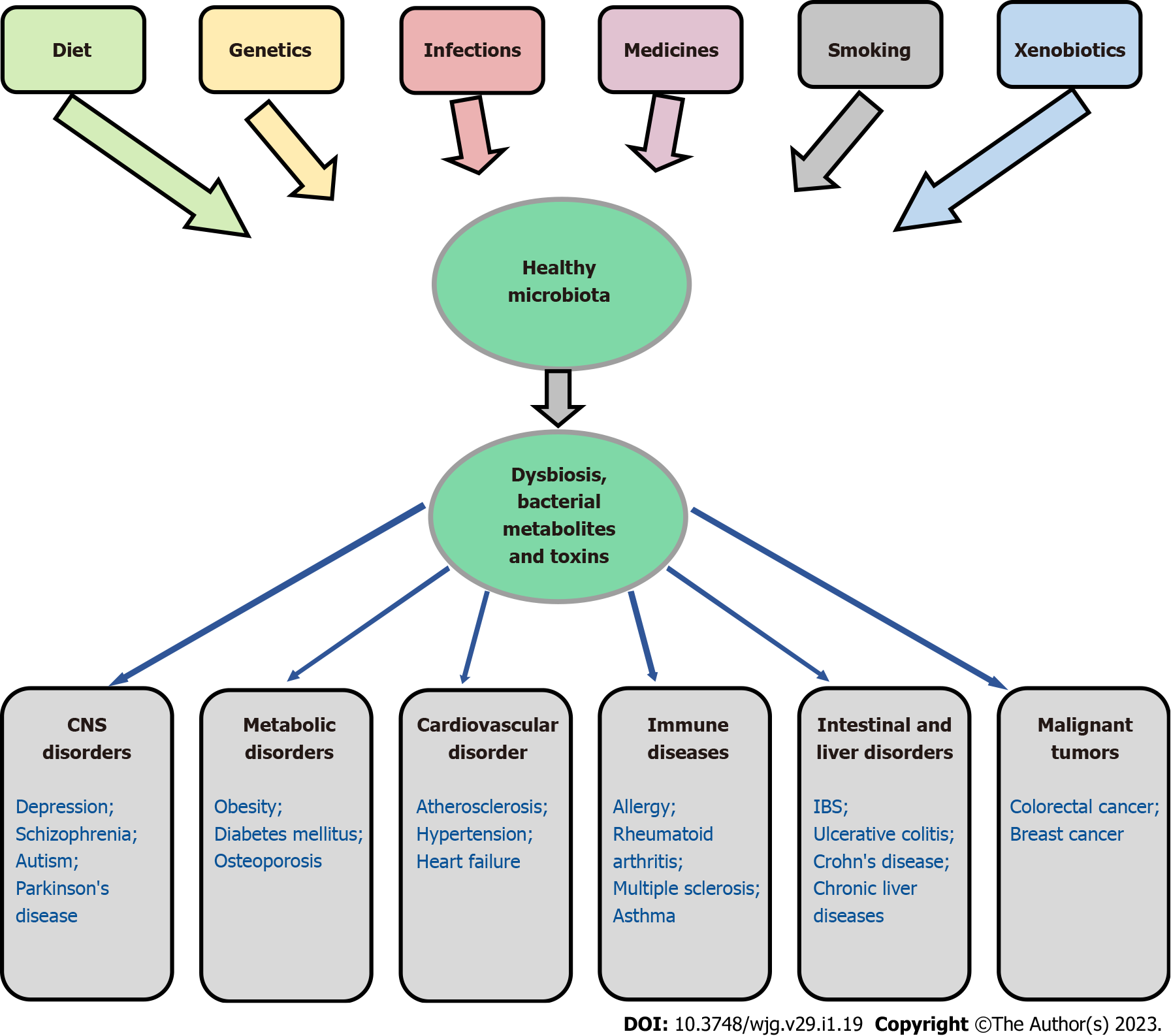
According to the results of numerous studies, in many diseases, including inflammatory bowel diseases[113,220-224], chronic liver diseases[225-227], obesity[228,229], diabetes mellitus[230,231], osteoporosis[41,232], cardiovascular[233,234] and oncological diseases[212,213,235], a decrease in the microbiota diversity and an overgrowth of pathogenic and conditionally pathogenic flora are observed[50,236,237]. Interestingly, in CRC, Fusobacterium nucleatum was found not only in the primary tumor but also in metastatic lesions in the liver[238] and lung[239]. In addition, it has been noted that dysbiosis to a certain extent can influence the development of depression, bipolar depression and schizophrenia[240], as well as autism[241] and Parkinson's disease[242,243]. Figure 1 demonstrates the association of gut dysbiosis with various human diseases.
It should be emphasized that the diseases associated with dysbacteriosis are multifactorial in nature[50,236,237,244] and are associated with bacterial invasion through the physical and chemical barriers of the gastrointestinal tract[50]. It is hypothesized that diet and other environmental influences may change the microbiome and thus provoke an unstable basis of genetic predisposition, which may lead to disease development, at least in some patients. Dysbiosis may be related to heredity, use of antibiotics, proton pump inhibitors, certain types of chemotherapy, advanced age, diet, and other factors[245,246]. At the same time, it should be noted that numerous studies have not revealed typical changes in the microbiome for a particular pathology[247]. A detailed analysis of host metabolism (metabolic index) and habitual diet (including the consumption of plant and animal foods, and fermented milk probiotics, such as yogurt) allowed Asnicar et al[248] to establish consistent gut microbiome signatures, segregating favourable and unfavourable taxa with multiple measures of both dietary intake and cardiometabolic health. However, we believe the issue cannot be considered definitively resolved, since it is not completely clear what is primary: A violation of the microbiome that then contributes to the development of diseases or disorder can cause changes in the microbiome. These questions require further research.
Considering that intestinal dysbiosis plays an important role in the development of a number of diseases, normalizing the microbiome seems to be a promising direction for their treatment. There are several approaches to solve this problem.
It is well known that a Mediterranean diet is rich in vegetables, fruits, whole grains and fish and thus creates favourable conditions for beneficial bacteria such as Firmicutes, which are involved in the production of butyrate necessary to maintain a healthy barrier between the colon and blood flow, preventing inflammation in the gut[64]. Unlike the Mediterranean diet, a “Western” high-fat, low-fiber diet contributes to colon inflammation and cancer[249,250].
Prebiotics are substrates that are selectively utilized by host microorganisms to provide health benefits[251]. The use of prebiotics, such as dietary fiber, reduces obesity and has anti-inflammatory and anticancer effects[85,250,252,253]. Dietary fiber intake is associated with a lower incidence of colon cancer, since fiber reduces the concentration of intestinal carcinogens due to increased stool mass, intestinal motility and production of butyrate, which maintain colonocyte health, increase apoptosis and inhibit cancer cell proliferation. A long-term fiber-rich diet increases the density of Firmicutes, which have the ability to mediate an immunomodulatory and anti-inflammatory immune response[64]. Moreover, dietary fiber physically interferes with fatty acid reabsorption and cholesterol absorption, thereby reducing the risk of obesity, diabetes, and atherosclerosis[254,255].
The mechanism of transplantation of fecal microbiota (FMT) is to restore the fermentation activity, pH and redox potential of the microbiome habitat in the respective niches and restore normal gas production and synthesis of SCFA[245,256]. FMT has been used in the treatment of nosocomial diarrhea caused by Clostridioides difficile[245] and other intestinal diseases[257-259]. Scibelli et al[260] demonstrated the effectiveness of using FMT in the treatment of rectal fistula in a patient with a colostomy who received intensive antibiotic therapy for a long time due to trauma. Impressive results have also been obtained in the treatment of Crohn's disease by FMT: nearly 60% of patients achieved a clinical response to treatment, and more than 20% of patients experienced sustained clinical remission, including 2 of 6 patients with perianal fistula[261]. The use of FMT has been shown to be beneficial in hepatic encephalitis, metabolic diseases, neuropsychiatric disorders, autoimmune diseases, allergic disorders, tumors, Parkinson's disease, multiple sclerosis, myoclonus dystonia, chronic fatigue syndrome, and idiopathic thrombocytopenic purpura[257,262-264].
The disadvantages of FMT are frequent side effects such as constipation, diarrhea, bloating and possible transmission of potential pathogens[245,265]. Given the enormous promise of microbiota transplantation, the search for new methods and ways to use it continues[266]. Attempts have been made to replenish only bacteria that had certain characteristics and whose number was reduced[245,267,268], but reproducibility and standardization of preparations used for transplantation have been proven to be a problem[267]. Zhang et al[269] first revealed that washed microbiota transplantation is safer, more precise and more quality-controllable than crude FMT by manual. Currently, there are a series of clinical trials conducted for SER-109, which is a consortium from several species of Firmicutes isolated from the stool of healthy human donors and encapsulated. The use of the medication reduced the risk of recurrence of nosocomial diarrhea caused by Clostridioides difficile from 41.3% in the placebo group to 11.1% in the treatment group[270].
Probiotics are live strains of carefully selected microorganisms that, when administered in adequate amounts, confer a health benefit on the host[271]. Probiotics can be easily and economically prepared and given to patients daily in the form of yogurt, drinks, cheese or capsules[272]. They contribute to the maintenance of intestinal barrier function, have immunomodulatory, metabolic and antiproliferative effects[273,274], and regulate DC maturation by producing tolerogenic DCs, which can reduce inflammation[275] and synthesize antimicrobial substances[276]. The ability of Bifidobacterium dentium and its secreted factors to suppress endoplasmic reticulum stress genes and promote MUC2 secretion, as well as to secrete the antioxidant γ-glutamylcysteine, which reduces the formation of ROS and suppresses NF-kB activation and IL-8 secretion, has been established[277].
Currently, Lactobacillus, Bifidobacterium and Saccharomyces are used to treat hospital-acquired diarrhea caused by Clostridioides difficile[278]. Oral use of Lactobacillus reuteri 6475 reduced the loss of total bone mineral density (BMD) in women aged 75 years to 80 years with low BMD[279]. Dietary supplementation with soluble corn fiber at doses of 10-20 g/d has also been associated with an increase in calcium absorption and a larger number of Clostridium and unclassified Clostridiaceae in feces[280]. These data suggest that both probiotics (live bacteria) and dietary supplements needed to feed the bacteria can be used as therapeutic agents to combat osteoporosis. It is possible that the use of their combination will be even more effective.
For therapeutic purposes, commensals (bacterial strains that are resistant to certain types of pathogenic bacteria), bacteriophages and fungi can be used. For example, CBM588 is a probiotic composed of the commensal Clostridium butyricum, which produces large amounts of butyrate and activates neutrophils and Th1 and Th17 cells[281]. Bacteriophages are viruses that can infect and multiply in pathogenic bacteria and eventually lyse them[245,282]. Yeasts such as Saccharomyces boulardii and Candida albicans, as well as fungal wall components such as β-glucans, can inhibit the growth of some enteric pathogens. Saccharomyces boulardii produces proteases or phosphatases that inactivate disease-causing toxins produced by gut bacteria and modulate multiple signaling pathways to suppress toxin-induced inflammation[245,283,284]. Because of the risk of fungemia, they should be used cautiously in debilitated patients[285].
The study of the influence of the gut microbiome on health and disease is one of the most relevant and interesting areas in modern science. The number of microorganisms inhabiting the human body is enormous, and their composition can vary significantly between individuals[248,286]. Dysbiosis has been found to play an important role in the development of a number of diseases, however, numerous studies have not revealed typical changes in the microbiome characteristic for a particular pathology[247]. It is likely that the impact of the microbiome and its metabolites on human health cannot be considered only in terms of health benefits or harms. Metabolites that perform important functions in the human body under certain conditions can have a negative impact on human health, while metabolites that are considered potentially dangerous can be beneficial[287].
As already noted, changes in the quantitative and qualitative microbiota composition can lead to an increase in the production of potentially toxic metabolites, such as secondary bile acids, TMAO and hydrogen sulfide, and an increase in the risk of developing intestinal, cardiovascular, neurological, oncological and other diseases. These changes may be related to diet, lifestyle, age, medications, and other factors. Thus, taking antibacterial drugs can increase sensitivity to viral infections[288], increase the risk of developing malignant neoplasms[289], and contribute to the development of resistance to chemotherapy drugs and immune checkpoint inhibitors in cancer patients[290]. Interestingly, some authors attribute an increase in the risk of malignant neoplasms with the use of antibiotics to a decrease in the synthesis of intestinal metabolites with antitumour activity, for example, LCA and cadaverine[289]. However, in some malignant neoplasms, the antitumour effect of antibacterial drugs was demonstrated. For example, in CRC, antibiotic therapy had a cytostatic effect due to the destruction of bacterial biofilms, the formation of which was associated with polyamines[291]. In an experiment, adding broad-spectrum antibiotics to drinking water for 3-4 wk reduced age-related oxidative stress and arterial dysfunction in mice[292]. It is believed that microbiota modulation using antibiotics, probiotics, fecal microbiota transplantation or nanotechnology can be effective in a variety of diseases, including enhancing the antitumour effect of chemotherapy drugs or immune checkpoint inhibitors[290].
