Published online Nov 14, 2022. doi: 10.3748/wjg.v28.i42.6002
Peer-review started: August 25, 2022
First decision: September 2, 2022
Revised: September 24, 2022
Accepted: October 27, 2022
Article in press: October 27, 2022
Published online: November 14, 2022
Processing time: 76 Days and 18.1 Hours
Gastrointestinal cancer (GIC) has high morbidity and mortality as one of the main causes of cancer death. Preoperative risk stratification is critical to guide patient management, but traditional imaging studies have difficulty predicting its biological behavior. The emerging field of radiomics allows the conversion of potential pathophysiological information in existing medical images that cannot be visually recognized into high-dimensional quantitative image features. Tumor lesion characterization, therapeutic response evaluation, and survival prediction can be achieved by analyzing the relationships between these features and clinical and genetic data. In recent years, the clinical application of radiomics to GIC has increased dramatically. In this editorial, we describe the latest progress in the application of radiomics to GIC and discuss the value of its potential clinical applications, as well as its limitations and future directions.
Core Tip: In this editorial, we summarize the latest advances of radiomics in the field of gastrointestinal cancer diagnosis and treatment. Radiomics has great potential in precision treatment decision-making for gastrointestinal cancer. However, radiomics studies have had relatively marked heterogeneity in their workflow. In the future, it will be necessary to establish and promote an imaging data acquisition protocol, standardize the research workflow, and conduct multicenter prospective studies on quality control.
- Citation: Mao Q, Zhou MT, Zhao ZP, Liu N, Yang L, Zhang XM. Role of radiomics in the diagnosis and treatment of gastrointestinal cancer. World J Gastroenterol 2022; 28(42): 6002-6016
- URL: https://www.wjgnet.com/1007-9327/full/v28/i42/6002.htm
- DOI: https://dx.doi.org/10.3748/wjg.v28.i42.6002
Gastrointestinal cancer (GIC) has high morbidity and mortality rates[1]. It causes approximately 5000000 new cases and 3540000 deaths worldwide each year, making it one of the main causes of cancer death[1]. Because of the high heterogeneity of these tumors, it is difficult to implement precision treatment[2]. Lambin et al[3] proposed the concept of radiomics in 2012. The emerging field of radiomics can convert potential pathophysiological information in existing medical images that cannot be recognized by the human eye into high-dimensional quantitative image features[2-4]. By analyzing the relationships between these features and clinical and genetic data, we can characterize tumor lesions, evaluate therapeutic responses, and predict survival. In recent years, research on the application of radiomics to GIC has grown dramatically. With this editorial, we aim to describe the latest advances of radiomics in the assessment of GIC and to explore the value of its potential clinical applications, its limitations, and its future directions.
Imaging modalities that can be used for radiomics analysis include computed tomography (CT), magnetic resonance imaging (MRI), and positron-emission tomography (PET). Since CT is the most commonly used staging method for esophageal cancer (EC) and gastric cancer (GC), most radiomics studies on EC and GC are based on CT images[5-9]. In contrast, as MRI is widely used for colorectal cancer (CRC) staging, most radiomics studies on CRC are based on MRI features[10-13].
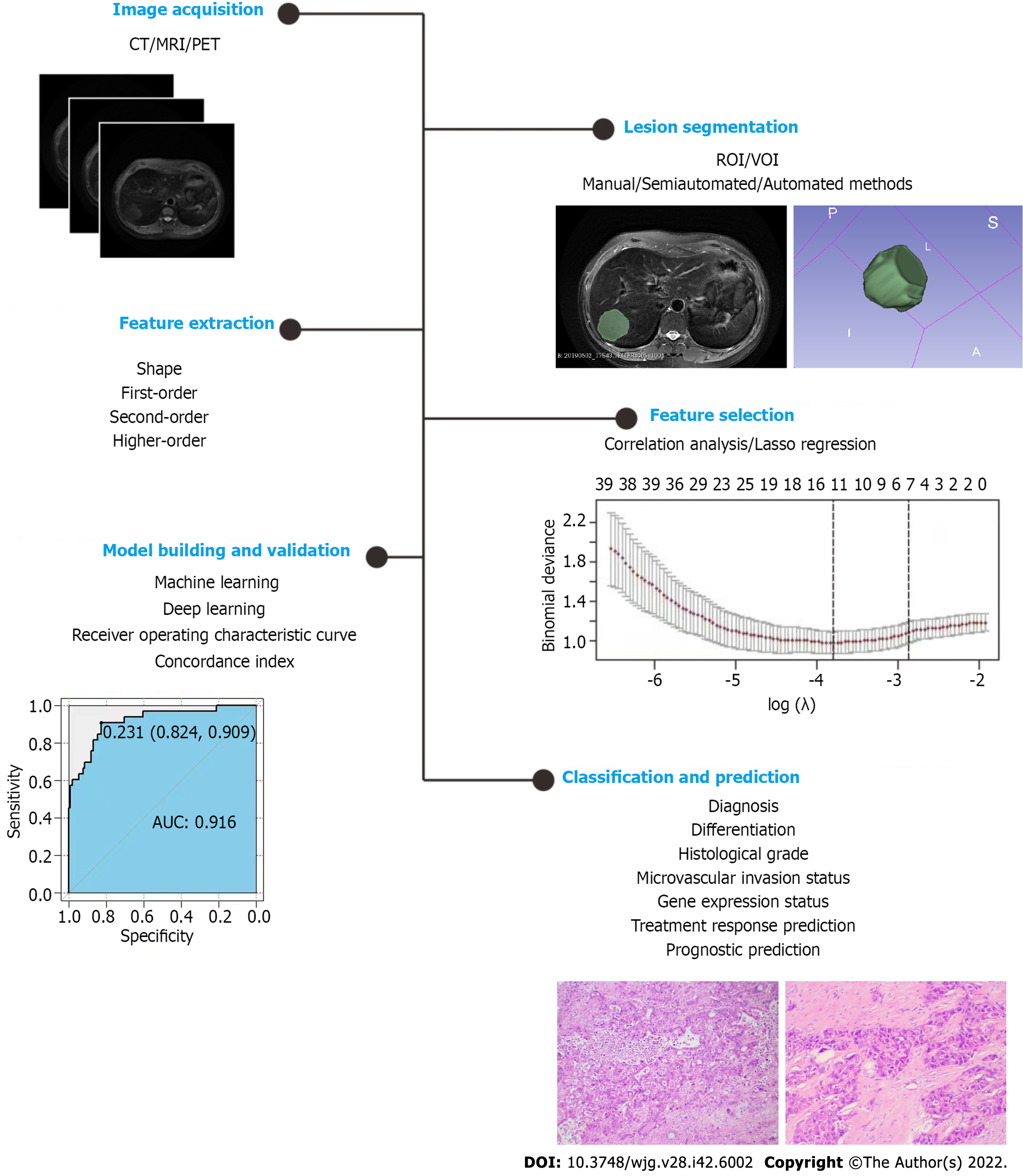
The workflow of radiomics usually includes image acquisition, lesion segmentation, feature extraction and selection, model building, and validation[14]. Lesion segmentation and feature extraction are the most essential steps. Manual, automatic, and semiautomatic segmentation methods are often used to segment the region of interest (ROI) or volume OI (VOI) (2D or 3D) in a target lesion, and manual segmentation is the most commonly used method (gold standard)[15]. After lesion segmen
Published studies have mainly investigated the predictive ability of radiomics in the staging, therapy response, and postoperative recurrence of EC[16-19].
Radiomic characteristics based on CT have good predictive potential for EC staging[20,21]. Yang et al[19] reported that CT radiomic characteristics were significantly correlated with the tumor (T) stage and tumor length of EC and showed good predictive performance for both; the area under the ROC curve (AUC), sensitivity, and specificitywere 0.86, 0.77 and 0.87, respectively, and 0.95, 0.92 and 0.91. Radiomic features also have good efficacy in predicting EC lymphatic metastasis[7,22-24]. Liu et al[20] suggested that baseline CT texture is a biomarker for the preoperative assessment of T, lymph node (N), and overall staging of esophageal squamous cell carcinoma (ESCC). Wu et al[25] established a model based on the radiomic characteristics of the late arterial phase of CT, which well distinguished early (I-II) and late (III-IV) ESCC, and the model’s efficacy was better than that of tumor volume.
Locally advanced EC often requires neoadjuvant chemoradiotherapy (NACRT)[26], whose treatment outcome is associated with tumor heterogeneity[27,28]. Radiomics can extract tumor heterogeneity data and has good application potential in improving the treatment stratification of patients. Radiomic characteristics are helpful for evaluating the response of EC to NAC or NACRT, distinguishing responders from nonresponders, for which it performs better than traditional parameters[29-32]. A prospective multicenter study[33] developed and validated a three-dimensional DL model applied to preprocessed CT images to predict the response of patients with locally advanced thoracic esophageal squamous cell carcinoma (TESCC) to concurrent chemoradiotherapy. The three-dimensional DL model achieved good predictive performance, with an AUC in the training cohort of 0.897 [95% confidence interval (CI): 0.840-0.959] and an AUC in the validation cohort of 0.833 (95%CI: 0.654-1.000). It is also feasible to use radiomics to predict the pathological complete response (pCR) of EC[34,35]. Patients with a pCR after NACRT have a higher overall survival (OS) rate[36,37], but nonresponders will not benefit from this therapy[38]. This information can provide guidance for personalized treatment of EC patients[28]. A CT-based radiomics study showed that a model that combined the intratumoral and peritumoral radiological characteristics could improve the predictive performance of the pCR of EC NACRT. In the test set, the AUC was 0.852 (95%CI: 0.753-0.951), the accuracy was 84.3%, the sensitivity was 90.3%, and the specificity was 79.5%[35]. Several studies of radiomics based on MRI or 18F-fluorodeoxyglucose (18F-FDG) PET also showed its efficacy in predicting the response to EC treatment[39-42]. The application of radiomics to immunotherapy has also achieved good response prediction value[43].
