Published online Apr 28, 2022. doi: 10.3748/wjg.v28.i16.1641
Peer-review started: January 18, 2022
First decision: February 7, 2022
Revised: February 9, 2022
Accepted: March 16, 2022
Article in press: March 16, 2022
Published online: April 28, 2022
Processing time: 96 Days and 3.3 Hours
Cancer has become the most life-threatening disease in the world. Mutations in and aberrant expression of genes encoding proteins and mutations in noncoding RNAs, especially long noncoding RNAs (lncRNAs), have significant effects in human cancers. LncRNAs have no protein-coding ability but function extensively in numerous physiological and pathological processes. Small nucleolar RNA host gene 3 (SNHG3) is a novel lncRNA and has been reported to be differentially expressed in various tumors, such as liver cancer, gastric cancer, and glioma. However, the interaction mechanisms for the regulation between SNHG3 and tumor progression are poorly understood. In this review, we summarize the results of SNHG3 studies in humans, animal models, and cells to underline the expression and role of SNHG3 in cancer. SNHG3 expression is upregulated in most tumors and is detrimental to patient prognosis. SNHG3 expression in lung adenocarcinoma remains controversial. Concurrently, SNHG3 affects oncogenes and tumor suppressor genes through various mechanisms, including competing endogenous RNA effects. A deeper understanding of the contribution of SNHG3 in clinical applications and tumor development may provide a new target for cancer diagnosis and treatment.
Core Tip: This review explores the differential expression of small nucleolar RNA host gene 3 (SNHG3) as a novel lncRNA in hepatocellular carcinoma as well as other tumours. SNHG3 is upregulated in most tumours and can influence tumourigenesis and progression through competing endogenous RNA effects and signalling pathways, thereby adversely affecting patient prognosis. Therefore, SNHG3 may become a new target for the diagnosis and treatment of many cancers, including hepatocellular carcinoma.
- Citation: Shan DD, Zheng QX, Wang J, Chen Z. Small nucleolar RNA host gene 3 functions as a novel biomarker in liver cancer and other tumour progression. World J Gastroenterol 2022; 28(16): 1641-1655
- URL: https://www.wjgnet.com/1007-9327/full/v28/i16/1641.htm
- DOI: https://dx.doi.org/10.3748/wjg.v28.i16.1641
Cancer has become one of the significant sources of mortality worldwide. Approximately 14.1 million individuals are newly diagnosed each year, and approximately 8.2 million deaths occurred in 2021 based on GLOBOCAN[1]. Despite considerable achievements and advances in clinical practices, cancer remains a dreadful illness with a poor prognosis and high health burden, which presents a global threat to health[2]. Cancer is a heterogeneous disease with substantial genotypic and phenotypic diversity[3]. Genomic regulation of the cancer genome plays a vital role in cancer initiation, progression, and metastasis[4,5]. It is of most importance to explore the intricate link underlying cancer development and progression.
Long noncoding RNAs (lncRNAs) are a class of noncoding RNAs (ncRNAs) with no protein-coding ability[6]. Evidence has shown that lncRNAs participate in genomic regulation at the transcriptional, translational, and epigenetic levels[6,7]. The regulatory functions of lncRNAs include activation and silencing of genes[8,9], recruitment of epigenetic regulator[10], modification of RNA interactions[11], transcriptional and posttranscriptional processes[12], mRNA decay[13], and protein recruitment[14]. LncRNA-associated regulatory functions are dynamically regulated in a cell-, tissue-, development- and distribution-specific manner[15]. LncRNAs in the cytoplasm may act as sponges, stabilizing mRNAs and regulating the mRNA translation process, thus modifying downstream target gene expression[16]. LncRNAs located in the nucleus may play “cis-acting” or “trans-acting” functions[17-19]. Recently, growing evidence demonstrates that lncRNAs are involved in various cancer functions and pathological processes, particularly in the initiation and progression of tumors[20]. LncRNAs regulate various malignant activities, including tumor progression, proliferation, apoptosis, migration, invasion, chromatin remodeling, metabolism[21-23].
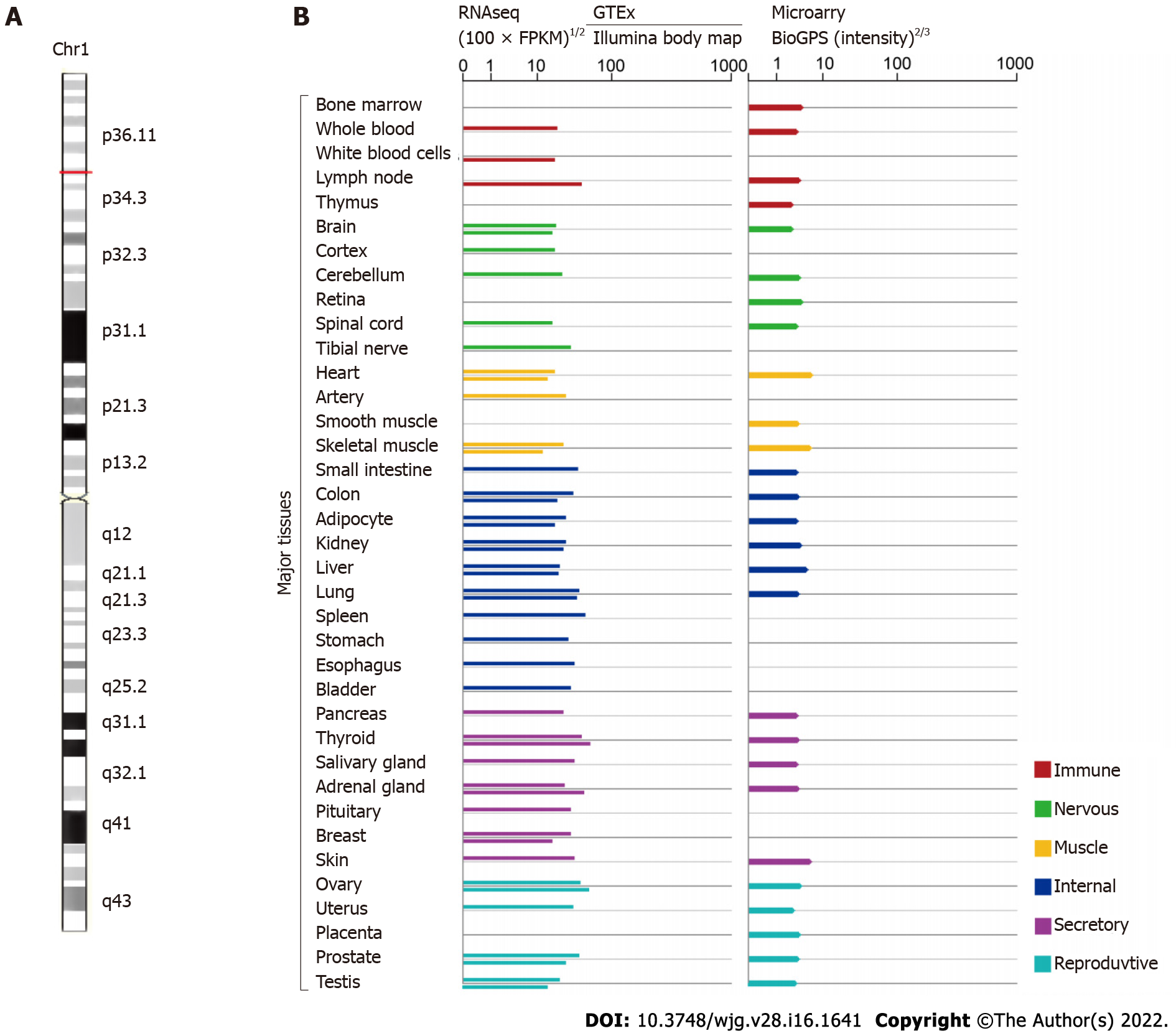
More recently, small nucleolar RNAs can be encoded by several lncRNAs called small RNA host genes, which are significantly differentially expressed in many diseases, including cancers[24]. Studies have unveiled that small nucleolar RNA host gene 3 (SNHG3), which is affiliated with the lncRNA class, plays an influential regulatory role in cancer initiation, development, progression, and tumor-associated microenvironment formation. LncRNA SNHG3 is located in 1p35.3, as shown in Figure 1 (GeneCards, http://www.genecards.org). SNHG3 is mainly localized to the nucleus, mitochondrion, and extracellular space. Although many studies on SNHG3 and tumors have been published, clinical studies and research remains limited. This paper reviews the upregulation of SNHG3 expression in most human tumors and its relationship with negative prognosis as well as the role of SNHG3 in the mechanism of tumorigenesis. Finally, it was concluded that SNHG3 can function as a biomarker in the diagnosis and prognosis of different cancers.
