Published online Jan 21, 2022. doi: 10.3748/wjg.v28.i3.365
Peer-review started: November 26, 2021
First decision: December 12, 2021
Revised: December 14, 2021
Accepted: January 11, 2022
Article in press: January 11, 2022
Published online: January 21, 2022
Processing time: 48 Days and 10.5 Hours
The gastric microbiota in patients with gastric cancer (GC) has received increasing attention, but the profiling of the gastric microbiome through the histological stages of gastric tumorigenesis remains poorly understood, especially for patients with Helicobacter pylori-negative GC (HPNGC).
To characterize microbial profiles of gastric mucosa and juice for HPNGC carcinogenesis and identify distinct taxa in precancerous lesions.
The 16S rRNA gene analysis was performed on gastric mucosa from 134 Helicobacter pylori-negative cases, including 56 superficial gastritis (SG), 9 atrophic gastritis (AG), 27 intestinal metaplasia (IM), 29 dysplasia (Dys), and 13 GC cases, to investigate differences in gastric microbial diversity and composition across the disease stages. In addition, paired gastric mucosa and juice samples from 18 SG, 18 IM, and 18 Dys samples were analyzed. α-Diversity was measured by Shannon and Chao1 indexes, and β-diversity was calculated using partial least squares discrimination analysis (PLS-DA). Differences in the microbial composition across disease stages in different sample types were assessed using the linear discriminant analysis effect size.
The diversity and composition of the bacterial microbiota in the gastric mucosa changed progressively across stages of gastric carcinogenesis. The diversity of the gastric mucosa microbiota was found to be significantly lower in the IM and Dys groups than in the SG group, and the patients with GC had the lowest bacterial community richness (P < 0.05). Patients with IM and those with Dys had similar gastric mucosa microbiota profiles with Ralstonia and Rhodococcus as the predominant genera. Microbial network analysis showed that there was increasing correlation strength between IM and Dys (|correlation threshold|≥ 0.5, P < 0.05). GC and its precancerous lesions have distinguishable bacterial taxa; our results identified HPNGC-associated bacteria Streptococcaceae and Lactobacillaceae (P < 0.05). Additionally, across precancerous lesion stages from AG to Dys in Helicobacter pylori-negative patients, Burkholderiaceae abundance continuously increased, while Streptococcaceae and Prevotellaceae abundance presented a continuous downward trend. Furthermore, the microbial diversity was higher in gastric juice (P < 0.001) than in the mucosa, while PLS-DA revealed a statistically significant difference between the two groups (ANOSIM, P = 0.001). A significant difference in the microbial structure was identified, with Proteobacteria being more prevalent in the gastric mucosa and Firmicutes being more abundant in gastric juice.
Our results provide insights into potential taxonomic biomarkers for HPNGC and its precancerous stages and assist in predicting the prognosis of IM and Dys based on the mucosal microbiota profile.
Core Tip: The gastric microbiome profile of Helicobacter pylori-negative precancerous lesions is poorly understood. This is the first study, to our knowledge, to compare the microbiota differences between paired gastric mucosa and gastric juice at different stages of gastric neoplastic progression. The findings revealed that the bacterial community of gastric juice differed from that of the gastric mucosa and that Helicobacter pylori-negative gastric cancer and precancerous lesions have distinct bacterial taxa. Patients with intestinal metaplasia and dysplasia had similar gastric mucosa microbiota profiles, with Ralstonia and Rhodococcus being the most predominant genera, which could aid in prognosis prediction.
- Citation: Sun QH, Zhang J, Shi YY, Zhang J, Fu WW, Ding SG. Microbiome changes in the gastric mucosa and gastric juice in different histological stages of Helicobacter pylori-negative gastric cancers. World J Gastroenterol 2022; 28(3): 365-380
- URL: https://www.wjgnet.com/1007-9327/full/v28/i3/365.htm
- DOI: https://dx.doi.org/10.3748/wjg.v28.i3.365
Gastric cancer (GC) is one of the most common tumor types. Despite its declining prevalence, GC is the sixth most prevalent cancer worldwide, accounting for 8.2% of all cancer-related fatalities[1]. Helicobacter pylori (H. pylori) infection is one of the major carcinogens associated with GC[2]. In the etiology of GC, H. pylori is the most important pathogen in the development of GC due to atrophic gastritis (AG), which mostly results in intestinal-type GC and non-AG, which primarily results in diffuse-type GC[3]. Gastric adenocarcinoma is a complex disease associated with several different risk factors. Approximately 30% of stomach malignancies are not caused by H. pylori infection[4]. Heterogeneity is influenced by factors such as demographic characteristics, lifestyle, excessive salt and nitrate diet, race, and genetic variables[5-9].
Even though H. pylori is recognized as a class I carcinogen by the International Agency for Research on Cancer because of its association with GC, an H. pylori-negative subgroup does exist[10]. The proportion of H. pylori-negative GC (HPNGC) among patients with GC varies from 0.7% to 47.8% in previous reports, and a possible poorer prognosis might exist in HPNGC[11-14].
It is gradually accepted that the stomach does indeed host a robust microbiota due to breakthroughs in PCR and metagenomics methods[15]. An increasing number of studies on the link between the gastric microbiota and GC have been spurred by these technological advancements. The majority of GC cases are the intestinal type of non-cardia GC, which develops from AG to intestinal metaplasia (IM) and to GC via predictable progression[16]. Gastric microbiota diversity has been characterized by the severity of phenotypes, including SG, AG, IM, and GC, in many studies[15,17-19].
However, it remains unclear whether there is a correlation between the diversity of gastric microbiota and the development of gastric carcinogenesis. There is currently no consensus on the relationship between microbiota diversity and GC development stage, despite the fact that several studies have used similar methods of data collection, exclusion criteria, molecular methods for analysis, and similar measures for diversity (via Shannon's diversity index or Chao1 richness estimator). The majority of studies investigating this problem have used gene sequencing on mucosal biopsy samples collected by upper endoscopy to examine the gastric microbiota of patients with conditions ranging from normal gastric mucosa to GC[17,20-22].
Until recently, although the gastric microbiota in patients with GC has received increasing attention, only a limited number of studies have focused on patients with HPNGC and research on gastric juice microbiota between precancerous disease progression has remained relatively scarce. Prioritizing patients with HPNGC and analyzing gastric juice samples will help fill the gap in our understanding of GC. Therefore, our study focused on H. pylori-negative patients and performed 16S rRNA gene analysis of gastric mucosal and juice samples to determine gastric microbiome dysbiosis across stages of HPNGC and the differences in bacterial communities between gastric mucosa and juice.
Patients were recruited from the Department of Gastroenterology of Peking University Third Hospital between September 2019 and October 2020 during upper gastroenterology endoscopic examination or endoscopic submucosal dissection[A1] due to precancerous mucosal lesions. Written informed consent was obtained from all subjects in this study. This study was performed in accordance with the Declaration of Helsinki of the World Medical Association and was approved by the Peking University Third Hospital Medical Ethics Committee (No. IRB00006761-M2017414).
The inclusion criteria were as follows: (1) Age > 18 years; and (2) biopsy specimens and gastric juice. The exclusion criteria were: (1) Present use of antibiotics, antacids, probiotics, and prebiotics or within the last month before gastroscopy; (2) Previous gastric surgery; (3) Use of immunosuppressants; (4) Comorbidity and complications with serious heart, liver, lung, kidney, blood, endocrine, nervous system, or autoimmune diseases; (5) Bile reflux gastritis, gastroesophageal reflux disease, gastroduodenal or esophagus ulcer, or colorectal cancer; (6) A positive test for human immunodeficiency virus or hepatitis B or C virus; and (7) pregnancy or lactation. Experienced endoscopists performed all endoscopic examinations and obtained biopsy specimens and gastric juice. Demographic information, medical history, medication use, and dietary habits were collected from all subjects.
Gastric mucosal biopsy samples of 1-2 mm were obtained using standard gastroscopic forceps. A biopsy for histologic examination was performed based on the disease condition and as needed. The gastric biopsy samples for histological examination were fixed in 10% formalin and placed in separate vials, which were labeled according to their topographic site. Additional mucosal biopsy specimens were taken from the gastric antrum for microbial analysis. The biopsy specimens for microbial analysis were immediately frozen in liquid nitrogen, transferred to the laboratory, and stored at −80°C until DNA extraction. Gastric juice was drained in a sterile drainage tube at the beginning of the endoscopy. Then, the mucous material was removed by centrifugation at 4000 rpm [A2] for 10 min at 4°C, and samples were stored at -80°C until DNA extraction.
Two pathologists reviewed the gastric mucosa specimens separately according to the criteria proposed by the Chinese Association of Gastric Cancer[23] and the Updated Sydney System[24]. The diagnosis and classification of dysplasia (Dys) were determined using the revised Vienna Classification System[25]. GC was confirmed to have gastric adenocarcinoma and was divided into diffuse, intestinal, and mixed types according to the Lauren Classification. Each biopsy was diagnosed as non-atrophic superficial gastritis (SG), chronic AG, IM, or Dys based on the most severe histology. Improved Warthin-Starry (W-S) silver staining was performed on each gastric mucosa specimen. Both positive 13C-urea breath test and positive W-S staining identified the specimens as H. pylori-positive; otherwise, they were preliminarily identified as negative.