Radiomics has also made progress in predicting the recurrence and prognosis of EC patients[44-47]. Tang et al[48] predicted the early recurrence of locally advanced ESCC after trimodal therapy based on enhanced CT radiomics. The results showed that in the training cohort, the AUCs of the radiomics model, the clinical model, and the combined model were 0.754, 0.679, and 0.821, respectively, and they were 0.646, 0.658, and 0.809 in the validation cohort; the combined model was the best. Qiu et al[49] developed and validated a prediction model based on radiomic features extracted from contrast-enhanced CT images to estimate the recurrence-free survival (RFS) of patients who achieved pCR through NACRT and surgery. The results showed that the radiomic characteristics were significantly correlated with RFS. In the training cohort and the validation cohort, compared with the nomograms of the radiomic characteristics and of clinical risk factors, the nomogram combining the radiomic characteristics and clinical risk factors had optimal performance. Other studies have shown that combining the radiomic characteristics of primary tumors and regional lymph nodes with clinical-pathological factors can improve OS prediction[50].
Other studies showed that CT-based radiomics features also had good predictive performance for classifying patients according to histological differentiation[51-53], the expression of programmed death-ligand 1, and CD8+ tumor-infiltrating lymphocytes of EC[5].
In recent years, some researchers have also explored the value of radiomics to the diagnosis and treatment of GC[9,54,55]. The CT radiomics model has high application value in the identification of GC[54,56-58]. Feng et al[59] used a transfer learning radiomics nomogram (TLRN) with whole-slide images of GC as the source domain data to distinguish Borrmann type IV GC from primary gastric lymphoma before surgery. The TLRN that integrated transfer learning radiomics signatures (TLRS), clinical factors, and CT subjective findings was developed through multiple logistic regression (LR). The results showed that the TLRN performed better than the clinical model and the TLRS. The AUCs of the internal and two external validation cohorts were 0.958 (95%CI: 0.883-0.991), 0.867 (95%CI: 0.794-0.922), and 0.921 (95%CI: 0.860-0.960), respectively[59]. Wang et al[60] reported that a DL radiomics model based on CT images had a potential role in the T staging of GC. For distinguishing T2 from T3/4 tumors, the AUCs of the arterial phase-based radiomics model in the training group and the test group were 0.899 (95%CI: 0.812-0.955) and 0.825 (95%CI: 0.718-0.904), respectively. The AUC of the radiomics model based on the portal vein phase in the training and testing cohorts was 0.843 (95%CI: 0.746-0.914) and 0.818 (95%CI: 0.711-0.899), respectively[60]. An important factor in the failure of GC treatment is lymph node metastasis (LNM) and cancer spread in the peritoneal cavity[61]. In GC, the most common metastatic sites are the distant lymph nodes (56%), liver (53%), and peritoneum (51%)[62]. Accurate assessment of LNM and preoperative N staging is critical for the accurate treatment of GC patients. Most studies have shown that CT-based radiomics models have good accuracy in predicting early GC lymph node and peritoneal metastasis before surgery[63-66]. A ML model based on preoperative 18F-FDG-PET/CT obtained similarly good results[67].
CT-based radiomic characteristics also perform well in predicting the response to NAC and radiotherapy in patients with advanced GC[68-71]. Jiang et al[72] showed that a DL CT signature could help to identify patients who might benefit from adjuvant chemotherapy for GC and improve prognostic prediction. A radiomics study based on 18F-FDG-PET signatures obtained similar results[73]. In addition, radiomics can be used to predict the histological grade of GC before surgery[74] and is useful for GC classification[75,76].
The application of radiomics to CRC has mainly focused on the evaluation of stage, neoadjuvant therapy outcome, and gene mutations[77,78].
Radiomics models are helpful for CRC staging[79-81]. LNM is an independent risk factor affecting the prognosis of CRC patients. Radiomics models can effectively predict LNM in CRC patients before surgery[82-85]. Liu et al[84] found that multiregional-based MRI radiomics combined with clinical data could improve the efficacy of predicting LNM. He et al[85] developed and tested five ML models based on the radiomic features of F-18-FDG-PET/CT and PET for their preoperative prediction of LNM in the CRC region: LR, support vector machine, random forest (RF), neural network, and extreme gradient boosting. The results showed that the LR (AUC 0.866, 95%CI: 0.808-0.925) and extreme gradient boosting models (AUC 0.903, 95%CI: 0.855-0.951) performed the best, outperforming F-18-FDG-PET/CT on both the training set and the test set[85]. Other studies have also shown that radiomics has a good ability to predict metastasis to distant organs, such as the liver and lung, as well as vascular and perineural invasion[86,87]. It is reported that the predictive power of CT-based radiomics for the preoperative staging of CRC. The results showed that the radiomic features were an independent predictor of CRC staging. CRC was successfully divided into stages I-II and III-IV in the training and validation datasets. The AUC in the training dataset was 0.792 (95%CI: 0.741-0.853), the sensitivity was 0.629, and the specificity was 0.874. The AUC in the validation dataset was 0.708 (95%CI: 0.698-0.718), the sensitivity was 0.611, and the specificity was 0.680[79].
Radiomics models have had excellent performance in noninvasively predicting the response to NAC and NACRT in patients with locally advanced CRC (including liver metastasis)[88-91]. They have also achieved good efficacy in predicting the response to CRC targeted therapy[77,92].
Mutations in the KRAS, NRAS, or BRAF gene indicate that CRC patients will lack a response to drugs targeting epidermal growth factor receptor. In 2016, the National Comprehensive Cancer Network guidelines recommended that all patients with suspected or confirmed metastatic CRC should be tested for KRAS/NRAS/BRAF mutations, but this requires pathological tissue specimens. It is gratifying that some radiogenomics studies have shown that the radiomic characteristics of CT and MRI may help to predict the genotype of CRC tumors before surgery[93-95]. Yang et al[96] reported that CT radiomic characteristics were associated with KRAS/NRAS/BRAF mutations. Another MRI radiomics study found a good correlation between quantitative features and gene mutations, while there was no correlation between qualitative features and gene mutations[97].
More recent studies have shown that radiomics can predict CRC histological grade before surgery[98,99].
The application of radiomics to hepatocellular carcinoma (HCC) involves differential diagnosis, determination of microvascular invasion (MVI) status, histological grade, gene expression status, and treatment response, and prognostic prediction[100-104].
Because HCC has a typical enhancement mode, dynamic contrast-enhanced CT, MRI, and ultrasound have played major roles in the diagnosis and differentiation of HCC[105-107]. However, it is sometimes difficult to distinguish some small nodules from atypical lesions[108-111]. Radiomics can achieve quantitative analysis of tumor biological behavior and heterogeneity, helping identify liver nodules[112-114]. Yasaka et al[115] investigated the performance of a DL method to distinguish liver masses on dynamic enhanced CT. There are five types of these masses: Type A, classic HCC; type B, malignant liver tumors other than HCC; type C, indeterminate masses or mass-like lesions, plus rare benign liver masses other than hemangiomas and cysts; type D, hemangiomas; type E, cysts. The median accuracy of the mass identification on the test set was 0.84. The AUC that distinguished the types A-B from types C-E was 0.92. Hamm et al[116] used a DL method to classify common liver lesions with typical imaging characteristics on multiphasic MRI, including a total of 494 liver lesions from six categories, which were divided into training (n = 434) and test groups (n = 60). Their DL system had an accuracy of 92%, a sensitivity of 92%, and a specificity of 98%. For HCC classification, the true-positive rate and false-positive rate were 93.5% and 1.6%, respectively, and the AUC was 0.992[116]. Other studies have reached similar conclusions[108,110].
The 5-year recurrence rate of HCC resection can reach 70%[103]. Pathological features such as histological grade and MVI of HCC were significantly correlated with postoperative recurrence and prognosis[117-120]. Histological grade, MVI status[121-125], and gene expression[113,126,127] in HCC can be successfully predicted by radiomics models before surgery. An MRI-based radiomics study showed that the AUCs of the MVI nomogram in the validation cohort using the RF algorithm and LR analysis were 0.920 (95%CI: 0.861-0.979) and 0.879 (95%CI: 0.820-0.938), respectively[123].
Radiomics models based on contrast-enhanced CT and MRI can predict the response of middle- and late-stage HCC to local treatment and systemic treatment[10,128-130]and the early recurrence and the prognosis after HCC resection[101,102,131,132]. Zhang et al[133] evaluated the effectiveness of predi
Some studies using radiomics to predict the occurrence of CRC liver metastases are particularly interesting[134-137]. Rao et al[137] retrospectively analyzed the primary staging CT data of 29 CRC patients. The patients were divided into three groups: The non-liver-metastasis group, the simultaneous liver metastasis (LM) group, and the metachronous LM group within 18 mo. Whole-liver texture analysis was performed on the liver parenchyma that was clearly disease-free on the portal vein image. The results showed that compared with those in nonmetastatic patients, the mean entropy 1.5 (E1.5)and E2.5 values of the whole liver in patients with synchronous metastasis were significantly increased, and the uniformity 1.5 (U1.5)and U2.5 values were significantly decreased. The AUCs for the diagnosis of synchronous metastasis based on E1.5, E2.5, U1.5, and U2.5 were 0.73-0.78[137]. Beckers et al[138] conducted a similar retrospective multicenter study. They included a total of 165 cases of CRC, which were also divided into the nonmetastasis group, the synchronous metastasis group, and the metastasis group (within 24 mo). Univariate analysis confirmed that U, sex, tumor site, nodal stage, and carcinoembryonic antigen (CEA) were potential predictive factors; multivariate analysis showed that U was still a factor predicting early metastasis; and none of the parameters could predict intermediate/late metastasis[138]. Other studies have shown no significant difference in CT texture parameters of liver parenchyma between CRC patients with and without liver metastasis[134,135]. The conclusions of these studies are inconsistent, so the prediction of LM of CRC based on the texture characteristics of the liver parenchyma requires further study. Recently, Liet al[139] investigated the efficacy of a radiomics model based on baseline CRC contrast-enhanced CT in predicting metachronous liver metastases in CRC patients. The AUC of the radiomics feature model was 0.78 ± 0.07, and the AUC of the clinical feature model was 0.79 ± 0.08. The model combining the two performed best, with AUCs of 0.79 ± 0.08 and 0.72 ± 0.07 in the internal and external validation cohorts, respectively. They believed that the radiomic characteristics of primary CRC lesions are often affected by fewer factors and are more stable; their radiomic characteristics have the potential to distinguish patients at risk of liver metastasis.