Growing evidence has demonstrated that SNHG3 acts as an oncogene and plays a critical regulatory role in the initiation and development of various cancers. SNHG3 exhibits characteristic oncogenic properties in multiple cancers. Studies have indicated that SNHG3 is significantly upregulated in hepatocellular carcinoma (HCC)[25,26] and other cancers, such as cervical cancer (CC)[27,28], and breast cancer (BC)[29-34]. Clear cell renal cell carcinoma (ccRCC)[35], colorectal cancer (CRC)[36-38], glioma[39,40], HCC[25,41,42], non-small cell lung cancer (NSCLC)[43-47], ovarian cancer (OC)[48-51], and papillary thyroid carcinoma (PTC)[52]. Different views exist on SNHG3 expression in lung adenocarcinoma, as shown in Table 1. Aberrant SNHG3 expression might represent a prognostic prediction value and therapeutic target in various cancers.
| Cancer types | Cases | Expression | Clinicopathologic parameters | PMID |
| Hepatocellular carcinoma | 47 pairs | Upregulated | Tumor stage, lymph node metastasis, distant metastasis | 31548493 |
| Hepatocellular carcinoma | 195 pairs | Upregulated | Tumor size, PVTT, relapse | 26373735 |
| Breast cancer | 30 pairs | Upregulated | 32495883 | |
| Breast cancer | 60 pairs | Upregulated | Histological grade, TNM stage, lymph node metastasis, ER, Her-2 | 31586299 |
| Breast cancer | 60 pairs | Upregulated | 32883233 | |
| Breast cancer | 48 pairs | Upregulated | 32945476 | |
| Cervical cancer | 88 pairs | Upregulated | FIGO stage, metastasis | 34816392 |
| Clear cell renal cell carcinoma | 36 pairs | Upregulated | Distant metastasis, T stage, pathological TNM stage, histologic grade | 31505165 |
| Colorectal cancer | 58 pairs | Upregulated | Tumor stage, distant metastasis | 32187965 |
| 50 pairs | Upregulated | 34661273 | ||
| Esophageal cancer | 384 pairs | Upregulated | 33596916 | |
| Gastric cancer | 60 pairs | Upregulated | 32930970 | |
| Glioma | 42 pairs | Upregulated | Tumor size, WHO grade | 33817254 |
| Glioma | 8 low-grade and 8 high-grade and 8 normal tissues | Upregulated | 34153159 | |
| Laryngeal carcinoma | 18 pairs | Upregulated | 31238052 | |
| Laryngeal carcinoma | 25 pairs | Upregulated | 32538668 | |
| Lung Adenocarcinoma | 65 pairs | Downregulated | TNM stage, lymph node metastasis | 32538668 |
| Non-small-cell lung cancer | 35 pairs | Upregulated | TNM stage, lymph node metastasis, tumor size | 34132359 |
| Non-small-cell lung cancer | 32 pairs | Upregulated | TNM stage, lymph node metastasis, tumor size | 31602642 |
| Non-small-cell lung cancer | 42 pairs | Upregulated | Stage, lymph node metastasis | 33177840 |
| Non-small-cell lung cancer | 15 pairs | Upregulated | 34032148 | |
| Oral squamous cell carcinoma | 30 pairs | Upregulated | 32989886 | |
| Osteosarcoma | 54 pairs | Upregulated | 30797154 | |
| Osteosarcoma | 127 pairs | Upregulated | 30791797 | |
| Ovarian cancer | 18 pairs | Upregulated | 29921511 | |
| Ovarian cancer | 96 pairs | Upregulated | FIGO stage, histological grade, lymph node metastasis | 33149611 |
| Ovarian cancer | 76 pairs | Upregulated | FIGO stage, lymph node metastasis | 29758922 |
| Ovarian cancer | 40 patients with OC and 19 patients with benign OC | Upregulated | 33552243 | |
| Papillary thyroid carcinoma | 72 pairs | Upregulated | TNM stage, lymph node metastasis | 32048306 |
| Prostate cancer | 50 pairs | Upregulated | 33416420 |
HCC is one of the most widespread cancerous malignancies, ranking third in cancer-related deaths[53]. Cures for HCC tend to be more effective when used at an early stage[54]. However, patients at this stage do not have specific symptoms or lack biomarkers for early diagnosis, which usually delays diagnosis. Accordingly, the development of biomarkers for early diagnosis and prognosis is urgent. Recent studies have shown that SNHG3 expression is upregulated in HCC tissues compared with normal tissues and positively correlates with tumor stage (P < 0.001), tumor size (P = 0.003), lymph node metastasis (P < 0.001), distant metastasis (P < 0.001), portal vein tumor thrombosis (P = 0.014), and relapse (P = 0.038)[25,26]. Based on the above evidence that SNHG3 influences HCC metastasis and growth, SNHG3 has the potential to be a reliable biomarker for the diagnosis and treatment of HCC.
BC accounts for 30% of all female tumors and is the second leading contributor to cancer deaths in women[55]. Early detection of BC can reduce mortality and improve patient survival[56-58]. The discovery of a new biomarker appears critical. Ma et al[31] reported elevated SNHG3 expression in BC tissues and correlated it with tumor malignancy. They also reported that SNHG3 expression was associated with clinicopathological features, such as histological grade (P = 0.016), tumor-node-metastasis (TNM) stage (P = 0.001), lymph node metastasis (P < 0.001), estrogen receptor (P = 0.009), and Her-2 (P = 0.001)[31]. ER and Her-2 are major molecular targets in the pathogenesis of BC and are associated with prognosis and treatment options for BC[59]. Therefore, SNHG3 plays a carcinogenic role in the progression and prognosis of BC. Therefore, SNHG3 may play an oncogenic role in the progression and prognosis of BC.
CC is the fourth most prevalent cancer among women worldwide in terms of incidence and mortality[53]. The survivability of patients with advanced CC is not desirable, and early diagnosis and treatment of CC improve its prognosis[60]. It is important to further explore the mechanisms of CC development to develop new therapeutic approaches[61]. The function of SNHG3 in CC has been recognized in several studies. Zhu et al[27] published that SNHG3 expression was obviously upregulated in CC tissues, and high SNHG3 expression affected International Federation of Gynecology and Obstetrics (FIGO) stage (P = 0.011) and metastasis (P = 0.018), implying that SNHG3 is a possible prognostic biomarker and a target for treatment in CC.
With high morbidity and mortality, ccRCC is the most common and serious type of renal cell carcinoma[62]. Using TCGA and GEO databases, Zhang et al[35] found that lncRNA SNHG3 expression was elevated in ccRCC and positively correlated with many clinicopathological parameters. They further quantified the relative expression of SNHG3 in ccRCC tissues compared with normal tissues using qRT–PCR and discovered that the relative expression of SNHG3 was increased in ccRCC tissues[35]. These results revealed that upregulated SNHG3 expression may play an essential role in the ccRCC pathway.
CRC is a dominant reason for morbidity and mortality worldwide, yet greater than half of patients have progressed to stage II/III at the time of diagnosis. The treatment of CRC typically consists of curative resection via surgery followed by adjuvant chemotherapy to reduce the risk of recurrence[63]. Therefore, it is critical to further elucidate the mechanisms of CRC and to pursue the development of effective targeted therapies. SNHG3 expression was remarkably upregulated in CRC tissues compared to nearby normal tissues and correlated with poor outcomes in CRC[36-38]. Moreover, Wen et al[36] explored the connection between SNHG3 and clinicopathological parameters, and the results reflected a close association with tumor stage (P = 0.006) and distant metastasis (P = 0.0033). Overall, SNHG3 promotes the progression of CRC, leading to an unfavorable prognosis.
Glioma is a fatal brain tumor that can be treated with radiation and surgery, but the median survival of patients is only approximately 14-17 mo[64]. Effective curative treatments for glioma are lacking. Thus, it is especially vital to determine the molecular mechanisms of glioma and identify novel strategies for treatment[65]. SNHG3 is highly expressed in glioma tissues compared to normal tissues[39,40]. Zhang et al[39] explored the relationship between SNHG3 expression and the clinicopathological features of GM patients, including tumor size (P = 0.0300) and World Health Organization classification (P = 0.0278). These studies indicated that SNHG3 offer new insights for the future treatment of glioma.
Lung cancer is one of the cancers with the highest mortality and morbidity rates worldwide[66], with NSCLC accounting for 85% of lung cancer cases[67]. Shi et al[44] revealed that high expression of lncRNA SNHG3 in NSCLC tissues was associated with TNM stage (P = 0.0053), lymph node metastasis (P = 0.0006), and tumor size (P = 0.0005). Thus, elevated SNHG3 expression is relevant to the lower overall survival (OS) rate of NSCLC patients. Liu et al[67] analyzed the TCGA database as well as the GSE19804 public database and found high SNHG3 expression in lung adenocarcinoma. These researchers used Kaplan–Meier plotter database analysis to determine that high SNHG3 expression decreases the OS time of lung adenocarcinoma patients. However, Kang et al[68] used quantitative polymerase chain reaction to show that SNHG3 expression levels were decreased in lung adenocarcinoma tissues, and Kaplan–Meier analysis demonstrated that patients with low SNHG3 expression levels had a short OS. In conclusion, SNHG3 is associated with unfavorable prognosis in lung cancer patients, but the expression and function of SNHG3 in lung adenocarcinoma still requires further research.
OC claims the lives of 151900 women worldwide each year and is one of the leading contributors to death in women[69]. Because there are no specific symptoms in the early stages, greater than 79% of OC patients reach stage III or IV[70]. The lack of effective biomarkers translates to a poor prognosis in OC patients. Recently, Hong et al[50] described high SNHG3 expression in OC tissues compared with normal tissues adjacent to cancer, and a high SNHG3 expression level was positively correlated with the FIGO stage (P = 0.007) and lymph node metastasis (P = 0.001) of OC patients. This finding suggests that high SNHG3 expression may be an individual prognostic factor affecting OS in OC patients.
PTC is the most common type of thyroid cancer, and the incidence of PTC is increasing[71]. The high rate of metastases in the cervical lymph nodes of PTC patients is closely related to the recurrence of PTC[72,73]. It seems relevant to study the mechanism of metastasis in PTC. Sui et al[52] analyzed 42 pairs of tissues and determined that SNHG3 expression was higher in PTC tissues compared with controls. High SNHG3 expression was positively correlated with TNM stage (P = 0.014) and lymph node metastasis (P = 0.0292) in advanced PTC. The above results suggest that SNHG3 could be a carcinogenic lncRNA in PTC.
