Published online Jan 7, 2023. doi: 10.3748/wjg.v29.i1.1
Peer-review started: September 10, 2022
First decision: September 29, 2022
Revised: October 11, 2022
Accepted: November 4, 2022
Article in press: November 4, 2022
Published online: January 7, 2023
Processing time: 115 Days and 15.4 Hours
Colorectal cancer (CRC) is one of the most common malignancies of the digestive tract, with the annual incidence and mortality increasing consistently. Oxaliplatin-based chemotherapy is a preferred therapeutic regimen for patients with advanced CRC. However, most patients will inevitably develop resistance to oxaliplatin. Many studies have reported that non-coding RNAs (ncRNAs), such as microRNAs, long non-coding RNAs, and circular RNAs, are extensively involved in cancer progression. Moreover, emerging evidence has revealed that ncRNAs mediate chemoresistance to oxaliplatin by transcriptional and post-transcriptional regulation, and by epigenetic modification. In this review, we summarize the mechanisms by which ncRNAs regulate the initiation and development of CRC chemoresistance to oxaliplatin. Furthermore, we investigate the clinical application of ncRNAs as promising biomarkers for liquid CRC biopsy. This review provides new insights into overcoming oxaliplatin resistance in CRC by targeting ncRNAs.
Core Tip: Oxaliplatin has served as a first-line chemotherapy option for colorectal cancer (CRC). However, owing to congenital or acquired resistance, treatment failure is common in some patients with CRC. Abundant evidence has revealed that non-coding RNAs (ncRNAs) are extensively involved in cancer progression, including drug resistance. Specifically, ncRNAs mediate resistance to oxaliplatin by mediating drug carriers, tumor microenvironment, resistance-related signaling pathways, and patterns of cell death. Importantly, we investigated the potential and clinical application values of these ncRNAs as liquid biopsy markers for CRC.
- Citation: Luo ZD, Wang YF, Zhao YX, Yu LC, Li T, Fan YJ, Zeng SJ, Zhang YL, Zhang Y, Zhang X. Emerging roles of non-coding RNAs in colorectal cancer oxaliplatin resistance and liquid biopsy potential. World J Gastroenterol 2023; 29(1): 1-18
- URL: https://www.wjgnet.com/1007-9327/full/v29/i1/1.htm
- DOI: https://dx.doi.org/10.3748/wjg.v29.i1.1
Colorectal cancer (CRC) is considered to be the leading cause of death associated with malignancy of the gastrointestinal tract, with approximately 1932 million new cases and 935000 deaths in 2020[1]. Currently, surgery is the preferred treatment option for early CRC patients; however, it has few clinical benefits for advanced patients due to high rates of postoperative metastasis and recurrence[2,3]. The annual survival rate for stage III CRC patients is about 30%-60%; this figure drops to just 10% for stage IV patients[4,5]. The combination of oxaliplatin, 5-fluorouracil, and leucovorin (FOLFOX) has significantly increased the overall survival of patients and now serves as a first-line standard regimen for metastatic CRC (mCRC)[6]. Oxaliplatin is a widely used third-generation platinum analog that functions by reacting with DNA to form hydration derivatives with intra- and inter-strand crosslinks eventually resulting in cell death[7]. However, oxaliplatin has not been satisfactory in improving the survival rate of some patients due to drug resistance[8]. Moreover, some toxic side effects persist after oxaliplatin treatment; these include peripheral neurotoxicity, which further hinders chemotherapy[9]. Fortunately, oxaliplatin-based chemotherapy retreatment strategies are being optimized to provide further personalized therapy for patients with mCRC[10]. Therefore, it is of great importance to understand the mechanism by which oxaliplatin resistance occurs, and to identify new biomarkers that can aid prediction of the outcomes for CRC patients treated with oxaliplatin-based therapy.
Although the processes of chemoresistance in CRC are intricate and inconclusive[11], ongoing research has revealed multiple oxaliplatin resistance mechanisms. Several studies have confirmed that chemoresistance is associated with the dysregulation of efflux proteins and drug-metabolizing enzymes, such as ABC transporters and GSTP1; these enzymes mediate drug uptake, transport, and toxicity[12,13]. Furthermore, accumulating evidence suggests that epithelial-mesenchymal transition (EMT)[14] and DNA damage repair[15] are driving factors that result in chemoresistance and tumor progression. Many studies have reported that various signaling pathways, such as the TGF-β/Smad[16], JNK/p38 MAPK[17], Wnt/β-catenin[18], and MEK/ERK/ELK1[19] pathways are closely associated with oxaliplatin resistance. As the main regulatory components of the tumor microenvironment (TME), cancer-associated fibroblasts (CAFs)[20] and tumor-associated macrophages[21] can modulate cancer chemotherapy resistance through complex mechanisms of crosstalk. Of note, emerging studies have confirmed that cell death mechanisms play a pivotal role in chemoresistance. It has been reported that apoptosis inhibition[22] and autophagy dysregulation[23] are underlying mechanisms leading to chemoresistance. Furthermore, numerous studies have identified that several additional death mechanisms can mediate CRC chemoresistance, including ferroptosis[24], pyroptosis[25], necroptosis[26], and others. However, the detailed molecular mechanisms by which these cell death modes lead to oxaliplatin resistance are unclear and require further investigation.
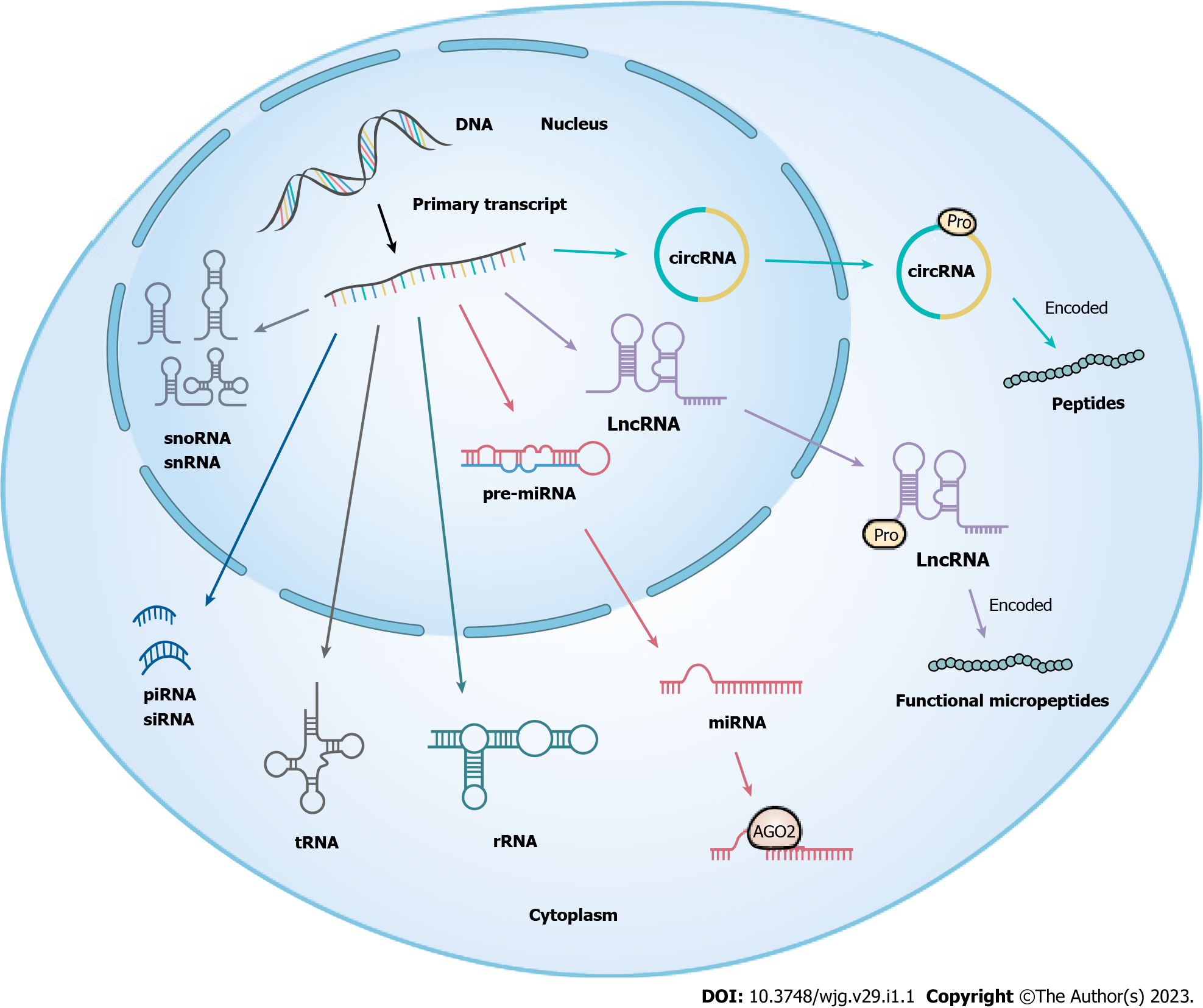
Non-coding RNAs (ncRNAs) refer to RNA molecules transcribed from genes that cannot encode proteins, accounting for more than 90% of human gene transcripts. ncRNAs can be divided into either small ncRNAs (< 200 nucleotides) or long non-coding RNAs (lncRNAs) (> 200 nucleotides) based on their length[27-29]. In the last decade, circular RNAs (circRNAs), a type of circular RNA generated in the splicing process of pre-mRNA, have emerged as a focus of research[30] (Figure 1). A growing number of studies have revealed that ncRNAs play critical roles in the occurrence and development of multiple human diseases via various mechanisms, such as epigenetics, transcription, and post-transcription[31,32]. Moreover, microRNA (miRNAs), lncRNAs, and circRNAs are three of the more widely studied types of ncRNAs and have been shown to mediate tumor progression by regulating tumor cell proliferation, aggression, metastasis, apoptosis, and drug resistance, amongst other pathways[33,34]. Drug resistance is one of the key factors in malignancy treatment failure, and whether this is mediated by ncRNAs has attracted increasing attention. In recent years, the rapid development of next-generation sequencing technologies has expanded the understanding of tumor pathogenesis and drug resistance mechanisms, and an increasing number of tumor chemoresistance-associated differential ncRNAs have been identified[35-37]; these developments have provided the expansion of open datasets of candidate ncRNAs for basic research and clinical application. Currently, the targeting of ncRNAs as biomarkers in the clinic is being actively facilitated[38]. As such, a systematic understanding of the underlying roles of ncRNAs in malignancies provides valuable insights into overcoming chemoresistance in CRC patients.
Although tissue biopsy is the gold standard for malignancy diagnosis, the availability of clinical samples is often limited, and subsequently, tumor heterogeneity may not be reflected in analysis and is therefore not suitable for longitudinal clinical monitoring[39]. As a noninvasive approach, liquid biopsy has been extensively applied for the real-time monitoring of variations in tumor dynamics in body fluids including blood, ascites, and others. With the development of novel molecular detection technologies, growing evidence suggests that circulating tumor cells, circulating tumor DNA, circulating free nucleic acids, tumor-educated platelet, exosomes, etc[40,41] have gradually become prominent liquid biopsy hallmarks. Owing to their stability and high abundance, circulating ncRNAs can provide substantial details regarding tumor biology and therapeutic efficacy, and have become candidate biomarkers for earlier diagnosis, therapeutic monitoring, and prognosis evaluation of malignancies[42,43].
In this study, we performed a systematic literature review to identify the latent mechanisms of ncRNAs (miRNAs, lncRNAs, and circRNAs) in CRC oxaliplatin resistance. In particular, exosomal ncRNAs, as one of the most promising biomarkers in liquid biopsies, are expected to improve diagnostic, therapeutic, and drug monitoring in CRC patients.
