Published online Nov 7, 2022. doi: 10.3748/wjg.v28.i41.5931
Peer-review started: June 29, 2022
First decision: August 19, 2022
Revised: August 31, 2022
Accepted: October 19, 2022
Article in press: October 19, 2022
Published online: November 7, 2022
Processing time: 127 Days and 14.1 Hours
Endoscopy artifacts are widespread in real capsule endoscopy (CE) images but not in high-quality standard datasets.
To improve the segmentation performance of polyps from CE images with artifacts based on ensemble learning.
We collected 277 polyp images with CE artifacts from 5760 h of videos from 480 patients at Guangzhou First People’s Hospital from January 2016 to December 2019. Two public high-quality standard external datasets were retrieved and used for the comparison experiments. For each dataset, we randomly segmented the data into training, validation, and testing sets for model training, selection, and testing. We compared the performance of the base models and the ensemble model in segmenting polyps from images with artifacts.
The performance of the semantic segmentation model was affected by artifacts in the sample images, which also affected the results of polyp detection by CE using a single model. The evaluation based on real datasets with artifacts and standard datasets showed that the ensemble model of all state-of-the-art models performed better than the best corresponding base learner on the real dataset with artifacts. Compared with the corresponding optimal base learners, the intersection over union (IoU) and dice of the ensemble learning model increased to different degrees, ranging from 0.08% to 7.01% and 0.61% to 4.93%, respectively. Moreover, in the standard datasets without artifacts, most of the ensemble models were slightly better than the base learner, as demonstrated by the IoU and dice increases ranging from -0.28% to 1.20% and -0.61% to 0.76%, respectively.
Ensemble learning can improve the segmentation accuracy of polyps from CE images with artifacts. Our results demonstrated an improvement in the detection rate of polyps with interference from artifacts.
Core Tip: Artificial intelligence has been widely used in capsule endoscopy to detect gastrointestinal polyps; however, it is often impaired by artifacts in clinical practice. At present, clear and high-quality images without artifacts are usually selected for research, which has not yet produced practical assistance regarding artifact interference. In this study, we demonstrated that ensemble learning can improve the segmentation performance of polyps under the interference of artifacts, which has a significant auxiliary role in the detection of polyps in clinical practice.
- Citation: Zhou JX, Yang Z, Xi DH, Dai SJ, Feng ZQ, Li JY, Xu W, Wang H. Enhanced segmentation of gastrointestinal polyps from capsule endoscopy images with artifacts using ensemble learning. World J Gastroenterol 2022; 28(41): 5931-5943
- URL: https://www.wjgnet.com/1007-9327/full/v28/i41/5931.htm
- DOI: https://dx.doi.org/10.3748/wjg.v28.i41.5931
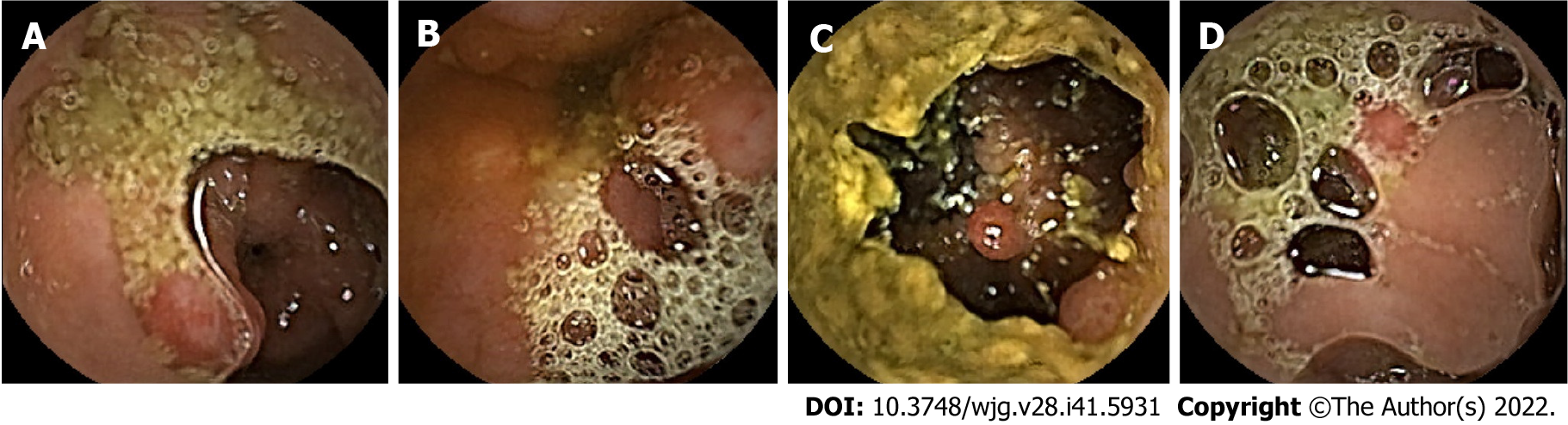
Colorectal cancer (CRC) is the second leading cause of death in the United States[1]. In China, an estimated 1101653 new cancer cases and 709529 cancer deaths from gastric cancer and CRC will occur in 2022, placing China first worldwide because of its large population[1]. Although other gastrointestinal lesions, such as erosions and ulcers, can also develop into cancers, most gastrointestinal cancers arise from precancerous polyps, which are the most common lesions found on endoscopy[2]. Therefore, early detection and removal of gastrointestinal polyps under endoscopy are critical for preventing gastrointestinal cancers[3-6]. Traditional gastroenteroscopy is widely used for the clinical assessment of gastrointestinal lesions. However, there are still some deficiencies, such as invasiveness and incomplete inspection of the site[7]. Additionally, some patients with small bowel diseases who have contraindications or are averse to undergoing gastroenteroscopy are more likely to use safer and non-invasive capsule endoscopy (CE) for visual examination of the digestive tract[8,9]. CE usually takes 8-12 h, which is not only time-consuming but also highly operator-dependent[10,11]. Otherwise, deep learning (DL) has greatly improved the sensitivity and specificity of CE for polyp detection while saving time[12]. Studies have indicated that for every 1% increase in the detection rate of colorectal adenoma, the risk of CRC can decrease by 3%[4]. However, inadequate intestinal cleansing can produce various artifacts, such as motion blur, specular reflections, bubbles, and debris (Figure 1), which can interfere with image reading, reduce the detection rate of polyps, cause patients to miss treatment, and increase the risk of tumor development[13,14]. In addition, high-quality and clear standard datasets that rarely appear in clinical practice are often used in these studies[15-17], and the intestinal lumen is often fully dilated in these images. This is significantly different from CE images with natural contraction of the intestinal lumen, which can present various artifacts (Figure 1). Therefore, these methods are often less effective in clinical practice. Hence, identifying gastrointestinal polyps and other lesions to the maximum extent when the gastrointestinal tract is insufficiently cleansed and dilated with interference factors, such as fecal residue, cloudy liquid, and bubbles in the lumen, is one of the biggest challenges in the application of artificial intelligence (AI) for CE in clinical practice and is of great concern to clinicians.
