Published online Jul 21, 2022. doi: 10.3748/wjg.v28.i27.3398
Peer-review started: February 7, 2022
First decision: April 10, 2022
Revised: April 18, 2022
Accepted: June 19, 2022
Article in press: June 19, 2022
Published online: July 21, 2022
Processing time: 160 Days and 22.5 Hours
Artificial intelligence (AI) is playing an increasingly important role in medicine, especially in the field of medical imaging. It can be used to diagnose diseases and predict certain statuses and possible events that may happen. Recently, more and more studies have confirmed the value of AI based on ultrasound in the evalua
Core Tip: Artificial intelligence (AI) is playing an increasingly important role in medicine, especially in the field of medical imaging. Currently, there is a need of a comprehensive review to introduce the application of AI based on ultrasound in diffuse and focal liver lesions. In this article, we introduce the application of AI in the assessment of liver fibrosis and nonalcoholic fatty liver and the differentiation of focal liver lesions. In addition, we discuss the performance of AI based on ultrasound in predicting curative effect, prognosis and microvascular invasion in hepatocellular carcinoma. Lastly, we illustrate the future prospect of AI in liver ultrasound.
- Citation: Cao LL, Peng M, Xie X, Chen GQ, Huang SY, Wang JY, Jiang F, Cui XW, Dietrich CF. Artificial intelligence in liver ultrasound. World J Gastroenterol 2022; 28(27): 3398-3409
- URL: https://www.wjgnet.com/1007-9327/full/v28/i27/3398.htm
- DOI: https://dx.doi.org/10.3748/wjg.v28.i27.3398
In the past several years, liver diseases have affected millions of lives and became one of the main causes of illness and death in the world[1]. It is reported that more than one-fifth of the Chinese population are affected by liver diseases, such as liver fibrosis, liver cancer and nonalcoholic fatty liver disease (NAFLD), contributing unambiguously to health loss. Therefore, paying more attention to liver diseases is of great significance.
Artificial intelligence (AI) is defined as the research of algorithms that enable machines to have the ability of reasoning and performing functions such as solving problems, recognizing object and word, inferring world states, and making decisions[2]. AI is a precise prediction technique that automates learning and recognizes patterns in data. Apart from this, AI has been extensively applied to medical diagnosis, especially in medical image analysis. This application mainly relies on deep learning, a subfield of machine learning. Deep learning is on the frontier of AI, which is based on deep neural networks (DNNs) with more than one hidden layer. Convolutional neural networks (CNNs) are a branch of DNNs that are particularly useful for recognizing images and have stimulated a large amount of interest from industry, academia and clinicians[3].
Compared to other medical imaging techniques, ultrasound is noninvasive, portable and can provide real-time imaging. In recent years, AI-powered ultrasound has become more developed and been implemented in clinical applications in order to reduce the subjectivity and improve the efficiency of ultrasound diagnosis[4]. Many studies have confirmed the value of AI in the evaluation of thyroid nodule, breast lesion and liver lesion classification by ultrasound. In addition to these applications, other AI applications in ultrasound have also been explored and achieved great progress.
In liver medical imaging, AI can make a quantitative assessment by recognizing imaging information automatically to aid physicians in making more precise and comprehensive imaging diagnoses[5]. This technique has been extensively applied to computed tomography (CT), positron emission tomography-CT, magnetic resonance imaging and ultrasound to diagnose liver lesions. For instance, deep learning based on CT and positron emission tomography-CT can be used to detect new liver tumors and metastatic liver malignancy, and to predict the primary origin of liver metastasis[6-8].
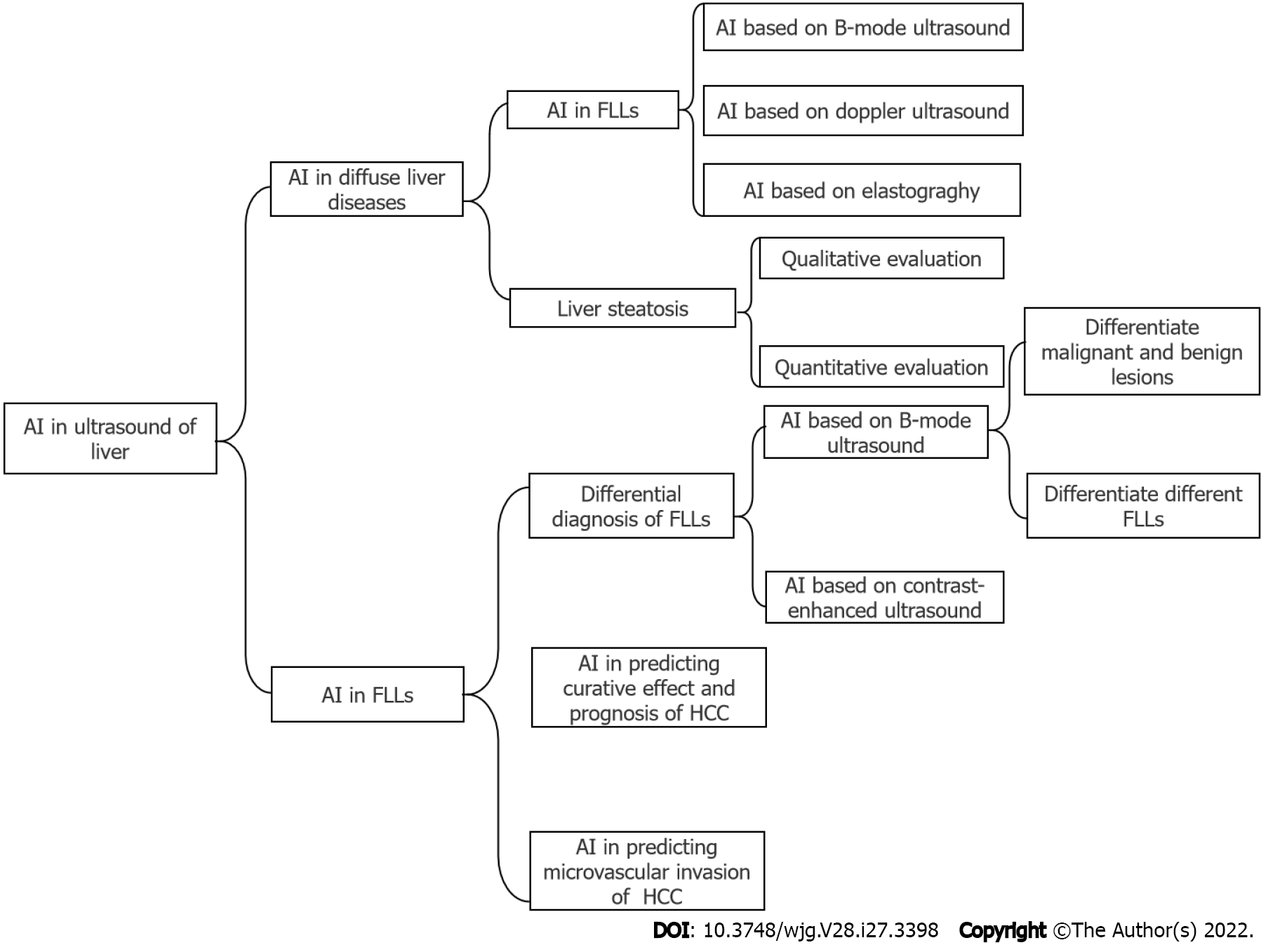
There are also many studies that have illustrated the application of AI in liver ultrasound, while a comprehensive review of AI in this field is lacking. In this review, we will introduce the application of AI based on ultrasound in diffuse liver diseases, including liver fibrosis and steatosis, and focal liver lesions (FLLs), including their differential diagnosis, prediction of a curative effect and prognosis and microvascular invasion (MVI) of hepatocellular carcinoma (HCC). The main structure of this review was illustrated in Figure 1.