MiRNAs are a class of small ncRNAs of approximately 22 nucleotides in length; they are produced by two RNase III proteins, Drosha and Dicer[44]. In general, miRNAs bind to the 3' untranslated region (UTR) of their target mRNAs resulting in cleavage or translational repression in the cytosol[45]. Intriguingly, numerous recent studies have demonstrated that some miRNAs activate gene transcription unconventionally in the nucleus by targeting enhancers, such miRNAs are known as nuclear activating miRNAs[46]. Moreover, enhancers (including super-enhancers) have been revealed to synergistically promote NamiRNA biogenesis and activate the expression of proximal genes[47,48]. Furthermore, studies have reported that the dysregulation of miRNAs may play an important role in CRC oxaliplatin resistance, as illustrated in Table 1.
| MiRNAs | Expression1 | Targets and pathways | Ref. |
| miR-135b-5p | ↑ | MUL1/ULK1 | [49] |
| miR-454-3p | ↑ | PTEN | [50] |
| miR-19a | ↑ | PTEN/PI3K/AKT | [51] |
| miR-543 | ↑ | PTEN/Akt/mTOR | [52] |
| miR-107 | ↑ | CAB39/AMPK/mTOR | [53] |
| miR-503-5p | ↑ | PUMA | [54] |
| miR-744 | ↑ | BIN1 | [55] |
| miR-5000-3p | ↑ | USP49 | [56] |
| miR-146b-5p | ↑ | WBSCR22 | [57] |
| miR-19b-3p | ↑ | SMAD4 | [58] |
| miR-1278 | ↓ | - | [59] |
| miR-506 | ↓ | Wnt/β-catenin | [60] |
| miR-200b-3p | ↓ | TUBB3 | [61] |
| miR-122 | ↓ | XIAP | [62] |
| miR-193a-5p | ↓ | CXCR4 | [63] |
| miR-483-3p | ↓ | FAM171B | [64] |
| miR-325 | ↓ | HSPA12B/PI3K/AKT/Bcl-2 | [65] |
| miR-195-5p | ↓ | - | [66] |
| miR-497-5p | ↓ | - | |
| miR-34a | ↓ | TGF-β/Smad4 | [68] |
| miR-27b-3p | ↓ | - | [69] |
MiR-135b-5p has been confirmed to be upregulated in the serum of CRC patients, mechanistically this has been shown to induce protective autophagy via the MUL1/ULK1 signaling pathway, a process that contributes to oxaliplatin resistance[49]. Another miRNA, miR-454-3p, is highly expressed in oxaliplatin-resistant CRC cells compared to oxaliplatin-sensitive cells and promoted oxaliplatin resistance by inhibiting PTEN expression and activation of the AKT pathway[50]. Similarly, targeting miR-19a with high expression in oxaliplatin-resistant CRC cell lines could promote oxaliplatin sensitization of resistant cells through activation of the PTEN/PI3K/AKT pathway[51]. Compounding these findings, it has been revealed that dichloroacetate, a pyruvate dehydrogenase kinase inhibitor, can enhance the chemosensitivity of oxaliplatin-resistant CRC cells through the miR-543/PTEN/Akt/ mTOR pathway and the miR-107/CAB39/AMPK/mTOR pathway[52,53]. In addition, an unbiased microRNA array demonstrated that miR-503-5p was up-regulated in oxaliplatin-resistant CRC cells; overexpression of miR-503-5p conferred resistance to oxaliplatin-induced apoptosis by inhibiting PUMA expression[54]. Further studies have revealed that by targeting BNI1, up-regulation of miR-744 enhanced the resistance of T84 and HCT116 cells to oxaliplatin[55]. Moreover, the oncogenic miR-5000-3p was upregulated in CRC tissues and oxaliplatin-resistant CRC cells, this miRNA negatively regulated USP9 to facilitate CRC chemoresistance[56]. Additionally, natural killer cells contributed to enhancing the sensitivity of oxaliplatin resistant CRC cells through the microRNA-146b-5p/WBSCR22 axis[57]. Remarkably, miR-19b-3p has been documented to be the most significantly upregulated candidate miRNA in colon cancer tissues, its expression correlates with tumorous histologic grading, staging, and poor prognosis in patients. Importantly, miR-19b-3p facilitated proliferation, curbed apoptosis, and induced oxaliplatin resistance in CRC cells by targeting SMAD4[58].
Interestingly, it has been demonstrated by some that tumor-suppressive miRNAs are associated with oxaliplatin resistance. Studies have identified that miR-1278 was significantly downregulated in CRC tissues, and that overexpression of miR-1278 inhibited CRC progression and enhanced oxaliplatin sensitivity through targeting of the KIF5B/BTG2 axis[59]. Similarly, miR-506 was weakly expressed in chemoresistant CRC cells, but its overexpression has been confirmed to impair the resistance of HCT116 cells to oxaliplatin via the Wnt/β-catenin pathway[60]. Moreover, miR-200b-3p was down-regulated in oxaliplatin-resistant CRC tissues and cells (HT29-OR and HCT116-OR), and studies revealed that miR-200b-3p reversed oxaliplatin resistance of CRC cells by targeting TUBB3[61]. Furthermore, miR-122, which was down-regulated in oxaliplatin-resistant SW480 and HT29 cells, has been demonstrated to be overexpressed in CRC cells, this could enhance the chemosensitivity of CRC by inhibiting the expression of XIAP[62]. As a tumor suppressor, miR-193a-5p can act directly upon CXCR4 to mitigate the chemosensitivity of CRC cells to 5-FU and oxaliplatin[63]. Using sequencing, Liang et al[64] discovered that miR-483-3p was negatively correlated with FAM171B expression, and targeting the miR-483-3p/FAM171B regulatory axis could enhance the sensitivity of CRC cells to oxaliplatin. Moreover, miR-325 mimics were observed to prevent the development of oxaliplatin resistance in CRC via interference with the HSPA12B/PI3K/AKT/Bcl-2 axis[65]. Finally, a proteomic analysis study reported that regulation of miR-195-5p and miR-497-5p is a potential strategy for alleviating oxaliplatin resistance in CRC cells[66].
As an additional factor, several groups showed that the interaction between ncRNAs and autophagy exerted significant roles in the therapeutic resistance of CRC[67]. Sun et al[68] demonstrated that miR-34a was expressed at low levels in oxaliplatin-resistant CRC patients and cells. Overexpression of miR-34a contributed to facilitating the sensitivity of CRC cells to oxaliplatin by regulating the TGF-β/Smad4 pathway resulting in the inhibition of macroautophagy. Furthermore, c-Myc has been shown to promote oxaliplatin resistance in CRC by integrating into the miR-27B promoter region and further mediating activation of the miR-27b-3p/ATG10 axis[69].
It is noteworthy that miR-181a could suppress the expression of BIRC6, a protein inhibitor of apoptosis, by directly targeting the 3'-UTR of BIRC6 mRNA[70]. Conversely, BIRC6 was observed to be significantly upregulated in acquired oxaliplatin-resistant CRC cells when compared to parental cells[71]. Considering that knockdown of BIRC6 helped enhance the chemosensitivity of CRC cells to oxaliplatin, targeting BIRC6 using miR-181a mimics may be a potential strategy to reverse oxaliplatin resistance in CRC.
LncRNAs are defined as a class of ncRNA molecules with transcript lengths of more than 200 nucleotides; they do not encode proteins due to their lack of an open reading frame[72]. LncRNAs can be classified into intergenic, intronic, sense, antisense, and bidirectional lncRNAs based on their genomic localization[73]. Alternatively, there are four categories of lncRNAs based on their functional mechanisms, including signal, decoy, guide, and scaffold lncRNAs[74,75]. It has been reported that lncRNAs can regulate transcription, translation, RNA stability, and alternative splicing[76-79]. It is noteworthy that numerous studies have revealed that aberrant lncRNAs (oncogenic and tumor suppressive lncRNAs) contribute to regulating the mechanism of oxaliplatin resistance through multiple cellular mechanisms, including but not limited to DNA damage repair, interference with drug influx and efflux, regulation of the cell cycle and apoptosis, and activation of signaling pathways[80,81]. Table 2 provides an overview of some aberrant lncRNAs and their target molecules in the context of oxaliplatin resistance in CRC.
| LncRNAs | Expression1 | Targets and pathways | Ref. |
| GIHCG | ↑ | - | [82] |
| lncARSR | ↑ | - | [83] |
| HOTAIR | ↑ | miR-1277-5p/ZEB1 | [85] |
| MALAT1 | ↑ | MALAT1/miR-218/EZH2 | [86] |
| ↑ | miR-324-3p/ADAM17 | [88] | |
| OIP5-AS1 | ↑ | miR-137 | [89] |
| Linc00152 | ↑ | miR-193a-3p/ERBB4/AKT | [90] |
| CASC15 | ↑ | miR-145/ABCC1 | [92] |
| CBR3-AS1 | ↑ | miR-145-5p | [93] |
| CRNDE | ↑ | miR-136/E2F1 | [94] |
| LINC00460 | ↑ | miR-149-5p/miR-150-5p/p53 | [95] |
| KCNQ1OT1 | ↑ | miR-34a/Atg4B | [96] |
| MIR155HG | ↑ | miR-650/Annexin A2 | [91] |
| lnc-RP11-536 K7.3 | ↑ | SOX2/HIF-1α/USP7 | [97] |
| TUG1 | ↑ | GATA6/BMP | [98] |
| LUCAT1 | ↑ | UBA52/RPL40/MDM2/p53 | [99] |
| PiHL | ↑ | EZH2/HMGA2/PI3K/Akt | [100] |
| ELFN1-AS1 | ↑ | EZH2/DNMT3a/MEIS1 | [102] |
| SNHG5 | ↑ | STAU1 | [103] |
| CCAT2 | ↑ | BOP1/AURKA | [104] |
| NBAT-1 | ↓ | WWC3/LATS1/YAP | [105] |
| MEG3 | ↓ | - | [106] |
| miR-141/PDCD4 | [107] | ||
| lnc-AP | ↓ | pep-AP/TALDO1 | [108] |
| lncRNA PVT1 | - | hsa-miR-297/GSTA2 | [109] |
Studies discovered that high expression of the lncRNA GIHCG promoted the proliferation, migration, and invasion of tumor cells, and enhanced their resistance to 5-FU and oxaliplatin[82]. Similarly, overexpression of the lncRNA ARSR reduced cell apoptosis and induced oxaliplatin resistance[83]. Notably, emerging evidence has revealed that lncRNAs could target mRNAs to promote chemoresistance[84]. For instance, upregulation of the lncRNA HOTAIR has been reported to facilitate EMT in a ZEB1-dependent manner by negatively regulating miR-1277-5p, a process that is involved in hypoxia-induced oxaliplatin resistance[85]. We previously discovered that metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) was overexpressed in CRC tissues compared to paired noncancerous tissues. Correspondingly, the overexpression of MALAT1 suppressed E-cadherin expression and promoted oxaliplatin-induced EMT by interacting with EZH2. Furthermore, upregulated MALAT1 further inhibited miR-218 expression, resulting in poor response to oxaliplatin-based chemotherapy in CRC patients[86]. Numerous studies have demonstrated that some endogenous transcripts (such as endogenous pseudogenes, lncRNAs, and circRNAs) can complement miRNAs in sequences and inhibit their expression, resulting in the upregulation of target gene expression. This phenomenon is called the miRNA “sponge” effect[87]. Analogously, Fan et al[88] demonstrated that MALAT1 could function as a “sponge” for miR-324-3p and increase the expression of ADAM17 to facilitate resistance to oxaliplatin in CRC cells.
Moreover, lncRNA Opa-interacting protein 5 antisense RNA 1 can complement the sites of miR-137 in sequence, sponging miR-137 and inhibiting its expression, thus conferring oxaliplatin resistance in CRC cells[89]. It was reported that Linc00152 increased ERB-B2 receptor tyrosine kinase 4 expression by acting as a “sponge” for miR-193a-3p resulting in the promoted phosphorylation of AKT at Thr308 and Ser47; this process mediated oxaliplatin resistance in CRC cells[90]. In addition, researchers have illustrated that the lncRNA MIR155HG promoted M2 macrophage polarization and enhanced oxaliplatin resistance in CRC cells by regulating the miR-650/Annexin A2 axis[91]. Similarly, other lncRNAs such as CASC15[92], CBR3-AS1[93], CRNDE[94], LINC00460[95], and KCNQ1OT1[96] have been reported to serve as “sponges” for miRNAs, thereby mediating oxaliplatin resistance in CRC.
Several research groups have revealed that lncRNAs could stabilize functional proteins, and mediate the chemoresistance of cancers. For example, lncRNA-RP11-536 K7.3 contributed to oxaliplatin resistance by recruiting SOX2 to activate deubiquitinase USP7 and stabilize the protein HIF-1α in CRC cells[97]. Furthermore, the lncRNA TUG1 induced oxaliplatin resistance in CRC stem cells and inhibited cell apoptosis by interacting with GATA6[98]. Moreover, two additional studies have further revealed that the lncRNAs LUCAT1 and PiHL mediated oxaliplatin resistance in CRC cells by interacting with UBA52 and EZH2 proteins, respectively[99,100].