Microbial genomic DNA was isolated using the E.Z.N.A® Soil DNA Kit (Omega Bio-tek, Norcross, GA, United States) according to the manufacturer’s instructions. The V3-V4 hypervariable regions of the bacterial 16S ribosomal RNA gene were amplified using the primers 338 F (5’-ACTCCTACGGGAGGCAGCAG-3’) and 806R (5’-GGACTACHVGGGTWTCTAAT-3’). The PCR cycling conditions were as follows: initial denaturation at 95°C for 3 min, followed by 27 cycles of denaturation at 95°C for 30 s, annealing at 55°C for 30 s, extension at 72°C for 45 s, and a final extension at 72°C for 10 min.
Raw reads of 16S rRNA gene sequences were de-multiplexed and quality-filtered using the Quantitative Insights Into Microbial Ecology (QIIME) platform[26]. Sequences were then clustered into OTUs based on 97% similarity. Using the Ribosomal Data Project Bayesian Classifier in QIIME, operational taxonomic units (OTUs) were assigned to phyla, classes, orders, families, and genera, and their relative abundances were calculated[27].
Bioinformatics analyses were performed using the Majorbio cloud platform. The read counts were normalized using the total sum normalization. Based on the normalized OTU abundance profile, microbial alpha diversity was measured using the Shannon and Chao1 indices. Alpha diversity indices were compared by one-way analysis of variance (ANOVA) followed by false discovery rate (FDR) correction. The dissimilarity of the microbial communities among groups was evaluated by partial least squares discrimination analysis (PLS-DA) using R software. Sample clustering in beta diversity analysis was tested using analysis of similarity (ANOSIM) using the vegan package in R software. Relative bacterial abundances were analyzed using the Kruskal-Wallis test with FDR correction for multiple testing. The key bacterial genera responsible for discrimination between different groups were identified using the linear discriminant analysis (LDA) effect size (LEfSe) algorithm. LDA > 3.5 and P < 0.05 indicated significantly enriched microbial communities[28]. The microbiome analyst platform was used to explore and visualize the associations between the core microbes. Heatmaps were generated according to the relative abundance of taxa using R software (http://www.R-project.org).
Spearman correlation analysis was performed to calculate the correlation coefficients (r values) between specific disease-related genera in the gastric mucosa. Two genera were connected by an edge if the correlation between them meets the P value (P < 0.05) and correlation threshold (|correlation threshold| ≥ 0.5) cut-off. The Kruskal–Wallis test was conducted to compare the interaction strengths between the different gastric lesion groups. Statistical significance was set at P < 0.05.
Statistical analysis was performed using GraphPad Prism 8 (GraphPad Software Inc., La Jolla, CA, United States). Data are presented as the mean ± SEM. ANOVA was used to compare differences among groups, followed by FDR correction for multiple comparisons. Statistical significance was set at P < 0.05. Correlation coefficients between disease phenotype parameters and alterations in microbial taxa were analyzed using Spearman’s correlation analysis.
A total of 183 patients were included, including 83 patients with SG, 21 with AG, 33 with IM, 34 with Dys, and 15 with GC according to the pathological report. Furthermore, samples with < 1% H. pylori relative abundance were grouped as H. pylori-negative, while those with > 1% H. pylori relative abundance were grouped as H. pylori-positive[18]. According to this standard, 56 SG, 9 AG, 27 IM, 29 Dys, and 13 GC were confirmed as H. pylori-negative and enrolled in our cohort. The demographic characteristics of the subjects are shown in Supplementary Table 1.
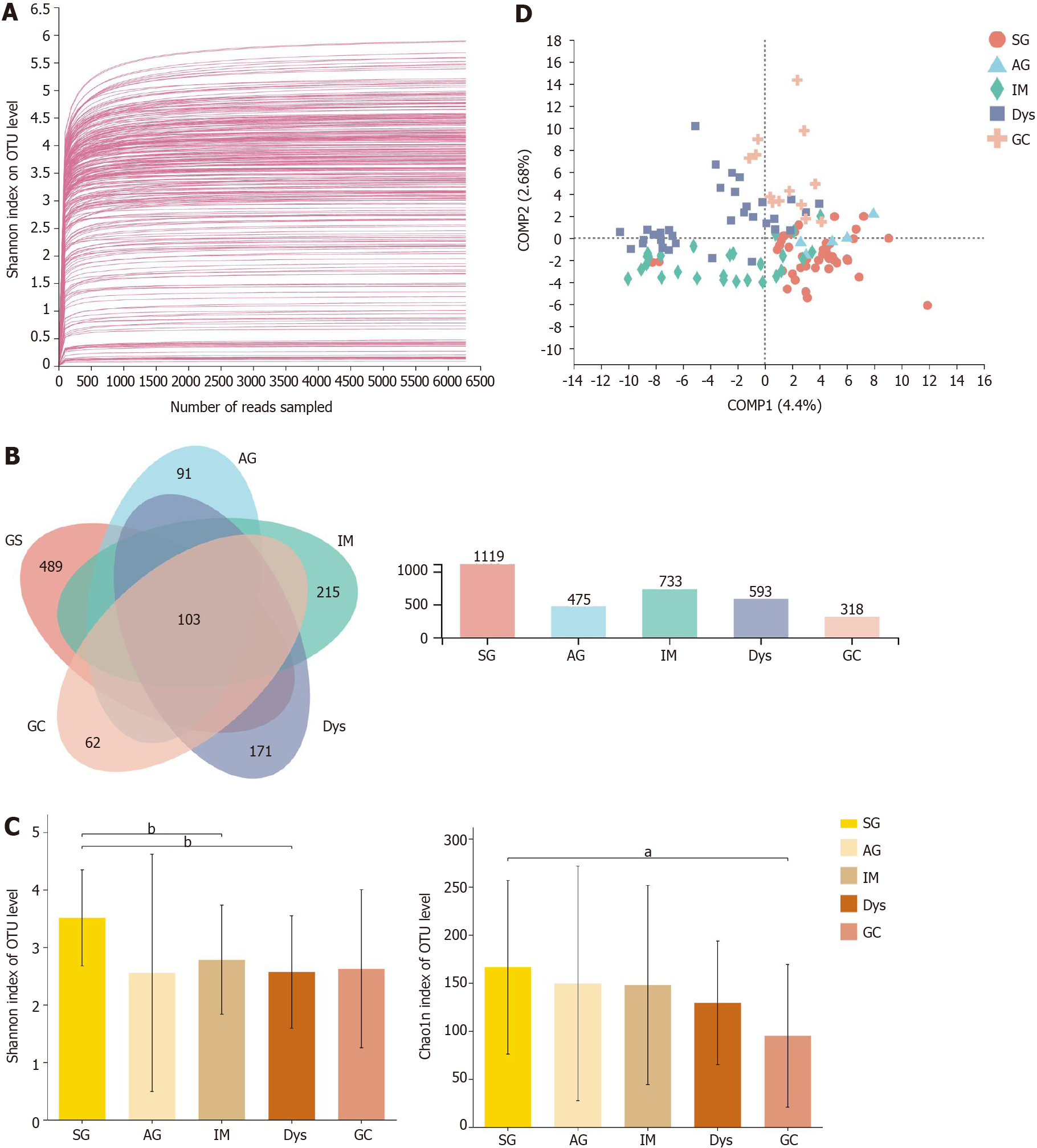
After sequencing and quality filtering, a total of 11699206 high-quality reads were generated from all samples. The average length of the sequences was 433 bp. The data were rarefied to 7234 sequences per sample to control for variations in sequencing efforts and clustered into 2296 OTUs at 97% sequence similarity. First, to test the sequencing depth, rarefaction curves were drawn, and the sequencing data volume was sufficient (Figure 1A). The generated Venn diagram showed that 103 OTUs were shared by five groups, with 489, 91, 215, 171, and 62 OTUs unique to the SG, AG, IM, Dys, and GC groups, respectively (Figure 1B). The Shannon and Chao1 indices were used to describe the α-diversity of the gastric bacterial community. The diversity and richness of the microbial community showed a declining trend across stages of gastric carcinogenesis, from SG, AG, IM, and Dys to GC. The diversity of microbiota was significantly higher in the SG group than in the IM and Dys groups (Shannon index, P = 0.003 and 0.001, respectively), and the richness of the microbiota was significantly higher in the SG group than in the GC group (Chao1 index, P = 0.027, Figure 1C). The β-diversity analysis with PLS-DA based on the OTU level revealed a pattern in which the samples were assigned into four separate groups (ANOSIM, P = 0.005; Figure 1D). Provoked by this interesting pattern, we conducted hierarchical clustering analysis at the genus level. IM samples were divided into two condensed groups, and the same result was applied to the Dys samples (Supplementary Figure 1). The IM and Dys samples were regrouped based on a hierarchical clustering tree plot. Subgroups IM-1 and Dys-1 had a similar microbiota composition with a high relative abundance of Ralstonia (Supplementary Figure 2).
