Published online Jul 7, 2022. doi: 10.3748/wjg.v28.i25.2890
Peer-review started: January 17, 2022
First decision: March 8, 2022
Revised: March 20, 2022
Accepted: May 14, 2022
Article in press: May 14, 2022
Published online: July 7, 2022
Processing time: 167 Days and 15 Hours
Non-alcoholic fatty liver disease (NAFLD) is the most common chronic liver disease worldwide and is strongly associated with metabolic deregulation. More recently, a significant impact of parental NAFLD in the offspring was demon
Core Tip: Non-alcoholic fatty liver disease (NAFLD) is a multifactorial disease and familial clustering has been described, although there is still some debate about this association. Among the factors that contribute to the disease in the offspring of NAFLD patients, genetic, epigenetic and environmental factors are the most plausible ones. In this review we describe the main genetic, environmental and epigenetic factors linked to NAFLD and the studies investigating the relation of NAFLD in parents and its offspring. Although there are many experimental studies in animals, there is still much to be elucidated regarding studies and interventions in human beings.
- Citation: Wajsbrot NB, Leite NC, Salles GF, Villela-Nogueira CA. Non-alcoholic fatty liver disease and the impact of genetic, epigenetic and environmental factors in the offspring. World J Gastroenterol 2022; 28(25): 2890-2899
- URL: https://www.wjgnet.com/1007-9327/full/v28/i25/2890.htm
- DOI: https://dx.doi.org/10.3748/wjg.v28.i25.2890
Although non-alcoholic fatty liver disease (NAFLD) is being replaced by metabolic dysfunction-associated fatty liver disease[1], studies concerning genetic and epigenetic factors in this new scenario are still scarce. This way, we will still adopt the nomenclature NAFLD when discussing the studies in this review.
NAFLD affects about 25% to 45% of the world's western population[2]. The spectrum of the disease includes simple steatosis, steatohepatitis with or without fibrosis, leading to cirrhosis, hepatic decompensation and hepatocellular carcinoma (HCC)[3]. NAFLD is currently the third indication for liver transplantation worldwide, and it will potentially be the leading indication in 2030[4].
Many cofactors have been recognized and related to NAFLD's high prevalence and severity. Metabolic syndrome, obesity and type 2 diabetes mellitus (T2DM) are the most relevant factors associated with progression from non-alcoholic fatty liver (NAFL) to non-alcoholic steatohepatitis (NASH) and fibrosis. Patients with T2DM have a higher prevalence of NAFLD, with a high prevalence of NASH and advanced fibrosis[5]. In a bidirectional relation, NAFLD also increases up to 5.5 times the risk of future development of T2DM and could be considered an early predictor of the disease[6]. Ethnicity also influences NAFLD prevalence, with Hispanics presenting a higher prevalence than Caucasians and African Americans, independently of metabolic factors. The genetic and environmental basis could be responsible for these findings in diverse ethnic groups[7]. Accordingly, the observation of NASH and cirrhosis familial clusters suggests a substantial hereditary influence on NAFLD progression[8]. Data from diverse epidemiological, familial aggregation and twin-cohorts studies, with a well-designed methodology, suggest that hepatic steatosis is highly heritable[9-12]. Some of these studies used magnetic resonance elastography to assess liver fibrosis or serum aminotransferase levels to infer hepatic steatosis. They demonstrated a high prevalence of NAFLD in family members of children with NAFLD, monozygotic and dizygotic twins, and first-degree family members of T2DM patients[11,13]. So far, the risk of hepatic steatosis and more severe disease in family members and children of patients with NAFLD is not fully understood, as well as the pathogenetic pathways involved in this process.
Genome-wide association studies have demonstrated the association of single nucleotide polymorphisms (SNP) with NAFLD. Patatin-like phospholipase-domain-containing 3 (rs738409 C>G encoding for PNPLA3 I148M), also known as adiponutrin gene, is located at chromosome 22 and was the first SNP described[14]. Although this is the most robust variant linked to NAFLD, additional genetic variants have been identified subsequently: Transmembrane 6 superfamily member 2 (TM6SF2)[15], glucokinase regulator (GCKR)[16], membrane-bound O-acyltransferase domain-containing 7 (MBOAT7)[17] and hydroxysteroid 17 β-dehydrogenase (HSD17B13)[18], among others. These variants have been associated with multiple pleiotropic effects, including a protective effect for NAFLD as seems to occur with the HSD17B13 polymorphism[18]. The different phenotypes resulting from these genes might partially explain the heritable component and metabolic profile of NAFLD patients and their offspring[9].
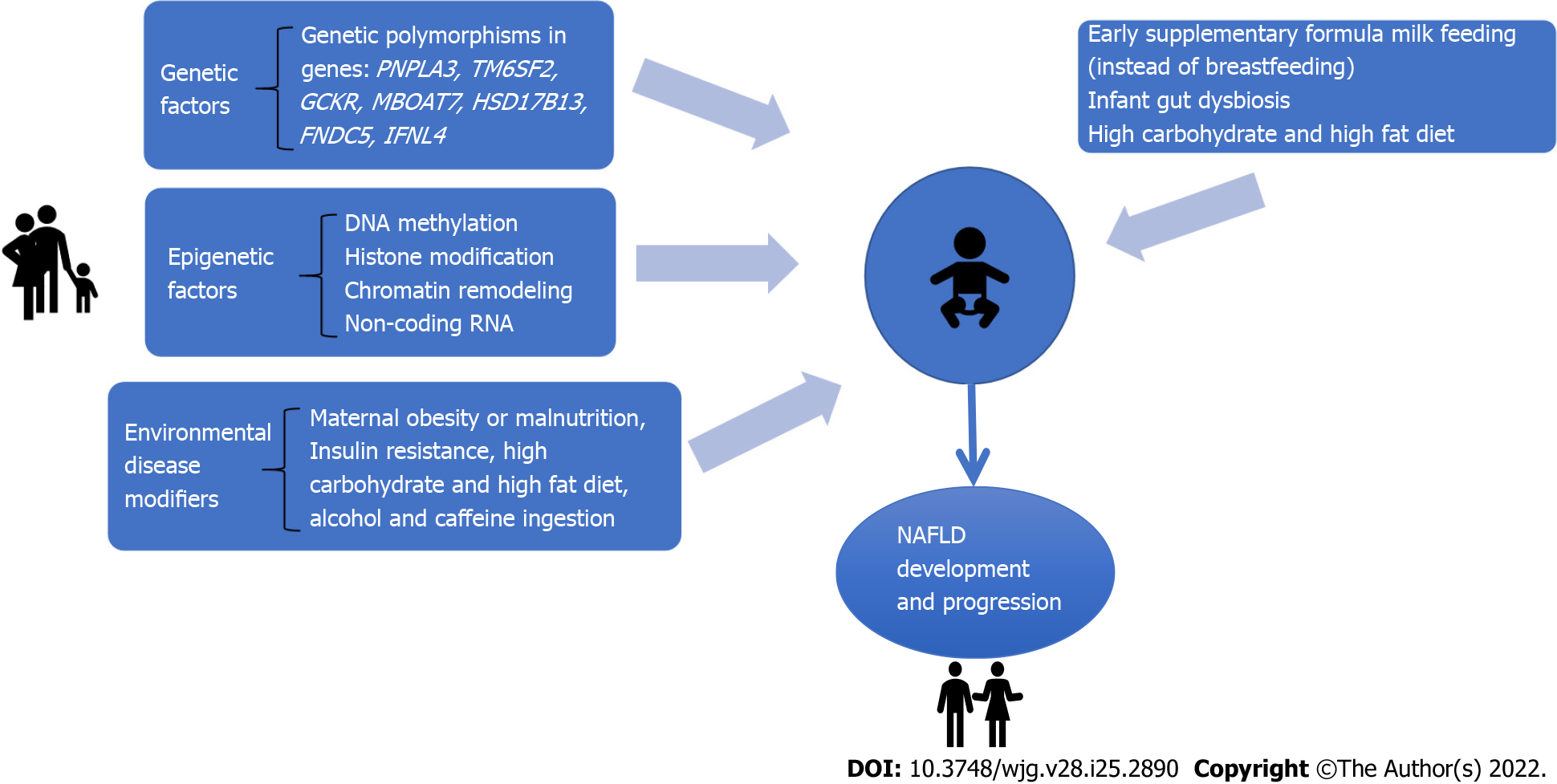