LncRNAs have also been demonstrated to be regulators of gene expression by enhancing the stability of mRNA through epigenetic mechanisms[101]. Studies have elucidated that the lncRNA ELFN1-AS1 suppressed myeloid ecotype virus insertion site 1 transcription and promoted oxaliplatin resistance by interacting with EZH2 and DNA methyltransferase 3 alpha after localization to the promoter region of MEIS1 gene[102]. In addition, overexpression of the lncRNA SNHG 5 contributed to the proliferation of CRC cells and their resistance to oxaliplatin-induced apoptosis by blocking the degradation of SPATS 2 by STAU 1[103]. Of note, it has been identified that in microsatellite stable CRC, overexpression of the lncRNA CCAT2 could induce chromosomal instability, and enhance 5-FU and oxaliplatin-resistance by the upregulation of ribosomal biogenesis factor and the activation of aurora kinase A[104].
Previously, several tumor suppressor lncRNAs were reported to be associated with CRC oxaliplatin resistance. For instance, the expression levels of NBAT-1 and MEG3 in CRC tissues were low compared with normal adjacent tissues, especially in oxaliplatin-resistant patients[105,106]. Mechanistically, NBAT-1 was revealed to inhibit the growth of oxaliplatin-resistant CRC cells by activating the WWC 3/LATS 1/YAP pathway after acting as a “sponge” for miR-454[105]. Similarly, MEG3 promoted the sensitivity of CRC cells to oxaliplatin by regulating the miR-141/PDCD4 axis[107]. It was further reported that lnc-AP is a lncRNA with coding potential, and is highly associated with oxaliplatin resistance in CRC; lnc-AP encodes the short peptide pep-AP that enhances sensitization of CRC cells to oxaliplatin via the pep-AP/TALDO1 pathway[108]. Intriguingly, an epidemiological investigation demonstrated that an rs2278176 CT/TT mutation in the lncRNA PVT1 increased the chemosensitivity of CRC patients to FOLFOX compared to CRC patients carrying distinct genotypes[109].
Of note, when compared to parental cells, the lncRNA CRNDE was significantly down-regulated in oxaliplatin-resistant cells and can be considered a predictor of oxaliplatin treatment response and tumor prognosis[110]. Interestingly, this lncRNA can be combined with arginine-rich splicing factor 6 to reduce its stability, knockdown of the latter can inhibit autophagy and increase the sensitivity of cancer cells to oxaliplatin by the alternative splicing of PICALM[111]. Given that up-regulated expression of CRNDE aids an increase in the sensitivity of cancer cells to oxaliplatin, we speculate that increasing CRNDE expression and decreasing the stability of SRSF6 may be a promising strategy to ameliorate CRC resistance to oxaliplatin.
Most circRNAs consist of covalently closed loop single-stranded RNAs produced by the back-splicing of exon precursor mRNAs that are more stable than their linear precursor gene[30]. Currently, circRNAs consist of four general categories: Exonic circRNAs, circular intronic RNAs, exon-intron circRNAs, and circRNA from other sources (such as antisense circular RNA or intergenic circular RNA) based on their structural domains and biogenesis features[112,113]. As vital biological regulators, circRNAs have been reported to widely participate in multiple oxaliplatin resistance-related mechanisms, such as apoptosis, autophagy, glycolysis, TME, EMT, DNA damage repair, etc[114]. Studies identified that hsa_circ_0040
The majority of circRNAs containing miRNA response elements (MREs) can regulate gene expression and signaling pathways by functioning as ceRNAs, mediating tumor chemoresistance[121] (Table 3). Studies have identified that circular RNA protein tyrosine kinase 2 was significantly up-regulated in CRC tissues compared to normal tissues, and its high levels of expression promoted CRC progression and oxaliplatin resistance by regulation of the miR-136-5p/YTHDF1 axis[122]. Similarly, circ_0032833 was highly expressed in FOLFOX-resistant CRC cells, and knock-down of circ_0032833 could promote apoptosis and partially enhance the sensitivity of CRC cells to 5-FU and oxaliplatin through the activity of miR-125-5p/MSI1[123]. Lai et al[124] also demonstrated that hsa_circ_0079662 functioned as a “sponge” for hsa-mir-324-5p and activated HOXA9 through activation of the TNF-α pathway; this process induced oxaliplatin-resistance in CRC cells. Furthermore, our previous research revealed that circHIPK3 was highly expressed in chemoresistance CRC patients, and this circRNA was strongly associated with tumor size, regional lymph node metastasis, and distant metastasis. The following functional investigation revealed that circHIPK3 facilitated oxaliplatin resistance, but not 5-FU resistance, by restraining autophagy-related cell death. Mechanistically, circHIPK3 acted as a ceRNA and a “sponge” for miR-637 resulting in activation of the STAT3/Bcl-2/Beclin1 pathway, and subsequent induction of oxaliplatin-resistance in CRC cells[125].
Intriguingly, oxaliplatin could directly induce the overexpression of some oncogenes, further exacerbating chemoresistance. Circular CCDC66, derived from exons 6 to 11, was highly expressed in oxaliplatin-resistant CRC tissues and cells, a phenomenon caused by oxaliplatin-triggered cell stress through phosphoinositide 3-kinase related kinases-mediated phosphorylation of DHX9[126]. Despite extensive literature, many circRNAs remain to be explored in the study of oxaliplatin-resistant CRC. Abu et al[127] identified differential expression of circular RNAs when analyzing chemosensitive and chemoresistant CRC cells using microarray; hsa_circ_32883 and hsa_circ_0338 were screened as promising candidates. Subsequently, their study also identified that exosome-derived circRNAs (hsa_circ_0032883, hsa_circ_0002039, and hsa_circ_0000338) may play important roles in the manifestation of chemoresistance[128].
Studies have reported that the expression of exosomal miR-21-5p from oxaliplatin-resistant cells was significantly elevated, and therefore levels of this miRNA could act as a predictor of chemotherapy response in CRC patients[129]. Interestingly, miR-21-5p can be sponged by some tumor-suppressive circRNAs, such as circDDX17[35] and circEPB41L2[130], thus resulting in activation of the downstream PTEN/AKT pathway. Importantly, previous studies have confirmed that targeting the PTEN/PI3K/AKT/mTOR pathway partially reversed oxaliplatin resistance in CRC[51,52]. Given this, we speculate that targeting these suppressive circRNAs might contribute to the sensitization of CRC cells to oxaliplatin via the miR-21-5p/PTEN/AKT axis. Furthermore, hsa_circ_0001955 and hsa_circ_0000977, potential upstream targets for chemoresistance-associated miRNAs[49,93], were demonstrated to be dysfunctional in CRC tissue, a finding that suggests these circRNAs may be involved in the progression of CRC chemoresistance in a ceRNA-dependent manner[131]. Currently, studies regarding the roles of circRNAs in oxaliplatin resistance are less commonly reported, the precise and comprehensive mechanisms of circRNAs in CRC chemoresistance need to be explored by further study.
Extracellular vesicles (EVs) are defined as a class of membranous vesicles that are released by cells to the extracellular matrix and play a key role in various physiological and pathological processes. According to the difference in their origin, size, content, and biological function, EVs can be roughly divided into three main subtypes—microbubbles, exosomes, and apoptotic bodies[132]. Exosomes are an emerging hallmark of liquid biopsy and have attracted much scientific attention of late, these small disc-shaped EVs have a diameter of approximately 40–150 nm and are enclosed by a lipid bilayer membrane[133]. Almost all cell types can secrete exosomes, and they also exist broadly within fluids including blood, urine, saliva, milk, ascites, cerebrospinal fluid, and others[40,134]. At present, ultracentrifugation remains the standard method for exosome extraction. However, with the emergence of novel technologies, exosome isolation and purification strategies are being continuously optimized, such as size exclusion chromatography, magnetic bead immune capture, and microfluidic-based chip technology techniques[135], these are aimed at efficiently obtaining exosomes with high abundance and quality. The characterization of exosomes has emerged as a prerequisite for their utilization, and the three basic characterization modes of morphological identification are particle size distribution and protein markers. However, more advanced features are gradually being included, in exosome characterization, such as the identification of the purity of exosomes and their uptake[135,136]. As signaling entities, numerous studies have demonstrated that exosomes exert biological effects in two ways: Firstly, exosomal membrane proteins or lipids act as ligands, directly activating receptors at the surface of target cells, generating cascade signal events and activating intracellular signaling pathways; Secondly, exosomes entrain and transport cellular signal-regulating molecules to target cells; these include DNA, lipids, proteins, and RNAs, which are extensively involved in tumorigenesis, metastasis, recurrence, and chemoresistance of cancers[137,138]. In particular, exosomal ncRNAs have received unprecedented appreciation in biomedical research and clinical application recently[139,140]. Several exosomal ncRNAs involved in CRC oxaliplatin resistance are listed in Table 4.
| Exosomal ncRNAs1 | Donor cells | Recipient cells | Targets and pathways | Ref. |
| miR-21↓ | THLG-293T/LG-293T cells | CRC cells | - | [156] |
| miR-208b↑ | CRC cells | T cells | PDCD4 | [145] |
| miR-92a-3p↑ | CAFs cells | CRC cells | FBXW7/MOAP1 | [20] |
| miR-46146↑ | CRC-R cells | CRC-S cells | PDCD10 | [146] |
| miR-1915-3p↓ | FHC cells | CRC cells | PFKFB3/USP2 | [147] |
| lncRNA H19↑ | CAFs cells | CRC cells | - | [141] |
| lncRNA CCAL↑ | CAFs cells | CRC cells | HuR/β-catenin | [142] |
| cricN4BP2L2 | CAFs cells | CRC cells | EIF4A3/PI3K/AKT/mTOR | [143] |
| ciRS-122↑ | CRC-R cells | CRC-S cells | miR-122/PKM2 | [148] |
| circ_0094343↓ | NCM460 cells | CRC cells | miR-766-5p/TRIM67 | [144] |
Increasing numbers of reports have revealed that exosomal ncRNAs can mediate chemoresistance via remodeling of the tumor microenvironment. Studies have identified that lncRNA H19 is highly expressed in CRC tissues compared to normal tissues. Moreover, lncRNA H19 derived from CAFs can act as a direct “sponge” for miR-141 and activate the β-catenin pathway, thus promoting stemness and oxaliplatin resistance[141]. Similarly, exosomal miR-92a-3p was transmitted from CAFs to cancer cells, promoting chemoresistance via the Wnt/β-catenin/apoptosis pathway[20]. Moreover, CAF-derived exosomes were revealed to deliver CRC-associated lncRNA to cancer cells, interacting with the mRNA of the stable protein human antigen R and increasing the expression of β-catenin; these processes promoted oxaliplatin resistance in CRC cells[142]. Qu et al[143] also found that exosomal cricN4BP2L2 secreted by CAFs can interact with EIF4A3 and modulate the PI3K/AKT/mTOR signaling pathway, thus promoting oxaliplatin resistance and the stemness of CRC cells. In addition, exosomal circ_0094343 derived from human colonic epithelial cells (NCM460 cells) was identified to be taken up by CRC cells in which it regulated cell apoptosis and glycolysis via the miR-766-5p/TRIM67 pathway, thereby enhancing the chemosensitivity of tumor cells[144].
Conversely, tumor cells have been revealed to generate exosomes in an autocrine or paracrine manner; they are taken up by themselves or delivered to the local microenvironment, thus exerting functional regulation. Ning et al[145] demonstrated that CRC cell-derived exosomal miR-208b could promote Treg expansion by directly inhibiting the expression of programmed cell death factor 4, resulting in promoted tumor progression and oxaliplatin resistance. Similarly, studies have revealed that exosomal miR-46146 conferred chemoresistance via the targeting of programmed cell death factor 10 in CRC cells[146]. Interestingly, one particular study revealed that miR-1915-3p was down-regulated in oxaliplatin-resistant CRC cells compared to oxaliplatin- sensitive cells and that EVs-derived miR-1915-3p increased the oxaliplatin sensitivity of drug-resistant cells by targeting PFKFB3/USP2[147]. Moreover, it has been reported that hsa_circ_0005963 can be transferred from oxaliplatin-resistant cells to sensitive cells by exosomes, directly acting as a miR-122 “sponge” and increasing the expression of PKM2, which promotes glycolysis and oxaliplatin-resistance in sensitive cells[148].
