Published online Oct 14, 2022. doi: 10.3748/wjg.v28.i38.5530
Peer-review started: July 1, 2022
First decision: July 13, 2022
Revised: August 12, 2022
Accepted: September 22, 2022
Article in press: September 22, 2022
Published online: October 14, 2022
Processing time: 103 Days and 0.8 Hours
Artificial intelligence (AI), especially deep learning, is gaining extensive attention for its excellent performance in medical image analysis. It can automatically make a quantitative assessment of complex medical images and help doctors to make more accurate diagnoses. In recent years, AI based on ultrasound has been shown to be very helpful in diffuse liver diseases and focal liver lesions, such as analyzing the severity of nonalcoholic fatty liver and the stage of liver fibrosis, identifying benign and malignant liver lesions, predicting the microvascular invasion of hepatocellular carcinoma, curative transarterial chemoembolization effect, and prognoses after thermal ablation. Moreover, AI based on endoscopic ultrasonography has been applied in some gastrointestinal diseases, such as distinguishing gastric mesenchymal tumors, detection of pancreatic cancer and intraductal papillary mucinous neoplasms, and predicting the preoperative tumor deposits in rectal cancer. This review focused on the basic technical knowledge about AI and the clinical application of AI in ultrasound of liver and gastroenterology diseases. Lastly, we discuss the challenges and future perspectives of AI.
Core Tip: Artificial intelligence (AI) based on ultrasound has been confirmed to be helpful in diagnosing diffuse liver diseases and focal liver lesions, such as analyzing the severity of nonalcoholic fatty liver and the stage of liver fibrosis, identifying benign and malignant liver lesions, predicting microvascular invasion of hepatocellular carcinoma, curative transarterial chemoembolization effect, and prognoses after thermal ablation. AI based on endoscopic ultrasonography has been applied in some gastrointestinal diseases. We focused on basic technical knowledge about AI and the aforementioned clinical application in the ultrasound of liver and gastroenterology. Additionally, we discuss the challenges and future perspectives of AI.
- Citation: Liu JQ, Ren JY, Xu XL, Xiong LY, Peng YX, Pan XF, Dietrich CF, Cui XW. Ultrasound-based artificial intelligence in gastroenterology and hepatology. World J Gastroenterol 2022; 28(38): 5530-5546
- URL: https://www.wjgnet.com/1007-9327/full/v28/i38/5530.htm
- DOI: https://dx.doi.org/10.3748/wjg.v28.i38.5530
Liver disease causes two million deaths per year in the world among which cirrhosis is the 11th leading cause of death in the world and liver cancer is the 16th leading cause of death[1]. The prevalence of nonalcoholic fatty liver disease (NAFLD) is 25.0% and is estimated to be 33.5% by 2030[2]. Gast
In clinical practice, many imaging techniques such as X-ray, computed tomography (CT), magnetic resonance imaging (MRI), and ultrasound have played a vital role in the detection and treatment of diseases[6]. Ultrasound, a noninvasive and real-time diagnostic technique, is the most commonly used method for detecting and diagnosing human digestive diseases[7]. However, the interpretation and analysis of ultrasound images depend deeply on the subjective judgment and experience of human experts. Radiologists may make mistakes due to exhaustion when dealing with a large number of images[8].
Artificial intelligence (AI) is defined as computer algorithms created by humans and improved with analogs of the thoughts, judgments, and reactions that take place in the human brain. In recent years, radiologists have increasingly embraced the aid of AI-powered diagnoses. AI can make a quantitative analysis by recognizing the information of images automatically and is widely applied in the medical images of ultrasound in diffuse liver diseases, focal liver lesions, PC, and colorectal cancer. In this review, we described the development of AI-based ultrasound in the aforementioned applications. In addition, we also discussed the future opportunities and challenges of AI-based ultrasound.
Currently, the algorithms of AI used in medical images mainly include traditional machine learning algorithms and deep learning.
Machine learning is described as a kind of data science that offers computers with the capacity to study without being programmed with specific rules[9]. It focuses on computer algorithms that are studied from the training model and give predictions on another model[10]. Machine learning depends primarily on the predefined characteristics that display the regular patterns inherent in models acquired from regions of interest with explicit parameters on the basis of expert experience. Then, other medical image features, such as various mass shape, size, and echo, can be quantified.
Radiomics, which belongs to traditional machine learning, is a popular field of study related to the acquisition and assessment of patterns within medical images, including CT, MRI, and ultrasound. These patterns include complicated patterns that are difficult to recognize or analyze by the human eye[11].
Deep learning is at the leading edge of AI and is developing rapidly. Deep learning is described as a group of artificial neural network (ANN) algorithms, which include many hidden layers. Namely, deep learning depends on a subset of algorithms that try to model high-level abstractions[12].
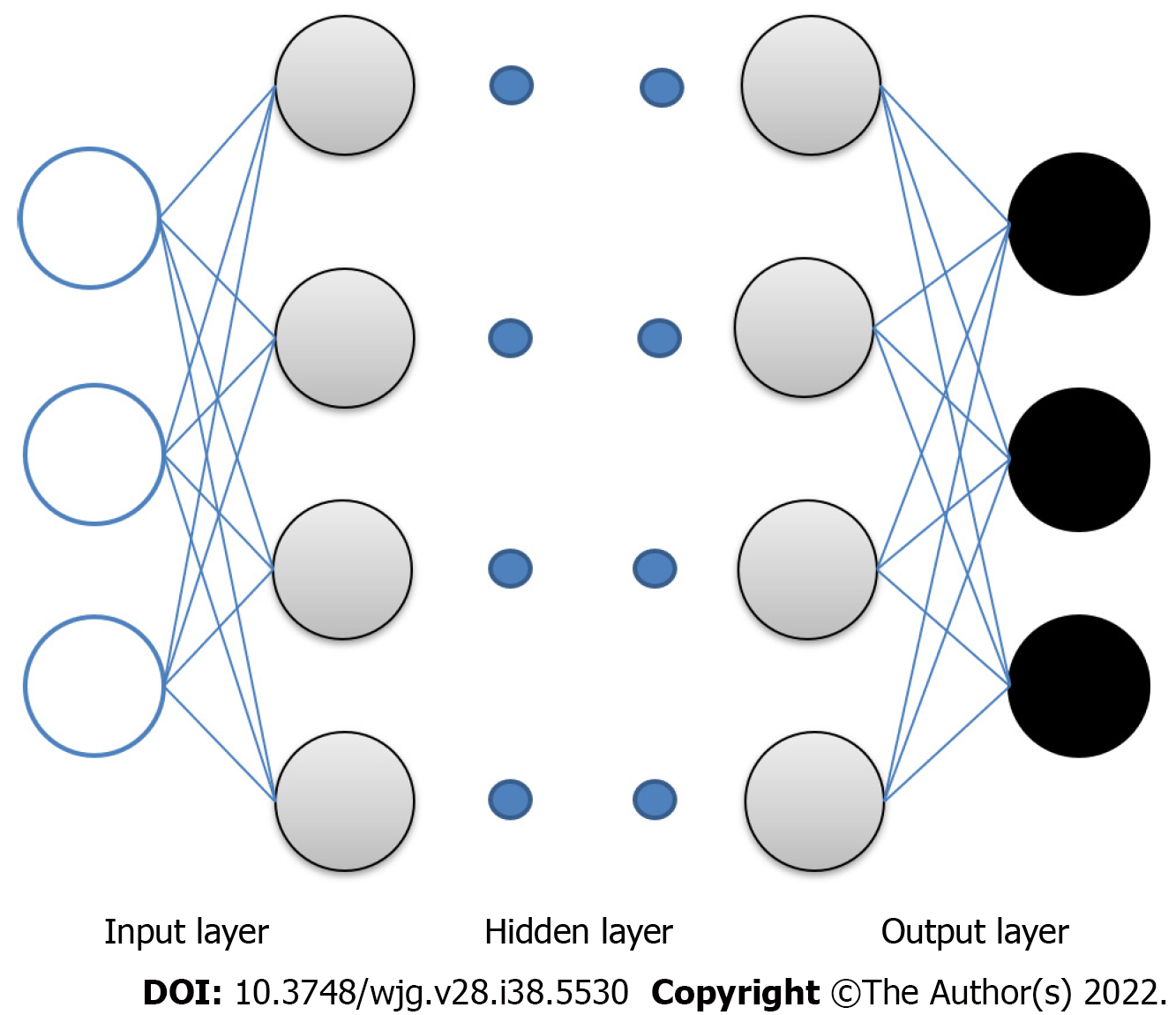
Recently, convolutional neural networks (CNNs) are the preferred type of deep learning architecture in the assessment of medical images[13]. CNNs consist of an input layer, multiple hidden layers, and an output layer (Figure 1). The hidden layers include convolutional layers, pooling layers, connected layers, and normalization layers. Convolutional layers and pooling layers can complete feature extraction and aggregation[9].
