Published online Jul 7, 2022. doi: 10.3748/wjg.v28.i25.2854
Peer-review started: January 14, 2022
First decision: March 9, 2022
Revised: March 23, 2022
Accepted: May 27, 2022
Article in press: May 27, 2022
Published online: July 7, 2022
Processing time: 170 Days and 14.6 Hours
Hepatocellular carcinoma (HCC) represents the primary carcinoma of the liver and the fourth leading cause of cancer-related deaths. The World Health Organization estimates an increase in cases in the coming years. The risk factors of HCC are multiple, and the incidence in different countries is closely related to the different risk factors to which the population is exposed. The molecular me
Core Tip: The molecular mechanisms that drive hepatocellular carcinoma tumorigenesis are extremely complex, and a univocal classification based on molecular features has not been defined. In the age of precision medicine, the study of hepatocellular carcinoma mutations is still a field worth investigating. Based on this, we wanted to analyze the possible correlations between molecular alterations and pathological features. Considering both the literature data and our personal experience, about 80% of hepatocellular carcinomas harbor mutations in at least one of the following genes, TERT, TP53, or CTNNB1, with different biological and clinical implications.
- Citation: Maloberti T, De Leo A, Sanza V, Gruppioni E, Altimari A, Riefolo M, Visani M, Malvi D, D’Errico A, Tallini G, Vasuri F, de Biase D. Correlation of molecular alterations with pathological features in hepatocellular carcinoma: Literature review and experience of an Italian center. World J Gastroenterol 2022; 28(25): 2854-2866
- URL: https://www.wjgnet.com/1007-9327/full/v28/i25/2854.htm
- DOI: https://dx.doi.org/10.3748/wjg.v28.i25.2854
Hepatocellular carcinoma (HCC) is one of the world-leading cancers, representing approximately 80% of the primary carcinomas of the liver[1] and the fourth most common cause of cancer-related deaths. The World Health Organization (WHO) estimated more than 905500 new HCC cases in 2020 worldwide[2], and based on its projection an increase of 58% is evaluated by 2040 with a total of 1400000 new cases and 1000000 deaths in 2030[2].
The etiological factors for HCC development are: (1) Infections, including hepatitis B virus (HBV) and hepatitis C virus (HCV), with or without coinfection of hepatitis delta virus; (2) Lifestyle risk factors and behaviors, such as alcohol addiction and smoking; (3) Environment, such as dietary toxins (e.g., aflatoxins, or aristolochic acid); (4) Underlying diseases, such as obesity, type 2 diabetes, nonalcoholic liver steatohepatitis/nonalcoholic fatty liver disease; and (5) Genetics, some single nucleotide polymorphisms are identified to be associated with HCC risk at different stages, from predisposition to risk factors to the severity of the chronic liver disease and its evolution to cirrhosis or to the malignant transformation and tumor progression[3,4]. For example, a single nucleotide polymorphism correlated with higher infection risk (MDM2 Promoter SNP309, MDM2 G-309T, rs2279744) has been associated with HCC patients with chronic hepatitis C[5].
The incidence of HCC in different countries varies considering the different risk factors mentioned above. In Eastern Asian countries and most African countries, the incidence of HCC is mostly due to aflatoxin exposure and HBV infection, except for Northern Africa where HCV infection is prevalent[6,7]. In traditional Chinese herbal medicines, practiced particularly in China, Vietnam, and Southeast Asia, plants containing aristolochic acid are commonly used. In this area, next-generation sequencing studies underlined that a fraction of HCCs harbored high rates of mutations matching a distinctive mutational signature of aristolochic acid exposure[8-10]. Moving to Western countries, the incidence of HCC is usually associated with HCV infection, dietary habits, and related metabolic diseases, such as nonalcoholic liver steatohepatitis and nonalcoholic fatty liver disease. In this area, the low incidence of HCC due to HBV/HCV infections can be explained considering the use of the vaccine for HBV and anti-viral treatments against HCV in contrast with the increased incidence of metabolic syndrome[11].
All the aforementioned risk factors lead to liver disease (cirrhosis or chronic inflammation) that causes an accumulation of genomic alterations driving HCC. In general, HCC arises during the progression of pre-existing chronic hepatitis, and in the vast majority (80%) of patients, HCC occurs in the setting of cirrhosis[12]. The development of HCC is a process characterized by a specific sequence of lesions, from regenerative nodules in cirrhosis, low-grade dysplastic nodules and high-grade dysplastic nodules to early and progressed HCC[13,14].
The molecular mechanisms driving HCC tumorigenesis are extremely complex. Understanding this multistep process, with underlying genetic alteration, is essential for prevention, diagnostic, prognostic, and therapeutic purposes. Considering the future perspective, a better knowledge of molecular mechanisms involved in HCC tumorigenesis would help for a correct classification of HCC, for improving patient outcomes, and to develop new therapeutic targets. The advent of NGS technologies may help in the comprehensive study of genetic alteration and the different pathways involved in the initiation and progression of HCC. In fact, the development of NGS multi-gene panels allows the parallel analysis of multiple markers giving a broad view of the genomic situation[15,16]. To date, this molecular landscape is crucial for therapeutic decision-making in other solid tumors[15]. The Cancer Genome Atlas Research Network investigated a total of 559 cases of HCC[17]. This study found that TERT, TP53, and CTNNB1 are the most frequently altered genes in HCCs; 77% of HCCs showed a mutation in at least one of these three genes[17]. Correlation data between HCC molecular signatures and etiological agents are shown in Table 1.
| Frequency mutation | Etiological factor | |||
| HBV | HCV | Non-viral | ||
| Driver gene | TERT | 50% | 61% | 65% |
| CTNNB1 | 15% | 30% | 39% | |
| TP53 | 10%-65 % | 24% | 16% | |
Bearing in mind all this evidence, the present review will discuss the main molecular mutations in HCC, with particular emphasis on the influence that these alterations have on HCC morphology and biological aggressiveness.
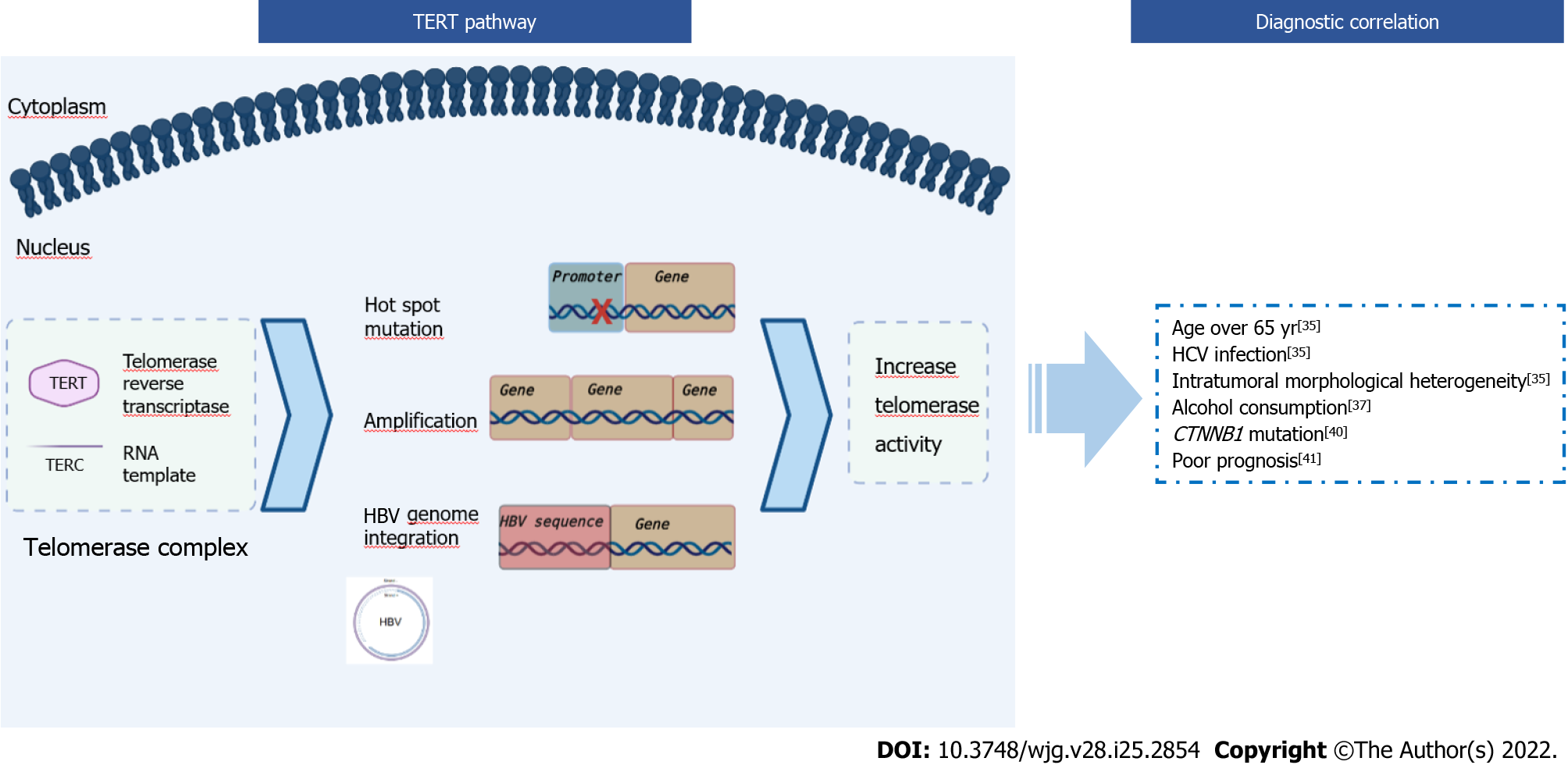
During cycles of genomic replication, the linear organization of chromosomes brings with it the problem of erosion of the 5’-terminus due to non-reproduction of the RNA primer binding site. Indeed, this erosion does not happen thanks to telomerase, constituted by telomerase reverse transcriptase (encoded by the TERT gene) and RNA template (encoded by the TERC gene). The telomerase complex adds nucleotides onto telomeres, preventing them from shortening. Telomeres are short tandem repeats of DNA (TTAGGG) coated by a protein complex known as Shelterin to protect the end of the chromosome where telomeres are located. Telomere synthesis is a controlled process activated in stem cells but deactivated in most somatic cells due to epigenetic silencing during the differentiation process. In the mature hepatocytes, the telomerase is not expressed[18,19]. The shortening of the telomeres exposes chromosomes to damage resulting in cellular senescence and is thought to be responsible for a sequence of events that drive to cancer[20].