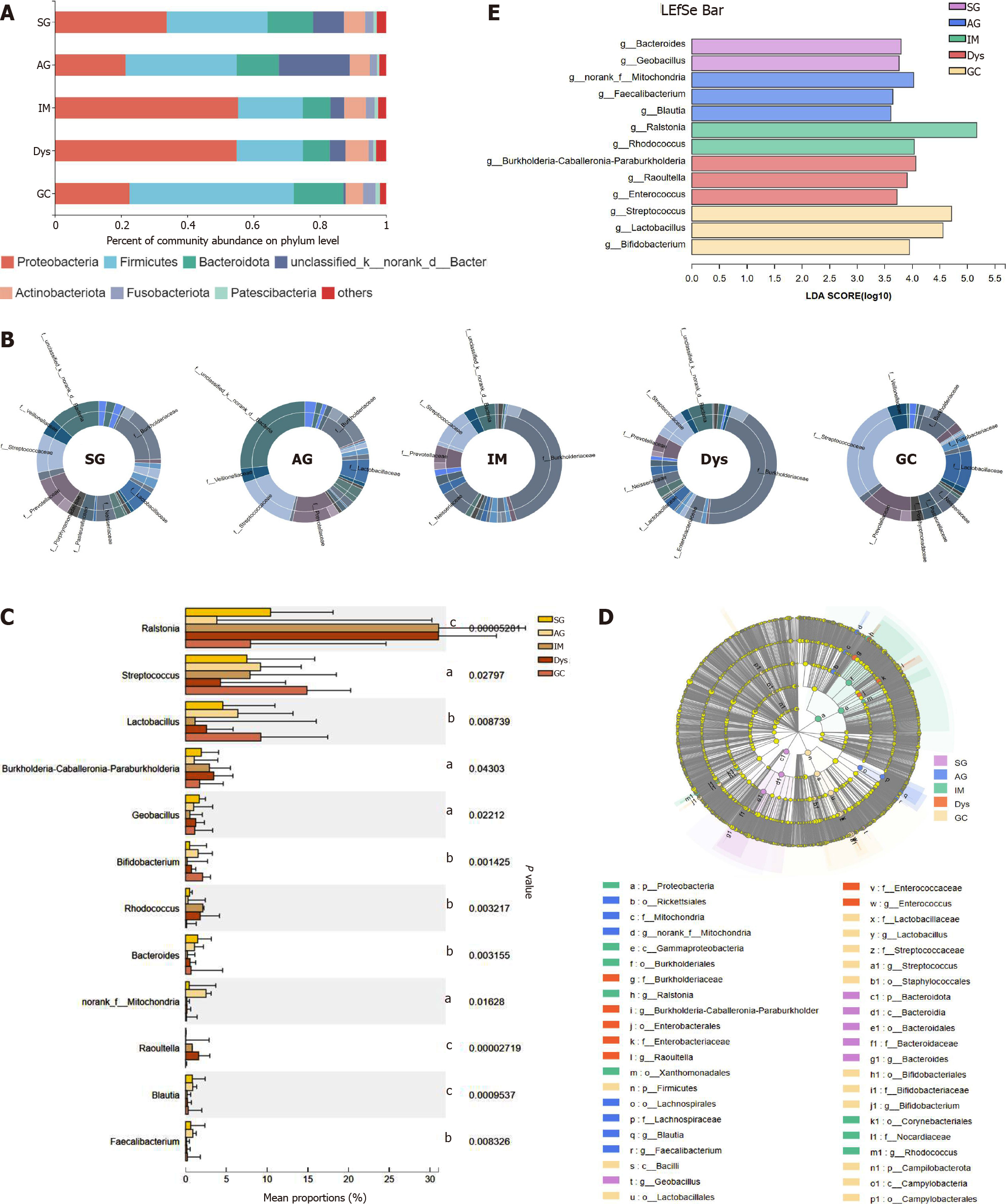
The differences in the gastric mucosa microbiota between each group were investigated at different taxonomic levels. The proportion of community abundance at the phylum level was calculated and is shown in Figure 2. Firmicutes, Bacteroidota, unclassified_k__norank_d__Bacteria, and Actinobacteria were the most predominant phyla, contributing to > 90% of the microbial composition of all groups. Both IM and Dys had higher abundances of Proteobacteria than the other disease stages (P < 0.001). The clusters of IM and Dys were close to each other, suggesting a similar gastric microbiota profile. Firmicutes was more abundant in patients with GC than in those with IM and Dys (P = 0.001 for both). Bacteroidetes was less abundant in the Dys group than in the SG group (P = 0.029) (Supplementary Table 2). The clusters of IM and Dys were close to each other, suggesting a similar gastric microbiota profile.
As shown by community analysis sunburst plots at the family level, Burkholderiaceae (12.44%), unclassified_k__norank_d__Bacteria (9.34%), Prevotellaceae (7.98%), and Streptococcaceae (7.54%) were more abundant in patients with SG. unclassified_k__nor-ank_d__Bacteria (26.29%), Prevotellaceae (9.42%), Streptococcaceae (9.21%), and Lactobacillaceae (6.42%) were the main communities in patients with AG. Burkholderiaceae (34.08%), Streptococcaceae (7.94%), Neisseriaceae (6.16%), and Prevotellaceae (5.34%) were more abundant in the IM group. Burkholderiaceae (34.59%), unclassified_k__nor-ank_d__Bacteria (4.73%), Prevotellaceae (4.52%), and Streptococcaceae (4.30%) were more abundant in patients with Dys. In the patients with GC, Streptococcaceae (23.92%), Prevotellaceae (11.11%), Lactobacillaceae (8.61%), and Burkholderiaceae (7.41%) were the dominant families. With the precancerous lesion stages from AG to Dys, Burkholderiaceae abundance continuously increased, while Streptococcaceae and Prevotellaceae presented a continuous trend of decline in abundance. Streptococcaceae and Lactobacillaceae abundance was significantly higher in the GC group than in the SG group (P < 0.05) (Figure 2B).
At the genus level, the top 12 genera that showed significant differences from each other were identified (Figure 2C). Taxonomic analysis indicated that the relative abundance of Ralstonia and Rhodococcus was significantly higher in patients with IM and Dys than in those with SG (Ralstonia: P = 0.008 and 0.004; Rhodococcus: P = 0.008 and 0.038, respectively). Streptococcus and Bifidobacterium abundance was significantly higher in patients with GC than in those with SG (P = 0.013 and 0.015, respectively). Raoultella abundance increased in patients with Dys, and norank_f__ mitochondria increased in patients with AG when compared to those with SG (P = 0.002 and 0.008, respectively) (Table 1).