For PC, the application of radiomics mainly focuses on identification, treatment response prediction, and prognostic prediction[140-142]. Many studies have focused on the diagnosis and differentiation of pancreatic ductal adenocarcinoma (PDAC)[143-146]. Chu et al[146] investigated the utility of CT radiomics in distinguishing PDAC from normal pancreas. In their retrospective casecontrol study, 190 PDAC patients and 190 healthy potential renal donors were included. The overall accuracy of RF binary classification was 99.2%, with an AUC of 99.9%; all PDAC cases were correctly classified. Park et al[145] confirmed that CT-based ML of radiomics features was helpful to distinguish between autoimmune pancreatitis and PDAC, with an overall accuracy of 95.2%. The radiomics model based on PET/CT also showed good performance in distinguishing benign autoimmune pancreatitis from malignant PDAC lesions[143,144].
Other studies have shown that radiomics can better predict the treatment response and prognosis of PC[142,147]. Simpson et al[141] evaluated the potential of MRI-based radiomics to predict the response to PC treatment. A total of 20 patients with unresected nonmetastatic PDAC were enrolled, all of whom received NAC followed by five rounds of MR-guided stereotactic body radiotherapy. Half of the 20 patients were defined as having histopathological tumor regression or tumor response based on an enhanced CT. The AUC of the model based on the RF algorithm was 0.81 (95%CI: 0.594-1.000); the adaptive least absolute shrinkage and selection operator (LASSO) algorithm achieved AUC of 0.81 (95%CI: 0.596-1.000). Xie et al[148] used a CT-based radiomics nomogram to predict the survival of patients with resected PDAC. The radiomics score developed based on CT imaging features was significantly correlated with disease-free survival (DFS) and OS in patients with PDAC. The radiomics nomogram showed good discrimination in both the training cohort and the validation cohort, being superior to the clinical model and the TNM staging system for survival estimation. The model integrating the radiomics score and clinical data had the best predictive performance, but there was no correlation between the radiomics score and recurrence pattern. Similar results were seen by Healy et al[149].
In this editorial, we summarize the results of the application of radiomics to the field of GIC diagnosis and treatment. These results show that radiomics has great potential for decision-making about precision treatments for GIC. Moreover, these results have important reference value for studies of other systemic tumors.
However, some limitations to the clinical application of radiomics remain[150,151]. The first key challenge is the use of different imaging techniques by different institutions and/or scanners. To ensure that the academic community can obtain high-quality radiological data resources, it is necessary to establish and promote certain imaging acquisition protocols[149]. Second, the current research uses different software and different feature selection methods, focuses on different feature sets, and applies different statistical and bioinformatic methods for data analysis and interpretation, which limit the reproducibility of radiomics models[152,153]. Future research workflows need to be standardized. Third, many relevant radiomics studies employ single-center retrospective datasets. A quality-controlled multicenter prospective study plan is ideal. In addition, the evidence level rating reflects the feasibility of incorporating radiomics research into clinical practice. Recently published guidelines and checklists aiming to improve the quality of radiomics studies, including the radiomics quality score, modified Quality Assessment of Diagnostic Accuracy Studies tool, image biomarker standardization initiativeguideline, and Transparent Reporting of a multivariable prediction model for Individual Prognosis or Diagnosis checklist, have been applied to radiomics evaluations[154-157]. These studies have shown that the current scientific and reporting quality of many radiomics studies is insufficient; feature reproducibility, open science categories, and clinical utility analyses need to be improved; and study objectives, blinding, sample sizes, and missing data must be reported[154-157]. In the future, radiomics studies should adhere to these guidelines to facilitate the translation of radiomics research into clinical practice[156].
Radiomics has great potential in precision treatment decision-making for gastrointestinal cancer. However, radiomics studies have had relatively marked heterogeneity in their workflow. In the future, it will be necessary to establish and promote an imaging data acquisition protocol, standardize the research workflow, and conduct multicenter prospective studies on quality control. In addition, the combination of radiomics with multiomics may lead to a major breakthrough in individualized medical treatment of tumors.
We thank MrTang Zhi and Miss Tao Yun-Yun for their contributions to graph creation.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Corresponding Author's Membership in Professional Societies: A member of the editorial board or peer reviewer of a BPG journal, 02832130.
Specialty type: Radiology, nuclear medicine and medical imaging
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Cannella R, Italy; Stabellini N, United States S-Editor: Chen YL L-Editor: A P-Editor: Chen YL
| 1. | Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75126] [Cited by in RCA: 64684] [Article Influence: 16171.0] [Reference Citation Analysis (177)] |
| 2. | Sexton RE, Al Hallak MN, Diab M, Azmi AS. Gastric cancer: a comprehensive review of current and future treatment strategies. Cancer Metastasis Rev. 2020;39:1179-1203. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 459] [Cited by in RCA: 461] [Article Influence: 92.2] [Reference Citation Analysis (0)] |
| 3. | Lambin P, Rios-Velazquez E, Leijenaar R, Carvalho S, van Stiphout RG, Granton P, Zegers CM, Gillies R, Boellard R, Dekker A, Aerts HJ. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer. 2012;48:441-446. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2415] [Cited by in RCA: 3857] [Article Influence: 296.7] [Reference Citation Analysis (2)] |
| 4. | Aerts HJ, Velazquez ER, Leijenaar RT, Parmar C, Grossmann P, Carvalho S, Bussink J, Monshouwer R, Haibe-Kains B, Rietveld D, Hoebers F, Rietbergen MM, Leemans CR, Dekker A, Quackenbush J, Gillies RJ, Lambin P. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun. 2014;5:4006. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2262] [Cited by in RCA: 3249] [Article Influence: 295.4] [Reference Citation Analysis (0)] |
| 5. | Wen Q, Yang Z, Zhu J, Qiu Q, Dai H, Feng A, Xing L. Pretreatment CT-Based Radiomics Signature as a Potential Imaging Biomarker for Predicting the Expression of PD-L1 and CD8+TILs in ESCC. Onco Targets Ther. 2020;13:12003-12013. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 37] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 6. | Gong J, Zhang W, Huang W, Liao Y, Yin Y, Shi M, Qin W, Zhao L. CT-based radiomics nomogram may predict local recurrence-free survival in esophageal cancer patients receiving definitive chemoradiation or radiotherapy: A multicenter study. Radiother Oncol. 2022;174:8-15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 33] [Reference Citation Analysis (0)] |
| 7. | Wu L, Yang X, Cao W, Zhao K, Li W, Ye W, Chen X, Zhou Z, Liu Z, Liang C. Multiple Level CT Radiomics Features Preoperatively Predict Lymph Node Metastasis in Esophageal Cancer: A Multicentre Retrospective Study. Front Oncol. 2019;9:1548. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 70] [Article Influence: 14.0] [Reference Citation Analysis (1)] |
| 8. | Rishi A, Zhang GG, Yuan Z, Sim AJ, Song EY, Moros EG, Tomaszewski MR, Latifi K, Pimiento JM, Fontaine JP, Mehta R, Harrison LB, Hoffe SE, Frakes JM. Pretreatment CT and 18 F-FDG PET-based radiomic model predicting pathological complete response and loco-regional control following neoadjuvant chemoradiation in oesophageal cancer. J Med Imaging Radiat Oncol. 2021;65:102-111. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 22] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 9. | Chen Q, Zhang L, Liu S, You J, Chen L, Jin Z, Zhang S, Zhang B. Radiomics in precision medicine for gastric cancer: opportunities and challenges. Eur Radiol. 2022;32:5852-5868. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 50] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 10. | Chen BY, Xie H, Li Y, Jiang XH, Xiong L, Tang XF, Lin XF, Li L, Cai PQ. MRI-Based Radiomics Features to Predict Treatment Response to Neoadjuvant Chemotherapy in Locally Advanced Rectal Cancer: A Single Center, Prospective Study. Front Oncol. 2022;12:801743. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 1] [Reference Citation Analysis (1)] |
| 11. | Mbanu P, Saunders MP, Mistry H, Mercer J, Malcomson L, Yousif S, Price G, Kochhar R, Renehan AG, van Herk M, Osorio EV. Clinical and radiomics prediction of complete response in rectal cancer pre-chemoradiotherapy. Phys Imaging Radiat Oncol. 2022;23:48-53. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 6] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 12. | Kekelidze M, D'Errico L, Pansini M, Tyndall A, Hohmann J. Colorectal cancer: current imaging methods and future perspectives for the diagnosis, staging and therapeutic response evaluation. World J Gastroenterol. 2013;19:8502-8514. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 115] [Cited by in RCA: 110] [Article Influence: 9.2] [Reference Citation Analysis (4)] |
| 13. | Qin Y, Zhu LH, Zhao W, Wang JJ, Wang H. Review of Radiomics- and Dosiomics-based Predicting Models for Rectal Cancer. Front Oncol. 2022;12:913683. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 11] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 14. | Miranda Magalhaes Santos JM, Clemente Oliveira B, Araujo-Filho JAB, Assuncao-Jr AN, de M Machado FA, Carlos Tavares Rocha C, Horvat JV, Menezes MR, Horvat N. State-of-the-art in radiomics of hepatocellular carcinoma: a review of basic principles, applications, and limitations. Abdom Radiol (NY). 2020;45:342-353. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 48] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 15. | Polan DF, Brady SL, Kaufman RA. Tissue segmentation of computed tomography images using a Random Forest algorithm: a feasibility study. Phys Med Biol. 2016;61:6553-6569. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 72] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 16. | Xie C, Yang P, Zhang X, Xu L, Wang X, Li X, Zhang L, Xie R, Yang L, Jing Z, Zhang H, Ding L, Kuang Y, Niu T, Wu S. Sub-region based radiomics analysis for survival prediction in oesophageal tumours treated by definitive concurrent chemoradiotherapy. EBioMedicine. 2019;44:289-297. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 111] [Article Influence: 18.5] [Reference Citation Analysis (0)] |