Reactivation of TERT expression has been observed in several cancers (e.g., melanomas, gliomas, poorly differentiated bladder cancer, anaplastic thyroid carcinomas, basal cell, squamous cell carcinomas) leading to a restoration of the telomerase activity[19] (Figure 1). This event avoids cellular senescence and leads cancer cells to acquire replicative immortality, a crucial feature in the progression of the neoplasm rather than in the transformation of the cells into malignant ones[21-24]. This upregulation of TERT in cancer can occur through several mechanisms, which are generally mutually exclusive: (1) Gene amplification, found in ovarian cancer, adrenocortical carcinoma, lung adenocarcinoma, and esophageal carcinoma[25]; (2) Gene rearrangements, found in high-risk neuroblastoma[26]; and (3) Gene mutations in hot-spot regions of the promoter region, found in melanomas, thyroid tumors, gliomas[19].
Alterations in the gene promoter region are the most common and most frequently detected, in particular C>T transition at chr5:1295228 (−124 or C228T) or chr5:1295250 (−146 or C250T). The C228T and C250T TERT mutations separately create an identical 11-base sequence that acts as a novel E-twenty-sis transcription factor binding site, causing TERT overexpression[21].
TERT and HCC: Telomere length and telomere expression play a key role in the pathogenesis of HCC. Several studies have found telomere shortening in cirrhotic tissue, independently of the etiology of the liver disease (e.g., alcohol abuse or viral hepatitis), suggesting that this event might represent a hallmark of liver senescence and chronic hepatitis[27-29]. In contrast to cirrhotic tissue, in 44%-59% of the HCC a reactivation of the TERT gene is observed[30]. Cellular senescence found in cirrhotic tissue followed by TERT reactivation is one of the mechanisms that may explain the development and progression of HCC in cirrhosis. In particular, with the accumulation of gene alterations senescence can induce neoplastic transformation, whereas subsequent telomerase activation can lead to a neoplastic progression (Figure 1).
Reactivation of TERT can also be caused by HBV infection (Figure 1). HBV is an enveloped virus with partially double-stranded DNA with the capacity to integrate its own genome into that of the host, leading to the deregulation of the gene involved. TERT promoter is the most frequent site of integration (38.5%) in HBV-related cancers, and the viral integration leads to TERT overexpression[31,32]. Intriguingly, TERT mutations have never been described in hepatocellular adenoma (HCA) in contrast to CTNNB1 (see “CTNNB1 and HCC” paragraph)[13,32-34].
TERT mutations in HCC have been statistically correlated with: (1) Age over 65 years (P = 0.018), HCV infection more than HBV (P = 0.048), and intratumoral morphological heterogeneity (P = 0.0001)[35,36]. In a study performed on 97 HCCs by Kwa et al[35], the histological patterns in the tumor areas were classified into four groups: early, well, moderate, and poor. In particular, regarding the morphological aspect in TERT mutated HCC they observed two or more histological patterns as opposed to TERT wild-type HCCs, which showed only a single dominant pattern[35]; (2) Alcohol consumption[32,37]. Schulze et al[32] performed a study on 243 surgically resected HCCs, and 60% of the alcohol-related HCCs had a mutation in the TERT gene promoter; (3) CTNNB1 mutations. Several studies have shown the association between CTNNB1 and TERT[38-40]. This correlation was demonstrated for the first time in a mice model in which it was observed that β-catenin binds the TERT promoter and participates in the control of its expression[40]; and (4) Poor prognosis, (P = 0.041). A study by Oh et al[41] on telomere length in HCC showed that telomere elongation was a poor prognostic factor, as it decreased overall survival (P = 0.044). Moreover, in the case of high telomerase activity the prognosis was unfavorable (P = 0.009)[41] (Figure 1).
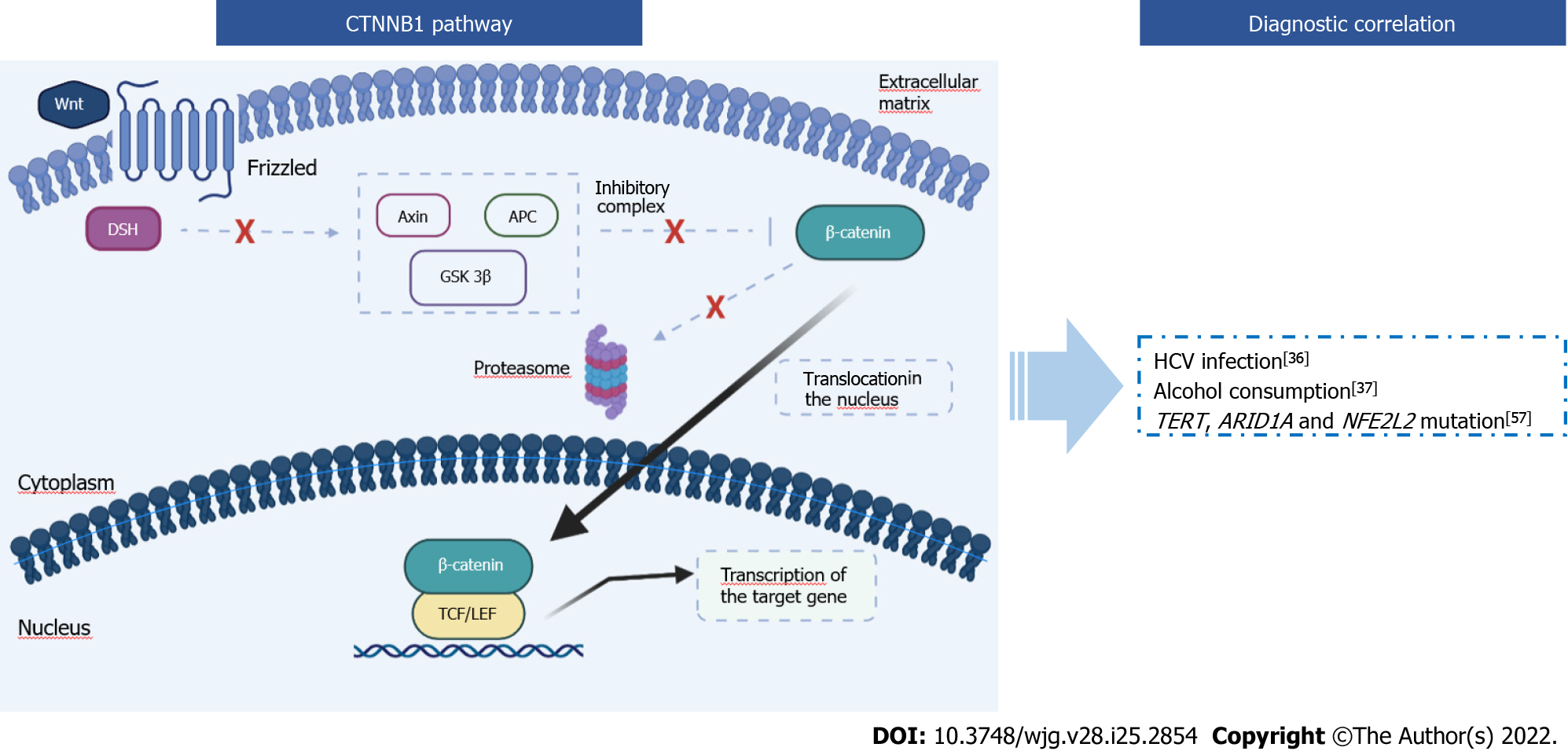
The CTNNB1 gene encodes β-catenin, a protein that performs several cellular functions. When interacting with the cadherin protein complex, β-catenin is important for the stabilization of the cytoskeleton and intracellular adhesions, but it also plays a role as a transcription factor in the canonical Wnt/β-catenin pathway. This pathway is involved in embryonic development, cellular homeostasis, and several diseases[42]. The cytoplasmic concentration of β-catenin is tightly controlled through its ubiquitination and proteasomal degradation. The phosphorylation required for this degradation mechanism is performed by glycogen synthase kinase 3 alpha and beta through the action of axin and the protein adenomatous polyposis coli (APC)[43,44]. In the cytoplasmic membrane, there are receptors for the Wnt molecules, called frizzled. The ligand-receptor complex triggers a cascade of cytoplasmic reactions, leading to the activation of the disheveled protein. This protein binds axin, preventing the bond between axin and glycogen synthase kinase 3[45]. This mechanism inhibits the proteasomal degradation of β-catenin. Given that CTNNB1 continues to be transcribed, the β-catenin cytoplasmic concentration increases.
Once all the β-catenin cytoplasmic binding sites are saturated, β-catenin protein is translocated into the nucleus. Here β-catenin interacts with many transcriptional factors, in particular with T-cell factor/lymphoid enhancing factor to promote the transcription of target genes, such as c-Myc, CyclinD-1, and Jun (Figure 2). Most of these gene targets encode for oncoproteins, leading to the activation of oncogenic mechanisms (e.g., uncontrolled growth or escape from apoptosis)[46]. For this reason, β-catenin is a molecule that may be involved in carcinogenesis and tumor progression of several cancers: HCC, lung cancer, brain and cerebellum cancer, breast cancer, colon cancer, leukemia, and others[47-49].
The Wnt pathway can also be activated by transforming growth factor-β. Dysregulation of its signaling pathway is associated with an invasive phenotype and plays a central role in inflammation, fibrogenesis, and immunomodulation in the HCC microenvironment[50,51].