Diffuse liver diseases display a failure in the metabolic and synthesis processes of the liver[14]. Liver biopsy is the gold standard for the diagnosis of fibrosis and NAFLD. However, liver biopsy is an invasive process that has many complications such as hemorrhage, biliary peritonitis, and pneumothorax[15]. In addition, liver biopsy is not feasible for the long-term management of patients with chronic liver diseases. Noninvasive liver imaging methods such as CT, MRI, and ultrasound have been extensively studied. Ultrasound is one of most common methods to diagnose liver diseases due to its noninvasiveness, inexpensive price, and real-time ability. Machine learning algorithms based on ultrasound have been applied for analysis of steatosis and the staging of liver fibrosis. Table 1 shows the application of ultrasound-based AI in diffuse liver disease.
| Ref. | Diseases: number of cases | Type of ultrasound | Algorithm of AI | Performance |
| Byra et al[21] | Severely obese patients: 55 | B-mode | CNN | Sensitivity: 100% |
| Specificity: 88% | ||||
| Accuracy: 96% | ||||
| AUC: 0.98 | ||||
| Fatty liver disease: 38 | ||||
| Biswas et al[22] | Normal patients: 27 | B-mode | Deep learning | Accuracy: 100% |
| Fatty liver disease: 36 | AUC: 1.0 | |||
| Han et al[24] | NAFLD: 140 | B-mode | CNN | Sensitivity: 97% |
| Specificity: 94% | ||||
| Accuracy: 96% | ||||
| Control: 64 | ||||
| AUC: 0.98 | ||||
| Yeh et al[28] | Postsurgical human liver samples: 20 | B-mode | SVM | F2 accuracy: 91% |
| F3 accuracy: 85% | ||||
| F4 accuracy: 81% | ||||
| F6 accuracy: 72% | ||||
| Zhang et al[29] | Liver fibrosis or cirrhosis: 239 | Duplex | ANN | Sensitivity: 95% |
| Specificity: 85% | ||||
| Training group: 179 | ||||
| Validation group: 60 | Accuracy: 88% | |||
| Gao et al[30] | S0: 4 | B-mode | ANN | S0 accuracy: 100% |
| S1: 16 | S1 accuracy: 90% | |||
| S2 accuracy: 70% | ||||
| S3 accuracy: 90% | ||||
| S2: 8 | S4 accuracy: 100% | |||
| S3: 5 | ||||
| S4: 4 | ||||
| Lee et al[31] | Patients: 3446 | B-mode | CNN | AUC: 0.86 |
| Internal validation set: 263 | ||||
| Internal test set: 266 | ||||
| External test set: 572 | ||||
| Gatos et al[34,35] | Chronic liver disease: 70 | Shear-wave elastography | SVM | Sensitivity: 94% |
| Healthy: 56 | Specificity: 81% | |||
| Accuracy: 87% | ||||
| Wang et al[36] | Liver fibrosis: 398 | Shear-wave elastography | Deep learning radiomic | F4 AUC: 0.97 |
| Training group: 266 | ||||
| Validation group: 132 | F3 AUC: 0.98 | |||
| F2 AUC: 0.85 | ||||
| Xue et al[38] | Liver fibrosis: 401 | Elastography | CNN by TL radiomics | S2 AUC: 0.95 |
| S3 AUC: 0.93 | ||||
| Patient without fibrosis: 65 | ||||
| S4 AUC: 0.93 |
Fatty liver diseases: An excess amount of fat in the liver cells is found in fatty liver diseases (FLD). The main causes of FLD include obesity, alcoholism, diabetes, nonalcoholic steatohepatitis, drugs, and toxins[16,17]. FLD is related to the growing risk of cirrhosis and liver cancer. The most common cause of FLD is NAFLD, which ranges in prevalence from 25% to 45%[18]. Several noninvasive imaging methods such as CT, MRI, and ultrasound can diagnose NAFLD[19]. Ultrasound is the cheapest diagnostic method with 93% sensitivity, while hepatic steatosis is greater than 33%[18].
Conventional ultrasound is commonly used for NAFLD evaluation, but its qualitative nature, doctor dependency, and unsatisfactory accuracy limits the application. Moreover, the ultrasound images of fatty liver and early cirrhosis have many common features, making it hard to distinguish the two diseases by the human eye[20].
In recent years, ultrasound-based AI has demonstrated high accuracy for detection of steatosis and represents excellent reproducibility and reliability.
Byra et al[21] created a CNN model to acquire features from B-mode ultrasound image. It was reported that they could assess the amount of steatosis present in the liver with the area under the receiver operating characteristic curve (AUC) of 0.98, and their approach may assist the doctors in automatically assessing the amount of fat in the liver clinically[21].
Biswas et al[22] revealed that a deep learning-based algorithm reached a superior performance for FLD identification and risk stratification with 100% accuracy and AUC of 1.0 when compared with a conventional machine learning system support vector machine (SVM) (accuracy: 82%, AUC: 0.79) and extreme learning machine (accuracy: 92%, AUC: 0.92).
Deep learning has also been applied to quantitatively evaluate NAFLD. The radiofrequency data of ultrasound displays much more information of hepatic microstructure than that of gray-scale B-mode images[23]. Han et al[24] developed a deep learning algorithm that used radiofrequency data for NAFLD assessment. The results revealed that the sensitivity, specificity, and positive predictive value (PPV) for NAFLD diagnosis were 97%, 94%, and 97%, respectively. They confirmed that the quantitative analysis of raw radiofrequency ultrasound signals showed the potential of identifying NAFLD and quantifying hepatic fat fraction[24].
Liver fibrosis and cirrhosis: Patients with chronic liver disease may have no clinical symptoms for an extended period, or it may develop to fibrosis and cirrhosis[25]. The activation of the resting hepatic stellate cell into an activated myofibroblast plays an important role in the progression of liver fibrosis. The activated myofibroblast expresses abundant a-smooth muscle actin and collagen[26].
Cirrhosis, which consists of various nodules and is harder than the normal liver, is the advanced period of fibrosis[27]. Liver fibrosis and early cirrhosis are confirmed to be partly reversible. Therefore, the precise diagnosis of liver fibrosis is vital for the treatment and management of chronic liver disease patients.
In clinical practice, liver biopsy is the gold standard for the diagnosis of liver fibrosis. Various noninvasive modalities such as ultrasound and elastography have been used as alternatives to liver biopsy. Some studies suggest that AI models based on ultrasound and elastography have great potential for the classification of liver fibrosis.
AI based on conventional ultrasound: AI based on conventional ultrasound has been applied to improve their performance for the diagnosis and grading of liver fibrosis.
Yeh et al[28] built an SVM model to analyze liver fibrosis. B-mode images of 20 fresh postsurgical human livers were used to assess ultrasound capacity in evaluating the stage of fibrosis. The study indicated the best classification accuracy of two, three, four, and six classes were 91%, 85%, 81%, and 72%, respectively[28]. The results confirmed that the SVM model may be suggested to assess diverse liver fibrosis stage.
Other than the B-mode ultrasound, duplex ultrasound has also been applied to diagnose liver fibrosis. Using an ANN model based on duplex ultrasound, Zhang et al[29] demonstrated that their model reached the accuracy, sensitivity, and specificity were 88.3%, 95.0%, and 85.0%, respectively. The ANN model included five ultrasonographic parameters: thickness of spleen, liver vein waveform, the hepatic parenchyma, liver artery pulsatile index, and hepatic damping index. The study suggested that their ANN model has the potential to diagnose liver fibrosis noninvasively[29].
Studies confirmed that radiomics show great performance in the grading of liver fibrosis. By the use of texture analysis to analyze ultrasound liver images, the study found the accuracies of S0-S4 were 100%, 90%, 70%, 90%, and 100%, respectively[30].
It was reported that deep learning has great potential for liver fibrosis evaluation. Lee et al[31] built a deep CNN and trained a four-class model (F0 vs F1 vs F23 vs F4) to predict METAVIR scores. They used 13608 ultrasound images of 3446 patients who accepted surgery, liver biopsy, or transient elastography to train the deep CNN model. The model achieved a higher AUC of 0.857 for the classification of cirrhosis compared with five radiologists (AUC range, 0.656-0.816; P < 0.05) using the external test set[31].
AI based on ultrasound elastography: ultrasound elastography has been performed to acquire quantitative assessment of liver tissue stiffness, which is related to the grades of fibrosis. These technologies include strain elastography and shear wave elastography (SWE)[32]. Recently, some studies confirmed that the AI based on SWE has great value to identify and stage liver fibrosis.
Compared to conventional radiomics, a multiparametric ultrasonic model using machine learning algorithms demonstrated better manifestation in fibrosis assessment[33]. By quantifying color information from SWE images, Gatos et al[34,35] created an SVM model that could differentiate patients with liver diseases from controls with accuracy, sensitivity, and specificity of 87.3%, 93.5% and 81.2%, respectively.
Deep learning has also been applied in the assessment of liver fibrosis. A multicenter study used deep learning radiomics on 2D-SWE ultrasound images for the classification of liver fibrosis[36]. 2D-SWE ultrasound images had higher AUCs of 0.97 for F4, 0.98 for ≥ F3, and 0.85 for ≥ F2 fibrosis when compared with standard 2D-SWE.
It is necessary to contain a large training dataset for deep learning. However, it is difficult and expensive to get abundant medical images in clinics. One method to solve this problem is the employment of transfer learning (TL), which can enhance the performance by TL from other areas to the ultrasound area[37]. A study developed a CNN model by TL radiomics to assess ultrasound images of gray-scale modality and elastogram modality for the grade of accurate liver fibrosis. TL in gray-scale modality and elastogram modality revealed much higher diagnostic accuracy of AUCs compared with non-TL. Multimodal gray-scale modality + elastogram modality was confirmed to be the most precise diagnostic model with AUCs of 0.930, 0.932, and 0.950 for classifying ≥ S2, ≥ S3, and S4, respectively. It was suggested that this TL model had excellent performance in liver fibrosis staging in clinical applications[38].
