Published online Jan 7, 2022. doi: 10.3748/wjg.v28.i1.108
Peer-review started: August 13, 2021
First decision: October 2, 2021
Revised: October 12, 2021
Accepted: December 25, 2021
Article in press: December 25, 2021
Published online: January 7, 2022
Processing time: 140 Days and 2.2 Hours
Colorectal cancer (CRC) is the third most common malignancy worldwide, with approximately 50% of patients developing colorectal cancer liver metastasis (CRLM) during the follow-up period. Management of CRLM is best achieved via a multidisciplinary approach and the diagnostic and therapeutic decision-making process is complex. In order to optimize patients’ survival and quality of life, there are several unsolved challenges which must be overcome. These primarily include a timely diagnosis and the identification of reliable prognostic factors. Furthermore, to allow optimal treatment options, a precision-medicine, personalized approach is required. The widespread digitalization of healthcare generates a vast amount of data and together with accessible high-performance computing, artificial intelligence (AI) technologies can be applied. By increasing diagnostic accuracy, reducing timings and costs, the application of AI could help mitigate the current shortcomings in CRLM management. In this review we explore the available evidence of the possible role of AI in all phases of the CRLM natural history. Radiomics analysis and convolutional neural networks (CNN) which combine computed tomography (CT) images with clinical data have been de
Core tip: The digitalization of healthcare generating huge amount of data set the ground for the progressive ubiquitous application of artificial intelligence (AI) technologies in healthcare. AI analyses can assist clinicians in all phases of colorectal liver metastases natural history: From predicting their occurrence, to increasing diagnostic accuracy or estimating recurrence risk after treatment and patient outcome. The implementation of AI resources supports the contemporary paradigm shift that sees healthcare focus moving from a generalized, disease-oriented to an individual, patient-centered, pre
- Citation: Rompianesi G, Pegoraro F, Ceresa CD, Montalti R, Troisi RI. Artificial intelligence in the diagnosis and management of colorectal cancer liver metastases. World J Gastroenterol 2022; 28(1): 108-122
- URL: https://www.wjgnet.com/1007-9327/full/v28/i1/108.htm
- DOI: https://dx.doi.org/10.3748/wjg.v28.i1.108
Colorectal cancer (CRC) is the most common gastrointestinal cancer, the third most frequently diagnosed malignancy (10.0%) overall, and the second highest cause of cancer-related deaths (9.4%), with incidences varying significantly worldwide[1,2]. CRC development is predominantly sporadic, with patient age, environmental and genetic factors associated with a significantly increased risk[3,4]. Over 20% of newly diagnosed CRC patients have distant metastases at presentation[5], with estimated 5-year survival dropping from 80%-90% in patients with local disease to a dismal 10%-15% in those with metastatic spread[6]. The liver is the preferential metastatic site, due to its anatomical proximity and the portal systemic circulation. This results in 25%-50% of CRC patients developing liver metastasis during the course of the disease[7,8]. In cases of synchronous resectable colorectal cancer liver metastasis (CRLM), the treat
The aim of this review is to summarize and analyze the available evidence on the application of AI technologies in the diagnosis and management of patients affected by CRLM.