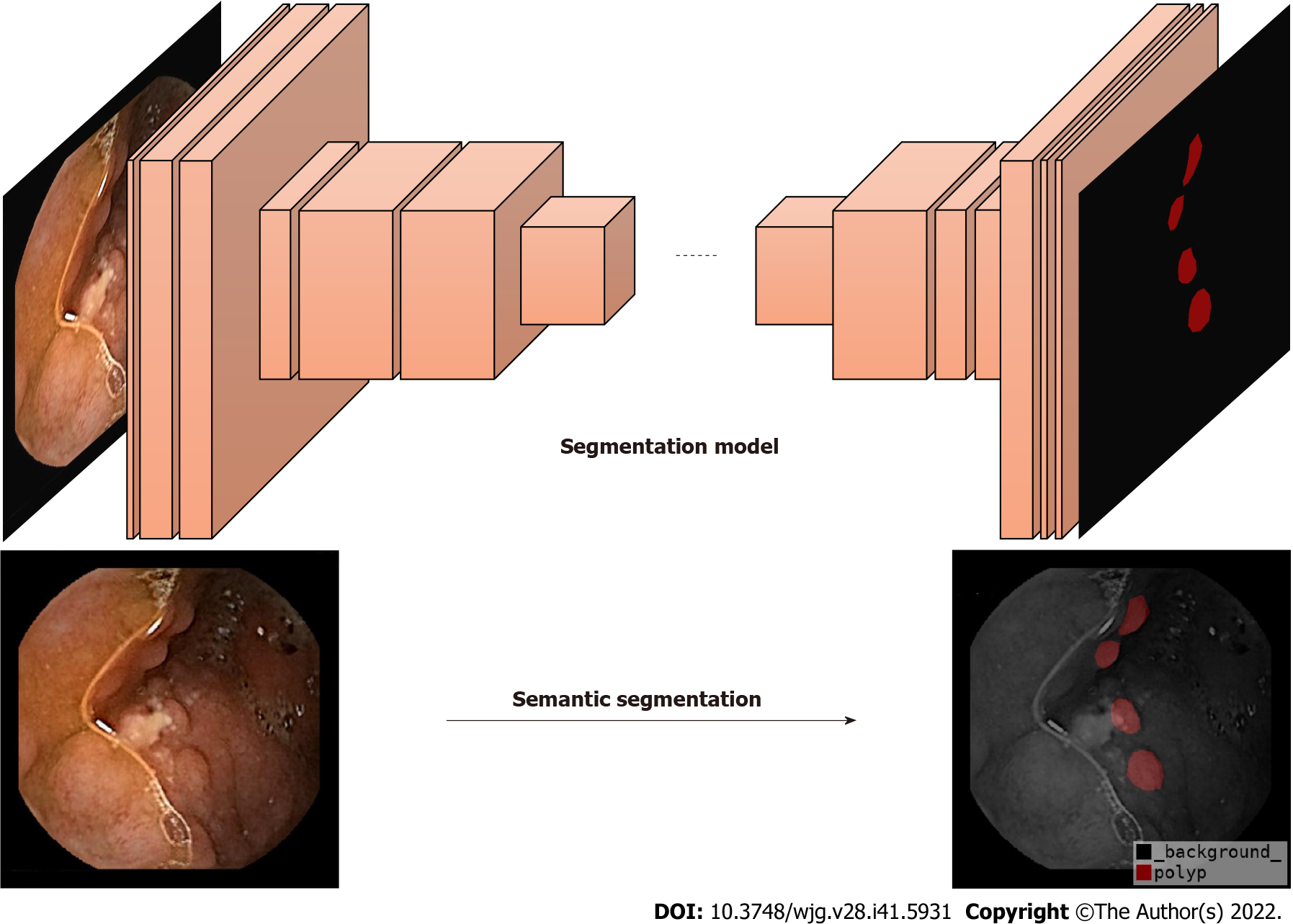
Currently, DL is a popular topic in the field of AI. It is based on the construction of computational models by simulating the neural network structure of the human brain[18]. Semantic segmentation is a part of DL algorithms that segments different objects according to each marked pixel in an image[19] (Figure 2). Some studies have proposed semantic segmentation models for medical images, such as SegNet[20], U-Net[21], Attention-UNet[22], Resnet-UNet[23], and HarDMSEG[24]. These studies have shown the significant superiority of various types of medical image semantic segmentation, as well as the feasibility of these models in tests with standard datasets. To improve the robustness of these models, researchers have begun to apply ensemble learning to medical image segmentation, not through a single model, but by combining several basic models to ensure the best prediction performance[25-27]. However, AI currently has limited ability to identify intestinal lesions with insufficient cleansing. For example, the detection rate of polyps in CE with a clean intestinal tract is significantly higher than that in CE with a dirty intestinal tract. In clinical practice, intestinal cleansing is not always performed well, and may not generate a clean image. Additionally, each patient has factors that can affect the identification by AI, such as insufficient intestinal distension, intestinal fecal residues, liquid residues, and air bubbles, resulting in the insufficient actual use of AI in clinical studies and low reliability.
In the present study, we combined semantic segmentation and ensemble learning methods for the first time to analyze CE images with artifacts. We then compared the performance of the ensemble and single models to further improve the detection rate of polyps. Our results demonstrate that ensemble learning can be used to reduce the influence of artifacts, which has a significant auxiliary role in the detection of polyps in clinical practice. To the best of our knowledge, this is the first study to propose the use of ensemble learning and semantic segmentation to reduce the negative impact of artifacts on model performance in clinical practice. Overall, our current findings have instructive significance for improving the analysis of medical images with artifacts in clinics.
This retrospective study was approved by the Ethics Committee of Guangzhou First People’s Hospital. All images were collected from videos of Ankon. This study has no conflicts of interest and did not receive any funding.
We collected 277 polyp CE images with artifacts selected from 5760 h of videos from 480 patients suffering from gastrointestinal disorders who received CE at Guangzhou First People’s Hospital from January 2016 to December 2019. The selection criteria for the experimental images were as follows: (1) The lumen on the picture was in a natural contraction state; (2) Images of the digestive tract with polyps; and (3) Artifacts in the lumen, such as feces, motion blur, specular reflections, bubbles, and debris. The polyps in these experimental images were verified for authenticity by using a large number of clear videos and photos containing the polyps or double-balloon enteroscopy. Additionally, to ensure the accuracy and rigor of the data annotation, the image data were obtained by an experienced gastroenterologist who watched the video recordings, extracted the frames where the polyps were captured through ES Navi, and annotated the pixel points of the polyp lesions using Labelme. Next, the annotated polyp profiles were carefully reviewed by two other experienced gastroenterologists. The processing time for each patient’s video was approximately 4-5 h. Before applying the dataset in the experiments, we cut off the black boxes of the images that typeset the patient’s name and other information to obtain 512 × 512 images.
The other class of data comprehended publicly available high-quality datasets with images that rarely have artifacts and included the CVC_Colon[16] dataset (created by the Computer Vision Center and Computer Science Department, Universitat Autònoma de Barcelona) and the CVC_Clinic[17] dataset (captured by the Hospital Clinic, Barcelona, Spain and labeled by the Computer Vision Center, Barcelona, Spain). CVC_Colon provided 380 colonoscopy images containing polyps with a frame size of 500 × 574 pixels. Similarly, the CVC-Clinic contained 612 still images with a size of 288 × 384 from 29 different sequences. Both datasets are frequently used in gastrointestinal endoscopic computer-assisted polyp detection studies, and several representative studies have used these datasets in their experiments.