It should be noted that despite the huge amount of research on the role of the microbiome in health and disease development, there are a number of issues that deserve further study. In particular, it is known that the microbiota affects the mood and behavior of a person, his physical activity, resistance to stress and diseases. At the same time, whether mental and physical health influence the human microbiome is not well understood. Many questions remain regarding the role of the gut microbiome in drug metabolism, the development of drug resistance and chemoresistance, and the role of the microbiome in cancer progression. Answers to these and other questions are still waiting for their researchers.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Russia
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): B, B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): E
P-Reviewer: Fu M, China; Łoniewski I, Poland; Luo ZW, China; Wan XH, China; Zhang F, China S-Editor: Gao CC L-Editor: A P-Editor: Gao CC
| 1. | Shreiner AB, Kao JY, Young VB. The gut microbiome in health and in disease. Curr Opin Gastroenterol. 2015;31:69-75. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 845] [Cited by in RCA: 1102] [Article Influence: 110.2] [Reference Citation Analysis (0)] |
| 2. | Milani C, Duranti S, Bottacini F, Casey E, Turroni F, Mahony J, Belzer C, Delgado Palacio S, Arboleya Montes S, Mancabelli L, Lugli GA, Rodriguez JM, Bode L, de Vos W, Gueimonde M, Margolles A, van Sinderen D, Ventura M. The First Microbial Colonizers of the Human Gut: Composition, Activities, and Health Implications of the Infant Gut Microbiota. Microbiol Mol Biol Rev. 2017;81. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1057] [Cited by in RCA: 1137] [Article Influence: 142.1] [Reference Citation Analysis (0)] |
| 3. | Barko PC, McMichael MA, Swanson KS, Williams DA. The Gastrointestinal Microbiome: A Review. J Vet Intern Med. 2018;32:9-25. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 329] [Cited by in RCA: 413] [Article Influence: 59.0] [Reference Citation Analysis (0)] |
| 4. | Fernández L, Langa S, Martín V, Maldonado A, Jiménez E, Martín R, Rodríguez JM. The human milk microbiota: origin and potential roles in health and disease. Pharmacol Res. 2013;69:1-10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 506] [Cited by in RCA: 533] [Article Influence: 41.0] [Reference Citation Analysis (0)] |
| 5. | Luo S, Zhu H, Zhang J, Wan D. The Pivotal Role of Microbiota in Modulating the Neuronal-Glial-Epithelial Unit. Infect Drug Resist. 2021;14:5613-5628. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 3] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 6. | Jha AR, Davenport ER, Gautam Y, Bhandari D, Tandukar S, Ng KM, Fragiadakis GK, Holmes S, Gautam GP, Leach J, Sherchand JB, Bustamante CD, Sonnenburg JL. Gut microbiome transition across a lifestyle gradient in Himalaya. PLoS Biol. 2018;16:e2005396. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 120] [Cited by in RCA: 118] [Article Influence: 16.9] [Reference Citation Analysis (0)] |
| 7. | Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, Knight R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A. 2010;107:11971-11975. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2922] [Cited by in RCA: 3164] [Article Influence: 210.9] [Reference Citation Analysis (0)] |
| 8. | Lundgren SN, Madan JC, Emond JA, Morrison HG, Christensen BC, Karagas MR, Hoen AG. Maternal diet during pregnancy is related with the infant stool microbiome in a delivery mode-dependent manner. Microbiome. 2018;6:109. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 130] [Cited by in RCA: 172] [Article Influence: 24.6] [Reference Citation Analysis (0)] |
| 9. | Yassour M, Vatanen T, Siljander H, Hämäläinen AM, Härkönen T, Ryhänen SJ, Franzosa EA, Vlamakis H, Huttenhower C, Gevers D, Lander ES, Knip M; DIABIMMUNE Study Group, Xavier RJ. Natural history of the infant gut microbiome and impact of antibiotic treatment on bacterial strain diversity and stability. Sci Transl Med. 2016;8:343ra81. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 553] [Cited by in RCA: 711] [Article Influence: 88.9] [Reference Citation Analysis (0)] |
| 10. | Lazar V, Ditu LM, Pircalabioru GG, Gheorghe I, Curutiu C, Holban AM, Picu A, Petcu L, Chifiriuc MC. Aspects of Gut Microbiota and Immune System Interactions in Infectious Diseases, Immunopathology, and Cancer. Front Immunol. 2018;9:1830. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 318] [Cited by in RCA: 341] [Article Influence: 48.7] [Reference Citation Analysis (0)] |
| 11. | Sender R, Fuchs S, Milo R. Revised Estimates for the Number of Human and Bacteria Cells in the Body. PLoS Biol. 2016;14:e1002533. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2328] [Cited by in RCA: 2907] [Article Influence: 323.0] [Reference Citation Analysis (0)] |
| 12. | Donaldson GP, Lee SM, Mazmanian SK. Gut biogeography of the bacterial microbiota. Nat Rev Microbiol. 2016;14:20-32. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1265] [Cited by in RCA: 1609] [Article Influence: 160.9] [Reference Citation Analysis (0)] |
| 13. | Ma J, Piao X, Mahfuz S, Long S, Wang J. The interaction among gut microbes, the intestinal barrier and short chain fatty acids. Anim Nutr. 2022;9:159-174. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 145] [Article Influence: 48.3] [Reference Citation Analysis (0)] |
| 14. | Chen Y, Wang X, Zhang C, Liu Z, Li C, Ren Z. Gut Microbiota and Bone Diseases: A Growing Partnership. Front Microbiol. 2022;13:877776. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 32] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 15. | Moszak M, Szulińska M, Bogdański P. You Are What You Eat-The Relationship between Diet, Microbiota, and Metabolic Disorders-A Review. Nutrients. 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 160] [Cited by in RCA: 224] [Article Influence: 44.8] [Reference Citation Analysis (0)] |
| 16. | Engevik MA, Luk B, Chang-Graham AL, Hall A, Herrmann B, Ruan W, Endres BT, Shi Z, Garey KW, Hyser JM, Versalovic J. Bifidobacterium dentium Fortifies the Intestinal Mucus Layer via Autophagy and Calcium Signaling Pathways. mBio. 2019;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 141] [Cited by in RCA: 174] [Article Influence: 29.0] [Reference Citation Analysis (0)] |
| 17. | Paone P, Cani PD. Mucus barrier, mucins and gut microbiota: the expected slimy partners? Gut. 2020;69:2232-2243. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 693] [Cited by in RCA: 933] [Article Influence: 186.6] [Reference Citation Analysis (0)] |
| 18. | Goto Y. Epithelial Cells as a Transmitter of Signals From Commensal Bacteria and Host Immune Cells. Front Immunol. 2019;10:2057. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 54] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 19. | Burgueño JF, Abreu MT. Epithelial Toll-like receptors and their role in gut homeostasis and disease. Nat Rev Gastroenterol Hepatol. 2020;17:263-278. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 144] [Cited by in RCA: 262] [Article Influence: 52.4] [Reference Citation Analysis (0)] |
| 20. | Gieryńska M, Szulc-Dąbrowska L, Struzik J, Mielcarska MB, Gregorczyk-Zboroch KP. Integrity of the Intestinal Barrier: The Involvement of Epithelial Cells and Microbiota-A Mutual Relationship. Animals (Basel). 2022;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 126] [Article Influence: 42.0] [Reference Citation Analysis (0)] |
| 21. | Gassler N. Paneth cells in intestinal physiology and pathophysiology. World J Gastrointest Pathophysiol. 2017;8:150-160. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 114] [Cited by in RCA: 98] [Article Influence: 12.3] [Reference Citation Analysis (2)] |
| 22. | Allaire JM, Crowley SM, Law HT, Chang SY, Ko HJ, Vallance BA. The Intestinal Epithelium: Central Coordinator of Mucosal Immunity. Trends Immunol. 2018;39:677-696. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 344] [Cited by in RCA: 595] [Article Influence: 85.0] [Reference Citation Analysis (0)] |
| 23. | Banerjee A, McKinley ET, von Moltke J, Coffey RJ, Lau KS. Interpreting heterogeneity in intestinal tuft cell structure and function. J Clin Invest. 2018;128:1711-1719. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 52] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 24. | Mabbott NA, Donaldson DS, Ohno H, Williams IR, Mahajan A. Microfold (M) cells: important immunosurveillance posts in the intestinal epithelium. Mucosal Immunol. 2013;6:666-677. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 423] [Cited by in RCA: 478] [Article Influence: 39.8] [Reference Citation Analysis (0)] |
| 25. | Turula H, Wobus CE. The Role of the Polymeric Immunoglobulin Receptor and Secretory Immunoglobulins during Mucosal Infection and Immunity. Viruses. 2018;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 127] [Cited by in RCA: 125] [Article Influence: 17.9] [Reference Citation Analysis (0)] |
| 26. | Knoop KA, Newberry RD. Goblet cells: multifaceted players in immunity at mucosal surfaces. Mucosal Immunol. 2018;11:1551-1557. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 140] [Cited by in RCA: 199] [Article Influence: 28.4] [Reference Citation Analysis (0)] |
| 27. | Kurashima Y, Tokuhara D, Kamioka M, Inagaki Y, Kiyono H. Intrinsic Control of Surface Immune and Epithelial Homeostasis by Tissue-Resident Gut Stromal Cells. Front Immunol. 2019;10:1281. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 9] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 28. | Yokoi Y, Nakamura K, Yoneda T, Kikuchi M, Sugimoto R, Shimizu Y, Ayabe T. Paneth cell granule dynamics on secretory responses to bacterial stimuli in enteroids. Sci Rep. 2019;9:2710. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 58] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 29. | Barreto E Barreto L, Rattes IC, da Costa AV, Gama P. Paneth cells and their multiple functions. Cell Biol Int. 2022;46:701-710. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 23] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 30. | Abokor AA, McDaniel GH, Golonka RM, Campbell C, Brahmandam S, Yeoh BS, Joe B, Vijay-Kumar M, Saha P. Immunoglobulin A, an Active Liaison for Host-Microbiota Homeostasis. Microorganisms. 2021;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 26] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 31. | Zhao Q, Maynard CL. Mucus, commensals, and the immune system. Gut Microbes. 2022;14:2041342. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 45] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 32. | Raya-Sandino A, Luissint AC, Kusters DHM, Narayanan V, Flemming S, Garcia-Hernandez V, Godsel LM, Green KJ, Hagen SJ, Conway DE, Parkos CA, Nusrat A. Regulation of intestinal epithelial intercellular adhesion and barrier function by desmosomal cadherin desmocollin-2. Mol Biol Cell. 2021;32:753-768. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 27] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 33. | Garcia MA, Nelson WJ, Chavez N. Cell-Cell Junctions Organize Structural and Signaling Networks. Cold Spring Harb Perspect Biol. 2018;10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 319] [Cited by in RCA: 325] [Article Influence: 46.4] [Reference Citation Analysis (1)] |
| 34. | Green KJ, Jaiganesh A, Broussard JA. Desmosomes: Essential contributors to an integrated intercellular junction network. F1000Res. 2019;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 64] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 35. | Schoenborn AA, von Furstenberg RJ, Valsaraj S, Hussain FS, Stein M, Shanahan MT, Henning SJ, Gulati AS. The enteric microbiota regulates jejunal Paneth cell number and function without impacting intestinal stem cells. Gut Microbes. 2019;10:45-58. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 46] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 36. | Paradis T, Bègue H, Basmaciyan L, Dalle F, Bon F. Tight Junctions as a Key for Pathogens Invasion in Intestinal Epithelial Cells. Int J Mol Sci. 2021;22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 146] [Article Influence: 36.5] [Reference Citation Analysis (0)] |
| 37. | Jackson DN, Theiss AL. Gut bacteria signaling to mitochondria in intestinal inflammation and cancer. Gut Microbes. 2020;11:285-304. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 96] [Cited by in RCA: 191] [Article Influence: 38.2] [Reference Citation Analysis (0)] |
| 38. | Haderer M, Neubert P, Rinner E, Scholtis A, Broncy L, Gschwendtner H, Kandulski A, Pavel V, Mehrl A, Brochhausen C, Schlosser S, Gülow K, Kunst C, Müller M. Novel pathomechanism for spontaneous bacterial peritonitis: disruption of cell junctions by cellular and bacterial proteases. Gut. 2022;71:580-592. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 32] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 39. | Bugge M, Bergstrom B, Eide OK, Solli H, Kjønstad IF, Stenvik J, Espevik T, Nilsen NJ. Surface Toll-like receptor 3 expression in metastatic intestinal epithelial cells induces inflammatory cytokine production and promotes invasiveness. J Biol Chem. 2017;292:15408-15425. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 32] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 40. | Hamonic G, Pasternak JA, Wilson HL. Recognizing conserved non-canonical localization patterns of toll-like receptors in tissues and across species. Cell Tissue Res. 2018;372:1-11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 17] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 41. | Bouskra D, Brézillon C, Bérard M, Werts C, Varona R, Boneca IG, Eberl G. Lymphoid tissue genesis induced by commensals through NOD1 regulates intestinal homeostasis. Nature. 2008;456:507-510. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 771] [Cited by in RCA: 804] [Article Influence: 47.3] [Reference Citation Analysis (0)] |
| 42. | Próchnicki T, Latz E. Inflammasomes on the Crossroads of Innate Immune Recognition and Metabolic Control. Cell Metab. 2017;26:71-93. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 190] [Cited by in RCA: 233] [Article Influence: 29.1] [Reference Citation Analysis (0)] |
| 43. | Uranga JA, Martínez V, Abalo R. Mast Cell Regulation and Irritable Bowel Syndrome: Effects of Food Components with Potential Nutraceutical Use. Molecules. 2020;25. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 39] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 44. | West AP, Brodsky IE, Rahner C, Woo DK, Erdjument-Bromage H, Tempst P, Walsh MC, Choi Y, Shadel GS, Ghosh S. TLR signalling augments macrophage bactericidal activity through mitochondrial ROS. Nature. 2011;472:476-480. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1023] [Cited by in RCA: 1267] [Article Influence: 90.5] [Reference Citation Analysis (0)] |