Although our understanding of genetic influence has exponentially increased in the past few years, it cannot thoroughly explain the high prevalence of NAFLD in family members of patients with the disease. Experimental studies have investigated different pathways related to NAFLD development in the offspring[9,19,20]. In this context, environmental and epigenetic mechanisms play an essential role in the occurrence and progression of NAFLD. Epigenetic factors involve mechanisms that affect and regulate gene expression without changes in DNA sequences[21]. Therefore, gene expression and cell phenotype related to NAFLD might depend on the genetic information encoded by DNA sequences and epigenetic factors[22]. Figure 1 shows the multifactorial mechanisms implicated in the offspring's NAFLD development. This review aims to discuss the impact of genetic, epigenetic and environment-related variables associated with NAFLD in the offspring.
Several studies have shown a solid familial clustering of NAFLD, particularly in coexisting metabolic traits[23-25]. Familial combined hyperlipidemia is the most frequent genetic dyslipidemia with a high risk of premature atherothrombotic cardiovascular disease. To assess whether liver steatosis is involved in the pathogenetic pathway of familial combined hyperlipidemia, Brouwers et al[23] studied family members with steatosis and twenty spouses. Fatty liver diagnosed by ultrasound was significantly more prevalent in familial combined hyperlipidemia probands (40%) and relatives (35%) compared with their spouses[23]. Moreover, the authors evaluated the correlations between indicators of fatty liver with plasma lipid levels. Liver steatosis and alanine aminotransferase levels correlated with triglyceride levels in all familial combined hyperlipidemia family members[23].
In the multigenerational Framingham Heart Study, a community-based study, individuals with at least one parent presenting hepatic steatosis had two-fold increased odds of having liver steatosis themselves than those without a parental history. More participants without metabolic diseases had liver steatosis if they had at least one parent with liver steatosis than those without any parent with steatosis. On the other hand, there was no difference in the prevalence of steatosis in patients with high cardiometabolic risk among participants with or without a parental history of liver steatosis. Based on these findings, this study suggested that a family history of liver steatosis was a significant risk factor for liver steatosis, but only in metabolically healthy participants[26]. This study goes against the previous one, which showed a higher prevalence of steatosis in those with familial hyperlipidemia. There was no investigation if the genetic aspects of those patients with familial hyperlipidemia could have influenced the higher prevalence of steatosis.
Schwimmer et al[11] evaluated 33 overweight children with biopsy-proven NAFLD and 11 overweight children without; NAFLD was significantly more observed in siblings and parents of the NAFLD children group. The correlation of liver fat fraction to body mass index (BMI) was more substantial in overweight children with NAFLD than without NAFLD, showing that there is likely an interaction between BMI and genetic factors on steatosis severity in families of children with NAFLD[11].
Similar to steatosis, hepatic fibrosis in NAFLD is also a heritable trait. Familial aggregation studies revealed a marked coexistence of advanced fibrosis or NAFLD cirrhosis among index patients and their first-degree relatives[24,25]. A cross-sectional analysis demonstrated that first-degree relatives of probands with NAFLD cirrhosis present a 12 times higher risk of advanced fibrosis compared with the relatives of non-NAFLD controls[25]. Interestingly, in another recent cross-sectional study of a prospective cohort comprising 156 twins and their families, the same authors identified a metabolite (3-4-hydroxyphenyl lactate) related to the abundance of several gut microbiota species in individuals with advanced fibrosis. Then, in their conclusions, they propose a link between genetics and microbiota composition concerning NAFLD heritability[27].
The potential genetic link of NAFLD regarding steatosis and fibrosis inheritance triggered the development of studies in twins to evaluate if both steatosis and fibrosis had a significant shared gene. The first study on twins regarding NAFLD inheritance included 60 monozygotic and dizygotic twins[13]. Both liver steatosis and fibrosis were non-invasively quantified by magnetic resonance imaging. The presence of hepatic steatosis by proton-density fat fraction magnetic resonance imaging (MRI) and fibrosis by magnetic resonance elastography correlated between monozygotic twins but not between dizygotic twins, providing evidence that both hepatic steatosis and fibrosis might be heritable traits as well[13].