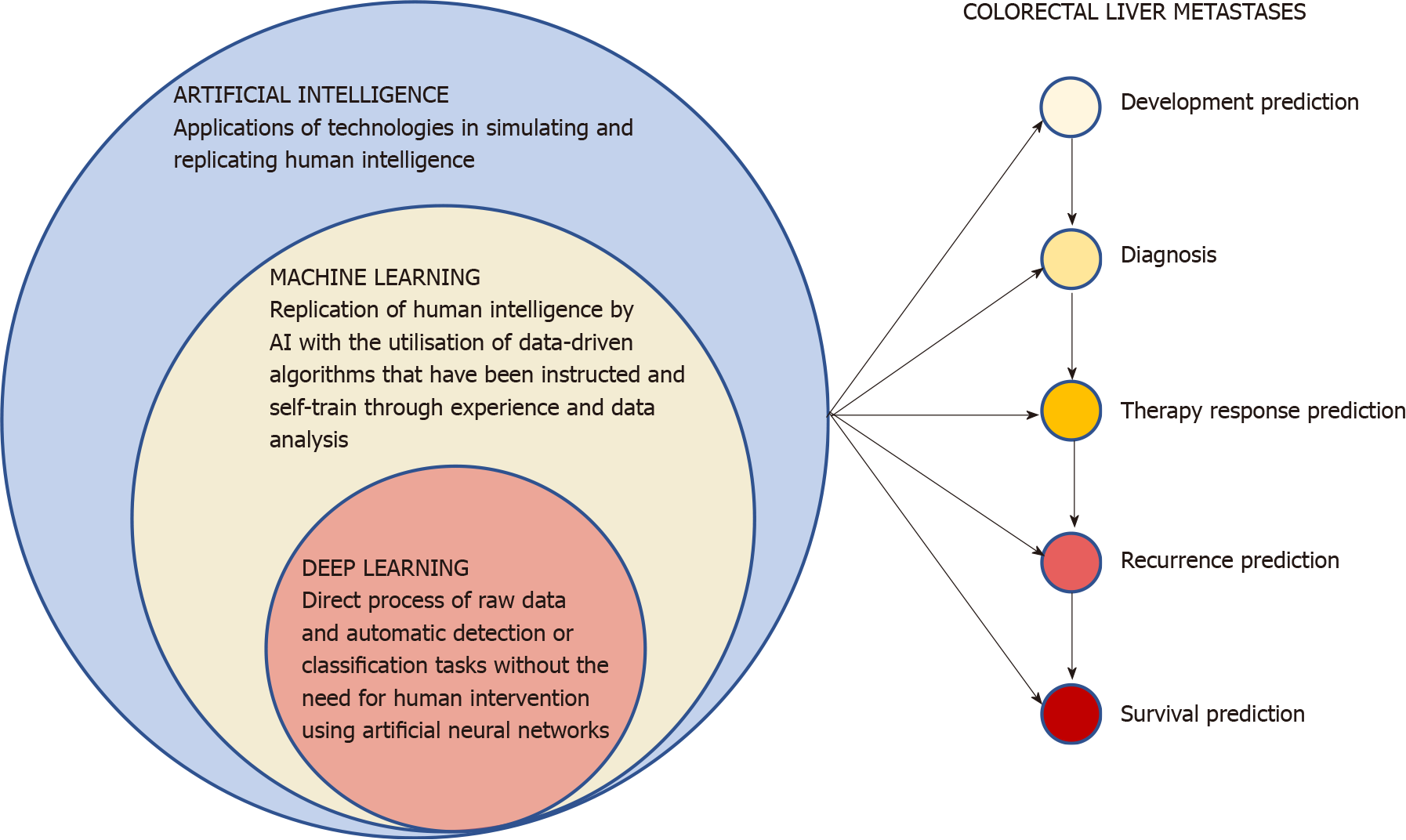
The term AI encompasses all the possible applications of technologies in simulating and replicating human intelligence[13]. These endless applications range from everyday life to finance and economics[14] or various medical fields, thanks to the advances in computational power and the collection and storage of large amounts of data in healthcare. After being adequately programmed and trained, AI has the potential to outperform clinicians in some tasks in terms of accuracy, speed of execution and reduced biases[15]. AI has therefore progressively demonstrated its potential across all human lifespan; from the optimization of embryo selection during in vitro fertilization[16] to the prediction of all-cause mortality[17]. The revolutionary potential of these technologies in healthcare has generated great interest in researchers, professionals and industries, with currently over 450 AI-based medical devices app
The replication of human intelligence by AI with the utilization of data-driven algorithms that have been instructed and self-train through experience and data analysis is generally defined as machine learning (ML)[13]. After been programmed, ML can find recurrent patterns in large amount of appropriately engineered data and progressively learn and independently improve performance accuracy without human intervention. The ML algorithms are generally classified in supervised learning (the most frequent one, which utilizes classified data), unsupervised learning (where algorithms can independently identify patterns in data without previous classification), semi-supervised learning (can use a combination of both labelled and unlabeled data) and reinforcement learning (uses estimated errors as proportional rewards or penalties to teach algorithms). Deep learning (DL) is a class of ML tech
A significant proportion of patients affected by CRC will develop CRLM during the follow-up period[21], but only about a quarter of them will be eligible for surgical resection and therefore potential cure[22]. Being able to identify the subgroup of patients at higher risk of CRLM development could allow the adoption of individualized and more intense screening protocols and adjuvant therapies.