Focal liver lesions (FLLs) are described as an abnormal part of the liver mainly coming from hepatocytes, biliary epithelium, and mesenchymal tissue[39]. Due to its cheap price, noninvasiveness, and real-time imaging, ultrasound is the preferred method for the diagnosis of FLLs. Based on this trend, the AI models using ultrasound images have more advantages over CT and MRI in routine clinical applications[40]. Table 2 shows the application of ultrasound-based AI in FLLs.
| Ref. | Diseases: number of cases | Type of ultrasound | Algorithm of AI | Performance |
| Xi et al[42] | Benign lesions: 300 | B-mode | CNN | All lesions |
| Accuracy: 84% | ||||
| Uncertain set of lesions | ||||
| Malignant lesions: 296 | Accuracy: 79% | |||
| Yang et al[43] | Benign tumor: 427 | B-mode | CNN | AUC for EV: 0.924 |
| Sensitivity: 86.5% | ||||
| Malignant tumor: 1786 | ||||
| Specificity: 85.5% | ||||
| Virmani et al[44] | HCC: 27 | B-mode | SVM | Accuracy of HCC: 91.6% |
| Sensitivity | ||||
| Metastatic liver tumor: 24 | HCC: 90% | |||
| Metastatic liver tumor: 93.3% | ||||
| Hwang et al[49] | Cyst: 29 | B-mode | ANN | Accuracy: 96% |
| Cyst vs hemangioma | ||||
| Cyst vs malignant | ||||
| Hemangioma: 37 | ||||
| Hemangioma vs malignant | ||||
| Malignant: 33 | ||||
| Schmauch et al[50] | Non-tumorous liver: 258 | B-mode | CNN | AUC |
| Hemangioma: 17 | FLL detection: 0.935 | |||
| Metastasis: 48 | ||||
| HCC: 6 | ||||
| FLL discrimination: 0.916 | ||||
| Cyst: 30 | ||||
| FNH: 8 | ||||
| Tiyarattanachai et al[51] | HCC: 2414 | B-mode | CNN | Detection rate: 87.0% |
| Cyst: 6600 | Sensitivity: 83.9% | |||
| Hemangioma: 5374 | ||||
| Specificity: 97.1% | ||||
| Focal fatty sparing: 5110 | ||||
| Focal fatty infiltration: 934 | ||||
| Gatos et al[47] | Benign FLL: 30 | CEUS | SVM | Accuracy: 90.3% |
| Sensitivity: 93.1% | ||||
| Malignant FLL: 22 | Specificity: 86.9% | |||
| Kondo et al[46] | Benign FLL: 31 | CEUS | SVM | Benign vs malignant |
| Accuracy: 91.8% | ||||
| Sensitivity: 94% | ||||
| Specificity: 87.1% | ||||
| Accuracy | ||||
| Malignant FLL: 67 | ||||
| Benign: 84.4% | ||||
| HCC: 87.7% | ||||
| Metastatic liver tumor: 85.7% | ||||
| Guo et al[48] | Benign FLL: 46 | CEUS | Deep canonical correlation analysis and multiple kernel learning | Accuracy: 90.4% |
| Sensitivity: 93.6% | ||||
| Malignant FLL: 47 | Specificity: 86.8% | |||
| Streba et al[52] | HCC: 41 | CEUS | ANN | Training accuracy: 94.5% |
| Hypervascular liver metastasis: 20 | Testing accuracy: 87.1% | |||
| Hypovascular liver metastasis: 12 | Sensitivity: 93.2% | |||
| Specificity: 89.7% | ||||
| Hemangioma: 16 | ||||
| Focal fatty changes: 23 | ||||
| Căleanu et al[53] | HCC: 30 | CEUS | Deep neural network | Accuracy: 88% |
| Hypervascular liver metastasis: 11 | ||||
| Hypovascular liver metastasis: 11 | ||||
| Hemangioma: 23 | ||||
| FNH: 16 | ||||
| Dong et al[56] | HCC: 322 | B-mode | Radiomics | AUC: 0.81 |
| Hu et al[57] | HCC: 482 | CEUS | Radiomics | AUC: 0.731 |
| Training cohort: 341 | ||||
| Validation cohort: 141 | ||||
| Zhang et al[58] | HCC: 313 | CEUS | Radiomics | AUC |
| Primary cohort: 192 | Primary dataset: 0.849 | |||
| Validation cohort: 121 | Validation dataset: 0.788 | |||
| Liu et al[63] | HCC: 130 | CEUS | Deep learning radiomics | AUC: 0.93 |
| Training cohort: 89 | ||||
| Validation cohort: 41 | ||||
| Ma et al[66] | HCC: 318 | CEUS | Radiomics | AUC: 0.89 |
| Training cohort: 255 | ||||
| Validation cohort: 63 | ||||
| Liu et al[69] | HCC: 419 | CEUS | Deep learning radiomics | C-index |
| RFA: 214 | RFA: 0.726 | |||
| SR: 0.741 | ||||
| SR: 205 |
The application of AI in the diagnosis of benign and malignant FLLs: Hepatocellular carcinoma (HCC) is the fifth most common malignancy worldwide and accounts for the second leading cause of cancer-related deaths[41]. It is vital to identify benign and malignant FLLs for patients in the early stage.
AI based on conventional ultrasound: deep learning based on B-mode ultrasound has been demonstrated to be helpful in the diagnosis of benign and malignant FLLs. A CNN model was used to distinguish benign and malignant FLLs and achieved a higher accuracy than two experts[42]. Yang et al[43] developed a multicenter study to improve the B-mode ultrasound diagnostic performance for FLLs. The CNN of ultrasound performed high sensitivity and specificity in detecting FLLs, and it may be helpful for less-experienced doctors to enhance their judgment in liver cancer diagnosis.
AI based on B-mode ultrasound images has also been applied for the diagnosis of primary or secondary malignant liver tumors. A study proposed machine learning for discriminating HCC and metastatic liver tumors using SVM. The results revealed a classification accuracy of 91.6% with a sensitivity of 90.0% for HCCs and 93.3% for metastatic liver tumors[44].
AI based on contrast-enhanced ultrasound (CEUS): Recently, CEUS has become a commonly used ultrasound modality for the detection of FLLs[45]. Many studies have indicated that CEUS images had better sensitivity and specificity for the differentiation of malignant and benign tumors compared with B-mode images. One of the advantages of CEUS is that the images can be analyzed quantitatively. Time intensity curve (TIC) is a common quantitative analysis tool for CEUS[46]. Recently, AI based on CEUS images was reported to have great performance for the discrimination of FLLs.
Gatos et al[47] created a pretrained SVM algorithm to distinguish benign and malignant FLLs. In this model, a complex segmentation method based on TIC was used to detect lesions and process contours of 52 CEUS images. The accuracy, sensitivity, and specificity were 90.3%, 93.1%, and 86.9%, respectively[47]. Another study using SVM revealed that the sensitivity, specificity, and accuracy of benign and malignant grading were 94.0%, 87.1%, and 91.8%, respectively, while the classification accuracy of HCC, metastatic liver tumor, and benign were 85.7%, 87.7%, and 84.4%, respectively[46].
In addition to TIC, extracting features except TICs from a region of interest on CEUS images and videos was also applied in AI. A two-stage multiview learning framework, which was the integration of deep canonical correlation analysis and multiple kernel learning for CEUS-based computer-aided diagnosis, was proposed to identify liver tumors. The deep canonical correlation analysis-multiple kernel learning framework achieved performance for discriminating benign from malignant liver tumors with the accuracy, sensitivity, and specificity of 90.4%, 93.6%, and 86.8%, respectively[48].
The application of AI for the differential diagnosis of FLLs: With the development of AI, AI based on B-mode ultrasound images has great performance on the diagnosis of different FLLs. Hwang et al[49] extracted hybrid textural features from ultrasound images and used an ANN to diagnose FLLs. They indicated that the model revealed enormous potential with the diagnosis accuracy of over 96% among all FLLs groups (hemangioma vs malignant, cyst vs hemangioma, and cyst vs malignant)[49].
Deep learning was also applied in the distinction of different FLLs. Schmauch et al[50] created an algorithm that simultaneously detected and characterized FLLs. Although the amount of training data was relatively small, the average AUC of FLL detection and characterization was 0.935 and 0.916, respectively.
A CNN model was developed and validated for tumor detection and 6-class discrimination (HCC, focal fatty sparing, focal fatty infiltration, hemangiomas, and cysts)[51]. This model reached 87.0% detection rate, 83.9% sensitivity, and 97.1% specificity in the internal evaluation. In external validation groups, the model achieved 75.0% detection rate, 84.9% sensitivity, and 97.1% specificity.
CEUS also had excellent potential for AI to distinguish different FLLs. An ANN was applied to study the role of TIC analysis parameters of 4-class discrimination of liver tumors. The neural network had 94.45% training accuracy and 87.12% testing accuracy. The automatic classification process registered 93.2% sensitivity and 89.7% specificity[52].
Căleanu et al[53] reported the 5-class classification of liver tumors using deep neural networks with an accuracy of 88%. In this study, deep neural network algorithms were compared with state-of-the-art architectures, and a novel leave-one-patient-out evaluation procedure was presented.
All these studies indicated that AI based on conventional ultrasound and CEUS played a vital role in the detection and distinction of FLLs.
The application of AI in the management of HCC patients: Because of the development of new treatments, the management of HCC patients has become much more complicated. Radiomics can offer accurate assessment of great numbers of image features from medical images. These features that are difficult to detect by the human eye can be detected by machine learning or deep learning. AI models based on radiomics has also been reported to be applicable for the management of HCC, such as the prediction of microvascular invasion (MVI), curative transarterial chemoembolization (TACE) effect, recurrence after thermal ablation, and prognosis.
Predicting MVI: MVI is described as the invasion of tumor cells within a vascular space lined by endothelium. It has been proven that MVI is a predictor of early recurrence of HCC and poor survival outcomes[54]. The only way to confirm MVI is via histopathology after surgery. Patients with HCC can receive a great benefit when MVI is identified noninvasively and accurately before surgery[55]. The application of AI based on gray-scale ultrasound images and CEUS indicated good performance in predicting preoperative MVI.
A study indicated that the radiological features of gray-scale ultrasound images of gross tumoral area predicted preoperative MVI of HCC with an AUC of 0.81[56]. A CEUS-based radiomics score was built for preoperative prediction of MVI in HCC[57]. The radiomics nomogram revealed great potential in the detection of MVI with an AUC of 0.731 compared with the clinical nomogram with an AUC of 0.634. It was indicated that the radiomics data based on ultrasound was a single predictor of MVI in HCC. Our group created a radiomics model based on CEUS to evaluate MVI of HCC patients before surgery. The model revealed a better detection in the primary group with an AUC of 0.849 vs 0.690 as well as the validation group with an AUC of 0.788 vs 0.661 when compared with the clinical model. We confirmed that the portal venous phase, delay phase, tumor size, rad-score, and alpha-fetoprotein level were single predictors related to MVI[58].
Predicting curative TACE effect: Pathways participating in important cancer-related progression, such as cell proliferation and angiogenesis, are major goals for the treatment of HCC patients. Additionally, transcription factors and cell cycle regulators are also considered to be interesting for anti-HCC drugs[59].
TACE is a widely used first-line therapy for HCC patients diagnosed at the intermediate stage. The tumor response to the first TACE treatment is highly different and obviously related to the subsequent therapies as well as the patients’ survival[60]. Hence, the exact prediction of HCC responses after the first TACE treatment is vital for patients.
The prediction of tumor responses to TACE heavily depends on MRI and serological biomarkers[61,62]. But these methods achieved unsatisfactory accuracy of prediction. The application of AI based on both B-mode ultrasound and CEUS demonstrated better prediction efficacy.