| 45. | Matarrese P, Falzano L, Fabbri A, Gambardella L, Frank C, Geny B, Popoff MR, Malorni W, Fiorentini C. Clostridium difficile toxin B causes apoptosis in epithelial cells by thrilling mitochondria. Involvement of ATP-sensitive mitochondrial potassium channels. J Biol Chem. 2007;282:9029-9041. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 94] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 46. | Rao R. Oxidative stress-induced disruption of epithelial and endothelial tight junctions. Front Biosci. 2008;13:7210-7226. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 215] [Cited by in RCA: 234] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 47. | Sardelli L, Pacheco DP, Ziccarelli A, Tunesi M, Caspani O, Fusari A, Briatico Vangosa F, Giordano C, Petrini P. Towards bioinspired in vitro models of intestinal mucus. RSC Adv. 2019;9:15887-15899. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 44] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 48. | Nyström EEL, Martinez-Abad B, Arike L, Birchenough GMH, Nonnecke EB, Castillo PA, Svensson F, Bevins CL, Hansson GC, Johansson MEV. An intercrypt subpopulation of goblet cells is essential for colonic mucus barrier function. Science. 2021;372. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 186] [Article Influence: 46.5] [Reference Citation Analysis (0)] |
| 49. | Herath M, Hosie S, Bornstein JC, Franks AE, Hill-Yardin EL. The Role of the Gastrointestinal Mucus System in Intestinal Homeostasis: Implications for Neurological Disorders. Front Cell Infect Microbiol. 2020;10:248. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 131] [Article Influence: 26.2] [Reference Citation Analysis (0)] |
| 50. | Inczefi O, Bacsur P, Resál T, Keresztes C, Molnár T. The Influence of Nutrition on Intestinal Permeability and the Microbiome in Health and Disease. Front Nutr. 2022;9:718710. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 48] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 51. | Johansson ME, Larsson JM, Hansson GC. The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host-microbial interactions. Proc Natl Acad Sci U S A. 2011;108 Suppl 1:4659-4665. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 848] [Cited by in RCA: 1012] [Article Influence: 67.5] [Reference Citation Analysis (0)] |
| 52. | Bergstrom K, Xia L. The barrier and beyond: Roles of intestinal mucus and mucin-type O-glycosylation in resistance and tolerance defense strategies guiding host-microbe symbiosis. Gut Microbes. 2022;14:2052699. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 38] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 53. | Ambort D, Johansson ME, Gustafsson JK, Nilsson HE, Ermund A, Johansson BR, Koeck PJ, Hebert H, Hansson GC. Calcium and pH-dependent packing and release of the gel-forming MUC2 mucin. Proc Natl Acad Sci U S A. 2012;109:5645-5650. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 218] [Cited by in RCA: 259] [Article Influence: 19.9] [Reference Citation Analysis (0)] |
| 54. | Bergstrom K, Shan X, Casero D, Batushansky A, Lagishetty V, Jacobs JP, Hoover C, Kondo Y, Shao B, Gao L, Zandberg W, Noyovitz B, McDaniel JM, Gibson DL, Pakpour S, Kazemian N, McGee S, Houchen CW, Rao CV, Griffin TM, Sonnenburg JL, McEver RP, Braun J, Xia L. Proximal colon-derived O-glycosylated mucus encapsulates and modulates the microbiota. Science. 2020;370:467-472. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 154] [Article Influence: 30.8] [Reference Citation Analysis (0)] |
| 55. | Van der Sluis M, De Koning BA, De Bruijn AC, Velcich A, Meijerink JP, Van Goudoever JB, Büller HA, Dekker J, Van Seuningen I, Renes IB, Einerhand AW. Muc2-deficient mice spontaneously develop colitis, indicating that MUC2 is critical for colonic protection. Gastroenterology. 2006;131:117-129. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1057] [Cited by in RCA: 1216] [Article Influence: 64.0] [Reference Citation Analysis (1)] |
| 56. | Velcich A, Yang W, Heyer J, Fragale A, Nicholas C, Viani S, Kucherlapati R, Lipkin M, Yang K, Augenlicht L. Colorectal cancer in mice genetically deficient in the mucin Muc2. Science. 2002;295:1726-1729. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 681] [Cited by in RCA: 716] [Article Influence: 31.1] [Reference Citation Analysis (0)] |
| 57. | Jakobsson HE, Rodríguez-Piñeiro AM, Schütte A, Ermund A, Boysen P, Bemark M, Sommer F, Bäckhed F, Hansson GC, Johansson ME. The composition of the gut microbiota shapes the colon mucus barrier. EMBO Rep. 2015;16:164-177. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 388] [Cited by in RCA: 501] [Article Influence: 45.5] [Reference Citation Analysis (0)] |
| 58. | Yao D, Dai W, Dong M, Dai C, Wu S. MUC2 and related bacterial factors: Therapeutic targets for ulcerative colitis. EBioMedicine. 2021;74:103751. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 137] [Article Influence: 34.3] [Reference Citation Analysis (1)] |
| 59. | Birchenough GM, Nyström EE, Johansson ME, Hansson GC. A sentinel goblet cell guards the colonic crypt by triggering Nlrp6-dependent Muc2 secretion. Science. 2016;352:1535-1542. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 413] [Cited by in RCA: 439] [Article Influence: 48.8] [Reference Citation Analysis (0)] |
| 60. | Cornick S, Kumar M, Moreau F, Gaisano H, Chadee K. VAMP8-mediated MUC2 mucin exocytosis from colonic goblet cells maintains innate intestinal homeostasis. Nat Commun. 2019;10:4306. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 66] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 61. | Okumura R, Kurakawa T, Nakano T, Kayama H, Kinoshita M, Motooka D, Gotoh K, Kimura T, Kamiyama N, Kusu T, Ueda Y, Wu H, Iijima H, Barman S, Osawa H, Matsuno H, Nishimura J, Ohba Y, Nakamura S, Iida T, Yamamoto M, Umemoto E, Sano K, Takeda K. Lypd8 promotes the segregation of flagellated microbiota and colonic epithelia. Nature. 2016;532:117-121. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 153] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 62. | Bergström JH, Birchenough GM, Katona G, Schroeder BO, Schütte A, Ermund A, Johansson ME, Hansson GC. Gram-positive bacteria are held at a distance in the colon mucus by the lectin-like protein ZG16. Proc Natl Acad Sci U S A. 2016;113:13833-13838. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 128] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 63. | Schroeder BO. Fight them or feed them: how the intestinal mucus layer manages the gut microbiota. Gastroenterol Rep (Oxf). 2019;7:3-12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 211] [Cited by in RCA: 307] [Article Influence: 51.2] [Reference Citation Analysis (0)] |
| 64. | Yang J, Yu J. The association of diet, gut microbiota and colorectal cancer: what we eat may imply what we get. Protein Cell. 2018;9:474-487. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 207] [Cited by in RCA: 203] [Article Influence: 29.0] [Reference Citation Analysis (0)] |
| 65. | Cheng HY, Ning MX, Chen DK, Ma WT. Interactions Between the Gut Microbiota and the Host Innate Immune Response Against Pathogens. Front Immunol. 2019;10:607. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 77] [Cited by in RCA: 146] [Article Influence: 24.3] [Reference Citation Analysis (0)] |
| 66. | Tyler CJ, McCarthy NE, Lindsay JO, Stagg AJ, Moser B, Eberl M. Antigen-Presenting Human γδ T Cells Promote Intestinal CD4(+) T Cell Expression of IL-22 and Mucosal Release of Calprotectin. J Immunol. 2017;198:3417-3425. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 45] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 67. | Abt MC, Buffie CG, Sušac B, Becattini S, Carter RA, Leiner I, Keith JW, Artis D, Osborne LC, Pamer EG. TLR-7 activation enhances IL-22-mediated colonization resistance against vancomycin-resistant enterococcus. Sci Transl Med. 2016;8:327ra25. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 78] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 68. | Semin I, Ninnemann J, Bondareva M, Gimaev I, Kruglov AA. Interplay Between Microbiota, Toll-Like Receptors and Cytokines for the Maintenance of Epithelial Barrier Integrity. Front Med (Lausanne). 2021;8:644333. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 26] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 69. | Price AE, Shamardani K, Lugo KA, Deguine J, Roberts AW, Lee BL, Barton GM. A Map of Toll-like Receptor Expression in the Intestinal Epithelium Reveals Distinct Spatial, Cell Type-Specific, and Temporal Patterns. Immunity. 2018;49:560-575.e6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 242] [Cited by in RCA: 253] [Article Influence: 36.1] [Reference Citation Analysis (0)] |
| 70. | Vaishnava S, Yamamoto M, Severson KM, Ruhn KA, Yu X, Koren O, Ley R, Wakeland EK, Hooper LV. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science. 2011;334:255-258. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1124] [Cited by in RCA: 1072] [Article Influence: 76.6] [Reference Citation Analysis (0)] |
| 71. | Nava GM, Friedrichsen HJ, Stappenbeck TS. Spatial organization of intestinal microbiota in the mouse ascending colon. ISME J. 2011;5:627-638. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 184] [Cited by in RCA: 201] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 72. | Cullen TW, Schofield WB, Barry NA, Putnam EE, Rundell EA, Trent MS, Degnan PH, Booth CJ, Yu H, Goodman AL. Gut microbiota. Antimicrobial peptide resistance mediates resilience of prominent gut commensals during inflammation. Science. 2015;347:170-175. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 263] [Cited by in RCA: 307] [Article Influence: 30.7] [Reference Citation Analysis (0)] |
| 73. | Garcia-Gutierrez E, Mayer MJ, Cotter PD, Narbad A. Gut microbiota as a source of novel antimicrobials. Gut Microbes. 2019;10:1-21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 140] [Cited by in RCA: 169] [Article Influence: 24.1] [Reference Citation Analysis (0)] |
| 74. | Umu ÖC, Bäuerl C, Oostindjer M, Pope PB, Hernández PE, Pérez-Martínez G, Diep DB. The Potential of Class II Bacteriocins to Modify Gut Microbiota to Improve Host Health. PLoS One. 2016;11:e0164036. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 74] [Cited by in RCA: 95] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 75. | McDole JR, Wheeler LW, McDonald KG, Wang B, Konjufca V, Knoop KA, Newberry RD, Miller MJ. Goblet cells deliver luminal antigen to CD103+ dendritic cells in the small intestine. Nature. 2012;483:345-349. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 597] [Cited by in RCA: 708] [Article Influence: 54.5] [Reference Citation Analysis (0)] |
| 76. | Mu Q, Kirby J, Reilly CM, Luo XM. Leaky Gut As a Danger Signal for Autoimmune Diseases. Front Immunol. 2017;8:598. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 266] [Cited by in RCA: 380] [Article Influence: 47.5] [Reference Citation Analysis (0)] |
| 77. | Sellin ME, Müller AA, Felmy B, Dolowschiak T, Diard M, Tardivel A, Maslowski KM, Hardt WD. Epithelium-intrinsic NAIP/NLRC4 inflammasome drives infected enterocyte expulsion to restrict Salmonella replication in the intestinal mucosa. Cell Host Microbe. 2014;16:237-248. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 253] [Cited by in RCA: 315] [Article Influence: 31.5] [Reference Citation Analysis (0)] |
| 78. | Mörbe UM, Jørgensen PB, Fenton TM, von Burg N, Riis LB, Spencer J, Agace WW. Human gut-associated lymphoid tissues (GALT); diversity, structure, and function. Mucosal Immunol. 2021;14:793-802. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 223] [Article Influence: 55.8] [Reference Citation Analysis (0)] |
| 79. | Dietert RR. Microbiome First Medicine in Health and Safety. Biomedicines. 2021;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 10] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 80. | Bhattacharya T, Ghosh TS, Mande SS. Global Profiling of Carbohydrate Active Enzymes in Human Gut Microbiome. PLoS One. 2015;10:e0142038. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 75] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 81. | Zafar H, Saier MH Jr. Gut Bacteroides species in health and disease. Gut Microbes. 2021;13:1-20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 510] [Cited by in RCA: 693] [Article Influence: 173.3] [Reference Citation Analysis (0)] |
| 82. | Sommer F, Bäckhed F. The gut microbiota--masters of host development and physiology. Nat Rev Microbiol. 2013;11:227-238. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2011] [Cited by in RCA: 2390] [Article Influence: 199.2] [Reference Citation Analysis (0)] |
| 83. | Shah T, Baloch Z, Shah Z, Cui X, Xia X. The Intestinal Microbiota: Impacts of Antibiotics Therapy, Colonization Resistance, and Diseases. Int J Mol Sci. 2021;22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 65] [Cited by in RCA: 49] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 84. | Schoeler M, Caesar R. Dietary lipids, gut microbiota and lipid metabolism. Rev Endocr Metab Disord. 2019;20:461-472. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 372] [Cited by in RCA: 734] [Article Influence: 122.3] [Reference Citation Analysis (0)] |
| 85. | Zeng H, Umar S, Rust B, Lazarova D, Bordonaro M. Secondary Bile Acids and Short Chain Fatty Acids in the Colon: A Focus on Colonic Microbiome, Cell Proliferation, Inflammation, and Cancer. Int J Mol Sci. 2019;20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 150] [Cited by in RCA: 318] [Article Influence: 53.0] [Reference Citation Analysis (0)] |
| 86. | Hernández MAG, Canfora EE, Jocken JWE, Blaak EE. The Short-Chain Fatty Acid Acetate in Body Weight Control and Insulin Sensitivity. Nutrients. 2019;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 162] [Cited by in RCA: 367] [Article Influence: 61.2] [Reference Citation Analysis (1)] |
| 87. | González-Bosch C, Boorman E, Zunszain PA, Mann GE. Short-chain fatty acids as modulators of redox signaling in health and disease. Redox Biol. 2021;47:102165. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 133] [Article Influence: 33.3] [Reference Citation Analysis (0)] |