Activating mutations found in the CTNNB1 gene are generally substitutions or in-frame deletions in hotspot regions that encode for the part of the protein that acts as a domain for the APC/AXIN1/ glycogen synthase kinase 3B complex. Thus, β-catenin is not degraded by the proteosome and then uncontrollably activates the transcription of oncogenes[52].
CTNNB1 and HCC: In HCCs, CTNNB1 mutations are among the most encountered genetic alterations, with a frequency of 20%-40%[32]. Regarding therapy, CTNNB1 mutations induce resistance to immune checkpoint inhibitors (anti-PD-1/PD-L1 inhibitors and anti-CTLA4)[53]. Another important aspect concerns HCA. According to the literature, 5%-10% of HCA are subject to malignant transformation, but the most recent WHO guidelines considered CTNNB1-mutated HCAs as a specific subtype, with a higher risk for malignant transformation that could lead to the development of HCC. In HCA CTNNB1 mutations are identified in 11%-43%[54-56].
CTNNB1 mutations are not the only alterations found in HCCs, regarding Wnt/β-catenin pathway. In fact, mutations in Axin and APC in HCCs have been detected in 6%-15% and 2%-4%, respectively[57]. Generally, mutations in CTNNB1, Axin, and APC are mutually exclusive[57].
HCCs with mutated CTNNB1 are statistically correlated to: (1) HCV infection[36]. For example, in a study on 22 HCV-related HCCs, an association between HCV infection and activation of the Wnt signaling pathway caused by the β-catenin mutation was found in 41% of cases[36], while according to the WHO, 30% of HCCs caused by HCV harbors a mutation in the CTNNB1 gene; (2) Alcohol consumption. Schulze et al[37] studied 243 surgically resected HCCs, and 37% of the alcohol-related HCCs harbored a mutation in the CTNNB1 gene; (3) TERT mutation[38,39]. Correlations between CTNNB1 and TERT have been described in the “TERT and HCC” paragraph; and (4) ARID1A mutations (P = 0.05) and NFE2L2 mutation (P = 0.015) associations with CTNNB1 were demonstrated in a study performed on 125 HCCs[57] (Figure 2).
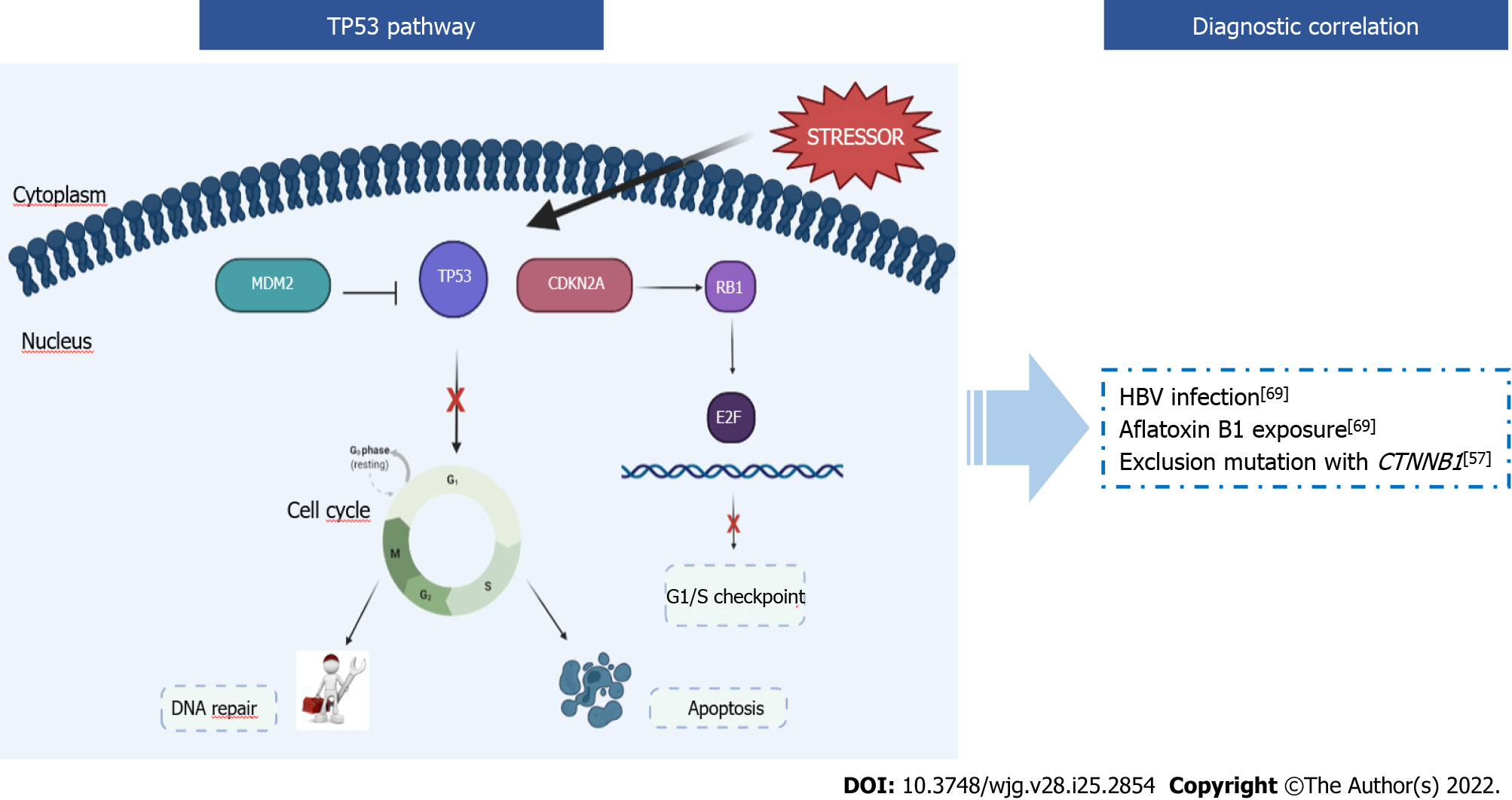
The TP53 gene encodes for the p53 protein, which owes its name to its molecular mass (53 kDa). p53 is called “the guardian of the genome” because it is an oncosuppressor that regulates the cell cycle, apoptosis, and genomic stability by preventing genomic mutations. The p53 pathway is crucial in cellular mechanisms as it interacts with other signal transduction pathways (e.g., retinoblastoma pathway, Wnt-β-catenin, cyclin-cdk). Plenty of positive and negative autoregulatory feedback mechanisms act on p53 functions[58,59]. The activation of p53 occurs in response to many different stressors, both intrinsic and extrinsic to the cell (e.g., gamma or UV radiation, oxidative stress, osmotic shock) that put faithful duplication of genetic material at risk[60]. The key event for p53 activation is the phosphorylation of the N-terminal domain by protein kinases. This event leads to the accumulation of p53 in the stressed cells through an increase in the half-life of the protein and an increase in efficiency as a transcription factor. After this activation, p53 initiates a program that blocks the cell cycle, leads the cell to senescence, and then to apoptosis[61] (Figure 3).
In unstressed cells, cytoplasmic levels of p53 are kept in check through its degradation. The Mdm2 protein binds p53, transports it from the nucleus to the cytosol, and acts as a ubiquitin ligase so that ubiquitin binds to p53 leading to proteasome degradation[62]. If the TP53 gene is altered, the p53 protein cannot function properly, driving tumorigenesis and tumor progression. As early as 1990, TP53 was defined as the most frequently mutated gene in human cancers. TP53 mutations remain among the most frequent and most significant in more common human cancers, although the frequency of mutations is highly variable depending on the type of cancer: from 90% in the ovary, 50%-80% in the lung up to less than 5% in the cervix[63]. Individuals affected by Li Fraumeni syndrome carry a mutated allele of TP53, and this syndrome predisposes to the development of several types of cancers[64].
TP53 and HCC: Approximately 15%-40% of HCCs carry mutations in the TP53 gene, with a higher frequency in advanced tumors[65]. Intriguingly, a specific TP53 mutation is significantly associated with dietary intake of aflatoxin B1, a mycotoxin produced by Aspergillus fungi. Exposure to aflatoxin B1 induces the transversion G → T at the TP53 codon 249, leading to the p.R249S (c.747G>T) substitution. This mutation could then be considered a mutational signature of exposure to aflatoxin B1 in HCC[66-68].
HCCs with mutated TP53 are statistically correlated to: (1) HBV infection and aflatoxin B1 exposure[68,69]. Lunn et al[69] conducted a population-based study on 110 HCCs. The relative risk (RR) that they obtained for HBV infection (RR=17.0), aflatoxin B1 exposure (RR=17.4), and the two risk agents together (RR=67.7) confirmed the correlation between these agents and HCC development. Exposure to aflatoxin B1 induces the p.R249S substitution in the TP53 gene, and HBV infection causes integration of the viral genome into that of the host, promoting mutations in genes crucial for cellular regulation, such as TP53. For these reasons HBV infection and aflatoxin B1 promote a high rate of mutagenesis in HCC[68,69]; and (2) TP53 alterations were usually exclusive from CTNNB1 mutations (P = 0.0001) but not from AXIN1 and APC[57] (Figure 3).
Over the years, many groups have tried to classify HCCs according to a molecular basis, but a univocal classification has been never reached.
Kabashima et al[70] grouped different classifications, starting from that of Shimada et al[71], and grouped HCCs into three molecular subtypes (MS1, MS2, MS3). The groups identified by the different studies are only fairly overlapping with each other. These classifications were based on clinical, molecular, and immunological features[70,71].
Regarding gene alterations, some correlations were found between TP53 and CTNNB1. TP53-mutated HCCs were classified into the MS1 group, which correlated with unfavorable prognosis, viral infection, high serum alpha-fetoprotein levels, vascular invasion and proliferation, extensive mitotic activity resulting in chromosomal instability, and stem cell-like properties.
CTNNB1-mutated HCCs were placed into the MS2 group, correlating with aberrant activation of Wnt/ β-Catenin pathway, which could explain the high rate of methylation in CpG islands present in this group, as the constitutively active β-catenin protein recruits a DNMT1 methyltransferase. Another feature associated with the MS2 group is the immunosuppressive phenotype. Moreover, this class is considered non-proliferative/less progressive than others.