When using these datasets, we cropped or padded the edges of the images for two reasons. First, black edges or information, such as patient and time on the edges of the images, have no effect on the polyp region segmentation. Second and the main reason is that when we cross-sectionally compared various base learners in our experiments, the convolution and pooling designs of some of them were found to be more suitable for images whose length and width were both divisible by powers of two. Therefore, to minimize the changes in the hyperparameters of these base learners, we cropped or padded the input images to match the model hyperparameter design. Finally, the GZ_Capcam dataset contains 277 images of size 512 × 512, the CVC_Clinic contains 612 images of size 288 × 384, and the CVC_Colon dataset contains 380 images of size 512 × 576; all images are eight-bit three-channel color images[17]. All images used in this study contained at least one polyp class, including the standard datasets.
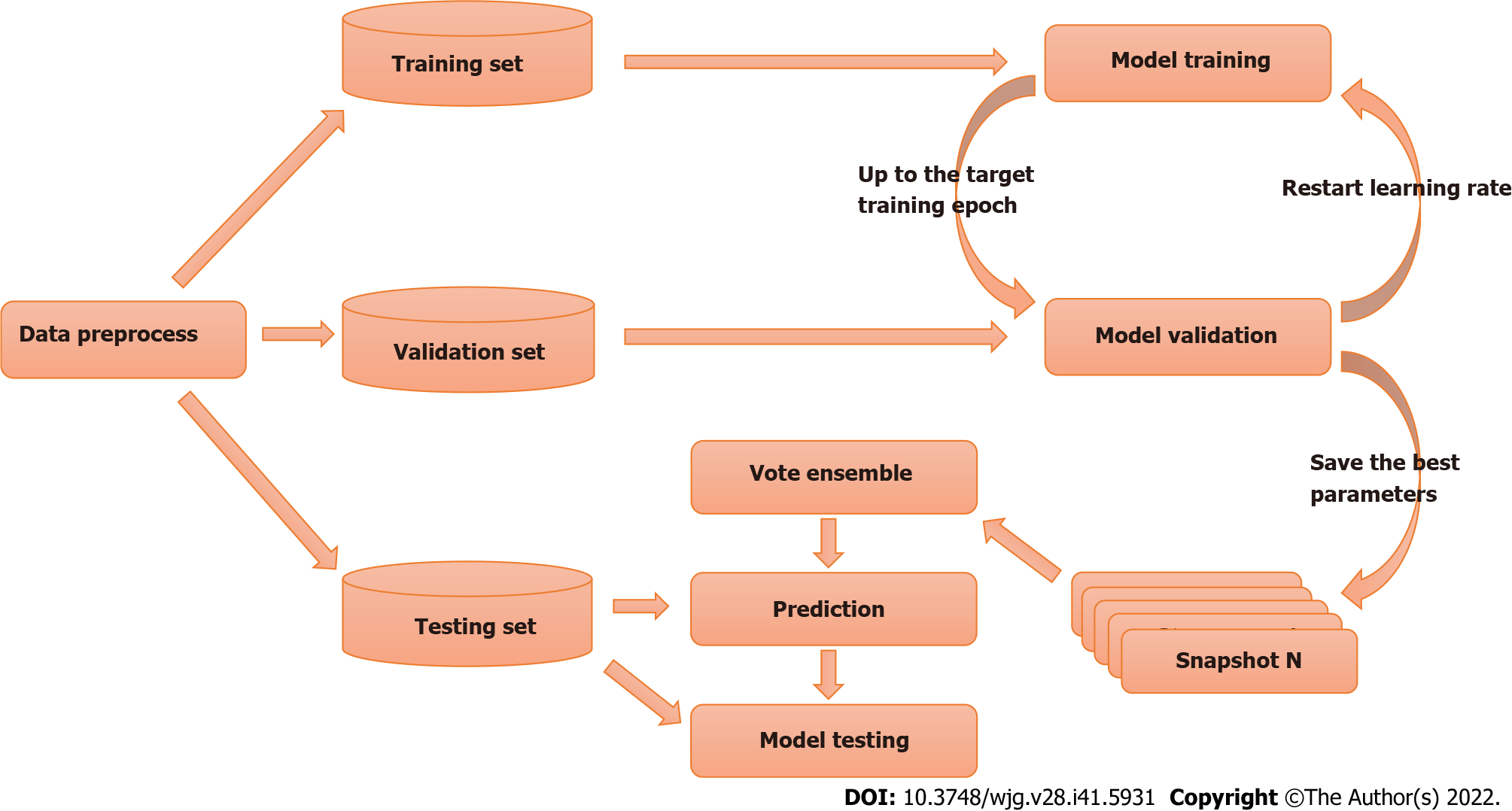
In supervised learning problems, we always expect to obtain models that perform well and are stable in all aspects; however, owing to the presence of randomness, the trained models are not always ideal, and the models obtained always have prediction preferences. The main goal of ensemble learning is to combine weak models to build a more integrated and comprehensive model that integrates the strengths of weak models. The snapshot ensemble method is a type of ensemble learning for DL models and was used in the present study[28].
In the DL method, the model parameters are adjusted according to the gradient of the objective function, as shown in Formula 1:
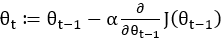
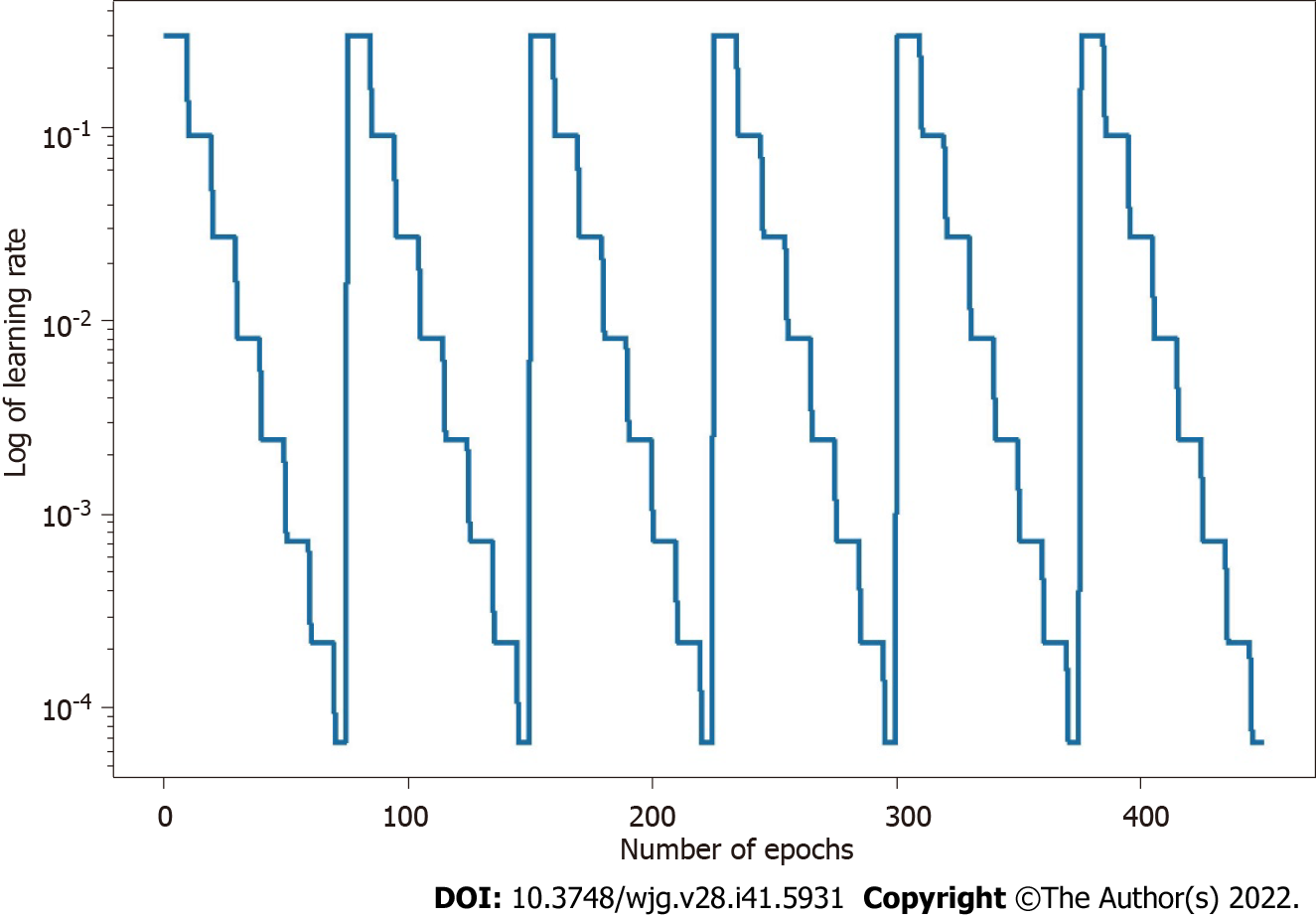
The parameters of the model take a step in the direction of the gradient descent at each iteration, and the size of the step depends on both the size of the current gradient and the learning rate, as shown in Formula 1, where θt denotes the model parameters in time step t and α denotes the learning rate. Usually, to speed up convergence and prevent DL models from repeatedly jumping at different local optima during training, the learning rate decays as the number of iterations increases, eventually