An AI-based radiomics was established and validated to predict the personalized responses of HCC to the first TACE session. The deep learning radiomics-based CEUS model showed better performance compared with the machine learning radiomics-based B-mode model and machine learning radiomics-based time intensity curve of CEUS model with AUCs of 0.93, 0.80, and 0.81, respectively[63]. They suggested that the deep learning-based radiomics could benefit TACE candidates in clinical work.
Predicting recurrence after thermal ablation: Thermal ablation has been confirmed to be an available therapy for early-stage HCC patients who are unsuitable for operation or recurrence after surgery[64]. In addition, the recent 2-year recurrence rates of HCC patients who underwent thermal ablation were reported as 2%-18%[65]. The accurate preoperative prediction of thermal ablation outcomes is of great importance for HCC patients. Compared with other imaging modalities, CEUS is radiation-free and has better temporal resolution when revealing the blood supply of the tumor. The application of AI based on CEUS could be performed for the preoperative prediction of thermal ablation outcomes.
A radiomics model was created to predict the early and late recurrence of HCC patients who accepted thermal ablation[66]. The combined model including CEUS, ultrasound radiomics, and clinical factors showed better performance for early recurrence with an AUC of 0.89 and for late recurrence prediction with a C-index of 0.77.
Predicting the prognoses: Surgical resection (SR) and radiofrequency ablation (RFA) are common curative strategies for HCC patients diagnosed at the early stage[64]. Some studies have compared the long-term survival of RFA and SR for early-stage HCC patients[67,68]. However, the conclusions were sharply different. Hence, it is necessary to find useful predictive means to select the optimal patients who are suitable for RFA or SR before surgery. AI models based on CEUS had great performance for the prediction of progression-free survival (PFS).
A deep learning-based radiomics from CEUS images was built to predict the PFS of SR and RFA for HCC patients. Both SR and RFA models achieved high prediction accuracy of 2-year PFS. They also identified that a higher average probability of 2-year PFS may be acquired while some RFA and SR patients exchange their choices[69]. By utilizing conventional ultrasound images and CEUS, these AI prediction models can be applied in the individualized management of HCC patients.
The majority of gastric mesenchymal tumors are occasionally found during routine esophagogastroduodenoscopy examinations. The incidence of gastric mesenchymal tumors is uncertain, but the prevalence of subepithelial tumors identified under endoscopy in Korea was reported as 1.7%[70]. Most gastric mesenchymal tumors are gastrointestinal stromal tumors (GISTs), which may metastasize to the liver and peritoneum after surgery[71,72]. Hence, distinguishing GISTs from benign mesenchymal tumors such as leiomyomas or schwannomas is of great importance in clinic practice. Endoscopic ultrasonography (EUS) is a common method to assess gastric mesenchymal tumors. It helps doctors evaluate the detailed size, shape, origin, and border of the lesions[73-75]. But the interpretation of EUS images by endoscopists is subjective and has poor interobserver agreement. Recently, EUS image interpretation using AI has developed rapidly and is applied to distinguish GISTs from benign mesenchymal tumors.
A convolutional neural network computer-aided diagnosis (CNN-CAD) model based on EUS images was developed to assess gastric mesenchymal tumors. They reported the model distinguished GISTs from non-GIST tumors with 83.0% sensitivity, 75.5% specificity, and 79.2% accuracy[76]. The CNN-CAD model had the potential to provide diagnostic assistance to endoscopists in the future.
EUS is currently a common tool to diagnose pancreatic diseases in clinical practice. However, the specificity for the diagnosis of pancreatic diseases using EUS images is low and deeply depends on the subjective judgment of endoscopists. Studies have confirmed that AI based on EUS improves their performance for the diagnosis of pancreatic diseases. Recently, AI using EUS images has been applied in the differential diagnosis of PC, distinguishing intraductal papillary mucinous neoplasms (IPMNs) and detecting pancreatic segmentation.
Pancreatic cancer: PC is relatively uncommon, with an incidence of 8-12 per 100000 per year. PC is attributed to hereditary germline or somatic acquired mutations in some genes such as tumor suppressor genes and cell cycle genes. These mutations are also associated with the progression and metastasis of PC. Moreover, shortened telomerase, cell turnover, and genomic instability have an important role in the development of PC[77].
The early diagnosis and surgery of PC, especially for lesions less than 1 cm, can achieve long-term prognoses with a 5-year survival rate of 80.4%[78]. However, PC is most frequently detected at an advanced stage, and the 5-year survival rate remains as low as 3%-15%[79]. Hence, early detection is vital for the treatment of PC patients. Studies have reported that AI based on EUS has great performance for the diagnosis of PC.
AI based on B-mode EUS: AI models based on B-mode EUS have been applied to improve their performance for the diagnosis of PC. Norton et al[80] first reported the use of CAD utilizing EUS images in pancreatic diseases in 2001. The study included 14 patients with focal chronic pancreatitis and 21 patients with PC. They showed the diagnostic sensitivity of the two diseases was 89%, and the overall accuracy was 80%[80]. However, this study cannot be referred to as AI-CAD in current applications as the number of patients was limited and the resolution of images were very low.
With the development of AI, ANN and SVM presented good performance in the diagnosis of PC[81-83]. Das et al[81] developed an ANN model to distinguish chronic pancreatitis from PC. The results achieved 93% sensitivity, 92% specificity, 87% PPV, 96% negative predictive value (NPV), and 0.93 AUC[81]. By using a multilayered neural network, the study confirmed the first machine learning results for the EUS images of the pancreas. But the sample size was small and lacked pathological evidence in the chronic pancreatitis and normal pancreas groups.
By selecting better texture features that included multifractal dimensional features, a quantitative measure of fractality (self-similarity), and complexity from EUS images, a SVM prediction model was created to identify PC and non-PC patients[83]. The model reached 97.98% accuracy, 94.32% sensitivity, 99.45% specificity, 98.65% PPV, and 97.77% NPV. The study demonstrated that SVM using EUS images is a useful tool for diagnosing PC and pancreatic diseases.
It was reported that AI was also applied for the age-dependent pancreatic changes on EUS images of PC cases. Ozkan et al[84] suggested a high-performance CAD model applying ANN to discriminate PC and noncancer patients in three age groups. In the under 40-year-old group, the accuracy, sensitivity and specificity were 92.0%, 87.5%, and 94.1%, respectively. In the 40-year-old to 60-year-old group, the accuracy, sensitivity, and specificity were 88.5%, 85.7%, and 91.7%, respectively. In the > 60-year-old group, the accuracy, sensitivity, and specificity were 91.7%, 93.3%, and 88.9%, respectively. The total performance of this model showed the accuracy, sensitivity, and specificity were 87.5%, 83.3%, and 93.3%, respectively.
Besides machine learning, deep learning has been applied to B-mode EUS images for analysis of PC. A CNN model using EUS images was developed for the detection of PC[85]. The sensitivity, specificity, PPV, and NPV were 90.2%, 74.9%, 80.1%, and 88.7%, respectively. The CNN model included six normalization layers, seven convolution layers, four max-pooling layers, and six activation layers. The EUS-CNN application was first reported to have the potential to detect PC from EUS images.
AI based on EUS elastography: Real-time EUS elastography can provide more information about the features of pancreatic masses by the use of strain assessment. It was reported that EUS elastography has been applied in the differential diagnosis of pancreatic lesions. However, the accuracy and reproducibility were unstable[86,87].
The application of AI improves their performance in the diagnosis of PC. A prospective, blinded, multicentric study using EUS elastography by ANN was performed in focal pancreatic lesions[88]. They demonstrated the sensitivity, specificity, PPV, and NPV values for the diagnosis of PC were 87.59%, 82.94%, 96.25%, and 57.22%, respectively. The study suggested that the ANN model may provide fast and accurate diagnoses in the clinical.
AI based on contrast-enhanced EUS: Contrast-enhanced EUS has been used to enhance the detection of pancreatic lesions[89]. AI based on contrast-enhanced EUS has great performance for the diagnosis of PC. An ANN model based on the TIC analysis from contrast-enhanced EUS images was designed to diagnose PC and chronic pancreatitis. The study reached 94.64% sensitivity, 94.44% specificity, 97.24% PPV, and 89.47% NPV[90]. The study suggested that the model could provide additional diagnostic value to CEUS interpretation and EUS fine needle aspiration results.
IPMNs: IPMNs are considered to be precursor lesions of pancreatic adenocarcinoma. Early surgical resection of IPMNs can provide a survival benefit for patients[91]. EUS is often used to assess the malignancy of IPMNs in clinics. Several predictive techniques were used to diagnose the malignancy of IPMNs with no satisfactory results (70%-80%)[92,93].
Compared with human diagnosis and conventional EUS features, AI via deep learning algorithms was confirmed to be a more exact and objective way for the differential diagnosis of malignant IPMNs. Kuwahara et al[94] performed a predictive CNN model using EUS images to detect malignant IPMNs. The model reached 95.7% sensitivity, 94.0% accuracy, and 92.6% specificity. The accuracy was higher compared with the diagnosis of a radiologist (56.0%). The author suggested that the application of AI can evaluate malignant IPMNs before surgery.
Pancreatic segmentation: AI using EUS images has also been applied in pancreatic segmentation. A deep learning-based classification system was created to utilize the “station approach” in EUS of pancreas[95]. The system obtained 90.0% accuracy in classification and 0.770 and 0.813 in blood vessel and pancreas segmentation, respectively. The results were similar to that of EUS experts. Thus, this study revealed that AI has the feasibility to detect the station and segmentation of the pancreas.
Colorectal cancer is the third most common cancer worldwide and accounts for the second leading cause of cancer-related deaths. Moreover, a growing number of patients diagnosed with rectal cancer are under 50-years-old[96]. Colorectal cancer is attributed to gene mutations of epithelial cells, such as oncogenes, tumor suppressor genes, and DNA repair genes. The specific molecular mechanisms implicated in this type of cancer may include the instability of chromosomes and microsatellites[97].
Recently, some researchers studied tumor deposits (TDs) of rectal cancer. TDs are described as focal aggregates of adenocarcinoma located in the surrounding fat of the colon or rectum. They are discontinuous with the primary tumor and unrelated to a lymph node[98,99].