There are a variety of diffuse liver diseases that can be asymptomatic or cause severe liver dysfunction, and many of them may lead to cirrhosis, hepatic carcinoma, and death. We will introduce the applications of AI based on ultrasound in two common diffuse liver diseases, i.e. liver fibrosis and steatosis.
Liver fibrosis is the early step of cirrhosis and an important pathological basis of HCC[9]. Therefore, the early detection and prevention of liver fibrosis is essential in the clinical setting. However, although liver biopsy is the gold standard for classifying liver fibrosis using the Metavir score[10] or New Inuyama classification[11] to distribute the score ranging from F0 (no fibrosis) to F4 (cirrhosis). The use of tissue examination for the assessment of liver fibrosis is controversial because liver biopsy is invasive and liver fibrosis is not equably distributed throughout the liver. There are an increasing number of credible, noninvasive and available approaches being widely applied in clinical practice. Recently, a large number of noninvasive techniques have been used to prevent adverse outcomes through the application of AI based on ultrasound.
AI based on B-mode ultrasound: As early as 20 years ago, AI was used to assist the diagnosis of liver fibrosis. Badawi et al[12] creatively proposed an approach that employed fuzzy reasoning techniques to identify diffuse liver diseases automatically by using digital quantitative features measured from the ultrasound images. They extracted parameters only from B-mode images, and the results revealed that this approach had higher specificity and sensitivity for the diagnosis of liver fibrosis than the statistical classification techniques, which had a certain effect but could not help much.
Apart from this, a novel deep multi-scale texture network based entirely on B-mode ultrasound images that was proposed recently seems to be more convenient[13]. The area under the receiver operating characteristic curve of this approach was 0.92 for significant fibrosis (≥ F2) and 0.89 for cirrhosis (F4) in the validation group, which outperformed the ultrasonographers and three serum biomarkers during diagnosis. Although it cannot be used for liver fibrosis staging now, it has excellent potential in the future workflow.
AI based on Doppler ultrasound: On the basis of grey-scale parameters from B-mode images, Doppler parameters of intrahepatic blood vasculature were added as essential parameters. Eventually, five ultrasonographic variables, including the liver parenchyma, thickness of the spleen, the hepatic vein waveform, hepatic artery pulsatile index and damping index, were selected as the input neurons. A data optimization procedure was used in an artificial neural network for the diagnosis of liver fibrosis, which achieved an area under the curve of 0.92[14]. Although this model proved to predict liver cirrhosis accurately, it still could not provide a specific grading.
AI based on elastography: In recent years, with the development of ultrasound, studies have proposed computer-aided techniques based on elastography that is of great importance in ultrasound images to identify and stage liver fibrosis. Real-time tissue elastography (RTE) is one of the recently developed elastography techniques. In a study, 11 image features were extracted directly from the RTE software that was installed in the ultrasound system to quantify the patterns of the RTE images[15]. Then, the data were processed and divided among four classical classifiers. The results showed that the performance of the adopted classifiers was much better than the previous liver fibrosis index method, which predicted the stage of fibrosis using RTE images and multiple regression analyses. The good performance in this study demonstrated that machine learning had the potential to perform as powerful tools for staging liver fibrosis.
Nowadays, most applications of AI in evaluating the stage of liver fibrosis are based on shear wave elastography. An automated approach including the image quality check, region of interest (ROI) selection and CNN classification based on shear wave elastography showed a more accurate detection of ≥ F2 fibrosis levels than a previously published baseline approach, with an area under curve of 0.89 vs 0.74[16]. The deep learning radiomics also presented the potential diagnostic performance in chronic hepatitis B patients compared with two-dimensional shear wave elastography[17]. AI could help stage liver fibrosis more accurately with the assistance of elastography.
Hepatic steatosis, characterized by the accumulation of fat droplets in hepatocytes, can develop into nonalcoholic fibrosis, steatohepatitis, cirrhosis, and even HCC[18,19]. Early detection and treatment may halt or reverse NAFLD progression[19]. As a consequence, there is a critical need to develop noninvasive imaging methods to assess hepatic steatosis. Noninvasive liver imaging methods including CT, magnetic resonance imaging and ultrasound have been extensively investigated[20].
Ultrasound is the first-line examination for identifying liver steatosis. It shows an enlarged liver with a greater number of echoes caused by fat droplets, and the liver is brighter and more hyperechoic compared with the right kidney. The image is qualitative and relies on the subjective judgement of the operator, which definitely leads to variable results and low reproducibility[21]. To overcome the observer bias, a series of quantitative and semi-quantitative parameters, including attenuation and backscatter coefficients, the hepato-renal index (HRI) and ultrasound envelope statistic parametric imaging (known as speckle statistics), have been implemented, some of which represent excellent reproducibility and reliability[21-24]. At present, almost all the studies published have concentrated on NAFLD.
It was reported that the detection of moderate and severe steatosis based on ultrasound has an 84.8% sensitivity and a 93.6% specificity, while mild steatosis has an even lower sensitivity[25]. Recently, some researchers applied AI to improve the ultrasound detection rate of NAFLD, and the results were promising. Table 1 shows the studies using AI based on ultrasound to identify steatosis. The studies showed that AI had tremendous potential in helping diagnose liver steatosis. Some studies attempted to optimize CNN models. In the future, classifying the degree of liver steatosis with the assistance of the AI could be a trend.