causing the model to fall into a certain local optimum and not jump out. The core idea of the snapshot ensemble method is to restart the learning rate when it decays to less than a certain threshold so that the model jumps out of the current local optimum and finds a new local optimum nearby and converges, and Formula 2 and Figure 3 show the specific changes in the learning rate:
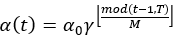
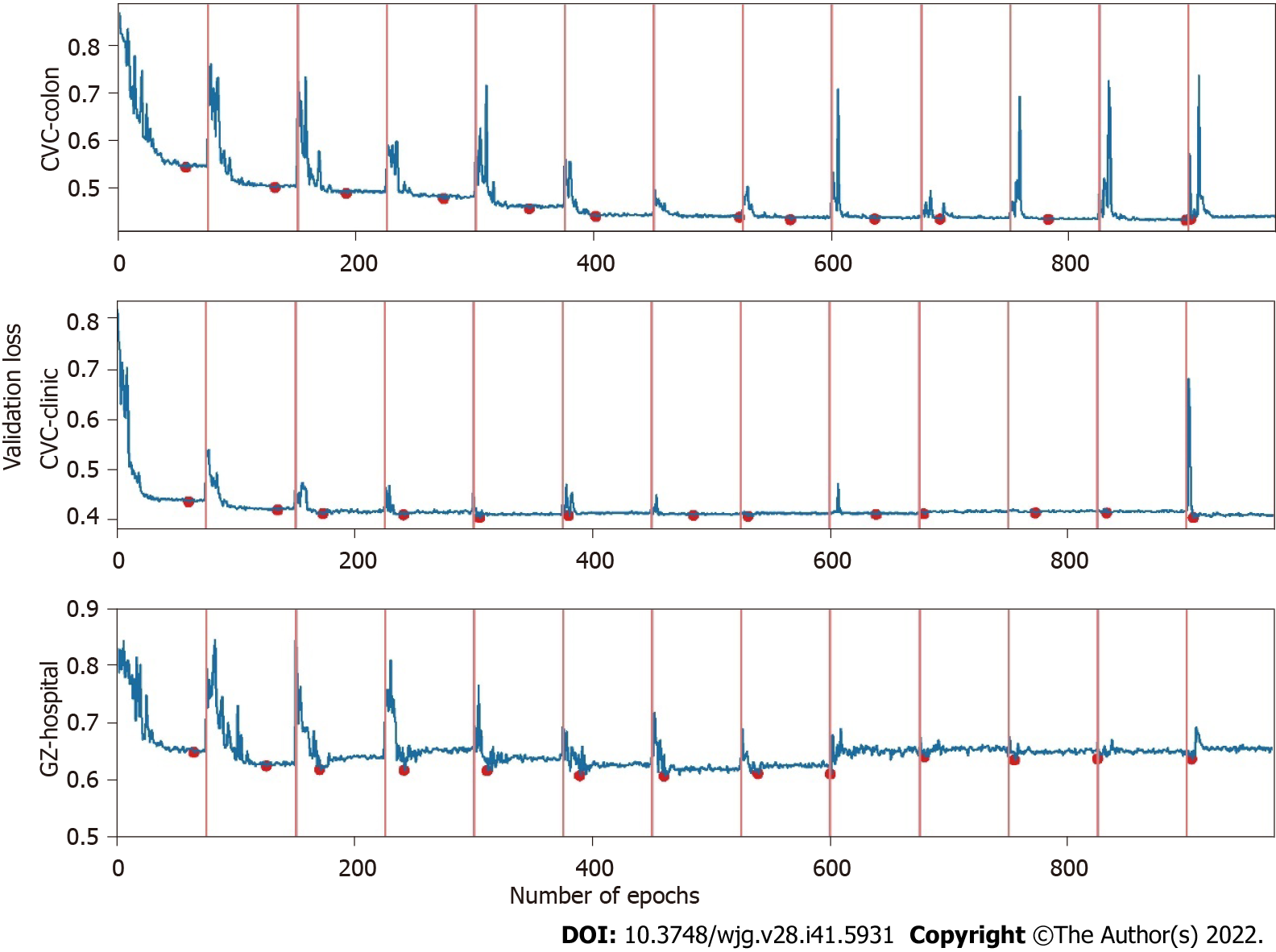
Where α0, γ, M, and T represent the initial learning rate, learning rate decay rate, number of epochs per learning rate decay, and number of epochs per learning rate restart cycle, respectively. In the snapshot ensemble method, the model that is at the local optimum before each restart learning rate is recorded as a weak model, and in the end, the prediction results of multiple weak models are integrated by ensemble voting. In the process of training, we set the number of learning rate restart cycles to 13, the learning rate decay rate to 0.3, and the number of epochs per learning rate decay to 10 and perform a total of 75 epochs in each cycle, i.e., 0.3 for γ, 10 for M, and 75 for T in Formula 2. In other words, the learning rate is reduced to 0.3 of the previous value every 10 epochs of training and reverts to the initial learning rate setting of 0.3 after 75 epochs. The model parameters that perform best on the validation set are retained in these 75 epochs as the parameters of the weak model. The entire training process lasted for 13 cycles, that is, we ended up with 13 weak learners. In the integration phase of weak models, we selected three, five, and seven weak learners with the best performance on the validation set and obtained the prediction results of the ensemble model by vote ensemble. All computational processes, including data pre-processing, model training, validation, and testing, were performed through Python programming. We built the model using PyTorch, and all experiments were based on an NVIDIA Titan V GPU. Figure 4 shows the change in validation loss in the experiment with the UNet model on the CVC_Colon dataset. The light pink line indicates the epochs of the restart learning rate, and the red points indicate the epochs of preserving the weak models.