| Relative abundance (%) | |||||||
| SG | AG | IM | Dys | GC | One-way ANOVA, | P value | |
| g__Ralstonia | 10.43 | 3.84 | 31.05 | 31.04 | 7.97 | 0.000 | 0.008b; |
| g__Streptococcus | 7.52 | 9.19 | 7.91 | 4.28 | 14.90 | 0.028 | 0.013d |
| g__Prevotella | 5.81 | 7.83 | 3.82 | 3.40 | 5.70 | 0.111 | |
| g__Lactobacillus | 4.56 | 6.42 | 1.16 | 2.57 | 9.23 | 0.009 | |
| g__Neisseria | 3.80 | 1.05 | 6.03 | 2.81 | 2.36 | 0.538 | |
| g__Veillonella | 3.15 | 3.00 | 2.31 | 1.49 | 4.53 | 0.497 | |
| g__Burkholderia-Caballeronia-Paraburkholderia | 1.93 | 1.06 | 2.93 | 3.45 | 1.72 | 0.043 | |
| g__Haemophilus | 2.95 | 0.78 | 2.08 | 1.32 | 2.33 | 0.540 | |
| g__Alloprevotella | 2.05 | 1.54 | 1.51 | 1.06 | 1.39 | 0.373 | |
| g__Acinetobacter | 1.74 | 0.96 | 1.12 | 1.78 | 1.93 | 0.716 | |
| g__Actinomyces | 1.42 | 2.80 | 1.04 | 0.92 | 1.22 | 0.364 | |
| g__Escherichia-Shigella | 1.86 | 1.12 | 1.11 | 1.70 | 1.47 | 0.193 | |
| g__Fusobacterium | 1.85 | 0.96 | 1.57 | 0.97 | 1.29 | 0.308 | |
| g__Pseudomonas | 1.56 | 0.47 | 0.88 | 1.65 | 1.70 | 0.128 | |
| g__Porphyromonas | 2.14 | 0.53 | 1.38 | 0.78 | 1.32 | 0.259 | |
| g__Geobacillus | 1.68 | 0.99 | 0.52 | 1.25 | 1.12 | 0.022 | |
| g__Bifidobacterium | 0.50 | 1.53 | 0.18 | 0.72 | 2.09 | 0.001 | 0.015d |
| g__Rhodococcus | 0.50 | 0.29 | 2.11 | 1.79 | 0.11 | 0.003 | 0.008b; |
| g__Gemella | 0.93 | 1.18 | 0.63 | 0.32 | 1.39 | 0.119 | |
| g__Delftia | 1.07 | 0.35 | 0.95 | 0.90 | 1.04 | 0.609 | |
| g__Granulicatella | 0.86 | 0.45 | 1.08 | 0.52 | 1.15 | 0.659 | |
| g__Bacteroides | 1.49 | 1.08 | 0.22 | 0.54 | 0.66 | 0.003 | |
| g__Leptotrichia | 0.66 | 0.73 | 1.02 | 0.49 | 0.65 | 0.689 | |
| g__norank_f__Mitochondria | 0.44 | 2.51 | 0.15 | 0.16 | 0.10 | 0.016 | 0.008a |
| g__Rothia | 0.77 | 0.70 | 1.00 | 0.28 | 0.56 | 0.762 | |
| g__unclassified_p__Proteobacteria | 0.49 | 1.03 | 0.64 | 0.28 | 0.12 | 0.195 | |
| g__Raoultella | 0.03 | 0.02 | 0.81 | 1.60 | 0.00 | 0.000 | 0.002c |
| g__Sphingomonas | 0.54 | 0.17 | 0.33 | 0.56 | 0.81 | 0.742 | |
| g__Blautia | 0.83 | 0.86 | 0.18 | 0.23 | 0.32 | 0.001 | |
| g__Clostridium_sensu_stricto_1 | 0.24 | 0.43 | 0.30 | 0.81 | 0.54 | 0.292 | |
| g__TM7x | 0.57 | 0.48 | 0.63 | 0.28 | 0.26 | 0.269 | |
| g__Corynebacterium | 0.64 | 0.16 | 0.24 | 0.64 | 0.45 | 0.777 | |
| g__norank_f__norank_o__Chloroplast | 0.63 | 0.53 | 0.33 | 0.22 | 0.30 | 0.924 | |
| g__Faecalibacterium | 0.62 | 0.88 | 0.11 | 0.16 | 0.21 | 0.008 | |
LEfSe analysis was used to identify the most relevant taxa responsible for the differences among disease stages. An LDA cutoff score of 3.5 was used to estimate the discriminatory impact of each community on the phylogenetic distribution. A total of 42 taxa were identified as key participants in the five groups (Figure 2D). Figure 2E shows the most relevant taxa responsible for the differences among disease stages at the genus level, with Bacteroides and Geobacillus identified in the SG group; Faecalibacterium, Blautia, and norank_f__Mitochondria in the AG group; Rhodococcus and Ralstonia in the IM group; Enterococcus, Burkholderia-Caballeronia-Paraburkholderia, and Raoultella in the Dys group; and Lactobacillus, Bifidobacterium, and Streptococcus in the GC group (Figure 2D).
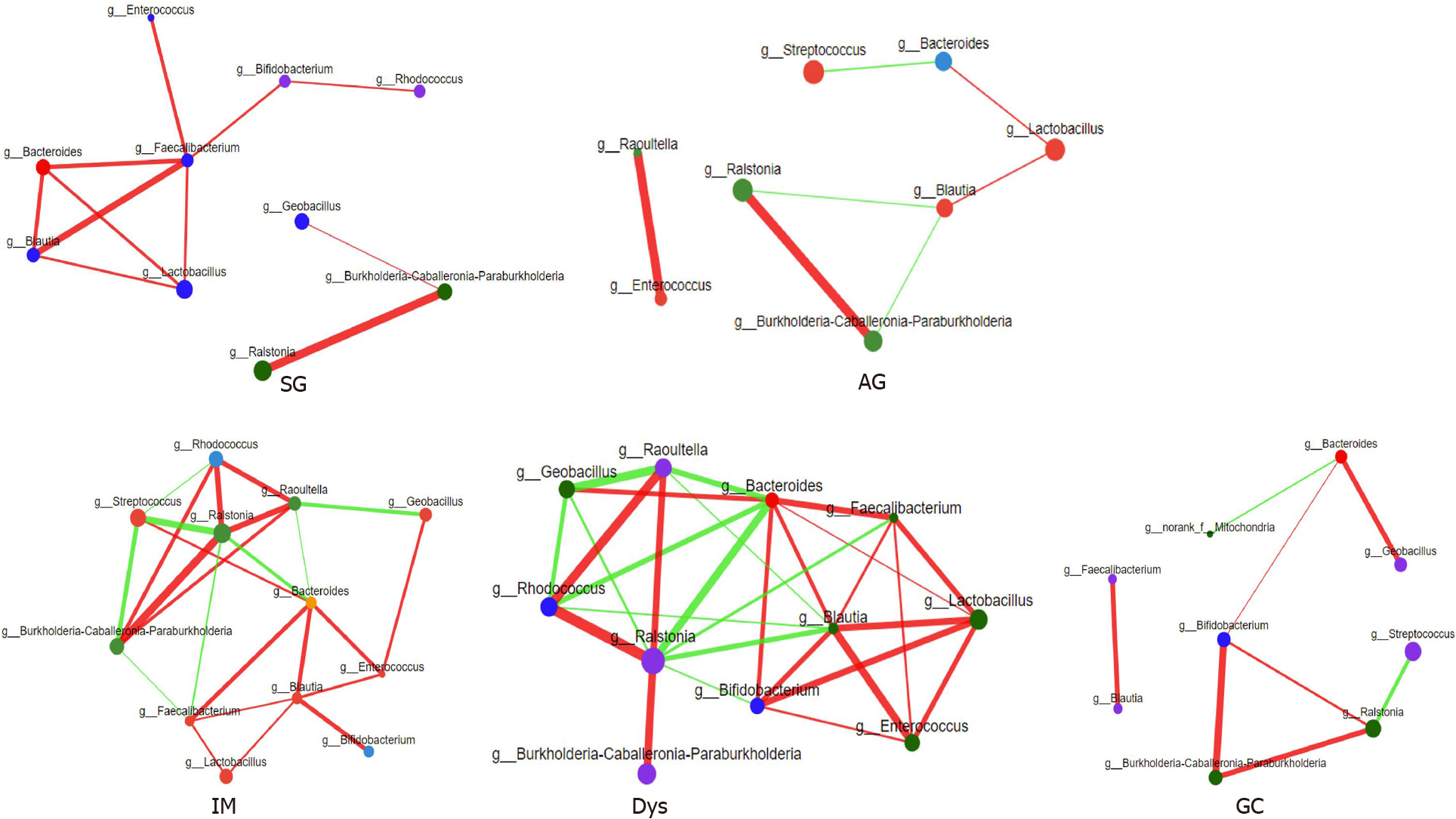
The relative abundance of the same 13 genera (the most relevant taxa responsible for the differences among disease stages are presented in Figure 2D) was compared among different gastric lesion groups. A network diagram was drawn based on the correlation between the genera to reflect the interactions between samples. The sizes of the nodes in the figure indicate the abundance of genera. The red color indicates a positive correlation, and green indicates a negative correlation. The thicker the line, the stronger the correlation between the genera. These results were used to visualize and identify possible associations among the important taxa. The IM and Dys groups had more complex interactions than the SG and GC groups (Figure 3). The transitivity, diameter, and average shortest path length of the different histological stages are shown in Supplementary Table 3.