| Task | Reference standard | Sample size | Method | Results | Ref. |
| Fatty liver disease diagnosis | Liver biopsy | 55 patients with severe obesity, 38 of whom had fatty liver disease | Deep learning with B-mode image ultrasound | Sensitivity: 100%; specificity: 88%; accuracy: 96%; AUC: 0.98 | [26] |
| Fatty liver disease diagnosis | Radiologist qualitative score | 157 ultrasound liver images from unknown number of participants | Deep learning with B-mode image ultrasound | Sensitivity: 95%; specificity: 85%; accuracy: 90.6%; AUC: 0.96 | [28] |
| NAFLD assessment | MRI proton density fat fraction | 204 participants, 140 of whom had NAFLD, 64 control participants | One-dimensional CNNs | Sensitivity: 97%; specificity: 94%; accuracy: 96%; AUC: 0.98 | [31] |
| NAFLD assessment | MRI proton density fat fraction | 135 adult participants with known or suspected NAFLD | Transfer learning with a pretrained CNN by four ultrasound views of liver routinely obtained | SCC: 0.81; AUC: 0.91 (PDFF ≥ 5%) | [27] |
| NAFLD assessment | Liver biopsy | 295 subjects, 198 mild fatty liver, one moderate degree of fatty liver | DCNN-based organ segmentation with Gaussian mixture modeling for automated quantification of the HRI | ICC of two radiologists and DCNN were 0.919, 0.916, 0.734 | [33] |
| The severity of fatty liver | Abdominal ultrasound | 21855 B-mode ultrasound images, 2070 patients with different severities from none to severe fatty liver | Pretrained CNN models with B-mode ultrasound images | The areas under the receiver operating characteristic curves were 0.974 (mild steatosis vs others), 0.971 (moderate steatosis vs others), 0.981 (severe steatosis vs others), 0.985 (any severity vs normal) and 0.996 (moderate-to-severe steatosis clinically abnormal vs normal-to-mild steatosis clinically normal) | [29] |
Qualitative evaluation: Deep learning has been applied to qualitatively evaluate NAFLD. An approach of assessing fatty liver disease by utilizing deep learning based on CNNs with B-mode images was proposed[26]. The study recruited 135 participants with known or suspected NAFLD to investigate the function of four liver views (three views in the transverse plane, including hepatic veins at the confluence with the inferior vena cava, right portal vein and right posterior portal vein, and one view in the sagittal plane, i.e. the liver and kidney view) in the assessment[27]. The study assessed attention maps for liver assessment based on CNNs, which illustrated that the available image features provided by each view could assess liver fat. Unlike the previous study, magnetic resonance imaging proton density fat fraction was used as a reference standard, which was not precise enough compared with liver biopsy.
A novel framework combining transfer learning with fine-tuning was proposed[28]. Although this study revealed that the new framework outperformed CNN, this conclusion was not entirely convincing because the radiologists’ qualitative scores were the reference standard. This framework was also utilized in other studies and achieved a good performance.
With the development of deep learning, Chou et al[29] established two-class, three-class and four-class prediction models to classify the severity of steatosis by using B-mode ultrasound images from 2070 patients. Although liver biopsy is the gold standard, the deep learning model could select eligible patients for a liver biopsy by evaluating the severity of fatty liver preliminarily, which would reduce unnecessary tests.
Different deep learning algorithms tend to have different performances. The combined deep learning algorithm based on B-mode images had an area under the receiver operating characteristic curve of 0.9999 and accuracy of 0.9864 compared with other algorithms[30]. Therefore, in future studies selecting an optimal algorithm is important.
Quantitative evaluation: AI has also been applied to quantitatively evaluate NAFLD. Radiofrequency signals may negate the loss or change of data during translation to B-mode ultrasound images. A study acquired the diagnosis and fat fraction of NAFLD by inputting the original data based on one-dimensional algorithms[31]. The investigators obtained a 97% sensitivity and 94% specificity, and the positive predictive value was up to 97%, which demonstrated that the utilization of original ultrasound radiofrequency could be applied in diagnosing NAFLD and to quantifying liver fat fraction. Similarly, in an animal experiment, a CNN model based on radiofrequency signals was shown to have a better performance than the traditional quantitative ultrasound when classifying steatosis[32].
An HRI model based on CNNs was also tested for NAFLD evaluation. Cha et al[33] reported that an automated approach had no significant difference in hepatic measurements and HRI calculations compared with experienced radiologists, which indicated that the aid of deep learning could reduce a radiologist’s workload and improve the residents’ diagnostic accuracy. In this study, an automated HRI calculation algorithm was used, including liver and kidney segmentation, kidney ROI extraction, liver ROI extraction, and calculation of the HRI.
HCC is the most conventional original malignant FLL, which is the sixth most common cancer in human beings as well as the fourth cause of cancer-related deaths in the world[1]. Hence, early accurate differential diagnosis of malignant and benign FLL is important for the management and prognosis of patients[34].
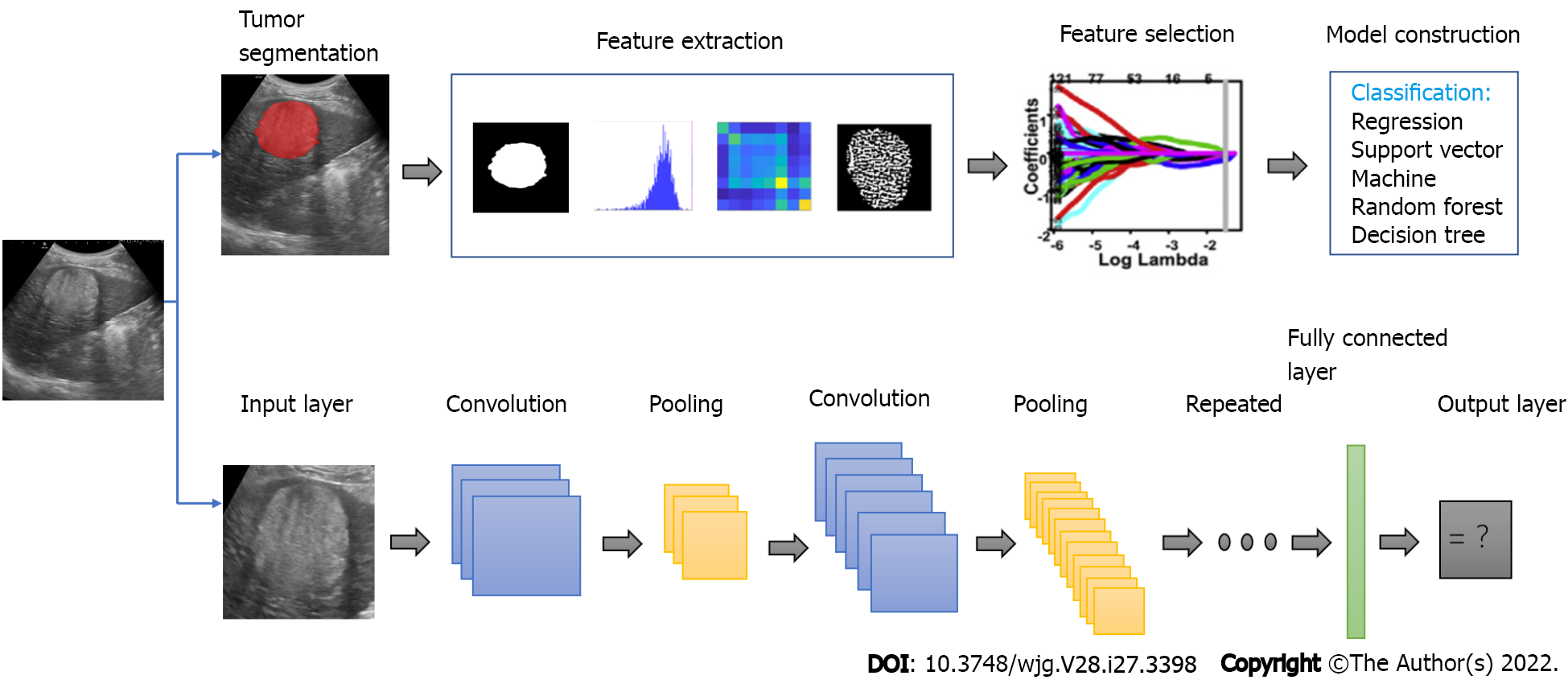
Ultrasound is the first-line imaging modality to identify FLLs in the clinical workflow. The development of AI provides a new method to improve the accuracy of ultrasound in diagnosing FLLs. Compared with radiologists viewing anatomical images, AI can better reflect monolithic tumor morphology as well as capture both granular and radiological patterns in a specific task, which is difficult by normal human vision[35]. Figure 2 illustrates the flowchart of the application of deep learning and radiomics in FLLs. Studies have confirmed that the application of AI can improve the diagnostic performance of ultrasound for FLLs (Table 2).