| 17. | Shi Z, Zhang Z, Liu Z, Zhao L, Ye Z, Dekker A, Wee L. Methodological quality of machine learning-based quantitative imaging analysis studies in esophageal cancer: a systematic review of clinical outcome prediction after concurrent chemoradiotherapy. Eur J Nucl Med Mol Imaging. 2022;49:2462-2481. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 14] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 18. | Peng H, Yang Q, Xue T, Chen Q, Li M, Duan S, Cai B, Feng F. Computed tomography-based radiomics analysis to predict lymphovascular invasion in esophageal squamous cell carcinoma. Br J Radiol. 2022;95:20210918. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 14] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 19. | Yang M, Hu P, Li M, Ding R, Wang Y, Pan S, Kang M, Kong W, Du D, Wang F. Computed Tomography-Based Radiomics in Predicting T Stage and Length of Esophageal Squamous Cell Carcinoma. Front Oncol. 2021;11:722961. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 20] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 20. | Liu S, Zheng H, Pan X, Chen L, Shi M, Guan Y, Ge Y, He J, Zhou Z. Texture analysis of CT imaging for assessment of esophageal squamous cancer aggressiveness. J Thorac Dis. 2017;9:4724-4732. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 29] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 21. | Kawahara D, Murakami Y, Tani S, Nagata Y. A prediction model for degree of differentiation for resectable locally advanced esophageal squamous cell carcinoma based on CT images using radiomics and machine-learning. Br J Radiol. 2021;94:20210525. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 11] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 22. | Li X, Liu Q, Hu B, Xu J, Huang C, Liu F. A computed tomography-based clinical-radiomics model for prediction of lymph node metastasis in esophageal carcinoma. J Cancer Res Ther. 2021;17:1665-1671. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 23. | Zhang C, Shi Z, Kalendralis P, Whybra P, Parkinson C, Berbee M, Spezi E, Roberts A, Christian A, Lewis W, Crosby T, Dekker A, Wee L, Foley KG. Prediction of lymph node metastases using pre-treatment PET radiomics of the primary tumour in esophageal adenocarcinoma: an external validation study. Br J Radiol. 2021;94:20201042. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 19] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 24. | Qu J, Shen C, Qin J, Wang Z, Liu Z, Guo J, Zhang H, Gao P, Bei T, Wang Y, Liu H, Kamel IR, Tian J, Li H. The MR radiomic signature can predict preoperative lymph node metastasis in patients with esophageal cancer. Eur Radiol. 2019;29:906-914. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 65] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 25. | Wu L, Wang C, Tan X, Cheng Z, Zhao K, Yan L, Liang Y, Liu Z, Liang C. Radiomics approach for preoperative identification of stages I-II and III-IV of esophageal cancer. Chin J Cancer Res. 2018;30:396-405. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 58] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 26. | Babic B, Fuchs HF, Bruns CJ. [Neoadjuvant chemoradiotherapy or chemotherapy for locally advanced esophageal cancer? Chirurg. 2020;91:379-383. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 16] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 27. | Secrier M, Li X, de Silva N, Eldridge MD, Contino G, Bornschein J, MacRae S, Grehan N, O'Donovan M, Miremadi A, Yang TP, Bower L, Chettouh H, Crawte J, Galeano-Dalmau N, Grabowska A, Saunders J, Underwood T, Waddell N, Barbour AP, Nutzinger B, Achilleos A, Edwards PA, Lynch AG, Tavaré S, Fitzgerald RC; Oesophageal Cancer Clinical and Molecular Stratification (OCCAMS) Consortium. Mutational signatures in esophageal adenocarcinoma define etiologically distinct subgroups with therapeutic relevance. Nat Genet. 2016;48:1131-1141. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 296] [Cited by in RCA: 291] [Article Influence: 32.3] [Reference Citation Analysis (0)] |
| 28. | Yang Z, He B, Zhuang X, Gao X, Wang D, Li M, Lin Z, Luo R. CT-based radiomic signatures for prediction of pathologic complete response in esophageal squamous cell carcinoma after neoadjuvant chemoradiotherapy. J Radiat Res. 2019;60:538-545. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 65] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 29. | Hou Z, Li S, Ren W, Liu J, Yan J, Wan S. Radiomic analysis in T2W and SPAIR T2W MRI: predict treatment response to chemoradiotherapy in esophageal squamous cell carcinoma. J Thorac Dis. 2018;10:2256-2267. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 33] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 30. | Jin X, Zheng X, Chen D, Jin J, Zhu G, Deng X, Han C, Gong C, Zhou Y, Liu C, Xie C. Prediction of response after chemoradiation for esophageal cancer using a combination of dosimetry and CT radiomics. Eur Radiol. 2019;29:6080-6088. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 63] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 31. | Hu Y, Xie C, Yang H, Ho JWK, Wen J, Han L, Lam KO, Wong IYH, Law SYK, Chiu KWH, Vardhanabhuti V, Fu J. Computed tomography-based deep-learning prediction of neoadjuvant chemoradiotherapy treatment response in esophageal squamous cell carcinoma. Radiother Oncol. 2021;154:6-13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 98] [Article Influence: 24.5] [Reference Citation Analysis (0)] |
| 32. | Luo HS, Huang SF, Xu HY, Li XY, Wu SX, Wu DH. A nomogram based on pretreatment CT radiomics features for predicting complete response to chemoradiotherapy in patients with esophageal squamous cell cancer. Radiat Oncol. 2020;15:249. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 37] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 33. | Li X, Gao H, Zhu J, Huang Y, Zhu Y, Huang W, Li Z, Sun K, Liu Z, Tian J, Li B. 3D Deep Learning Model for the Pretreatment Evaluation of Treatment Response in Esophageal Carcinoma: A Prospective Study (ChiCTR2000039279). Int J Radiat Oncol Biol Phys. 2021;111:926-935. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 24] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 34. | Kao YS, Hsu Y. A Meta-Analysis for Using Radiomics to Predict Complete Pathological Response in Esophageal Cancer Patients Receiving Neoadjuvant Chemoradiation. In Vivo. 2021;35:1857-1863. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 25] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 35. | Hu Y, Xie C, Yang H, Ho JWK, Wen J, Han L, Chiu KWH, Fu J, Vardhanabhuti V. Assessment of Intratumoral and Peritumoral Computed Tomography Radiomics for Predicting Pathological Complete Response to Neoadjuvant Chemoradiation in Patients With Esophageal Squamous Cell Carcinoma. JAMA Netw Open. 2020;3:e2015927. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 117] [Article Influence: 23.4] [Reference Citation Analysis (0)] |
| 36. | Donahue JM, Nichols FC, Li Z, Schomas DA, Allen MS, Cassivi SD, Jatoi A, Miller RC, Wigle DA, Shen KR, Deschamps C. Complete pathologic response after neoadjuvant chemoradiotherapy for esophageal cancer is associated with enhanced survival. Ann Thorac Surg. 2009;87:392-8; discussion 398. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 226] [Cited by in RCA: 247] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 37. | Lin JW, Hsu CP, Yeh HL, Chuang CY, Lin CH. The impact of pathological complete response after neoadjuvant chemoradiotherapy in locally advanced squamous cell carcinoma of esophagus. J Chin Med Assoc. 2018;81:18-24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 24] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 38. | Dittrick GW, Weber JM, Shridhar R, Hoffe S, Melis M, Almhanna K, Barthel J, McLoughlin J, Karl RC, Meredith KL. Pathologic nonresponders after neoadjuvant chemoradiation for esophageal cancer demonstrate no survival benefit compared with patients treated with primary esophagectomy. Ann Surg Oncol. 2012;19:1678-1684. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 51] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 39. | Murakami Y, Kawahara D, Tani S, Kubo K, Katsuta T, Imano N, Takeuchi Y, Nishibuchi I, Saito A, Nagata Y. Predicting the Local Response of Esophageal Squamous Cell Carcinoma to Neoadjuvant Chemoradiotherapy by Radiomics with a Machine Learning Method Using 18F-FDG PET Images. Diagnostics (Basel). 2021;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 13] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 40. | Qu J, Ma L, Lu Y, Wang Z, Guo J, Zhang H, Yan X, Liu H, Kamel IR, Qin J, Li H. DCE-MRI radiomics nomogram can predict response to neoadjuvant chemotherapy in esophageal cancer. Discov Oncol. 2022;13:3. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 12] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 41. | Li Y, Beck M, Päßler T, Lili C, Hua W, Mai HD, Amthauer H, Biebl M, Thuss-Patience PC, Berger J, Stromberger C, Tinhofer I, Kruppa J, Budach V, Hofheinz F, Lin Q, Zschaeck S. A FDG-PET radiomics signature detects esophageal squamous cell carcinoma patients who do not benefit from chemoradiation. Sci Rep. 2020;10:17671. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 42. | Beukinga RJ, Poelmann FB, Kats-Ugurlu G, Viddeleer AR, Boellaard R, De Haas RJ, Plukker JTM, Hulshoff JB. Prediction of Non-Response to Neoadjuvant Chemoradiotherapy in Esophageal Cancer Patients with 18F-FDG PET Radiomics Based Machine Learning Classification. Diagnostics (Basel). 2022;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 9] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 43. | Zhu Y, Yao W, Xu BC, Lei YY, Guo QK, Liu LZ, Li HJ, Xu M, Yan J, Chang DD, Feng ST, Zhu ZH. Predicting response to immunotherapy plus chemotherapy in patients with esophageal squamous cell carcinoma using non-invasive Radiomic biomarkers. BMC Cancer. 2021;21:1167. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 27] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 44. | Luo HS, Chen YY, Huang WZ, Wu SX, Huang SF, Xu HY, Xue RL, Du ZS, Li XY, Lin LX, Huang HC. Development and validation of a radiomics-based model to predict local progression-free survival after chemo-radiotherapy in patients with esophageal squamous cell cancer. Radiat Oncol. 2021;16:201. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 33] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 45. | Yan T, Liu L, Yan Z, Peng M, Wang Q, Zhang S, Wang L, Zhuang X, Liu H, Ma Y, Wang B, Cui Y. A Radiomics Nomogram for Non-Invasive Prediction of Progression-Free Survival in Esophageal Squamous Cell Carcinoma. Front Comput Neurosci. 2022;16:885091. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 46. | Cao B, Mi K, Dai W, Liu T, Xie T, Li Q, Lang J, Han Y, Peng L, Wang Q. Prognostic and incremental value of computed tomography-based radiomics from tumor and nodal regions in esophageal squamous cell carcinoma. Chin J Cancer Res. 2022;34:71-82. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 11] [Reference Citation Analysis (0)] |