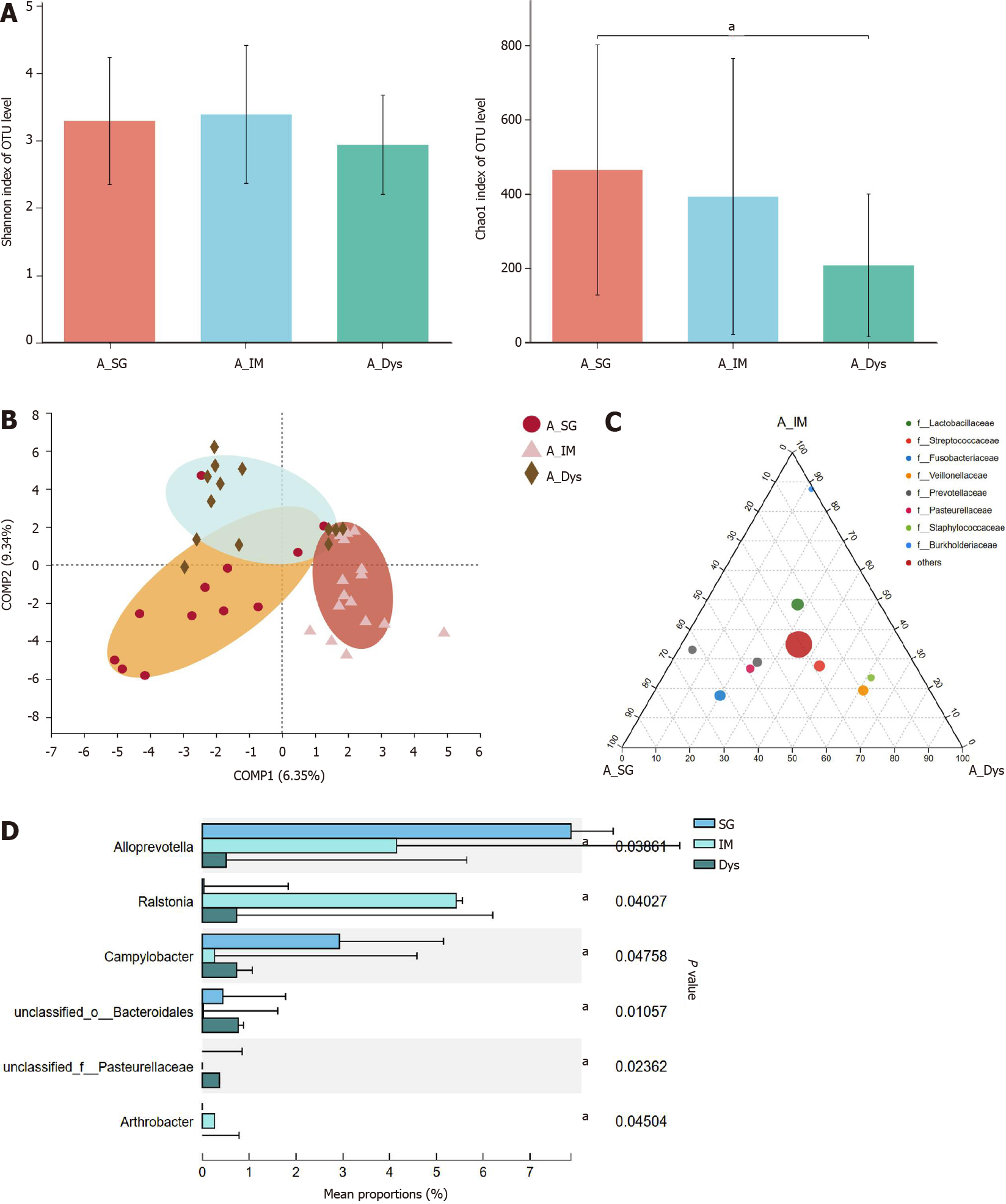
Paired-gastric juice and mucosa from 18 SG, 18 IM, and 18 Dys patients were analyzed. In the first step, we analyzed the gastric juice from patients with SG, IM, and Dys. The richness of the microbiota was significantly higher in the SG group than in the Dys group (Chao1 index, P = 0.025), but there were no significant differences in the diversity of microbiota between these groups (Figure 4A). Although β-diversity analysis with PLS-DA based on the OTU level revealed a pattern with three clusters, ANOSIM showed that the clusters for the three groups were not significantly different (P = 0.230; Figure 4B). As shown by ternary analysis at the family level, Burkholderiaceae was more abundant in the IM group, Fusobacteriaceae and Prevotellaceae were more abundant in the SG group, and Veillonellaceae and Staphylococcaceae were more abundant in the Dys group (Figure 4C). At the genus level, taxonomic analysis indicated that the relative abundance of Alloprevotella in Dys was significantly decreased, while that in IM and Campylobacter abundance in SG were significantly increased (P < 0.05, Figure 4D).
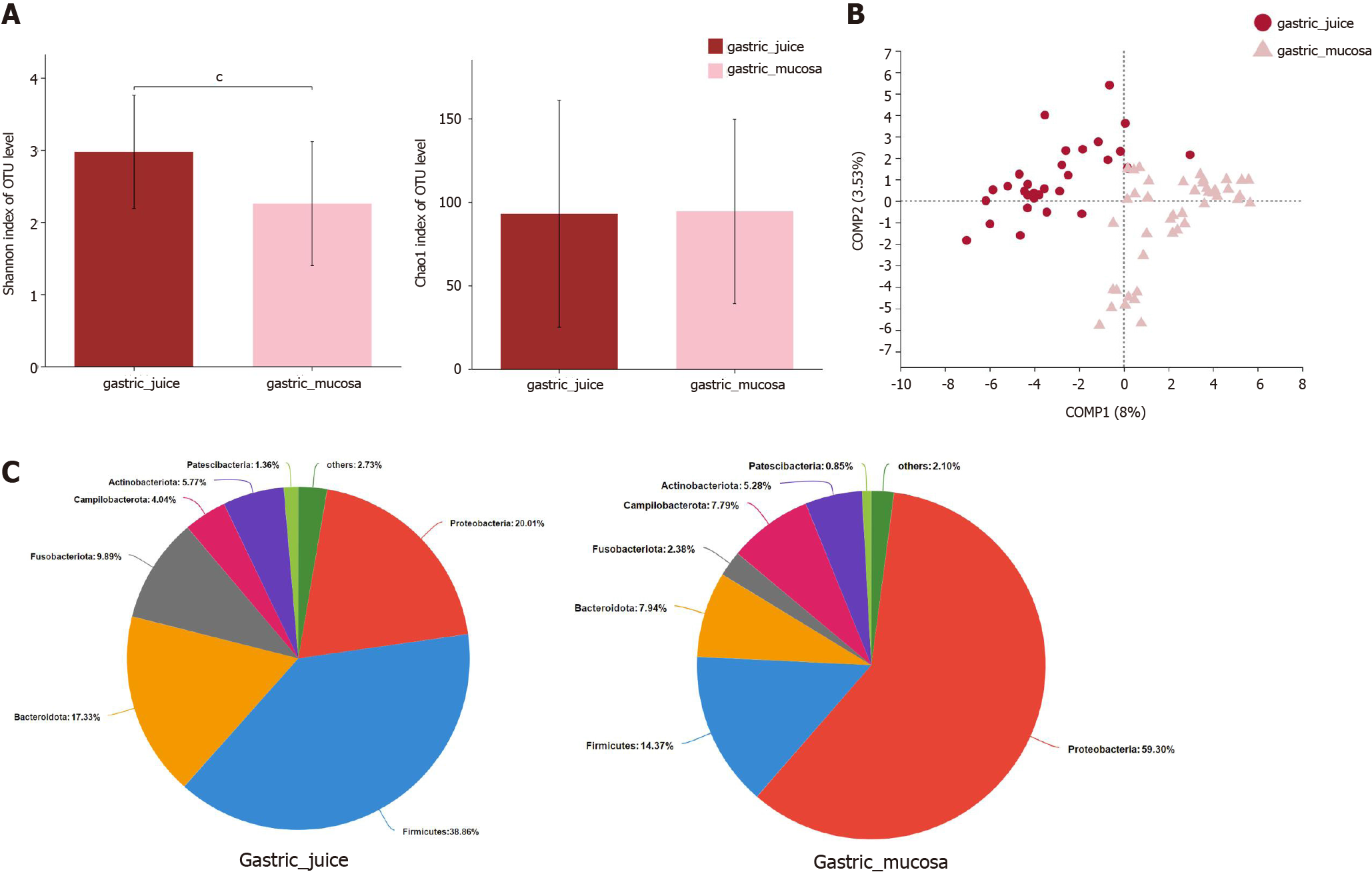
Next, the microbial α-diversity and β-diversity were measured to analyze the differences in the microbiota structure between the gastric mucosa and juice. We found that the microbial community diversity was significantly higher in gastric juice (P < 0.001), while there was no significant difference in microbial community richness between the two groups (Figure 5A). PLS-DA at the OTU level revealed a statistically significant separation of the groups (ANOSIM, P = 0.001; Figure 5B), suggesting different microbial community structures. Proteobacteria (59.30%), Firmicutes (14.37%), and Bacteroidetes (7.94%) were three of the most predominant phyla in the gastric mucosa, while Firmicutes (38.86%), Proteobacteria (20.01%), and Bacteroidetes (17.33%) were the top three most abundant phyla in gastric juice (Figure 5C).