Researchers have elucidated the high expression of SNHG3 in acute myeloid leukemia (AML)[74], esophageal cancer[75], gastric cancer (GC)[76-78], laryngeal carcinoma[79,80], oral squamous cell carcinoma[81,82], osteosarcoma[83,84], and prostate cancer[85,86], but have not yet explored the relationship between SNHG3 and clinicopathological features. Subsequent investigations into the biological mechanisms of SNHG3 in other diseases will further support the potential of SNHG3 as a new diagnostic and therapeutic biomarker.
SNHG3 may function as an oncogene and has the potential to become a novel potential therapeutic target for many cancers. Several studies have used animal models to explore the effect of SNHG3 in vivo. Zhao et al[25] assessed the role of SNHG3 in HCC growth in vivo. They injected stable SNHG3-depleted HepG2 cells subcutaneously into the right dorsum of female BALB/c nude mice and measured the volume of tumor. The results showed that SNHG3depleted tumors grew significantly slower than that of controls. HCC growth was inhibited in vivo due to the knockdown of the SNHG3 gene[25]. Similarly, SNHG3 gene knockdown inhibited the growth of other tumors, such as BC[34], CC[27], ccRCC[35], CRC[36,37], GC[77], glioma[40], laryngeal carcinoma[80], NSCLC[45,46], OC[49], PTC[52], prostate cancer[85]. Overall, SNHG3 promoted the progression of the abovementioned tumors.
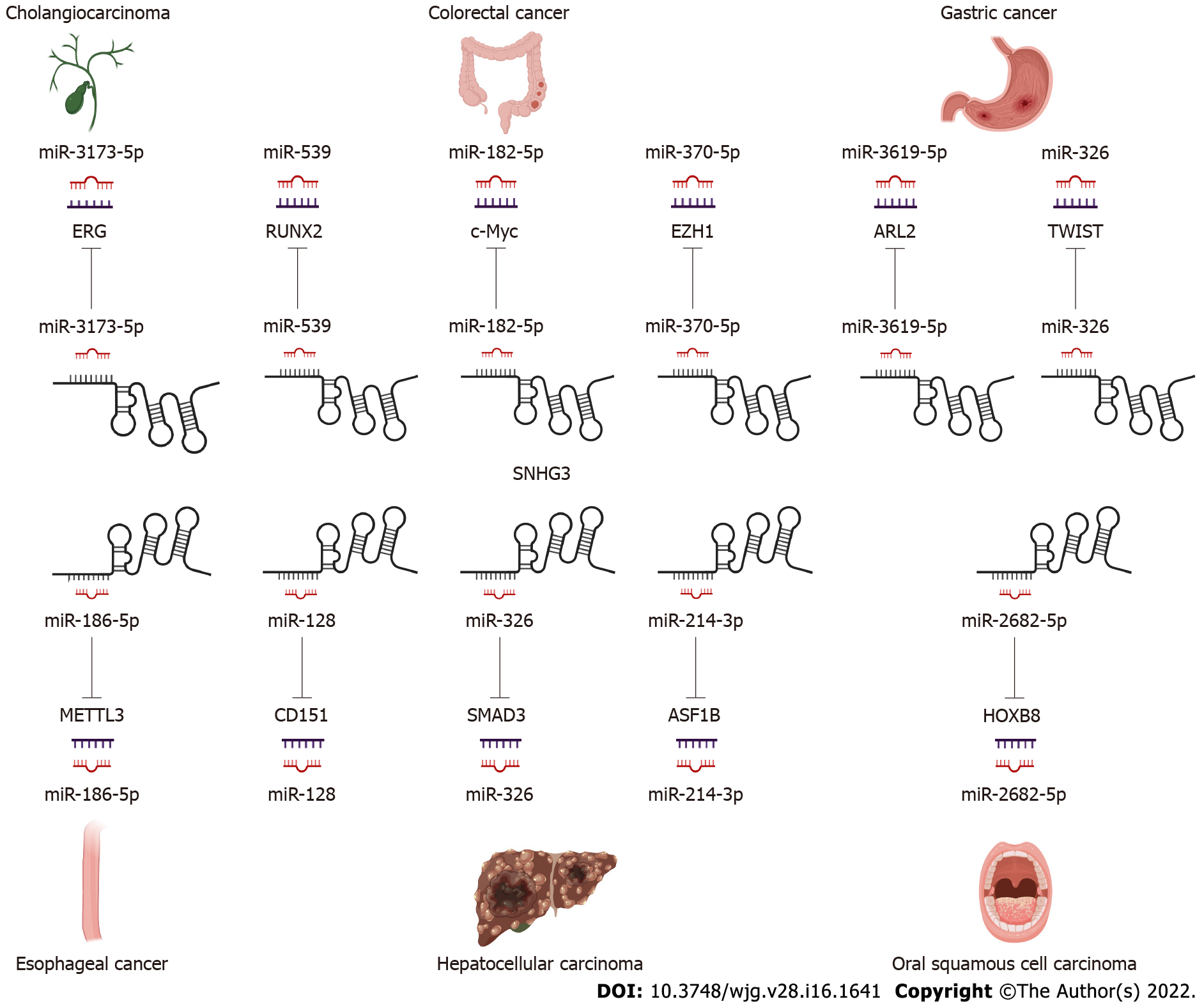
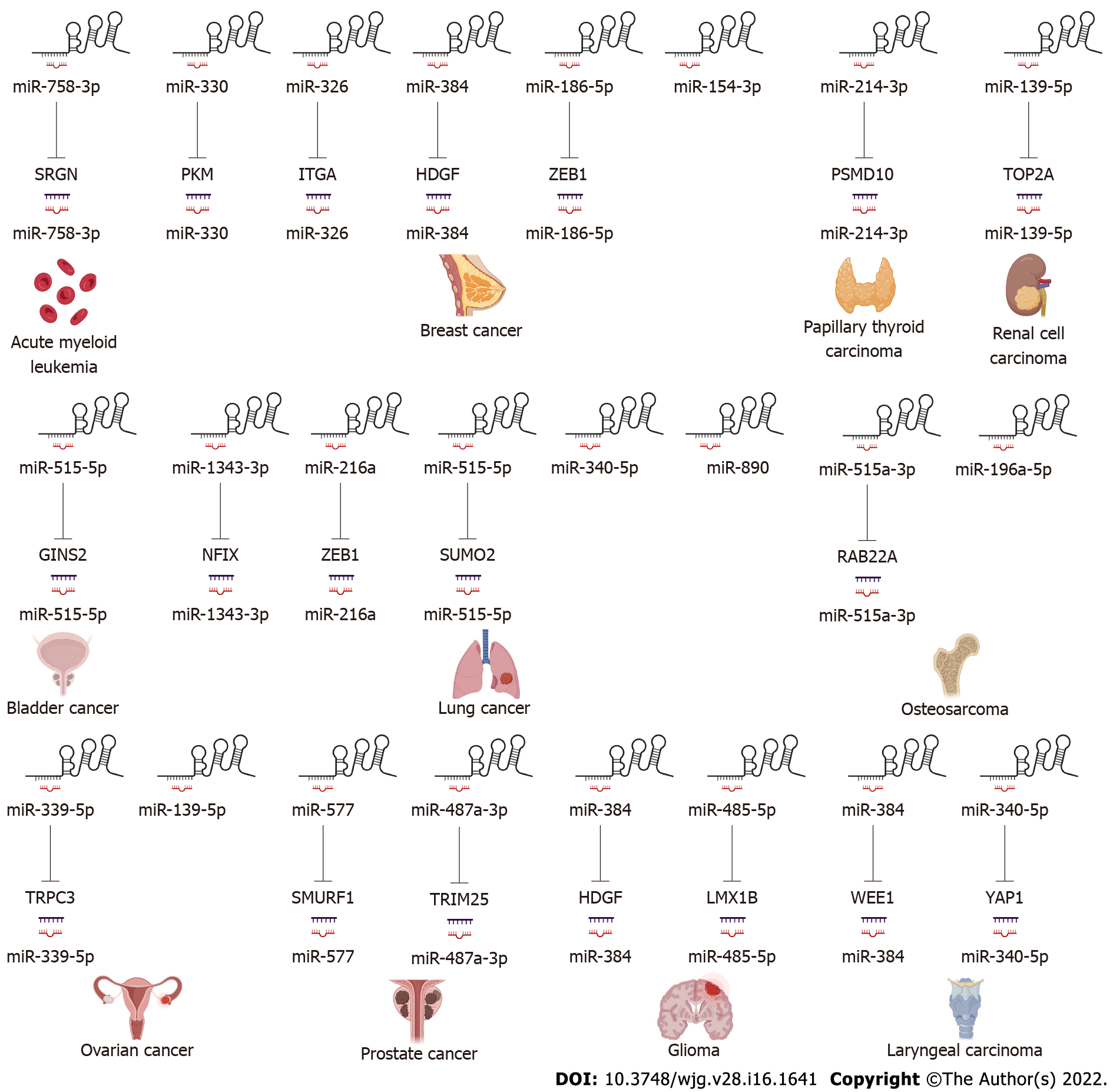
In the following section, we will explore the biological function of SNHG3 in different cell lines. Many works have shown that SNHG3 affects cancer cell proliferation, epithelial-mesenchymal transition (EMT)[25,41,44,47,85-87], apoptosis, invasion, migration, and metabolism[29,48] specifically through the action of different molecular mechanisms. In HCC, SNHG3 affects cell proliferation, apoptosis, metastasis and invasion mainly by means of competing endogenous RNA (ceRNA) and EMT signaling pathway, as shown in Figure 2 and Table 2.