Liquid biopsy, in which diseases are evaluated via sampling biological fluids, can avoid the influence of tissue heterogeneity on tumor molecular typing to some extent. More and more evidence show that abundant exosomes are enriched in body fluids and participate in many physiological and pathological processes. As an emerging hallmark of liquid biopsy, exosomes are of substantial value in cancer diagnosis, monitoring, and prediction[136,140]. Studies found that the ratio of exosomal miRNAs is an effective predictor of tumor response in peritoneal metastatic patients after repeated intraperitoneal chemotherapy, as patients with high ratios of miR-223-3p/miR-29b-3p or miR-21-5p/miR-29b-3p had inferior survival outcomes compared to patients with low ratios[149]. By analogy, exosomes derived miR-21-5p, miR-1246, miR-1229-5p, and miR-96-5p were highly expressed in CRC patients and cancer cell lines resistant to 5-FU and oxaliplatin compared with those sensitive to chemotherapy. The area under the curve (AUC) of four combinations of exosomal miRNAs was 0.804, which is expected to be an effective predictor of chemotherapy[129]. In addition, researchers found that plasma-derived exosomal miRNA-125b was highly expressed in patients with progressive disease (PD) compared with the healthy control group or patients with stable disease. Importantly, some patients with PD who respond to modified fluorouracil, leucovorin, and oxaliplatin (mFOLFOX6) have obvious differences in the expression of plasma exosomal miRNA-125b before and after chemotherapy, and patients with low expression of exosomal miRNA-125b have higher progression-free survival (PFS)[150]. Furthermore, it was reported that miR-208b was highly expressed in the serum of FOLFOX-resistant CRC patients and the AUC of serum miRNA-208b was 0.771 [95% confidence interval (CI): 0.688-0.855], which is better than that of serum CEA of 0.493 (95%CI: 0.385-0.601). Their research suggests that this miRNA can be regarded as a promising liquid biopsy indicator to predict FOLFOX sensitivity in cancer patients[145]. Encouragingly, a panel of plasma exosomal miR-17-5p and miR-185-5p were successfully used to predict FOLFOX4/FOLFIRI responses in patients with advanced CRC[151].
A large number of reports have revealed the prospects for the application of exosomes to overcome chemoresistance, their strong stability, intrinsic biocompatibility, low immunogenicity, and natural targeting ability making them favorable targets[152,153]. Novel research has exhibited an engineered exosome encapsulating oxaliplatin and PGM5 Antisense RNA 1 that were delivered into CRC cells, and these effectively relieved chemoresistance and inhibited tumor progression[154]. Furthermore, Pi et al[155] constructed engineered EVs enwrapped in folate nanoparticles and survivin siRNA, which significantly inhibited tumor growth in CRC xenograft mice. Similarly, miR-21 inhibitors have been co-incorporated into exosomes, resulting in enhanced chemosensitivity of CRC cells[156]. Taken together, these findings suggest that exogenous delivery of ncRNAs could serve as a novel therapeutic target for CRC.
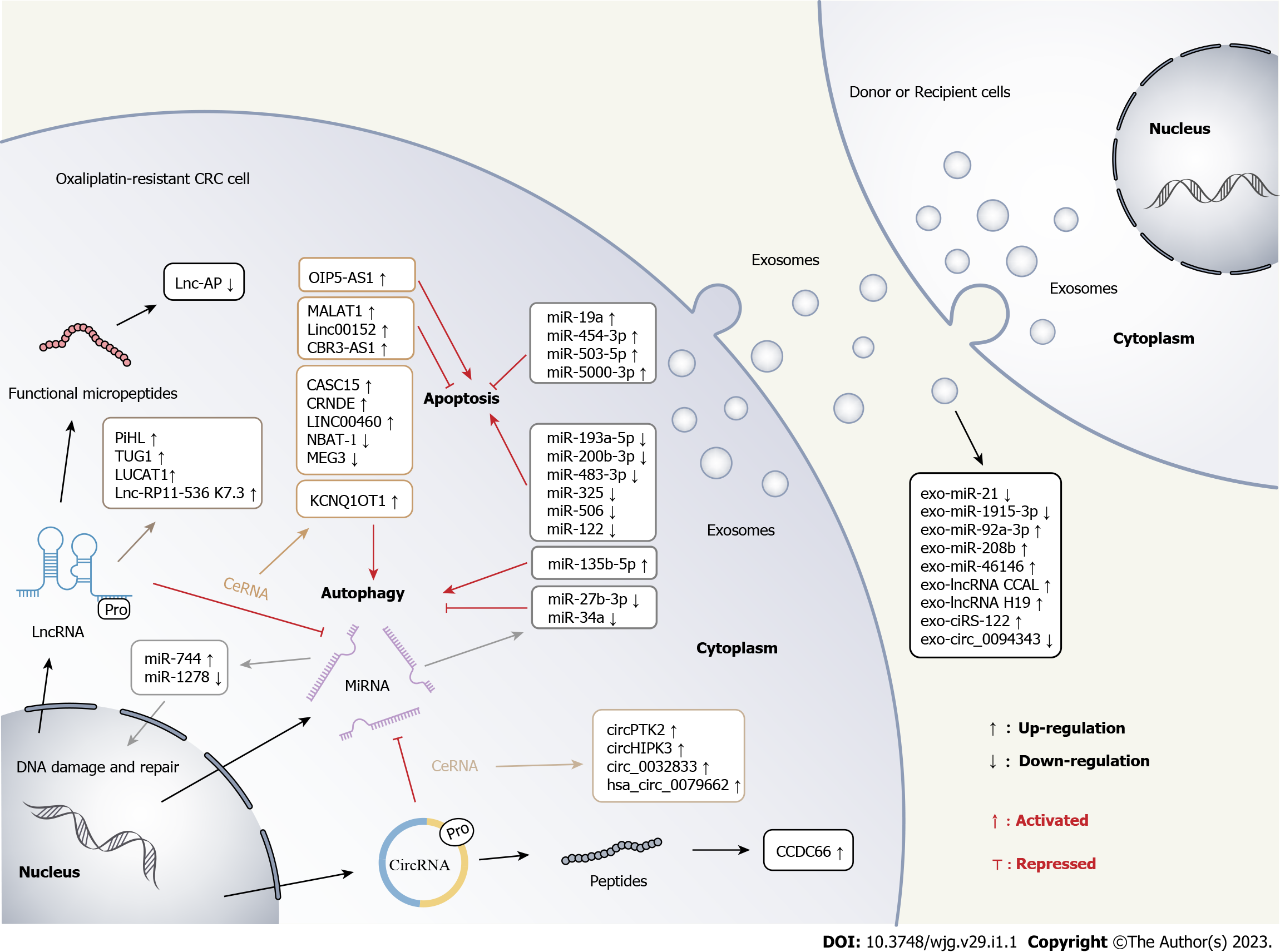
A multitude of emerging studies have confirmed that dysfunctional ncRNAs contribute to the development of malignancies, including chemoresistance. In this review, we systematically summarized the multiple mechanisms by which ncRNAs function in CRC oxaliplatin resistance and discussed the biomedical prospects of exosomal ncRNAs as hallmarks for fluid biopsy and therapeutic targets. Figure 2 schematically illustrates the multiple regulatory roles of ncRNAs in CRC oxaliplatin resistance. Owing to the differential expression patterns of ncRNAs in cancer patients, it is considered they may be used as biomarkers for early detection, tumor staging, and clinical outcome. Although many mechanisms by which ncRNAs exert their biological effects remain to be deciphered, targeting these ncRNAs has emerged as a promising strategy to ameliorate chemoresistance.
Nevertheless, the integration and application of ncRNAs in clinical practice are far from the existing research fervor and are currently being tested in experimental studies; there are still many challenges to be overcome before ncRNAs can be extensively implemented in clinics. Firstly, the results of existing research are confounding due to differing nomenclature being utilized for circRNAs. The circRNAs labeled using different naming standards make the subsequent genomic localization cumbersome. Secondly, the expression of ncRNAs in biological fluids is relatively low, and therefore accurate detection and quantification of these molecules is a prerequisite for the development of biomarkers. Furthermore, due to differences in the lengths and mechanisms by which ncRNAs function in distinct tumor types, it is a challenge to select appropriate targets for these multitudinous candidates. Exosomes may be a better intermediary for delivering ncRNAs therapies; this could be achieved in an endogenous or exogenous manner. However, it is crucial to increase the loading of ncRNAs delivered by endogenous exosomes and ensure the safety of engineered exosomes with complex structures. Measurements of the efficacy of exosomal ncRNAs in clinical applications cannot be limited to laboratory data alone. A series of large validation cohorts will be required to obtain reliable predictive models and accurate therapeutic doses. In the future, clinical translational trials utilizing target ncRNAs are expected to overcome oxaliplatin resistance and thus improve the clinical outcomes of CRC patients.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Imai Y, Japan; Jeong KY, South Korea; Manojlovic N, Serbia S-Editor: Liu GL L-Editor: Webster JR P-Editor: Liu GL
| 1. | Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75126] [Cited by in RCA: 64583] [Article Influence: 16145.8] [Reference Citation Analysis (176)] |
| 2. | Dekker E, Tanis PJ, Vleugels JLA, Kasi PM, Wallace MB. Colorectal cancer. Lancet. 2019;394:1467-1480. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1570] [Cited by in RCA: 3016] [Article Influence: 502.7] [Reference Citation Analysis (3)] |
| 3. | Biller LH, Schrag D. Diagnosis and Treatment of Metastatic Colorectal Cancer: A Review. JAMA. 2021;325:669-685. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 398] [Cited by in RCA: 1432] [Article Influence: 358.0] [Reference Citation Analysis (0)] |
| 4. | Kanemitsu Y, Shitara K, Mizusawa J, Hamaguchi T, Shida D, Komori K, Ikeda S, Ojima H, Ike H, Shiomi A, Watanabe J, Takii Y, Yamaguchi T, Katsumata K, Ito M, Okuda J, Hyakudomi R, Shimada Y, Katayama H, Fukuda H; JCOG Colorectal Cancer Study Group. Primary Tumor Resection Plus Chemotherapy Versus Chemotherapy Alone for Colorectal Cancer Patients With Asymptomatic, Synchronous Unresectable Metastases (JCOG1007; iPACS): A Randomized Clinical Trial. J Clin Oncol. 2021;39:1098-1107. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 136] [Article Influence: 34.0] [Reference Citation Analysis (0)] |
| 5. | Grothey A, Sobrero AF, Shields AF, Yoshino T, Paul J, Taieb J, Souglakos J, Shi Q, Kerr R, Labianca R, Meyerhardt JA, Vernerey D, Yamanaka T, Boukovinas I, Meyers JP, Renfro LA, Niedzwiecki D, Watanabe T, Torri V, Saunders M, Sargent DJ, Andre T, Iveson T. Duration of Adjuvant Chemotherapy for Stage III Colon Cancer. N Engl J Med. 2018;378:1177-1188. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 639] [Cited by in RCA: 685] [Article Influence: 97.9] [Reference Citation Analysis (0)] |
| 6. | Hecht JR, Lonardi S, Bendell J, Sim HW, Macarulla T, Lopez CD, Van Cutsem E, Muñoz Martin AJ, Park JO, Greil R, Wang H, Hozak RR, Gueorguieva I, Lin Y, Rao S, Ryoo BY. Randomized Phase III Study of FOLFOX Alone or With Pegilodecakin as Second-Line Therapy in Patients With Metastatic Pancreatic Cancer That Progressed After Gemcitabine (SEQUOIA). J Clin Oncol. 2021;39:1108-1118. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 79] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 7. | Martinez-Balibrea E, Martínez-Cardús A, Ginés A, Ruiz de Porras V, Moutinho C, Layos L, Manzano JL, Bugés C, Bystrup S, Esteller M, Abad A. Tumor-Related Molecular Mechanisms of Oxaliplatin Resistance. Mol Cancer Ther. 2015;14:1767-1776. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 160] [Cited by in RCA: 236] [Article Influence: 23.6] [Reference Citation Analysis (0)] |
| 8. | Van der Jeught K, Xu HC, Li YJ, Lu XB, Ji G. Drug resistance and new therapies in colorectal cancer. World J Gastroenterol. 2018;24:3834-3848. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 448] [Cited by in RCA: 413] [Article Influence: 59.0] [Reference Citation Analysis (5)] |
| 9. | Kang L, Tian Y, Xu S, Chen H. Oxaliplatin-induced peripheral neuropathy: clinical features, mechanisms, prevention and treatment. J Neurol. 2021;268:3269-3282. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 108] [Article Influence: 21.6] [Reference Citation Analysis (0)] |
| 10. | Mauri G, Gori V, Bonazzina E, Amatu A, Tosi F, Bencardino K, Ruggieri L, Patelli G, Arena S, Bardelli A, Siena S, Sartore-Bianchi A. Oxaliplatin retreatment in metastatic colorectal cancer: Systematic review and future research opportunities. Cancer Treat Rev. 2020;91:102112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 39] [Article Influence: 7.8] [Reference Citation Analysis (1)] |
| 11. | Wei L, Wang X, Lv L, Zheng Y, Zhang N, Yang M. The emerging role of noncoding RNAs in colorectal cancer chemoresistance. Cell Oncol (Dordr). 2019;42:757-768. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 67] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 12. | Ghanbarian M, Afgar A, Yadegarazari R, Najafi R, Teimoori-Toolabi L. Through oxaliplatin resistance induction in colorectal cancer cells, increasing ABCB1 level accompanies decreasing level of miR-302c-5p, miR-3664-5p and miR-129-5p. Biomed Pharmacother. 2018;108:1070-1080. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 44] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 13. | Cui J, Li G, Yin J, Li L, Tan Y, Wei H, Liu B, Deng L, Tang J, Chen Y, Yi L. GSTP1 and cancer: Expression, methylation, polymorphisms and signaling (Review). Int J Oncol. 2020;56:867-878. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 41] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 14. | Sreekumar R, Al-Saihati H, Emaduddin M, Moutasim K, Mellone M, Patel A, Kilic S, Cetin M, Erdemir S, Navio MS, Lopez MA, Curtis N, Yagci T, Primrose JN, Price BD, Berx G, Thomas GJ, Tulchinsky E, Mirnezami A, Sayan AE. The ZEB2-dependent EMT transcriptional programme drives therapy resistance by activating nucleotide excision repair genes ERCC1 and ERCC4 in colorectal cancer. Mol Oncol. 2021;15:2065-2083. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 17] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 15. | Xiao Y, Lin FT, Lin WC. ACTL6A promotes repair of cisplatin-induced DNA damage, a new mechanism of platinum resistance in cancer. Proc Natl Acad Sci U S A. 2021;118. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 52] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 16. | Liu J, Miao X, Xiao B, Huang J, Tao X, Zhang J, Zhao H, Pan Y, Wang H, Gao G, Xiao GG. Obg-Like ATPase 1 Enhances Chemoresistance of Breast Cancer via Activation of TGF-β/Smad Axis Cascades. Front Pharmacol. 2020;11:666. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 17. | Kwak AW, Park JW, Lee SO, Lee JY, Seo JH, Yoon G, Lee MH, Choi JS, Shim JH. Isolinderalactone sensitizes oxaliplatin-resistance colorectal cancer cells through JNK/p38 MAPK signaling pathways. Phytomedicine. 2022;105:154383. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 25] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 18. | Liu X, Su K, Sun X, Jiang Y, Wang L, Hu C, Zhang C, Lu M, Du X, Xing B. Sec62 promotes stemness and chemoresistance of human colorectal cancer through activating Wnt/β-catenin pathway. J Exp Clin Cancer Res. 2021;40:132. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 89] [Cited by in RCA: 125] [Article Influence: 31.3] [Reference Citation Analysis (0)] |