The Radiomics Intelligent Analysis Toolkit-based analysis platform built by Li et al[23] allowed the construction of individualized nomograms able to combine ma
Prompt diagnosis of CRLM at an early stage gives patients the best chances of effective treatment and a superior outcome. One of the key steps in the diagnostic process is tumor segmentation, with nodule volume being a better predictor than diameter[26]. This process is usually done manually but requires a significant expertise, is operator-dependent and time-consuming. In this setting, semiautomatic tumor segmentation methods based on texture analysis have been developed[26] in order to take full advantage of AI’s unique potential to increase sensitivity and specificity of metastatic tumor detection[27].
Starting with a manual tumor/nontumor class prediction voxel classification, a defor
A challenging scenario that can occur in 16%-26% of patients with CRC is when the staging CT scan shows small hypoattenuating hepatic nodules defined as too small to characterize. Further imaging such as magnetic resonance (MR), repeat CT after a time interval, or performing a biopsy can delay treatment, increase costs, remain incon
Despite CT imaging being the most widely used modality in detecting metastatic liver tumors, it can still miss up to 25% of CRLM[35] and MR has progressively gained an established role thanks to the high sensitivity and specificity and absence of ionizing radiations[36,37]. AI utilizing CNN for liver segmentation and CRLM detection could assist radiologists in this complex task and potentially reduce the manual liver lesion detection failure rate of 5%-13%[38]. The CRLM detection method developed by Jansen et al[38] is based on a fully CNN with an automatic liver segmentation and the analysis of both dynamic contrast-enhanced and diffusion-weighted MR images in 121 patients. It resulted in an impressive a high sensitivity of 99.8% and a low number of false positives.
Interestingly, a ML model has been used by Steenhuis et al[39] to analyze data from a retrospective cohort of 62 patients following curative CRC resection to detect CRLM development or local recurrence. The volatile organic compounds (VOCs) from patients’ exhaled air are gaseous products of metabolism known to be altered by pathological processes, such as abnormal cell growth, necrosis or intestinal micro
The applications of AI and ML in diagnosing CRLM have been extended to histopathological examination in order to rapidly and accurately identify CRLM tissue. A probe electrospray ionization-mass spectrometry and ML model was able to distinguish CRLM (103 samples) from noncancer liver parenchyma (80 control samples) with an accuracy rate of 99.5% and a AUROC of 0.9999[41]. CRLM patients are a heterogeneous group with considerable variations, including histopathological growth patterns (HGPs) and corresponding microvasculature[42]. The two predominant types of HGPs are the desmoplastic and replacement, with the pushing and mixed types being far less common. Once accurately determined by analyzing the interface between the tumor cells and the nearby normal liver, HGPs can represent a useful prognostic and predictive biomarker for response to therapy and overall survival[43-46]. The MR-based radiomics model developed by Han et al[47] aims at preoperatively identifying HGP of CRLM with an AUC of 0.906 in the internal validation cohort when the analysis is performed on the tumor-liver interface zone.
Surgical resection offers patients presenting with synchronous or metachronous CRLM the only potential for cure and a superior long-term survival[48] but unfortunately only a fraction of newly diagnosed patients are suitable for surgery. Liver-directed ablative therapies have progressively gained a role in treating nonsurgical candidates with acceptable safety and efficacy profiles[49]. In spite of this, recurrence after CRLM treatment represents a major problem, with an overall risk of local or distant tumor development after surgical resection or ablation as high as 70%-80%, with early recurrences being associated with a poorer prognosis[50,51]. Chemotherapy is of paramount importance in determining outcome of patients with either resectable or unresectable CRLM[8] and can convert up to one third of initially unresectable patients to receive potentially curative treatment[52].
A reliable assessment of response to chemotherapy is of paramount importance for the personalized treatment decision-making process to determine eligibility for surgery, or the need for second-line treatments[53]. Discriminating responsive from unresponsive nodules or new lesions on the CT scan often represents a challenging task for radi
In order to predict early local tumor progression after ablation treatment of up to five nodules per patient with a maximum diameter of 30 mm, Taghavi et al[59] developed a ML-based radiomics analysis of the pretreatment CT scan combined with patients’ clinical features that showed a concordance index in the validation cohort of 0.79 (95%CI: 0.78-0.80).