| Cancer types | Assessed cancer cell lines | Expression | Related genes and pathways | Biological significance | PMID |
| Hepatocellular carcinoma | PLC/PRF/5, Hep3B, HepG2, MHCC97L, Huh7, SMMC-7721, HCCLM3 | Up | miR-128, CD151 | Invasion, EMT, sorafenib resistance | 30132868 |
| Hepatocellular carcinoma | HepG2, HCCLM3 | Up | miR-326, SMAD3, ZEB1 | Proliferation, migration, EMT, anti-apoptosis | 31548493 |
| Hepatocellular carcinoma | HepG2, HuH-7 | Up | miR-214-3p, ASF1B | 34336642 | |
| Acute myeloid leukemia | HL-60,THP-1,KG-1, NB4, MOLM-14 | Up | miR-758-3p, SRGN | Proliferation, anti-apoptosis | 31452272 |
| Bladder cancer | 5637,T24 | Up | miR-515-5p, GINS2 | Proliferation, migration, invasion, EMT | 32596993 |
| Breast cancer | MCF-7, MD-MBA-453 | Up | miR-330, PKM | Proliferation, mitochondrial metabolism | 31956955 |
| Breast cancer | MDA-MB-231, BT-549, MDA-MB-468 | Up | miR-326, ITGA5 Vav2/Rac1 | Viability, migration, invasion, anti-apoptosis | 32495883 |
| Breast cancer | MDA-MB-231, MCF-7 | Up | miR-384, HDGF | Proliferation, migration, invasion | 31586299 |
| Breast cancer | MCF-7, MDA-MB-231, HCC1937, BT474, SKBr-3 | Up | miR-154-3p, Notch | Proliferation, migration, invasion | 32883233 |
| Breast cancer | MDA-MB-231, MCF-7 | Up | miR-186-5p, ZEB1 | Proliferation, migration, invasion | 33594311 |
| Breast cancer | MCF-7, MDA-MB-231, MDA-MB-468, BT-474 | Up | miR-326 | Proliferation, migration, invasion | 32945476 |
| Cervical cancer | C33A, HeLa, SiHa, CaSki and HCC94 | Up | YAP1 | Proliferation, migration, invasion | 34816392 |
| Cervical cancer | SiHa | Up | Proliferation, migration, invasion | 34238747 | |
| Cholangiocarcinoma | HCCC9810, QBC939, RBE, HUCCT1 | Up | miR-3173–5p, ERG | Proliferation, migration, invasion | 34647226 |
| Clear cell renal cell carcinoma | ACHN, A498, Caki-1 | Up | miR-139-5p, TOP2A | Proliferation, migration, invasion | 31505165 |
| Colorectal cancer | HCT116, LoVo, SW480, and SW620 | Up | miR-539, RUNX2 | Proliferation, migration, invasion | 32187965 |
| Colorectal cancer | HT29, HCT116, SW480, and LoVo | Up | miR-182-5p, c-Myc | Proliferation, migration, invasion | 28731158 |
| Colorectal cancer | SW480, SW620, HCT8, HT29 | Up | miR-370-5p, EZH1 | Proliferation, migration, invasion | 34661273 |
| Esophageal cancer | KYSE-150, Eca-9706 | Up | miR-186-5p, METTL3, m6A | 33596916 | |
| Gastric cancer | MGC-803, AGS, BGC-823, SGC-7901, MKN-45, HGC-27 | Up | EZH2, MED18 | Proliferation, migration, invasion | 31534128 |
| Gastric cancer | SGC7901, BGC823 | Up | IL-6, STAT3, SNHG3, miR-3619-5p, ARL2 | Proliferation | 32930970 |
| Gastric cancer | HGC-27, GC9811-P | Up | miR-326, TWIST | Proliferation, migration, invasion | 34257656 |
| Glioma | A172, SHG44 | Up | miR-384, HDGF | Proliferation, migration, invasion, anti-apoptosis | 33817254 |
| Glioma | U-87-MG, U-251-MG | Up | miR-485-5p, LMX1B | Proliferation, migration, invasion | 34153159 |
| Laryngeal carcinoma | 16HBE, TU212, TU686 | Up | miR-384, WEE1 | Proliferation, migration, invasion | 31238052 |
| Laryngeal carcinoma | TU177, AMC-HN-8 | Up | Wnt/β-catenin, miR-340-5p, YAP1 | Proliferation, migration, anti-apoptosis, mitochondrial metabolism | 32538668 |
| Lung adenocarcinoma | A549, H1299 | Up | Proliferation, cell cycle,anti-apoptosis | 30154938 | |
| Lung adenocarcinoma | A549, H1299, H1975 | Down | miR-890 | Anti-proliferation, anti-migration, anti-invasion, apoptosis | 34306585 |
| Non-small-cell lung cancer | H1299, H358, A549, H1975 | Up | miR-13433p, NFIX | Proliferation, migration, invasion | 34132359 |
| Non-small-cell lung cancer | CMT-167, LLC, CMT-170, CMT-181 | Up | TGF-β pathway, IL-6, JAK2, STAT3 pathway | Proliferation, migration, EMT | 31602642 |
| Non-small-cell lung cancer | SPC-A1, A549, NCI-H23, NCI-H460 | Up | miR-340-5p | Lymph node infiltration, distant metastases | 33225676 |
| Non-small-cell lung cancer | A549, H322, H1299, GLC-82, SPC-A1 | Up | miR-216a, ZEB1 | Proliferation, migration, invasion | 33177840 |
| Non-small-cell lung cancer | A549, HCC827, H2170, H520 | Up | miR-515-5p, SUMO2 | Proliferation, migration, invasion, EMT | 34032148 |
| Oral squamous cell carcinoma | SCC4, CAL-27 | Up | miR-2682-5p, HOXB8 | Proliferation, migration | 32989886 |
| Oral squamous cell carcinoma | SCC-15, SCC-9, CAL-27, HN5 | Up | Wnt/β-catenin, NFYC | Proliferation, migration | 31762107 |
| Osteosarcoma | Saos2, MG63, U2OS, HOS | Up | miR-151a-3p, RAB22A | Proliferation, migration, invasion | 30797154 |
| Osteosarcoma | MG-63, 143B, HOS, SW-1353, Saos-2, U-2OS | Up | miR-196a-5p | Proliferation, colony formation | 30791797 |
| Ovarian cancer | SK-OV3, TOV-21G, and OVCAR-3 | Up | Energy metabolism | 29921511 | |
| Ovarian cancer | SKOV3, HeyA8, A2780 | Up | miR-339-5p, TRPC3 | Proliferation, anti-apoptosis | 33149611 |
| Ovarian cancer | SKOV3, 60OVCAR3, A2780, ES2 | Up | GSK3β/β-catenin | Proliferation, migration | 29758922 |
| Ovarian cancer | A2780, SKOV3, OVCAR3, OV90 | Up | miR-139-5p, Notch1 | Proliferation, migration | 33552243 |
| Papillary thyroid carcinoma | BCPAP, TPC-1 | Up | miR-214-3p, PSMD10 | Migration, invasion, proliferation, and colony formation | 32048306 |
| Prostate cancer | PC3, DU145, 22RV1, NCaP | Up | miR-577, SMURF1 | Proliferation, migration, EMT, anti-apoptosis | 32248648 |
| Prostate cancer | PC3, 22RV1, DU145, LNCaP | Up | miR-487a-3p,TRIM25 | Viability, migration, invasion, EMT | 33416420 |
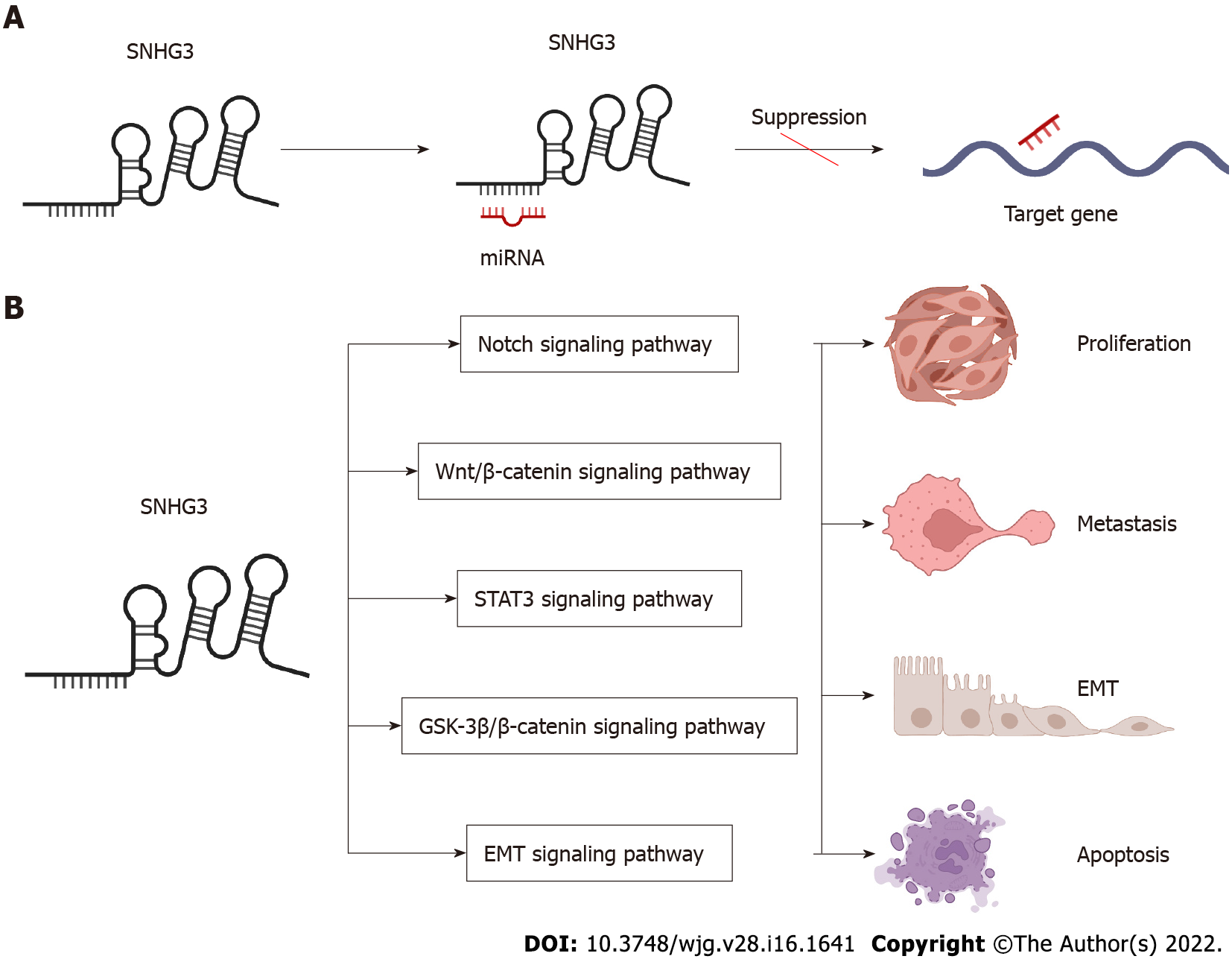