| Modality and task | Approach | Target disease: number of the case | Performance | Ref. |
| Classifying different FLLs based on B-mode | ANN | Cyst: 29; hemangioma: 37; malignant tumor: 33 | Cyst vs hemangioma accuracy: 99.7%; cyst vs malignant tumor accuracy: 98.7%; hemangioma vs malignant tumor accuracy: 96.1% | [40] |
| Differentiating benign and malignant lesions based on B-mode | CNN | Benign lesions: 300; malignant lesions: 296 | All lesion accuracy: 84%; uncertain set of lesion accuracy: 79% | [37] |
| Classifying different FLLs based on B-mode | ANN (sparse autoencoder) | Normal liver: 16; cyst: 44; hemangioma: 18; HCC: 30 | overall accuracy: 97.2%; overall sensitivity: 98%; overall specificity: 95.7% | [41] |
| Differentiating benign and malignant lesions based on B-mode | FSVM | training set; DS1: benign lesions: 132, malignant lesions: 68; DS2: malignant liver cancer: 50, hepatocellular adenoma: 150, hemangioma: 35, focal nodular hyperplasia: 145, lipoma: 70 | DS1: accuracy: 97%, sensitivity: 100%, specificity: 95.5%, AUC: 0.984; DS2: accuracy: 95.1%, sensitivity: 92.0%, specificity: 95.5%, AUC: 0.971 | [36] |
| Classifying different FLLs based on B-mode | CNN | Non-tumorous liver: 258, hemangioma: 17, HCC: 6, cyst: 30, focal nodular hyperplasia: 8 | AUC for tumor detection: 0.935; AUC for tumor discrimination (mean): 0.916 | [42] |
| Diagnosing HCC based on B-mode | CNN | Malignant tumor: 1786; benign tumor: 427 | AUC for EV: 0.924 | [38] |
| Differentiating benign and malignant lesions based on B-mode | CNN | HCC: 6; cyst: 6600; hemangioma: 5374; focal fatty sparing: 5110; focal fatty infiltration: 934 | IV: overall sensitivity: 83.9%; overall specificity: 97.1%; HCC detection rate: 85.3%; EV: overall sensitivity: 84.9%; overall specificity: 97.1%; HCC detection rate: 78.3% | [39] |
| Classifying different FLLs based on CEUS | ANN | hemangioma: 16; focal fatty liver: 23; HCC: 41; metastatic tumor: 32 (hypervascular: 20 hypovascular: 12) | Accuracy: 94.5%; sensitivity: 93.2%; specificity: 89.7% | [47] |
| Differentiating benign and malignant lesions based on CEUS | Deep belief networks | HCC: 6; hemangioma: 10; liver abscess: 4; metastases: 3; focal fatty sparing: 3 | Accuracy: 83.4%; sensitivity: 83.3%; specificity: 87.5% | [59] |
| Differentiating benign and malignant lesions based on CEUS | SVM | Benign tumor: 30; malignant tumor: 22 | Accuracy: 90.3%; sensitivity: 93.1%; specificity: 86.9% | [45] |
| Differentiating benign and malignant lesions based on CEUS | SVM | Benign tumor, HCC or metastatic tumor: 98 | Benign vs malignant accuracy: 91.8%, sensitivity: 93.1%, specificity: 86.9%; benign vs HCC vs metastatic carcinoma: accuracy: 85.7%; sensitivity: 84.4%; specificity: 87.7% | [46] |
| Differentiating benign and malignant lesions based on CEUS | Deep canonical correlation analysis + multiple kernel learning | Benign tumor: 46; malignant tumor: 47 | Accuracy: 90.4%; sensitivity: 93.6%; specificity: 86.9% | [43] |
| Differentiating benign and malignant lesions based on CEUS | 3D-CNN | HCC: 2110; focal nodular hyperplasia: 2310 | Accuracy: 93.1%; sensitivity: 94.5%; specificity: 93.6% | [44] |
| Differentiating benign and malignant lesions based on CEUS | Deep neural network | Focal nodular hyperplasia: 16; HCC: 30; hemangioma: 23; hypervascular metastasis: 11; hypovascular metastasis: 11 | Top accuracy: 88% | [48] |
| Differentiating benign and malignant lesions based on CEUS | CNN | Development set: malignant tumor: 281, benign tumor: 82; testing set: malignant tumor: 164, benign tumor: 47 | Accuracy: 91.0%; sensitivity: 92.7%; specificity: 85.1%; AUC: 0.934 | [49] |
AI based on B-mode ultrasound: AI has been widely used in differentiating malignant and benign FLLs based on B-mode ultrasound. Gray level co-occurrence matrix could be used in extracting features from B-mode images, which has been used in differentiating malignant and benign FLLs combined with a fuzzy support vector machine[36]. This study achieved an area under the curve of 0.984 and 0.971 in database 1 and database 2, respectively, which confirmed the feasibility of AI in this field.
With the development of AI, deep learning plays a more vital role in the differential diagnosis of FLLs. A CNN with ResNet50 was utilized to recognize benign from malignant solid liver lesions through ultrasonography. The performance was comparable to that of expert radiologists[37]. However, this study did not involve other information such as clinical factors. In another (multicenter) study, after adding seven clinical factors, a higher accuracy, sensitivity and specificity was obtained, compared with those from radiologists with 15 years of experience. The area under the curve for recognizing malignant from benign lesions reached up to 0.924 in the external validation cohort[38].
However, the aforementioned studies only included common FLLs. Increasing the types of FLLs may confuse the diagnosis and reduce the accuracy. Similar to the previous study, a multicenter study estimating internal validation and external validation cohorts had a larger volume of training data and involved more varieties of FLLs, including cysts, HCC, hemangiomas, focal fatty infiltration and focal fatty sparing[39]. Although they obtained a lower sensitivity due to more types of diseases, the performance in the external validation cohorts was still satisfactory. In addition, they utilized videos as training materials to achieve real-time analysis in the future workflow. This novel approach would offer great convenience to radiologists in helping differentiate FLLs.