The MS3 group is not associated with molecular signatures, but only with metabolic disease-associated tumors[51,72-74].
The TERT gene is rarely found in these classifications. However, it should be considered that the most frequent mutations in TERT fall in a promoter region usually not covered by the exome sequencing studies. The Cancer Genome Atlas 2017 classification detected TERT mutations in HCC and included HCC-TERT samples in iCluster2 and iCluster3[17]. In 2019, further classification was drafted by Yang et al[75] that divides HCCs into 3 groups (C1, C2, C3), overlapping with a previous study performed by Hoshida et al[51]: C1→S3, C2→S1, C3→S2.
In 2017, Calderaro et al[76] discriminated HCCs based on the presence of TP53 or CTNNB1 mutations, considering that these are two genes that mutate in a mutually exclusive manner and together comprise 57% of HCCs. In this study, CTNNB1-mutated HCCs were described as larger than the CTNNB1-wildtype HCCs but characterized by a lower tumor grade, with microtrabecular and pseudoglandular patterns of growth, without inflammatory infiltrates and with the presence of cholestasis[76]. Conversely, TP53-mutated HCCs were described as poorly differentiated tumors, with large multinucleated and pleomorphic cells, solid pattern of growth, frequent vascular invasion, and angiogenesis[76].
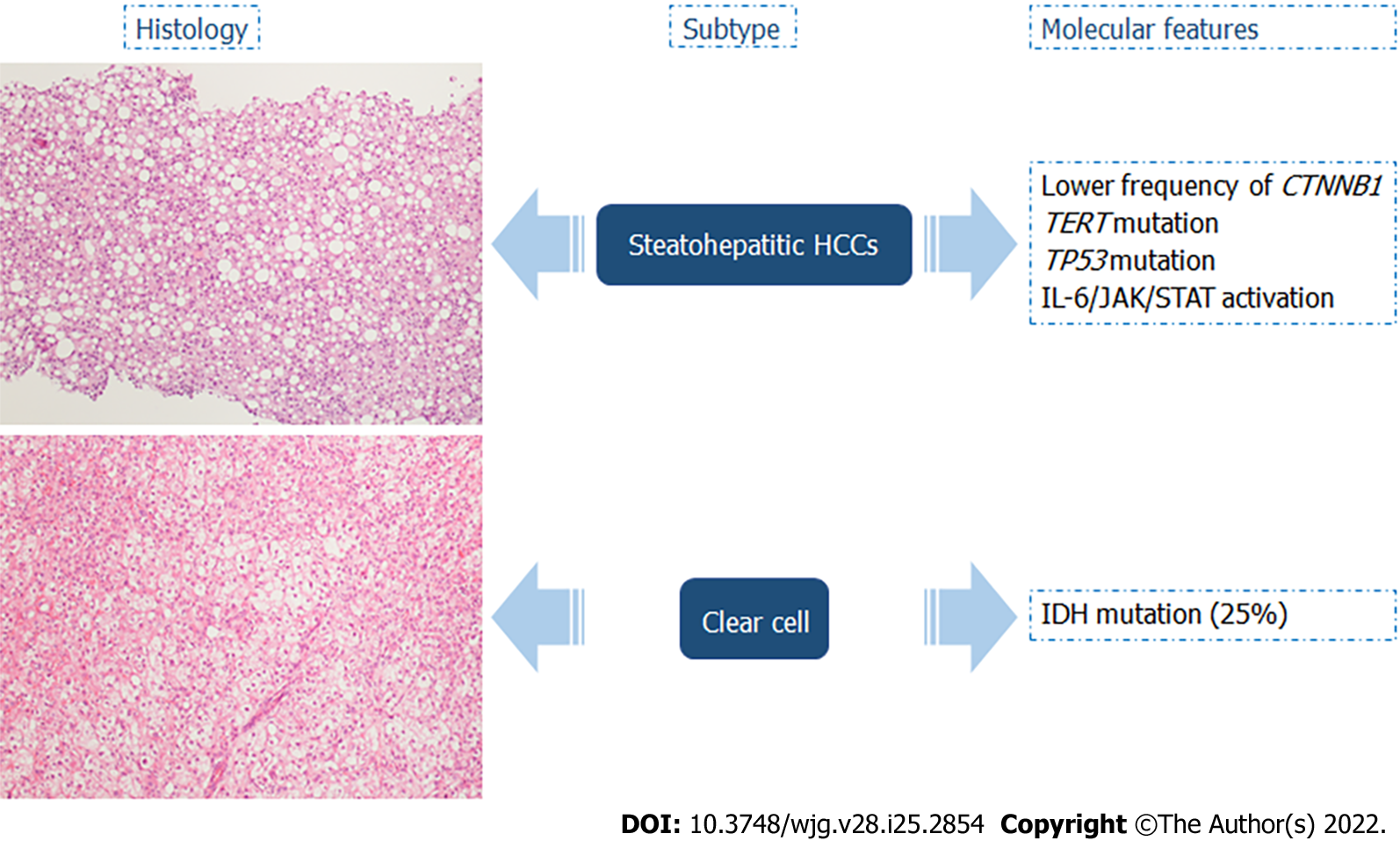
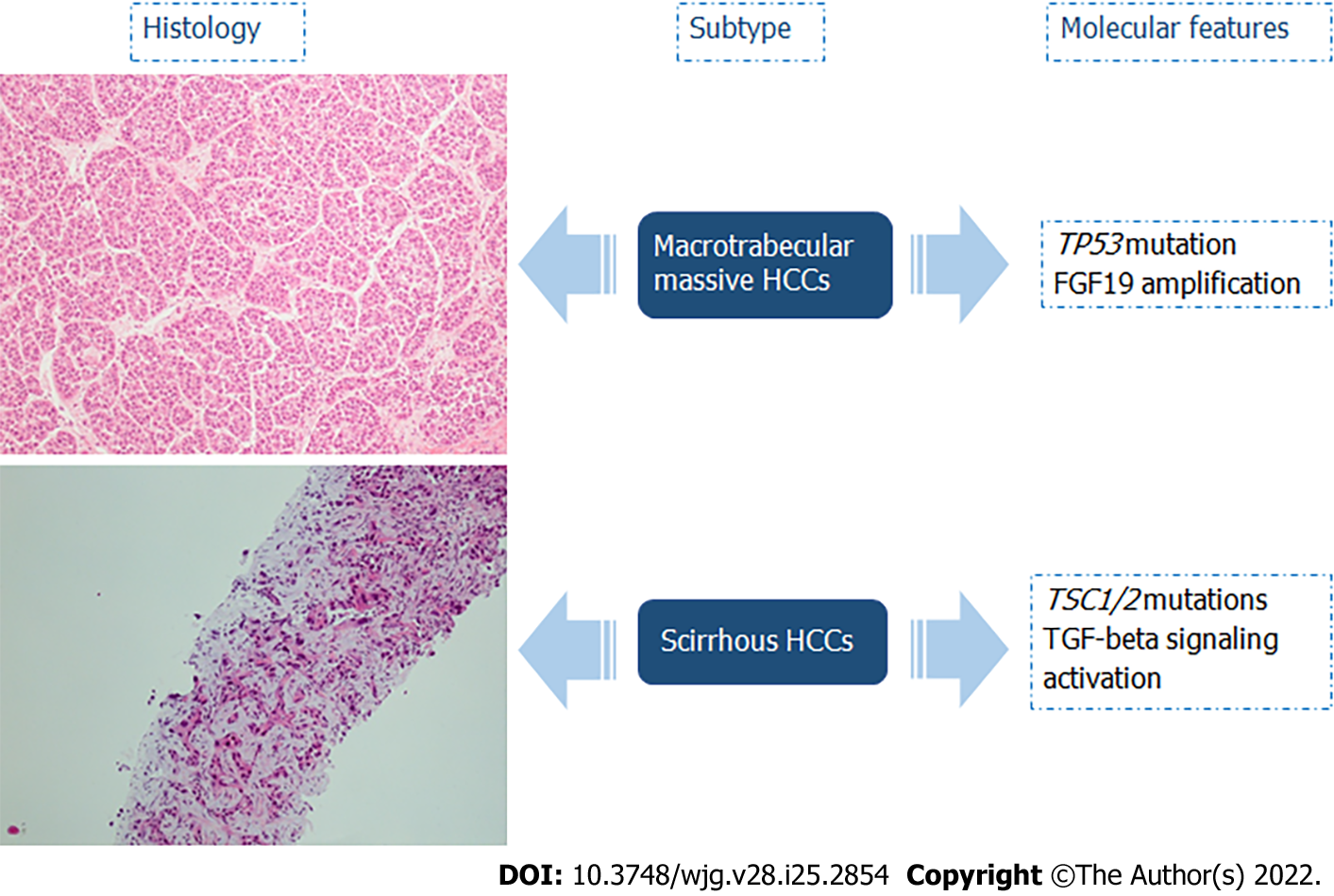
Selected subtypes of HCCs recognized by the most recent WHO Classification of Tumors were described to have specific molecular alterations (Figures 4 and 5)[14]: (1) Up to 63% of steatohepatitic HCCs were associated with nonalcoholic fatty liver disease. This type of HCC is characterized by the following histologic features: macrovesicular steatosis, Mallory-Denk bodies, ballooning of tumoral hepatocytes, inflammation, and trabecular or pericellular fibrosis. With regard to key molecular features, steatohepatitic HCCs were significantly associated with a lower frequency of CTNNB1 mutations, higher rate of mutations in TERT and TP53, IL-6/JAK/STAT activation, high level of C-reactive protein, and serum amyloid A positive at immunohistochemistry[14,77-80]; (2) Clear cell HCCs are considered a well-differentiated type, characterized by a cytoplasmic clearing, due to accumulation of glycogen, lipopolysaccharides, mucopolysaccharides, or cytoplasmic vesicles. IDH mutations were identified in 25% of clear cell HCCs, and these alterations were significantly associated with a worse prognosis. Moreover, IDH mutations are also found in intrahepatic cholangiocarcinoma, a tumor with a significantly worse prognosis than HCC[14,80-83]; (3) Macrotrabecular massive HCCs are frequently larger than 50 mm with vascular invasion, correlated high alpha-fetoprotein serum levels, high expression of angiopoietin 2, and vascular endothelial growth factor A. At the histological level, this subgroup is characterized by massive trabeculae surrounded by vascular spaces and coated by immature endothelial cells. On the molecular side, TP53 mutations and FGF19 amplifications have been detected. Macrotrabecular massive HCC is an aggressive phenotype associated with a worse prognosis[14,80,84,85]; and (5) Scirrhous HCCs develop in the non-cirrhotic liver, and they are characterized by hyaline stroma, intratumoral fibrosis with thin trabecular pattern growth (due to this characteristic is easily confusable radiologically to cholangiocarcinoma), or the lymphoepithelioma-like subtype, consisting of dense intratumor lymphocytic infiltration. Scirrhous HCCs may exhibit TSC1/TSC2 mutations and transforming growth factor-β signaling activation. Regarding prognosis, scirrhous HCCs are an aggressive subgroup, often with invasion of the portal vein, but as far as long-term follow-up is concerned, the prognosis is similar or sometimes better to conventional HCCs[76,86,87].