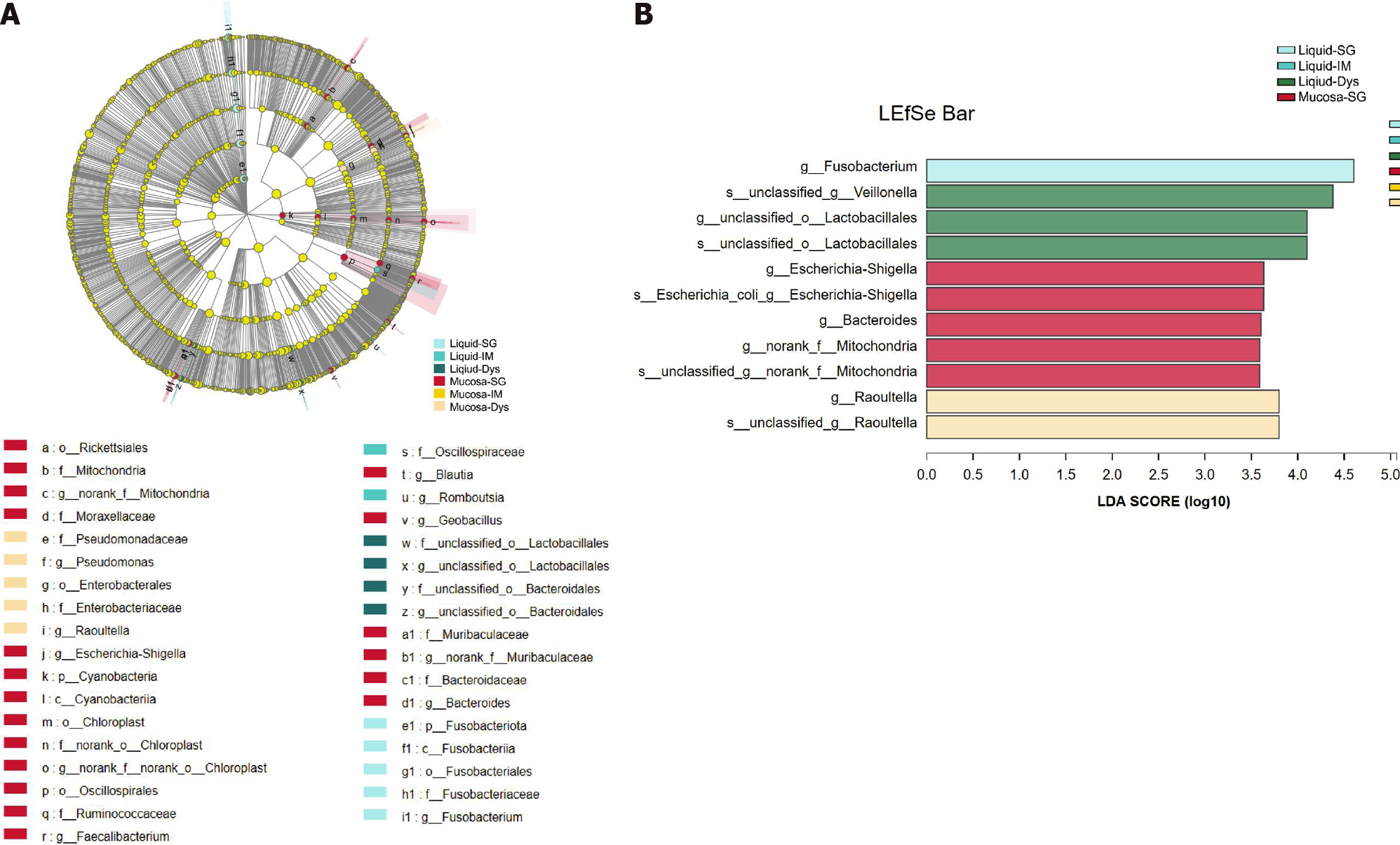
To assess the microbiota characteristics of different stomach microhabitats, we compared pairs of gastric juice and mucosa samples for each disease stage from patients with SG, IM, and Dys. LEfSe analysis was applied to identify the most relevant taxa responsible for the differences between gastric liquid and mucosa among the disease stages (Figure 6A). We focused on bacterial taxa with different abundances at the genus and species levels. In the gastric juice of patients with Dys, enrichment in the genera unclassified_o__Lactobacillales and Veillonella was observed. In the gastric mucosa group of patients with Dys, the enriched genera were Raoultella and Bacteroides. The SG-enriched genera in the gastric mucosa were Escherichia-Shigella and norank_f_Mitochondria (Figure 6B).
In recent years, many researchers and clinicians have explored the role of the microbiome in various disease processes, which has resulted in a significant surge in the number of studies on this topic[29]. Although the severely acidic conditions of the stomach have formerly hampered research into the gastric microbiota, studies on the gastric microbiota have risen over the past decade owing to the development of modern PCR techniques and metagenomic analyses. The majority of research has compared the gastric mucosal microbiota of GC to that of SG or healthy controls without distinguishing non-H. pylori-infected individuals from H. pylori-infected ones[30-32]. Additionally, the number of studies examining the gastric microbiota utilizing gastric juice samples is still limited, and well-designed comparative studies analyzing the link between the mucosal and luminal microbiota are even rarer. To bridge these gaps, we studied the mucosal bacterial community from SG, AG, IM, Dys, and GC in H. pylori-negative patients as well as the bacterial composition of gastric juice and its deviations from the mucosal microbiota.
In this study, we discovered that the α-diversity of the gastric mucosa microbiota was significantly lower in the IM and Dys groups than in the SG group using the Shannon index, and that the bacterial community richness was lowest in the patients with GC, which is supported by earlier research results[19,30,32].
Our study's findings on microbial β-diversity revealed that the SG, IM, Dys, and GC groups could be distinguished from each other, which is in agreement with the results of previous studies showing that there is a shift in the composition of the microbial community along different histological phases of stomach neoplastic progression[18,33]. Notably, the AG group was intermingled with the SG group in the β-diversity plot (Figure 1D), indicating that the microbial profile of AG may be comparable to that of SG. Another possibility is that a comprehensive assessment of the microbiome in AG is not feasible owing to the sample size constraint. Another interesting finding was that patients with IM were dispersed throughout the SG and Dys as a bridge in the PLS-DA (Figure 1D). Similarly, the distribution region of β-diversity in the Dys group exhibited distinct characteristics: one portion heavily overlapped with the GC group, while the other part was near the IM group. Based on the above findings, we further plotted hierarchical clustering trees and found that there were two distinct clusters in the IM and Dys groups. We then investigated the bacterial composition of patients with IM and Dys and discovered that both groups could be divided into two sub
As precancerous lesions, IM and Dys have been considered intermediate stages between cancer and gastritis, and consecutive alterations in the microbiota may play a role in the progression of mucosal precancerous lesions. In clinical practice, it is challenging for digestive endoscopists to choose the appropriate interval and frequency of endoscopic follow-up for patients with IM or Dys. According to our findings, one possible solution to this problem is to use the microbiota profile of the gastric mucosa to determine whether a patient has a more cancer- or gastritis-like microbiota, allowing cancer-like patients to undergo more rigorous and frequent endoscopic monitoring or magnifying endoscopy because they may be at a higher risk of cancer. It might be more reasonable to assess gastric lesions by pathological reports combined with gastric microbiota profiles. Further research with a larger sample size is needed to validate this theory.
Streptococcus was found to be significantly more abundant in patients with GC than in those with SG, which is supported by the findings of multiple studies that used mucosal samples[33,34]. Interestingly, several investigations have indicated a large increase in Streptococcus abundance in malignant tissues compared to normal tissues from the same patients with GC, and similar findings have also been drawn using fecal samples, which made us more certain that Streptococcus should play an essential role throughout the process of gastric cancerization[35-37]. One plausible explanation is that Streptococcus antigens might induce cancer, since Streptococcus bovis has previously been shown to have such an effect in two studies using animal models[38,39].
In our study, the relative abundance of Alloprevotella was shown to decline significantly lower in patients with Dys than in those with SG at the genus level (P < 0.05, Figure 4D). This result is in-line with that of a recent study, which indicated that Alloprevotella levels are significantly lower in the IM/DYS group than in the normal/SG group[40]. Alloprevotella is known to have anti-inflammatory properties, which may explain this outcome to some degree[41,42]. In addition, Ralstonia abundance was found in our study to be significantly increased in the IM and Dys groups compared to in the SG group, which is consistent with the results of an earlier study[43]. Ralstonia has been shown to play a role in the initiation of inflammation, which explains why there was an increase in relative abundance[44]. In addition, Ralstonia and Helicobacter were verified as the top two genera of discriminant abundance in the stomachs of patients with GC, which warrants deeper analysis of the association between these two genera and GC[45].
In most clinical trials, intragastric bacterial overgrowth is examined using gastric juice culture and rarely via gastric mucosal tissue. Gastric juice samples are easier to collect, generally non-invasive compared to mucosal tissues, and exhibit integrated properties. They have been used to characterize the gastric microbiota in some studies[46,47]. In general, gastric juice samples include a combination of mucosal microbes and luminal communities[48], which have not been previously assessed in patients with GC. It has been demonstrated that oral or fecal commensal flora are usually found in the gastric juice of patients with GC[22], which indicates that there might be differences between the microbiota in gastric juice and mucosa. With respect to the influence of sample type, it was demonstrated in our study that the alterations of microbiota in gastric mucosa and gastric juice showed a discrepancy despite several earlier studies showing that microbial communities of different anatomical gastric positions are similar[18,20,36], which illustrated that gastric sample type may be a factor influencing research results, and that juice and mucosal samples should be treated separately. Future studies are still needed to confirm the differences between the mucosal microbiota and the gastral cavity microbiota.