It was reported that a patient who is TD-positive has more malignant tumors, with decreased disease-free survival and overall survival[100]. However, TDs are often diagnosed by pathology only after surgery. Hence, the noninvasive preoperative prediction of TDs is important for rectal cancer patients. EUS is currently a common tool to detect rectal masses. Recently, ultrasound-based radiomics have been applied to predict the status of TDs.
Chen et al[101] developed an ANN system using ultrasound radiomics and clinical factors to predict TDs. Endorectal ultrasound and SWE examinations were conducted for 127 patients with rectal cancer. The accuracy was 75.0% in the validation group. The model reached 72.7% sensitivity, 75.9% specificity, and 0.743 AUC. The study suggested that ultrasound-based radiomics has the potential for the prediction of TDs before treatment. Table 3 shows the application of ultrasound-based AI in gastrointestinal disease.
| Ref. | Diseases: number of cases | Type of ultrasound | Algorithm of AI | Performance |
| Kim et al[76] | GISTs: 125 | B-mode EUS | CNN | Sensitivity: 83.0% |
| Leiomyomas: 33 | Specificity: 75.5% | |||
| Accuracy: 79.2% | ||||
| Schwannomas: 21 | ||||
| Norton et al[80] | Chronic pancreatitis: 14 | B-mode EUS | Basic neural network | Sensitivity: 89% |
| Pancreatic cancer: 21 | Accuracy: 80% | |||
| Das et al[81] | Chronic pancreatitis: 12 | B-mode EUS | ANN | Sensitivity: 93% |
| Pancreatic cancer: 22 | ||||
| Specificity: 92% | ||||
| Normal patient: 22 | ||||
| AUC: 0.93 | ||||
| Zhu et al[82] | Chronic pancreatitis: 126 | B-mode EUS | SVM | Sensitivity: 96.25% |
| Specificity: 93.38% | ||||
| Accuracy: 94.2% | ||||
| Pancreatic cancer: 262 | ||||
| Zhang et al[83] | Pancreatic cancer: 153 | B-mode EUS | SVM | Sensitivity: 94.32% |
| Specificity: 99.45% | ||||
| Normal patient: 63 | ||||
| Accuracy: 97.98% | ||||
| Ozkan et al[84] | Pancreatic cancer: 202 | B-mode EUS | ANN | Sensitivity: 83.3% |
| Specificity: 93.3% | ||||
| Normal patient: 130 | Accuracy: 87.5% | |||
| Tonozuka et al[85] | Chronic pancreatitis: 34 | B-mode EUS | CNN | Sensitivity: 90.2% |
| Pancreatic cancer: 76 | ||||
| Normal patient: 29 | ||||
| Specificity: 74.9% | ||||
| Săftoiu et al[88] | Chronic pancreatitis: 47 | EUS elastography | ANN | Sensitivity: 87.59% |
| Specificity: 82.94% | ||||
| Pancreatic cancer: 211 | ||||
| Săftoiu et al[90] | Chronic pancreatitis: 55 | Contrast-enhanced EUS | ANN | Sensitivity: 94.64% |
| Pancreatic cancer: 122 | Specificity: 94.44% | |||
| Kuwahara et al[94] | IPMN: 50 | B-mode EUS | CNN | Sensitivity: 95.7% |
| Specificity: 92.6% | ||||
| Accuracy: 94.0% | ||||
| Zhang et al[95] | Training: 291 | B-mode EUS | CNN | Accuracy: 90.0% |
| Testing: 181 | ||||
| Chen et al[101] | Rectal cancer: 127 | Endorectal ultrasound | ANN | Sensitivity: 72.7% |
| Specificity: 75.9% | ||||
| Shear-wave elastography | ||||
| AUC: 0.743 |
In recent years, AI models using ultrasound images have developed rapidly. They can offer a more precise and efficient diagnosis and ease the burden of doctors. AI based on ultrasound has been confirmed to be helpful in diffuse liver diseases and FLLs, such as assessing the severity of NAFLD and the grade of liver fibrosis, distinguishing benign and malignant liver lesions, predicting the MVI of HCC, curative TACE effect, and prognoses after thermal ablation. In addition, AI based on EUS has great performance in gastrointestinal diseases, such as distinguishing gastric mesenchymal tumors, differential diagnosis of PC, distinguishing IPMNs, and predicting the status of TDs in rectal cancer.
However, the application of AI based on ultrasound in clinical practice has some limitations. The main reason may be due to the high variability between radiologists in ultrasound image acquisition and interpretation[102]. Hence, it is necessary to unify the ultrasonic image acquisition process as well as the standard of ultrasonic data measurement during the ultrasound examination.
In addition, some studies of AI-powered ultrasound were retrospective and trained on limited data offered by a single hospital with potential data selection bias, and the amount of data in the training set was not enough. Abundant multicenter prospective studies should assure the efficiency and stability of these AI models. Additionally, deep learning needs a large number of images, so it is necessary to establish an abundant database with common collaborative efforts.
In addition, the application of AI based on EUS has some limitations. The number of EUS examinations is overwhelmingly low compared to other examinations such as endoscopy and CT, especially in gastrointestinal diseases.
In the future, AI based on ultrasound may be used to develop highly accurate and more efficient models for more digestive diseases such as peptic ulcers, stomach neoplasms, inflammatory bowel disease, and so on. These models may heavily reduce the workload for doctors by automatic identification of disease on radiologic and histopathologic images. Moreover, the application of AI can enable building individual management for patients as well as predicting disease progression and complications in clinics. Additionally, AI may improve distance teaching by remote monitoring and enhance medical services in undeveloped areas.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Amin A, United Arab Emirates; Surani S, United States S-Editor: Gong ZM L-Editor: Filipodia P-Editor: Gong ZM
| 1. | Asrani SK, Devarbhavi H, Eaton J, Kamath PS. Burden of liver diseases in the world. J Hepatol. 2019;70:151-171. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1382] [Cited by in RCA: 2297] [Article Influence: 382.8] [Reference Citation Analysis (0)] |
| 2. | Araújo AR, Rosso N, Bedogni G, Tiribelli C, Bellentani S. Global epidemiology of non-alcoholic fatty liver disease/non-alcoholic steatohepatitis: What we need in the future. Liver Int. 2018;38 Suppl 1:47-51. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 225] [Cited by in RCA: 281] [Article Influence: 40.1] [Reference Citation Analysis (0)] |
| 3. | Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87-108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18694] [Cited by in RCA: 21373] [Article Influence: 2137.3] [Reference Citation Analysis (3)] |
| 4. | Peery AF, Dellon ES, Lund J, Crockett SD, McGowan CE, Bulsiewicz WJ, Gangarosa LM, Thiny MT, Stizenberg K, Morgan DR, Ringel Y, Kim HP, DiBonaventura MD, Carroll CF, Allen JK, Cook SF, Sandler RS, Kappelman MD, Shaheen NJ. Burden of gastrointestinal disease in the United States: 2012 update. Gastroenterology. 2012;143:1179-1187.e3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1355] [Cited by in RCA: 1467] [Article Influence: 112.8] [Reference Citation Analysis (1)] |
| 5. | Yadav D, Lowenfels AB. The epidemiology of pancreatitis and pancreatic cancer. Gastroenterology. 2013;144:1252-1261. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1151] [Cited by in RCA: 1347] [Article Influence: 112.3] [Reference Citation Analysis (0)] |
| 6. | Brody H. Medical imaging. Nature. 2013;502:S81. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 25] [Article Influence: 2.1] [Reference Citation Analysis (3)] |
| 7. | Tchelepi H, Ralls PW, Radin R, Grant E. Sonography of diffuse liver disease. J Ultrasound Med. 2002;21:1023-32; quiz 1033. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 180] [Cited by in RCA: 168] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 8. | Vicas C, Lupsor M, Badea R, Nedevschi S. Usefulness of textural analysis as a tool for noninvasive liver fibrosis staging. J Med Ultrason (2001). 2011;38:105-117. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 9. | Wang S, Summers RM. Machine learning and radiology. Med Image Anal. 2012;16:933-951. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 451] [Cited by in RCA: 352] [Article Influence: 27.1] [Reference Citation Analysis (0)] |
| 10. | Chen H, Sung JJY. Potentials of AI in medical image analysis in Gastroenterology and Hepatology. J Gastroenterol Hepatol. 2021;36:31-38. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 24] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 11. | Mayerhoefer ME, Materka A, Langs G, Häggström I, Szczypiński P, Gibbs P, Cook G. Introduction to Radiomics. J Nucl Med. 2020;61:488-495. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 211] [Cited by in RCA: 1018] [Article Influence: 203.6] [Reference Citation Analysis (0)] |
| 12. | LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521:436-444. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36149] [Cited by in RCA: 20062] [Article Influence: 2006.2] [Reference Citation Analysis (0)] |
| 13. | Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, van der Laak JAWM, van Ginneken B, Sánchez CI. A survey on deep learning in medical image analysis. Med Image Anal. 2017;42:60-88. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5573] [Cited by in RCA: 4942] [Article Influence: 617.8] [Reference Citation Analysis (0)] |
| 14. | Ros PR, Mortele KJ. Diffuse liver disease. Clin Liver Dis. 2002;6:181-201. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 15] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 15. | Piccinino F, Sagnelli E, Pasquale G, Giusti G. Complications following percutaneous liver biopsy. A multicentre retrospective study on 68,276 biopsies. J Hepatol. 1986;2:165-173. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 850] [Cited by in RCA: 804] [Article Influence: 20.6] [Reference Citation Analysis (0)] |
| 16. | Farrell GC, Larter CZ. Nonalcoholic fatty liver disease: from steatosis to cirrhosis. Hepatology. 2006;43:S99-S112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1756] [Cited by in RCA: 1822] [Article Influence: 95.9] [Reference Citation Analysis (0)] |
| 17. | Sriraam N, Roopa J, Saranya M, Dhanalakshmi M. Performance evaluation of computer aided diagnostic tool (CAD) for detection of ultrasonic based liver disease. J Med Syst. 2009;33:267-274. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 4] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 18. | Rinella ME. Nonalcoholic fatty liver disease: a systematic review. JAMA. 2015;313:2263-2273. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1508] [Cited by in RCA: 1758] [Article Influence: 175.8] [Reference Citation Analysis (0)] |