AI could also be used in the classification of FLLs. In order to optimize feature sets, a hybrid textural feature extraction system was proposed by Hwang et al[40]. In their preliminary study, a high accuracy was observed in classifying cysts vs hemangiomas and cysts vs malignant lesions. However, when classifying hemangiomas vs malignant lesions by extracting multiple ROI, the accuracy was only 80%. The proposed approach exhibited a better accuracy in all classification groups by quantifying the key features in ultrasound images, especially in classifying hemangioma vs malignant, with an accuracy of 96.13%.
Later, a sparse autoencoder system based on deep learning was proposed in diagnosing cysts, hemangiomas and malignant lesions, and it outperformed the three progressive techniques, including K-enarest neighbor, multi-support vector machine and naive Bayes, with an overall accuracy of 97.2%[41].
These two studies[40,41] focused on three kinds of FLLs. An algorithm that could simultaneously detect and characterize FLLs based on deep learning was proposed in diagnosing HCC, focal nodular hyperplasia, cysts, hemangiomas and metastasis, and it achieved an average area under the curve of 0.916[42]. That study yielded promising results by using a small amount of data. Larger databases would increase the accuracy of this model.
AI based on contrast-enhanced ultrasound: It is reported that contrast-enhanced ultrasound (CEUS) images had better sensitivity and specificity for differentiating between malignant and benign tumors compared with B-mode images, which indicated CEUS had a superior diagnostic performance. Combining AI with CEUS could differentiate benign and malignant FLLs and classify different kinds of malignant lesions.
AI could be used to differentiate malignant and benign FLLs based on three-phase CEUS images. A two-stage multiple view learning that represented the integration of deep canonical correlation analysis and multiple kernel learning was used to fuse the characteristics of three-phase patterns in CEUS, presenting an accuracy of 90.41%[43]. The proposed algorithm had both a low computational complex and a high predictive accuracy. For multiview CEUS images, utilizing a multimodal feature fusion algorithm is necessary.
Compared with deep canonical correlation analysis-multiple kernel learning, the use of a three-dimensioned CNN, which integrated the relationship between two temporally adjacent frames to extract features spatially and temporally, achieved a higher accuracy of 93.1%, sensitivity of 94.5% and specificity of 93.6%[44]. However, this algorithm still needs to be validated.
These two studies above[43,44] exploited heterogeneous visual morphology to describe the difference between liver masses. Apart from this method, time-intensity curve (TIC), which represents the contrast intensity constantly and generates the fitted curve of enhanced intensity during the process, was used in many studies.
Support vector machine[45,46] and deep learning[47,48] based on TICs have presented good performances in differentiating FLLs. A support vector machine-based image analysis system was used for FLL classification and presented an area under the curve of 0.89[45]. An artificial neural network diagnostic system based on TICs was shown to have a similar accuracy and specificity in classifying five different liver tumors 10 years ago[47]. Later, deep learning became more mature, and TICs of the arterial and the portal vein phases of CEUS videos were extracted on the basis of the deep belief networks, a kind of neural network that was composed of layers of Boltzmann machines, to analyze the extracted TICs[48]. The accuracy of this deep learning method for classifying benign from malignant lesions was 83.36%. A novel evaluation procedure named ‘leave-one-patient-out’ and custom DNNs were creatively presented. That study involved various types of liver lesions and compared the custom DNN designs with the state-of-the-art architectures and obtained a maximal accuracy of 88% by utilizing the proposed evaluation procedure in both pretrained and trained-from-scratch models. This novel approach has a magnificent prospect for development, and it is worth further investigation.
AI based on CEUS was shown to assist in clinical settings as the reference and improve the performance of residents in the differentiation of benign and malignant FLLs[49]. In the future, it will likely play a supporting role in clinical work. However, AI based on TICs tends to complicate the calculation because generating TICs is a time-consuming process. Therefore, developing new approaches to extract features from CEUS images is important.
It was reported that the Edmondson-Steiner grade is a vital preoperation predictor of tumor survival and recurrence after undergoing surgical resection[50,51]. Because preoperative pathological differentiation grade can only be obtained by an invasive biopsy[52], it is necessary to explore a noninvasive method to predict therapeutic effect, recurrence and metastasis to achieve personalized treatment.
Some studies[53-55] demonstrated the superiority of AI based on CEUS in predicting a curative effect and prognosis in HCC. Although the results revealed a better performance of AI models compared with a single clinical or ultrasound model, a better performance may be obtained by adding clinical factors in the future studies.
Predicting curative effect of HCC: Transarterial chemoembolization (TACE) is the first-line therapy in patients who are diagnosed with mid-stage HCC, and the response to the first TACE treatment is related to the subsequent curative effect and survival. Therefore, it is necessary to predict the personalized responses to the first TACE treatment. A deep learning radiomics-based CEUS model, machine learning radiomics-based B-mode image model and machine learning radiomics-based TIC of the CEUS model were established to achieve this function[53]. These models presented a better performance compared with the hepatoma arterial embolization prognostic score based on three indexes concerning liver function and tumor load, which will be of great benefit in selecting both first treatment and subsequent therapies after the first TACE treatment.
Predicting prognosis of patients with HCC: Radiofrequency ablation and surgical resection are recommended for early-stage HCC. Deep learning could also be used to predict the progression-free survival of these two therapies in HCC patients[54]. Two models based on these two kinds of therapies provided a satisfactory prediction accuracy and calibration of 2-year progression-free survival. In another study[55], a three dimensional-CNN model, which avoided missing information from CEUS images compared with extracting features from four-phase images, was used. It was observed that predicting prognosis of different treatments in advance and timely swapping of treatment would increase the 2-year progression-free survival, which could contribute to a better prognosis.
MVI has shown to be the independent predictor of recurrence and poor outcomes of HCC. Therefore, making a noninvasive and accurate preoperative identification of MVI would be of great significance for HCC patients. The application of AI in predicting MVI achieved good performance based on gray-scale ultrasound images and CEUS.
The radiomics score based on ultrasound of HCC was established and shown to be an independent predictor of MVI[56]. The performance of the clinical nomogram improved significantly with the aid of a radiomics score, which demonstrated the important role of this technique.
Features of the peritumoral area have been shown to be more accurate recently[56]. The radiomic signatures from the gross and peritumoral region showed the best performance compared with the gross-tumoral region and the peritumoral region[57]. The area under the curves were 0.726 based on features of the gross and peritumoral region and 0.744 after the incorporation of essential clinical information.
These studies[56,57] mentioned above have confirmed the application of AI based on gray-scale ultrasound, and AI could be applied to CEUS in predicting MVI as well. Zhang et al[58] extracted radiomics features from the B-mode, artery phase, portal venous phase and delay phase images of preoperative CEUS to construct four radiomics scores based on the primary dataset. Then, they used four radiomic scores and clinical factors for multivariate logistic regression analysis, which demonstrated that the portal venous phase and delay phase radiomics score, tumor size and alpha-fetoprotein level were independent risk factors in predicting MVI. The radiomics nomogram based on these four predictors indicated a better discrimination and a good calibration compared with the clinical model (based on tumor size and alpha-fetoprotein level) in both the primary dataset (area under the curve: 0.849 vs 0.690) and the validation dataset (area under the curve: 0.788 vs 0.661). This study developed a new noninvasive predictive nomogram based on CEUS that could provide useful information in predicting MVI preoperatively, thus facilitating the choice of a more appropriate surgical option.