| 88. | Martinez-Guryn K, Hubert N, Frazier K, Urlass S, Musch MW, Ojeda P, Pierre JF, Miyoshi J, Sontag TJ, Cham CM, Reardon CA, Leone V, Chang EB. Small Intestine Microbiota Regulate Host Digestive and Absorptive Adaptive Responses to Dietary Lipids. Cell Host Microbe. 2018;23:458-469.e5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 247] [Cited by in RCA: 424] [Article Influence: 60.6] [Reference Citation Analysis (0)] |
| 89. | Kim S, Covington A, Pamer EG. The intestinal microbiota: Antibiotics, colonization resistance, and enteric pathogens. Immunol Rev. 2017;279:90-105. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 508] [Cited by in RCA: 465] [Article Influence: 58.1] [Reference Citation Analysis (0)] |
| 90. | Chandrakesan P, Jakkula LU, Ahmed I, Roy B, Anant S, Umar S. Differential effects of β-catenin and NF-κB interplay in the regulation of cell proliferation, inflammation and tumorigenesis in response to bacterial infection. PLoS One. 2013;8:e79432. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 18] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 91. | Sommer F, Nookaew I, Sommer N, Fogelstrand P, Bäckhed F. Site-specific programming of the host epithelial transcriptome by the gut microbiota. Genome Biol. 2015;16:62. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 99] [Cited by in RCA: 118] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 92. | Frei R, Akdis M, O'Mahony L. Prebiotics, probiotics, synbiotics, and the immune system: experimental data and clinical evidence. Curr Opin Gastroenterol. 2015;31:153-158. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 150] [Cited by in RCA: 166] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 93. | Takiishi T, Fenero CIM, Câmara NOS. Intestinal barrier and gut microbiota: Shaping our immune responses throughout life. Tissue Barriers. 2017;5:e1373208. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 248] [Cited by in RCA: 620] [Article Influence: 77.5] [Reference Citation Analysis (1)] |
| 94. | Ticinesi A, Lauretani F, Tana C, Nouvenne A, Ridolo E, Meschi T. Exercise and immune system as modulators of intestinal microbiome: implications for the gut-muscle axis hypothesis. Exerc Immunol Rev. 2019;25:84-95. [PubMed] |
| 95. | Rowland I, Gibson G, Heinken A, Scott K, Swann J, Thiele I, Tuohy K. Gut microbiota functions: metabolism of nutrients and other food components. Eur J Nutr. 2018;57:1-24. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 993] [Cited by in RCA: 1594] [Article Influence: 199.3] [Reference Citation Analysis (0)] |
| 96. | Pan WH, Sommer F, Falk-Paulsen M, Ulas T, Best P, Fazio A, Kachroo P, Luzius A, Jentzsch M, Rehman A, Müller F, Lengauer T, Walter J, Künzel S, Baines JF, Schreiber S, Franke A, Schultze JL, Bäckhed F, Rosenstiel P. Exposure to the gut microbiota drives distinct methylome and transcriptome changes in intestinal epithelial cells during postnatal development. Genome Med. 2018;10:27. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 116] [Cited by in RCA: 112] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 97. | Huang R, Xiang J, Zhou P. Vitamin D, gut microbiota, and radiation-related resistance: a love-hate triangle. J Exp Clin Cancer Res. 2019;38:493. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 36] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 98. | Tian Y, Zuo L, Guo Q, Li J, Hu Z, Zhao K, Li C, Li X, Zhou J, Zhou Y, Li XA. Potential role of fecal microbiota in patients with constipation. Therap Adv Gastroenterol. 2020;13:1756284820968423. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 44] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 99. | Yang W, Yu T, Huang X, Bilotta AJ, Xu L, Lu Y, Sun J, Pan F, Zhou J, Zhang W, Yao S, Maynard CL, Singh N, Dann SM, Liu Z, Cong Y. Intestinal microbiota-derived short-chain fatty acids regulation of immune cell IL-22 production and gut immunity. Nat Commun. 2020;11:4457. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 415] [Cited by in RCA: 642] [Article Influence: 128.4] [Reference Citation Analysis (0)] |
| 100. | He J, Zhang P, Shen L, Niu L, Tan Y, Chen L, Zhao Y, Bai L, Hao X, Li X, Zhang S, Zhu L. Short-Chain Fatty Acids and Their Association with Signalling Pathways in Inflammation, Glucose and Lipid Metabolism. Int J Mol Sci. 2020;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 127] [Cited by in RCA: 535] [Article Influence: 107.0] [Reference Citation Analysis (0)] |
| 101. | Raveschot C, Coutte F, Frémont M, Vaeremans M, Dugersuren J, Demberel S, Drider D, Dhulster P, Flahaut C, Cudennec B. Probiotic Lactobacillus strains from Mongolia improve calcium transport and uptake by intestinal cells in vitro. Food Res Int. 2020;133:109201. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 42] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 102. | Lucas S, Omata Y, Hofmann J, Böttcher M, Iljazovic A, Sarter K, Albrecht O, Schulz O, Krishnacoumar B, Krönke G, Herrmann M, Mougiakakos D, Strowig T, Schett G, Zaiss MM. Short-chain fatty acids regulate systemic bone mass and protect from pathological bone loss. Nat Commun. 2018;9:55. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 243] [Cited by in RCA: 466] [Article Influence: 66.6] [Reference Citation Analysis (0)] |
| 103. | Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, Fukuda S, Saito T, Narushima S, Hase K, Kim S, Fritz JV, Wilmes P, Ueha S, Matsushima K, Ohno H, Olle B, Sakaguchi S, Taniguchi T, Morita H, Hattori M, Honda K. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500:232-236. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1875] [Cited by in RCA: 2134] [Article Influence: 177.8] [Reference Citation Analysis (2)] |
| 104. | Ye X, Wang R, Bhattacharya R, Boulbes DR, Fan F, Xia L, Adoni H, Ajami NJ, Wong MC, Smith DP, Petrosino JF, Venable S, Qiao W, Baladandayuthapani V, Maru D, Ellis LM. Fusobacterium Nucleatum Subspecies Animalis Influences Proinflammatory Cytokine Expression and Monocyte Activation in Human Colorectal Tumors. Cancer Prev Res (Phila). 2017;10:398-409. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 110] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 105. | Silva YP, Bernardi A, Frozza RL. The Role of Short-Chain Fatty Acids From Gut Microbiota in Gut-Brain Communication. Front Endocrinol (Lausanne). 2020;11:25. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 780] [Cited by in RCA: 1582] [Article Influence: 316.4] [Reference Citation Analysis (0)] |
| 106. | Mirzaei R, Bouzari B, Hosseini-Fard SR, Mazaheri M, Ahmadyousefi Y, Abdi M, Jalalifar S, Karimitabar Z, Teimoori A, Keyvani H, Zamani F, Yousefimashouf R, Karampoor S. Role of microbiota-derived short-chain fatty acids in nervous system disorders. Biomed Pharmacother. 2021;139:111661. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 177] [Cited by in RCA: 158] [Article Influence: 39.5] [Reference Citation Analysis (0)] |
| 107. | Trzeciak P, Herbet M. Role of the Intestinal Microbiome, Intestinal Barrier and Psychobiotics in Depression. Nutrients. 2021;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 79] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 108. | Neufeld KM, Kang N, Bienenstock J, Foster JA. Reduced anxiety-like behavior and central neurochemical change in germ-free mice. Neurogastroenterol Motil. 2011;23:255-264, e119. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 887] [Cited by in RCA: 931] [Article Influence: 66.5] [Reference Citation Analysis (0)] |
| 109. | Braniste V, Al-Asmakh M, Kowal C, Anuar F, Abbaspour A, Tóth M, Korecka A, Bakocevic N, Ng LG, Kundu P, Gulyás B, Halldin C, Hultenby K, Nilsson H, Hebert H, Volpe BT, Diamond B, Pettersson S. The gut microbiota influences blood-brain barrier permeability in mice. Sci Transl Med. 2014;6:263ra158. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1139] [Cited by in RCA: 1692] [Article Influence: 169.2] [Reference Citation Analysis (0)] |
| 110. | Ratajczak W, Rył A, Mizerski A, Walczakiewicz K, Sipak O, Laszczyńska M. Immunomodulatory potential of gut microbiome-derived short-chain fatty acids (SCFAs). Acta Biochim Pol. 2019;66:1-12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 202] [Article Influence: 33.7] [Reference Citation Analysis (0)] |
| 111. | Vich Vila A, Collij V, Sanna S, Sinha T, Imhann F, Bourgonje AR, Mujagic Z, Jonkers DMAE, Masclee AAM, Fu J, Kurilshikov A, Wijmenga C, Zhernakova A, Weersma RK. Impact of commonly used drugs on the composition and metabolic function of the gut microbiota. Nat Commun. 2020;11:362. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 228] [Cited by in RCA: 470] [Article Influence: 94.0] [Reference Citation Analysis (0)] |
| 112. | Feng W, Liu J, Ao H, Yue S, Peng C. Targeting gut microbiota for precision medicine: Focusing on the efficacy and toxicity of drugs. Theranostics. 2020;10:11278-11301. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 70] [Article Influence: 14.0] [Reference Citation Analysis (1)] |
| 113. | Mars RAT, Yang Y, Ward T, Houtti M, Priya S, Lekatz HR, Tang X, Sun Z, Kalari KR, Korem T, Bhattarai Y, Zheng T, Bar N, Frost G, Johnson AJ, van Treuren W, Han S, Ordog T, Grover M, Sonnenburg J, D'Amato M, Camilleri M, Elinav E, Segal E, Blekhman R, Farrugia G, Swann JR, Knights D, Kashyap PC. Longitudinal Multi-omics Reveals Subset-Specific Mechanisms Underlying Irritable Bowel Syndrome. Cell. 2020;183:1137-1140. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 45] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 114. | Ait-Belgnaoui A, Colom A, Braniste V, Ramalho L, Marrot A, Cartier C, Houdeau E, Theodorou V, Tompkins T. Probiotic gut effect prevents the chronic psychological stress-induced brain activity abnormality in mice. Neurogastroenterol Motil. 2014;26:510-520. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 257] [Cited by in RCA: 311] [Article Influence: 28.3] [Reference Citation Analysis (0)] |
| 115. | Duncan SH, Barcenilla A, Stewart CS, Pryde SE, Flint HJ. Acetate utilization and butyryl coenzyme A (CoA):acetate-CoA transferase in butyrate-producing bacteria from the human large intestine. Appl Environ Microbiol. 2002;68:5186-5190. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 500] [Cited by in RCA: 506] [Article Influence: 22.0] [Reference Citation Analysis (0)] |
| 116. | Vital M, Howe AC, Tiedje JM. Revealing the bacterial butyrate synthesis pathways by analyzing (meta)genomic data. mBio. 2014;5:e00889. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 562] [Cited by in RCA: 822] [Article Influence: 74.7] [Reference Citation Analysis (1)] |
| 117. | O'Keefe SJ. Diet, microorganisms and their metabolites, and colon cancer. Nat Rev Gastroenterol Hepatol. 2016;13:691-706. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 521] [Cited by in RCA: 768] [Article Influence: 85.3] [Reference Citation Analysis (0)] |
| 118. | Perego S, Sansoni V, Banfi G, Lombardi G. Sodium butyrate has anti-proliferative, pro-differentiating, and immunomodulatory effects in osteosarcoma cells and counteracts the TNFα-induced low-grade inflammation. Int J Immunopathol Pharmacol. 2018;32:394632017752240. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 21] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 119. | Melo AD, Silveira H, Bortoluzzi C, Lara LJ, Garbossa CA, Preis G, Costa LB, Rostagno MH. Intestinal alkaline phosphatase and sodium butyrate may be beneficial in attenuating LPS-induced intestinal inflammation. Genet Mol Res. 2016;15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 20] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 120. | Simeoli R, Mattace Raso G, Pirozzi C, Lama A, Santoro A, Russo R, Montero-Melendez T, Berni Canani R, Calignano A, Perretti M, Meli R. An orally administered butyrate-releasing derivative reduces neutrophil recruitment and inflammation in dextran sulphate sodium-induced murine colitis. Br J Pharmacol. 2017;174:1484-1496. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 98] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 121. | Zhang H, Du M, Yang Q, Zhu MJ. Butyrate suppresses murine mast cell proliferation and cytokine production through inhibiting histone deacetylase. J Nutr Biochem. 2016;27:299-306. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 54] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 122. | Matheus VA, Monteiro L, Oliveira RB, Maschio DA, Collares-Buzato CB. Butyrate reduces high-fat diet-induced metabolic alterations, hepatic steatosis and pancreatic beta cell and intestinal barrier dysfunctions in prediabetic mice. Exp Biol Med (Maywood). 2017;242:1214-1226. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 116] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 123. | Gershon MD, Tack J. The serotonin signaling system: from basic understanding to drug development for functional GI disorders. Gastroenterology. 2007;132:397-414. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 988] [Cited by in RCA: 1125] [Article Influence: 62.5] [Reference Citation Analysis (0)] |
| 124. | Arnoldussen IAC, Wiesmann M, Pelgrim CE, Wielemaker EM, van Duyvenvoorde W, Amaral-Santos PL, Verschuren L, Keijser BJF, Heerschap A, Kleemann R, Wielinga PY, Kiliaan AJ. Butyrate restores HFD-induced adaptations in brain function and metabolism in mid-adult obese mice. Int J Obes (Lond). 2017;41:935-944. [PubMed] [DOI] [Full Text] |
| 125. | Wang HB, Wang PY, Wang X, Wan YL, Liu YC. Butyrate enhances intestinal epithelial barrier function via up-regulation of tight junction protein Claudin-1 transcription. Dig Dis Sci. 2012;57:3126-3135. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 375] [Cited by in RCA: 549] [Article Influence: 42.2] [Reference Citation Analysis (0)] |
| 126. | Huang C, Song P, Fan P, Hou C, Thacker P, Ma X. Dietary Sodium Butyrate Decreases Postweaning Diarrhea by Modulating Intestinal Permeability and Changing the Bacterial Communities in Weaned Piglets. J Nutr. 2015;145:2774-2780. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 155] [Cited by in RCA: 194] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 127. | Zeng H, Briske-Anderson M. Prolonged butyrate treatment inhibits the migration and invasion potential of HT1080 tumor cells. J Nutr. 2005;135:291-295. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 51] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 128. | Ragsdale SW, Pierce E. Acetogenesis and the Wood-Ljungdahl pathway of CO(2) fixation. Biochim Biophys Acta. 2008;1784:1873-1898. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1030] [Cited by in RCA: 779] [Article Influence: 45.8] [Reference Citation Analysis (0)] |