Following the same rationale, Cui et al[10] investigated a prospective cohort of community-dwelling monozygotic and dizygotic twin pairs living in Southern California, using non-invasive proton-density fat fraction MRI and magnetic resonance elastography to assess steatosis and fibrosis. They investigated if individuals prone to genetic susceptibilities to steatosis and fibrosis also had genetic susceptibilities to metabolic variables such as arterial hypertension, dyslipidemia, insulin resistance and diabetes mellitus. The authors have shown that hepatic steatosis and fibrosis have statistically and clinically significant shared genetic determination and metabolic traits such as high-density lipoprotein, triglycerides, insulin resistance, and glycosylated hemoglobin[10]. In another study, the same cohort of twins was evaluated regarding the metabolites of the gut microbiome and its effect on steatosis and liver fibrosis compared to a biopsy-proven NAFLD cohort. This proof of concept study provided a link between the gut-microbiome and 3-lactate that shared gene-effect with hepatic steatosis and fibrosis[27]. Hence, the heritage of NAFLD might relate to multiple factors like a genetic inheritance that could directly affect steatosis and fibrosis and heritable traits of the gut microbiome inherited, or even be influenced by a shared lifestyle in the probands and its parents.
Genetic polymorphisms are involved in NAFLD expression regarding its relation with liver steatosis, advanced stages of fibrosis, and even a possible protective effect for disease progression[9,18,28]. However, studies evaluating their impact on the offspring of patients with NAFLD are scarce.
As previously described, PNPLA3 rs738409 C>G variant is associated with hepatic steatosis and severity of NAFLD, progression to cirrhosis and HCC, resulting in a worse prognosis[14]. PNPLA3 encodes a triacylglycerol lipase, and this variant promotes hepatic triglyceride accumulation by restricting substrate access to the catalytic dyad, thus inhibiting triglyceride hydrolysis in the cell[14].
TM6SF2 function is related to regulating cholesterol synthesis and secretion of lipoproteins. Individuals who carry the SNP rs58542926 C>T, which encodes the E167K amino acidic substitution, have a higher risk of NAFLD and histological disease severity. However, there is still a strong debate if it has a protective effect on coronary artery disease. A large study with 60801 patients with coronary artery disease compared to 123504 healthy individuals described a protective effect of the T variant of TM6SF2 on this disease and found an equivalent, although modest effect for the G variant of PNPLA3, that was more intense in the recessive model (genotype GG). At last, an exome study including more than 300000 individuals showed that both TM6SF2 and PNPLA3 polymorphisms induce a protective effect on coronary artery disease and an increased risk of NAFLD[29]. So far, there is no study regarding the evaluation of the impact of TM6SF2 in the offspring of NAFLD patients.
In young adolescents, the rs1260326 C>T variant in GCKR was significantly associated with de novo lipogenesis in those with TT genotype. Another variant in GCKR, the rs780094 A>G, was also associated with NAFLD in a meta-analysis involving 2091 cases and 3003 controls[30].
MBOAT7 was first studied in alcohol abusers and was related to a higher risk of cirrhosis. It encodes a protein involved in the re-acylation of phospholipids as a component of the phospholipid-remodeling pathway, known as the land cycle. Subsequently, the rs641738 C>T variant in this gene was associated with increased hepatic fat, more severe liver damage and fibrosis in NAFLD individuals of European descent; moreover, it has been demonstrated that the T allele may predispose to HCC in patients without cirrhosis[31].
Recently, three polymorphisms have been identified as protective against advanced stages of NAFLD. Results from exome-sequence data from 46455 individuals have shown an association of rs72613567:TA in HSD17B13, a variant with an adenine insertion, with lower levels of aminotransferases and reduced risk of chronic liver disease, including NASH[18]. Pirola et al[32] demonstrated the effect of this variant on a Hispanic population submitted to liver biopsy, investigating its association with histological parameters of NAFLD. They identified a lower risk of ballooning degeneration, lobular inflammation and liver fibrosis, mediated by reduced enzyme activity in converting retinol to retinoic acid, suggesting a protective effect in inflammation and fibrosis[32]. Di Sessa et al[33] evaluated 685 obese children (mean age 10.56 ± 2.94 years) and demonstrated that carriers of the HSD17B13 A allele had a lower percentage of liver steatosis on ultrasound imaging and lower serum aminotransferases levels[33].
Petta et al[34] evaluated the role of irisin, a myokine encoded by the fibronectin type III domain-containing protein 5 gene (FNDC5), in NAFLD patients. The variant rs3480 A>G was not associated with the severity of steatosis and NASH but was correlated with a lower prevalence of clinically significant fibrosis (F2-F4), showing a protective effect against fibrosis. They also found that irisin is expressed in human activated hepatic stellate cells, promoting profibrogenic actions and collagen synthesis. Thus, the FNDC5 genotype might affect hepatic fibrogenesis by modulating irisin secretion[34].
The genetic polymorphisms associated with NAFLD, their functions and effects are summarized in Table 1.
| Gene | Variant (s) | Function | Phenotype |
| PNPLA3 | rs738409 C>G | Triglyceride hydrolysis | ↑ NAFLD, NASH, fibrosis, HCC |
| TM6SF2 | rs58542926 C>T | Lipoproteins secretion | ↑ NAFLD, NASH, fibrosis |
| GCKR | rs1260326 C>Trs780094 A>G | De novo lipogenesis regulation | ↑ NAFLD, NASH, fibrosis |
| MBOAT7 | rs641738 C>T | Phospholipid metabolism | ↑ NAFLD and fibrosis |
| HSD17B13 | rs72613567:TA | Conversion of retinol to retinoic acid | ↓ NASH and fibrosis |
| FNDC5 | rs3480 A>G | Hepatic fibrogenesis | ↓ fibrosis |
Concerning NAFLD and family inheritance, the PNPLA3 polymorphism was the only one studied. Overweight and obese children with NAFLD confirmed by histology were evaluated regarding the role of lifetime exposures in association with a genetic predisposition, parental obesity, economic income, programming during fetal life, being breastfed or not, and later biomarkers of dietary habits and lifestyle, correlating with fibrosis. In this study, 75% of the children had fibrosis, independently associated with PNPLA3-GG genotype, parental obesity, not being breastfed, vitamin D levels (< 20 mg/dL) and fructose consumption. Notably, a high socioeconomic maternal occupation was related to less severe fibrosis[35]. These findings reinforce the multifactorial impact of NAFLD inheritance. Recently, Jain et al[36] studied 51 patients with NAFLD and their parents compared to 50 individuals without NAFLD and their parents as a control group. They observed that parents of the NAFLD group had a higher frequency of GG genotype when compared to parents of those without NAFLD (15% vs 5%)[36]. In this study, no other factors except for PNPLA3 polymorphism were evaluated.