| 19. | Ma J, Zeng S, Zhang Y, Deng G, Qu Y, Guo C, Yin L, Han Y, Cai C, Li Y, Wang G, Bonkovsky HL, Shen H. BMP4 promotes oxaliplatin resistance by an induction of epithelial-mesenchymal transition via MEK1/ERK/ELK1 signaling in hepatocellular carcinoma. Cancer Lett. 2017;411:117-129. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 63] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 20. | Hu JL, Wang W, Lan XL, Zeng ZC, Liang YS, Yan YR, Song FY, Wang FF, Zhu XH, Liao WJ, Liao WT, Ding YQ, Liang L. CAFs secreted exosomes promote metastasis and chemotherapy resistance by enhancing cell stemness and epithelial-mesenchymal transition in colorectal cancer. Mol Cancer. 2019;18:91. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 240] [Cited by in RCA: 558] [Article Influence: 93.0] [Reference Citation Analysis (0)] |
| 21. | Yin Y, Yao S, Hu Y, Feng Y, Li M, Bian Z, Zhang J, Qin Y, Qi X, Zhou L, Fei B, Zou J, Hua D, Huang Z. The Immune-microenvironment Confers Chemoresistance of Colorectal Cancer through Macrophage-Derived IL6. Clin Cancer Res. 2017;23:7375-7387. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 208] [Cited by in RCA: 193] [Article Influence: 24.1] [Reference Citation Analysis (0)] |
| 22. | Rubio MF, Lira MC, Rosa FD, Sambresqui AD, Salazar Güemes MC, Costas MA. RAC3 influences the chemoresistance of colon cancer cells through autophagy and apoptosis inhibition. Cancer Cell Int. 2017;17:111. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 23] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 23. | Zhang H, Lu B. The Roles of ceRNAs-Mediated Autophagy in Cancer Chemoresistance and Metastasis. Cancers (Basel). 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 34] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 24. | Yang C, Zhang Y, Lin S, Liu Y, Li W. Suppressing the KIF20A/NUAK1/Nrf2/GPX4 signaling pathway induces ferroptosis and enhances the sensitivity of colorectal cancer to oxaliplatin. Aging (Albany NY). 2021;13:13515-13534. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 109] [Article Influence: 27.3] [Reference Citation Analysis (0)] |
| 25. | Hou X, Xia J, Feng Y, Cui L, Yang Y, Yang P, Xu X. USP47-Mediated Deubiquitination and Stabilization of TCEA3 Attenuates Pyroptosis and Apoptosis of Colorectal Cancer Cells Induced by Chemotherapeutic Doxorubicin. Front Pharmacol. 2021;12:713322. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 26] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 26. | Lan H, Liu Y, Liu J, Wang X, Guan Z, Du J, Jin K. Tumor-Associated Macrophages Promote Oxaliplatin Resistance via METTL3-Mediated m6A of TRAF5 and Necroptosis in Colorectal Cancer. Mol Pharm. 2021;18:1026-1037. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 72] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 27. | Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, Xue C, Marinov GK, Khatun J, Williams BA, Zaleski C, Rozowsky J, Röder M, Kokocinski F, Abdelhamid RF, Alioto T, Antoshechkin I, Baer MT, Bar NS, Batut P, Bell K, Bell I, Chakrabortty S, Chen X, Chrast J, Curado J, Derrien T, Drenkow J, Dumais E, Dumais J, Duttagupta R, Falconnet E, Fastuca M, Fejes-Toth K, Ferreira P, Foissac S, Fullwood MJ, Gao H, Gonzalez D, Gordon A, Gunawardena H, Howald C, Jha S, Johnson R, Kapranov P, King B, Kingswood C, Luo OJ, Park E, Persaud K, Preall JB, Ribeca P, Risk B, Robyr D, Sammeth M, Schaffer L, See LH, Shahab A, Skancke J, Suzuki AM, Takahashi H, Tilgner H, Trout D, Walters N, Wang H, Wrobel J, Yu Y, Ruan X, Hayashizaki Y, Harrow J, Gerstein M, Hubbard T, Reymond A, Antonarakis SE, Hannon G, Giddings MC, Ruan Y, Wold B, Carninci P, Guigó R, Gingeras TR. Landscape of transcription in human cells. Nature. 2012;489:101-108. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3924] [Cited by in RCA: 3987] [Article Influence: 306.7] [Reference Citation Analysis (0)] |
| 28. | ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57-74. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14554] [Cited by in RCA: 12945] [Article Influence: 995.8] [Reference Citation Analysis (0)] |
| 29. | Ali SA, Peffers MJ, Ormseth MJ, Jurisica I, Kapoor M. The non-coding RNA interactome in joint health and disease. Nat Rev Rheumatol. 2021;17:692-705. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 142] [Article Influence: 35.5] [Reference Citation Analysis (0)] |
| 30. | Liu CX, Chen LL. Circular RNAs: Characterization, cellular roles, and applications. Cell. 2022;185:2016-2034. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 559] [Article Influence: 186.3] [Reference Citation Analysis (0)] |
| 31. | Kaikkonen MU, Lam MT, Glass CK. Non-coding RNAs as regulators of gene expression and epigenetics. Cardiovasc Res. 2011;90:430-440. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 466] [Cited by in RCA: 467] [Article Influence: 33.4] [Reference Citation Analysis (0)] |
| 32. | Eidem TM, Kugel JF, Goodrich JA. Noncoding RNAs: Regulators of the Mammalian Transcription Machinery. J Mol Biol. 2016;428:2652-2659. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 41] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 33. | Slack FJ, Chinnaiyan AM. The Role of Non-coding RNAs in Oncology. Cell. 2019;179:1033-1055. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 521] [Cited by in RCA: 1109] [Article Influence: 221.8] [Reference Citation Analysis (0)] |
| 34. | Yan H, Bu P. Non-coding RNA in cancer. Essays Biochem. 2021;65:625-639. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 386] [Article Influence: 96.5] [Reference Citation Analysis (0)] |
| 35. | Li XN, Wang ZJ, Ye CX, Zhao BC, Li ZL, Yang Y. RNA sequencing reveals the expression profiles of circRNA and indicates that circDDX17 acts as a tumor suppressor in colorectal cancer. J Exp Clin Cancer Res. 2018;37:325. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 133] [Cited by in RCA: 187] [Article Influence: 26.7] [Reference Citation Analysis (0)] |
| 36. | Zhu KP, Zhang CL, Ma XL, Hu JP, Cai T, Zhang L. Analyzing the Interactions of mRNAs and ncRNAs to Predict Competing Endogenous RNA Networks in Osteosarcoma Chemo-Resistance. Mol Ther. 2019;27:518-530. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 153] [Article Influence: 25.5] [Reference Citation Analysis (0)] |
| 37. | Li Y, Zhao J, Yu S, Wang Z, He X, Su Y, Guo T, Sheng H, Chen J, Zheng Q, Li Y, Guo W, Cai X, Shi G, Wu J, Wang L, Wang P, Huang S. Extracellular Vesicles Long RNA Sequencing Reveals Abundant mRNA, circRNA, and lncRNA in Human Blood as Potential Biomarkers for Cancer Diagnosis. Clin Chem. 2019;65:798-808. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 204] [Article Influence: 34.0] [Reference Citation Analysis (0)] |
| 38. | Chen B, Dragomir MP, Yang C, Li Q, Horst D, Calin GA. Targeting non-coding RNAs to overcome cancer therapy resistance. Signal Transduct Target Ther. 2022;7:121. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 254] [Article Influence: 84.7] [Reference Citation Analysis (0)] |
| 39. | Overman MJ, Modak J, Kopetz S, Murthy R, Yao JC, Hicks ME, Abbruzzese JL, Tam AL. Use of research biopsies in clinical trials: are risks and benefits adequately discussed? J Clin Oncol. 2013;31:17-22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 206] [Cited by in RCA: 247] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 40. | Yu W, Hurley J, Roberts D, Chakrabortty SK, Enderle D, Noerholm M, Breakefield XO, Skog JK. Exosome-based liquid biopsies in cancer: opportunities and challenges. Ann Oncol. 2021;32:466-477. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 328] [Cited by in RCA: 555] [Article Influence: 138.8] [Reference Citation Analysis (0)] |
| 41. | Zhou H, Zhu L, Song J, Wang G, Li P, Li W, Luo P, Sun X, Wu J, Liu Y, Zhu S, Zhang Y. Liquid biopsy at the frontier of detection, prognosis and progression monitoring in colorectal cancer. Mol Cancer. 2022;21:86. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 152] [Article Influence: 50.7] [Reference Citation Analysis (1)] |
| 42. | Anfossi S, Babayan A, Pantel K, Calin GA. Clinical utility of circulating non-coding RNAs - an update. Nat Rev Clin Oncol. 2018;15:541-563. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 256] [Cited by in RCA: 349] [Article Influence: 58.2] [Reference Citation Analysis (0)] |
| 43. | Winkle M, El-Daly SM, Fabbri M, Calin GA. Noncoding RNA therapeutics - challenges and potential solutions. Nat Rev Drug Discov. 2021;20:629-651. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 884] [Cited by in RCA: 1021] [Article Influence: 255.3] [Reference Citation Analysis (0)] |
| 44. | Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15:509-524. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3368] [Cited by in RCA: 4118] [Article Influence: 374.4] [Reference Citation Analysis (1)] |
| 45. | Lu TX, Rothenberg ME. MicroRNA. J Allergy Clin Immunol. 2018;141:1202-1207. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 853] [Cited by in RCA: 1818] [Article Influence: 227.3] [Reference Citation Analysis (0)] |
| 46. | Liang Y, Zou Q, Yu W. Steering Against Wind: A New Network of NamiRNAs and Enhancers. Genomics Proteomics Bioinformatics. 2017;15:331-337. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 23] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 47. | Suzuki HI, Young RA, Sharp PA. Super-Enhancer-Mediated RNA Processing Revealed by Integrative MicroRNA Network Analysis. Cell. 2017;168:1000-1014.e15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 226] [Cited by in RCA: 214] [Article Influence: 26.8] [Reference Citation Analysis (0)] |