| 47. | Chu F, Liu Y, Liu Q, Li W, Jia Z, Wang C, Wang Z, Lu S, Li P, Zhang Y, Liao Y, Xu M, Yao X, Wang S, Liu C, Zhang H, Yan X, Kamel IR, Sun H, Yang G, Qu J. Development and validation of MRI-based radiomics signatures models for prediction of disease-free survival and overall survival in patients with esophageal squamous cell carcinoma. Eur Radiol. 2022;32:5930-5942. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 21] [Reference Citation Analysis (0)] |
| 48. | Tang S, Ou J, Liu J, Wu YP, Wu CQ, Chen TW, Zhang XM, Li R, Tang MJ, Yang LQ, Tan BG, Lu FL, Hu J. Application of contrast-enhanced CT radiomics in prediction of early recurrence of locally advanced oesophageal squamous cell carcinoma after trimodal therapy. Cancer Imaging. 2021;21:38. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 23] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 49. | Qiu Q, Duan J, Deng H, Han Z, Gu J, Yue NJ, Yin Y. Development and Validation of a Radiomics Nomogram Model for Predicting Postoperative Recurrence in Patients With Esophageal Squamous Cell Cancer Who Achieved pCR After Neoadjuvant Chemoradiotherapy Followed by Surgery. Front Oncol. 2020;10:1398. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 53] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 50. | Lu N, Zhang WJ, Dong L, Chen JY, Zhu YL, Zhang SH, Fu JH, Yin SH, Li ZC, Xie CM. Dual-region radiomics signature: Integrating primary tumor and lymph node computed tomography features improves survival prediction in esophageal squamous cell cancer. Comput Methods Programs Biomed. 2021;208:106287. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 24] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 51. | Cheng L, Wu L, Chen S, Ye W, Liu Z, Liang C. [CT-based radiomics analysis for evaluating the differentiation degree of esophageal squamous carcinoma]. Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2019;44:251-256. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 52. | Foy JJ, Armato SG 3rd, Al-Hallaq HA. Effects of variability in radiomics software packages on classifying patients with radiation pneumonitis. J Med Imaging (Bellingham). 2020;7:014504. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 12] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 53. | Anthony GJ, Cunliffe A, Castillo R, Pham N, Guerrero T, Armato SG 3rd, Al-Hallaq HA. Incorporation of pre-therapy 18 F-FDG uptake data with CT texture features into a radiomics model for radiation pneumonitis diagnosis. Med Phys. 2017;44:3686-3694. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 33] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 54. | Feng B, Huang L, Li C, Quan Y, Chen Y, Xue H, Chen Q, Sun S, Li R, Long W. A Heterogeneity Radiomic Nomogram for Preoperative Differentiation of Primary Gastric Lymphoma From Borrmann Type IV Gastric Cancer. J Comput Assist Tomogr. 2021;45:191-202. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 4] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 55. | Lan Q, Guan X, Lu S, Yuan W, Jiang Z, Lin H, Long L. Radiomics in addition to computed tomography-based body composition nomogram may improve the prediction of postoperative complications in gastric cancer patients. Ann Nutr Metab. 2022;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 4] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 56. | Deng J, Tan Y, Gu Q, Rong P, Wang W, Liu S. [Application of CT-based radiomics in differentiating primary gastric lymphoma from Borrmann type IV gastric cancer]. Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2019;44:257-263. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 57. | Ma Z, Fang M, Huang Y, He L, Chen X, Liang C, Huang X, Cheng Z, Dong D, Xie J, Tian J, Liu Z. CT-based radiomics signature for differentiating Borrmann type IV gastric cancer from primary gastric lymphoma. Eur J Radiol. 2017;91:142-147. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 89] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 58. | Wang R, Liu H, Liang P, Zhao H, Li L, Gao J. Radiomics analysis of CT imaging for differentiating gastric neuroendocrine carcinomas from gastric adenocarcinomas. Eur J Radiol. 2021;138:109662. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 14] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 59. | Feng B, Huang L, Liu Y, Chen Y, Zhou H, Yu T, Xue H, Chen Q, Zhou T, Kuang Q, Yang Z, Chen X, Peng Z, Long W. A Transfer Learning Radiomics Nomogram for Preoperative Prediction of Borrmann Type IV Gastric Cancer From Primary Gastric Lymphoma. Front Oncol. 2021;11:802205. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 16] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 60. | Wang Y, Liu W, Yu Y, Liu JJ, Jiang L, Xue HD, Lei J, Jin Z, Yu JC. Prediction of the Depth of Tumor Invasion in Gastric Cancer: Potential Role of CT Radiomics. Acad Radiol. 2020;27:1077-1084. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 28] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 61. | Coccolini F, Cotte E, Glehen O, Lotti M, Poiasina E, Catena F, Yonemura Y, Ansaloni L. Intraperitoneal chemotherapy in advanced gastric cancer. Meta-analysis of randomized trials. Eur J Surg Oncol. 2014;40:12-26. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 143] [Cited by in RCA: 179] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 62. | Verstegen MH, Harker M, van de Water C, van Dieren J, Hugen N, Nagtegaal ID, Rosman C, van der Post RS. Metastatic pattern in esophageal and gastric cancer: Influenced by site and histology. World J Gastroenterol. 2020;26:6037-6046. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 26] [Cited by in RCA: 55] [Article Influence: 11.0] [Reference Citation Analysis (7)] |
| 63. | Gao X, Ma T, Cui J, Zhang Y, Wang L, Li H, Ye Z. A radiomics-based model for prediction of lymph node metastasis in gastric cancer. Eur J Radiol. 2020;129:109069. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 28] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 64. | Gao X, Ma T, Cui J, Zhang Y, Wang L, Li H, Ye Z. A CT-based Radiomics Model for Prediction of Lymph Node Metastasis in Early Stage Gastric Cancer. Acad Radiol. 2021;28:e155-e164. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 30] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 65. | Wang X, Li C, Fang M, Zhang L, Zhong L, Dong D, Tian J, Shan X. Integrating No.3 lymph nodes and primary tumor radiomics to predict lymph node metastasis in T1-2 gastric cancer. BMC Med Imaging. 2021;21:58. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 11] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 66. | Dong D, Fang MJ, Tang L, Shan XH, Gao JB, Giganti F, Wang RP, Chen X, Wang XX, Palumbo D, Fu J, Li WC, Li J, Zhong LZ, De Cobelli F, Ji JF, Liu ZY, Tian J. Deep learning radiomic nomogram can predict the number of lymph node metastasis in locally advanced gastric cancer: an international multicenter study. Ann Oncol. 2020;31:912-920. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 269] [Article Influence: 53.8] [Reference Citation Analysis (1)] |
| 67. | Liu Q, Li J, Xin B, Sun Y, Feng D, Fulham MJ, Wang X, Song S. 18F-FDG PET/CT Radiomics for Preoperative Prediction of Lymph Node Metastases and Nodal Staging in Gastric Cancer. Front Oncol. 2021;11:723345. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 24] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 68. | Wang W, Peng Y, Feng X, Zhao Y, Seeruttun SR, Zhang J, Cheng Z, Li Y, Liu Z, Zhou Z. Development and Validation of a Computed Tomography-Based Radiomics Signature to Predict Response to Neoadjuvant Chemotherapy for Locally Advanced Gastric Cancer. JAMA Netw Open. 2021;4:e2121143. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 74] [Article Influence: 18.5] [Reference Citation Analysis (0)] |
| 69. | Hou Z, Yang Y, Li S, Yan J, Ren W, Liu J, Wang K, Liu B, Wan S. Radiomic analysis using contrast-enhanced CT: predict treatment response to pulsed low dose rate radiotherapy in gastric carcinoma with abdominal cavity metastasis. Quant Imaging Med Surg. 2018;8:410-420. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 34] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 70. | Chen Y, Xu W, Li YL, Liu W, Sah BK, Wang L, Xu Z, Wels M, Zheng Y, Yan M, Zhang H, Ma Q, Zhu Z, Li C. CT-Based Radiomics Showing Generalization to Predict Tumor Regression Grade for Advanced Gastric Cancer Treated With Neoadjuvant Chemotherapy. Front Oncol. 2022;12:758863. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 12] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 71. | Chen Y, Yuan F, Wang L, Li E, Xu Z, Wels M, Yao W, Zhang H. Evaluation of dual-energy CT derived radiomics signatures in predicting outcomes in patients with advanced gastric cancer after neoadjuvant chemotherapy. Eur J Surg Oncol. 2022;48:339-347. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 19] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 72. | Jiang Y, Jin C, Yu H, Wu J, Chen C, Yuan Q, Huang W, Hu Y, Xu Y, Zhou Z, Fisher GA Jr, Li G, Li R. Development and Validation of a Deep Learning CT Signature to Predict Survival and Chemotherapy Benefit in Gastric Cancer: A Multicenter, Retrospective Study. Ann Surg. 2021;274:e1153-e1161. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 101] [Article Influence: 20.2] [Reference Citation Analysis (0)] |