LncRNAs and microRNAs (miRNAs) are ncRNAs. LncRNAs compete with miRNAs by acting as sponges for miRNAs to reduce the activity or expression of miRNAs[88]. These lncRNAs are called ceRNAs. Numerous studies have found that SNHG3 can function as a ceRNA by competitively binding to various miRNAs, including miR-758-3p[74], miR-515-5p[87], miR-330[29], miR-326[25,30,34,78], miR-384[31,39,79], miR-154-3p[32], miR-186-5p[33,75], miR3173–5p[89], miR-139-5p[35], miR-539[36], miR-182-5p[37], miR-370-5p[38], miR-3619-5p[76], miR-485-5p[40], miR-128[41], miR-214-3p[42,52], miR-340-5p[45,80], miR-890[68], miR-1343-3p[43], miR-216a[46], miR-515-5p[47], miR-2682-5p[81], miRNA-151a-3p[83], miR-196a-5p[84], miR-339-5p[49], miR-139-5p[51], miR-577[85], and miR-487a-3p[86]. In HCC SNHG3 plays its role as a ceRNA mainly by binding to miR-128[41], miR-326[25] and miR-214-3p[42]. SNHG3 acts as a sponge to bind to miRNAs, subsequently blocking the effects of miRNAs on their downstream target mRNAs. Thus, SNHG3 regulates the expression of oncogenes or tumor suppressor genes, such as SRGN[74], PKM[29], HDGF[31], and c-Myc[37], ultimately affecting cancer cell proliferation, apoptosis, metastasis, metabolism and EMT, as shown in Figures 3 and 4.
The EMT process has been shown to be critical in cancer[90,91]. Shi et al[44] showed that human lung cancer cells overexpressing the SNHG3 gene exhibited increased expression of mesenchymal markers (N-calmodulin and waveform protein) and reduced expression of epithelial cell markers (E-calmodulin) while also promoting cancer cell proliferation and metastasis through the TGF-β pathway. In addition, Zhao et al[25] learned that SNHG3 promotes cancer cell migration by upregulating the expression of ZEB1, a key transcription factor of EMT in HCC cells. The same conclusion was reached in BC[33] and NSCLC[46]. The above achievements point to the potential application of SNHG3 in the tumor EMT signaling pathway.
The Notch signaling pathway is a strongly conserved cellular signaling system in most multicellular organisms and is required for a variety of cellular processes, including stem cell functions, cell proliferation, differentiation, and cell death. Several lncRNAs have been proven to participate in the Notch signaling pathway. Zhang et al[51] demonstrated that SNHG3 promotes OC proliferation and migration by regulating Notch1. In addition, Jiang et al[32], suggested that SNHG3 promotes BC cell proliferation and metastasis through upregulation of the Notch signaling pathway. These studies indicate that SNHG3 plays a crucial part in Notch pathways, which have the potential to develop novel therapeutic targets for cancer therapy.
Aberrant activation of Wnt/β-catenin signaling was found in some tumors, and Wnt/β-catenin signaling can regulate oncogenes, such as c-Myc and Bcl-2, leading to tumorigenesis and cell proliferation. SNHG3 may upregulate the Wnt/β-catenin pathway by regulating miR-340-5p and YAP1 in laryngeal cancer cells[80]. In addition, SNHG3 may also promote the proliferation and metastasis of oral squamous carcinoma cells by upregulating the NFYC and Wnt/β-catenin pathways[82]. SNHG3 is involved in the Wnt/β-catenin signaling pathway that mediates tumor development and could represent a new tool for tumor therapy.
Tyrosine kinase signaling delivered by STAT3 is frequently activated in cancer cells, and the STAT3 signaling pathway plays an important role in cancer progression, where it can be activated by cytokines, such as IL-6[92], and growth factors[93,94]. STAT3 is phosphorylated by receptor-associated JAK and thus enters the nucleus to act as a transcriptional activator, regulating oncogene expression[95]. The impacts of STAT3 signaling in suppressing tumor immune surveillance have also been reported[96].
Sun et al[76] demonstrated that SNHG3 was overexpressed in GC tissues, and cellular experiments revealed that IL-6-activated STAT3 positively regulates SNHG3 and that SNHG3 promotes stem cell-like properties in GC cells. Shi et al[44] found that the IL-6/JAK2/STAT3 pathway activated SNHG3 in NSCLC and promoted cell proliferation and migration. This finding leads to the conclusion that the STAT3 signaling pathway involved in SNHG3 is a novel mechanism of carcinogenesis, suggesting that SNHG3 may represent a biomarker for the treatment of these carcinomas.
GSK-3β can negatively regulate β-catenin signaling, which is also implicated in cell proliferation[97]. Research has illustrated that GSK-3β/β-catenin signaling is, for example, one of the downstream drivers of SNHG3 in OC[50], and high expression of SNHG3 fosters cell proliferation and invasion via GSK-3β/β-catenin.
SNHG3 can revitalize multiple signaling pathways to promote human tumorigenesis, such as the Notch signaling pathway, Wnt/β-catenin signaling pathway, STAT3 signaling pathway, GSK-3β/β-catenin signaling pathway, and EMT signaling pathway. Thus, SNHG3 may become a target in the treatment of tumors.
LncRNAs function as multifunctional signaling modulators, facilitating tumor initiation, progression, and metastasis by regulating tumor cell proliferation, migration, apoptosis, cell cycle, drug resistance, epithelial-mesenchymal transition, metabolic reprogramming, and immune response[98-100]. Compelling studies have suggested that lncRNAs can act as diagnostic indices, prognostic biomarkers, and therapeutic targets for diseases[101,102]. These reports indicate that SNHG3 expression is upregulated in tumor tissues, such as HCC, GC, CC, PTC, and AML, compared to adjacent normal tissues. However, the expression of SNHG3 in lung adenocarcinoma has shown different results and needs to be further explored. At the same time, SNHG3 has been associated with various clinicopathological parameters such as staging and distant metastasis. In addition, SNHG3 has been shown to promote tumour development in in vivo experiments, and at the cellular level, SNHG3 plays a significant role in promoting tumour cell proliferation, migration and metastasis, disrupting the cell cycle and inhibiting apoptosis in a variety of cancers through a variety of signalling pathways, resulting in a poor prognosis for patients.In short, these findings implied that SNHG3 might function as a new target in the diagnosis and treatment of tumours.
In summary, a better understanding of the function of SNHG3 in the clinicopathological features and mechanisms of tumor development may help to improve the efficiency and targeting of treatment. Further studies on SNHG3 and its regulatory mechanism may pave the way to improve prevention, cancer diagnosis, and treatment based on patients' biological and pathological characteristics.