In addition to the genetic information encoded by DNA sequences, epigenetic modifications increase or inhibit the expression of specific genes and affect chromatin structure without modifying nucleotide sequence. Epigenetics implies inheritable changes in the expression of genes, but they can also be acquired and may occur in response to environmental factors, such as nutrition, contributing to disease risk and severity[37]. These alterations can be transferred to the next generation and, in this way, may modify metabolic and NAFLD risk in the offspring. As epigenetic changes can be inheritable and modulated by environmental stimuli, they are considered reversible and could offer new individualized prevention and therapy[37]. So far, the impact of maternal and/or paternal risk factors on the clinical phenotypes of the offspring and the underlying epigenetic mechanisms has not been fully elucidated[37].
Epigenetic phenomena include four regulatory mechanisms: Modification in DNA methylation, covalent histone modification, chromatin remodeling, and RNA-based mechanisms, such as non-coding RNA. DNA methylation is the most studied[22,38].
Some experimental studies, most of them in mice, tried to elucidate the mechanisms involved in the inheritance of NAFLD and the external factors that could modulate NAFLD development in the offspring through epigenetic factors. It has been shown that many factors during pregnancy may activate lipogenic and inflammatory pathways leading to NAFLD in the progeny[19]. Many authors have studied the impact of breastfeeding, maternal obesity and diet before or during pregnancy in animal models.
Oben et al[19] demonstrated that maternal obesity before and throughout pregnancy and lactation could be linked to dysmetabolism in the offspring of female mice. Offspring of obese dams showed a dysmetabolic pattern related to insulin resistance and NAFLD phenotype. Moreover, the offspring of lean dams fed by obese dams presented increased body weight and higher insulin levels and cytokines such as leptin, interleukin-6 and tumor necrosis factor-alpha. Raised levels of leptin were also observed in the breast milk of obese mice compared to lean ones. They proposed that a modified pathway over hypothalamic appetite nuclei signaling by maternal breast milk and neonatal adipose tissue-derived leptin in the early postnatal period was the mechanism behind these findings.
Considering the hypothesis that diet during and after pregnancy might also be involved in NAFLD in the post-weaning period, Pruis et al[39] observed that a maternal western-type diet during pregnancy could stimulate metabolic programming or phenotype induction, leading to NAFLD development.
Another study[40] suggested that modifying the diet during pregnancy could benefit the offspring by preventing a disrupted liver lipid profile[61]. When pregnant mice were fed either with a high fat-slow digestive diet or a rapid digestive diet, the offspring of the high fat-rapid digestive diet showed an abnormal liver lipid profile. However, it was not observed in their counterparts born from high fat-slow digestive diet fed-mice.
The relationship between obesity in pregnancy and circadian cycle deregulation might affect metabolic pathways related to NAFLD in adults. Mouralidarane et al[20] suggested that, in addition to an obesogenic post-weaning diet, obesity in the mother might lead to NAFLD by disrupting the liver's canonical metabolic rhythmicity gene expression. It implicates the role of abnormal circadian rhythm in the genesis of NAFLD, and alterations in this system during critical developmental periods might be responsible for the onset of the disease later in adulthood.
Another issue that might be considered regarding further development of NAFLD after birth is ethanol exposure[41]. Shen et al[41] developed a rat model of intrauterine growth retardation by prenatal ethanol exposure. These models were fed with normal and high-fat diets. Enhanced liver expression of the insulin growth factor-1 pathway, gluconeogenesis, lipid synthesis and diminished expression of lipid output were accompanied in prenatal ethanol exposure female offspring fed with a high-fat diet.
Oliveira et al[42] studied Wistar rats fed with a standard diet and a high-fat diet. Rats born from mice fed with a standard diet were not affected by changes in liver morphology, as did the offspring of high-fat-fed rats. Therefore, the study concluded that fructose intake during adolescence hastens NAFLD onset and reveals a differentiated hepatic response to metabolic insult, depending on the maternal diet. Notwithstanding, Nicolás-Toledo et al[43] showed that sucrose intake in adulthood increases fat content only in female rat offspring of dams fed with a low-protein diet during pregnancy, reinforcing the influence of maternal diet in the offspring[43]. Of note, regarding specific epigenetic mechanisms, Suter et al[44] have described that epigenetic changes to histones may act as a molecular memory of intrauterine exposure, rendering the risk of adult disease. The genome-wide epigenetic modifications in the fetal liver of susceptible offspring were analyzed, concluding that a maternal high-fat diet is associated with functional alterations to fetal hepatic histones, some of which may persist up to five weeks of age[44].
Another study by Wei et al[45] connected NAFLD with epigenetic methylation of specific genes in fathers that can be transmitted from gametes to embryos across generations. They have shown that even paternal diet patterns and prediabetes increase the risk of diabetes in the offspring through gametic epigenetic alterations such as different methylation of genes in the sperm of prediabetic fathers.
All these experimental studies in animal models have revealed that maternal obesity and parental diet during pregnancy or lactation may significantly influence NAFLD and lipid dysmetabolism in the offspring, either by environmental factors or through epigenetic factors, some yet to be better specified, mainly concerning environmental factors. Hence, cofactors as alcohol and fructose intake, among others not yet identified, may activate lipogenic and inflammatory pathways that can lead to NAFLD in the offspring.
Studies in mothers and newborns: Animal studies confirmed that disruptions during early deve
In prior studies, it was demonstrated that low birth weight babies exhibit an altered postnatal metabolism after developing an adaptative response to a suboptimal fetal environment[46,47]. Although the mechanism is not entirely understood, exposure to excessive and deficient nutrition during the prenatal period may induce a nutritional mismatch between metabolic efficiency and energy expenditure, increasing the risk of future cardiometabolic diseases. If confirmed, an early and straightforward nutritional intervention might prevent the further development of metabolic diseases in adulthood.
Modi et al[48] evaluated 105 healthy mother-neonate pairs. They measured neonatal adipose tissue content by whole-body MRI and intrahepatic lipid content by a proton magnetic resonance spectroscopy. They have demonstrated that infant adiposity, particularly abdominal adipose tissue and intra hepatocellular lipid correlated with increased maternal BMI. Recently, Bedogni et al[49] studied the prevalence and risk factors associated with bright liver in 391 1-year-old toddlers born from healthy mothers. The PNPLA3 I148M variant and maternal weight gain during pregnancy were related to the presence of bright liver in the ultrasonography[49]. Thus, interestingly, the authors suggested a potential gene-environment interchange between PNPLA3 and maternal environmental factors contributing to the risk of fatty liver disease at this earlier age, reinforcing the multifactorial inheritance of NAFLD.