| 129. | Jugder BE, Kamareddine L, Watnick PI. Microbiota-derived acetate activates intestinal innate immunity via the Tip60 histone acetyltransferase complex. Immunity. 2021;54:1683-1697.e3. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 49] [Article Influence: 12.3] [Reference Citation Analysis (1)] |
| 130. | Okamoto T, Morino K, Ugi S, Nakagawa F, Lemecha M, Ida S, Ohashi N, Sato D, Fujita Y, Maegawa H. Microbiome potentiates endurance exercise through intestinal acetate production. Am J Physiol Endocrinol Metab. 2019;316:E956-E966. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 144] [Article Influence: 24.0] [Reference Citation Analysis (0)] |
| 131. | Zheng H, Xu P, Jiang Q, Xu Q, Zheng Y, Yan J, Ji H, Ning J, Zhang X, Li C, Zhang L, Li Y, Li X, Song W, Gao H. Depletion of acetate-producing bacteria from the gut microbiota facilitates cognitive impairment through the gut-brain neural mechanism in diabetic mice. Microbiome. 2021;9:145. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 98] [Article Influence: 24.5] [Reference Citation Analysis (0)] |
| 132. | Galuppo B, Cline G, Van Name M, Shabanova V, Wagner D, Kien CL, Santoro N. Colonic Fermentation and Acetate Production in Youth with and without Obesity. J Nutr. 2021;151:3292-3298. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 3] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 133. | Hetzel M, Arslandemir C, König HH, Buck AK, Nüssle K, Glatting G, Gabelmann A, Hetzel J, Hombach V, Schirrmeister H. F-18 NaF PET for detection of bone metastases in lung cancer: accuracy, cost-effectiveness, and impact on patient management. J Bone Miner Res. 2003;18:2206-2214. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 141] [Cited by in RCA: 109] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 134. | Koh A, De Vadder F, Kovatcheva-Datchary P, Bäckhed F. From Dietary Fiber to Host Physiology: Short-Chain Fatty Acids as Key Bacterial Metabolites. Cell. 2016;165:1332-1345. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2533] [Cited by in RCA: 4170] [Article Influence: 463.3] [Reference Citation Analysis (0)] |
| 135. | Tong LC, Wang Y, Wang ZB, Liu WY, Sun S, Li L, Su DF, Zhang LC. Propionate Ameliorates Dextran Sodium Sulfate-Induced Colitis by Improving Intestinal Barrier Function and Reducing Inflammation and Oxidative Stress. Front Pharmacol. 2016;7:253. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 129] [Cited by in RCA: 227] [Article Influence: 25.2] [Reference Citation Analysis (0)] |
| 136. | Huang T, Shi H, Xu Y, Ji L. The gut microbiota metabolite propionate ameliorates intestinal epithelial barrier dysfunction-mediated Parkinson's disease via the AKT signaling pathway. Neuroreport. 2021;32:244-251. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 22] [Article Influence: 5.5] [Reference Citation Analysis (1)] |
| 137. | Bajic D, Niemann A, Hillmer AK, Mejias-Luque R, Bluemel S, Docampo M, Funk MC, Tonin E, Boutros M, Schnabl B, Busch DH, Miki T, Schmid RM, van den Brink MRM, Gerhard M, Stein-Thoeringer CK. Gut Microbiota-Derived Propionate Regulates the Expression of Reg3 Mucosal Lectins and Ameliorates Experimental Colitis in Mice. J Crohns Colitis. 2020;14:1462-1472. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 80] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 138. | Chambers ES, Byrne CS, Morrison DJ, Murphy KG, Preston T, Tedford C, Garcia-Perez I, Fountana S, Serrano-Contreras JI, Holmes E, Reynolds CJ, Roberts JF, Boyton RJ, Altmann DM, McDonald JAK, Marchesi JR, Akbar AN, Riddell NE, Wallis GA, Frost GS. Dietary supplementation with inulin-propionate ester or inulin improves insulin sensitivity in adults with overweight and obesity with distinct effects on the gut microbiota, plasma metabolome and systemic inflammatory responses: a randomised cross-over trial. Gut. 2019;68:1430-1438. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 174] [Cited by in RCA: 283] [Article Influence: 47.2] [Reference Citation Analysis (0)] |
| 139. | Dupraz L, Magniez A, Rolhion N, Richard ML, Da Costa G, Touch S, Mayeur C, Planchais J, Agus A, Danne C, Michaudel C, Spatz M, Trottein F, Langella P, Sokol H, Michel ML. Gut microbiota-derived short-chain fatty acids regulate IL-17 production by mouse and human intestinal γδ T cells. Cell Rep. 2021;36:109332. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 170] [Article Influence: 42.5] [Reference Citation Analysis (0)] |
| 140. | Haghikia A, Zimmermann F, Schumann P, Jasina A, Roessler J, Schmidt D, Heinze P, Kaisler J, Nageswaran V, Aigner A, Ceglarek U, Cineus R, Hegazy AN, van der Vorst EPC, Döring Y, Strauch CM, Nemet I, Tremaroli V, Dwibedi C, Kränkel N, Leistner DM, Heimesaat MM, Bereswill S, Rauch G, Seeland U, Soehnlein O, Müller DN, Gold R, Bäckhed F, Hazen SL, Haghikia A, Landmesser U. Propionate attenuates atherosclerosis by immune-dependent regulation of intestinal cholesterol metabolism. Eur Heart J. 2022;43:518-533. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 187] [Article Influence: 46.8] [Reference Citation Analysis (0)] |
| 141. | Scheiman J, Luber JM, Chavkin TA, MacDonald T, Tung A, Pham LD, Wibowo MC, Wurth RC, Punthambaker S, Tierney BT, Yang Z, Hattab MW, Avila-Pacheco J, Clish CB, Lessard S, Church GM, Kostic AD. Meta-omics analysis of elite athletes identifies a performance-enhancing microbe that functions via lactate metabolism. Nat Med. 2019;25:1104-1109. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 290] [Cited by in RCA: 519] [Article Influence: 86.5] [Reference Citation Analysis (0)] |
| 142. | Zhang L, Chu J, Hao W, Zhang J, Li H, Yang C, Yang J, Chen X, Wang H. Gut Microbiota and Type 2 Diabetes Mellitus: Association, Mechanism, and Translational Applications. Mediators Inflamm. 2021;2021:5110276. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 78] [Article Influence: 19.5] [Reference Citation Analysis (0)] |
| 143. | Jacobson A, Lam L, Rajendram M, Tamburini F, Honeycutt J, Pham T, Van Treuren W, Pruss K, Stabler SR, Lugo K, Bouley DM, Vilches-Moure JG, Smith M, Sonnenburg JL, Bhatt AS, Huang KC, Monack D. A Gut Commensal-Produced Metabolite Mediates Colonization Resistance to Salmonella Infection. Cell Host Microbe. 2018;24:296-307.e7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 334] [Cited by in RCA: 338] [Article Influence: 48.3] [Reference Citation Analysis (0)] |
| 144. | Chen LM, Bao CH, Wu Y, Liang SH, Wang D, Wu LY, Huang Y, Liu HR, Wu HG. Tryptophan-kynurenine metabolism: a link between the gut and brain for depression in inflammatory bowel disease. J Neuroinflammation. 2021;18:135. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 113] [Article Influence: 28.3] [Reference Citation Analysis (0)] |
| 145. | Clarke G, Grenham S, Scully P, Fitzgerald P, Moloney RD, Shanahan F, Dinan TG, Cryan JF. The microbiome-gut-brain axis during early life regulates the hippocampal serotonergic system in a sex-dependent manner. Mol Psychiatry. 2013;18:666-673. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1121] [Cited by in RCA: 1314] [Article Influence: 109.5] [Reference Citation Analysis (0)] |
| 146. | Marx W, McGuinness AJ, Rocks T, Ruusunen A, Cleminson J, Walker AJ, Gomes-da-Costa S, Lane M, Sanches M, Diaz AP, Tseng PT, Lin PY, Berk M, Clarke G, O'Neil A, Jacka F, Stubbs B, Carvalho AF, Quevedo J, Soares JC, Fernandes BS. The kynurenine pathway in major depressive disorder, bipolar disorder, and schizophrenia: a meta-analysis of 101 studies. Mol Psychiatry. 2021;26:4158-4178. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 178] [Article Influence: 44.5] [Reference Citation Analysis (0)] |
| 147. | Dehhaghi M, Kazemi Shariat Panahi H, Guillemin GJ. Microorganisms, Tryptophan Metabolism, and Kynurenine Pathway: A Complex Interconnected Loop Influencing Human Health Status. Int J Tryptophan Res. 2019;12:1178646919852996. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 84] [Cited by in RCA: 152] [Article Influence: 25.3] [Reference Citation Analysis (0)] |
| 148. | Jamshed L, Debnath A, Jamshed S, Wish JV, Raine JC, Tomy GT, Thomas PJ, Holloway AC. An Emerging Cross-Species Marker for Organismal Health: Tryptophan-Kynurenine Pathway. Int J Mol Sci. 2022;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 26] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 149. | Cervenka I, Agudelo LZ, Ruas JL. Kynurenines: Tryptophan's metabolites in exercise, inflammation, and mental health. Science. 2017;357. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 489] [Cited by in RCA: 881] [Article Influence: 125.9] [Reference Citation Analysis (0)] |
| 150. | Hajsl M, Hlavackova A, Broulikova K, Sramek M, Maly M, Dyr JE, Suttnar J. Tryptophan Metabolism, Inflammation, and Oxidative Stress in Patients with Neurovascular Disease. Metabolites. 2020;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 57] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 151. | Bhattarai Y, Williams BB, Battaglioli EJ, Whitaker WR, Till L, Grover M, Linden DR, Akiba Y, Kandimalla KK, Zachos NC, Kaunitz JD, Sonnenburg JL, Fischbach MA, Farrugia G, Kashyap PC. Gut Microbiota-Produced Tryptamine Activates an Epithelial G-Protein-Coupled Receptor to Increase Colonic Secretion. Cell Host Microbe. 2018;23:775-785.e5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 153] [Cited by in RCA: 289] [Article Influence: 48.2] [Reference Citation Analysis (0)] |
| 152. | Konopelski P, Ufnal M. Indoles - Gut Bacteria Metabolites of Tryptophan with Pharmacotherapeutic Potential. Curr Drug Metab. 2018;19:883-890. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 64] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 153. | Ridlon JM, Harris SC, Bhowmik S, Kang DJ, Hylemon PB. Consequences of bile salt biotransformations by intestinal bacteria. Gut Microbes. 2016;7:22-39. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 532] [Cited by in RCA: 733] [Article Influence: 81.4] [Reference Citation Analysis (0)] |
| 154. | Jia W, Xie G, Jia W. Bile acid-microbiota crosstalk in gastrointestinal inflammation and carcinogenesis. Nat Rev Gastroenterol Hepatol. 2018;15:111-128. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1355] [Cited by in RCA: 1240] [Article Influence: 177.1] [Reference Citation Analysis (0)] |
| 155. | Fang Y, Yan C, Zhao Q, Xu J, Liu Z, Gao J, Zhu H, Dai Z, Wang D, Tang D. The roles of microbial products in the development of colorectal cancer: a review. Bioengineered. 2021;12:720-735. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 83] [Cited by in RCA: 78] [Article Influence: 19.5] [Reference Citation Analysis (0)] |
| 156. | Cao W, Tian W, Hong J, Li D, Tavares R, Noble L, Moss SF, Resnick MB. Expression of bile acid receptor TGR5 in gastric adenocarcinoma. Am J Physiol Gastrointest Liver Physiol. 2013;304:G322-G327. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 69] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 157. | Vaughn BP, Kaiser T, Staley C, Hamilton MJ, Reich J, Graiziger C, Singroy S, Kabage AJ, Sadowsky MJ, Khoruts A. A pilot study of fecal bile acid and microbiota profiles in inflammatory bowel disease and primary sclerosing cholangitis. Clin Exp Gastroenterol. 2019;12:9-19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 66] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 158. | Liang H, Estes MK, Zhang H, Du G, Zhou Y. Bile acids target proteolipid nano-assemblies of EGFR and phosphatidic acid in the plasma membrane for stimulation of MAPK signaling. PLoS One. 2018;13:e0198983. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 10] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 159. | Shant J, Cheng K, Marasa BS, Wang JY, Raufman JP. Akt-dependent NF-kappaB activation is required for bile acids to rescue colon cancer cells from stress-induced apoptosis. Exp Cell Res. 2009;315:432-450. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 43] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 160. | Pai R, Tarnawski AS, Tran T. Deoxycholic acid activates beta-catenin signaling pathway and increases colon cell cancer growth and invasiveness. Mol Biol Cell. 2004;15:2156-2163. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 143] [Cited by in RCA: 139] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 161. | Cheng K, Raufman JP. Bile acid-induced proliferation of a human colon cancer cell line is mediated by transactivation of epidermal growth factor receptors. Biochem Pharmacol. 2005;70:1035-1047. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 116] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 162. | Ward JBJ, Lajczak NK, Kelly OB, O'Dwyer AM, Giddam AK, Ní Gabhann J, Franco P, Tambuwala MM, Jefferies CA, Keely S, Roda A, Keely SJ. Ursodeoxycholic acid and lithocholic acid exert anti-inflammatory actions in the colon. Am J Physiol Gastrointest Liver Physiol. 2017;312:G550-G558. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117] [Cited by in RCA: 190] [Article Influence: 23.8] [Reference Citation Analysis (0)] |
| 163. | West AC, Jenkins BJ. Inflammatory and non-inflammatory roles for Toll-like receptors in gastrointestinal cancer. Curr Pharm Des. 2015;21:2968-2977. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 18] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 164. | Mikó E, Vida A, Kovács T, Ujlaki G, Trencsényi G, Márton J, Sári Z, Kovács P, Boratkó A, Hujber Z, Csonka T, Antal-Szalmás P, Watanabe M, Gombos I, Csoka B, Kiss B, Vígh L, Szabó J, Méhes G, Sebestyén A, Goedert JJ, Bai P. Lithocholic acid, a bacterial metabolite reduces breast cancer cell proliferation and aggressiveness. Biochim Biophys Acta Bioenerg. 2018;1859:958-974. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 138] [Article Influence: 19.7] [Reference Citation Analysis (0)] |
| 165. | Kim EK, Cho JH, Kim E, Kim YJ. Ursodeoxycholic acid inhibits the proliferation of colon cancer cells by regulating oxidative stress and cancer stem-like cell growth. PLoS One. 2017;12:e0181183. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 43] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 166. | Pearson T, Caporaso JG, Yellowhair M, Bokulich NA, Padi M, Roe DJ, Wertheim BC, Linhart M, Martinez JA, Bilagody C, Hornstra H, Alberts DS, Lance P, Thompson PA. Effects of ursodeoxycholic acid on the gut microbiome and colorectal adenoma development. Cancer Med. 2019;8:617-628. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 81] [Cited by in RCA: 72] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 167. | Carlström M, Moretti CH, Weitzberg E, Lundberg JO. Microbiota, diet and the generation of reactive nitrogen compounds. Free Radic Biol Med. 2020;161:321-325. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 22] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 168. | Gatarek P, Kaluzna-Czaplinska J. Trimethylamine N-oxide (TMAO) in human health. EXCLI J. 2021;20:301-319. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 77] [Reference Citation Analysis (0)] |