The authors gratefully acknowledge all the people that have made this study.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Biochemistry and molecular biology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Dalili S, Iran; Pang Y, China S-Editor: Zhang H L-Editor: A P-Editor: Zhang H
| 1. | Sharma Y, Miladi M, Dukare S, Boulay K, Caudron-Herger M, Groß M, Backofen R, Diederichs S. A pan-cancer analysis of synonymous mutations. Nat Commun. 2019;10:2569. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 86] [Cited by in RCA: 137] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 2. | Nuevo-Tapioles C, Santacatterina F, Stamatakis K, Núñez de Arenas C, Gómez de Cedrón M, Formentini L, Cuezva JM. Coordinate β-adrenergic inhibition of mitochondrial activity and angiogenesis arrest tumor growth. Nat Commun. 2020;11:3606. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 83] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 3. | Ni Y, Schmidt KR, Werner BA, Koenig JK, Guldner IH, Schnepp PM, Tan X, Jiang L, Host M, Sun L, Howe EN, Wu J, Littlepage LE, Nakshatri H, Zhang S. Death effector domain-containing protein induces vulnerability to cell cycle inhibition in triple-negative breast cancer. Nat Commun. 2019;10:2860. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 17] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 4. | Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150:12-27. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1920] [Cited by in RCA: 2277] [Article Influence: 175.2] [Reference Citation Analysis (0)] |
| 5. | Paull EO, Aytes A, Jones SJ, Subramaniam PS, Giorgi FM, Douglass EF, Tagore S, Chu B, Vasciaveo A, Zheng S, Verhaak R, Abate-Shen C, Alvarez MJ, Califano A. A modular master regulator landscape controls cancer transcriptional identity. Cell. 2021;184:334-351.e20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 84] [Article Influence: 21.0] [Reference Citation Analysis (0)] |
| 6. | Statello L, Guo CJ, Chen LL, Huarte M. Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol. 2021;22:96-118. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3257] [Cited by in RCA: 3036] [Article Influence: 759.0] [Reference Citation Analysis (0)] |
| 7. | Yao RW, Wang Y, Chen LL. Cellular functions of long noncoding RNAs. Nat Cell Biol. 2019;21:542-551. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 693] [Cited by in RCA: 1041] [Article Influence: 173.5] [Reference Citation Analysis (0)] |
| 8. | Dong H, Hu J, Zou K, Ye M, Chen Y, Wu C, Chen X, Han M. Activation of LncRNA TINCR by H3K27 acetylation promotes Trastuzumab resistance and epithelial-mesenchymal transition by targeting MicroRNA-125b in breast Cancer. Mol Cancer. 2019;18:3. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 155] [Cited by in RCA: 170] [Article Influence: 28.3] [Reference Citation Analysis (0)] |
| 9. | Li L, van Breugel PC, Loayza-Puch F, Ugalde AP, Korkmaz G, Messika-Gold N, Han R, Lopes R, Barbera EP, Teunissen H, de Wit E, Soares RJ, Nielsen BS, Holmstrøm K, Martínez-Herrera DJ, Huarte M, Louloupi A, Drost J, Elkon R, Agami R. LncRNA-OIS1 regulates DPP4 activation to modulate senescence induced by RAS. Nucleic Acids Res. 2018;46:4213-4227. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 43] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 10. | Wu H, Qin W, Lu S, Wang X, Zhang J, Sun T, Hu X, Li Y, Chen Q, Wang Y, Zhao H, Piao H, Zhang R, Wei M. Long noncoding RNA ZFAS1 promoting small nucleolar RNA-mediated 2'-O-methylation via NOP58 recruitment in colorectal cancer. Mol Cancer. 2020;19:95. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 96] [Article Influence: 19.2] [Reference Citation Analysis (0)] |
| 11. | Shen SN, Li K, Liu Y, Yang CL, He CY, Wang HR. Down-regulation of long noncoding RNA PVT1 inhibits esophageal carcinoma cell migration and invasion and promotes cell apoptosis via microRNA-145-mediated inhibition of FSCN1. Mol Oncol. 2019;13:2554-2573. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 62] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 12. | Stojic L, Niemczyk M, Orjalo A, Ito Y, Ruijter AE, Uribe-Lewis S, Joseph N, Weston S, Menon S, Odom DT, Rinn J, Gergely F, Murrell A. Transcriptional silencing of long noncoding RNA GNG12-AS1 uncouples its transcriptional and product-related functions. Nat Commun. 2016;7:10406. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 69] [Cited by in RCA: 66] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 13. | Gong C, Maquat LE. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3' UTRs via Alu elements. Nature. 2011;470:284-288. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1072] [Cited by in RCA: 1004] [Article Influence: 71.7] [Reference Citation Analysis (0)] |
| 14. | McHugh CA, Chen CK, Chow A, Surka CF, Tran C, McDonel P, Pandya-Jones A, Blanco M, Burghard C, Moradian A, Sweredoski MJ, Shishkin AA, Su J, Lander ES, Hess S, Plath K, Guttman M. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature. 2015;521:232-236. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 866] [Cited by in RCA: 859] [Article Influence: 85.9] [Reference Citation Analysis (0)] |
| 15. | Sun Q, Tripathi V, Yoon JH, Singh DK, Hao Q, Min KW, Davila S, Zealy RW, Li XL, Polycarpou-Schwarz M, Lehrmann E, Zhang Y, Becker KG, Freier SM, Zhu Y, Diederichs S, Prasanth SG, Lal A, Gorospe M, Prasanth KV. MIR100 host gene-encoded lncRNAs regulate cell cycle by modulating the interaction between HuR and its target mRNAs. Nucleic Acids Res. 2018;46:10405-10416. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 58] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 16. | Rashid F, Shah A, Shan G. Long Non-coding RNAs in the Cytoplasm. Genomics Proteomics Bioinformatics. 2016;14:73-80. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 216] [Cited by in RCA: 287] [Article Influence: 31.9] [Reference Citation Analysis (0)] |
| 17. | Gil N, Ulitsky I. Regulation of gene expression by cis-acting long non-coding RNAs. Nat Rev Genet. 2020;21:102-117. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 276] [Cited by in RCA: 479] [Article Influence: 79.8] [Reference Citation Analysis (0)] |
| 18. | Kopp F, Mendell JT. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell. 2018;172:393-407. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1719] [Cited by in RCA: 2635] [Article Influence: 439.2] [Reference Citation Analysis (0)] |
| 19. | Tuunanen TH, Tervo TM. Excimer laser phototherapeutic keratectomy for corneal diseases: a follow-up study. CLAO J. 1995;21:67-72. [PubMed] |
| 20. | Chen W, Yang J, Fang H, Li L, Sun J. Relevance Function of Linc-ROR in the Pathogenesis of Cancer. Front Cell Dev Biol. 2020;8:696. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 33] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 21. | Atianand MK, Caffrey DR, Fitzgerald KA. Immunobiology of Long Noncoding RNAs. Annu Rev Immunol. 2017;35:177-198. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 274] [Cited by in RCA: 364] [Article Influence: 45.5] [Reference Citation Analysis (0)] |
| 22. | Atianand MK, Fitzgerald KA. Long non-coding RNAs and control of gene expression in the immune system. Trends Mol Med. 2014;20:623-631. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 157] [Cited by in RCA: 189] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 23. | Mowel WK, Kotzin JJ, McCright SJ, Neal VD, Henao-Mejia J. Control of Immune Cell Homeostasis and Function by lncRNAs. Trends Immunol. 2018;39:55-69. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 109] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 24. | Yang W, Zhang K, Li L, Ma K, Hong B, Gong Y, Gong K. Discovery and validation of the prognostic value of the lncRNAs encoding snoRNAs in patients with clear cell renal cell carcinoma. Aging (Albany NY). 2020;12:4424-4444. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 32] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 25. | Zhao Q, Wu C, Wang J, Li X, Fan Y, Gao S, Wang K. LncRNA SNHG3 Promotes Hepatocellular Tumorigenesis by Targeting miR-326. Tohoku J Exp Med. 2019;249:43-56. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 39] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 26. | Zhang T, Cao C, Wu D, Liu L. SNHG3 correlates with malignant status and poor prognosis in hepatocellular carcinoma. Tumour Biol. 2016;37:2379-2385. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 81] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 27. | Zhu H, Zhu C, Feng X, Luo Y. Long noncoding RNA SNHG3 promotes malignant phenotypes in cervical cancer cells via association with YAP1. Hum Cell. 2022;35:320-332. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 28. | Sun Z, Hu J, Hu K, Tang M, Sun S, Fang Y, Yu H, Zhang Y. [Role of long noncoding RNA SNHG3 in regulating proliferation, migration and invasion of cervical cancer SiHa cells]. Nan Fang Yi Ke Da Xue Xue Bao. 2021;41:931-936. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 29. | Li Y, Zhao Z, Liu W, Li X. SNHG3 Functions as miRNA Sponge to Promote Breast Cancer Cells Growth Through the Metabolic Reprogramming. Appl Biochem Biotechnol. 2020;191:1084-1099. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 76] [Cited by in RCA: 100] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 30. | Wang P, Liu GZ, Wang JF, Du YY. SNHG3 silencing suppresses the malignant development of triple-negative breast cancer cells by regulating miRNA-326/integrin α5 axis and inactivating Vav2/Rac1 signaling pathway. Eur Rev Med Pharmacol Sci. 2020;24:5481-5492. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |
| 31. | Ma Q, Qi X, Lin X, Li L, Chen L, Hu W. LncRNA SNHG3 promotes cell proliferation and invasion through the miR-384/hepatoma-derived growth factor axis in breast cancer. Hum Cell. 2020;33:232-242. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 48] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 32. | Jiang H, Li X, Wang W, Dong H. Long non-coding RNA SNHG3 promotes breast cancer cell proliferation and metastasis by binding to microRNA-154-3p and activating the notch signaling pathway. BMC Cancer. 2020;20:838. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 26] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 33. | Wan Q, Tang M, Sun SL, Hu J, Sun ZJ, Fang YT, He TC, Zhang Y. SNHG3 promotes migration, invasion, and epithelial-mesenchymal transition of breast cancer cells through the miR-186-5p/ZEB1 axis. Am J Transl Res. 2021;13:585-600. [PubMed] |