In conclusion, AI can provide great assistance in the evaluation of diffuse liver diseases (including liver fibrosis and liver steatosis) and FLLs. First, it could be applied to identify and stage liver fibrosis on the basis of B-mode ultrasound, Doppler ultrasound, and elastography. Second, the application of deep learning could be used to make qualitative evaluation based entirely on B-mode images and quantitative evaluation based on radiofrequency signals and HRI, which would improve the ultrasound detection rate of NAFLD. Third, AI has the ability to differentiate malignant FLLs from benign FLLs as well as classify different kinds of FLLs with a better performance compared with clinical indexes. Fourth, the curative effect and prognosis of HCC treatment can be predicted, and an optimal personalized treatment can be chosen. Last, AI based on B-mode ultrasound and CEUS could predict MVI of HCC preoperatively, which could be helpful for more appropriate surgical planning. These applications had good specificity, accuracy and a comparable or even better performance compared with experts in the diagnosis and differentiation of diffuse and focal liver lesions.
There are also some limitations in the applications of AI using ultrasound. First, it is difficult to prepare a large-scale dataset, especially for medical images. Second, although deep learning is the widest used algorithm and has good performance in various studies, its interpretability and generalization is low. Third, the input data may vary from different equipment and operators, which would influence the performance of AI. Last, because a large amount of data is needed to train and validate the established algorithms, the conclusions of many single-center studies are not convincing. Therefore, researchers are expected to conduct more multicenter studies and incorporate more samples, as much as possible. At the same time, optimizing algorithms and creating standards for medical images are also necessary. Despite medical images, researchers could also build the database containing important clinical factors to establish a more comprehensive AI model for future work.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Hussain J, Oman; Li L, China S-Editor: Fan JR L-Editor: A P-Editor: Fan JR
| 1. | Xiao J, Wang F, Wong NK, He J, Zhang R, Sun R, Xu Y, Liu Y, Li W, Koike K, He W, You H, Miao Y, Liu X, Meng M, Gao B, Wang H, Li C. Global liver disease burdens and research trends: Analysis from a Chinese perspective. J Hepatol. 2019;71:212-221. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 239] [Cited by in RCA: 378] [Article Influence: 63.0] [Reference Citation Analysis (1)] |
| 2. | Bellman RE. An Introduction to Artificial Intelligence: Can Computers Think? Spring: 1978. |
| 3. | Chan HP, Samala RK, Hadjiiski LM, Zhou C. Deep Learning in Medical Image Analysis. Adv Exp Med Biol. 2020;1213:3-21. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 292] [Cited by in RCA: 328] [Article Influence: 65.6] [Reference Citation Analysis (0)] |
| 4. | Akkus Z, Cai J, Boonrod A, Zeinoddini A, Weston AD, Philbrick KA, Erickson BJ. A Survey of Deep-Learning Applications in Ultrasound: Artificial Intelligence-Powered Ultrasound for Improving Clinical Workflow. J Am Coll Radiol. 2019;16:1318-1328. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 161] [Article Influence: 32.2] [Reference Citation Analysis (0)] |
| 5. | Ambinder EP. A history of the shift toward full computerization of medicine. J Oncol Pract. 2005;1:54-56. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 26] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 6. | Vivanti R, Szeskin A, Lev-Cohain N, Sosna J, Joskowicz L. Automatic detection of new tumors and tumor burden evaluation in longitudinal liver CT scan studies. Int J Comput Assist Radiol Surg. 2017;12:1945-1957. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 50] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 7. | Preis O, Blake MA, Scott JA. Neural network evaluation of PET scans of the liver: a potentially useful adjunct in clinical interpretation. Radiology. 2011;258:714-721. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 18] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 8. | Ben-Cohen A, Klang E, Diamant I, Rozendorn N, Raskin SP, Konen E, Amitai MM, Greenspan H. CT Image-based Decision Support System for Categorization of Liver Metastases Into Primary Cancer Sites: Initial Results. Acad Radiol. 2017;24:1501-1509. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 28] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 9. | Yoo S, Wang W, Wang Q, Fiel MI, Lee E, Hiotis SP, Zhu J. A pilot systematic genomic comparison of recurrence risks of hepatitis B virus-associated hepatocellular carcinoma with low- and high-degree liver fibrosis. BMC Med. 2017;15:214. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 63] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 10. | Bedossa P, Poynard T. An algorithm for the grading of activity in chronic hepatitis C. The METAVIR Cooperative Study Group. Hepatology. 1996;24:289-293. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2860] [Cited by in RCA: 3082] [Article Influence: 106.3] [Reference Citation Analysis (0)] |
| 11. | Yamada G. [Histopathological characteristics and clinical significance of New Inuyama Classification in chronic hepatitis B]. Nihon Rinsho. 2004;62 Suppl 8:290-292. [PubMed] |
| 12. | Badawi AM, Derbala AS, Youssef AM. Fuzzy logic algorithm for quantitative tissue characterization of diffuse liver diseases from ultrasound images. Int J Med Inform. 1999;55:135-147. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 39] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 13. | Ruan D, Shi Y, Jin L, Yang Q, Yu W, Ren H, Zheng W, Chen Y, Zheng N, Zheng M. An ultrasound image-based deep multi-scale texture network for liver fibrosis grading in patients with chronic HBV infection. Liver Int. 2021;41:2440-2454. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 14] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 14. | Zhang L, Li QY, Duan YY, Yan GZ, Yang YL, Yang RJ. Artificial neural network aided non-invasive grading evaluation of hepatic fibrosis by duplex ultrasonography. BMC Med Inform Decis Mak. 2012;12:55. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 22] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 15. | Chen Y, Luo Y, Huang W, Hu D, Zheng RQ, Cong SZ, Meng FK, Yang H, Lin HJ, Sun Y, Wang XY, Wu T, Ren J, Pei SF, Zheng Y, He Y, Hu Y, Yang N, Yan H. Machine-learning-based classification of real-time tissue elastography for hepatic fibrosis in patients with chronic hepatitis B. Comput Biol Med. 2017;89:18-23. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 60] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 16. | Brattain LJ, Telfer BA, Dhyani M, Grajo JR, Samir AE. Objective Liver Fibrosis Estimation from Shear Wave Elastography. Annu Int Conf IEEE Eng Med Biol Soc. 2018;2018:1-5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 16] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 17. | Wang K, Lu X, Zhou H, Gao Y, Zheng J, Tong M, Wu C, Liu C, Huang L, Jiang T, Meng F, Lu Y, Ai H, Xie XY, Yin LP, Liang P, Tian J, Zheng R. Deep learning Radiomics of shear wave elastography significantly improved diagnostic performance for assessing liver fibrosis in chronic hepatitis B: a prospective multicentre study. Gut. 2019;68:729-741. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 226] [Cited by in RCA: 354] [Article Influence: 59.0] [Reference Citation Analysis (1)] |