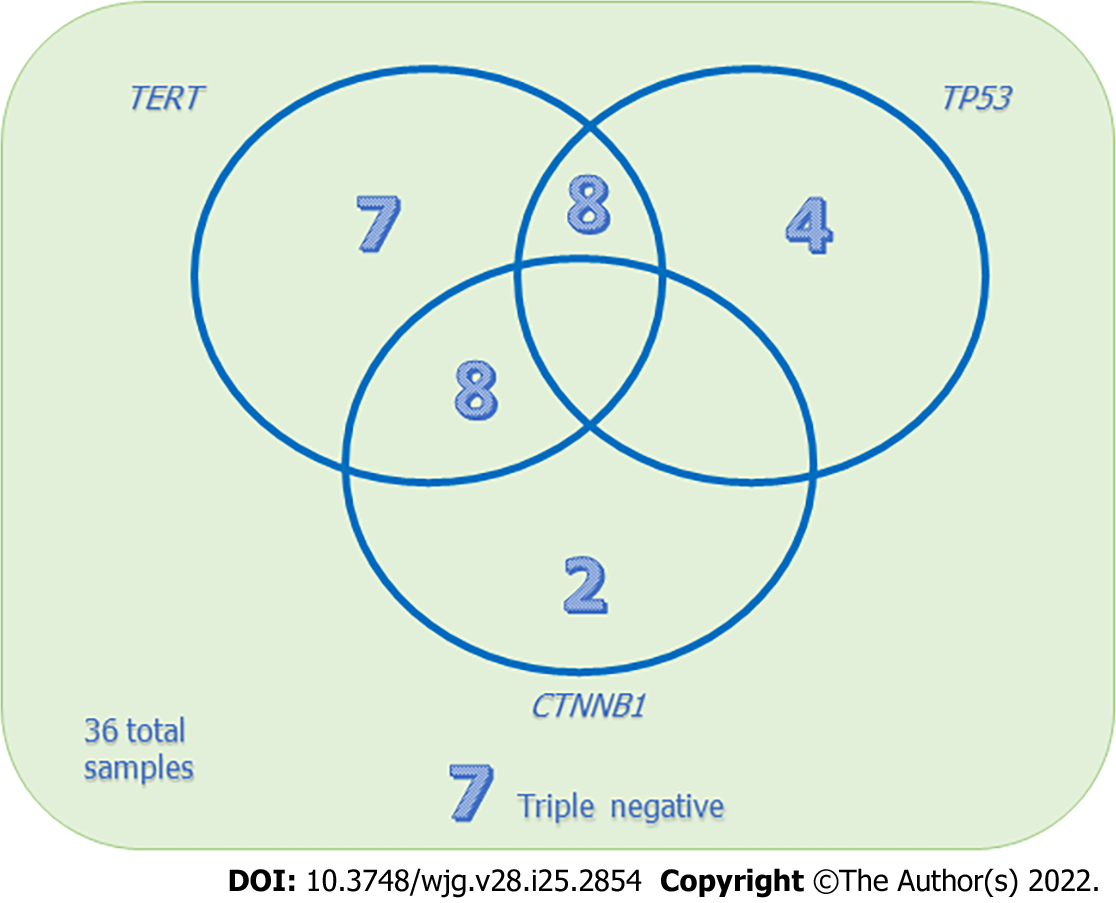
Our preliminary results focused on 36 prospectively enrolled patients, all resected for HCC and selected for NGS by means of a laboratory-developed multi-gene panel Gene-Studio S5 sequencer, which comprises specific target regions including TERT[15]. We detected single mutations in the TERT promoter in 7 (19.4%) cases, in TP53 in 4 (11.1%) cases, and in CTNNB1 in 2 (5.6%) cases. TERT and CTNNB1 coexistent mutations were observed in 8 (22.2%) cases, while TERT and TP53 were in 8 (22.2%) cases. In 7 (19.4%) cases no mutations in these three genes were detected (Figure 6).
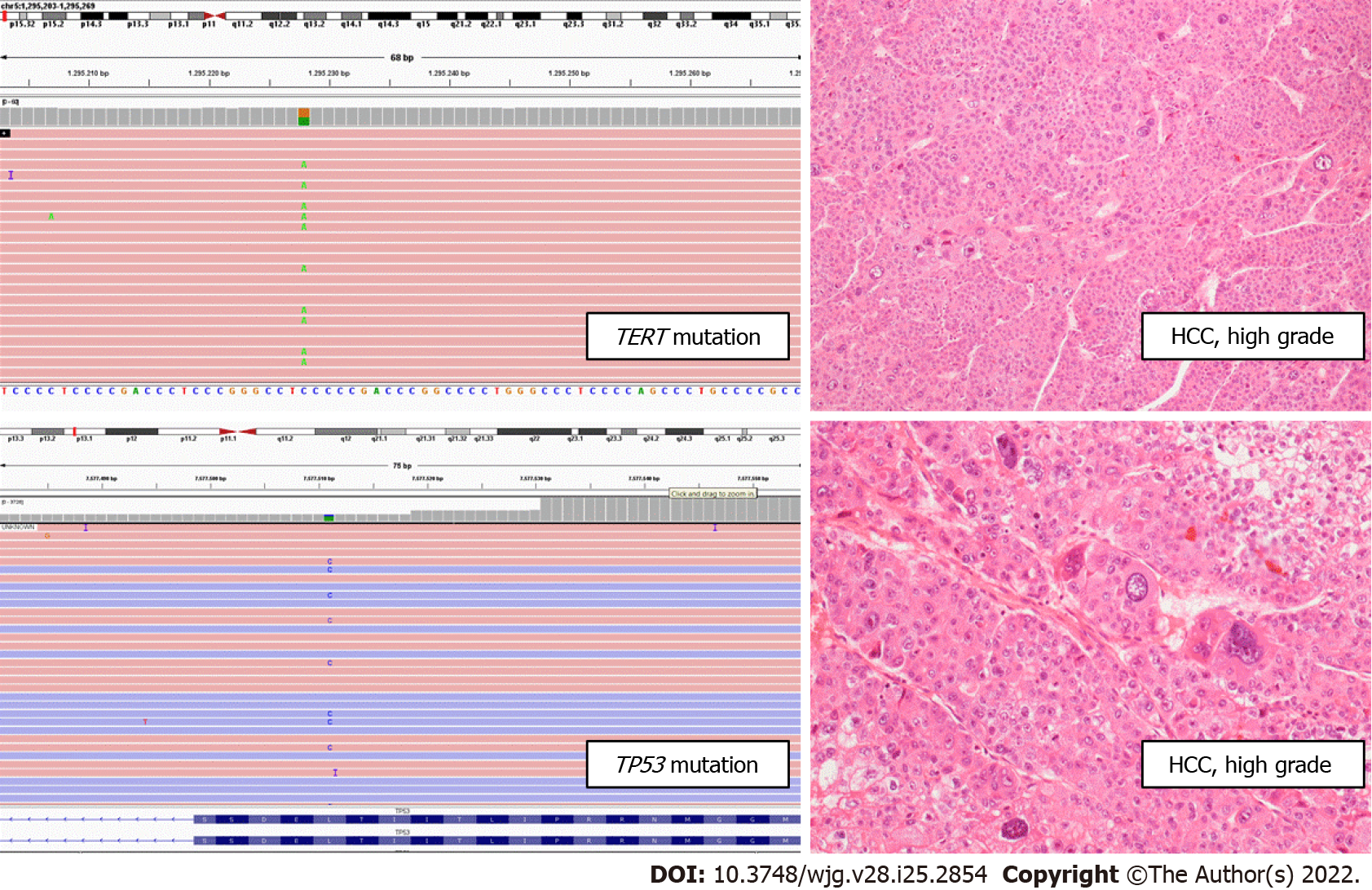
In line with a previous study by Calderaro et al[76], we observed a trend of TERT-mutated HCC towards a macrotrabecular or solid architecture. Moreover, the presence of TERT promoter mutations in combination with TP53 mutation correlated with high-grade HCC (P = 0.011; Figure 7).
Interestingly, no correlations were found between mutations and tumor dimensions. This evidence leads us to hypothesize that the presence of TERT promoter mutations, alone or in combination with TP53 alteration, correlates with a morphological progression in HCC, in terms of a higher tumor grade and an architecture more related to aggressive behavior (solid, macrotrabecular) but not of a dimensional evolution.
Most of the HCC in non-cirrhotic livers of our series showed no mutations or harbored only a CTNNB1 mutation (P = 0.031), as a countercheck of the correlation between tumor progression and mutations. The validation of these results on a larger series as well as with post-surgical follow-up might indicate that small HCC may have an aggressive behavior from a molecular and morphological point of view, despite their dimensions.