| 48. | Odame E, Chen Y, Zheng S, Dai D, Kyei B, Zhan S, Cao J, Guo J, Zhong T, Wang L, Li L, Zhang H. Enhancer RNAs: transcriptional regulators and workmates of NamiRNAs in myogenesis. Cell Mol Biol Lett. 2021;26:4. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 14] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 49. | Wang H, Wang X, Zhang H, Deng T, Liu R, Liu Y, Li H, Bai M, Ning T, Wang J, Ge S, Ba Y. The HSF1/miR-135b-5p axis induces protective autophagy to promote oxaliplatin resistance through the MUL1/ULK1 pathway in colorectal cancer. Oncogene. 2021;40:4695-4708. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 46] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 50. | Qian XL, Zhou F, Xu S, Jiang J, Chen ZP, Wang SK, Zuo Y, Ni C. MiR-454-3p Promotes Oxaliplatin Resistance by Targeting PTEN in Colorectal Cancer. Front Oncol. 2021;11:638537. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 20] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 51. | Zhang Y, Liu X, Zhang J, Xu Y, Shao J, Hu Y, Shu P, Cheng H. Inhibition of miR-19a partially reversed the resistance of colorectal cancer to oxaliplatin via PTEN/PI3K/AKT pathway. Aging (Albany NY). 2020;12:5640-5650. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 29] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 52. | Liang Y, Zhu D, Zhu L, Hou Y, Hou L, Huang X, Li L, Wang Y, Zou H, Wu T, Yao M, Wang J, Meng X. Dichloroacetate Overcomes Oxaliplatin Chemoresistance in Colorectal Cancer through the miR-543/PTEN/Akt/mTOR Pathway. J Cancer. 2019;10:6037-6047. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 37] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 53. | Liang Y, Zhu D, Hou L, Wang Y, Huang X, Zhou C, Zhu L, Li L, Gu Y, Luo M, Wang J, Meng X. MiR-107 confers chemoresistance to colorectal cancer by targeting calcium-binding protein 39. Br J Cancer. 2020;122:705-714. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 30] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 54. | Xu K, Chen G, Qiu Y, Yuan Z, Li H, Yuan X, Sun J, Xu J, Liang X, Yin P. miR-503-5p confers drug resistance by targeting PUMA in colorectal carcinoma. Oncotarget. 2017;8:21719-21732. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 50] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 55. | Zhou Y, He A, Zhang L, Yi G. MiR-744 mediates the Oxaliplatin chemoresistance in colorectal cancer through inhibiting BIN1. Neoplasma. 2020;67:296-303. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 11] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 56. | Zhuang YY, Zhong W, Xia ZS, Lin SZ, Chan MC, Jiang K, Li WF, Xu XY. miR-5000-3p confers oxaliplatin resistance by targeting ubiquitin-specific peptidase 49 in colorectal cancer. Cell Death Discov. 2021;7:129. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 13] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 57. | Zhao H, Su W, Kang Q, Xing Z, Lin X, Wu Z. Natural killer cells inhibit oxaliplatin-resistant colorectal cancer by repressing WBSCR22 via upregulating microRNA-146b-5p. Am J Cancer Res. 2018;8:824-834. [PubMed] |
| 58. | Jiang T, Ye L, Han Z, Liu Y, Yang Y, Peng Z, Fan J. miR-19b-3p promotes colon cancer proliferation and oxaliplatin-based chemoresistance by targeting SMAD4: validation by bioinformatics and experimental analyses. J Exp Clin Cancer Res. 2017;36:131. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 75] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 59. | Lin W, Zou H, Mo J, Jin C, Jiang H, Yu C, Jiang Z, Yang Y, He B, Wang K. Micro1278 Leads to Tumor Growth Arrest, Enhanced Sensitivity to Oxaliplatin and Vitamin D and Inhibits Metastasis via KIF5B, CYP24A1, and BTG2, Respectively. Front Oncol. 2021;11:637878. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 10] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 60. | Zhou H, Lin C, Zhang Y, Zhang X, Zhang C, Zhang P, Xie X, Ren Z. miR-506 enhances the sensitivity of human colorectal cancer cells to oxaliplatin by suppressing MDR1/P-gp expression. Cell Prolif. 2017;50. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 82] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 61. | Wu YZ, Lin HY, Zhang Y, Chen WF. miR-200b-3p mitigates oxaliplatin resistance via targeting TUBB3 in colorectal cancer. J Gene Med. 2020;22:e3178. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 22] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 62. | Hua Y, Zhu Y, Zhang J, Zhu Z, Ning Z, Chen H, Liu L, Chen Z, Meng Z. miR-122 Targets X-Linked Inhibitor of Apoptosis Protein to Sensitize Oxaliplatin-Resistant Colorectal Cancer Cells to Oxaliplatin-Mediated Cytotoxicity. Cell Physiol Biochem. 2018;51:2148-2159. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 20] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 63. | Azar MRMH, Aghazadeh H, Mohammed HN, Sara MRS, Hosseini A, Shomali N, Tamjidifar R, Tarzi S, Mansouri M, Sarand SP, Marofi F, Akbari M, Xu H, Shotorbani SS. miR-193a-5p as a promising therapeutic candidate in colorectal cancer by reducing 5-FU and Oxaliplatin chemoresistance by targeting CXCR4. Int Immunopharmacol. 2021;92:107355. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 40] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 64. | Liang H, Xu Y, Zhang Q, Yang Y, Mou Y, Gao Y, Chen R, Chen C, Dai P. MiR-483-3p regulates oxaliplatin resistance by targeting FAM171B in human colorectal cancer cells. Artif Cells Nanomed Biotechnol. 2019;47:725-736. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 31] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 65. | Zhang L, Chen H, Song Y, Gu Q, Zhang L, Xie Q, Xu J, Zhang M. MiR-325 Promotes Oxaliplatin-Induced Cytotoxicity Against Colorectal Cancer Through the HSPA12B/PI3K/AKT/Bcl-2 Pathway. Dig Dis Sci. 2021;66:2651-2660. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 12] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 66. | Poel D, Boyd LNC, Beekhof R, Schelfhorst T, Pham TV, Piersma SR, Knol JC, Jimenez CR, Verheul HMW, Buffart TE. Proteomic Analysis of miR-195 and miR-497 Replacement Reveals Potential Candidates that Increase Sensitivity to Oxaliplatin in MSI/P53wt Colorectal Cancer Cells. Cells. 2019;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 20] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 67. | Chen L, He M, Zhang M, Sun Q, Zeng S, Zhao H, Yang H, Liu M, Ren S, Meng X, Xu H. The Role of non-coding RNAs in colorectal cancer, with a focus on its autophagy. Pharmacol Ther. 2021;226:107868. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 78] [Article Influence: 19.5] [Reference Citation Analysis (0)] |
| 68. | Sun C, Wang FJ, Zhang HG, Xu XZ, Jia RC, Yao L, Qiao PF. miR-34a mediates oxaliplatin resistance of colorectal cancer cells by inhibiting macroautophagy via transforming growth factor-β/Smad4 pathway. World J Gastroenterol. 2017;23:1816-1827. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 69] [Cited by in RCA: 86] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 69. | Sun W, Li J, Zhou L, Han J, Liu R, Zhang H, Ning T, Gao Z, Liu B, Chen X, Ba Y. The c-Myc/miR-27b-3p/ATG10 regulatory axis regulates chemoresistance in colorectal cancer. Theranostics. 2020;10:1981-1996. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 109] [Article Influence: 21.8] [Reference Citation Analysis (0)] |
| 70. | Liu XY, Zhang FR, Shang JY, Liu YY, Lv XF, Yuan JN, Zhang TT, Li K, Lin XC, Liu X, Lei Q, Fu XD, Zhou JG, Liang SJ. Renal inhibition of miR-181a ameliorates 5-fluorouracil-induced mesangial cell apoptosis and nephrotoxicity. Cell Death Dis. 2018;9:610. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 39] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 71. | Van Houdt WJ, Emmink BL, Pham TV, Piersma SR, Verheem A, Vries RG, Fratantoni SA, Pronk A, Clevers H, Borel Rinkes IH, Jimenez CR, Kranenburg O. Comparative proteomics of colon cancer stem cells and differentiated tumor cells identifies BIRC6 as a potential therapeutic target. Mol Cell Proteomics. 2011;10:M111.011353. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 72] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 72. | Chen LL. Linking Long Noncoding RNA Localization and Function. Trends Biochem Sci. 2016;41:761-772. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 587] [Cited by in RCA: 783] [Article Influence: 87.0] [Reference Citation Analysis (0)] |
| 73. | Connerty P, Lock RB, de Bock CE. Long Non-coding RNAs: Major Regulators of Cell Stress in Cancer. Front Oncol. 2020;10:285. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 42] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 74. | Gao N, Li Y, Li J, Gao Z, Yang Z, Liu H, Fan T. Long Non-Coding RNAs: The Regulatory Mechanisms, Research Strategies, and Future Directions in Cancers. Front Oncol. 2020;10:598817. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 168] [Cited by in RCA: 205] [Article Influence: 41.0] [Reference Citation Analysis (0)] |
| 75. | Rai MI, Alam M, Lightfoot DA, Gurha P, Afzal AJ. Classification and experimental identification of plant long non-coding RNAs. Genomics. 2019;111:997-1005. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 62] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 76. | Sun Q, Hao Q, Prasanth KV. Nuclear Long Noncoding RNAs: Key Regulators of Gene Expression. Trends Genet. 2018;34:142-157. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 427] [Cited by in RCA: 435] [Article Influence: 62.1] [Reference Citation Analysis (0)] |
| 77. | Postepska-Igielska A, Giwojna A, Gasri-Plotnitsky L, Schmitt N, Dold A, Ginsberg D, Grummt I. LncRNA Khps1 Regulates Expression of the Proto-oncogene SPHK1 via Triplex-Mediated Changes in Chromatin Structure. Mol Cell. 2015;60:626-636. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 175] [Cited by in RCA: 247] [Article Influence: 27.4] [Reference Citation Analysis (0)] |
| 78. | Xu M, Chen X, Lin K, Zeng K, Liu X, Pan B, Xu X, Xu T, Hu X, Sun L, He B, Pan Y, Sun H, Wang S. The long noncoding RNA SNHG1 regulates colorectal cancer cell growth through interactions with EZH2 and miR-154-5p. Mol Cancer. 2018;17:141. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 152] [Cited by in RCA: 234] [Article Influence: 33.4] [Reference Citation Analysis (0)] |
| 79. | Lan Z, Yao X, Sun K, Li A, Liu S, Wang X. The Interaction Between lncRNA SNHG6 and hnRNPA1 Contributes to the Growth of Colorectal Cancer by Enhancing Aerobic Glycolysis Through the Regulation of Alternative Splicing of PKM. Front Oncol. 2020;10:363. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 67] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 80. | Chen QN, Wei CC, Wang ZX, Sun M. Long non-coding RNAs in anti-cancer drug resistance. Oncotarget. 2017;8:1925-1936. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 138] [Cited by in RCA: 155] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 81. | Hu Y, Zhu QN, Deng JL, Li ZX, Wang G, Zhu YS. Emerging role of long non-coding RNAs in cisplatin resistance. Onco Targets Ther. 2018;11:3185-3194. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 62] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 82. | Jiang X, Li Q, Zhang S, Song C, Zheng P. Long noncoding RNA GIHCG induces cancer progression and chemoresistance and indicates poor prognosis in colorectal cancer. Onco Targets Ther. 2019;12:1059-1070. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 30] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 83. | Li T, Jin X, Dong J, Deng H. Long noncoding RNA ARSR is associated with a poor prognosis in patients with colorectal cancer. J Gene Med. 2020;22:e3241. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 84. | Yamamura S, Imai-Sumida M, Tanaka Y, Dahiya R. Interaction and cross-talk between non-coding RNAs. Cell Mol Life Sci. 2018;75:467-484. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 220] [Cited by in RCA: 241] [Article Influence: 34.4] [Reference Citation Analysis (0)] |
| 85. | Weng X, Liu H, Ruan J, Du M, Wang L, Mao J, Cai Y, Lu X, Chen W, Huang Y, Zhi X, Shan J. HOTAIR/miR-1277-5p/ZEB1 axis mediates hypoxia-induced oxaliplatin resistance via regulating epithelial-mesenchymal transition in colorectal cancer. Cell Death Discov. 2022;8:310. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 27] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 86. | Li P, Zhang X, Wang H, Wang L, Liu T, Du L, Yang Y, Wang C. MALAT1 Is Associated with Poor Response to Oxaliplatin-Based Chemotherapy in Colorectal Cancer Patients and Promotes Chemoresistance through EZH2. Mol Cancer Ther. 2017;16:739-751. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 154] [Cited by in RCA: 211] [Article Influence: 26.4] [Reference Citation Analysis (0)] |