The systematic comparative analysis of quantitative imaging biomarkers based on the geometric and radiomics analysis of the liver tumor burden by Mühlberg et al[60], performed on a retrospective cohort of 103 patients with CRLM with automated segmentation of baseline contrast-enhanced CT images, showed that the tumor burden score (TBS) had the best discriminative performance for 1-year survival (AUC: 0.70; 95%CI: 0.56-0.90). The TBS[61] is calculated combining tumor number and maximum diameter through the Pythagorean theorem [TBS2 = (maximum tumor diameter)2 + (number of liver lesions)2]. An ML method has been used by Hao et al[62] to analyze whole-genome methylation data to predict cancer versus normal tissue of four common tumors (including 29 of 30 CRLMs) with > 95% accuracy and patient prognosis and survival through DNA methylation analysis.
Anti-epidermal growth factor receptor (EGFR) therapies are an effective option for RAS wild-type mutational status CRLM, but there is a need for reliable biomarkers that can estimate the balance between risks and clinical benefits of such therapies in individual patients[63]. Dercle et al[64] developed an AI model that through ML could create a signature that evaluated a change in tumor phenotype on interval CT scan images (baseline to 8 wk). The resultant model was able to successfully predict both sensitivity to anti-EGFR therapy (0.80; 95%CI: 0.69-0.94) and overall survival (P < 0.05).
The ANN model constructed by Spelt et al[65] retrospectively analyzed a single-center cohort of 241 patients who underwent liver resection for CRLM. Six of the 28 potential risk variables (age, preoperative chemotherapy, size of largest metastasis, hemorrhagic complications, preoperative CEA level and number of metastases) were selected by the ANN model to predict survival more accurately than the Cox regression model, with C-index of 0.72 versus 0.66. Paredes et al[66] in 2020 published the results of their ML recurrence-free prediction model for patients with CRLM undergoing curative-intent resection using clinical, pathological and morphological tumor characteristics with genetic Kirsten rat sarcoma 2 viral oncogene homolog information. The model, built on the analysis of 1406 multi-institutional patients undergoing liver resection, showed a discriminative ability to predict the recurrence risk at 1, 3 and 5 years (AUROC of 0.693, 0.669 and 0.669, respectively) more accurate than the ones of Fong[67] and Vauthey[68] scores.
In spite of AI’s clear potential there remain several unresolved issues and limitations. These include the potential for artefacts in radiomics analyses to affect the results, the ethical and legal considerations, the definition of minimal accuracy rates and safe
| Author | Study design | AI model type | Data source | Total sample size/training cohort/validation cohort | AUC training/AUC validation | Sensitivity/specificity | PPV/NPV | Accuracy |
| CRLM development | ||||||||
| Li et al[23] (2020) | Retrospective; Single center | Radiomics/ML | CT images ± clinical data | 100/NA/80 | 0.90/0.906 | 81%/84% | 85%/79% | NA |
| Taghavi et al[24] (2021) | Retrospective; Multicenter | Radiomics/ML | CT images ± clinical data | 91/70/21 | 0.952-0.683-0.954/0.862-0.713-0.864 | NA/NA | NA/NA | NA |
| Lee et al[25] (2020) | Retrospective; Single center | Radiomics/CNN | CT images ± clinical data | 2019/1413/606 | NA/0.6062-0.7093-0.7474 | NA/NA | NA/NA | NA |
| Diagnosis | ||||||||
| Vorontsov et al[26] (2017) | Retrospective; Single center | Radiomics/CNN | CT images | 40/32/8 | NA/NA | 84%/92% | NA/NA | 88% |