| 34. | Zhang H, Wei N, Zhang W, Shen L, Ding R, Li Q, Li S, Du Y. lncRNA SNHG3 promotes breast cancer progression by acting as a miR326 sponge. Oncol Rep. 2020;44:1502-1510. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 8] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 35. | Zhang C, Qu Y, Xiao H, Xiao W, Liu J, Gao Y, Li M. LncRNA SNHG3 promotes clear cell renal cell carcinoma proliferation and migration by upregulating TOP2A. Exp Cell Res. 2019;384:111595. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 42] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 36. | Dacheng W, Songhe L, Weidong J, Shutao Z, Jingjing L, Jiaming Z. LncRNA SNHG3 promotes the growth and metastasis of colorectal cancer by regulating miR-539/RUNX2 axis. Biomed Pharmacother. 2020;125:110039. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 39] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 37. | Huang W, Tian Y, Dong S, Cha Y, Li J, Guo X, Yuan X. The long non-coding RNA SNHG3 functions as a competing endogenous RNA to promote malignant development of colorectal cancer. Oncol Rep. 2017;38:1402-1410. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 76] [Cited by in RCA: 93] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 38. | Zhang Y, Li L, Lu KX, Yu LB, Meng J, Liu CY. LncRNA SNHG3 is responsible for the deterioration of colorectal carcinoma through regulating the miR-370-5p/EZH1 axis. Eur Rev Med Pharmacol Sci. 2021;25:6131-6137. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 39. | Zhang X, Zheng W, Jiang W, Lin R, Xing C. Long non-coding RNA SNHG3 accelerates progression in glioma by modulating miR-384/HDGF axis. Open Life Sci. 2020;15:654-664. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 40. | Guo X, Zheng J, Yu MJ, Piao HZ, Zhao HY. Long noncoding RNA SNHG3 promotes glioma tumorigenesis by sponging miR-485-5p to upregulate LMX1B expression. Kaohsiung J Med Sci. 2021;37:851-862. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 8] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 41. | Zhang PF, Wang F, Wu J, Wu Y, Huang W, Liu D, Huang XY, Zhang XM, Ke AW. LncRNA SNHG3 induces EMT and sorafenib resistance by modulating the miR-128/CD151 pathway in hepatocellular carcinoma. J Cell Physiol. 2019;234:2788-2794. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 143] [Cited by in RCA: 145] [Article Influence: 24.2] [Reference Citation Analysis (0)] |
| 42. | Zhan T, Gao X, Wang G, Li F, Shen J, Lu C, Xu L, Li Y, Zhang J. Construction of Novel lncRNA-miRNA-mRNA Network Associated With Recurrence and Identification of Immune-Related Potential Regulatory Axis in Hepatocellular Carcinoma. Front Oncol. 2021;11:626663. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 33] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 43. | Zhao L, Song X, Guo Y, Ding N, Wang T, Huang L. Long noncoding RNA SNHG3 promotes the development of nonsmall cell lung cancer via the miR13433p/NFIX pathway. Int J Mol Med. 2021;48. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 44. | Shi J, Li J, Yang S, Hu X, Chen J, Feng J, Shi T, He Y, Mei Z, He W, Xie J, Li S, Jie Z, Tu S. LncRNA SNHG3 is activated by E2F1 and promotes proliferation and migration of non-small-cell lung cancer cells through activating TGF-β pathway and IL-6/JAK2/STAT3 pathway. J Cell Physiol. 2020;235:2891-2900. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 68] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 45. | He WW, Ma HT, Guo X, Wu WM, Gao EJ, Zhao YH. lncRNA SNHG3 accelerates the proliferation and invasion of non-small cell lung cancer by downregulating miR-340-5p. J Biol Regul Homeost Agents. 2020;34:2017-2027. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 46. | Zhao S, Gao X, Zhong C, Li Y, Wang M, Zang S. SNHG3 Knockdown Suppresses Proliferation, Migration and Invasion, and Promotes Apoptosis in Non-Small Cell Lung Cancer Through Regulating miR-216a/ZEB1 Axis. Onco Targets Ther. 2020;13:11327-11336. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 12] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 47. | Li Y, Gao L, Zhang C, Meng J. LncRNA SNHG3 Promotes Proliferation and Metastasis of Non-Small-Cell Lung Cancer Cells Through miR-515-5p/SUMO2 Axis. Technol Cancer Res Treat. 2021;20:15330338211019376. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 14] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 48. | Li N, Zhan X. The lncRNA SNHG3 regulates energy metabolism of ovarian cancer by an analysis of mitochondrial proteomes. Gynecol Oncol. 2018;150:343-354. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 74] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 49. | Liu EL, Zhou YX, Li J, Zhang DH, Liang F. Long-Chain Non-Coding RNA SNHG3 Promotes the Growth of Ovarian Cancer Cells by Targeting miR-339-5p/TRPC3 Axis. Onco Targets Ther. 2020;13:10959-10971. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 9] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 50. | Hong L, Chen W, Wu D, Wang Y. Upregulation of SNHG3 expression associated with poor prognosis and enhances malignant progression of ovarian cancer. Cancer Biomark. 2018;22:367-374. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 39] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 51. | Zhang L, Li G, Wang X, Zhang Y, Huang X, Wu H. lncRNA SNHG3 acts as oncogene in ovarian cancer through miR-139-5p and Notch1. Oncol Lett. 2021;21:122. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 52. | Sui G, Zhang B, Fei D, Wang H, Guo F, Luo Q. The lncRNA SNHG3 accelerates papillary thyroid carcinoma progression via the miR-214-3p/PSMD10 axis. J Cell Physiol. 2020;235:6615-6624. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 25] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 53. | Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53206] [Cited by in RCA: 55827] [Article Influence: 7975.3] [Reference Citation Analysis (132)] |
| 54. | Ren Z, Li A, Jiang J, Zhou L, Yu Z, Lu H, Xie H, Chen X, Shao L, Zhang R, Xu S, Zhang H, Cui G, Sun R, Wen H, Lerut JP, Kan Q, Li L, Zheng S. Gut microbiome analysis as a tool towards targeted non-invasive biomarkers for early hepatocellular carcinoma. Gut. 2019;68:1014-1023. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 542] [Cited by in RCA: 509] [Article Influence: 84.8] [Reference Citation Analysis (0)] |
| 55. | Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer Statistics, 2021. CA Cancer J Clin. 2021;71:7-33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8287] [Cited by in RCA: 11936] [Article Influence: 2984.0] [Reference Citation Analysis (4)] |
| 56. | Teh YC, Tan GH, Taib NA, Rahmat K, Westerhout CJ, Fadzli F, See MH, Jamaris S, Yip CH. Opportunistic mammography screening provides effective detection rates in a limited resource healthcare system. BMC Cancer. 2015;15:405. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 33] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 57. | O'Mahony M, Comber H, Fitzgerald T, Corrigan MA, Fitzgerald E, Grunfeld EA, Flynn MG, Hegarty J. Interventions for raising breast cancer awareness in women. Cochrane Database Syst Rev. 2017;2:CD011396. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 32] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 58. | Coleman C. Early Detection and Screening for Breast Cancer. Semin Oncol Nurs. 2017;33:141-155. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 133] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 59. | Waks AG, Winer EP. Breast Cancer Treatment: A Review. JAMA. 2019;321:288-300. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1800] [Cited by in RCA: 2855] [Article Influence: 475.8] [Reference Citation Analysis (0)] |
| 60. | Frumovitz M, Querleu D, Gil-Moreno A, Morice P, Jhingran A, Munsell MF, Macapinlac HA, Leblanc E, Martinez A, Ramirez PT. Lymphadenectomy in locally advanced cervical cancer study (LiLACS): Phase III clinical trial comparing surgical with radiologic staging in patients with stages IB2-IVA cervical cancer. J Minim Invasive Gynecol. 2014;21:3-8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 69] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 61. | Wang L, Zhao Y, Wang Y, Wu X. The Role of Galectins in Cervical Cancer Biology and Progression. Biomed Res Int. 2018;2018:2175927. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 33] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 62. | Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7-34. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13300] [Cited by in RCA: 15472] [Article Influence: 2578.7] [Reference Citation Analysis (2)] |
| 63. | Glaire MA, Domingo E, Sveen A, Bruun J, Nesbakken A, Nicholson G, Novelli M, Lawson K, Oukrif D, Kildal W, Danielsen HE, Kerr R, Kerr D, Tomlinson I, Lothe RA, Church DN. Tumour-infiltrating CD8+ lymphocytes and colorectal cancer recurrence by tumour and nodal stage. Br J Cancer. 2019;121:474-482. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 41] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 64. | Reifenberger G, Wirsching HG, Knobbe-Thomsen CB, Weller M. Advances in the molecular genetics of gliomas - implications for classification and therapy. Nat Rev Clin Oncol. 2017;14:434-452. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 338] [Cited by in RCA: 467] [Article Influence: 51.9] [Reference Citation Analysis (0)] |
| 65. | Riabovol OO, Tsymbal DO, Minchenko DO, Lebid-Biletska KM, Sliusar MY, Rudnytska OV, Minchenko OH. Effect of glucose deprivation on the expression of genes encoding glucocorticoid receptor and some related factors in ERN1-knockdown U87 glioma cells. Endocr Regul. 2019;53:237-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 66. | Youlden DR, Cramb SM, Baade PD. The International Epidemiology of Lung Cancer: geographical distribution and secular trends. J Thorac Oncol. 2008;3:819-831. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 499] [Cited by in RCA: 566] [Article Influence: 33.3] [Reference Citation Analysis (0)] |