| 18. | Loomba R, Sanyal AJ. The global NAFLD epidemic. Nat Rev Gastroenterol Hepatol. 2013;10:686-690. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1127] [Cited by in RCA: 1330] [Article Influence: 110.8] [Reference Citation Analysis (0)] |
| 19. | Friedman SL, Neuschwander-Tetri BA, Rinella M, Sanyal AJ. Mechanisms of NAFLD development and therapeutic strategies. Nat Med. 2018;24:908-922. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1376] [Cited by in RCA: 2918] [Article Influence: 416.9] [Reference Citation Analysis (1)] |
| 20. | Lăpădat AM, Jianu IR, Ungureanu BS, Florescu LM, Gheonea DI, Sovaila S, Gheonea IA. Non-invasive imaging techniques in assessing non-alcoholic fatty liver disease: a current status of available methods. J Med Life. 2017;10:19-26. [PubMed] |
| 21. | Lee DH. Imaging evaluation of non-alcoholic fatty liver disease: focused on quantification. Clin Mol Hepatol. 2017;23:290-301. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 63] [Cited by in RCA: 106] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 22. | Ballestri S, Nascimbeni F, Lugari S, Lonardo A, Francica G. A critical appraisal of the use of ultrasound in hepatic steatosis. Expert Rev Gastroenterol Hepatol. 2019;13:667-681. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 41] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 23. | Pirmoazen AM, Khurana A, El Kaffas A, Kamaya A. Quantitative ultrasound approaches for diagnosis and monitoring hepatic steatosis in nonalcoholic fatty liver disease. Theranostics. 2020;10:4277-4289. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 76] [Cited by in RCA: 106] [Article Influence: 21.2] [Reference Citation Analysis (0)] |
| 24. | Xia MF, Yan HM, He WY, Li XM, Li CL, Yao XZ, Li RK, Zeng MS, Gao X. Standardized ultrasound hepatic/renal ratio and hepatic attenuation rate to quantify liver fat content: an improvement method. Obesity (Silver Spring). 2012;20:444-452. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 112] [Cited by in RCA: 112] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 25. | Kotronen A, Peltonen M, Hakkarainen A, Sevastianova K, Bergholm R, Johansson LM, Lundbom N, Rissanen A, Ridderstråle M, Groop L, Orho-Melander M, Yki-Järvinen H. Prediction of non-alcoholic fatty liver disease and liver fat using metabolic and genetic factors. Gastroenterology. 2009;137:865-872. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 490] [Cited by in RCA: 601] [Article Influence: 37.6] [Reference Citation Analysis (0)] |
| 26. | Byra M, Styczynski G, Szmigielski C, Kalinowski P, Michałowski Ł, Paluszkiewicz R, Ziarkiewicz-Wróblewska B, Zieniewicz K, Sobieraj P, Nowicki A. Transfer learning with deep convolutional neural network for liver steatosis assessment in ultrasound images. Int J Comput Assist Radiol Surg. 2018;13:1895-1903. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 206] [Cited by in RCA: 154] [Article Influence: 22.0] [Reference Citation Analysis (0)] |
| 27. | Byra M, Han A, Boehringer AS, Zhang YN, O'Brien WD Jr, Erdman JW Jr, Loomba R, Sirlin CB, Andre M. Liver Fat Assessment in Multiview Sonography Using Transfer Learning With Convolutional Neural Networks. J Ultrasound Med. 2022;41:175-184. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 30] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 28. | Reddy DS, Bharath R, Rajalakshmi P. A Novel Computer-Aided Diagnosis Framework Using Deep Learning for Classification of Fatty Liver Disease in Ultrasound Imaging. Int Confer e-Health Network App Ser. 2018;1-5. |
| 29. | Chou TH, Yeh HJ, Chang CC, Tang JH, Kao WY, Su IC, Li CH, Chang WH, Huang CK, Sufriyana H, Su EC. Deep learning for abdominal ultrasound: A computer-aided diagnostic system for the severity of fatty liver. J Chin Med Assoc. 2021;84:842-850. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 27] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 30. | Zamanian H, Mostaar A, Azadeh P, Ahmadi M. Implementation of Combinational Deep Learning Algorithm for Non-alcoholic Fatty Liver Classification in Ultrasound Images. J Biomed Phys Eng. 2021;11:73-84. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 29] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 31. | Han A, Byra M, Heba E, Andre MP, Erdman JW Jr, Loomba R, Sirlin CB, O'Brien WD Jr. Noninvasive Diagnosis of Nonalcoholic Fatty Liver Disease and Quantification of Liver Fat with Radiofrequency Ultrasound Data Using One-dimensional Convolutional Neural Networks. Radiology. 2020;295:342-350. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 85] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 32. | Nguyen TN, Podkowa AS, Park TH, Miller RJ, Do MN, Oelze ML. Use of a convolutional neural network and quantitative ultrasound for diagnosis of fatty liver. Ultrasound Med Biol. 2021;47:556-568. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 17] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 33. | Cha DI, Kang TW, Min JH, Joo I, Sinn DH, Ha SY, Kim K, Lee G, Yi J. Deep learning-based automated quantification of the hepatorenal index for evaluation of fatty liver by ultrasonography. Ultrasonography. 2021;40:565-574. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 14] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 34. | Amarapurkar D, Han KH, Chan HL, Ueno Y; Asia-Pacific Working Party on Prevention of Hepatocellular Carcinoma. Application of surveillance programs for hepatocellular carcinoma in the Asia-Pacific Region. J Gastroenterol Hepatol. 2009;24:955-961. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 51] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 35. | Kumar V, Gu Y, Basu S, Berglund A, Eschrich SA, Schabath MB, Forster K, Aerts HJ, Dekker A, Fenstermacher D, Goldgof DB, Hall LO, Lambin P, Balagurunathan Y, Gatenby RA, Gillies RJ. Radiomics: the process and the challenges. Magn Reson Imaging. 2012;30:1234-1248. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1505] [Cited by in RCA: 1555] [Article Influence: 119.6] [Reference Citation Analysis (0)] |
| 36. | Xian GM. An identification method of malignant and benign liver tumors from ultrasonography based on GLCM texture features and fuzzy SVM. Expert System App. 2010;37:6737-6741. [DOI] [Full Text] |
| 37. | Xi IL, Wu J, Guan J, Zhang PJ, Horii SC, Soulen MC, Zhang Z, Bai HX. Deep learning for differentiation of benign and malignant solid liver lesions on ultrasonography. Abdom Radiol (NY). 2021;46:534-543. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 27] [Article Influence: 6.8] [Reference Citation Analysis (1)] |