| 73. | Jiang Y, Yuan Q, Lv W, Xi S, Huang W, Sun Z, Chen H, Zhao L, Liu W, Hu Y, Lu L, Ma J, Li T, Yu J, Wang Q, Li G. Radiomic signature of 18F fluorodeoxyglucose PET/CT for prediction of gastric cancer survival and chemotherapeutic benefits. Theranostics. 2018;8:5915-5928. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 110] [Cited by in RCA: 107] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 74. | Huang J, Yao H, Li Y, Dong M, Han C, He L, Huang X, Xia T, Yi Z, Wang H, Zhang Y, He J, Liang C, Liu Z. Development and validation of a CT-based radiomics nomogram for preoperative prediction of tumor histologic grade in gastric adenocarcinoma. Chin J Cancer Res. 2021;33:69-78. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 75. | Wang XX, Ding Y, Wang SW, Dong D, Li HL, Chen J, Hu H, Lu C, Tian J, Shan XH. Intratumoral and peritumoral radiomics analysis for preoperative Lauren classification in gastric cancer. Cancer Imaging. 2020;20:83. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 38] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 76. | Sun Z, Jin L, Zhang S, Duan S, Xing W, Hu S. Preoperative prediction for lauren type of gastric cancer: A radiomics nomogram analysis based on CT images and clinical features. J Xray Sci Technol. 2021;29:675-686. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 2] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 77. | Zhao Y, Yang J, Luo M, Yang Y, Guo X, Zhang T, Hao J, Yao Y, Ma X. Contrast-Enhanced CT-based Textural Parameters as Potential Prognostic Factors of Survival for Colorectal Cancer Patients Receiving Targeted Therapy. Mol Imaging Biol. 2021;23:427-435. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 8] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 78. | Vicini S, Bortolotto C, Rengo M, Ballerini D, Bellini D, Carbone I, Preda L, Laghi A, Coppola F, Faggioni L. A narrative review on current imaging applications of artificial intelligence and radiomics in oncology: focus on the three most common cancers. Radiol Med. 2022;127:819-836. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 79. | Liang C, Huang Y, He L, Chen X, Ma Z, Dong D, Tian J, Liang C, Liu Z. The development and validation of a CT-based radiomics signature for the preoperative discrimination of stage I-II and stage III-IV colorectal cancer. Oncotarget. 2016;7:31401-31412. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 101] [Cited by in RCA: 129] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 80. | Lin X, Zhao S, Jiang H, Jia F, Wang G, He B, Ma X, Li J, Shi Z. A radiomics-based nomogram for preoperative T staging prediction of rectal cancer. Abdom Radiol (NY). 2021;46:4525-4535. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 15] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 81. | Dou Y, Liu Y, Kong X, Yang S. T staging with functional and radiomics parameters of computed tomography in colorectal cancer patients. Medicine (Baltimore). 2022;101:e29244. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 82. | Li J, Zhou Y, Wang X, Zhou M, Chen X, Luan K. An MRI-based multi-objective radiomics model predicts lymph node status in patients with rectal cancer. Abdom Radiol (NY). 2021;46:1816-1824. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 26] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 83. | Li C, Yin J. Radiomics Based on T2-Weighted Imaging and Apparent Diffusion Coefficient Images for Preoperative Evaluation of Lymph Node Metastasis in Rectal Cancer Patients. Front Oncol. 2021;11:671354. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 23] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 84. | Liu X, Yang Q, Zhang C, Sun J, He K, Xie Y, Zhang Y, Fu Y, Zhang H. Multiregional-Based Magnetic Resonance Imaging Radiomics Combined With Clinical Data Improves Efficacy in Predicting Lymph Node Metastasis of Rectal Cancer. Front Oncol. 2020;10:585767. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 37] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 85. | He J, Wang Q, Zhang Y, Wu H, Zhou Y, Zhao S. Preoperative prediction of regional lymph node metastasis of colorectal cancer based on 18F-FDG PET/CT and machine learning. Ann Nucl Med. 2021;35:617-627. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 29] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 86. | Huang Y, He L, Dong D, Yang C, Liang C, Chen X, Ma Z, Huang X, Yao S, Tian J, Liu Z. Individualized prediction of perineural invasion in colorectal cancer: development and validation of a radiomics prediction model. Chin J Cancer Res. 2018;30:40-50. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 52] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 87. | Zhang K, Ren Y, Xu S, Lu W, Xie S, Qu J, Wang X, Shen B, Pang P, Cai X, Sun J. A clinical-radiomics model incorporating T2-weighted and diffusion-weighted magnetic resonance images predicts the existence of lymphovascular invasion / perineural invasion in patients with colorectal cancer. Med Phys. 2021;48:4872-4882. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 88. | Siriwardena AK, Mason JM, Mullamitha S, Hancock HC, Jegatheeswaran S. Management of colorectal cancer presenting with synchronous liver metastases. Nat Rev Clin Oncol. 2014;11:446-459. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 126] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 89. | Ayez N, Burger JW, van der Pool AE, Eggermont AM, Grunhagen DJ, de Wilt JH, Verhoef C. Long-term results of the "liver first" approach in patients with locally advanced rectal cancer and synchronous liver metastases. Dis Colon Rectum. 2013;56:281-287. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 47] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 90. | Liu Z, Zhang XY, Shi YJ, Wang L, Zhu HT, Tang Z, Wang S, Li XT, Tian J, Sun YS. Radiomics Analysis for Evaluation of Pathological Complete Response to Neoadjuvant Chemoradiotherapy in Locally Advanced Rectal Cancer. Clin Cancer Res. 2017;23:7253-7262. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 275] [Cited by in RCA: 407] [Article Influence: 50.9] [Reference Citation Analysis (0)] |
| 91. | Wang J, Chen J, Zhou R, Gao Y, Li J. Machine learning-based multiparametric MRI radiomics for predicting poor responders after neoadjuvant chemoradiotherapy in rectal Cancer patients. BMC Cancer. 2022;22:420. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 15] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 92. | Giannini V, Rosati S, Defeudis A, Balestra G, Vassallo L, Cappello G, Mazzetti S, De Mattia C, Rizzetto F, Torresin A, Sartore-Bianchi A, Siena S, Vanzulli A, Leone F, Zagonel V, Marsoni S, Regge D. Radiomics predicts response of individual HER2-amplified colorectal cancer liver metastases in patients treated with HER2-targeted therapy. Int J Cancer. 2020;147:3215-3223. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 27] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 93. | Zhang Z, Shen L, Wang Y, Wang J, Zhang H, Xia F, Wan J, Zhang Z. MRI Radiomics Signature as a Potential Biomarker for Predicting KRAS Status in Locally Advanced Rectal Cancer Patients. Front Oncol. 2021;11:614052. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 11] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 94. | Zhang G, Chen L, Liu A, Pan X, Shu J, Han Y, Huan Y, Zhang J. Comparable Performance of Deep Learning-Based to Manual-Based Tumor Segmentation in KRAS/NRAS/BRAF Mutation Prediction With MR-Based Radiomics in Rectal Cancer. Front Oncol. 2021;11:696706. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 12] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 95. | Xue T, Peng H, Chen Q, Li M, Duan S, Feng F. Preoperative prediction of KRAS mutation status in colorectal cancer using a CT-based radiomics nomogram. Br J Radiol. 2022;95:20211014. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 13] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 96. | Yang L, Dong D, Fang M, Zhu Y, Zang Y, Liu Z, Zhang H, Ying J, Zhao X, Tian J. Can CT-based radiomics signature predict KRAS/NRAS/BRAF mutations in colorectal cancer? Eur Radiol. 2018;28:2058-2067. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 175] [Article Influence: 25.0] [Reference Citation Analysis (0)] |
| 97. | Horvat N, Veeraraghavan H, Pelossof RA, Fernandes MC, Arora A, Khan M, Marco M, Cheng CT, Gonen M, Golia Pernicka JS, Gollub MJ, Garcia-Aguillar J, Petkovska I. Radiogenomics of rectal adenocarcinoma in the era of precision medicine: A pilot study of associations between qualitative and quantitative MRI imaging features and genetic mutations. Eur J Radiol. 2019;113:174-181. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 33] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 98. | Golia Pernicka JS, Gagniere J, Chakraborty J, Yamashita R, Nardo L, Creasy JM, Petkovska I, Do RRK, Bates DDB, Paroder V, Gonen M, Weiser MR, Simpson AL, Gollub MJ. Radiomics-based prediction of microsatellite instability in colorectal cancer at initial computed tomography evaluation. Abdom Radiol (NY). 2019;44:3755-3763. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 74] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 99. | Li Z, Chen F, Zhang S, Ma X, Xia Y, Shen F, Lu Y, Shao C. The feasibility of MRI-based radiomics model in presurgical evaluation of tumor budding in locally advanced rectal cancer. Abdom Radiol (NY). 2022;47:56-65. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 19] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 100. | Gong XQ, Tao YY, Wu YK, Liu N, Yu X, Wang R, Zheng J, Huang XH, Li JD, Yang G, Wei XQ, Yang L, Zhang XM. Progress of MRI Radiomics in Hepatocellular Carcinoma. Front Oncol. 2021;11:698373. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 31] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 101. | He Y, Hu B, Zhu C, Xu W, Ge Y, Hao X, Dong B, Chen X, Dong Q, Zhou X. A Novel Multimodal Radiomics Model for Predicting Prognosis of Resected Hepatocellular Carcinoma. Front Oncol. 2022;12:745258. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 102. | Liu Q, Li J, Liu F, Yang W, Ding J, Chen W, Wei Y, Li B, Zheng L. A radiomics nomogram for the prediction of overall survival in patients with hepatocellular carcinoma after hepatectomy. Cancer Imaging. 2020;20:82. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 45] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 103. | Zhang W, Zhang B, Chen XP. Adjuvant treatment strategy after curative resection for hepatocellular carcinoma. Front Med. 2021;15:155-169. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 61] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 104. | Lee S, Kim SH, Lee JE, Sinn DH, Park CK. Preoperative gadoxetic acid-enhanced MRI for predicting microvascular invasion in patients with single hepatocellular carcinoma. J Hepatol. 2017;67:526-534. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 205] [Cited by in RCA: 349] [Article Influence: 43.6] [Reference Citation Analysis (0)] |