| 67. | Liu L, Ni J, He X. Upregulation of the Long Noncoding RNA SNHG3 Promotes Lung Adenocarcinoma Proliferation. Dis Markers. 2018;2018:5736716. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 36] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 68. | Kang B, Qiu C, Zhang Y. The Effect of lncRNA SNHG3 Overexpression on Lung Adenocarcinoma by Regulating the Expression of miR-890. J Healthc Eng. 2021;2021:1643788. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 7] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 69. | Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87-108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18694] [Cited by in RCA: 21368] [Article Influence: 2136.8] [Reference Citation Analysis (3)] |
| 70. | Torre LA, Trabert B, DeSantis CE, Miller KD, Samimi G, Runowicz CD, Gaudet MM, Jemal A, Siegel RL. Ovarian cancer statistics, 2018. CA Cancer J Clin. 2018;68:284-296. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2132] [Cited by in RCA: 2366] [Article Influence: 338.0] [Reference Citation Analysis (0)] |
| 71. | Seib CD, Sosa JA. Evolving Understanding of the Epidemiology of Thyroid Cancer. Endocrinol Metab Clin North Am. 2019;48:23-35. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 175] [Cited by in RCA: 307] [Article Influence: 51.2] [Reference Citation Analysis (0)] |
| 72. | Wang X, Lei J, Wei T, Zhu J, Li Z. Clinicopathological characteristics and recurrence risk of papillary thyroid microcarcinoma in the elderly. Cancer Manag Res. 2019;11:2371-2377. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 12] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 73. | Zheng X, Peng C, Gao M, Zhi J, Hou X, Zhao J, Wei X, Chi J, Li D, Qian B. Risk factors for cervical lymph node metastasis in papillary thyroid microcarcinoma: a study of 1,587 patients. Cancer Biol Med. 2019;16:121-130. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 68] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 74. | Peng L, Zhang Y, Xin H. lncRNA SNHG3 facilitates acute myeloid leukemia cell growth via the regulation of miR-758-3p/SRGN axis. J Cell Biochem. 2020;121:1023-1031. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 33] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 75. | Zhang M, Bai M, Wang L, Lu N, Wang J, Yan R, Cui M, Yan H, Zhang L. Targeting SNHG3/miR-186-5p reverses the increased m6A level caused by platinum treatment through regulating METTL3 in esophageal cancer. Cancer Cell Int. 2021;21:114. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 24] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 76. | Sun B, Han Y, Cai H, Huang H, Xuan Y. Long non-coding RNA SNHG3, induced by IL-6/STAT3 transactivation, promotes stem cell-like properties of gastric cancer cells by regulating the miR-3619-5p/ARL2 axis. Cell Oncol (Dordr). 2021;44:179-192. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 22] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 77. | Xuan Y, Wang Y. Long non-coding RNA SNHG3 promotes progression of gastric cancer by regulating neighboring MED18 gene methylation. Cell Death Dis. 2019;10:694. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 74] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 78. | Rao J, Fu J, Meng C, Huang J, Qin X, Zhuang S. LncRNA SNHG3 Promotes Gastric Cancer Cells Proliferation, Migration, and Invasion by Targeting miR-326. J Oncol. 2021;2021:9935410. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 20] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 79. | Wang L, Su K, Wu H, Li J, Song D. LncRNA SNHG3 regulates laryngeal carcinoma proliferation and migration by modulating the miR-384/WEE1 axis. Life Sci. 2019;232:116597. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 37] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 80. | Kang R, Yao DF, Xu GZ, Zhou YH. The knockdown of SNHG3 inhibits the progression of laryngeal squamous cell carcinoma by miR-340-5p/YAP1 axis and Wnt/β-catenin pathway. Neoplasma. 2020;67:1094-1105. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 17] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 81. | Lu N, Yin Y, Yao Y, Zhang P. SNHG3/miR-2682-5p/HOXB8 promotes cell proliferation and migration in oral squamous cell carcinoma. Oral Dis. 2021;27:1161-1170. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 16] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 82. | Liu Z, Tao H. Small nucleolar RNA host gene 3 facilitates cell proliferation and migration in oral squamous cell carcinoma via targeting nuclear transcription factor Y subunit gamma. J Cell Biochem. 2020;121:2150-2158. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 18] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 83. | Zheng S, Jiang F, Ge D, Tang J, Chen H, Yang J, Yao Y, Yan J, Qiu J, Yin Z, Ni Y, Zhao L, Chen X, Li H, Yang L. LncRNA SNHG3/miRNA-151a-3p/RAB22A axis regulates invasion and migration of osteosarcoma. Biomed Pharmacother. 2019;112:108695. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 110] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 84. | Chen J, Wu Z, Zhang Y. LncRNA SNHG3 promotes cell growth by sponging miR-196a-5p and indicates the poor survival in osteosarcoma. Int J Immunopathol Pharmacol. 2019;33:2058738418820743. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 39] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 85. | Li T, Xing Y, Yang F, Sun Y, Zhang S, Wang Q, Zhang W. LncRNA SNHG3 sponges miR-577 to up-regulate SMURF1 expression in prostate cancer. Cancer Med. 2020;9:3852-3862. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 40] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 86. | Yu L, Ren Y. Long Noncoding RNA Small Nucleolar RNA Host Gene 3 Mediates Prostate Cancer Migration, Invasion, and Epithelial-Mesenchymal Transition by Sponging miR-487a-3p to Regulate TRIM25. Cancer Biother Radiopharm. 2021;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 87. | Dai G, Huang C, Yang J, Jin L, Fu K, Yuan F, Zhu J, Xue B. LncRNA SNHG3 promotes bladder cancer proliferation and metastasis through miR-515-5p/GINS2 axis. J Cell Mol Med. 2020;24:9231-9243. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 55] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 88. | Zhang H, Lu B. The Roles of ceRNAs-Mediated Autophagy in Cancer Chemoresistance and Metastasis. Cancers (Basel). 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 34] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 89. | Sun ZP, Tan ZG, Peng C, Yi WM. LncRNA SNHG3 Facilitates the Malignant Phenotype of Cholangiocarcinoma Cells via the miR-3173-5p/ERG Axis. J Gastrointest Surg. 2021;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 15] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 90. | Li Z, Chen Y, An T, Liu P, Zhu J, Yang H, Zhang W, Dong T, Jiang J, Zhang Y, Jiang M, Yang X. Nuciferine inhibits the progression of glioblastoma by suppressing the SOX2-AKT/STAT3-Slug signaling pathway. J Exp Clin Cancer Res. 2019;38:139. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 52] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 91. | Scanlon CS, Van Tubergen EA, Inglehart RC, D'Silva NJ. Biomarkers of epithelial-mesenchymal transition in squamous cell carcinoma. J Dent Res. 2013;92:114-121. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 197] [Cited by in RCA: 244] [Article Influence: 18.8] [Reference Citation Analysis (0)] |
| 92. | Heinrich PC, Behrmann I, Müller-Newen G, Schaper F, Graeve L. Interleukin-6-type cytokine signalling through the gp130/Jak/STAT pathway. Biochem J. 1998;334 ( Pt 2):297-314. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1532] [Cited by in RCA: 1649] [Article Influence: 61.1] [Reference Citation Analysis (0)] |
| 93. | Miscia S, Marchisio M, Grilli A, Di Valerio V, Centurione L, Sabatino G, Garaci F, Zauli G, Bonvini E, Di Baldassarre A. Tumor necrosis factor alpha (TNF-alpha) activates Jak1/Stat3-Stat5B signaling through TNFR-1 in human B cells. Cell Growth Differ. 2002;13:13-18. [PubMed] |
| 94. | Park OK, Schaefer TS, Nathans D. In vitro activation of Stat3 by epidermal growth factor receptor kinase. Proc Natl Acad Sci U S A. 1996;93:13704-13708. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 147] [Cited by in RCA: 159] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 95. | Yang L, Lin S, Xu L, Lin J, Zhao C, Huang X. Novel activators and small-molecule inhibitors of STAT3 in cancer. Cytokine Growth Factor Rev. 2019;49:10-22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 97] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 96. | Miklossy G, Hilliard TS, Turkson J. Therapeutic modulators of STAT signalling for human diseases. Nat Rev Drug Discov. 2013;12:611-629. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 286] [Cited by in RCA: 349] [Article Influence: 29.1] [Reference Citation Analysis (0)] |
| 97. | Soda M, Willert K, Kaushansky K, Geddis AE. Inhibition of GSK-3beta promotes survival and proliferation of megakaryocytic cells through a beta-catenin-independent pathway. Cell Signal. 2008;20:2317-2323. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 19] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 98. | Hu Q, Egranov SD, Lin C, Yang L. Long noncoding RNA loss in immune suppression in cancer. Pharmacol Ther. 2020;213:107591. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 43] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 99. | Sun H, Huang Z, Sheng W, Xu MD. Emerging roles of long non-coding RNAs in tumor metabolism. J Hematol Oncol. 2018;11:106. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 69] [Cited by in RCA: 70] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 100. | Zhang L, Xu X, Su X. Noncoding RNAs in cancer immunity: functions, regulatory mechanisms, and clinical application. Mol Cancer. 2020;19:48. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 73] [Cited by in RCA: 72] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 101. | Liu K, Gao L, Ma X, Huang JJ, Chen J, Zeng L, Ashby CR Jr, Zou C, Chen ZS. Long non-coding RNAs regulate drug resistance in cancer. Mol Cancer. 2020;19:54. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 69] [Cited by in RCA: 140] [Article Influence: 28.0] [Reference Citation Analysis (0)] |
| 102. | Zhang X, Xie K, Zhou H, Wu Y, Li C, Liu Y, Liu Z, Xu Q, Liu S, Xiao D, Tao Y. Role of non-coding RNAs and RNA modifiers in cancer therapy resistance. Mol Cancer. 2020;19:47. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 139] [Cited by in RCA: 180] [Article Influence: 36.0] [Reference Citation Analysis (0)] |