| 169. | Janeiro MH, Ramírez MJ, Milagro FI, Martínez JA, Solas M. Implication of Trimethylamine N-Oxide (TMAO) in Disease: Potential Biomarker or New Therapeutic Target. Nutrients. 2018;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 257] [Cited by in RCA: 436] [Article Influence: 62.3] [Reference Citation Analysis (0)] |
| 170. | Chhibber-Goel J, Gaur A, Singhal V, Parakh N, Bhargava B, Sharma A. The complex metabolism of trimethylamine in humans: endogenous and exogenous sources. Expert Rev Mol Med. 2016;18:e8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 63] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 171. | Senchukova M, Tomchuk O, Shurygina E, Letuta S, Alidzhanov E, Nikiyan H, Razdobreev D. Calcium Carbonate Nanoparticles Can Activate the Epithelial⁻Mesenchymal Transition in an Experimental Gastric Cancer Model. Biomedicines. 2019;7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 2] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 172. | Millard HR, Musani SK, Dibaba DT, Talegawkar SA, Taylor HA, Tucker KL, Bidulescu A. Dietary choline and betaine; associations with subclinical markers of cardiovascular disease risk and incidence of CVD, coronary heart disease and stroke: the Jackson Heart Study. Eur J Nutr. 2018;57:51-60. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 73] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 173. | Senthong V, Wang Z, Li XS, Fan Y, Wu Y, Tang WH, Hazen SL. Intestinal Microbiota-Generated Metabolite Trimethylamine-N-Oxide and 5-Year Mortality Risk in Stable Coronary Artery Disease: The Contributory Role of Intestinal Microbiota in a COURAGE-Like Patient Cohort. J Am Heart Assoc. 2016;5. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 145] [Cited by in RCA: 204] [Article Influence: 22.7] [Reference Citation Analysis (0)] |
| 174. | Yang S, Li X, Yang F, Zhao R, Pan X, Liang J, Tian L, Liu L, Xing Y, Wu M. Gut Microbiota-Dependent Marker TMAO in Promoting Cardiovascular Disease: Inflammation Mechanism, Clinical Prognostic, and Potential as a Therapeutic Target. Front Pharmacol. 2019;10:1360. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 114] [Cited by in RCA: 226] [Article Influence: 37.7] [Reference Citation Analysis (0)] |
| 175. | Kuo CH, Liu CH, Wang JH, Hsu BG. Serum Trimethylamine N-Oxide Levels Correlate with Metabolic Syndrome in Coronary Artery Disease Patients. Int J Environ Res Public Health. 2022;19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 176. | Randrianarisoa E, Lehn-Stefan A, Wang X, Hoene M, Peter A, Heinzmann SS, Zhao X, Königsrainer I, Königsrainer A, Balletshofer B, Machann J, Schick F, Fritsche A, Häring HU, Xu G, Lehmann R, Stefan N. Relationship of Serum Trimethylamine N-Oxide (TMAO) Levels with early Atherosclerosis in Humans. Sci Rep. 2016;6:26745. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 177] [Cited by in RCA: 234] [Article Influence: 26.0] [Reference Citation Analysis (0)] |
| 177. | Chou RH, Chen CY, Chen IC, Huang HL, Lu YW, Kuo CS, Chang CC, Huang PH, Chen JW, Lin SJ. Trimethylamine N-Oxide, Circulating Endothelial Progenitor Cells, and Endothelial Function in Patients with Stable Angina. Sci Rep. 2019;9:4249. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 90] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 178. | Yao ME, Liao PD, Zhao XJ, Wang L. Trimethylamine-N-oxide has prognostic value in coronary heart disease: a meta-analysis and dose-response analysis. BMC Cardiovasc Disord. 2020;20:7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 68] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 179. | Wiese GN, Biruete A, Moorthi RN, Moe SM, Lindemann SR, Hill Gallant KM. Plant-Based Diets, the Gut Microbiota, and Trimethylamine N-Oxide Production in Chronic Kidney Disease: Therapeutic Potential and Methodological Considerations. J Ren Nutr. 2021;31:121-131. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 18] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 180. | Zixin Y, Lulu C, Xiangchang Z, Qing F, Binjie Z, Chunyang L, Tai R, Dongsheng O. TMAO as a potential biomarker and therapeutic target for chronic kidney disease: A review. Front Pharmacol. 2022;13:929262. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 48] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 181. | Bae S, Ulrich CM, Neuhouser ML, Malysheva O, Bailey LB, Xiao L, Brown EC, Cushing-Haugen KL, Zheng Y, Cheng TY, Miller JW, Green R, Lane DS, Beresford SA, Caudill MA. Plasma choline metabolites and colorectal cancer risk in the Women's Health Initiative Observational Study. Cancer Res. 2014;74:7442-7452. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 200] [Cited by in RCA: 190] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 182. | Liu X, Liu H, Yuan C, Zhang Y, Wang W, Hu S, Liu L, Wang Y. Preoperative serum TMAO level is a new prognostic marker for colorectal cancer. Biomark Med. 2017;11:443-447. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 43] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 183. | Ma G, Pan B, Chen Y, Guo C, Zhao M, Zheng L, Chen B. Trimethylamine N-oxide in atherogenesis: impairing endothelial self-repair capacity and enhancing monocyte adhesion. Biosci Rep. 2017;37. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 113] [Cited by in RCA: 187] [Article Influence: 23.4] [Reference Citation Analysis (0)] |
| 184. | Liu Y, Dai M. Trimethylamine N-Oxide Generated by the Gut Microbiota Is Associated with Vascular Inflammation: New Insights into Atherosclerosis. Mediators Inflamm. 2020;2020:4634172. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 122] [Article Influence: 24.4] [Reference Citation Analysis (0)] |
| 185. | Seldin MM, Meng Y, Qi H, Zhu W, Wang Z, Hazen SL, Lusis AJ, Shih DM. Trimethylamine N-Oxide Promotes Vascular Inflammation Through Signaling of Mitogen-Activated Protein Kinase and Nuclear Factor-κB. J Am Heart Assoc. 2016;5. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 395] [Cited by in RCA: 650] [Article Influence: 72.2] [Reference Citation Analysis (0)] |
| 186. | Chen K, Zheng X, Feng M, Li D, Zhang H. Gut Microbiota-Dependent Metabolite Trimethylamine N-Oxide Contributes to Cardiac Dysfunction in Western Diet-Induced Obese Mice. Front Physiol. 2017;8:139. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 115] [Cited by in RCA: 185] [Article Influence: 23.1] [Reference Citation Analysis (0)] |
| 187. | Yang S, Dai H, Lu Y, Li R, Gao C, Pan S. Trimethylamine N-Oxide Promotes Cell Proliferation and Angiogenesis in Colorectal Cancer. J Immunol Res. 2022;2022:7043856. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 39] [Reference Citation Analysis (0)] |
| 188. | Xu R, Wang Q, Li L. A genome-wide systems analysis reveals strong link between colorectal cancer and trimethylamine N-oxide (TMAO), a gut microbial metabolite of dietary meat and fat. BMC Genomics. 2015;16 Suppl 7:S4. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 91] [Cited by in RCA: 138] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 189. | Chan CWH, Law BMH, Waye MMY, Chan JYW, So WKW, Chow KM. Trimethylamine-N-oxide as One Hypothetical Link for the Relationship between Intestinal Microbiota and Cancer - Where We Are and Where Shall We Go? J Cancer. 2019;10:5874-5882. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 65] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 190. | Wallace JL, Motta JP, Buret AG. Hydrogen sulfide: an agent of stability at the microbiome-mucosa interface. Am J Physiol Gastrointest Liver Physiol. 2018;314:G143-G149. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 77] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 191. | Khattak S, Rauf MA, Khan NH, Zhang QQ, Chen HJ, Muhammad P, Ansari MA, Alomary MN, Jahangir M, Zhang CY, Ji XY, Wu DD. Hydrogen Sulfide Biology and Its Role in Cancer. Molecules. 2022;27. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 64] [Article Influence: 21.3] [Reference Citation Analysis (0)] |
| 192. | Mottawea W, Chiang CK, Mühlbauer M, Starr AE, Butcher J, Abujamel T, Deeke SA, Brandel A, Zhou H, Shokralla S, Hajibabaei M, Singleton R, Benchimol EI, Jobin C, Mack DR, Figeys D, Stintzi A. Altered intestinal microbiota-host mitochondria crosstalk in new onset Crohn's disease. Nat Commun. 2016;7:13419. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 213] [Cited by in RCA: 310] [Article Influence: 34.4] [Reference Citation Analysis (0)] |
| 193. | Motta JP, Flannigan KL, Agbor TA, Beatty JK, Blackler RW, Workentine ML, Da Silva GJ, Wang R, Buret AG, Wallace JL. Hydrogen sulfide protects from colitis and restores intestinal microbiota biofilm and mucus production. Inflamm Bowel Dis. 2015;21:1006-1017. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 115] [Cited by in RCA: 145] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 194. | Liu Y, Liao R, Qiang Z, Yang W, Cao J. Exogenous H2S promotes ion channel reconstruction to regulate colonic motility in rats with dinitrobenzene sulfonic acid-induced colitis. Ann Transl Med. 2022;10:681. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 195. | Zhen Y, Pan W, Hu F, Wu H, Feng J, Zhang Y, Chen J. Exogenous hydrogen sulfide exerts proliferation/anti-apoptosis/angiogenesis/migration effects via amplifying the activation of NF-κB pathway in PLC/PRF/5 hepatoma cells. Int J Oncol. 2015;46:2194-2204. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 56] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 196. | Zhen Y, Wu Q, Ding Y, Zhang W, Zhai Y, Lin X, Weng Y, Guo R, Zhang Y, Feng J, Lei Y, Chen J. Exogenous hydrogen sulfide promotes hepatocellular carcinoma cell growth by activating the STAT3-COX-2 signaling pathway. Oncol Lett. 2018;15:6562-6570. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 13] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 197. | Dong Q, Yang B, Han JG, Zhang MM, Liu W, Zhang X, Yu HL, Liu ZG, Zhang SH, Li T, Wu DD, Ji XY, Duan SF. A novel hydrogen sulfide-releasing donor, HA-ADT, suppresses the growth of human breast cancer cells through inhibiting the PI3K/AKT/mTOR and Ras/Raf/MEK/ERK signaling pathways. Cancer Lett. 2019;455:60-72. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 68] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 198. | Ma Y, Yan Z, Deng X, Guo J, Hu J, Yu Y, Jiao F. Anticancer effect of exogenous hydrogen sulfide in cisplatinresistant A549/DDP cells. Oncol Rep. 2018;39:2969-2977. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 13] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 199. | Ijssennagger N, van der Meer R, van Mil SWC. Sulfide as a Mucus Barrier-Breaker in Inflammatory Bowel Disease? Trends Mol Med. 2016;22:190-199. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 142] [Cited by in RCA: 208] [Article Influence: 23.1] [Reference Citation Analysis (0)] |
| 200. | Rong F, Wang T, Zhou Q, Peng H, Yang J, Fan Q, Li P. Intelligent polymeric hydrogen sulfide delivery systems for therapeutic applications. Bioact Mater. 2023;19:198-216. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 27] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 201. | Liu J, Mesfin FM, Hunter CE, Olson KR, Shelley WC, Brokaw JP, Manohar K, Markel TA. Recent Development of the Molecular and Cellular Mechanisms of Hydrogen Sulfide Gasotransmitter. Antioxidants (Basel). 2022;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 16] [Reference Citation Analysis (0)] |
| 202. | Murray Stewart T, Dunston TT, Woster PM, Casero RA Jr. Polyamine catabolism and oxidative damage. J Biol Chem. 2018;293:18736-18745. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 135] [Cited by in RCA: 177] [Article Influence: 25.3] [Reference Citation Analysis (0)] |
| 203. | Sagar NA, Tarafdar S, Agarwal S, Tarafdar A, Sharma S. Polyamines: Functions, Metabolism, and Role in Human Disease Management. Med Sci (Basel). 2021;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 72] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 204. | Hanus M, Parada-Venegas D, Landskron G, Wielandt AM, Hurtado C, Alvarez K, Hermoso MA, López-Köstner F, De la Fuente M. Immune System, Microbiota, and Microbial Metabolites: The Unresolved Triad in Colorectal Cancer Microenvironment. Front Immunol. 2021;12:612826. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 120] [Article Influence: 30.0] [Reference Citation Analysis (0)] |
| 205. | Tofalo R, Cocchi S, Suzzi G. Polyamines and Gut Microbiota. Front Nutr. 2019;6:16. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 163] [Cited by in RCA: 167] [Article Influence: 27.8] [Reference Citation Analysis (0)] |
| 206. | Pugin B, Barcik W, Westermann P, Heider A, Wawrzyniak M, Hellings P, Akdis CA, O'Mahony L. A wide diversity of bacteria from the human gut produces and degrades biogenic amines. Microb Ecol Health Dis. 2017;28:1353881. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 83] [Cited by in RCA: 103] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 207. | Kibe R, Kurihara S, Sakai Y, Suzuki H, Ooga T, Sawaki E, Muramatsu K, Nakamura A, Yamashita A, Kitada Y, Kakeyama M, Benno Y, Matsumoto M. Upregulation of colonic luminal polyamines produced by intestinal microbiota delays senescence in mice. Sci Rep. 2014;4:4548. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 136] [Cited by in RCA: 184] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 208. | Kurihara S. Polyamine metabolism and transport in gut microbes. Biosci Biotechnol Biochem. 2022;86:957-966. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 12] [Reference Citation Analysis (0)] |
| 209. | Ramos-Molina B, Queipo-Ortuño MI, Lambertos A, Tinahones FJ, Peñafiel R. Dietary and Gut Microbiota Polyamines in Obesity- and Age-Related Diseases. Front Nutr. 2019;6:24. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 75] [Cited by in RCA: 130] [Article Influence: 21.7] [Reference Citation Analysis (0)] |
| 210. | Kovács T, Mikó E, Vida A, Sebő É, Toth J, Csonka T, Boratkó A, Ujlaki G, Lente G, Kovács P, Tóth D, Árkosy P, Kiss B, Méhes G, Goedert JJ, Bai P. Cadaverine, a metabolite of the microbiome, reduces breast cancer aggressiveness through trace amino acid receptors. Sci Rep. 2019;9:1300. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 72] [Cited by in RCA: 124] [Article Influence: 20.7] [Reference Citation Analysis (0)] |