In a large study, Ayonrinde et al[50] investigated the relation of maternal factors and infant nutrition with the future development of NAFLD in adolescents aged 17 years. They concluded that average pre-gestational BMI, breastfeeding for at least six months and avoiding early supplementary formula milk feeding reduce the risk of NAFLD diagnosis by liver ultrasound[50]. Additionally, more extended maintenance of breastfeeding resulted in multiple benefits on maternal metabolism and a lower risk of NAFLD in mid-life[51-53].
The Healthy Start study examined a cohort of 951 mothers from different ethnicities[54]. Similar to others, they found that maternal BMI was correlated to increased neonatal adiposity. It has also been demonstrated that increased maternal insulin resistance and fasting glucose levels contribute to this association. Excessive insulin resistance during pregnancy activates placental inflammatory pathways and affects the fetus indirectly by increasing placental nutrient transfer capacity[55].
Still regarding insulin-glucose metabolism, elevated blood glucose and insulin concentrations exacerbate de novo lipogenesis, resulting in increased intrahepatic lipids. Additionally, reduced glucose and pyruvate consumption in parallel with increased triglyceride concentrations and excess fatty acids incompletely oxidized can impair mitochondrial function and gene expression, limiting mitochondrial biogenesis and leading to NAFLD[55].
Peroxisome proliferator-activated receptor γcoactivator 1 (PGC1) gene is a transcriptional coactivator that participates in mitochondrial biogenesis and function and hypermethylation PGC1 promoter was associated with decreased mitochondrial DNA content and insulin resistance in NAFLD patients[56]. In a cross-sectional analysis, Gemma et al[57] noticed a positive correlation between maternal BMI and methylation of the PGC1 gene in the umbilical cord of their babies[57]. Based on their findings, the authors speculated that PGC1 might be a promising candidate gene involved in metabolic programming by epigenetic regulation[57]. DNA methylation in regulatory regions of different genes participates in NAFLD development and progression. Other epigenetic mechanisms affecting NAFLD pathogenesis include histone modification and microRNA (miRNA)-mediated processes. Notably, circulating miRNAs have been associated with the presence and heritability of NAFLD in a population study in 40 pairs of twins. Serum miR-331-3p and miR-30c were identified among the 21 miRs that differed between NAFLD and non-NAFLD individuals. These miRNAs are highly inheritable and correlate with each other suggesting a common pathway related to NAFLD[58].
Although shreds of evidence support that high pre-pregnancy BMI in the mothers may lead to significant modifications in the infant gut microbiome[59], few studies link maternal obesity and infant dysbiosis with NAFLD risk in later life. The neonatal gut microbiome can be essential for later homeostasis, and disruption of this early process may increase the risk of future metabolic diseases[60]. Emerging data provides evidence that the gut-liver axis is a fundamental element in the onset and progression of NAFLD. Gut microbiota dysbiosis may contribute to NAFLD by increasing concentrations of bacteria-derived endotoxins, pro-inflammatory cytokines, amino-acid metabolites, short-chain fatty acids and bile acids, all of which might exert effects that promote macrophage programming and activation, favoring liver injury[61].
The interplay among multiple genetic, epigenetic and environmental factors determine an individual's susceptibility to NAFLD. Current evidence points to genetic polymorphisms as pleiotropic tools that lead to diverse traits and phenotypes, including typical metabolic profiles in parents and their offspring. Importantly, epigenetic markers can also be transferred to successors by transgenerational epigenetic inheritance. Current studies in mothers and their offspring, although still small, show a direct effect of these factors and their related outcome, NAFLD. Future studies may clarify what interventions are essential for preventing this complex disease in the perinatal or postnatal period to reach the better liver and metabolic-related outcomes in the upcoming adult population.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Corresponding Author's Membership in Professional Societies: American Association for the Study of Liver Diseases, No. 114358; European Association for the study of Liver Diseases, No. 60371.
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Brazil
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Lara Riegos JC, Mexico; Sporea I, Romania A-Editor: Maslennikov R S-Editor: Fan JR L-Editor: A P-Editor: Fan JR
| 1. | Kawaguchi T, Tsutsumi T, Nakano D, Torimura T. MAFLD: Renovation of clinical practice and disease awareness of fatty liver. Hepatol Res. 2021;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 67] [Article Influence: 22.3] [Reference Citation Analysis (1)] |
| 2. | Bellentani S, Marino M. Epidemiology and natural history of non-alcoholic fatty liver disease (NAFLD). Ann Hepatol. 2009;8 Suppl 1:S4-S8. [PubMed] |
| 3. | Angulo P. Nonalcoholic fatty liver disease. N Engl J Med. 2002;346:1221-1231. [PubMed] |
| 4. | Shaker M, Tabbaa A, Albeldawi M, Alkhouri N. Liver transplantation for nonalcoholic fatty liver disease: new challenges and new opportunities. World J Gastroenterol. 2014;20:5320-5330. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 77] [Cited by in RCA: 92] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 5. | Leite NC, Villela-Nogueira CA, Pannain VL, Bottino AC, Rezende GF, Cardoso CR, Salles GF. Histopathological stages of nonalcoholic fatty liver disease in type 2 diabetes: prevalences and correlated factors. Liver Int. 2011;31:700-706. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 130] [Cited by in RCA: 131] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 6. | Shibata M, Kihara Y, Taguchi M, Tashiro M, Otsuki M. Nonalcoholic fatty liver disease is a risk factor for type 2 diabetes in middle-aged Japanese men. Diabetes Care. 2007;30:2940-2944. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 175] [Cited by in RCA: 184] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 7. | Wagenknecht LE, Scherzinger AL, Stamm ER, Hanley AJ, Norris JM, Chen YD, Bryer-Ash M, Haffner SM, Rotter JI. Correlates and heritability of nonalcoholic fatty liver disease in a minority cohort. Obesity (Silver Spring). 2009;17:1240-1246. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 89] [Cited by in RCA: 90] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 8. | Adibi A, Janghorbani M, Shayganfar S, Amini M. First-degree relatives of patients with type 2 diabetes mellitus and risk of non-alcoholic Fatty liver disease. Rev Diabet Stud. 2007;4:236-241. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 11] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 9. | Eslam M, George J. Genetic contributions to NAFLD: leveraging shared genetics to uncover systems biology. Nat Rev Gastroenterol Hepatol. 2020;17:40-52. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 239] [Cited by in RCA: 217] [Article Influence: 43.4] [Reference Citation Analysis (0)] |