To show that the ensemble classification is effective in improving the segmentation in comparison with the single model when dealing with medical images with artifacts, and to illustrate the generality of its enhancement effect, we used five existing state-of-the-art (SOTA) segmentation models as base learners; SegNet[20], which is proposed to solve the deep network model of image semantic segmentation for autonomous driving or intelligent robots, and is mainly based on full convolutional networks; U-Net[21], which performs well on neuron structure segmentation datasets with only a small number of annotations, and is a basic solution for medical image analysis of small datasets; Attention-UNet[22], which is an improved model based on U-Net, and achieves performance beyond that of the U-Net model for semantic segmentation of human organs on abdominal three-dimensional computed tomography scans; ResNet-UNet[23], which is also an improved version of U-Net, and gets outstanding performance on the public challenge of identifying pneumothorax diseases on chest X-rays; and HarDMSEG[24], which is an efficient image segmentation model, and achieves SOTA level in terms of both computational efficiency and analytical accuracy, in comparison experiments to illustrate that ensemble learning method improves their analysis capability in the face of images with artifacts.
First, we randomly divided the experimental and public data into training (195 images), validation (41 images), and testing (41 images) sets. The model was trained using the training set, and the best model was selected for the final test on the validation set to ensure that the model did not overfit the final test data. Finally, we tested the model using the testing set. For each model and dataset pair, multiple cycles of the learning rate restart were performed during the training phase. The best-performing model, evaluated using the validation set data in each restart learning rate cycle, was retained as a weak model. Finally, every weak model and the strong model comprehending several of the best weak models were evaluated using the validation set and tested using the testing set. Figure 5 shows the overall experimental design.
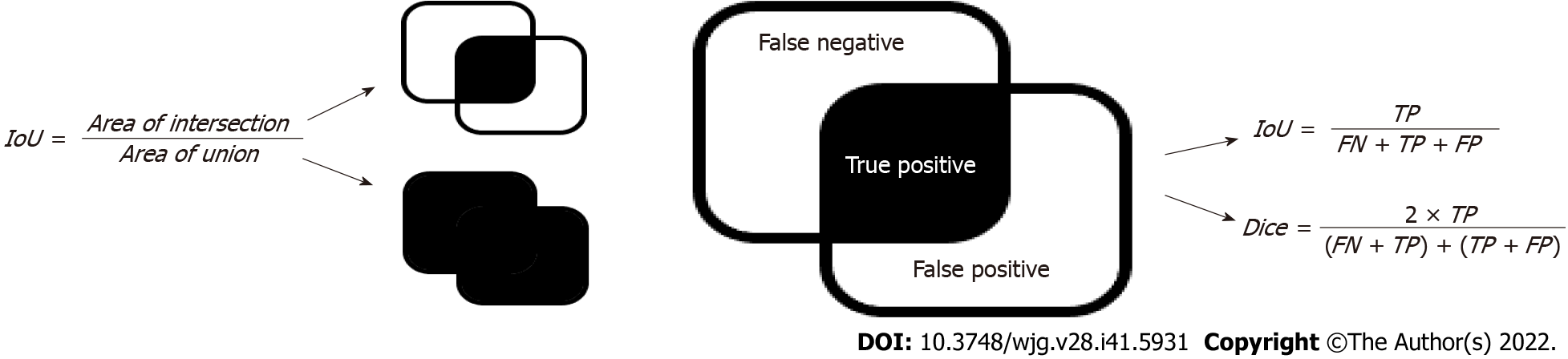
The intersection over union (IoU) and dice coefficients (Figure 6) are the most widely used metrics for semantic segmentation problems[29-31]. Both metrics measure the similarity between the sets of real and predicted regions. The calculation process is illustrated in Figure 6, where the area of intersection denotes the number of pixels in the intersection between the prediction area and ground truth, and the area of the union denotes that of the union. These two metrics were used to assess the performance of the segmentation models.
In summary, we performed two sets of comparison experiments using SOTA base models for the two types of datasets. In the first experiment, we compared the performance of the ensemble learning model with that of single models on a dataset with artifacts. In the second experiment, we compared the performance of the single models with that of the ensemble model on high-quality datasets without artifacts. Finally, we compared the differences between the improvements of the ensemble learning method for datasets with and without artifacts.
First, we compared the performance of the ensemble learning model with that of single models on CE images for all the five aforementioned base learners. A total of 41 images from the test dataset of GZ_Capcam were used for the final test. These test images were used only in the final testing phase to avoid data leakage and the consequent erroneous evaluation of the models. To illustrate that the ensemble learning model improves the performance of the single model on the artifact-infected dataset, we replicated all base learners mentioned in the previous section to illustrate the robustness of the conclusions in this study.
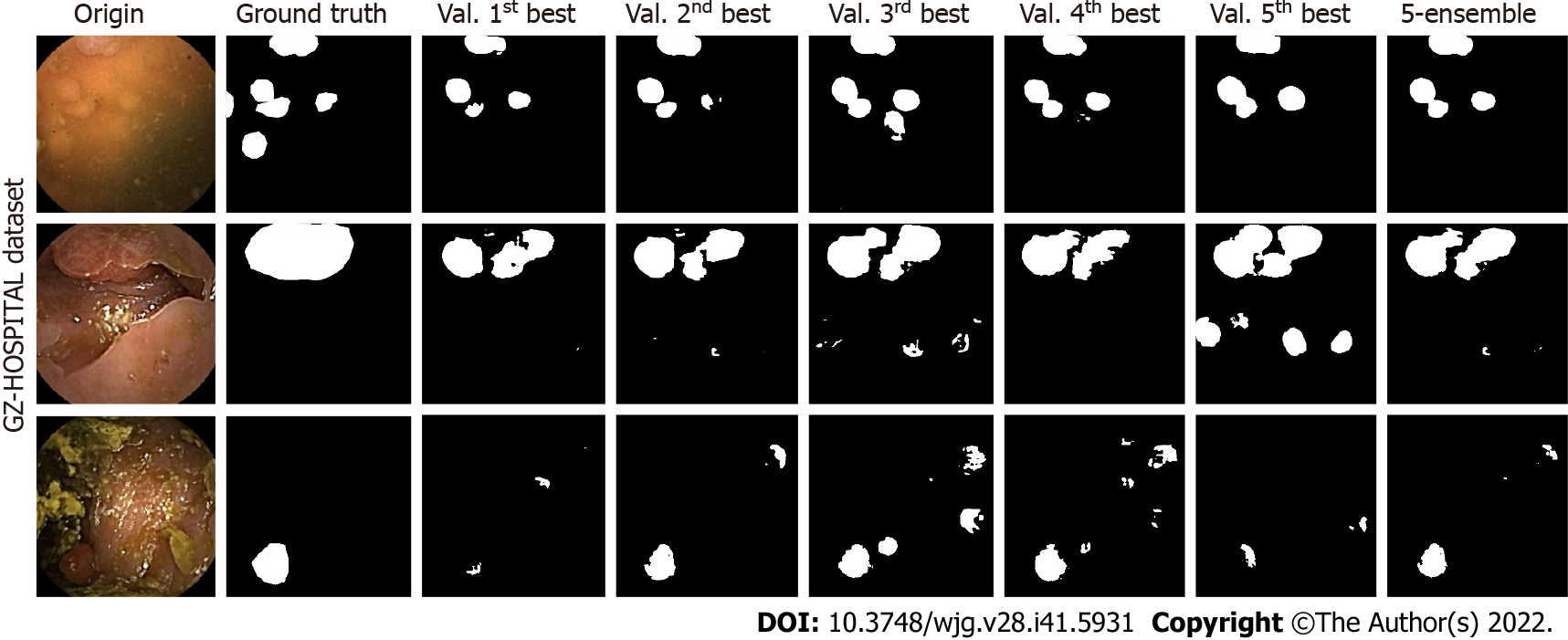
For U-Net, three test samples in the GZ-Hospital dataset were selected to compare the performances of the single and ensemble models (Figure 7). We can see that the semantic segmentation model was affected by different noises, such as stains, blurs, and light-dark variations in the sample images, leading to results that were not always clear. However, the performance of the ensemble model often met or exceeded the best results of a single model, indicating that a model constructed based on ensemble learning can effectively mitigate the effects of artifacts on the performance of the semantic segmentation model.