| 211. | Casero RA Jr, Murray Stewart T, Pegg AE. Polyamine metabolism and cancer: treatments, challenges and opportunities. Nat Rev Cancer. 2018;18:681-695. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 352] [Cited by in RCA: 559] [Article Influence: 79.9] [Reference Citation Analysis (0)] |
| 212. | Yang Y, Misra BB, Liang L, Bi D, Weng W, Wu W, Cai S, Qin H, Goel A, Li X, Ma Y. Integrated microbiome and metabolome analysis reveals a novel interplay between commensal bacteria and metabolites in colorectal cancer. Theranostics. 2019;9:4101-4114. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 92] [Cited by in RCA: 203] [Article Influence: 33.8] [Reference Citation Analysis (0)] |
| 213. | Avuthu N, Guda C. Meta-Analysis of Altered Gut Microbiota Reveals Microbial and Metabolic Biomarkers for Colorectal Cancer. Microbiol Spectr. 2022;10:e0001322. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 59] [Article Influence: 19.7] [Reference Citation Analysis (0)] |
| 214. | Huang CY, Fang YJ, Abulimiti A, Yang X, Li L, Liu KY, Zhang X, Feng XL, Chen YM, Zhang CX. Dietary Polyamines Intake and Risk of Colorectal Cancer: A Case-Control Study. Nutrients. 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 13] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 215. | Nogacka AM, Gómez-Martín M, Suárez A, González-Bernardo O, de Los Reyes-Gavilán CG, González S. Xenobiotics Formed during Food Processing: Their Relation with the Intestinal Microbiota and Colorectal Cancer. Int J Mol Sci. 2019;20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 49] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 216. | Kobets T, Smith BPC, Williams GM. Food-Borne Chemical Carcinogens and the Evidence for Human Cancer Risk. Foods. 2022;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 27] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 217. | Abu-Ghazaleh N, Chua WJ, Gopalan V. Intestinal microbiota and its association with colon cancer and red/processed meat consumption. J Gastroenterol Hepatol. 2021;36:75-88. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 82] [Article Influence: 20.5] [Reference Citation Analysis (0)] |
| 218. | Zhang J, Empl MT, Schwab C, Fekry MI, Engels C, Schneider M, Lacroix C, Steinberg P, Sturla SJ. Gut Microbial Transformation of the Dietary Imidazoquinoxaline Mutagen MelQx Reduces Its Cytotoxic and Mutagenic Potency. Toxicol Sci. 2017;159:266-276. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 22] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 219. | Zhang J, Lacroix C, Wortmann E, Ruscheweyh HJ, Sunagawa S, Sturla SJ, Schwab C. Gut microbial beta-glucuronidase and glycerol/diol dehydratase activity contribute to dietary heterocyclic amine biotransformation. BMC Microbiol. 2019;19:99. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 48] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 220. | Saulnier DM, Riehle K, Mistretta TA, Diaz MA, Mandal D, Raza S, Weidler EM, Qin X, Coarfa C, Milosavljevic A, Petrosino JF, Highlander S, Gibbs R, Lynch SV, Shulman RJ, Versalovic J. Gastrointestinal microbiome signatures of pediatric patients with irritable bowel syndrome. Gastroenterology. 2011;141:1782-1791. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 559] [Cited by in RCA: 501] [Article Influence: 35.8] [Reference Citation Analysis (0)] |
| 221. | Pittayanon R, Lau JT, Yuan Y, Leontiadis GI, Tse F, Surette M, Moayyedi P. Gut Microbiota in Patients With Irritable Bowel Syndrome-A Systematic Review. Gastroenterology. 2019;157:97-108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 303] [Cited by in RCA: 460] [Article Influence: 76.7] [Reference Citation Analysis (0)] |
| 222. | Hugerth LW, Andreasson A, Talley NJ, Forsberg AM, Kjellström L, Schmidt PT, Agreus L, Engstrand L. No distinct microbiome signature of irritable bowel syndrome found in a Swedish random population. Gut. 2020;69:1076-1084. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 66] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 223. | Hirano A, Umeno J, Okamoto Y, Shibata H, Ogura Y, Moriyama T, Torisu T, Fujioka S, Fuyuno Y, Kawarabayasi Y, Matsumoto T, Kitazono T, Esaki M. Comparison of the microbial community structure between inflamed and non-inflamed sites in patients with ulcerative colitis. J Gastroenterol Hepatol. 2018;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 90] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 224. | Gevers D, Kugathasan S, Denson LA, Vázquez-Baeza Y, Van Treuren W, Ren B, Schwager E, Knights D, Song SJ, Yassour M, Morgan XC, Kostic AD, Luo C, González A, McDonald D, Haberman Y, Walters T, Baker S, Rosh J, Stephens M, Heyman M, Markowitz J, Baldassano R, Griffiths A, Sylvester F, Mack D, Kim S, Crandall W, Hyams J, Huttenhower C, Knight R, Xavier RJ. The treatment-naive microbiome in new-onset Crohn's disease. Cell Host Microbe. 2014;15:382-392. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1945] [Cited by in RCA: 2341] [Article Influence: 212.8] [Reference Citation Analysis (0)] |
| 225. | Mouzaki M, Comelli EM, Arendt BM, Bonengel J, Fung SK, Fischer SE, McGilvray ID, Allard JP. Intestinal microbiota in patients with nonalcoholic fatty liver disease. Hepatology. 2013;58:120-127. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 496] [Cited by in RCA: 555] [Article Influence: 46.3] [Reference Citation Analysis (0)] |
| 226. | Wei X, Yan X, Zou D, Yang Z, Wang X, Liu W, Wang S, Li X, Han J, Huang L, Yuan J. Abnormal fecal microbiota community and functions in patients with hepatitis B liver cirrhosis as revealed by a metagenomic approach. BMC Gastroenterol. 2013;13:175. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 88] [Cited by in RCA: 106] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 227. | Boursier J, Mueller O, Barret M, Machado M, Fizanne L, Araujo-Perez F, Guy CD, Seed PC, Rawls JF, David LA, Hunault G, Oberti F, Calès P, Diehl AM. The severity of nonalcoholic fatty liver disease is associated with gut dysbiosis and shift in the metabolic function of the gut microbiota. Hepatology. 2016;63:764-775. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 763] [Cited by in RCA: 1045] [Article Influence: 116.1] [Reference Citation Analysis (0)] |
| 228. | Liu BN, Liu XT, Liang ZH, Wang JH. Gut microbiota in obesity. World J Gastroenterol. 2021;27:3837-3850. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 98] [Cited by in RCA: 283] [Article Influence: 70.8] [Reference Citation Analysis (22)] |
| 229. | Zeng Q, Li D, He Y, Li Y, Yang Z, Zhao X, Liu Y, Wang Y, Sun J, Feng X, Wang F, Chen J, Zheng Y, Yang Y, Sun X, Xu X, Wang D, Kenney T, Jiang Y, Gu H, Zhou K, Li S, Dai W. Discrepant gut microbiota markers for the classification of obesity-related metabolic abnormalities. Sci Rep. 2019;9:13424. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 111] [Cited by in RCA: 263] [Article Influence: 43.8] [Reference Citation Analysis (0)] |
| 230. | Zhou Z, Sun B, Yu D, Zhu C. Gut Microbiota: An Important Player in Type 2 Diabetes Mellitus. Front Cell Infect Microbiol. 2022;12:834485. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 134] [Article Influence: 44.7] [Reference Citation Analysis (0)] |
| 231. | Tang SS, Liang CH, Liu YL, Wei W, Deng XR, Shi XY, Wang LM, Zhang LJ, Yuan HJ. Intermittent hypoxia is involved in gut microbial dysbiosis in type 2 diabetes mellitus and obstructive sleep apnea-hypopnea syndrome. World J Gastroenterol. 2022;28:2320-2333. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 10] [Cited by in RCA: 27] [Article Influence: 9.0] [Reference Citation Analysis (4)] |
| 232. | Ozaki D, Kubota R, Maeno T, Abdelhakim M, Hitosugi N. Association between gut microbiota, bone metabolism, and fracture risk in postmenopausal Japanese women. Osteoporos Int. 2021;32:145-156. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 75] [Article Influence: 18.8] [Reference Citation Analysis (0)] |
| 233. | Long D, Mao C, Zhang X, Liu Y, Shangguan X, Zou M, Zhu Y, Wang X. Coronary heart disease and gut microbiota: A bibliometric and visual analysis from 2002 to 2022. Front Cardiovasc Med. 2022;9:949859. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 12] [Reference Citation Analysis (0)] |
| 234. | Yu H, Li L, Deng Y, Zhang G, Jiang M, Huang H, Li C, Lv Z, Zhou Y, Liu X. The relationship between the number of stenotic coronary arteries and the gut microbiome in coronary heart disease patients. Front Cell Infect Microbiol. 2022;12:903828. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 235. | Huo RX, Wang YJ, Hou SB, Wang W, Zhang CZ, Wan XH. Gut mucosal microbiota profiles linked to colorectal cancer recurrence. World J Gastroenterol. 2022;28:1946-1964. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 36] [Cited by in RCA: 34] [Article Influence: 11.3] [Reference Citation Analysis (4)] |
| 236. | Bajaj JS, Khoruts A. Microbiota changes and intestinal microbiota transplantation in liver diseases and cirrhosis. J Hepatol. 2020;72:1003-1027. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 129] [Article Influence: 25.8] [Reference Citation Analysis (1)] |
| 237. | Novakovic M, Rout A, Kingsley T, Kirchoff R, Singh A, Verma V, Kant R, Chaudhary R. Role of gut microbiota in cardiovascular diseases. World J Cardiol. 2020;12:110-122. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 144] [Cited by in RCA: 117] [Article Influence: 23.4] [Reference Citation Analysis (8)] |
| 238. | Bullman S, Pedamallu CS, Sicinska E, Clancy TE, Zhang X, Cai D, Neuberg D, Huang K, Guevara F, Nelson T, Chipashvili O, Hagan T, Walker M, Ramachandran A, Diosdado B, Serna G, Mulet N, Landolfi S, Ramon Y Cajal S, Fasani R, Aguirre AJ, Ng K, Élez E, Ogino S, Tabernero J, Fuchs CS, Hahn WC, Nuciforo P, Meyerson M. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science. 2017;358:1443-1448. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 649] [Cited by in RCA: 1051] [Article Influence: 131.4] [Reference Citation Analysis (0)] |
| 239. | Chen S, Su T, Zhang Y, Lee A, He J, Ge Q, Wang L, Si J, Zhuo W. Fusobacterium nucleatum promotes colorectal cancer metastasis by modulating KRT7-AS/KRT7. Gut Microbes. 2020;11:511-525. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 159] [Article Influence: 31.8] [Reference Citation Analysis (0)] |
| 240. | Dickerson F, Severance E, Yolken R. The microbiome, immunity, and schizophrenia and bipolar disorder. Brain Behav Immun. 2017;62:46-52. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 169] [Cited by in RCA: 181] [Article Influence: 22.6] [Reference Citation Analysis (0)] |
| 241. | van De Sande MM, van Buul VJ, Brouns FJ. Autism and nutrition: the role of the gut-brain axis. Nutr Res Rev. 2014;27:199-214. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 74] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 242. | Gerhardt S, Mohajeri MH. Changes of Colonic Bacterial Composition in Parkinson's Disease and Other Neurodegenerative Diseases. Nutrients. 2018;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 194] [Cited by in RCA: 201] [Article Influence: 28.7] [Reference Citation Analysis (0)] |
| 243. | Minato T, Maeda T, Fujisawa Y, Tsuji H, Nomoto K, Ohno K, Hirayama M. Progression of Parkinson's disease is associated with gut dysbiosis: Two-year follow-up study. PLoS One. 2017;12:e0187307. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 155] [Cited by in RCA: 189] [Article Influence: 23.6] [Reference Citation Analysis (0)] |
| 244. | Li X, He C, Li N, Ding L, Chen H, Wan J, Yang X, Xia L, He W, Xiong H, Shu X, Zhu Y, Lu N. The interplay between the gut microbiota and NLRP3 activation affects the severity of acute pancreatitis in mice. Gut Microbes. 2020;11:1774-1789. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 100] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 245. | Zhang Y, Saint Fleur A, Feng H. The development of live biotherapeutics against Clostridioides difficile infection towards reconstituting gut microbiota. Gut Microbes. 2022;14:2052698. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 17] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 246. | Ianiro G, Mullish BH, Kelly CR, Kassam Z, Kuijper EJ, Ng SC, Iqbal TH, Allegretti JR, Bibbò S, Sokol H, Zhang F, Fischer M, Costello SP, Keller JJ, Masucci L, van Prehn J, Quaranta G, Quraishi MN, Segal J, Kao D, Satokari R, Sanguinetti M, Tilg H, Gasbarrini A, Cammarota G. Reorganisation of faecal microbiota transplant services during the COVID-19 pandemic. Gut. 2020;69:1555-1563. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 116] [Cited by in RCA: 116] [Article Influence: 23.2] [Reference Citation Analysis (0)] |
| 247. | Yu D, Meng X, de Vos WM, Wu H, Fang X, Maiti AK. Implications of Gut Microbiota in Complex Human Diseases. Int J Mol Sci. 2021;22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 26] [Article Influence: 6.5] [Reference Citation Analysis (1)] |
| 248. | Asnicar F, Berry SE, Valdes AM, Nguyen LH, Piccinno G, Drew DA, Leeming E, Gibson R, Le Roy C, Khatib HA, Francis L, Mazidi M, Mompeo O, Valles-Colomer M, Tett A, Beghini F, Dubois L, Bazzani D, Thomas AM, Mirzayi C, Khleborodova A, Oh S, Hine R, Bonnett C, Capdevila J, Danzanvilliers S, Giordano F, Geistlinger L, Waldron L, Davies R, Hadjigeorgiou G, Wolf J, Ordovás JM, Gardner C, Franks PW, Chan AT, Huttenhower C, Spector TD, Segata N. Microbiome connections with host metabolism and habitual diet from 1,098 deeply phenotyped individuals. Nat Med. 2021;27:321-332. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 551] [Cited by in RCA: 563] [Article Influence: 140.8] [Reference Citation Analysis (0)] |
| 249. | O'Neill AM, Burrington CM, Gillaspie EA, Lynch DT, Horsman MJ, Greene MW. High-fat Western diet-induced obesity contributes to increased tumor growth in mouse models of human colon cancer. Nutr Res. 2016;36:1325-1334. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 54] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 250. | Zeng H, Lazarova DL, Bordonaro M. Mechanisms linking dietary fiber, gut microbiota and colon cancer prevention. World J Gastrointest Oncol. 2014;6:41-51. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 183] [Cited by in RCA: 165] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 251. | Gibson GR, Hutkins R, Sanders ME, Prescott SL, Reimer RA, Salminen SJ, Scott K, Stanton C, Swanson KS, Cani PD, Verbeke K, Reid G. Expert consensus document: The International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nat Rev Gastroenterol Hepatol. 2017;14:491-502. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2256] [Cited by in RCA: 3168] [Article Influence: 396.0] [Reference Citation Analysis (0)] |