| 87. | Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet. 2016;17:272-283. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1116] [Cited by in RCA: 1589] [Article Influence: 176.6] [Reference Citation Analysis (0)] |
| 88. | Fan C, Yuan Q, Liu G, Zhang Y, Yan M, Sun Q, Zhu C. Long non-coding RNA MALAT1 regulates oxaliplatin-resistance via miR-324-3p/ADAM17 axis in colorectal cancer cells. Cancer Cell Int. 2020;20:473. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 32] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 89. | Liang J, Tian XF, Yang W. Effects of long non-coding RNA Opa-interacting protein 5 antisense RNA 1 on colon cancer cell resistance to oxaliplatin and its regulation of microRNA-137. World J Gastroenterol. 2020;26:1474-1489. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 10] [Cited by in RCA: 14] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 90. | Yue B, Cai D, Liu C, Fang C, Yan D. Linc00152 Functions as a Competing Endogenous RNA to Confer Oxaliplatin Resistance and Holds Prognostic Values in Colon Cancer. Mol Ther. 2016;24:2064-2077. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 155] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 91. | Zhou L, Li J, Liao M, Zhang Q, Yang M. LncRNA MIR155HG induces M2 macrophage polarization and drug resistance of colorectal cancer cells by regulating ANXA2. Cancer Immunol Immunother. 2022;71:1075-1091. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 49] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 92. | Gao R, Fang C, Xu J, Tan H, Li P, Ma L. LncRNA CACS15 contributes to oxaliplatin resistance in colorectal cancer by positively regulating ABCC1 through sponging miR-145. Arch Biochem Biophys. 2019;663:183-191. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 75] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 93. | Xie L, Cui G, Li T. Long Noncoding RNA CBR3-AS1 Promotes Stem-like Properties and Oxaliplatin Resistance of Colorectal Cancer by Sponging miR-145-5p. J Oncol. 2022;2022:2260211. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 7] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 94. | Gao H, Song X, Kang T, Yan B, Feng L, Gao L, Ai L, Liu X, Yu J, Li H. Long noncoding RNA CRNDE functions as a competing endogenous RNA to promote metastasis and oxaliplatin resistance by sponging miR-136 in colorectal cancer. Onco Targets Ther. 2017;10:205-216. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 77] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 95. | Meng X, Sun W, Yu J, Zhou Y, Gu Y, Han J, Zhou L, Jiang X, Wang C. LINC00460-miR-149-5p/miR-150-5p-Mutant p53 Feedback Loop Promotes Oxaliplatin Resistance in Colorectal Cancer. Mol Ther Nucleic Acids. 2020;22:1004-1015. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 45] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 96. | Li Y, Li C, Li D, Yang L, Jin J, Zhang B. lncRNA KCNQ1OT1 enhances the chemoresistance of oxaliplatin in colon cancer by targeting the miR-34a/ATG4B pathway. Onco Targets Ther. 2019;12:2649-2660. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 95] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 97. | Li Q, Sun H, Luo D, Gan L, Mo S, Dai W, Liang L, Yang Y, Xu M, Li J, Zheng P, Li X, Li Y, Wang Z. Lnc-RP11-536 K7.3/SOX2/HIF-1α signaling axis regulates oxaliplatin resistance in patient-derived colorectal cancer organoids. J Exp Clin Cancer Res. 2021;40:348. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 54] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 98. | Sun J, Zhou H, Bao X, Wu Y, Jia H, Zhao H, Liu G. lncRNA TUG1 Facilitates Colorectal Cancer Stem Cell Characteristics and Chemoresistance by Enhancing GATA6 Protein Stability. Stem Cells Int. 2021;2021:1075481. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 99. | Zhou Q, Hou Z, Zuo S, Zhou X, Feng Y, Sun Y, Yuan X. LUCAT1 promotes colorectal cancer tumorigenesis by targeting the ribosomal protein L40-MDM2-p53 pathway through binding with UBA52. Cancer Sci. 2019;110:1194-1207. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 80] [Cited by in RCA: 82] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 100. | Deng X, Kong F, Li S, Jiang H, Dong L, Xu X, Zhang X, Yuan H, Xu Y, Chu Y, Peng H, Guan M. A KLF4/PiHL/EZH2/HMGA2 regulatory axis and its function in promoting oxaliplatin-resistance of colorectal cancer. Cell Death Dis. 2021;12:485. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 55] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 101. | Quinodoz S, Guttman M. Long noncoding RNAs: an emerging link between gene regulation and nuclear organization. Trends Cell Biol. 2014;24:651-663. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 235] [Cited by in RCA: 259] [Article Influence: 23.5] [Reference Citation Analysis (0)] |
| 102. | Li Y, Gan Y, Liu J, Li J, Zhou Z, Tian R, Sun R, Xiao Q, Li Y, Lu P, Peng Y, Shu G, Yin G. Downregulation of MEIS1 mediated by ELFN1-AS1/EZH2/DNMT3a axis promotes tumorigenesis and oxaliplatin resistance in colorectal cancer. Signal Transduct Target Ther. 2022;7:87. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 101] [Article Influence: 33.7] [Reference Citation Analysis (0)] |
| 103. | Damas ND, Marcatti M, Côme C, Christensen LL, Nielsen MM, Baumgartner R, Gylling HM, Maglieri G, Rundsten CF, Seemann SE, Rapin N, Thézenas S, Vang S, Ørntoft T, Andersen CL, Pedersen JS, Lund AH. SNHG5 promotes colorectal cancer cell survival by counteracting STAU1-mediated mRNA destabilization. Nat Commun. 2016;7:13875. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 131] [Cited by in RCA: 172] [Article Influence: 19.1] [Reference Citation Analysis (0)] |
| 104. | Chen B, Dragomir MP, Fabris L, Bayraktar R, Knutsen E, Liu X, Tang C, Li Y, Shimura T, Ivkovic TC, De Los Santos MC, Anfossi S, Shimizu M, Shah MY, Ling H, Shen P, Multani AS, Pardini B, Burks JK, Katayama H, Reineke LC, Huo L, Syed M, Song S, Ferracin M, Oki E, Fromm B, Ivan C, Bhuvaneshwar K, Gusev Y, Mimori K, Menter D, Sen S, Matsuyama T, Uetake H, Vasilescu C, Kopetz S, Parker-Thornburg J, Taguchi A, Hanash SM, Girnita L, Slaby O, Goel A, Varani G, Gagea M, Li C, Ajani JA, Calin GA. The Long Noncoding RNA CCAT2 Induces Chromosomal Instability Through BOP1-AURKB Signaling. Gastroenterology. 2020;159:2146-2162.e33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 90] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 105. | Li C, Li X. Antitumor Activity of lncRNA NBAT-1 via Inhibition of miR-4504 to Target to WWC3 in Oxaliplatin-Resistant Colorectal Carcinoma. J Healthc Eng. 2022;2022:9121554. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 106. | Li L, Shang J, Zhang Y, Liu S, Peng Y, Zhou Z, Pan H, Wang X, Chen L, Zhao Q. MEG3 is a prognostic factor for CRC and promotes chemosensitivity by enhancing oxaliplatin-induced cell apoptosis. Oncol Rep. 2017;38:1383-1392. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 64] [Cited by in RCA: 60] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 107. | Wang H, Li H, Zhang L, Yang D. Overexpression of MEG3 sensitizes colorectal cancer cells to oxaliplatin through regulation of miR-141/PDCD4 axis. Biomed Pharmacother. 2018;106:1607-1615. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 60] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 108. | Wang X, Zhang H, Yin S, Yang Y, Yang H, Yang J, Zhou Z, Li S, Ying G, Ba Y. lncRNA-encoded pep-AP attenuates the pentose phosphate pathway and sensitizes colorectal cancer cells to Oxaliplatin. EMBO Rep. 2022;23:e53140. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 46] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 109. | Wu S, Yang X, Tang W, Familiari G, Relucenti M, Aschner M, Li X, Chen R. Chemotherapeutic Risk lncRNA-PVT1 SNP Sensitizes Metastatic Colorectal Cancer to FOLFOX Regimen. Front Oncol. 2022;12:808889. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 8] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 110. | Sun F, Liang W, Qian J. The identification of CRNDE, H19, UCA1 and HOTAIR as the key lncRNAs involved in oxaliplatin or irinotecan resistance in the chemotherapy of colorectal cancer based on integrative bioinformatics analysis. Mol Med Rep. 2019;20:3583-3596. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 22] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 111. | Zhang F, Wang H, Yu J, Yao X, Yang S, Li W, Xu L, Zhao L. LncRNA CRNDE attenuates chemoresistance in gastric cancer via SRSF6-regulated alternative splicing of PICALM. Mol Cancer. 2021;20:6. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 141] [Article Influence: 35.3] [Reference Citation Analysis (0)] |
| 112. | Liu J, Liu T, Wang X, He A. Circles reshaping the RNA world: from waste to treasure. Mol Cancer. 2017;16:58. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 219] [Cited by in RCA: 330] [Article Influence: 41.3] [Reference Citation Analysis (0)] |
| 113. | Huang A, Zheng H, Wu Z, Chen M, Huang Y. Circular RNA-protein interactions: functions, mechanisms, and identification. Theranostics. 2020;10:3503-3517. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 607] [Cited by in RCA: 586] [Article Influence: 117.2] [Reference Citation Analysis (0)] |
| 114. | Wang J, Zhang Y, Liu L, Yang T, Song J. Circular RNAs: new biomarkers of chemoresistance in cancer. Cancer Biol Med. 2021;. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 21] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 115. | Mao G, Zhou B, Xu W, Jiao N, Wu Z, Li J, Liu Y. Hsa_circ_0040809 regulates colorectal cancer development by upregulating methyltransferase DNMT1 via targeting miR-515-5p. J Gene Med. 2021;23:e3388. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 9] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 116. | Li Z, Yao H, Wang S, Li G, Gu X. CircTADA2A suppresses the progression of colorectal cancer via miR-374a-3p/KLF14 axis. J Exp Clin Cancer Res. 2020;39:160. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 45] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 117. | Zhou J, Wang L, Sun Q, Chen R, Zhang C, Yang P, Tan Y, Peng C, Wang T, Jin C, Ji J, Jin K, Sun Y. Hsa_circ_0001666 suppresses the progression of colorectal cancer through the miR-576-5p/PCDH10 axis. Clin Transl Med. 2021;11:e565. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 73] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 118. | Zhang N, Zhang X, Xu W, Mu Z. CircRNA_103948 inhibits autophagy in colorectal cancer in a ceRNA manner. Ann N Y Acad Sci. 2021;1503:88-101. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 119. | Feng J, Li Z, Li L, Xie H, Lu Q, He X. Hypoxiainduced circCCDC66 promotes the tumorigenesis of colorectal cancer via the miR3140/autophagy pathway. Int J Mol Med. 2020;46:1973-1982. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 30] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 120. | Chen Z, Ren R, Wan D, Wang Y, Xue X, Jiang M, Shen J, Han Y, Liu F, Shi J, Kuang Y, Li W, Zhi Q. Hsa_circ_101555 functions as a competing endogenous RNA of miR-597-5p to promote colorectal cancer progression. Oncogene. 2019;38:6017-6034. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 72] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 121. | Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384-388. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4631] [Cited by in RCA: 6044] [Article Influence: 503.7] [Reference Citation Analysis (0)] |