The results for the GZ_Capcam dataset are presented in Table 1, which includes images rich in artifacts. The IoU and dice metrics were calculated, as previously described. The performances of the single and ensemble models on the test set are presented in Supplementary Table 1. The results for all five basic learners on the CE dataset showed that the ensemble model outperformed the single models. Compared with single models, specifically, on the dataset with artifacts, the ensemble learning models with SegNet, U-Net, Attention-UNet, Resnet-UNet, and HarDMSEG as the base learners improved the detection by 0.08%, 7.01%, 3.88%, 5.13%, and 2.22%, respectively, using the IoU metric, and 1.71%, 4.93%, 1.40%, 2.86%, and 0.61%, respectively, using the dice metric. Overall, the ensemble model outperformed the single models. The performance of a truly single model, that is, a model obtained from a single training validation, was consistently worse than that of the ensemble model, as shown in the results for the weak models excluding the best one.
| Model | SegNet | U-Net | Attention-UNet | ResNet-UNet | HarDMSEG | |||||
| Metric | IoU | Dice | IoU | Dice | IoU | Dice | IoU | Dice | IoU | Dice |
| Snap_max | 0.341 | 0.454 | 0.379 | 0.507 | 0.361 | 0.502 | 0.414 | 0.522 | 0.538 | 0.649 |
| Ens_max | 0.341 | 0.462 | 0.406 | 0.532 | 0.375 | 0.509 | 0.435 | 0.537 | 0.550 | 0.653 |
| Improve | 0.000 | 0.008 | 0.027 | 0.025 | 0.014 | 0.007 | 0.021 | 0.015 | 0.012 | 0.004 |
| Improve (%) | 0.08 | 1.71 | 7.01 | 4.93 | 3.88 | 1.40 | 5.13 | 2.86 | 2.22 | 0.61 |
Similarly, we checked the performance of the single and ensemble models using standard datasets (Figure 8, Tables 2 and 3). The performances of the single and ensemble models on the test set are presented in Supplementary Tables 2 and 3. By comparing the results presented in Figures 7 and 8, we found that the ensemble learning method can improve the robustness of the semantic segmentation model when the dataset is affected by artifacts.
| Model | SegNet | UNet | Attention-UNet | ResNet-UNet | HarDMSEG | |||||
| IoU | Dice | IoU | Dice | IoU | Dice | IoU | Dice | IoU | Dice | |
| Snap_max | 0.700 | 0.780 | 0.713 | 0.788 | 0.754 | 0.830 | 0.747 | 0.819 | 0.840 | 0.901 |
| Ens_max | 0.702 | 0.779 | 0.711 | 0.783 | 0.752 | 0.829 | 0.750 | 0.816 | 0.840 | 0.901 |
| Improve | 0.002 | -0.001 | -0.002 | -0.005 | -0.002 | -0.002 | 0.003 | -0.002 | 0.000 | 0.000 |
| Improve (%) | 0.30 | -0.08 | -0.28 | -0.61 | -0.22 | -0.22 | 0.42 | -0.26 | 0.03 | -0.01 |
| Model | SegNet | UNet | Attention-UNet | ResNet-UNet | HarDMSEG | |||||
| IoU | Dice | IoU | Dice | IoU | Dice | IoU | Dice | IoU | Dice | |
| Snap_max | 0.814 | 0.890 | 0.816 | 0.884 | 0.836 | 0.900 | 0.823 | 0.884 | 0.845 | 0.893 |
| Ens_max | 0.815 | 0.890 | 0.826 | 0.891 | 0.838 | 0.898 | 0.824 | 0.884 | 0.844 | 0.892 |
| Improve | 0.001 | 0.001 | 0.010 | 0.007 | 0.002 | -0.001 | 0.001 | 0.000 | -0.001 | -0.001 |
| Improve (%) | 0.13 | 0.07 | 1.20 | 0.76 | 0.23 | -0.14 | 0.08 | 0.02 | -0.08 | -0.06 |
In the present study, we demonstrated that current computer-aided medical image analysis methods performed poorly in the presence of artifacts that were previously ignored. Nevertheless, almost every patient presents with insufficient intestinal cleansing. Thus, we used ensemble learning to improve the existing AI models and enhance their robustness in dealing with images with artifacts. Previous studies have extensively analyzed and concluded that integrated learning methods improve the robustness of medical image classification in a credible manner[32]. By improving the segmentation performance of the model, we can separate polyps more accurately from surrounding tissues, which can improve the detection probability of polyps and aid in monitoring the size of polyps in patients with unresectable polyps[33-36]. Semantic segmentation provides pixel-level classification and clearer polyp boundaries, which are also crucial in surgical procedures or radiofrequency ablation, and is expected to be used for real-time detection of polyp boundaries in surgical resection under gastroenteroscopy to assist polyp resection[33,36]. More in-depth studies have shown that the noise immunity of single models is weaker than that of integrated learning models[37], and clinical images, such as the CE images used in this study, are not always perfect in terms of image quality.
We used CE image datasets as samples, mainly because CE is an increasingly widely used and safe form of endoscopy but also has many artifacts[15,38]. The ensemble learning approach was tested for 15 pairs, consisting of three datasets and five SOTA segmentation models. The results showed that for CE images with various artifacts, ensemble learning improved the analytical performance of AI models. Herein, we demonstrated that ensemble learning can reduce the influence of artifacts on the semantic segmentation of CE images, which might also apply to other medical images.
In general, artifacts are prevalent in medical images and seriously challenge the performance of existing computer-aided diagnosis (CAD) models; therefore, this study discusses the enhancement of ensemble learning methods for CAD models to analyze images with artifacts, mainly using CE images as an example in the experiments. In addition, our experiments did not involve the injection of a priori knowledge of gastroenterology; in other words, the use of the ensemble learning approach mentioned in this paper does not imply any additional workload or workflow reordering. The only additional cost associated with the method is the computational resources. Additionally, from the perspective of DL, better model performance often relies on more model parameters and computational resources. Methods that already use DL models can easily apply ensemble learning methods to improve model performance without the need for additional workflow tuning. It is worth mentioning that, although the ensemble learning approach can improve the robustness of CAD image analysis models, misuse may lead to a less-than-expected improvement in the model’s effectiveness, mainly because the essence of ensemble learning is to reduce model variance, and when the variance of a single model is already very low, the improvement brought by ensemble learning may be very limited.