The molecular signature of a tumor is becoming increasingly important in the approach of patients with different types of cancers, on diagnostic, prognostic, and predictive grounds. In the age of precision medicine, the study of HCC mutations is still a field that is worth investigating. Considering both the literature data and our personal experience, about 80% of HCCs harbor mutations in at least one gene among TERT, TP53, or CTNNB1, with different biological and clinical implications.
In the near future, a deeper analysis of these three genes is surely desirable since a molecular characterization of HCC would open up the possibility of personalized therapies, as has happened for other cancers (e.g., lung adenocarcinomas, melanomas, gastrointestinal stromal tumors, colorectal adenocarcinomas). Moreover, the evidence of a tight correlation between the mutational profile and the HCC morphology is likely to imply an increasing integrative approach between anatomic pathology and molecular laboratories.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Italy
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Castro-Gil M, Mexico; Citores MJ, Spain; Yang L, China A-Editor: Zhou SM, China S-Editor: Ma YJ L-Editor: Filipodia P-Editor: Ma YJ
| 1. | Harris PS, Hansen RM, Gray ME, Massoud OI, McGuire BM, Shoreibah MG. Hepatocellular carcinoma surveillance: An evidence-based approach. World J Gastroenterol. 2019;25:1550-1559. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 156] [Cited by in RCA: 151] [Article Influence: 25.2] [Reference Citation Analysis (4)] |
| 2. | World Health Organization. Cancer Today. Available from: https://gco.iarc.fr/today/home. |
| 3. | Nahon P, Zucman-Rossi J. Single nucleotide polymorphisms and risk of hepatocellular carcinoma in cirrhosis. J Hepatol. 2012;57:663-674. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 119] [Cited by in RCA: 132] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 4. | Bidkhori G, Benfeitas R, Klevstig M, Zhang C, Nielsen J, Uhlen M, Boren J, Mardinoglu A. Metabolic network-based stratification of hepatocellular carcinoma reveals three distinct tumor subtypes. Proc Natl Acad Sci U S A. 2018;115:E11874-E11883. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 158] [Article Influence: 22.6] [Reference Citation Analysis (0)] |
| 5. | Dharel N, Kato N, Muroyama R, Moriyama M, Shao RX, Kawabe T, Omata M. MDM2 promoter SNP309 is associated with the risk of hepatocellular carcinoma in patients with chronic hepatitis C. Clin Cancer Res. 2006;12:4867-4871. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 76] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 6. | Park JW, Chen M, Colombo M, Roberts LR, Schwartz M, Chen PJ, Kudo M, Johnson P, Wagner S, Orsini LS, Sherman M. Global patterns of hepatocellular carcinoma management from diagnosis to death: the BRIDGE Study. Liver Int. 2015;35:2155-2166. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 569] [Cited by in RCA: 939] [Article Influence: 93.9] [Reference Citation Analysis (0)] |
| 7. | Wild CP, Miller JD, Groopman JD. Mycotoxin Control in Low- And Middle-Income Countries. In: Wild CP, Miller JD, Groopman JD, editors. Mycotoxin Control in Low- and Middle-Income Countries. Lyon (FR), 2015. |
| 8. | Ng AWT, Poon SL, Huang MN, Lim JQ, Boot A, Yu W, Suzuki Y, Thangaraju S, Ng CCY, Tan P, Pang ST, Huang HY, Yu MC, Lee PH, Hsieh SY, Chang AY, Teh BT, Rozen SG. Aristolochic acids and their derivatives are widely implicated in liver cancers in Taiwan and throughout Asia. Sci Transl Med. 2017;9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 207] [Cited by in RCA: 256] [Article Influence: 36.6] [Reference Citation Analysis (0)] |
| 9. | Arlt VM, Stiborova M, Schmeiser HH. Aristolochic acid as a probable human cancer hazard in herbal remedies: a review. Mutagenesis. 2002;17:265-277. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 364] [Cited by in RCA: 356] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 10. | Yang JD, Hainaut P, Gores GJ, Amadou A, Plymoth A, Roberts LR. A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat Rev Gastroenterol Hepatol. 2019;16:589-604. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2184] [Cited by in RCA: 2889] [Article Influence: 481.5] [Reference Citation Analysis (17)] |
| 11. | Baecker A, Liu X, La Vecchia C, Zhang ZF. Worldwide incidence of hepatocellular carcinoma cases attributable to major risk factors. Eur J Cancer Prev. 2018;27:205-212. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 181] [Article Influence: 30.2] [Reference Citation Analysis (0)] |
| 12. | Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma. Lancet. 2012;379:1245-1255. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3249] [Cited by in RCA: 3594] [Article Influence: 276.5] [Reference Citation Analysis (4)] |
| 13. | Zucman-Rossi J, Villanueva A, Nault JC, Llovet JM. Genetic Landscape and Biomarkers of Hepatocellular Carcinoma. Gastroenterology. 2015;149:1226-1239.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 744] [Cited by in RCA: 951] [Article Influence: 95.1] [Reference Citation Analysis (0)] |
| 14. | Tumours WCo. Digestive System Tumours. 5th Edition. Lyon: IARC, 2019. |
| 15. | de Biase D, Acquaviva G, Visani M, Sanza V, Argento CM, De Leo A, Maloberti T, Pession A, Tallini G. Molecular Diagnostic of Solid Tumor Using a Next Generation Sequencing Custom-Designed Multi-Gene Panel. Diagnostics (Basel). 2020;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 51] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 16. | Malvi D, de Biase D, Fittipaldi S, Grillini M, Visani M, Pession A, D'Errico A, Vasuri F. Immunomorphology and molecular biology of mixed primary liver cancers: is Nestin a marker of intermediate-cell carcinoma? Histopathology. 2020;76:265-274. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 21] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 17. | Cancer Genome Atlas Research Network. ; Cancer Genome Atlas Research Network. Comprehensive and Integrative Genomic Characterization of Hepatocellular Carcinoma. Cell. 2017;169:1327-1341.e23. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1578] [Cited by in RCA: 1727] [Article Influence: 215.9] [Reference Citation Analysis (1)] |
| 18. | Nandakumar J, Cech TR. Finding the end: recruitment of telomerase to telomeres. Nat Rev Mol Cell Biol. 2013;14:69-82. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 282] [Cited by in RCA: 281] [Article Influence: 23.4] [Reference Citation Analysis (0)] |
| 19. | Heidenreich B, Kumar R. TERT promoter mutations in telomere biology. Mutat Res Rev Mutat Res. 2017;771:15-31. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 143] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 20. | Mason PJ, Perdigones N. Telomere biology and translational research. Transl Res. 2013;162:333-342. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 35] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 21. | Colebatch AJ, Dobrovic A, Cooper WA. TERT gene: its function and dysregulation in cancer. J Clin Pathol. 2019;72:281-284. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 68] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 22. | Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646-674. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51728] [Cited by in RCA: 47059] [Article Influence: 3361.4] [Reference Citation Analysis (5)] |
| 23. | Dratwa M, Wysoczańska B, Łacina P, Kubik T, Bogunia-Kubik K. TERT-Regulation and Roles in Cancer Formation. Front Immunol. 2020;11:589929. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 189] [Article Influence: 37.8] [Reference Citation Analysis (0)] |
| 24. | Esopi D, Graham MK, Brosnan-Cashman JA, Meyers J, Vaghasia A, Gupta A, Kumar B, Haffner MC, Heaphy CM, De Marzo AM, Meeker AK, Nelson WG, Wheelan SJ, Yegnasubramanian S. Pervasive promoter hypermethylation of silenced TERT alleles in human cancers. Cell Oncol (Dordr). 2020;43:847-861. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 15] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 25. | Barthel FP, Wei W, Tang M, Martinez-Ledesma E, Hu X, Amin SB, Akdemir KC, Seth S, Song X, Wang Q, Lichtenberg T, Hu J, Zhang J, Zheng S, Verhaak RG. Systematic analysis of telomere length and somatic alterations in 31 cancer types. Nat Genet. 2017;49:349-357. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 322] [Cited by in RCA: 460] [Article Influence: 57.5] [Reference Citation Analysis (0)] |
| 26. | Peifer M, Hertwig F, Roels F, Dreidax D, Gartlgruber M, Menon R, Krämer A, Roncaioli JL, Sand F, Heuckmann JM, Ikram F, Schmidt R, Ackermann S, Engesser A, Kahlert Y, Vogel W, Altmüller J, Nürnberg P, Thierry-Mieg J, Thierry-Mieg D, Mariappan A, Heynck S, Mariotti E, Henrich KO, Gloeckner C, Bosco G, Leuschner I, Schweiger MR, Savelyeva L, Watkins SC, Shao C, Bell E, Höfer T, Achter V, Lang U, Theissen J, Volland R, Saadati M, Eggert A, de Wilde B, Berthold F, Peng Z, Zhao C, Shi L, Ortmann M, Büttner R, Perner S, Hero B, Schramm A, Schulte JH, Herrmann C, O'Sullivan RJ, Westermann F, Thomas RK, Fischer M. Telomerase activation by genomic rearrangements in high-risk neuroblastoma. Nature. 2015;526:700-704. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 393] [Cited by in RCA: 438] [Article Influence: 43.8] [Reference Citation Analysis (0)] |