| 122. | Jiang Z, Hou Z, Liu W, Yu Z, Liang Z, Chen S. Circular RNA protein tyrosine kinase 2 (circPTK2) promotes colorectal cancer proliferation, migration, invasion and chemoresistance. Bioengineered. 2022;13:810-823. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 37] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 123. | Li S, Zheng S. Down-Regulation of Circ_0032833 Sensitizes Colorectal Cancer to 5-Fluorouracil and Oxaliplatin Partly Depending on the Regulation of miR-125-5p and MSI1. Cancer Manag Res. 2020;12:11257-11269. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 21] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 124. | Lai M, Liu G, Li R, Bai H, Zhao J, Xiao P, Mei J. Hsa_circ_0079662 induces the resistance mechanism of the chemotherapy drug oxaliplatin through the TNF-α pathway in human colon cancer. J Cell Mol Med. 2020;24:5021-5027. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 46] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 125. | Zhang Y, Li C, Liu X, Wang Y, Zhao R, Yang Y, Zheng X, Zhang Y, Zhang X. circHIPK3 promotes oxaliplatin-resistance in colorectal cancer through autophagy by sponging miR-637. EBioMedicine. 2019;48:277-288. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 96] [Cited by in RCA: 106] [Article Influence: 17.7] [Reference Citation Analysis (1)] |
| 126. | Lin YC, Yu YS, Lin HH, Hsiao KY. Oxaliplatin-Induced DHX9 Phosphorylation Promotes Oncogenic Circular RNA CCDC66 Expression and Development of Chemoresistance. Cancers (Basel). 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 50] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 127. | Abu N, Hon KW, Jeyaraman S, Yahaya A, Abdullah NM, Mustangin M, Sulaiman SA, Jamal R, Ab-Mutalib NS. Identification of differentially expressed circular RNAs in chemoresistant colorectal cancer. Epigenomics. 2019;11:875-884. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 43] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 128. | Hon KW, Ab-Mutalib NS, Abdullah NMA, Jamal R, Abu N. Extracellular Vesicle-derived circular RNAs confers chemoresistance in Colorectal cancer. Sci Rep. 2019;9:16497. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 91] [Cited by in RCA: 108] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 129. | Jin G, Liu Y, Zhang J, Bian Z, Yao S, Fei B, Zhou L, Yin Y, Huang Z. A panel of serum exosomal microRNAs as predictive markers for chemoresistance in advanced colorectal cancer. Cancer Chemother Pharmacol. 2019;84:315-325. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 93] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 130. | Jiang Z, Hou Z, Li L, Liu W, Yu Z, Chen S. Exosomal circEPB41L2 serves as a sponge for miR-21-5p and miR-942-5p to suppress colorectal cancer progression by regulating the PTEN/AKT signalling pathway. Eur J Clin Invest. 2021;51:e13581. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 31] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 131. | Ding B, Yao M, Fan W, Lou W. Whole-transcriptome analysis reveals a potential hsa_circ_0001955/hsa_circ_0000977-mediated miRNA-mRNA regulatory sub-network in colorectal cancer. Aging (Albany NY). 2020;12:5259-5279. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 62] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 132. | Van Niel G, D'Angelo G, Raposo G. Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol. 2018;19:213-228. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3060] [Cited by in RCA: 5614] [Article Influence: 802.0] [Reference Citation Analysis (0)] |
| 133. | Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200:373-383. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4900] [Cited by in RCA: 6158] [Article Influence: 513.2] [Reference Citation Analysis (0)] |
| 134. | Kalluri R. The biology and function of exosomes in cancer. J Clin Invest. 2016;126:1208-1215. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 944] [Cited by in RCA: 1418] [Article Influence: 157.6] [Reference Citation Analysis (0)] |
| 135. | Zhu L, Sun HT, Wang S, Huang SL, Zheng Y, Wang CQ, Hu BY, Qin W, Zou TT, Fu Y, Shen XT, Zhu WW, Geng Y, Lu L, Jia HL, Qin LX, Dong QZ. Isolation and characterization of exosomes for cancer research. J Hematol Oncol. 2020;13:152. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 323] [Cited by in RCA: 324] [Article Influence: 64.8] [Reference Citation Analysis (0)] |
| 136. | Yu D, Li Y, Wang M, Gu J, Xu W, Cai H, Fang X, Zhang X. Exosomes as a new frontier of cancer liquid biopsy. Mol Cancer. 2022;21:56. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 335] [Cited by in RCA: 485] [Article Influence: 161.7] [Reference Citation Analysis (0)] |
| 137. | Kalluri R, LeBleu VS. The biology, function, and biomedical applications of exosomes. Science. 2020;367. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6920] [Cited by in RCA: 6553] [Article Influence: 1310.6] [Reference Citation Analysis (0)] |
| 138. | Yokoi A, Ochiya T. Exosomes and extracellular vesicles: Rethinking the essential values in cancer biology. Semin Cancer Biol. 2021;74:79-91. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 99] [Article Influence: 24.8] [Reference Citation Analysis (0)] |
| 139. | He C, Zheng S, Luo Y, Wang B. Exosome Theranostics: Biology and Translational Medicine. Theranostics. 2018;8:237-255. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 700] [Cited by in RCA: 915] [Article Influence: 130.7] [Reference Citation Analysis (0)] |
| 140. | Baassiri A, Nassar F, Mukherji D, Shamseddine A, Nasr R, Temraz S. Exosomal Non Coding RNA in LIQUID Biopsies as a Promising Biomarker for Colorectal Cancer. Int J Mol Sci. 2020;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 75] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 141. | Ren J, Ding L, Zhang D, Shi G, Xu Q, Shen S, Wang Y, Wang T, Hou Y. Carcinoma-associated fibroblasts promote the stemness and chemoresistance of colorectal cancer by transferring exosomal lncRNA H19. Theranostics. 2018;8:3932-3948. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 296] [Cited by in RCA: 558] [Article Influence: 79.7] [Reference Citation Analysis (0)] |
| 142. | Deng X, Ruan H, Zhang X, Xu X, Zhu Y, Peng H, Kong F, Guan M. Long noncoding RNA CCAL transferred from fibroblasts by exosomes promotes chemoresistance of colorectal cancer cells. Int J Cancer. 2020;146:1700-1716. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 161] [Article Influence: 26.8] [Reference Citation Analysis (0)] |
| 143. | Qu Z, Yang KD, Luo BH, Zhang F. CAFs-secreted exosomal cricN4BP2L2 promoted colorectal cancer stemness and chemoresistance by interacting with EIF4A3. Exp Cell Res. 2022;418:113266. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 27] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 144. | Li C, Li X. Exosome-Derived Circ_0094343 Promotes Chemosensitivity of Colorectal Cancer Cells by Regulating Glycolysis via the miR-766-5p/TRIM67 Axis. Contrast Media Mol Imaging. 2022;2022:2878557. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 16] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 145. | Ning T, Li J, He Y, Zhang H, Wang X, Deng T, Liu R, Li H, Bai M, Fan Q, Zhu K, Ying G, Ba Y. Exosomal miR-208b related with oxaliplatin resistance promotes Treg expansion in colorectal cancer. Mol Ther. 2021;29:2723-2736. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 110] [Article Influence: 27.5] [Reference Citation Analysis (0)] |
| 146. | Xu Y, Zhu M. Novel exosomal miR-46146 transfer oxaliplatin chemoresistance in colorectal cancer. Clin Transl Oncol. 2020;22:1105-1116. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 29] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 147. | Xiao Z, Liu Y, Li Q, Liu Q, Luo Y, Wei S. EVs delivery of miR-1915-3p improves the chemotherapeutic efficacy of oxaliplatin in colorectal cancer. Cancer Chemother Pharmacol. 2021;88:1021-1031. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 22] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 148. | Wang X, Zhang H, Yang H, Bai M, Ning T, Deng T, Liu R, Fan Q, Zhu K, Li J, Zhan Y, Ying G, Ba Y. Exosome-delivered circRNA promotes glycolysis to induce chemoresistance through the miR-122-PKM2 axis in colorectal cancer. Mol Oncol. 2020;14:539-555. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 294] [Cited by in RCA: 400] [Article Influence: 80.0] [Reference Citation Analysis (0)] |
| 149. | Ohzawa H, Kimura Y, Saito A, Yamaguchi H, Miyato H, Sakuma Y, Horie H, Hosoya Y, Lefor AK, Sata N, Kitayama J. Ratios of miRNAs in Peritoneal Exosomes are Useful Biomarkers to Predict Tumor Response to Intraperitoneal Chemotherapy in Patients with Peritoneal Metastases from Gastric Cancer. Ann Surg Oncol. 2020;27:5057-5064. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 150. | Yagi T, Iinuma H, Hayama T, Matsuda K, Nozawa K, Tsukamoto M, Shimada R, Akahane T, Tsuchiya T, Ozawa T, Hashiguchi Y. Plasma exosomal microRNA-125b as a monitoring biomarker of resistance to mFOLFOX6-based chemotherapy in advanced and recurrent colorectal cancer patients. Mol Clin Oncol. 2019;11:416-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 28] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 151. | Sur D, Balacescu L, Cainap SS, Visan S, Pop L, Burz C, Havasi A, Buiga R, Cainap C, Irimie A, Balacescu O. Predictive Efficacy of MiR-125b-5p, MiR-17-5p, and MiR-185-5p in Liver Metastasis and Chemotherapy Response Among Advanced Stage Colorectal Cancer Patients. Front Oncol. 2021;11:651380. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 16] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 152. | Kim H, Kim EH, Kwak G, Chi SG, Kim SH, Yang Y. Exosomes: Cell-Derived Nanoplatforms for the Delivery of Cancer Therapeutics. Int J Mol Sci. 2020;22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 101] [Article Influence: 20.2] [Reference Citation Analysis (0)] |
| 153. | Zhang Y, Liu Q, Zhang X, Huang H, Tang S, Chai Y, Xu Z, Li M, Chen X, Liu J, Yang C. Recent advances in exosome-mediated nucleic acid delivery for cancer therapy. J Nanobiotechnology. 2022;20:279. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 182] [Article Influence: 60.7] [Reference Citation Analysis (0)] |
| 154. | Hui B, Lu C, Wang J, Xu Y, Yang Y, Ji H, Li X, Xu L, Tang W, Wang K, Gu Y. Engineered exosomes for co-delivery of PGM5-AS1 and oxaliplatin to reverse drug resistance in colon cancer. J Cell Physiol. 2022;237:911-933. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 49] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 155. | Pi F, Binzel DW, Lee TJ, Li Z, Sun M, Rychahou P, Li H, Haque F, Wang S, Croce CM, Guo B, Evers BM, Guo P. Nanoparticle orientation to control RNA loading and ligand display on extracellular vesicles for cancer regression. Nat Nanotechnol. 2018;13:82-89. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 275] [Cited by in RCA: 372] [Article Influence: 53.1] [Reference Citation Analysis (0)] |
| 156. | Liang G, Zhu Y, Ali DJ, Tian T, Xu H, Si K, Sun B, Chen B, Xiao Z. Engineered exosomes for targeted co-delivery of miR-21 inhibitor and chemotherapeutics to reverse drug resistance in colon cancer. J Nanobiotechnology. 2020;18:10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 182] [Cited by in RCA: 433] [Article Influence: 86.6] [Reference Citation Analysis (0)] |