| 19. | Acharya UR, Raghavendra U, Fujita H, Hagiwara Y, Koh JE, Jen Hong T, Sudarshan VK, Vijayananthan A, Yeong CH, Gudigar A, Ng KH. Automated characterization of fatty liver disease and cirrhosis using curvelet transform and entropy features extracted from ultrasound images. Comput Biol Med. 2016;79:250-258. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 68] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 20. | Mathiesen UL, Franzén LE, Aselius H, Resjö M, Jacobsson L, Foberg U, Frydén A, Bodemar G. Increased liver echogenicity at ultrasound examination reflects degree of steatosis but not of fibrosis in asymptomatic patients with mild/moderate abnormalities of liver transaminases. Dig Liver Dis. 2002;34:516-522. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 246] [Cited by in RCA: 262] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 21. | Byra M, Styczynski G, Szmigielski C, Kalinowski P, Michałowski Ł, Paluszkiewicz R, Ziarkiewicz-Wróblewska B, Zieniewicz K, Sobieraj P, Nowicki A. Transfer learning with deep convolutional neural network for liver steatosis assessment in ultrasound images. Int J Comput Assist Radiol Surg. 2018;13:1895-1903. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 206] [Cited by in RCA: 154] [Article Influence: 22.0] [Reference Citation Analysis (0)] |
| 22. | Biswas M, Kuppili V, Edla DR, Suri HS, Saba L, Marinhoe RT, Sanches JM, Suri JS. Symtosis: A liver ultrasound tissue characterization and risk stratification in optimized deep learning paradigm. Comput Methods Programs Biomed. 2018;155:165-177. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 119] [Article Influence: 17.0] [Reference Citation Analysis (1)] |
| 23. | Oelze ML, Mamou J. Review of Quantitative Ultrasound: Envelope Statistics and Backscatter Coefficient Imaging and Contributions to Diagnostic Ultrasound. IEEE Trans Ultrason Ferroelectr Freq Control. 2016;63:336-351. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 256] [Cited by in RCA: 225] [Article Influence: 25.0] [Reference Citation Analysis (0)] |
| 24. | Han A, Byra M, Heba E, Andre MP, Erdman JW Jr, Loomba R, Sirlin CB, ’'Brien WD Jr. Noninvasive Diagnosis of Nonalcoholic Fatty Liver Disease and Quantification of Liver Fat with Radiofrequency Ultrasound Data Using One-dimensional Convolutional Neural Networks. Radiology. 2020;295:342-350. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 85] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 25. | Ishak K, Baptista A, Bianchi L, Callea F, De Groote J, Gudat F, Denk H, Desmet V, Korb G, MacSween RN. Histological grading and staging of chronic hepatitis. J Hepatol. 1995;22:696-699. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3521] [Cited by in RCA: 3784] [Article Influence: 126.1] [Reference Citation Analysis (1)] |
| 26. | Amin A, Mahmoud-Ghoneim D. Texture analysis of liver fibrosis microscopic images: a study on the effect of biomarkers. Acta Biochim Biophys Sin (Shanghai). 2011;43:193-203. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 47] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 27. | Bharti P, Mittal D, Ananthasivan R. Computer-aided Characterization and Diagnosis of Diffuse Liver Diseases Based on Ultrasound Imaging: A Review. Ultrason Imaging. 2017;39:33-61. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 25] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 28. | Yeh WC, Huang SW, Li PC. Liver fibrosis grade classification with B-mode ultrasound. Ultrasound Med Biol. 2003;29:1229-1235. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 54] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 29. | Zhang L, Li QY, Duan YY, Yan GZ, Yang YL, Yang RJ. Artificial neural network aided non-invasive grading evaluation of hepatic fibrosis by duplex ultrasonography. BMC Med Inform Decis Mak. 2012;12:55. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 22] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 30. | Gao S, Peng Y, Guo H, Liu W, Gao T, Xu Y, Tang X. Texture analysis and classification of ultrasound liver images. Biomed Mater Eng. 2014;24:1209-1216. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 22] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 31. | Lee JH, Joo I, Kang TW, Paik YH, Sinn DH, Ha SY, Kim K, Choi C, Lee G, Yi J, Bang WC. Deep learning with ultrasonography: automated classification of liver fibrosis using a deep convolutional neural network. Eur Radiol. 2020;30:1264-1273. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 86] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 32. | Gatos I, Tsantis S, Spiliopoulos S, Karnabatidis D, Theotokas I, Zoumpoulis P, Loupas T, Hazle JD, Kagadis GC. Temporal stability assessment in shear wave elasticity images validated by deep learning neural network for chronic liver disease fibrosis stage assessment. Med Phys. 2019;46:2298-2309. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 42] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 33. | Li W, Huang Y, Zhuang BW, Liu GJ, Hu HT, Li X, Liang JY, Wang Z, Huang XW, Zhang CQ, Ruan SM, Xie XY, Kuang M, Lu MD, Chen LD, Wang W. Multiparametric ultrasomics of significant liver fibrosis: A machine learning-based analysis. Eur Radiol. 2019;29:1496-1506. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 62] [Cited by in RCA: 96] [Article Influence: 13.7] [Reference Citation Analysis (1)] |
| 34. | Gatos I, Tsantis S, Spiliopoulos S, Karnabatidis D, Theotokas I, Zoumpoulis P, Loupas T, Hazle JD, Kagadis GC. A new computer aided diagnosis system for evaluation of chronic liver disease with ultrasound shear wave elastography imaging. Med Phys. 2016;43:1428-1436. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 20] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 35. | Gatos I, Tsantis S, Spiliopoulos S, Karnabatidis D, Theotokas I, Zoumpoulis P, Loupas T, Hazle JD, Kagadis GC. A Machine-Learning Algorithm Toward Color Analysis for Chronic Liver Disease Classification, Employing Ultrasound Shear Wave Elastography. Ultrasound Med Biol. 2017;43:1797-1810. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 73] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 36. | Wang K, Lu X, Zhou H, Gao Y, Zheng J, Tong M, Wu C, Liu C, Huang L, Jiang T, Meng F, Lu Y, Ai H, Xie XY, Yin LP, Liang P, Tian J, Zheng R. Deep learning Radiomics of shear wave elastography significantly improved diagnostic performance for assessing liver fibrosis in chronic hepatitis B: a prospective multicentre study. Gut. 2019;68:729-741. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 226] [Cited by in RCA: 354] [Article Influence: 59.0] [Reference Citation Analysis (1)] |
| 37. | Liu S, Wang Y, Yang X, Lei B, Liu L, Li SX, Ni D, Wang T. Deep Learning in Medical Ultrasound Analysis: A Review. Engineering. 2019;5:261-275. [DOI] [Full Text] |
| 38. | Xue LY, Jiang ZY, Fu TT, Wang QM, Zhu YL, Dai M, Wang WP, Yu JH, Ding H. Transfer learning radiomics based on multimodal ultrasound imaging for staging liver fibrosis. Eur Radiol. 2020;30:2973-2983. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 79] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 39. | Bruix J, Sherman M; American Association for the Study of Liver Diseases. Management of hepatocellular carcinoma: an update. Hepatology. 2011;53:1020-1022. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5972] [Cited by in RCA: 6573] [Article Influence: 469.5] [Reference Citation Analysis (1)] |
| 40. | Nishida N, Kudo M. Artificial Intelligence in Medical Imaging and Its Application in Sonography for the Management of Liver Tumor. Front Oncol. 2020;10:594580. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 24] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 41. | Heimbach JK, Kulik LM, Finn RS, Sirlin CB, Abecassis MM, Roberts LR, Zhu AX, Murad MH, Marrero JA. AASLD guidelines for the treatment of hepatocellular carcinoma. Hepatology. 2018;67:358-380. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2107] [Cited by in RCA: 3031] [Article Influence: 433.0] [Reference Citation Analysis (3)] |
| 42. | Xi IL, Wu J, Guan J, Zhang PJ, Horii SC, Soulen MC, Zhang Z, Bai HX. Deep learning for differentiation of benign and malignant solid liver lesions on ultrasonography. Abdom Radiol (NY). 2021;46:534-543. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 27] [Article Influence: 6.8] [Reference Citation Analysis (1)] |
| 43. | Yang Q, Wei J, Hao X, Kong D, Yu X, Jiang T, Xi J, Cai W, Luo Y, Jing X, Yang Y, Cheng Z, Wu J, Zhang H, Liao J, Zhou P, Song Y, Zhang Y, Han Z, Cheng W, Tang L, Liu F, Dou J, Zheng R, Yu J, Tian J, Liang P. Improving B-mode ultrasound diagnostic performance for focal liver lesions using deep learning: A multicentre study. eBioMedicine. 2020;56:102777. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 71] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 44. | Virmani J, Kumar V, Kalra N, Khandelwal N. Characterization of primary and secondary malignant liver lesions from B-mode ultrasound. J Digit Imaging. 2013;26:1058-1070. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 35] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 45. | Claudon M, Cosgrove D, Albrecht T, Bolondi L, Bosio M, Calliada F, Correas JM, Darge K, Dietrich C, ’'Onofrio M, Evans DH, Filice C, Greiner L, Jäger K, Jong Nd, Leen E, Lencioni R, Lindsell D, Martegani A, Meairs S, Nolsøe C, Piscaglia F, Ricci P, Seidel G, Skjoldbye B, Solbiati L, Thorelius L, Tranquart F, Weskott HP, Whittingham T. Guidelines and good clinical practice recommendations for contrast enhanced ultrasound (CEUS)–- update 2008. Ultraschall Med. 2008;29:28-44. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 556] [Cited by in RCA: 499] [Article Influence: 29.4] [Reference Citation Analysis (0)] |