| 27. | Kitada T, Seki S, Kawakita N, Kuroki T, Monna T. Telomere shortening in chronic liver diseases. Biochem Biophys Res Commun. 1995;211:33-39. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 112] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 28. | Wiemann SU, Satyanarayana A, Tsahuridu M, Tillmann HL, Zender L, Klempnauer J, Flemming P, Franco S, Blasco MA, Manns MP, Rudolph KL. Hepatocyte telomere shortening and senescence are general markers of human liver cirrhosis. FASEB J. 2002;16:935-942. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 374] [Cited by in RCA: 382] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 29. | Carulli L, Anzivino C. Telomere and telomerase in chronic liver disease and hepatocarcinoma. World J Gastroenterol. 2014;20:6287-6292. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 42] [Cited by in RCA: 36] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 30. | Quaas A, Oldopp T, Tharun L, Klingenfeld C, Krech T, Sauter G, Grob TJ. Frequency of TERT promoter mutations in primary tumors of the liver. Virchows Arch. 2014;465:673-677. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 52] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 31. | Jang JW, Kim HS, Kim JS, Lee SK, Han JW, Sung PS, Bae SH, Choi JY, Yoon SK, Han DJ, Kim TM, Roberts LR. Distinct Patterns of HBV Integration and TERT Alterations between in Tumor and Non-Tumor Tissue in Patients with Hepatocellular Carcinoma. Int J Mol Sci. 2021;22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 15] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 32. | Schulze K, Nault JC, Villanueva A. Genetic profiling of hepatocellular carcinoma using next-generation sequencing. J Hepatol. 2016;65:1031-1042. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 167] [Cited by in RCA: 224] [Article Influence: 24.9] [Reference Citation Analysis (0)] |
| 33. | Lee SE, Chang SH, Kim WY, Lim SD, Kim WS, Hwang TS, Han HS. Frequent somatic TERT promoter mutations and CTNNB1 mutations in hepatocellular carcinoma. Oncotarget. 2016;7:69267-69275. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 37] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 34. | Nault JC, Zucman-Rossi J. TERT promoter mutations in primary liver tumors. Clin Res Hepatol Gastroenterol. 2016;40:9-14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 71] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 35. | Kwa WT, Effendi K, Yamazaki K, Kubota N, Hatano M, Ueno A, Masugi Y, Sakamoto M. Telomerase reverse transcriptase (TERT) promoter mutation correlated with intratumoral heterogeneity in hepatocellular carcinoma. Pathol Int. 2020;70:624-632. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 36. | Huang H, Fujii H, Sankila A, Mahler-Araujo BM, Matsuda M, Cathomas G, Ohgaki H. Beta-catenin mutations are frequent in human hepatocellular carcinomas associated with hepatitis C virus infection. Am J Pathol. 1999;155:1795-1801. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 211] [Cited by in RCA: 205] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 37. | Schulze K, Imbeaud S, Letouzé E, Alexandrov LB, Calderaro J, Rebouissou S, Couchy G, Meiller C, Shinde J, Soysouvanh F, Calatayud AL, Pinyol R, Pelletier L, Balabaud C, Laurent A, Blanc JF, Mazzaferro V, Calvo F, Villanueva A, Nault JC, Bioulac-Sage P, Stratton MR, Llovet JM, Zucman-Rossi J. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat Genet. 2015;47:505-511. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1006] [Cited by in RCA: 1340] [Article Influence: 134.0] [Reference Citation Analysis (0)] |
| 38. | Nault JC, Mallet M, Pilati C, Calderaro J, Bioulac-Sage P, Laurent C, Laurent A, Cherqui D, Balabaud C, Zucman-Rossi J. High frequency of telomerase reverse-transcriptase promoter somatic mutations in hepatocellular carcinoma and preneoplastic lesions. Nat Commun. 2013;4:2218. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 414] [Cited by in RCA: 511] [Article Influence: 46.5] [Reference Citation Analysis (1)] |
| 39. | Pinyol R, Tovar V, Llovet JM. TERT promoter mutations: gatekeeper and driver of hepatocellular carcinoma. J Hepatol. 2014;61:685-687. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 41] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 40. | Hoffmeyer K, Raggioli A, Rudloff S, Anton R, Hierholzer A, Del Valle I, Hein K, Vogt R, Kemler R. Wnt/β-catenin signaling regulates telomerase in stem cells and cancer cells. Science. 2012;336:1549-1554. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 387] [Cited by in RCA: 413] [Article Influence: 31.8] [Reference Citation Analysis (0)] |
| 41. | Oh BK, Kim H, Park YN, Yoo JE, Choi J, Kim KS, Lee JJ, Park C. High telomerase activity and long telomeres in advanced hepatocellular carcinomas with poor prognosis. Lab Invest. 2008;88:144-152. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 73] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 42. | Shang S, Hua F, Hu ZW. The regulation of β-catenin activity and function in cancer: therapeutic opportunities. Oncotarget. 2017;8:33972-33989. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 429] [Cited by in RCA: 486] [Article Influence: 60.8] [Reference Citation Analysis (0)] |
| 43. | Xing Y, Clements WK, Kimelman D, Xu W. Crystal structure of a beta-catenin/axin complex suggests a mechanism for the beta-catenin destruction complex. Genes Dev. 2003;17:2753-2764. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 184] [Cited by in RCA: 199] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 44. | Liu J, Xing Y, Hinds TR, Zheng J, Xu W. The third 20 amino acid repeat is the tightest binding site of APC for beta-catenin. J Mol Biol. 2006;360:133-144. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 66] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 45. | Fiedler M, Mendoza-Topaz C, Rutherford TJ, Mieszczanek J, Bienz M. Dishevelled interacts with the DIX domain polymerization interface of Axin to interfere with its function in down-regulating β-catenin. Proc Natl Acad Sci U S A. 2011;108:1937-1942. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 190] [Cited by in RCA: 172] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 46. | Nishida N, Goel A. Genetic and epigenetic signatures in human hepatocellular carcinoma: a systematic review. Curr Genomics. 2011;12:130-137. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 133] [Cited by in RCA: 152] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 47. | Kobayashi M, Honma T, Matsuda Y, Suzuki Y, Narisawa R, Ajioka Y, Asakura H. Nuclear translocation of beta-catenin in colorectal cancer. Br J Cancer. 2000;82:1689-1693. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 62] [Cited by in RCA: 88] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 48. | Khramtsov AI, Khramtsova GF, Tretiakova M, Huo D, Olopade OI, Goss KH. Wnt/beta-catenin pathway activation is enriched in basal-like breast cancers and predicts poor outcome. Am J Pathol. 2010;176:2911-2920. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 343] [Cited by in RCA: 418] [Article Influence: 27.9] [Reference Citation Analysis (0)] |
| 49. | Gekas C, D'Altri T, Aligué R, González J, Espinosa L, Bigas A. β-Catenin is required for T-cell leukemia initiation and MYC transcription downstream of Notch1. Leukemia. 2016;30:2002-2010. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 47] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 50. | Chen J, Gingold JA, Su X. Immunomodulatory TGF-β Signaling in Hepatocellular Carcinoma. Trends Mol Med. 2019;25:1010-1023. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 195] [Article Influence: 32.5] [Reference Citation Analysis (0)] |
| 51. | Hoshida Y, Nijman SM, Kobayashi M, Chan JA, Brunet JP, Chiang DY, Villanueva A, Newell P, Ikeda K, Hashimoto M, Watanabe G, Gabriel S, Friedman SL, Kumada H, Llovet JM, Golub TR. Integrative transcriptome analysis reveals common molecular subclasses of human hepatocellular carcinoma. Cancer Res. 2009;69:7385-7392. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 956] [Cited by in RCA: 942] [Article Influence: 58.9] [Reference Citation Analysis (0)] |
| 52. | de La Coste A, Romagnolo B, Billuart P, Renard CA, Buendia MA, Soubrane O, Fabre M, Chelly J, Beldjord C, Kahn A, Perret C. Somatic mutations of the beta-catenin gene are frequent in mouse and human hepatocellular carcinomas. Proc Natl Acad Sci U S A. 1998;95:8847-8851. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 835] [Cited by in RCA: 840] [Article Influence: 31.1] [Reference Citation Analysis (0)] |
| 53. | Pinyol R, Sia D, Llovet JM. Immune Exclusion-Wnt/CTNNB1 Class Predicts Resistance to Immunotherapies in HCC. Clin Cancer Res. 2019;25:2021-2023. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 167] [Article Influence: 27.8] [Reference Citation Analysis (0)] |
| 54. | Nault JC, Couchy G, Balabaud C, Morcrette G, Caruso S, Blanc JF, Bacq Y, Calderaro J, Paradis V, Ramos J, Scoazec JY, Gnemmi V, Sturm N, Guettier C, Fabre M, Savier E, Chiche L, Labrune P, Selves J, Wendum D, Pilati C, Laurent A, De Muret A, Le Bail B, Rebouissou S, Imbeaud S; GENTHEP Investigators, Bioulac-Sage P, Letouzé E, Zucman-Rossi J. Molecular Classification of Hepatocellular Adenoma Associates With Risk Factors, Bleeding, and Malignant Transformation. Gastroenterology. 2017;152:880-894.e6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 225] [Cited by in RCA: 269] [Article Influence: 33.6] [Reference Citation Analysis (0)] |
| 55. | Shen XY, Hu XG, Kim YB, Kim MN, Hong SY, Kim BW, Wang HJ. Molecular classification of hepatocellular adenoma: A single-center experience. Ann Hepatobiliary Pancreat Surg. 2019;23:109-114. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 1] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 56. | Rebouissou S, Franconi A, Calderaro J, Letouzé E, Imbeaud S, Pilati C, Nault JC, Couchy G, Laurent A, Balabaud C, Bioulac-Sage P, Zucman-Rossi J. Genotype-phenotype correlation of CTNNB1 mutations reveals different ß-catenin activity associated with liver tumor progression. Hepatology. 2016;64:2047-2061. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 173] [Cited by in RCA: 237] [Article Influence: 26.3] [Reference Citation Analysis (0)] |
| 57. | Guichard C, Amaddeo G, Imbeaud S, Ladeiro Y, Pelletier L, Maad IB, Calderaro J, Bioulac-Sage P, Letexier M, Degos F, Clément B, Balabaud C, Chevet E, Laurent A, Couchy G, Letouzé E, Calvo F, Zucman-Rossi J. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat Genet. 2012;44:694-698. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 962] [Cited by in RCA: 1148] [Article Influence: 88.3] [Reference Citation Analysis (0)] |