| 38. | Yang Q, Wei J, Hao X, Kong D, Yu X, Jiang T, Xi J, Cai W, Luo Y, Jing X, Yang Y, Cheng Z, Wu J, Zhang H, Liao J, Zhou P, Song Y, Zhang Y, Han Z, Cheng W, Tang L, Liu F, Dou J, Zheng R, Yu J, Tian J, Liang P. Improving B-mode ultrasound diagnostic performance for focal liver lesions using deep learning: A multicentre study. EBioMedicine. 2020;56:102777. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 71] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 39. | Tiyarattanachai T, Apiparakoon T, Marukatat S, Sukcharoen S, Geratikornsupuk N, Anukulkarnkusol N, Mekaroonkamol P, Tanpowpong N, Sarakul P, Rerknimitr R, Chaiteerakij R. Development and validation of artificial intelligence to detect and diagnose liver lesions from ultrasound images. PLoS One. 2021;16:e0252882. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 22] [Article Influence: 5.5] [Reference Citation Analysis (1)] |
| 40. | Hwang YN, Lee JH, Kim GY, Jiang YY, Kim SM. Classification of focal liver lesions on ultrasound images by extracting hybrid textural features and using an artificial neural network. Biomed Mater Eng. 2015;26 Suppl 1:S1599-S1611. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 28] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 41. | Hassan TM, Elmogy M, Sallam ES. Diagnosis of Focal Liver Diseases Based on Deep Learning Technique for Ultrasound Images. J Sci Engi. 2017;42:3127-3140. [RCA] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 60] [Article Influence: 7.5] [Reference Citation Analysis (1)] |
| 42. | Schmauch B, Herent P, Jehanno P, Dehaene O, Saillard C, Aubé C, Luciani A, Lassau N, Jégou S. Diagnosis of focal liver lesions from ultrasound using deep learning. Diagn Interv Imaging. 2019;100:227-233. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 104] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 43. | Guo LH, Wang D, Qian YY, Zheng X, Zhao CK, Li XL, Bo XW, Yue WW, Zhang Q, Shi J, Xu HX. A two-stage multi-view learning framework based computer-aided diagnosis of liver tumors with contrast enhanced ultrasound images. Clin Hemorheol Microcirc. 2018;69:343-354. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 72] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 44. | Pan F, Huang Q, Li X. Classification of liver tumors with CEUS based on 3D-CNN. Int Confer Adv Rob Mechat. 2019;845-849. |
| 45. | Gatos I, Tsantis S, Spiliopoulos S, Skouroliakou A, Theotokas I, Zoumpoulis P, Hazle JD, Kagadis GC. A new automated quantification algorithm for the detection and evaluation of focal liver lesions with contrast-enhanced ultrasound. Med Phys. 2015;42:3948-3959. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 42] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 46. | Kondo S, Takagi K, Nishida M, Iwai T, Kudo Y, Ogawa K, Kamiyama T, Shibuya H, Kahata K, Shimizu C. Computer-Aided Diagnosis of Focal Liver Lesions Using Contrast-Enhanced Ultrasonography With Perflubutane Microbubbles. IEEE Trans Med Imaging. 2017;36:1427-1437. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 54] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 47. | Streba CT, Ionescu M, Gheonea DI, Sandulescu L, Ciurea T, Saftoiu A, Vere CC, Rogoveanu I. Contrast-enhanced ultrasonography parameters in neural network diagnosis of liver tumors. World J Gastroenterol. 2012;18:4427-4434. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 53] [Cited by in RCA: 46] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 48. | Căleanu CD, Sîrbu CL, Simion G. Deep Neural Architectures for Contrast Enhanced Ultrasound (CEUS) Focal Liver Lesions Automated Diagnosis. Sensors (Basel). 2021;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 22] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 49. | Hu HT, Wang W, Chen LD, Ruan SM, Chen SL, Li X, Lu MD, Xie XY, Kuang M. Artificial intelligence assists identifying malignant vs benign liver lesions using contrast-enhanced ultrasound. J Gastroenterol Hepatol. 2021;36:2875-2883. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 40] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 50. | Zhou L, Rui JA, Zhou WX, Wang SB, Chen SG, Qu Q. Edmondson-Steiner grade: A crucial predictor of recurrence and survival in hepatocellular carcinoma without microvascular invasio. Pathol Res Pract. 2017;213:824-830. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 73] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 51. | Zhou L, Rui JA, Wang SB, Chen SG, Qu Q, Chi TY, Wei X, Han K, Zhang N, Zhao HT. Factors predictive for long-term survival of male patients with hepatocellular carcinoma after curative resection. J Surg Oncol. 2007;95:298-303. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 33] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 52. | Court CM, Harlander-Locke MP, Markovic D, French SW, Naini BV, Lu DS, Raman SS, Kaldas FM, Zarrinpar A, Farmer DG, Finn RS, Sadeghi S, Tomlinson JS, Busuttil RW, Agopian VG. Determination of hepatocellular carcinoma grade by needle biopsy is unreliable for liver transplant candidate selection. Liver Transpl. 2017;23:1123-1132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 31] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 53. | Liu D, Liu F, Xie X, Su L, Liu M, Kuang M, Huang G, Wang Y, Zhou H, Wang K, Lin M, Tian J. Accurate prediction of responses to transarterial chemoembolization for patients with hepatocellular carcinoma by using artificial intelligence in contrast-enhanced ultrasound. Eur Radiol. 2020;30:2365-2376. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 96] [Article Influence: 19.2] [Reference Citation Analysis (0)] |
| 54. | Liu F, Liu D, Wang K, Xie X, Su L, Kuang M, Huang G, Peng B, Wang Y, Lin M, Tian J. Deep Learning Radiomics Based on Contrast-Enhanced Ultrasound Might Optimize Curative Treatments for Very-Early or Early-Stage Hepatocellular Carcinoma Patients. Liver Cancer. 2020;9:397-413. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 103] [Article Influence: 20.6] [Reference Citation Analysis (0)] |
| 55. | Wang W, Wu SS, Zhang JC, Xian MF, Huang H, Li W, Zhou ZM, Zhang CQ, Wu TF, Li X, Xu M, Xie XY, Kuang M, Lu MD, Hu HT. Preoperative Pathological Grading of Hepatocellular Carcinoma Using Ultrasomics of Contrast-Enhanced Ultrasound. Acad Radiol. 2021;28:1094-1101. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 22] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 56. | Hu HT, Wang Z, Huang XW, Chen SL, Zheng X, Ruan SM, Xie XY, Lu MD, Yu J, Tian J, Liang P, Wang W, Kuang M. Ultrasound-based radiomics score: a potential biomarker for the prediction of microvascular invasion in hepatocellular carcinoma. Eur Radiol. 2019;29:2890-2901. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 141] [Article Influence: 20.1] [Reference Citation Analysis (0)] |
| 57. | Dong Y, Zhou L, Xia W, Zhao XY, Zhang Q, Jian JM, Gao X, Wang WP. Preoperative Prediction of Microvascular Invasion in Hepatocellular Carcinoma: Initial Application of a Radiomic Algorithm Based on Grayscale Ultrasound Images. Front Oncol. 2020;10:353. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 28] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 58. | Zhang D, Wei Q, Wu GG, Zhang XY, Lu WW, Lv WZ, Liao JT, Cui XW, Ni XJ, Dietrich CF. Preoperative Prediction of Microvascular Invasion in Patients With Hepatocellular Carcinoma Based on Radiomics Nomogram Using Contrast-Enhanced Ultrasound. Front Oncol. 2021;11:709339. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 24] [Article Influence: 6.0] [Reference Citation Analysis (0)] |