From the experimental results, although the ensemble learning approach improves the performance of the segmentation model on the dataset with artifacts, there are still false-positive and false-negative cases. On the one hand, the main reason for false-negative cases was that the model confuses normal-color polyps with normal gastrointestinal folds or confuses abnormal-color polyps with artifacts, such as yellow bubbles. However, the main cause of false-positive cases was that some artifacts or normal folds had a high similarity with polyps in the image, which led the model to misidentify them as polyps. Overall, the main reason for segmentation errors is that the color and texture are highly confusing, and we will further attempt to improve the ability of the model to distinguish polyps, normal tissues, and artifacts in a subsequent study.
In clinical practice, video frames can be completely infested with artifacts, making the content of the image simply unrecognizable. Therefore, the appearance of these frames is inevitable in clinical practice. In the present study, we confirmed the authenticity of polyps in pictures with artifacts by using more images, videos, and other inspection methods. Thus, we solved the dilemma of applying AI to these medical images. However, our study has some limitations. For example, the images were insufficient and did not involve lesions other than polyps.
We believe that the direction of feature AI for CE imaging research lies in making existing computer models better serve clinical diagnosis in a practical sense rather than letting these methods stay in the laboratory. CE is commonly used to examine digestive diseases. In addition to polyps, many digestive diseases can be detected using CE. Thus, AI for CE imaging can be considered to enrich the diagnosis, localization, and grading of more forms of the disease, such as ulcers and erosions, to assist doctors in more refined disease research and diagnosis. In the future, we will validate the ensemble learning method in clinical practice to demonstrate that it can improve the detection rate of polyps in CE in the clinic and evaluate the potential of this method for other types of medical images or lesions[39].
Ensemble learning can improve the semantic segmentation performance of AI models on CE images with artifacts.
Artificial intelligence (AI)-assisted capsule endoscopy (CE) can improve the detection rate of gastrointestinal polyps and reduce the incidence of gastrointestinal cancer.
Most previous studies ignored the serious impact of the existence of a large number of artifacts in the real world on the detection ability of existing AI models for polyps in CE images.
In this study, semantic segmentation and ensemble learning methods were combined to analyze polyp images of CE with artifacts, proving that ensemble learning methods can better solve the impact of artifacts in the real world.
This study retrospectively analyzed CE images of patients at our research center from January 2016 to December 2019. Polyp images with artifacts were selected and randomly divided into a training set (195 images), a validation set (41 images), and a test set (41 images). Further validation was performed on two public datasets with good background quality.
Compared with the corresponding optimal base model, intersection over union and dice are improved by 0.08%-7.01% and 0.61%-4.93%, respectively. For public datasets with good background quality, the segmentation performance of most ensemble learning models was better than that of a single model.
The ensemble learning method can improve the performance of semantic segmentation of polyps in CE images with artifacts.
We will validate other digestive tract lesions and other medical images and perform real-time detection during endoscopic and surgical procedures.
The authors would like to acknowledge Ai-Hua Lin for assistance with statistical analysis.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Lee YJ, South Korea; Naganuma H, Japan S-Editor: Wang JJ L-Editor: Wang TQ P-Editor: Wang JJ
| 1. | Xia C, Dong X, Li H, Cao M, Sun D, He S, Yang F, Yan X, Zhang S, Li N, Chen W. Cancer statistics in China and United States, 2022: profiles, trends, and determinants. Chin Med J (Engl). 2022;135:584-590. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1790] [Cited by in RCA: 2164] [Article Influence: 721.3] [Reference Citation Analysis (1)] |
| 2. | Nguyen LH, Goel A, Chung DC. Pathways of Colorectal Carcinogenesis. Gastroenterology. 2020;158:291-302. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 344] [Cited by in RCA: 303] [Article Influence: 60.6] [Reference Citation Analysis (0)] |
| 3. | Zhou JX, Li JY, Liu KJ, Liang YX, Wang H. Study on the promotion of lipopolysaccharide from shewanella algae on the occurrence and development of colorectal adenoma. Gut. 2021;70:A60-A63. [DOI] [Full Text] |
| 4. | Corley DA, Jensen CD, Marks AR, Zhao WK, Lee JK, Doubeni CA, Zauber AG, de Boer J, Fireman BH, Schottinger JE, Quinn VP, Ghai NR, Levin TR, Quesenberry CP. Adenoma detection rate and risk of colorectal cancer and death. N Engl J Med. 2014;370:1298-1306. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1251] [Cited by in RCA: 1550] [Article Influence: 140.9] [Reference Citation Analysis (0)] |
| 5. | Abdelrahim M, Saiga H, Maeda N, Hossain E, Ikeda H, Bhandari P. Automated sizing of colorectal polyps using computer vision. Gut. 2022;71:7-9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 16] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 6. | Wang WJ, Zhou YL, He J, Feng ZQ, Zhang L, Lai XB, Zhou JX, Wang H. Characterizing the composition of intestinal microflora by 16S rRNA gene sequencing. World J Gastroenterol. 2020;26:614-626. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 11] [Cited by in RCA: 17] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 7. | Pennazio M. Enteroscopy and capsule endoscopy. Endoscopy. 2006;38:1079-1086. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 6] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 8. | Ding Z, Shi H, Zhang H, Meng L, Fan M, Han C, Zhang K, Ming F, Xie X, Liu H, Liu J, Lin R, Hou X. Gastroenterologist-Level Identification of Small-Bowel Diseases and Normal Variants by Capsule Endoscopy Using a Deep-Learning Model. Gastroenterology. 2019;157:1044-1054.e5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 248] [Cited by in RCA: 206] [Article Influence: 34.3] [Reference Citation Analysis (0)] |
| 9. | Ertem FU, Hartman D, Dubner HM. Unexpected Cause of Obscure Gastrointestinal Bleeding: Primary Small-Intestinal Angiosarcoma. Am J Gastroenterol. 2021;116:1579. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 10. | Trasolini R, Byrne MF. Artificial intelligence and deep learning for small bowel capsule endoscopy. Dig Endosc. 2021;33:290-297. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 22] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 11. | Melson J, Trikudanathan G, Abu Dayyeh BK, Bhutani MS, Chandrasekhara V, Jirapinyo P, Krishnan K, Kumta NA, Pannala R, Parsi MA, Sethi A, Trindade AJ, Watson RR, Maple JT, Lichtenstein DR. Video capsule endoscopy. Gastrointest Endosc. 2021;93:784-796. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 23] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 12. | Hassan C, Wallace MB, Sharma P, Maselli R, Craviotto V, Spadaccini M, Repici A. New artificial intelligence system: first validation study versus experienced endoscopists for colorectal polyp detection. Gut. 2020;69:799-800. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 147] [Cited by in RCA: 122] [Article Influence: 24.4] [Reference Citation Analysis (0)] |