| 10. | Cui J, Chen CH, Lo MT, Schork N, Bettencourt R, Gonzalez MP, Bhatt A, Hooker J, Shaffer K, Nelson KE, Long MT, Brenner DA, Sirlin CB, Loomba R; For The Genetics Of Nafld In Twins Consortium. Shared genetic effects between hepatic steatosis and fibrosis: A prospective twin study. Hepatology. 2016;64:1547-1558. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 65] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 11. | Schwimmer JB, Celedon MA, Lavine JE, Salem R, Campbell N, Schork NJ, Shiehmorteza M, Yokoo T, Chavez A, Middleton MS, Sirlin CB. Heritability of nonalcoholic fatty liver disease. Gastroenterology. 2009;136:1585-1592. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 381] [Cited by in RCA: 358] [Article Influence: 22.4] [Reference Citation Analysis (0)] |
| 12. | Dickson I. NAFLD: Increased familial risk of fibrosis in NAFLD. Nat Rev Gastroenterol Hepatol. 2017;14:450. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 13. | Loomba R, Schork N, Chen CH, Bettencourt R, Bhatt A, Ang B, Nguyen P, Hernandez C, Richards L, Salotti J, Lin S, Seki E, Nelson KE, Sirlin CB, Brenner D; Genetics of NAFLD in Twins Consortium. Heritability of Hepatic Fibrosis and Steatosis Based on a Prospective Twin Study. Gastroenterology. 2015;149:1784-1793. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 230] [Cited by in RCA: 302] [Article Influence: 30.2] [Reference Citation Analysis (0)] |
| 14. | Romeo S, Kozlitina J, Xing C, Pertsemlidis A, Cox D, Pennacchio LA, Boerwinkle E, Cohen JC, Hobbs HH. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2008;40:1461-1465. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2701] [Cited by in RCA: 2603] [Article Influence: 153.1] [Reference Citation Analysis (0)] |
| 15. | Kozlitina J, Smagris E, Stender S, Nordestgaard BG, Zhou HH, Tybjærg-Hansen A, Vogt TF, Hobbs HH, Cohen JC. Exome-wide association study identifies a TM6SF2 variant that confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2014;46:352-356. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 724] [Cited by in RCA: 932] [Article Influence: 84.7] [Reference Citation Analysis (0)] |
| 16. | Yang J, Trépo E, Nahon P, Cao Q, Moreno C, Letouzé E, Imbeaud S, Bayard Q, Gustot T, Deviere J, Bioulac-Sage P, Calderaro J, Ganne-Carrié N, Laurent A, Blanc JF, Guyot E, Sutton A, Ziol M, Zucman-Rossi J, Nault JC. A 17-Beta-Hydroxysteroid Dehydrogenase 13 Variant Protects From Hepatocellular Carcinoma Development in Alcoholic Liver Disease. Hepatology. 2019;70:231-240. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 64] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 17. | Basyte-Bacevice V, Skieceviciene J, Valantiene I, Sumskiene J, Petrenkiene V, Kondrackiene J, Petrauskas D, Lammert F, Kupcinskas J. TM6SF2 and MBOAT7 Gene Variants in Liver Fibrosis and Cirrhosis. Int J Mol Sci. 2019;20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 33] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 18. | Abul-Husn NS, Cheng X, Li AH, Xin Y, Schurmann C, Stevis P, Liu Y, Kozlitina J, Stender S, Wood GC, Stepanchick AN, Still MD, McCarthy S, O'Dushlaine C, Packer JS, Balasubramanian S, Gosalia N, Esopi D, Kim SY, Mukherjee S, Lopez AE, Fuller ED, Penn J, Chu X, Luo JZ, Mirshahi UL, Carey DJ, Still CD, Feldman MD, Small A, Damrauer SM, Rader DJ, Zambrowicz B, Olson W, Murphy AJ, Borecki IB, Shuldiner AR, Reid JG, Overton JD, Yancopoulos GD, Hobbs HH, Cohen JC, Gottesman O, Teslovich TM, Baras A, Mirshahi T, Gromada J, Dewey FE. A Protein-Truncating HSD17B13 Variant and Protection from Chronic Liver Disease. N Engl J Med. 2018;378:1096-1106. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 445] [Cited by in RCA: 611] [Article Influence: 87.3] [Reference Citation Analysis (0)] |
| 19. | Oben JA, Mouralidarane A, Samuelsson AM, Matthews PJ, Morgan ML, McKee C, Soeda J, Fernandez-Twinn DS, Martin-Gronert MS, Ozanne SE, Sigala B, Novelli M, Poston L, Taylor PD. Maternal obesity during pregnancy and lactation programs the development of offspring non-alcoholic fatty liver disease in mice. J Hepatol. 2010;52:913-920. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 261] [Cited by in RCA: 236] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 20. | Mouralidarane A, Soeda J, Sugden D, Bocianowska A, Carter R, Ray S, Saraswati R, Cordero P, Novelli M, Fusai G, Vinciguerra M, Poston L, Taylor PD, Oben JA. Maternal obesity programs offspring non-alcoholic fatty liver disease through disruption of 24-h rhythms in mice. Int J Obes (Lond). 2015;39:1339-1348. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 41] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 21. | Meaney MJ. Epigenetics and the biological definition of gene x environment interactions. Child Dev. 2010;81:41-79. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 770] [Cited by in RCA: 586] [Article Influence: 39.1] [Reference Citation Analysis (0)] |
| 22. | Chen L, Huang W, Wang L, Zhang Z, Zhang F, Zheng S, Kong D. The effects of epigenetic modification on the occurrence and progression of liver diseases and the involved mechanism. Expert Rev Gastroenterol Hepatol. 2020;14:259-270. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 16] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 23. | Brouwers MC, Cantor RM, Kono N, Yoon JL, van der Kallen CJ, Bilderbeek-Beckers MA, van Greevenbroek MM, Lusis AJ, de Bruin TW. Heritability and genetic loci of fatty liver in familial combined hyperlipidemia. J Lipid Res. 2006;47:2799-2807. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 51] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 24. | Struben VM, Hespenheide EE, Caldwell SH. Nonalcoholic steatohepatitis and cryptogenic cirrhosis within kindreds. Am J Med. 2000;108:9-13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 205] [Cited by in RCA: 195] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 25. | Caussy C, Soni M, Cui J, Bettencourt R, Schork N, Chen CH, Ikhwan MA, Bassirian S, Cepin S, Gonzalez MP, Mendler M, Kono Y, Vodkin I, Mekeel K, Haldorson J, Hemming A, Andrews B, Salotti J, Richards L, Brenner DA, Sirlin CB, Loomba R; Familial NAFLD Cirrhosis Research Consortium. Nonalcoholic fatty liver disease with cirrhosis increases familial risk for advanced fibrosis. J Clin Invest. 