It is a pity that only gastric juice samples in SG, IM, and Dys groups were available when samples were collected with a lack of data from the GC group. Another shortcoming is that the sample size of the AG groups was relatively small, which might not reflect the bacterial composition to the fullest. It has also been shown that tea drinking as well as fresh vegetable and fruit intake might play a role in slowing carcinogenic progression[49], which might have some influence on gastric microbiota. A detailed dietary questionnaire would ensure more rigorous and well-founded results. In-depth research on the pathogenic mechanisms of non-H. pylori bacteria in gastric carcinogenesis will be strongly desired in the future.
Our study showed a shift in the gastric microbial community structure along the SG-AG-IM-Dys-GC stages in the H. pylori-negative stages. The diversity and composition of the gastric mucosal microbiota altered gradually across the stages of gastric neoplastic progression. Patients with IM and Dys had similar gastric mucosa microbiota profiles, and their potential to be indicators of IM and Dys prognosis needs to be verified in further studies. Our findings also revealed that the bacterial community of gastric juice differed from that of the gastric mucosa, and that HPNGC and its precancerous lesions have distinct bacterial taxa. Streptococcaceae and Lactobacillaceae were enriched in HPNGC. In addition, from AG to Dys, Burkholderiaceae abundance increased continuously, while Streptococcaceae and Prevotellaceae presented a continuous downward trend in abundance, which suggested that Burkholderiaceae, Streptococcaceae, and Prevotellaceae might play different roles in the carcinogenesis of HPNGC.
The gastric microbiome through the histological stages of gastric tumorigenesis remains poorly understood, especially for the Helicobacter pylori-negative gastric cancer (HPNGC).
To get a better knowledge of gastric microbiota and to identify microbial indicators at different histological stages of gastric tumorigenesis.
To identify distinct taxa in precancerous lesions and describe microbial profiles of gastric mucosa and juice for HPNGC carcinogenesis.
We designed a clinical cohort study and utilized the 16S rRNA gene sequencing analysis.
Our study showed a change in the gastric microbial community structure along the precancerous lesions in the Helicobacter pylori-negative stages. Patients with intestinal metaplasia and dysplasia had similar gastric mucosa microbiota profiles, and their potential to be indicators for prognosis. Our findings revealed that the bacterial community of gastric juice differed from that of the gastric mucosa, and that HPNGC and its precancerous lesions have distinct bacterial taxa.
Using the gastric microbiota profile, we were able to identify possible taxonomic biomarkers for HPNGC and its precancerous phases, as well as help predict prognoses for intestinal metaplasia and dysplasia.
Our research revealed the core pathogenic bacteria in Helicobacter pylori-negative precancerous lesions, allowing for further investigation of the pathogenic process.
STROBE Statement: The guidelines of the STROBE Statement have been adopted.
Provenance and peer review: Unsolicited article; externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Huh YM, Hwang GS, Shin CM S-Editor: Liu JH L-Editor: A P-Editor: Liu JH
| 1. | Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53206] [Cited by in RCA: 55814] [Article Influence: 7973.4] [Reference Citation Analysis (132)] |
| 2. | Plummer M, Franceschi S, Vignat J, Forman D, de Martel C. Global burden of gastric cancer attributable to Helicobacter pylori. Int J Cancer. 2015;136:487-490. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 643] [Cited by in RCA: 696] [Article Influence: 69.6] [Reference Citation Analysis (0)] |
| 3. | Conteduca V, Sansonno D, Lauletta G, Russi S, Ingravallo G, Dammacco F. H. pylori infection and gastric cancer: state of the art (review). Int J Oncol. 2013;42:5-18. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 135] [Cited by in RCA: 162] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 4. | de Martel C, Ferlay J, Franceschi S, Vignat J, Bray F, Forman D, Plummer M. Global burden of cancers attributable to infections in 2008: a review and synthetic analysis. Lancet Oncol. 2012;13:607-615. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1583] [Cited by in RCA: 1718] [Article Influence: 132.2] [Reference Citation Analysis (1)] |
| 5. | Guilford P, Humar B, Blair V. Hereditary diffuse gastric cancer: translation of CDH1 germline mutations into clinical practice. Gastric Cancer. 2010;13:1-10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 113] [Cited by in RCA: 105] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 6. | Kamineni A, Williams MA, Schwartz SM, Cook LS, Weiss NS. The incidence of gastric carcinoma in Asian migrants to the United States and their descendants. Cancer Causes Control. 1999;10:77-83. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 78] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 7. | van Loon AJ, Botterweck AA, Goldbohm RA, Brants HA, van den Brandt PA. Nitrate intake and gastric cancer risk: results from the Netherlands cohort study. Cancer Lett. 1997;114:259-261. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 21] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 8. | Wang XQ, Terry PD, Yan H. Review of salt consumption and stomach cancer risk: epidemiological and biological evidence. World J Gastroenterol. 2009;15:2204-2213. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 114] [Cited by in RCA: 122] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 9. | Hamilton JP, Meltzer SJ. A review of the genomics of gastric cancer. Clin Gastroenterol Hepatol. 2006;4:416-425. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 78] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 10. | Wu MS, Hung HW, Wang JT, Tseng CC, Shun CT, Wang HP, Lee WJ, Lin JT. Helicobacter pylori-seronegative gastric carcinoma: a subset of gastric carcinoma with distinct clinicopathologic features. Hepatogastroenterology. 1998;45:2432-2436. [PubMed] |
| 11. | Marrelli D, Pedrazzani C, Berardi A, Corso G, Neri A, Garosi L, Vindigni C, Santucci A, Figura N, Roviello F. Negative Helicobacter pylori status is associated with poor prognosis in patients with gastric cancer. Cancer. 2009;115:2071-2080. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 80] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 12. | Meimarakis G, Winter H, Assmann I, Kopp R, Lehn N, Kist M, Stolte M, Jauch KW, Hatz RA. Helicobacter pylori as a prognostic indicator after curative resection of gastric carcinoma: a prospective study. Lancet Oncol. 2006;7:211-222. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 83] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 13. | Yoon H, Kim N, Lee HS, Shin CM, Park YS, Lee DH, Jung HC, Song IS. Helicobacter pylori-negative gastric cancer in South Korea: incidence and clinicopathologic characteristics. Helicobacter. 2011;16:382-388. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 72] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 14. | Qiu HB, Zhang LY, Keshari RP, Wang GQ, Zhou ZW, Xu DZ, Wang W, Zhan YQ, Li W. Relationship between H.Pylori infection and clinicopathological features and prognosis of gastric cancer. BMC Cancer. 2010;10:374. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 47] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 15. | Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, Mende DR, Li J, Xu J, Li S, Li D, Cao J, Wang B, Liang H, Zheng H, Xie Y, Tap J, Lepage P, Bertalan M, Batto JM, Hansen T, Le Paslier D, Linneberg A, Nielsen HB, Pelletier E, Renault P, Sicheritz-Ponten T, Turner K, Zhu H, Yu C, Jian M, Zhou Y, Li Y, Zhang X, Qin N, Yang H, Wang J, Brunak S, Doré J, Guarner F, Kristiansen K, Pedersen O, Parkhill J, Weissenbach J; MetaHIT Consortium, Bork P, Ehrlich SD, Wang J. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59-65. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9101] [Cited by in RCA: 7837] [Article Influence: 522.5] [Reference Citation Analysis (4)] |
| 16. | Park YH, Kim N. Review of atrophic gastritis and intestinal metaplasia as a premalignant lesion of gastric cancer. J Cancer Prev. 2015;20:25-40. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 164] [Cited by in RCA: 206] [Article Influence: 20.6] [Reference Citation Analysis (0)] |
| 17. | Wang L, Zhou J, Xin Y, Geng C, Tian Z, Yu X, Dong Q. Bacterial overgrowth and diversification of microbiota in gastric cancer. Eur J Gastroenterol Hepatol. 2016;28:261-266. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 171] [Cited by in RCA: 158] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 18. | Coker OO, Dai Z, Nie Y, Zhao G, Cao L, Nakatsu G, Wu WK, Wong SH, Chen Z, Sung JJY, Yu J. Mucosal microbiome dysbiosis in gastric carcinogenesis. Gut. 2018;67:1024-1032. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 304] [Cited by in RCA: 476] [Article Influence: 68.0] [Reference Citation Analysis (0)] |
| 19. | Park CH, Lee AR, Lee YR, Eun CS, Lee SK, Han DS. Evaluation of gastric microbiome and metagenomic function in patients with intestinal metaplasia using 16S rRNA gene sequencing. Helicobacter. 2019;24:e12547. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 74] [Cited by in RCA: 65] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 20. | Bik EM, Eckburg PB, Gill SR, Nelson KE, Purdom EA, Francois F, Perez-Perez G, Blaser MJ, Relman DA. Molecular analysis of the bacterial microbiota in the human stomach. Proc Natl Acad Sci U S A. 2006;103:732-737. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 730] [Cited by in RCA: 779] [Article Influence: 41.0] [Reference Citation Analysis (0)] |