| 105. | Zhou W, Jian W, Cen X, Zhang L, Guo H, Liu Z, Liang C, Wang G. Prediction of Microvascular Invasion of Hepatocellular Carcinoma Based on Contrast-Enhanced MR and 3D Convolutional Neural Networks. Front Oncol. 2021;11:588010. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 20] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 106. | Feng ST, Jia Y, Liao B, Huang B, Zhou Q, Li X, Wei K, Chen L, Li B, Wang W, Chen S, He X, Wang H, Peng S, Chen ZB, Tang M, Chen Z, Hou Y, Peng Z, Kuang M. Preoperative prediction of microvascular invasion in hepatocellular cancer: a radiomics model using Gd-EOB-DTPA-enhanced MRI. Eur Radiol. 2019;29:4648-4659. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149] [Cited by in RCA: 148] [Article Influence: 24.7] [Reference Citation Analysis (0)] |
| 107. | Ariff B, Lloyd CR, Khan S, Shariff M, Thillainayagam AV, Bansi DS, Khan SA, Taylor-Robinson SD, Lim AK. Imaging of liver cancer. World J Gastroenterol. 2009;15:1289-1300. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 80] [Cited by in RCA: 76] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 108. | Mokrane FZ, Lu L, Vavasseur A, Otal P, Peron JM, Luk L, Yang H, Ammari S, Saenger Y, Rousseau H, Zhao B, Schwartz LH, Dercle L. Radiomics machine-learning signature for diagnosis of hepatocellular carcinoma in cirrhotic patients with indeterminate liver nodules. Eur Radiol. 2020;30:558-570. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 125] [Article Influence: 25.0] [Reference Citation Analysis (0)] |
| 109. | Abdelrahman AS, Madkour SS, Ekladious MEY. Interrater reliability and agreement of the liver imaging reporting and data system (LI-RADS) v2018 for the evaluation of hepatic lesions. Pol J Radiol. 2022;87:e316-e324. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 110. | Zhong X, Guan T, Tang D, Li J, Lu B, Cui S, Tang H. Differentiation of small (≤ 3 cm) hepatocellular carcinomas from benign nodules in cirrhotic liver: the added additive value of MRI-based radiomics analysis to LI-RADS version 2018 algorithm. BMC Gastroenterol. 2021;21:155. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 111. | Zhong X, Tang H, Guan T, Lu B, Zhang C, Tang D, Li J, Cui S. Added Value of Quantitative Apparent Diffusion Coefficients for Identifying Small Hepatocellular Carcinoma from Benign Nodule Categorized as LI-RADS 3 and 4 in Cirrhosis. J Clin Transl Hepatol. 2022;10:34-41. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 6] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 112. | Hectors SJ, Wagner M, Bane O, Besa C, Lewis S, Remark R, Chen N, Fiel MI, Zhu H, Gnjatic S, Merad M, Hoshida Y, Taouli B. Quantification of hepatocellular carcinoma heterogeneity with multiparametric magnetic resonance imaging. Sci Rep. 2017;7:2452. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 77] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 113. | Hectors SJ, Lewis S, Besa C, King MJ, Said D, Putra J, Ward S, Higashi T, Thung S, Yao S, Laface I, Schwartz M, Gnjatic S, Merad M, Hoshida Y, Taouli B. MRI radiomics features predict immuno-oncological characteristics of hepatocellular carcinoma. Eur Radiol. 2020;30:3759-3769. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 113] [Article Influence: 22.6] [Reference Citation Analysis (0)] |
| 114. | Kurucay M, Kloth C, Kaufmann S, Nikolaou K, Bösmüller H, Horger M, Thaiss WM. Multiparametric imaging for detection and characterization of hepatocellular carcinoma using gadoxetic acid-enhanced MRI and perfusion-CT: which parameters work best? Cancer Imaging. 2017;17:18. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 10] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 115. | Yasaka K, Akai H, Abe O, Kiryu S. Deep Learning with Convolutional Neural Network for Differentiation of Liver Masses at Dynamic Contrast-enhanced CT: A Preliminary Study. Radiology. 2018;286:887-896. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 293] [Cited by in RCA: 394] [Article Influence: 49.3] [Reference Citation Analysis (0)] |
| 116. | Hamm CA, Wang CJ, Savic LJ, Ferrante M, Schobert I, Schlachter T, Lin M, Duncan JS, Weinreb JC, Chapiro J, Letzen B. Deep learning for liver tumor diagnosis part I: development of a convolutional neural network classifier for multi-phasic MRI. Eur Radiol. 2019;29:3338-3347. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117] [Cited by in RCA: 207] [Article Influence: 34.5] [Reference Citation Analysis (0)] |
| 117. | Hui TCH, Chuah TK, Low HM, Tan CH. Predicting early recurrence of hepatocellular carcinoma with texture analysis of preoperative MRI: a radiomics study. Clin Radiol. 2018;73:1056.e11-1056.e16. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 53] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 118. | Zhang Z, Jiang H, Chen J, Wei Y, Cao L, Ye Z, Li X, Ma L, Song B. Hepatocellular carcinoma: radiomics nomogram on gadoxetic acid-enhanced MR imaging for early postoperative recurrence prediction. Cancer Imaging. 2019;19:22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 97] [Cited by in RCA: 101] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 119. | Shi G, Han X, Wang Q, Ding Y, Liu H, Zhang Y, Dai Y. Evaluation of Multiple Prognostic Factors of Hepatocellular Carcinoma with Intra-Voxel Incoherent Motions Imaging by Extracting the Histogram Metrics. Cancer Manag Res. 2020;12:6019-6031. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 17] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 120. | Zhou Y, Sun SW, Liu QP, Xu X, Zhang Y, Zhang YD. TED: Two-stage expert-guided interpretable diagnosis framework for microvascular invasion in hepatocellular carcinoma. Med Image Anal. 2022;82:102575. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 121. | Ma X, Wei J, Gu D, Zhu Y, Feng B, Liang M, Wang S, Zhao X, Tian J. Preoperative radiomics nomogram for microvascular invasion prediction in hepatocellular carcinoma using contrast-enhanced CT. Eur Radiol. 2019;29:3595-3605. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 163] [Article Influence: 27.2] [Reference Citation Analysis (1)] |
| 122. | Xu X, Zhang HL, Liu QP, Sun SW, Zhang J, Zhu FP, Yang G, Yan X, Zhang YD, Liu XS. Radiomic analysis of contrast-enhanced CT predicts microvascular invasion and outcome in hepatocellular carcinoma. J Hepatol. 2019;70:1133-1144. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 278] [Cited by in RCA: 495] [Article Influence: 82.5] [Reference Citation Analysis (0)] |
| 123. | Chong HH, Yang L, Sheng RF, Yu YL, Wu DJ, Rao SX, Yang C, Zeng MS. Multi-scale and multi-parametric radiomics of gadoxetate disodium-enhanced MRI predicts microvascular invasion and outcome in patients with solitary hepatocellular carcinoma ≤ 5 cm. Eur Radiol. 2021;31:4824-4838. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 149] [Article Influence: 37.3] [Reference Citation Analysis (1)] |
| 124. | Xu T, Ren L, Liao M, Zhao B, Wei R, Zhou Z, He Y, Zhang H, Chen D, Chen H, Liao W. Preoperative Radiomics Analysis of Contrast-Enhanced CT for Microvascular Invasion and Prognosis Stratification in Hepatocellular Carcinoma. J Hepatocell Carcinoma. 2022;9:189-201. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 2] [Article Influence: 0.7] [Reference Citation Analysis (1)] |
| 125. | Brancato V, Garbino N, Salvatore M, Cavaliere C. MRI-Based Radiomic Features Help Identify Lesions and Predict Histopathological Grade of Hepatocellular Carcinoma. Diagnostics (Basel). 2022;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 14] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 126. | Segal E, Sirlin CB, Ooi C, Adler AS, Gollub J, Chen X, Chan BK, Matcuk GR, Barry CT, Chang HY, Kuo MD. Decoding global gene expression programs in liver cancer by noninvasive imaging. Nat Biotechnol. 2007;25:675-680. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 400] [Cited by in RCA: 443] [Article Influence: 24.6] [Reference Citation Analysis (0)] |
| 127. | Gu D, Xie Y, Wei J, Li W, Ye Z, Zhu Z, Tian J, Li X. MRI-Based Radiomics Signature: A Potential Biomarker for Identifying Glypican 3-Positive Hepatocellular Carcinoma. J Magn Reson Imaging. 2020;52:1679-1687. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 43] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 128. | Wang F, Tan BF, Poh SS, Siow TR, Lim FLWT, Yip CSP, Wang MLC, Nei W, Tan HQ. Predicting outcomes for locally advanced rectal cancer treated with neoadjuvant chemoradiation with CT-based radiomics. Sci Rep. 2022;12:6167. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 14] [Reference Citation Analysis (0)] |
| 129. | Shahzadi I, Zwanenburg A, Lattermann A, Linge A, Baldus C, Peeken JC, Combs SE, Diefenhardt M, Rödel C, Kirste S, Grosu AL, Baumann M, Krause M, Troost EGC, Löck S. Analysis of MRI and CT-based radiomics features for personalized treatment in locally advanced rectal cancer and external validation of published radiomics models. Sci Rep. 2022;12:10192. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 18] [Reference Citation Analysis (0)] |