| 252. | McNabney SM, Henagan TM. Short Chain Fatty Acids in the Colon and Peripheral Tissues: A Focus on Butyrate, Colon Cancer, Obesity and Insulin Resistance. Nutrients. 2017;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 361] [Cited by in RCA: 346] [Article Influence: 43.3] [Reference Citation Analysis (0)] |
| 253. | Makki K, Deehan EC, Walter J, Bäckhed F. The Impact of Dietary Fiber on Gut Microbiota in Host Health and Disease. Cell Host Microbe. 2018;23:705-715. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 898] [Cited by in RCA: 1542] [Article Influence: 220.3] [Reference Citation Analysis (0)] |
| 254. | Gunness P, Michiels J, Vanhaecke L, De Smet S, Kravchuk O, Van de Meene A, Gidley MJ. Reduction in circulating bile acid and restricted diffusion across the intestinal epithelium are associated with a decrease in blood cholesterol in the presence of oat β-glucan. FASEB J. 2016;30:4227-4238. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 57] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 255. | Gunness P, Williams BA, Gerrits WJ, Bird AR, Kravchuk O, Gidley MJ. Circulating triglycerides and bile acids are reduced by a soluble wheat arabinoxylan via modulation of bile concentration and lipid digestion rates in a pig model. Mol Nutr Food Res. 2016;60:642-651. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 40] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 256. | Brown JR, Flemer B, Joyce SA, Zulquernain A, Sheehan D, Shanahan F, O'Toole PW. Changes in microbiota composition, bile and fatty acid metabolism, in successful faecal microbiota transplantation for Clostridioides difficile infection. BMC Gastroenterol. 2018;18:131. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 77] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 257. | Hassouneh R, Bajaj JS. Gut Microbiota Modulation and Fecal Transplantation: An Overview on Innovative Strategies for Hepatic Encephalopathy Treatment. J Clin Med. 2021;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 33] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 258. | Costello SP, Hughes PA, Waters O, Bryant RV, Vincent AD, Blatchford P, Katsikeros R, Makanyanga J, Campaniello MA, Mavrangelos C, Rosewarne CP, Bickley C, Peters C, Schoeman MN, Conlon MA, Roberts-Thomson IC, Andrews JM. Effect of Fecal Microbiota Transplantation on 8-Week Remission in Patients With Ulcerative Colitis: A Randomized Clinical Trial. JAMA. 2019;321:156-164. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 414] [Cited by in RCA: 613] [Article Influence: 102.2] [Reference Citation Analysis (0)] |
| 259. | Caldeira LF, Borba HH, Tonin FS, Wiens A, Fernandez-Llimos F, Pontarolo R. Fecal microbiota transplantation in inflammatory bowel disease patients: A systematic review and meta-analysis. PLoS One. 2020;15:e0238910. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 70] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 260. | Scibelli N, Singh P, Raynor K. Intestinal Dysbiosis Disguised as a Rectal Fistula Treated With Autologous Fecal Microbiota Transplantation. Cureus. 2021;13:e14115. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 261. | Xiang L, Ding X, Li Q, Wu X, Dai M, Long C, He Z, Cui B, Zhang F. Efficacy of faecal microbiota transplantation in Crohn's disease: a new target treatment? Microb Biotechnol. 2020;13:760-769. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 56] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 262. | Xu MQ, Cao HL, Wang WQ, Wang S, Cao XC, Yan F, Wang BM. Fecal microbiota transplantation broadening its application beyond intestinal disorders. World J Gastroenterol. 2015;21:102-111. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 161] [Cited by in RCA: 158] [Article Influence: 15.8] [Reference Citation Analysis (2)] |
| 263. | Vendrik KEW, Ooijevaar RE, de Jong PRC, Laman JD, van Oosten BW, van Hilten JJ, Ducarmon QR, Keller JJ, Kuijper EJ, Contarino MF. Fecal Microbiota Transplantation in Neurological Disorders. Front Cell Infect Microbiol. 2020;10:98. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 110] [Cited by in RCA: 263] [Article Influence: 52.6] [Reference Citation Analysis (0)] |
| 264. | Chen D, Wu J, Jin D, Wang B, Cao H. Fecal microbiota transplantation in cancer management: Current status and perspectives. Int J Cancer. 2019;145:2021-2031. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 113] [Cited by in RCA: 212] [Article Influence: 30.3] [Reference Citation Analysis (0)] |
| 265. | DeFilipp Z, Bloom PP, Torres Soto M, Mansour MK, Sater MRA, Huntley MH, Turbett S, Chung RT, Chen YB, Hohmann EL. Drug-Resistant E. coli Bacteremia Transmitted by Fecal Microbiota Transplant. N Engl J Med. 2019;381:2043-2050. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 575] [Cited by in RCA: 825] [Article Influence: 137.5] [Reference Citation Analysis (0)] |
| 266. | Wang X, Zhao J, Feng Y, Feng Z, Ye Y, Liu L, Kang G, Cao X. Evolutionary Insights Into Microbiota Transplantation in Inflammatory Bowel Disease. Front Cell Infect Microbiol. 2022;12:916543. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 267. | Rode AA, Chehri M, Krogsgaard LR, Heno KK, Svendsen AT, Ribberholt I, Helms M, Engberg J, Schønning K, Tvede M, Andersen CØ, Jensen US, Petersen AM, Bytzer P. Randomised clinical trial: a 12-strain bacterial mixture vs faecal microbiota transplantation vs vancomycin for recurrent Clostridioides difficile infections. Aliment Pharmacol Ther. 2021;53:999-1009. [PubMed] |
| 268. | Ghimire S, Roy C, Wongkuna S, Antony L, Maji A, Keena MC, Foley A, Scaria J. Identification of Clostridioides difficile-Inhibiting Gut Commensals Using Culturomics, Phenotyping, and Combinatorial Community Assembly. mSystems. 2020;5. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 46] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 269. | Zhang T, Lu G, Zhao Z, Liu Y, Shen Q, Li P, Chen Y, Yin H, Wang H, Marcella C, Cui B, Cheng L, Ji G, Zhang F. Washed microbiota transplantation vs. manual fecal microbiota transplantation: clinical findings, animal studies and in vitro screening. Protein Cell. 2020;11:251-266. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 194] [Cited by in RCA: 188] [Article Influence: 37.6] [Reference Citation Analysis (0)] |
| 270. | Khanna S, Pardi DS, Kelly CR, Kraft CS, Dhere T, Henn MR, Lombardo MJ, Vulic M, Ohsumi T, Winkler J, Pindar C, McGovern BH, Pomerantz RJ, Aunins JG, Cook DN, Hohmann EL. A Novel Microbiome Therapeutic Increases Gut Microbial Diversity and Prevents Recurrent Clostridium difficile Infection. J Infect Dis. 2016;214:173-181. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 281] [Cited by in RCA: 246] [Article Influence: 27.3] [Reference Citation Analysis (0)] |
| 271. | Hill C, Guarner F, Reid G, Gibson GR, Merenstein DJ, Pot B, Morelli L, Canani RB, Flint HJ, Salminen S, Calder PC, Sanders ME. Expert consensus document. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat Rev Gastroenterol Hepatol. 2014;11:506-514. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4055] [Cited by in RCA: 5516] [Article Influence: 501.5] [Reference Citation Analysis (2)] |
| 272. | Shen NT, Maw A, Tmanova LL, Pino A, Ancy K, Crawford CV, Simon MS, Evans AT. Timely Use of Probiotics in Hospitalized Adults Prevents Clostridium difficile Infection: A Systematic Review With Meta-Regression Analysis. Gastroenterology. 2017;152:1889-1900.e9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 177] [Cited by in RCA: 199] [Article Influence: 24.9] [Reference Citation Analysis (0)] |
| 273. | Dos Reis SA, da Conceição LL, Siqueira NP, Rosa DD, da Silva LL, Peluzio MD. Review of the mechanisms of probiotic actions in the prevention of colorectal cancer. Nutr Res. 2017;37:1-19. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 125] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 274. | Commane D, Hughes R, Shortt C, Rowland I. The potential mechanisms involved in the anti-carcinogenic action of probiotics. Mutat Res. 2005;591:276-289. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 131] [Cited by in RCA: 118] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 275. | Baradaran Ghavami S, Asadzadeh Aghdaei H, Sorrentino D, Shahrokh S, Farmani M, Ashrafian F, Dore MP, Keshavarz Azizi Raftar S, Mobin Khoramjoo S, Zali MR. Probiotic-Induced Tolerogenic Dendritic Cells: A Novel Therapy for Inflammatory Bowel Disease? Int J Mol Sci. 2021;22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 276. | Sanders ME, Merenstein DJ, Reid G, Gibson GR, Rastall RA. Probiotics and prebiotics in intestinal health and disease: from biology to the clinic. Nat Rev Gastroenterol Hepatol. 2019;16:605-616. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 570] [Cited by in RCA: 1050] [Article Influence: 175.0] [Reference Citation Analysis (0)] |
| 277. | Engevik MA, Herrmann B, Ruan W, Engevik AC, Engevik KA, Ihekweazu F, Shi Z, Luck B, Chang-Graham AL, Esparza M, Venable S, Horvath TD, Haidacher SJ, Hoch KM, Haag AM, Schady DA, Hyser JM, Spinler JK, Versalovic J. Bifidobacterium dentium-derived y-glutamylcysteine suppresses ER-mediated goblet cell stress and reduces TNBS-driven colonic inflammation. Gut Microbes. 2021;13:1-21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 61] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 278. | Lau CS, Chamberlain RS. Probiotics are effective at preventing Clostridium difficile-associated diarrhea: a systematic review and meta-analysis. Int J Gen Med. 2016;9:27-37. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 80] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 279. | Nilsson AG, Sundh D, Bäckhed F, Lorentzon M. Lactobacillus reuteri reduces bone loss in older women with low bone mineral density: a randomized, placebo-controlled, double-blind, clinical trial. J Intern Med. 2018;284:307-317. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 121] [Cited by in RCA: 200] [Article Influence: 28.6] [Reference Citation Analysis (0)] |
| 280. | Whisner CM, Martin BR, Nakatsu CH, Story JA, MacDonald-Clarke CJ, McCabe LD, McCabe GP, Weaver CM. Soluble Corn Fiber Increases Calcium Absorption Associated with Shifts in the Gut Microbiome: A Randomized Dose-Response Trial in Free-Living Pubertal Females. J Nutr. 2016;146:1298-1306. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 116] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 281. | Hayashi A, Nagao-Kitamoto H, Kitamoto S, Kim CH, Kamada N. The Butyrate-Producing Bacterium Clostridium butyricum Suppresses Clostridioides difficile Infection via Neutrophil- and Antimicrobial Cytokine-Dependent but GPR43/109a-Independent Mechanisms. J Immunol. 2021;206:1576-1585. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 56] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 282. | Nale JY, Spencer J, Hargreaves KR, Buckley AM, Trzepiński P, Douce GR, Clokie MR. Bacteriophage Combinations Significantly Reduce Clostridium difficile Growth In Vitro and Proliferation In Vivo. Antimicrob Agents Chemother. 2016;60:968-981. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 122] [Cited by in RCA: 181] [Article Influence: 18.1] [Reference Citation Analysis (0)] |
| 283. | Enaud R, Prevel R, Ciarlo E, Beaufils F, Wieërs G, Guery B, Delhaes L. The Gut-Lung Axis in Health and Respiratory Diseases: A Place for Inter-Organ and Inter-Kingdom Crosstalks. Front Cell Infect Microbiol. 2020;10:9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 372] [Cited by in RCA: 465] [Article Influence: 93.0] [Reference Citation Analysis (0)] |
| 284. | Markey L, Shaban L, Green ER, Lemon KP, Mecsas J, Kumamoto CA. Pre-colonization with the commensal fungus Candida albicans reduces murine susceptibility to Clostridium difficile infection. Gut Microbes. 2018;9:497-509. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 58] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 285. | Cassone M, Serra P, Mondello F, Girolamo A, Scafetti S, Pistella E, Venditti M. Outbreak of Saccharomyces cerevisiae subtype boulardii fungemia in patients neighboring those treated with a probiotic preparation of the organism. J Clin Microbiol. 2003;41:5340-5343. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 147] [Cited by in RCA: 133] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 286. | El-Sayed A, Aleya L, Kamel M. Microbiota's role in health and diseases. Environ Sci Pollut Res Int. 2021;28:36967-36983. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 75] [Article Influence: 18.8] [Reference Citation Analysis (0)] |
| 287. | Zhang B, Wang X, Xia R, Li C. Gut microbiota in coronary artery disease: a friend or foe? Biosci Rep. 2020;40. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 288. | Li N, Ma WT, Pang M, Fan QL, Hua JL. The Commensal Microbiota and Viral Infection: A Comprehensive Review. Front Immunol. 2019;10:1551. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 134] [Cited by in RCA: 182] [Article Influence: 30.3] [Reference Citation Analysis (0)] |
| 289. | Mikó E, Kovács T, Sebő É, Tóth J, Csonka T, Ujlaki G, Sipos A, Szabó J, Méhes G, Bai P. Microbiome-Microbial Metabolome-Cancer Cell Interactions in Breast Cancer-Familiar, but Unexplored. Cells. 2019;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 125] [Cited by in RCA: 141] [Article Influence: 23.5] [Reference Citation Analysis (0)] |
| 290. | Cheng WY, Wu CY, Yu J. The role of gut microbiota in cancer treatment: friend or foe? Gut. 2020;69:1867-1876. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 117] [Cited by in RCA: 235] [Article Influence: 47.0] [Reference Citation Analysis (0)] |
| 291. | Johnson CH, Dejea CM, Edler D, Hoang LT, Santidrian AF, Felding BH, Ivanisevic J, Cho K, Wick EC, Hechenbleikner EM, Uritboonthai W, Goetz L, Casero RA Jr, Pardoll DM, White JR, Patti GJ, Sears CL, Siuzdak G. Metabolism links bacterial biofilms and colon carcinogenesis. Cell Metab. 2015;21:891-897. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 237] [Cited by in RCA: 270] [Article Influence: 27.0] [Reference Citation Analysis (0)] |
| 292. | Brunt VE, Gioscia-Ryan RA, Richey JJ, Zigler MC, Cuevas LM, Gonzalez A, Vázquez-Baeza Y, Battson ML, Smithson AT, Gilley AD, Ackermann G, Neilson AP, Weir T, Davy KP, Knight R, Seals DR. Suppression of the gut microbiome ameliorates age-related arterial dysfunction and oxidative stress in mice. J Physiol. 2019;597:2361-2378. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 134] [Article Influence: 22.3] [Reference Citation Analysis (0)] |