2017;127:2697-2704. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 132] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 26. | Long MT, Gurary EB, Massaro JM, Ma J, Hoffmann U, Chung RT, Benjamin EJ, Loomba R. Parental non-alcoholic fatty liver disease increases risk of non-alcoholic fatty liver disease in offspring. Liver Int. 2019;39:740-747. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 32] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 27. | Caussy C, Hsu C, Lo MT, Liu A, Bettencourt R, Ajmera VH, Bassirian S, Hooker J, Sy E, Richards L, Schork N, Schnabl B, Brenner DA, Sirlin CB, Chen CH, Loomba R; Genetics of NAFLD in Twins Consortium. Link between gut-microbiome derived metabolite and shared gene-effects with hepatic steatosis and fibrosis in NAFLD. Hepatology. 2018;68:918-932. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 139] [Cited by in RCA: 154] [Article Influence: 22.0] [Reference Citation Analysis (0)] |
| 28. | Anstee QM, Day CP. The Genetics of Nonalcoholic Fatty Liver Disease: Spotlight on PNPLA3 and TM6SF2. Semin Liver Dis. 2015;35:270-290. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 119] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 29. | Simons N, Isaacs A, Koek GH, Kuč S, Schaper NC, Brouwers MCGJ. PNPLA3, TM6SF2, and MBOAT7 Genotypes and Coronary Artery Disease. Gastroenterology. 2017;152:912-913. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 76] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 30. | Zain SM, Mohamed Z, Mohamed R. Common variant in the glucokinase regulatory gene rs780094 and risk of nonalcoholic fatty liver disease: a meta-analysis. J Gastroenterol Hepatol. 2015;30:21-27. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 64] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 31. | Donati B, Dongiovanni P, Romeo S, Meroni M, McCain M, Miele L, Petta S, Maier S, Rosso C, De Luca L, Vanni E, Grimaudo S, Romagnoli R, Colli F, Ferri F, Mancina RM, Iruzubieta P, Craxi A, Fracanzani AL, Grieco A, Corradini SG, Aghemo A, Colombo M, Soardo G, Bugianesi E, Reeves H, Anstee QM, Fargion S, Valenti L. MBOAT7 rs641738 variant and hepatocellular carcinoma in non-cirrhotic individuals. Sci Rep. 2017;7:4492. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 203] [Cited by in RCA: 196] [Article Influence: 24.5] [Reference Citation Analysis (0)] |
| 32. | Pirola CJ, Garaycoechea M, Flichman D, Arrese M, San Martino J, Gazzi C, Castaño GO, Sookoian S. Splice variant rs72613567 prevents worst histologic outcomes in patients with nonalcoholic fatty liver disease. J Lipid Res. 2019;60:176-185. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 107] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 33. | Di Sessa A, Umano GR, Cirillo G, Passaro AP, Verde V, Cozzolino D, Guarino S, Marzuillo P, Miraglia Del Giudice E. Pediatric non-alcoholic fatty liver disease and kidney function: Effect of HSD17B13 variant. World J Gastroenterol. 2020;26:5474-5483. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 19] [Cited by in RCA: 22] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 34. | Petta S, Valenti L, Svegliati-Baroni G, Ruscica M, Pipitone RM, Dongiovanni P, Rychlicki C, Ferri N, Cammà C, Fracanzani AL, Pierantonelli I, Di Marco V, Meroni M, Giordano D, Grimaudo S, Maggioni M, Cabibi D, Fargion S, Craxì A. Fibronectin Type III Domain-Containing Protein 5 rs3480 A>G Polymorphism, Irisin, and Liver Fibrosis in Patients With Nonalcoholic Fatty Liver Disease. J Clin Endocrinol Metab. 2017;102:2660-2669. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 44] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 35. | Mosca A, De Cosmi V, Parazzini F, Raponi M, Alisi A, Agostoni C, Nobili V. The Role of Genetic Predisposition, Programing During Fetal Life, Family Conditions, and Post-natal Diet in the Development of Pediatric Fatty Liver Disease. J Pediatr. 2019;211:72-77.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 22] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 36. | Jain S, Thanage R, Panchal F, Rathi PM, Munshi R, Udgirkar SS, Contractor QQ, Chandnani SJ, Sujit NP, Debnath P, Singh A. Screening of Family Members of Nonalcoholic Fatty Liver Disease Patients can Detect Undiagnosed Nonalcoholic Fatty Liver Disease Among Them: Is There a Genetic Link? J Clin Exp Hepatol. 2021;11:466-474. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 3] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 37. | Jonas W, Schürmann A. Genetic and epigenetic factors determining NAFLD risk. Mol Metab. 2021;50:101111. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 107] [Article Influence: 21.4] [Reference Citation Analysis (0)] |
| 38. | Botello-Manilla AE, Chávez-Tapia NC, Uribe M, Nuño-Lámbarri N. Genetics and epigenetics purpose in nonalcoholic fatty liver disease. Expert Rev Gastroenterol Hepatol. 2020;14:733-748. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 13] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 39. | Pruis MG, Lendvai A, Bloks VW, Zwier MV, Baller JF, de Bruin A, Groen AK, Plösch T. Maternal western diet primes non-alcoholic fatty liver disease in adult mouse offspring. Acta Physiol (Oxf). 2014;210:215-227. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 70] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 40. | Salto R, Manzano M, Girón MD, Cano A, Castro A, Vílchez JD, Cabrera E, López-Pedrosa JM. A Slow-Digesting Carbohydrate Diet during Rat Pregnancy Protects Offspring from Non-Alcoholic Fatty Liver Disease Risk through the Modulation of the Carbohydrate-Response Element and Sterol Regulatory Element Binding Proteins. Nutrients. 2019;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 41. | Shen L, Liu Z, Gong J, Zhang L, Wang L, Magdalou J, Chen L, Wang H. Prenatal ethanol exposure programs an increased susceptibility of non-alcoholic fatty liver disease in female adult offspring rats. Toxicol Appl Pharmacol. 2014;274:263-273. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 82] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 42. | Oliveira LS, Caetano B, Miranda RA, Souza AFP, Cordeiro A, Woyames J, Andrade CBV, Atella GC, Takiya CM, Fortunato RS, Trevenzoli IH, Souza LL, Pazos-Moura CC. Differentiated Hepatic Response to Fructose Intake during Adolescence Reveals the Increased Susceptibility to Non-Alcoholic Fatty Liver Disease of Maternal High-Fat Diet Male Rat Offspring. Mol Nutr Food Res. 2020;64:e1900838. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 15] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 43. | Nicolás-Toledo L, Cervantes-Rodríguez M, Cuevas-Romero E, Corona-Quintanilla DL, Pérez-Sánchez E, Zambrano E, Castelán F, Rodríguez-Antolín J. Hitting a triple in the non-alcoholic fatty liver disease field: sucrose intake in adulthood increases fat content in the female but not in the male rat offspring of dams fed a gestational low-protein diet. J Dev Orig Health Dis. 2018;9:151-159. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 6] [Article Influence: 0.8] [Reference Citation Analysis (1)] |