| 130. | Bonomo P, Socarras Fernandez J, Thorwarth D, Casati M, Livi L, Zips D, Gani C. Simulation CT-based radiomics for prediction of response after neoadjuvant chemo-radiotherapy in patients with locally advanced rectal cancer. Radiat Oncol. 2022;17:84. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 3] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 131. | Zhang L, Hu J, Hou J, Jiang X, Guo L, Tian L. Radiomics-based model using gadoxetic acid disodium-enhanced MR images: associations with recurrence-free survival of patients with hepatocellular carcinoma treated by surgical resection. Abdom Radiol (NY). 2021;46:3845-3854. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 20] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 132. | Zhu HB, Zheng ZY, Zhao H, Zhang J, Zhu H, Li YH, Dong ZY, Xiao LS, Kuang JJ, Zhang XL, Liu L. Radiomics-based nomogram using CT imaging for noninvasive preoperative prediction of early recurrence in patients with hepatocellular carcinoma. Diagn Interv Radiol. 2020;26:411-419. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 27] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 133. | Zhang Z, Chen J, Jiang H, Wei Y, Zhang X, Cao L, Duan T, Ye Z, Yao S, Pan X, Song B. Gadoxetic acid-enhanced MRI radiomics signature: prediction of clinical outcome in hepatocellular carcinoma after surgical resection. Ann Transl Med. 2020;8:870. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 25] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 134. | Lee SJ, Zea R, Kim DH, Lubner MG, Deming DA, Pickhardt PJ. CT texture features of liver parenchyma for predicting development of metastatic disease and overall survival in patients with colorectal cancer. Eur Radiol. 2018;28:1520-1528. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 32] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 135. | Beckers RCJ, Beets-Tan RGH, Schnerr RS, Maas M, da Costa Andrade LA, Beets GL, Dejong CH, Houwers JB, Lambregts DMJ. Whole-volume vs. segmental CT texture analysis of the liver to assess metachronous colorectal liver metastases. Abdom Radiol (NY). 2017;42:2639-2645. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 10] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 136. | Ganeshan B, Miles KA, Young RC, Chatwin CR. Texture analysis in non-contrast enhanced CT: impact of malignancy on texture in apparently disease-free areas of the liver. Eur J Radiol. 2009;70:101-110. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 108] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 137. | Rao SX, Lambregts DM, Schnerr RS, van Ommen W, van Nijnatten TJ, Martens MH, Heijnen LA, Backes WH, Verhoef C, Zeng MS, Beets GL, Beets-Tan RG. Whole-liver CT texture analysis in colorectal cancer: Does the presence of liver metastases affect the texture of the remaining liver? United European Gastroenterol J. 2014;2:530-538. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 51] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 138. | Beckers RCJ, Lambregts DMJ, Schnerr RS, Maas M, Rao SX, Kessels AGH, Thywissen T, Beets GL, Trebeschi S, Houwers JB, Dejong CH, Verhoef C, Beets-Tan RGH. Whole liver CT texture analysis to predict the development of colorectal liver metastases-A multicentre study. Eur J Radiol. 2017;92:64-71. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 34] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 139. | Li Y, Gong J, Shen X, Li M, Zhang H, Feng F, Tong T. Assessment of Primary Colorectal Cancer CT Radiomics to Predict Metachronous Liver Metastasis. Front Oncol. 2022;12:861892. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 15] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 140. | Bian Y, Jiang H, Ma C, Cao K, Fang X, Li J, Wang L, Zheng J, Lu J. Performance of CT-based radiomics in diagnosis of superior mesenteric vein resection margin in patients with pancreatic head cancer. Abdom Radiol (NY). 2020;45:759-773. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 18] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 141. | Simpson G, Spieler B, Dogan N, Portelance L, Mellon EA, Kwon D, Ford JC, Yang F. Predictive value of 0.35 T magnetic resonance imaging radiomic features in stereotactic ablative body radiotherapy of pancreatic cancer: A pilot study. Med Phys. 2020;47:3682-3690. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 38] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 142. | Parr E, Du Q, Zhang C, Lin C, Kamal A, McAlister J, Liang X, Bavitz K, Rux G, Hollingsworth M, Baine M, Zheng D. Radiomics-Based Outcome Prediction for Pancreatic Cancer Following Stereotactic Body Radiotherapy. Cancers (Basel). 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 43] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 143. | Zhang Y, Cheng C, Liu Z, Wang L, Pan G, Sun G, Chang Y, Zuo C, Yang X. Radiomics analysis for the differentiation of autoimmune pancreatitis and pancreatic ductal adenocarcinoma in 18 F-FDG PET/CT. Med Phys. 2019;46:4520-4530. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 55] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 144. | Liu Z, Li M, Zuo C, Yang Z, Yang X, Ren S, Peng Y, Sun G, Shen J, Cheng C. Radiomics model of dual-time 2-[18F]FDG PET/CT imaging to distinguish between pancreatic ductal adenocarcinoma and autoimmune pancreatitis. Eur Radiol. 2021;31:6983-6991. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 27] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 145. | Park S, Chu LC, Hruban RH, Vogelstein B, Kinzler KW, Yuille AL, Fouladi DF, Shayesteh S, Ghandili S, Wolfgang CL, Burkhart R, He J, Fishman EK, Kawamoto S. Differentiating autoimmune pancreatitis from pancreatic ductal adenocarcinoma with CT radiomics features. Diagn Interv Imaging. 2020;101:555-564. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 75] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 146. | Chu LC, Park S, Kawamoto S, Fouladi DF, Shayesteh S, Zinreich ES, Graves JS, Horton KM, Hruban RH, Yuille AL, Kinzler KW, Vogelstein B, Fishman EK. Utility of CT Radiomics Features in Differentiation of Pancreatic Ductal Adenocarcinoma From Normal Pancreatic Tissue. AJR Am J Roentgenol. 2019;213:349-357. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 132] [Article Influence: 22.0] [Reference Citation Analysis (0)] |
| 147. | Tomaszewski MR, Latifi K, Boyer E, Palm RF, El Naqa I, Moros EG, Hoffe SE, Rosenberg SA, Frakes JM, Gillies RJ. Delta radiomics analysis of Magnetic Resonance guided radiotherapy imaging data can enable treatment response prediction in pancreatic cancer. Radiat Oncol. 2021;16:237. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 31] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 148. | Xie T, Wang X, Li M, Tong T, Yu X, Zhou Z. Pancreatic ductal adenocarcinoma: a radiomics nomogram outperforms clinical model and TNM staging for survival estimation after curative resection. Eur Radiol. 2020;30:2513-2524. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 64] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 149. | Healy GM, Salinas-Miranda E, Jain R, Dong X, Deniffel D, Borgida A, Hosni A, Ryan DT, Njeze N, McGuire A, Conlon KC, Dodd JD, Ryan ER, Grant RC, Gallinger S, Haider MA. Pre-operative radiomics model for prognostication in resectable pancreatic adenocarcinoma with external validation. Eur Radiol. 2022;32:2492-2505. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 26] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 150. | van Timmeren JE, Cester D, Tanadini-Lang S, Alkadhi H, Baessler B. Radiomics in medical imaging-"how-to" guide and critical reflection. Insights Imaging. 2020;11:91. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 588] [Cited by in RCA: 771] [Article Influence: 154.2] [Reference Citation Analysis (0)] |
| 151. | Verma V, Simone CB 2nd, Krishnan S, Lin SH, Yang J, Hahn SM. The Rise of Radiomics and Implications for Oncologic Management. J Natl Cancer Inst. 2017;109. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 97] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 152. | Lambin P, Leijenaar RTH, Deist TM, Peerlings J, de Jong EEC, van Timmeren J, Sanduleanu S, Larue RTHM, Even AJG, Jochems A, van Wijk Y, Woodruff H, van Soest J, Lustberg T, Roelofs E, van Elmpt W, Dekker A, Mottaghy FM, Wildberger JE, Walsh S. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol. 2017;14:749-762. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1825] [Cited by in RCA: 3569] [Article Influence: 446.1] [Reference Citation Analysis (0)] |
| 153. | Zwanenburg A, Vallières M, Abdalah MA, Aerts HJWL, Andrearczyk V, Apte A, Ashrafinia S, Bakas S, Beukinga RJ, Boellaard R, Bogowicz M, Boldrini L, Buvat I, Cook GJR, Davatzikos C, Depeursinge A, Desseroit MC, Dinapoli N, Dinh CV, Echegaray S, El Naqa I, Fedorov AY, Gatta R, Gillies RJ, Goh V, Götz M, Guckenberger M, Ha SM, Hatt M, Isensee F, Lambin P, Leger S, Leijenaar RTH, Lenkowicz J, Lippert F, Losnegård A, Maier-Hein KH, Morin O, Müller H, Napel S, Nioche C, Orlhac F, Pati S, Pfaehler EAG, Rahmim A, Rao AUK, Scherer J, Siddique MM, Sijtsema NM, Socarras Fernandez J, Spezi E, Steenbakkers RJHM, Tanadini-Lang S, Thorwarth D, Troost EGC, Upadhaya T, Valentini V, van Dijk LV, van Griethuysen J, van Velden FHP, Whybra P, Richter C, Löck S. The Image Biomarker Standardization Initiative: Standardized Quantitative Radiomics for High-Throughput Image-based Phenotyping. Radiology. 2020;295:328-338. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2090] [Cited by in RCA: 2320] [Article Influence: 464.0] [Reference Citation Analysis (0)] |
| 154. | Zhong J, Hu Y, Xing Y, Ge X, Ding D, Zhang H, Yao W. A systematic review of radiomics in pancreatitis: applying the evidence level rating tool for promoting clinical transferability. Insights Imaging. 2022;13:139. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 12] [Reference Citation Analysis (0)] |
| 155. | Zhong J, Hu Y, Ge X, Xing Y, Ding D, Zhang G, Zhang H, Yang Q, Yao W. A systematic review of radiomics in chondrosarcoma: assessment of study quality and clinical value needs handy tools. Eur Radiol. 2022;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 16] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 156. | Park CJ, Park YW, Ahn SS, Kim D, Kim EH, Kang SG, Chang JH, Kim SH, Lee SK. Quality of Radiomics Research on Brain Metastasis: A Roadmap to Promote Clinical Translation. Korean J Radiol. 2022;23:77-88. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 16] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 157. | Park JE, Kim D, Kim HS, Park SY, Kim JY, Cho SJ, Shin JH, Kim JH. Quality of science and reporting of radiomics in oncologic studies: room for improvement according to radiomics quality score and TRIPOD statement. Eur Radiol. 2020;30:523-536. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 182] [Article Influence: 30.3] [Reference Citation Analysis (0)] |