| 21. | Dicksved J, Lindberg M, Rosenquist M, Enroth H, Jansson JK, Engstrand L. Molecular characterization of the stomach microbiota in patients with gastric cancer and in controls. J Med Microbiol. 2009;58:509-516. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 211] [Cited by in RCA: 242] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 22. | Castaño-Rodríguez N, Goh KL, Fock KM, Mitchell HM, Kaakoush NO. Dysbiosis of the microbiome in gastric carcinogenesis. Sci Rep. 2017;7:15957. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 109] [Cited by in RCA: 182] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 23. | You WC, Blot WJ, Li JY, Chang YS, Jin ML, Kneller R, Zhang L, Han ZX, Zeng XR, Liu WD. Precancerous gastric lesions in a population at high risk of stomach cancer. Cancer Res. 1993;53:1317-1321. [PubMed] |
| 24. | Dixon MF, Genta RM, Yardley JH, Correa P. Classification and grading of gastritis. The updated Sydney System. International Workshop on the Histopathology of Gastritis, Houston 1994. Am J Surg Pathol. 1996;20:1161-1181. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3221] [Cited by in RCA: 3552] [Article Influence: 122.5] [Reference Citation Analysis (3)] |
| 25. | Dixon MF. Gastrointestinal epithelial neoplasia: Vienna revisited. Gut. 2002;51:130-131. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 445] [Cited by in RCA: 498] [Article Influence: 21.7] [Reference Citation Analysis (0)] |
| 26. | Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335-336. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29318] [Cited by in RCA: 23737] [Article Influence: 1582.5] [Reference Citation Analysis (0)] |
| 27. | Kuczynski J, Stombaugh J, Walters WA, González A, Caporaso JG, Knight R. Using QIIME to analyze 16S rRNA gene sequences from microbial communities. Curr Protoc Bioinformatics. 2011;Chapter 10:Unit 10.7.. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 391] [Cited by in RCA: 397] [Article Influence: 28.4] [Reference Citation Analysis (0)] |
| 28. | Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11842] [Cited by in RCA: 10310] [Article Influence: 736.4] [Reference Citation Analysis (0)] |
| 29. | Stewart OA, Wu F, Chen Y. The role of gastric microbiota in gastric cancer. Gut Microbes. 2020;11:1220-1230. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 125] [Article Influence: 25.0] [Reference Citation Analysis (0)] |
| 30. | Wang Z, Gao X, Zeng R, Wu Q, Sun H, Wu W, Zhang X, Sun G, Yan B, Wu L, Ren R, Guo M, Peng L, Yang Y. Changes of the Gastric Mucosal Microbiome Associated With Histological Stages of Gastric Carcinogenesis. Front Microbiol. 2020;11:997. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 94] [Article Influence: 18.8] [Reference Citation Analysis (0)] |
| 31. | Gantuya B, El Serag HB, Matsumoto T, Ajami NJ, Uchida T, Oyuntsetseg K, Bolor D, Yamaoka Y. Gastric mucosal microbiota in a Mongolian population with gastric cancer and precursor conditions. Aliment Pharmacol Ther. 2020;51:770-780. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 81] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 32. | Wu ZF, Zou K, Wu GN, Jin ZJ, Xiang CJ, Xu S, Wang YH, Wu XY, Chen C, Xu Z, Li WS, Yao XQ, Zhang JF, Liu FK. A Comparison of Tumor-Associated and Non-Tumor-Associated Gastric Microbiota in Gastric Cancer Patients. Dig Dis Sci. 2021;66:1673-1682. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 31] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 33. | Eun CS, Kim BK, Han DS, Kim SY, Kim KM, Choi BY, Song KS, Kim YS, Kim JF. Differences in gastric mucosal microbiota profiling in patients with chronic gastritis, intestinal metaplasia, and gastric cancer using pyrosequencing methods. Helicobacter. 2014;19:407-416. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 197] [Cited by in RCA: 220] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 34. | Gong J, Li L, Zuo X, Li Y. Change of the duodenal mucosa-associated microbiota is related to intestinal metaplasia. BMC Microbiol. 2019;19:275. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 19] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 35. | Liu X, Shao L, Liu X, Ji F, Mei Y, Cheng Y, Liu F, Yan C, Li L, Ling Z. Alterations of gastric mucosal microbiota across different stomach microhabitats in a cohort of 276 patients with gastric cancer. EBioMedicine. 2019;40:336-348. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 222] [Cited by in RCA: 208] [Article Influence: 34.7] [Reference Citation Analysis (0)] |
| 36. | Yu G, Torres J, Hu N, Medrano-Guzman R, Herrera-Goepfert R, Humphrys MS, Wang L, Wang C, Ding T, Ravel J, Taylor PR, Abnet CC, Goldstein AM. Molecular Characterization of the Human Stomach Microbiota in Gastric Cancer Patients. Front Cell Infect Microbiol. 2017;7:302. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 83] [Cited by in RCA: 118] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 37. | Qi YF, Sun JN, Ren LF, Cao XL, Dong JH, Tao K, Guan XM, Cui YN, Su W. Intestinal Microbiota Is Altered in Patients with Gastric Cancer from Shanxi Province, China. Dig Dis Sci. 2019;64:1193-1203. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 59] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 38. | Biarc J, Nguyen IS, Pini A, Gossé F, Richert S, Thiersé D, Van Dorsselaer A, Leize-Wagner E, Raul F, Klein JP, Schöller-Guinard M. Carcinogenic properties of proteins with pro-inflammatory activity from Streptococcus infantarius (formerly S.bovis). Carcinogenesis. 2004;25:1477-1484. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 154] [Cited by in RCA: 161] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 39. | Ellmerich S, Schöller M, Duranton B, Gossé F, Galluser M, Klein JP, Raul F. Promotion of intestinal carcinogenesis by Streptococcus bovis. Carcinogenesis. 2000;21:753-756. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 171] [Cited by in RCA: 177] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 40. | Guo Y, Zhang Y, Gerhard M, Gao JJ, Mejias-Luque R, Zhang L, Vieth M, Ma JL, Bajbouj M, Suchanek S, Liu WD, Ulm K, Quante M, Li ZX, Zhou T, Schmid R, Classen M, Li WQ, You WC, Pan KF. Effect of Helicobacter pylori on gastrointestinal microbiota: a population-based study in Linqu, a high-risk area of gastric cancer. Gut. 2020;69:1598-1607. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 112] [Cited by in RCA: 214] [Article Influence: 42.8] [Reference Citation Analysis (1)] |
| 41. | Meehan CJ, Beiko RG. A phylogenomic view of ecological specialization in the Lachnospiraceae, a family of digestive tract-associated bacteria. Genome Biol Evol. 2014;6:703-713. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 434] [Cited by in RCA: 546] [Article Influence: 49.6] [Reference Citation Analysis (0)] |
| 42. | Wei X, Tao J, Xiao S, Jiang S, Shang E, Zhu Z, Qian D, Duan J. Xiexin Tang improves the symptom of type 2 diabetic rats by modulation of the gut microbiota. Sci Rep. 2018;8:3685. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 169] [Cited by in RCA: 190] [Article Influence: 27.1] [Reference Citation Analysis (0)] |
| 43. | Wang L, Xin Y, Zhou J, Tian Z, Liu C, Yu X, Meng X, Jiang W, Zhao S, Dong Q. Gastric Mucosa-Associated Microbial Signatures of Early Gastric Cancer. Front Microbiol. 2020;11:1548. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 40] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 44. | Tejera D, Limongi G, Bertullo M, Cancela M. Ralstonia pickettii bacteremia in hemodialysis patients: a report of two cases. Rev Bras Ter Intensiva. 2016;28:195-198. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 14] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 45. | Tseng CH, Lin JT, Ho HJ, Lai ZL, Wang CB, Tang SL, Wu CY. Gastric microbiota and predicted gene functions are altered after subtotal gastrectomy in patients with gastric cancer. Sci Rep. 2016;6:20701. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 67] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 46. | Conlon MA, Bird AR. The impact of diet and lifestyle on gut microbiota and human health. Nutrients. 2014;7:17-44. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 950] [Cited by in RCA: 982] [Article Influence: 89.3] [Reference Citation Analysis (1)] |
| 47. | Morita E, Narikiyo M, Yano A, Nishimura E, Igaki H, Sasaki H, Terada M, Hanada N, Kawabe R. Different frequencies of Streptococcus anginosus infection in oral cancer and esophageal cancer. Cancer Sci. 2003;94:492-496. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 76] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 48. | Hu YL, Pang W, Huang Y, Zhang Y, Zhang CJ. The Gastric Microbiome Is Perturbed in Advanced Gastric Adenocarcinoma Identified Through Shotgun Metagenomics. Front Cell Infect Microbiol. 2018;8:433. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 89] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 49. | Crew KD, Neugut AI. Epidemiology of gastric cancer. World J Gastroenterol. 2006;12:354-362. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 1193] [Cited by in RCA: 1254] [Article Influence: 66.0] [Reference Citation Analysis (8)] |