| 13. | Van Gossum A, Munoz-Navas M, Fernandez-Urien I, Carretero C, Gay G, Delvaux M, Lapalus MG, Ponchon T, Neuhaus H, Philipper M, Costamagna G, Riccioni ME, Spada C, Petruzziello L, Fraser C, Postgate A, Fitzpatrick A, Hagenmuller F, Keuchel M, Schoofs N, Devière J. Capsule endoscopy versus colonoscopy for the detection of polyps and cancer. N Engl J Med. 2009;361:264-270. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 267] [Cited by in RCA: 224] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 14. | Namikawa K, Hirasawa T, Yoshio T, Fujisaki J, Ozawa T, Ishihara S, Aoki T, Yamada A, Koike K, Suzuki H, Tada T. Utilizing artificial intelligence in endoscopy: a clinician's guide. Expert Rev Gastroenterol Hepatol. 2020;14:689-706. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 17] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 15. | Pennazio M, Rondonotti E, Koulaouzidis A. Small Bowel Capsule Endoscopy: Normal Findings and Normal Variants of the Small Bowel. Gastrointest Endosc Clin N Am. 2017;27:29-50. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 16. | Bernal J, Sánchez J, Vilario F. Towards automatic polyp detection with a polyp appearance model. Pattern Recognit. 2012;45:3166-3182. [DOI] [Full Text] |
| 17. | Bernal J, Sánchez FJ, Fernández-Esparrach G, Gil D, Rodríguez C, Vilariño F. WM-DOVA maps for accurate polyp highlighting in colonoscopy: Validation vs. saliency maps from physicians. Comput Med Imaging Graph. 2015;43:99-111. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 336] [Cited by in RCA: 442] [Article Influence: 44.2] [Reference Citation Analysis (0)] |
| 18. | Klang E, Grinman A, Soffer S, Margalit Yehuda R, Barzilay O, Amitai MM, Konen E, Ben-Horin S, Eliakim R, Barash Y, Kopylov U. Automated Detection of Crohn's Disease Intestinal Strictures on Capsule Endoscopy Images Using Deep Neural Networks. J Crohns Colitis. 2021;15:749-756. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 56] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 19. | Cai L, Xu X, Zhang L, Foo CS. Exploring Spatial Diversity for Region-Based Active Learning. IEEE Trans Image Process. 2021;30:8702-8712. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 20. | Badrinarayanan V, Kendall A, Cipolla R. SegNet: A Deep Convolutional Encoder-Decoder Architecture for Image Segmentation. IEEE Trans Pattern Anal Mach Intell. 2017;39:2481-2495. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8315] [Cited by in RCA: 3600] [Article Influence: 450.0] [Reference Citation Analysis (0)] |
| 21. | Falk T, Mai D, Bensch R, Çiçek Ö, Abdulkadir A, Marrakchi Y, Böhm A, Deubner J, Jäckel Z, Seiwald K, Dovzhenko A, Tietz O, Dal Bosco C, Walsh S, Saltukoglu D, Tay TL, Prinz M, Palme K, Simons M, Diester I, Brox T, Ronneberger O. U-Net: deep learning for cell counting, detection, and morphometry. Nat Methods. 2019;16:67-70. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 801] [Cited by in RCA: 853] [Article Influence: 121.9] [Reference Citation Analysis (0)] |
| 22. | Lin H, Li Z, Yang Z, Wang Y. Variance-aware attention U-Net for multi-organ segmentation. Med Phys. 2021;48:7864-7876. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 19] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 23. | de Groof AJ, Struyvenberg MR, van der Putten J, van der Sommen F, Fockens KN, Curvers WL, Zinger S, Pouw RE, Coron E, Baldaque-Silva F, Pech O, Weusten B, Meining A, Neuhaus H, Bisschops R, Dent J, Schoon EJ, de With PH, Bergman JJ. Deep-Learning System Detects Neoplasia in Patients With Barrett's Esophagus With Higher Accuracy Than Endoscopists in a Multistep Training and Validation Study With Benchmarking. Gastroenterology. 2020;158:915-929.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 172] [Cited by in RCA: 220] [Article Influence: 44.0] [Reference Citation Analysis (0)] |
| 24. | Huang CH, Wu HY, Lin YL. HarDNet-MSEG: A Simple Encoder-Decoder Polyp Segmentation Neural Network that Achieves over 0.9 Mean Dice and 86 FPS. 2021 Preprint. Available from: arXiv:2101.07172. [DOI] [Full Text] |
| 25. | Liew WS, Tang TB, Lin CH, Lu CK. Automatic colonic polyp detection using integration of modified deep residual convolutional neural network and ensemble learning approaches. Comput Methods Programs Biomed. 2021;206:106114. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 27] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 26. | Paul A, Tang YX, Shen TC, Summers RM. Discriminative ensemble learning for few-shot chest x-ray diagnosis. Med Image Anal. 2021;68:101911. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 26] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 27. | Jiang Z, Xiao SR, Liu R. Dissecting and predicting different types of binding sites in nucleic acids based on structural information. Brief Bioinform. 2022;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 28. | Gao H, Li Y, Pleiss G, Zhuang L, Weinberger KQ. Snapshot Ensembles: Train 1, get M for free. ICLR 2017: Proceedings of the 5th International Conference on Learning Representations; 2017 Apr 24-26; Toulon, France. |
| 29. | Li L, Wei M, Liu B, Atchaneeyasakul K, Zhou F, Pan Z, Kumar SA, Zhang JY, Pu Y, Liebeskind DS, Scalzo F. Deep Learning for Hemorrhagic Lesion Detection and Segmentation on Brain CT Images. IEEE J Biomed Health Inform. 2021;25:1646-1659. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 56] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 30. | Li CC, Wu MY, Sun YC, Chen HH, Wu HM, Fang ST, Chung WY, Guo WY, Lu HH. Ensemble classification and segmentation for intracranial metastatic tumors on MRI images based on 2D U-nets. Sci Rep. 2021;11:20634. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 4] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 31. | Grøvik E, Yi D, Iv M, Tong E, Nilsen LB, Latysheva A, Saxhaug C, Jacobsen KD, Helland Å, Emblem KE, Rubin DL, Zaharchuk G. Handling missing MRI sequences in deep learning segmentation of brain metastases: a multicenter study. NPJ Digit Med. 2021;4:33. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 28] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 32. | Müller D, Soto-Rey I, Kramer F. An Analysis on Ensemble Learning optimized Medical Image Classification with Deep Convolutional Neural Networks. 2022 Preprint. Available from: arXiv:2201.11440. [DOI] [Full Text] |
| 33. | Jha D, Ali S, Tomar NK, Johansen HD, Johansen D, Rittscher J, Riegler MA, Halvorsen P. Real-Time Polyp Detection, Localization and Segmentation in Colonoscopy Using Deep Learning. IEEE Access. 2021;9:40496-40510. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 164] [Cited by in RCA: 91] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 34. | Mahmud T, Paul B, Fattah SA. PolypSegNet: A modified encoder-decoder architecture for automated polyp segmentation from colonoscopy images. Comput Biol Med. 2021;128:104119. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 31] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 35. | Yao Y, Gou S, Tian R, Zhang X, He S. Automated Classification and Segmentation in Colorectal Images Based on Self-Paced Transfer Network. Biomed Res Int. 2021;2021:6683931. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 8] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 36. | Shi L, Wang Y, Li Z, Qiumiao W. FRCNet: Feature Refining and Context-Guided Network for Efficient Polyp Segmentation. Front Bioeng Biotechnol. 2022;10:799541. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 2] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 37. | Strauss T, Hanselmann M, Junginger A, Ulmer H. Ensemble Methods as a Defense to Adversarial Perturbations Against Deep Neural Networks. 2017 Preprint. Available from: arXiv:1709.03423. [DOI] [Full Text] |
| 38. | Brooker J, Smith AC. Capsule endoscopy artifact: the afterimage. Gastrointest Endosc. 2007;66:820; discussion 821. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 39. | Huang TY, Zhan SQ, Chen PJ, Yang CW, Lu HH. Accurate diagnosis of endoscopic mucosal healing in ulcerative colitis using deep learning and machine learning. J Chin Med Assoc. 2021;84:678-681. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 14] [Article Influence: 3.5] [Reference Citation Analysis (0)] |