| 46. | Kondo S, Takagi K, Nishida M, Iwai T, Kudo Y, Ogawa K, Kamiyama T, Shibuya H, Kahata K, Shimizu C. Computer-Aided Diagnosis of Focal Liver Lesions Using Contrast-Enhanced Ultrasonography With Perflubutane Microbubbles. IEEE Trans Med Imaging. 2017;36:1427-1437. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 54] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 47. | Gatos I, Tsantis S, Spiliopoulos S, Skouroliakou A, Theotokas I, Zoumpoulis P, Hazle JD, Kagadis GC. A new automated quantification algorithm for the detection and evaluation of focal liver lesions with contrast-enhanced ultrasound. Med Phys. 2015;42:3948-3959. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 42] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 48. | Guo LH, Wang D, Qian YY, Zheng X, Zhao CK, Li XL, Bo XW, Yue WW, Zhang Q, Shi J, Xu HX. A two-stage multi-view learning framework based computer-aided diagnosis of liver tumors with contrast enhanced ultrasound images. Clin Hemorheol Microcirc. 2018;69:343-354. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 72] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 49. | Hwang YN, Lee JH, Kim GY, Jiang YY, Kim SM. Classification of focal liver lesions on ultrasound images by extracting hybrid textural features and using an artificial neural network. Biomed Mater Eng. 2015;26 Suppl 1:S1599-S1611. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 25] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 50. | Schmauch B, Herent P, Jehanno P, Dehaene O, Saillard C, Aubé C, Luciani A, Lassau N, Jégou S. Diagnosis of focal liver lesions from ultrasound using deep learning. Diagn Interv Imaging. 2019;100:227-233. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 104] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 51. | Tiyarattanachai T, Apiparakoon T, Marukatat S, Sukcharoen S, Geratikornsupuk N, Anukulkarnkusol N, Mekaroonkamol P, Tanpowpong N, Sarakul P, Rerknimitr R, Chaiteerakij R. Development and validation of artificial intelligence to detect and diagnose liver lesions from ultrasound images. pLoS One. 2021;16:e0252882. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 22] [Article Influence: 5.5] [Reference Citation Analysis (1)] |
| 52. | Streba CT, Ionescu M, Gheonea DI, Sandulescu L, Ciurea T, Saftoiu A, Vere CC, Rogoveanu I. Contrast-enhanced ultrasonography parameters in neural network diagnosis of liver tumors. World J Gastroenterol. 2012;18:4427-4434. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 53] [Cited by in RCA: 46] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 53. | Căleanu CD, Sîrbu CL, Simion G. Deep Neural Architectures for Contrast Enhanced Ultrasound (CEUS) Focal Liver Lesions Automated Diagnosis. Sensors (Basel). 2021;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 22] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 54. | Lim KC, Chow PK, Allen JC, Chia GS, Lim M, Cheow PC, Chung AY, Ooi LL, Tan SB. Microvascular invasion is a better predictor of tumor recurrence and overall survival following surgical resection for hepatocellular carcinoma compared to the Milan criteria. Ann Surg. 2011;254:108-113. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 284] [Cited by in RCA: 414] [Article Influence: 29.6] [Reference Citation Analysis (0)] |
| 55. | Banerjee S, Wang DS, Kim HJ, Sirlin CB, Chan MG, Korn RL, Rutman AM, Siripongsakun S, Lu D, Imanbayev G, Kuo MD. A computed tomography radiogenomic biomarker predicts microvascular invasion and clinical outcomes in hepatocellular carcinoma. Hepatology. 2015;62:792-800. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 203] [Cited by in RCA: 277] [Article Influence: 27.7] [Reference Citation Analysis (0)] |
| 56. | Dong Y, Zhou L, Xia W, Zhao XY, Zhang Q, Jian JM, Gao X, Wang WP. Preoperative Prediction of Microvascular Invasion in Hepatocellular Carcinoma: Initial Application of a Radiomic Algorithm Based on Grayscale Ultrasound Images. Front Oncol. 2020;10:353. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 49] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 57. | Hu HT, Wang Z, Huang XW, Chen SL, Zheng X, Ruan SM, Xie XY, Lu MD, Yu J, Tian J, Liang P, Wang W, Kuang M. Ultrasound-based radiomics score: a potential biomarker for the prediction of microvascular invasion in hepatocellular carcinoma. Eur Radiol. 2019;29:2890-2901. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 142] [Article Influence: 20.3] [Reference Citation Analysis (0)] |
| 58. | Zhang D, Wei Q, Wu GG, Zhang XY, Lu WW, Lv WZ, Liao JT, Cui XW, Ni XJ, Dietrich CF. Preoperative Prediction of Microvascular Invasion in Patients With Hepatocellular Carcinoma Based on Radiomics Nomogram Using Contrast-Enhanced Ultrasound. Front Oncol. 2021;11:709339. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 24] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 59. | Juaid N, Amin A, Abdalla A, Reese K, Alamri Z, Moulay M, Abdu S, Miled N. Anti-Hepatocellular Carcinoma Biomolecules: Molecular Targets Insights. Int J Mol Sci. 2021;22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 67] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 60. | Kim BK, Kim SU, Kim KA, Chung YE, Kim MJ, Park MS, Park JY, Kim DY, Ahn SH, Kim MD, Park SI, Won JY, Lee DY, Han KH. Complete response at first chemoembolization is still the most robust predictor for favorable outcome in hepatocellular carcinoma. J Hepatol. 2015;62:1304-1310. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 160] [Cited by in RCA: 162] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 61. | Loosen SH, Schulze-Hagen M, Leyh C, Benz F, Vucur M, Kuhl C, Trautwein C, Tacke F, Bruners P, Roderburg C, Luedde T. IL-6 and IL-8 Serum Levels Predict Tumor Response and Overall Survival after TACE for Primary and Secondary Hepatic Malignancies. Int J Mol Sci. 2018;19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 36] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 62. | Lahrsow M, Albrecht MH, Bickford MW, Vogl TJ. Predicting Treatment Response of Colorectal Cancer Liver Metastases to Conventional Lipiodol-Based Transarterial Chemoembolization Using Diffusion-Weighted MR Imaging: Value of Pretreatment Apparent Diffusion Coefficients (ADC) and ADC Changes Under Therapy. Cardiovasc Intervent Radiol. 2017;40:852-859. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 12] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 63. | Liu D, Liu F, Xie X, Su L, Liu M, Kuang M, Huang G, Wang Y, Zhou H, Wang K, Lin M, Tian J. Accurate prediction of responses to transarterial chemoembolization for patients with hepatocellular carcinoma by using artificial intelligence in contrast-enhanced ultrasound. Eur Radiol. 2020;30:2365-2376. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 96] [Article Influence: 19.2] [Reference Citation Analysis (0)] |
| 64. | European Association for the Study of the Liver. EASL Clinical Practice Guidelines: Management of hepatocellular carcinoma. J Hepatol. 2018;69:182-236. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5593] [Cited by in RCA: 6065] [Article Influence: 866.4] [Reference Citation Analysis (3)] |
| 65. | European Association For The Study Of The Liver. European Organisation For Research And Treatment Of Cancer. EASL-EORTC clinical practice guidelines: management of hepatocellular carcinoma. J Hepatol. 2012;56:908-943. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4059] [Cited by in RCA: 4521] [Article Influence: 347.8] [Reference Citation Analysis (2)] |
| 66. | Ma QP, He XL, Li K, Wang JF, Zeng QJ, Xu EJ, He XQ, Li SY, Kun W, Zheng RQ, Tian J. Dynamic Contrast-Enhanced Ultrasound Radiomics for Hepatocellular Carcinoma Recurrence Prediction After Thermal Ablation. Mol Imaging Biol. 2021;23:572-585. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 32] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 67. | Chen MS, Li JQ, Zheng Y, Guo RP, Liang HH, Zhang YQ, Lin XJ, Lau WY. A prospective randomized trial comparing percutaneous local ablative therapy and partial hepatectomy for small hepatocellular carcinoma. Ann Surg. 2006;243:321-328. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1100] [Cited by in RCA: 1104] [Article Influence: 58.1] [Reference Citation Analysis (0)] |
| 68. | Feng K, Yan J, Li X, Xia F, Ma K, Wang S, Bie P, Dong J. A randomized controlled trial of radiofrequency ablation and surgical resection in the treatment of small hepatocellular carcinoma. J Hepatol. 2012;57:794-802. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 471] [Cited by in RCA: 598] [Article Influence: 46.0] [Reference Citation Analysis (0)] |
| 69. | Liu F, Liu D, Wang K, Xie X, Su L, Kuang M, Huang G, Peng B, Wang Y, Lin M, Tian J. Deep Learning Radiomics Based on Contrast-Enhanced Ultrasound Might Optimize Curative Treatments for Very-Early or Early-Stage Hepatocellular Carcinoma Patients. Liver Cancer. 2020;9:397-413. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 103] [Article Influence: 20.6] [Reference Citation Analysis (0)] |
| 70. | Lee JH, Lee HL, Ahn YW, Lee KN, Jun DW, Lee OY, Han DS, Yoon BC, Choi HS. Prevalence of Gastric Subepithelial Tumors in Korea: A Single Center Experience. Korean J Gastroenterol. 2015;66:274-276. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 17] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 71. | Blay JY, Bonvalot S, Casali P, Choi H, Debiec-Richter M, Dei Tos AP, Emile JF, Gronchi A, Hogendoorn PC, Joensuu H, Le Cesne A, McClure J, Maurel J, Nupponen N, Ray-Coquard I, Reichardt P, Sciot R, Stroobants S, van Glabbeke M, van Oosterom A, Demetri GD; GIST consensus meeting panelists. Consensus meeting for the management of gastrointestinal stromal tumors. Report of the GIST Consensus Conference of 20-21 March 2004, under the auspices of ESMO. Ann Oncol. 2005;16:566-578. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 500] [Cited by in RCA: 487] [Article Influence: 24.4] [Reference Citation Analysis (0)] |
| 72. | Miettinen M, Sobin LH, Lasota J. Gastrointestinal stromal tumors of the stomach: a clinicopathologic, immunohistochemical, and molecular genetic study of 1765 cases with long-term follow-up. Am J Surg Pathol. 2005;29:52-68. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 852] [Cited by in RCA: 867] [Article Influence: 43.4] [Reference Citation Analysis (0)] |