| 58. | Harris SL, Levine AJ. The p53 pathway: positive and negative feedback loops. Oncogene. 2005;24:2899-2908. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1399] [Cited by in RCA: 1410] [Article Influence: 70.5] [Reference Citation Analysis (0)] |
| 59. | Toufektchan E, Toledo F. The Guardian of the Genome Revisited: p53 Downregulates Genes Required for Telomere Maintenance, DNA Repair, and Centromere Structure. Cancers (Basel). 2018;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 78] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 60. | Overholtzer M, Rao PH, Favis R, Lu XY, Elowitz MB, Barany F, Ladanyi M, Gorlick R, Levine AJ. The presence of p53 mutations in human osteosarcomas correlates with high levels of genomic instability. Proc Natl Acad Sci U S A. 2003;100:11547-11552. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 148] [Cited by in RCA: 145] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 61. | Jin S, Levine AJ. The p53 functional circuit. J Cell Sci. 2001;114:4139-4140. [PubMed] |
| 62. | Purvis JE, Karhohs KW, Mock C, Batchelor E, Loewer A, Lahav G. p53 dynamics control cell fate. Science. 2012;336:1440-1444. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 538] [Cited by in RCA: 572] [Article Influence: 44.0] [Reference Citation Analysis (0)] |
| 63. | Leroy B, Anderson M, Soussi T. TP53 mutations in human cancer: database reassessment and prospects for the next decade. Hum Mutat. 2014;35:672-688. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 204] [Cited by in RCA: 282] [Article Influence: 25.6] [Reference Citation Analysis (0)] |
| 64. | Gargallo P, Yáñez Y, Segura V, Juan A, Torres B, Balaguer J, Oltra S, Castel V, Cañete A. Li-Fraumeni syndrome heterogeneity. Clin Transl Oncol. 2020;22:978-988. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 16] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 65. | Lombardo D, Saitta C, Giosa D, Di Tocco FC, Musolino C, Caminiti G, Chines V, Franzè MS, Alibrandi A, Navarra G, Raimondo G, Pollicino T. Frequency of somatic mutations in TERT promoter, TP53 and CTNNB1 genes in patients with hepatocellular carcinoma from Southern Italy. Oncol Lett. 2020;19:2368-2374. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 20] [Reference Citation Analysis (0)] |
| 66. | Bressac B, Kew M, Wands J, Ozturk M. Selective G to T mutations of p53 gene in hepatocellular carcinoma from southern Africa. Nature. 1991;350:429-431. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 972] [Cited by in RCA: 872] [Article Influence: 25.6] [Reference Citation Analysis (0)] |
| 67. | Unsal H, Yakicier C, Marçais C, Kew M, Volkmann M, Zentgraf H, Isselbacher KJ, Ozturk M. Genetic heterogeneity of hepatocellular carcinoma. Proc Natl Acad Sci U S A. 1994;91:822-826. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 109] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 68. | Gouas D, Shi H, Hainaut P. The aflatoxin-induced TP53 mutation at codon 249 (R249S): biomarker of exposure, early detection and target for therapy. Cancer Lett. 2009;286:29-37. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 98] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 69. | Lunn RM, Zhang YJ, Wang LY, Chen CJ, Lee PH, Lee CS, Tsai WY, Santella RM. p53 mutations, chronic hepatitis B virus infection, and aflatoxin exposure in hepatocellular carcinoma in Taiwan. Cancer Res. 1997;57:3471-3477. [PubMed] |
| 70. | Kabashima A, Shimada S, Shimokawa M, Akiyama Y, Tanabe M, Tanaka S. Molecular and immunological paradigms of hepatocellular carcinoma: Special reference to therapeutic approaches. J Hepatobiliary Pancreat Sci. 2021;28:62-75. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 6] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 71. | Shimada S, Mogushi K, Akiyama Y, Furuyama T, Watanabe S, Ogura T, Ogawa K, Ono H, Mitsunori Y, Ban D, Kudo A, Arii S, Tanabe M, Wands JR, Tanaka S. Comprehensive molecular and immunological characterization of hepatocellular carcinoma. EBioMedicine. 2019;40:457-470. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 192] [Cited by in RCA: 179] [Article Influence: 29.8] [Reference Citation Analysis (0)] |
| 72. | Lee JS, Chu IS, Heo J, Calvisi DF, Sun Z, Roskams T, Durnez A, Demetris AJ, Thorgeirsson SS. Classification and prediction of survival in hepatocellular carcinoma by gene expression profiling. Hepatology. 2004;40:667-676. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 677] [Cited by in RCA: 689] [Article Influence: 32.8] [Reference Citation Analysis (0)] |
| 73. | Boyault S, Rickman DS, de Reyniès A, Balabaud C, Rebouissou S, Jeannot E, Hérault A, Saric J, Belghiti J, Franco D, Bioulac-Sage P, Laurent-Puig P, Zucman-Rossi J. Transcriptome classification of HCC is related to gene alterations and to new therapeutic targets. Hepatology. 2007;45:42-52. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 827] [Cited by in RCA: 926] [Article Influence: 51.4] [Reference Citation Analysis (0)] |
| 74. | Chiang DY, Villanueva A, Hoshida Y, Peix J, Newell P, Minguez B, LeBlanc AC, Donovan DJ, Thung SN, Solé M, Tovar V, Alsinet C, Ramos AH, Barretina J, Roayaie S, Schwartz M, Waxman S, Bruix J, Mazzaferro V, Ligon AH, Najfeld V, Friedman SL, Sellers WR, Meyerson M, Llovet JM. Focal gains of VEGFA and molecular classification of hepatocellular carcinoma. Cancer Res. 2008;68:6779-6788. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 570] [Cited by in RCA: 564] [Article Influence: 33.2] [Reference Citation Analysis (0)] |
| 75. | Yang C, Huang X, Liu Z, Qin W, Wang C. Metabolism-associated molecular classification of hepatocellular carcinoma. Mol Oncol. 2020;14:896-913. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 150] [Article Influence: 30.0] [Reference Citation Analysis (0)] |
| 76. | Calderaro J, Couchy G, Imbeaud S, Amaddeo G, Letouzé E, Blanc JF, Laurent C, Hajji Y, Azoulay D, Bioulac-Sage P, Nault JC, Zucman-Rossi J. Histological subtypes of hepatocellular carcinoma are related to gene mutations and molecular tumour classification. J Hepatol. 2017;67:727-738. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 373] [Cited by in RCA: 535] [Article Influence: 66.9] [Reference Citation Analysis (0)] |
| 77. | Salomao M, Yu WM, Brown RS Jr, Emond JC, Lefkowitch JH. Steatohepatitic hepatocellular carcinoma (SH-HCC): a distinctive histological variant of HCC in hepatitis C virus-related cirrhosis with associated NAFLD/NASH. Am J Surg Pathol. 2010;34:1630-1636. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 144] [Cited by in RCA: 163] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 78. | Ando S, Shibahara J, Hayashi A, Fukayama M. β-catenin alteration is rare in hepatocellular carcinoma with steatohepatitic features: immunohistochemical and mutational study. Virchows Arch. 2015;467:535-542. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 34] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 79. | Taniai M, Hashimoto E, Tobari M, Kodama K, Tokushige K, Yamamoto M, Takayama T, Sugitani M, Sano K, Kondo F, Fukusato T. Clinicopathological investigation of steatohepatitic hepatocellular carcinoma: A multicenter study using immunohistochemical analysis of adenoma-related markers. Hepatol Res. 2018;48:947-955. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 26] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 80. | El Jabbour T, Lagana SM, Lee H. Update on hepatocellular carcinoma: Pathologists' review. World J Gastroenterol. 2019;25:1653-1665. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 82] [Cited by in RCA: 99] [Article Influence: 16.5] [Reference Citation Analysis (4)] |
| 81. | Lee WS, Lee KW, Heo JS, Kim SJ, Choi SH, Kim YI, Joh JW. Comparison of combined hepatocellular and cholangiocarcinoma with hepatocellular carcinoma and intrahepatic cholangiocarcinoma. Surg Today. 2006;36:892-897. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 119] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 82. | Lee JH, Shin DH, Park WY, Shin N, Kim A, Lee HJ, Kim YK, Choi KU, Kim JY, Yang YI, Lee CH, Sol MY. IDH1 R132C mutation is detected in clear cell hepatocellular carcinoma by pyrosequencing. World J Surg Oncol. 2017;15:82. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 22] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 83. | Bannasch P, Ribback S, Su Q, Mayer D. Clear cell hepatocellular carcinoma: origin, metabolic traits and fate of glycogenotic clear and ground glass cells. Hepatobiliary Pancreat Dis Int. 2017;16:570-594. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 21] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 84. | Vasuri F, Fittipaldi S, Giunchi F, Monica M, Ravaioli M, Degiovanni A, Bonora S, Golfieri R, Bolondi L, Grigioni WF, Pasquinelli G, D'Errico-Grigioni A. Facing the enigma of the vascular network in hepatocellular carcinomas in cirrhotic and non-cirrhotic livers. J Clin Pathol. 2016;69:102-108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 85. | Ziol M, Poté N, Amaddeo G, Laurent A, Nault JC, Oberti F, Costentin C, Michalak S, Bouattour M, Francoz C, Pageaux GP, Ramos J, Decaens T, Luciani A, Guiu B, Vilgrain V, Aubé C, Derman J, Charpy C, Zucman-Rossi J, Barget N, Seror O, Ganne-Carrié N, Paradis V, Calderaro J. Macrotrabecular-massive hepatocellular carcinoma: A distinctive histological subtype with clinical relevance. Hepatology. 2018;68:103-112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 179] [Cited by in RCA: 173] [Article Influence: 24.7] [Reference Citation Analysis (1)] |
| 86. | Matsuura S, Aishima S, Taguchi K, Asayama Y, Terashi T, Honda H, Tsuneyoshi M. 'Scirrhous' type hepatocellular carcinomas: a special reference to expression of cytokeratin 7 and hepatocyte paraffin 1. Histopathology. 2005;47:382-390. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 55] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 87. | Li W, Gomez E, Zhang Z. Immunohistochemical expression of stromal cell-derived factor-1 (SDF-1) and CXCR4 ligand receptor system in hepatocellular carcinoma. J Exp Clin Cancer Res. 2007;26:527-533. [PubMed] |