| 44. | Suter MA, Ma J, Vuguin PM, Hartil K, Fiallo A, Harris RA, Charron MJ, Aagaard KM. In utero exposure to a maternal high-fat diet alters the epigenetic histone code in a murine model. Am J Obstet Gynecol. 2014;210:463.e1-463.e11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 51] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 45. | Wei Y, Yang CR, Wei YP, Zhao ZA, Hou Y, Schatten H, Sun QY. Paternally induced transgenerational inheritance of susceptibility to diabetes in mammals. Proc Natl Acad Sci U S A. 2014;111:1873-1878. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 302] [Cited by in RCA: 317] [Article Influence: 28.8] [Reference Citation Analysis (0)] |
| 46. | Barker DJ, Hales CN, Fall CH, Osmond C, Phipps K, Clark PM. Type 2 (non-insulin-dependent) diabetes mellitus, hypertension and hyperlipidaemia (syndrome X): relation to reduced fetal growth. Diabetologia. 1993;36:62-67. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1710] [Cited by in RCA: 1599] [Article Influence: 50.0] [Reference Citation Analysis (0)] |
| 47. | Sarr O, Yang K, Regnault TR. In utero programming of later adiposity: the role of fetal growth restriction. J Pregnancy. 2012;2012:134758. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 68] [Cited by in RCA: 80] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 48. | Modi N, Murgasova D, Ruager-Martin R, Thomas EL, Hyde MJ, Gale C, Santhakumaran S, Doré CJ, Alavi A, Bell JD. The influence of maternal body mass index on infant adiposity and hepatic lipid content. Pediatr Res. 2011;70:287-291. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 144] [Cited by in RCA: 124] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 49. | Bedogni G, De Matteis G, Fabrizi M, Alisi A, Crudele A, Pizzolante F, Signore F, Dallapiccola B, Nobili V, Manco M. Association of Bright Liver With the PNPLA3 I148M Gene Variant in 1-Year-Old Toddlers. J Clin Endocrinol Metab. 2019;104:2163-2170. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 3] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 50. | Ayonrinde OT, Oddy WH, Adams LA, Mori TA, Beilin LJ, de Klerk N, Olynyk JK. Infant nutrition and maternal obesity influence the risk of non-alcoholic fatty liver disease in adolescents. J Hepatol. 2017;67:568-576. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 93] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 51. | Gunderson EP, Jacobs DR Jr, Chiang V, Lewis CE, Feng J, Quesenberry CP Jr, Sidney S. Duration of lactation and incidence of the metabolic syndrome in women of reproductive age according to gestational diabetes mellitus status: a 20-Year prospective study in CARDIA (Coronary Artery Risk Development in Young Adults). Diabetes. 2010;59:495-504. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 179] [Cited by in RCA: 187] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 52. | Stuebe AM, Rich-Edwards JW, Willett WC, Manson JE, Michels KB. Duration of lactation and incidence of type 2 diabetes. JAMA. 2005;294:2601-2610. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 331] [Cited by in RCA: 314] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 53. | Ajmera V, Liu A, Bettencourt R, Dhar D, Richards L, Loomba R. The impact of genetic risk on liver fibrosis in non-alcoholic fatty liver disease as assessed by magnetic resonance elastography. Aliment Pharmacol Ther. 2021;54:68-77. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 24] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 54. | Shapiro AL, Schmiege SJ, Brinton JT, Glueck D, Crume TL, Friedman JE, Dabelea D. Testing the fuel-mediated hypothesis: maternal insulin resistance and glucose mediate the association between maternal and neonatal adiposity, the Healthy Start study. Diabetologia. 2015;58:937-941. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 45] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 55. | Dimasuay KG, Boeuf P, Powell TL, Jansson T. Placental Responses to Changes in the Maternal Environment Determine Fetal Growth. Front Physiol. 2016;7:12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 174] [Cited by in RCA: 191] [Article Influence: 21.2] [Reference Citation Analysis (0)] |
| 56. | Puigserver P, Spiegelman BM. Peroxisome proliferator-activated receptor-gamma coactivator 1 alpha (PGC-1 alpha): transcriptional coactivator and metabolic regulator. Endocr Rev. 2003;24:78-90. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1537] [Cited by in RCA: 1607] [Article Influence: 73.0] [Reference Citation Analysis (0)] |
| 57. | Gemma C, Sookoian S, Alvariñas J, García SI, Quintana L, Kanevsky D, González CD, Pirola CJ. Maternal pregestational BMI is associated with methylation of the PPARGC1A promoter in newborns. Obesity (Silver Spring). 2009;17:1032-1039. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 130] [Cited by in RCA: 116] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 58. | Zarrinpar A, Gupta S, Maurya MR, Subramaniam S, Loomba R. Serum microRNAs explain discordance of non-alcoholic fatty liver disease in monozygotic and dizygotic twins: a prospective study. Gut. 2016;65:1546-1554. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 75] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 59. | Galley JD, Bailey M, Kamp Dush C, Schoppe-Sullivan S, Christian LM. Maternal obesity is associated with alterations in the gut microbiome in toddlers. PLoS One. 2014;9:e113026. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 117] [Cited by in RCA: 135] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 60. | Soderborg TK, Clark SE, Mulligan CE, Janssen RC, Babcock L, Ir D, Young B, Krebs N, Lemas DJ, Johnson LK, Weir T, Lenz LL, Frank DN, Hernandez TL, Kuhn KA, D'Alessandro A, Barbour LA, El Kasmi KC, Friedman JE. The gut microbiota in infants of obese mothers increases inflammation and susceptibility to NAFLD. Nat Commun. 2018;9:4462. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 128] [Cited by in RCA: 214] [Article Influence: 30.6] [Reference Citation Analysis (0)] |
| 61. | Wesolowski SR, Kasmi KC, Jonscher KR, Friedman JE. Developmental origins of NAFLD: a womb with a clue. Nat Rev Gastroenterol Hepatol. 2017;14:81-96. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 174] [Article Influence: 21.8] [Reference Citation Analysis (0)] |