| 73. | Chak A, Canto MI, Rösch T, Dittler HJ, Hawes RH, Tio TL, Lightdale CJ, Boyce HW, Scheiman J, Carpenter SL, Van Dam J, Kochman ML, Sivak MV Jr. Endosonographic differentiation of benign and malignant stromal cell tumors. Gastrointest Endosc. 1997;45:468-473. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 230] [Cited by in RCA: 206] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 74. | Palazzo L, Landi B, Cellier C, Cuillerier E, Roseau G, Barbier JP. Endosonographic features predictive of benign and malignant gastrointestinal stromal cell tumours. Gut. 2000;46:88-92. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 244] [Cited by in RCA: 221] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 75. | Kim GH, Park DY, Kim S, Kim DH, Choi CW, Heo J, Song GA. Is it possible to differentiate gastric GISTs from gastric leiomyomas by EUS? World J Gastroenterol. 2009;15:3376-3381. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 66] [Cited by in RCA: 64] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 76. | Kim YH, Kim GH, Kim KB, Lee MW, Lee BE, Baek DH, Kim DH, Park JC. Application of A Convolutional Neural Network in The Diagnosis of Gastric Mesenchymal Tumors on Endoscopic Ultrasonography Images. J Clin Med. 2020;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 33] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 77. | Goral V. Pancreatic Cancer: Pathogenesis and Diagnosis. Asian Pac J Cancer Prev. 2015;16:5619-5624. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 174] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 78. | Egawa S, Toma H, Ohigashi H, Okusaka T, Nakao A, Hatori T, Maguchi H, Yanagisawa A, Tanaka M. Japan Pancreatic Cancer Registry; 30th year anniversary: Japan Pancreas Society. Pancreas. 2012;41:985-992. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 257] [Cited by in RCA: 307] [Article Influence: 23.6] [Reference Citation Analysis (0)] |
| 79. | Pereira SP, Oldfield L, Ney A, Hart PA, Keane MG, Pandol SJ, Li D, Greenhalf W, Jeon CY, Koay EJ, Almario CV, Halloran C, Lennon AM, Costello E. Early detection of pancreatic cancer. Lancet Gastroenterol Hepatol. 2020;5:698-710. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 254] [Cited by in RCA: 303] [Article Influence: 60.6] [Reference Citation Analysis (1)] |
| 80. | Norton ID, Zheng Y, Wiersema MS, Greenleaf J, Clain JE, Dimagno EP. Neural network analysis of EUS images to differentiate between pancreatic malignancy and pancreatitis. Gastrointest Endosc. 2001;54:625-629. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 76] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 81. | Das A, Nguyen CC, Li F, Li B. Digital image analysis of EUS images accurately differentiates pancreatic cancer from chronic pancreatitis and normal tissue. Gastrointest Endosc. 2008;67:861-867. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 81] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 82. | Zhu M, Xu C, Yu J, Wu Y, Li C, Zhang M, Jin Z, Li Z. Differentiation of pancreatic cancer and chronic pancreatitis using computer-aided diagnosis of endoscopic ultrasound (EUS) images: a diagnostic test. pLoS One. 2013;8:e63820. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 84] [Cited by in RCA: 74] [Article Influence: 6.2] [Reference Citation Analysis (2)] |
| 83. | Zhang MM, Yang H, Jin ZD, Yu JG, Cai ZY, Li ZS. Differential diagnosis of pancreatic cancer from normal tissue with digital imaging processing and pattern recognition based on a support vector machine of EUS images. Gastrointest Endosc. 2010;72:978-985. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 63] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 84. | Ozkan M, Cakiroglu M, Kocaman O, Kurt M, Yilmaz B, Can G, Korkmaz U, Dandil E, Eksi Z. Age-based computer-aided diagnosis approach for pancreatic cancer on endoscopic ultrasound images. Endosc Ultrasound. 2016;5:101-107. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 75] [Cited by in RCA: 67] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 85. | Tonozuka R, Itoi T, Nagata N, Kojima H, Sofuni A, Tsuchiya T, Ishii K, Tanaka R, Nagakawa Y, Mukai S. Deep learning analysis for the detection of pancreatic cancer on endosonographic images: a pilot study. J Hepatobiliary Pancreat Sci. 2021;28:95-104. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 79] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 86. | Iglesias-Garcia J, Larino-Noia J, Abdulkader I, Forteza J, Dominguez-Munoz JE. EUS elastography for the characterization of solid pancreatic masses. Gastrointest Endosc. 2009;70:1101-1108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 126] [Cited by in RCA: 124] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 87. | Iglesias-Garcia J, Larino-Noia J, Abdulkader I, Forteza J, Dominguez-Munoz JE. Quantitative endoscopic ultrasound elastography: an accurate method for the differentiation of solid pancreatic masses. Gastroenterology. 2010;139:1172-1180. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 202] [Cited by in RCA: 195] [Article Influence: 13.0] [Reference Citation Analysis (1)] |
| 88. | Săftoiu A, Vilmann P, Gorunescu F, Janssen J, Hocke M, Larsen M, Iglesias-Garcia J, Arcidiacono P, Will U, Giovannini M, Dietrich CF, Havre R, Gheorghe C, McKay C, Gheonea DI, Ciurea T; European EUS Elastography Multicentric Study Group. Efficacy of an artificial neural network-based approach to endoscopic ultrasound elastography in diagnosis of focal pancreatic masses. Clin Gastroenterol Hepatol. 2012;10:84-90.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149] [Cited by in RCA: 130] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 89. | Dietrich CF, Braden B, Hocke M, Ott M, Ignee A. Improvedharacterizationn of solitary solid pancreatic tumours using contrast enhanced transabdominal ultrasound. J Cancer Res Clin Oncol. 2008;134:635-643. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 59] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 90. | Săftoiu A, Vilmann P, Dietrich CF, Iglesias-Garcia J, Hocke M, Seicean A, Ignee A, Hassan H, Streba CT, Ioncică AM, Gheonea DI, Ciurea T. Quantitative contrast-enhanced harmonic EUS in differential diagnosis of focal pancreatic masses (with videos). Gastrointest Endosc. 2015;82:59-69. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 112] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 91. | Moris D, Damaskos C, Spartalis E, Papalampros A, Vernadakis S, Dimitroulis D, Griniatsos J, Felekouras E, Nikiteas N. Updates and Critical Evaluation on Novel Biomarkers for the Malignant Progression of Intraductal Papillary Mucinous Neoplasms of the Pancreas. Anticancer Res. 2017;37:2185-2194. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 29] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 92. | Shimizu Y, Hijioka S, Hirono S, Kin T, Ohtsuka T, Kanno A, Koshita S, Hanada K, Kitano M, Inoue H, Itoi T, Ueki T, Matsuo K, Yanagisawa A, Yamaue H, Sugiyama M, Okazaki K. New Model for Predicting Malignancy in Patients With Intraductal Papillary Mucinous Neoplasm. Ann Surg. 2020;272:155-162. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 52] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 93. | Shimizu Y, Yamaue H, Maguchi H, Yamao K, Hirono S, Osanai M, Hijioka S, Hosoda W, Nakamura Y, Shinohara T, Yanagisawa A. Predictors of malignancy in intraductal papillary mucinous neoplasm of the pancreas: analysis of 310 pancreatic resection patients at multiple high-volume centers. Pancreas. 2013;42:883-888. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 102] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 94. | Kuwahara T, Hara K, Mizuno N, Okuno N, Matsumoto S, Obata M, Kurita Y, Koda H, Toriyama K, Onishi S, Ishihara M, Tanaka T, Tajika M, Niwa Y. Usefulness of Deep Learning Analysis for the Diagnosis of Malignancy in Intraductal Papillary Mucinous Neoplasms of the Pancreas. Clin Transl Gastroenterol. 2019;10:1-8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 130] [Article Influence: 26.0] [Reference Citation Analysis (0)] |
| 95. | Zhang J, Zhu L, Yao L, Ding X, Chen D, Wu H, Lu Z, Zhou W, Zhang L, An P, Xu B, Tan W, Hu S, Cheng F, Yu H. Deep learning-based pancreas segmentation and station recognition system in EUS: development and validation of a useful training tool (with video). Gastrointest Endosc. 2020;92:874-885.e3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 70] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 96. | Keller DS, Berho M, Perez RO, Wexner SD, Chand M. The multidisciplinary management of rectal cancer. Nat Rev Gastroenterol Hepatol. 2020;17:414-429. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 202] [Article Influence: 40.4] [Reference Citation Analysis (0)] |
| 97. | Alzahrani SM, Al Doghaither HA, Al-Ghafari AB. General insight into cancer: An overview of colorectal cancer (Review). Mol Clin Oncol. 2021;15:271. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 98] [Article Influence: 24.5] [Reference Citation Analysis (0)] |
| 98. | Greene FL. Tumor deposits in colorectal cancer: a moving target. Ann Surg. 2012;255:214-215. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 13] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 99. | Tong LL, Gao P, Wang ZN, Song YX, Xu YY, Sun Z, Xing CZ, Xu HM. Is the seventh edition of the UICC/AJCC TNM staging system reasonable for patients with tumor deposits in colorectal cancer? Ann Surg. 2012;255:208-213. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 70] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 100. | Nagtegaal ID, Knijn N, Hugen N, Marshall HC, Sugihara K, Tot T, Ueno H, Quirke P. Tumor Deposits in Colorectal Cancer: Improving the Value of Modern Staging-A Systematic Review and Meta-Analysis. J Clin Oncol. 2017;35:1119-1127. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 196] [Article Influence: 21.8] [Reference Citation Analysis (1)] |
| 101. | Chen LD, Li W, Xian MF, Zheng X, Lin Y, Liu BX, Lin MX, Li X, Zheng YL, Xie XY, Lu MD, Kuang M, Xu JB, Wang W. Preoperative prediction of tumour deposits in rectal cancer by an artificial neural network-based US radiomics model. Eur Radiol. 2020;30:1969-1979. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 40] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 102. | Akkus Z, Cai J, Boonrod A, Zeinoddini A, Weston AD, Philbrick KA, Erickson BJ. A Survey of Deep-Learning Applications in Ultrasound: Artificial Intelligence-Powered Ultrasound for Improving Clinical Workflow. J Am Coll Radiol. 2019;16:1318-1328. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 161] [Article Influence: 32.2] [Reference Citation Analysis (0)] |