| Vorontsov et al[28] (2019) | Retrospective; Single center | Radiomics/CNN | CT images | 156/115/15 | NA/NA | 59%5/NA | 80%5/NA | NA |
| Ma et al[30] (2020) | Retrospective; Multicenter | CNN | CT images | 909/479/202 (2286) | NA/0.837-0.8446 | 82%6/74%5 | 75%6/81%6 | NA |
| Kim et al[31] (2021) | Retrospective; Single center | DL | CT images | 587/502/85 | NA/0.631 | 81.82%/22.22% | NA/NA | NA |
| Khalili et al[34] (2020) | Retrospective; Single center | CNN | CT images ± liver metastatic status | 199/150/49 | NA/0.84-0.957 | (81.5%-81.5%7)/(76.2%-96.4%7) | NA/NA | 78.3%; 90.6%6 |
| Jansen et al[38] (2019) | Retrospective; Single center | CNN | MRI images | 121/3341/861 | NA/NA | 99.8%/NA | NA/NA | NA |
| Steenhuis et al[39] (2020) | Retrospective; Single center | ML | VOCs | 62/NA/NA | NA/0.86 | 88%/75% | 72%/90% | 81% |
| Miller-Atkins et al[40] (2020) | Prospective; Single center | ML | VOCs | 296/284/NA | NA/NA | 51%/94% | NA/NA | 86% |
| Kiritani et al[41] (2021) | Retrospective; Single center | ML | Histologic markers | 183/NA/40 | NA/0.999 | 100%/99% | NA/NA | 99.5% |
| Han et al[47] (2020) | Retrospective; Single center | Radiomics/ML | MRI images ± clinical data | 107/611/311 | 0.9742-0.6593-0.9714/0.9122-0.6763-0.9094 | 95.2%2-57.1%3-95.2%4/80.0%2-70.0%3-70.0%4 | NA/NA | 90.3%2; 61.3%3; 87.1%4 |
| Chemotherapy response | ||||||||
| Maaref et al[54] (2020) | Retrospective; Single center | DL CNN | CT images | 202/70%/10% | 0.97/0.88 | 98%/54% | NA/NA | 91%8; 78%9 |
| Wei et al[55] (2021) | Retrospective; Single center | Radiomics/DL | CT images ± CEA | 192/144/48 | 0.90310-0.93511/0.82010-0.83011 | 90.9%/73.3% | 88.2%/78.6% | 85.4% |
| Giannini et al[57] (2020) | Retrospective; Multicenter | Radiomics/ML | CT images | 38/28/10 | NA/NA | 92%/86% | 96%/75% | NA |
| Nakanishi et al[58] (2021) | Retrospective; Single center | Radiomics | CT images | 42/941/321 | 0.8512/0.7792 | NA/NA | NA/NA | NA |
| Local ablative therapies efficacy | ||||||||
| Taghavi et al[59] (2021) | Retrospective; Single center | Radiomics/ML | CT images | 90/63/27 | NA/0.782-0.563-0.794 | NA/NA | NA/NA | NA |
| Survival prediction | ||||||||
| Mühlberg et al[60] (2021) | Retrospective; Single center | Radiomics/ML | CT images ± WLTB ± TBS | 103/NA/NA | NA/0.7012–0.7313-0.7614 | NA/NA | NA/NA | NA |
| Hao et al[62] (2017) | Retrospective; Multicenter | ML | DNA methylation | 17921/NA/8841 (7181,6) | NA/NA | NA/NA | NA/NA | 98.4% |
| Dercle et al[64] (2020) | Retrospective; Multicenter | ML | CT images | 667/438/229 | 0.83/0.80 | 80%/78% | NA/NA | NA |
| Spelt et al[65] (2013) | Retrospective; Single center | ANN | Clinical variables | 241/NA/NA | NA/NA | NA/NA | NA/NA | 72% |
| Paredes et al[66] (2020) | Retrospective; Multicenter | ML | Clinical variables | 1406/703/703 | 0.52715-0.52516-0.69317/0.52415-0.50116-0.64217 | NA/NA | NA/NA | NA |
The progressive widespread availability of high-performance computing, together with the accessibility to a large amount of data constantly generated as the result of the increase in the digitalization, set the ground for the ubiquitous implementation of AI technologies in contemporary healthcare. The fields of medical and surgical oncology have welcomed with enthusiasm the advent of augmented medicine with numerous studies investigating its potential, also given the high complexity and diversity of cancer patients. CRC makes no exception and still represents a leading cause of cancer-related death due to its high incidence, rapid progression potential and biological heterogeneity that advocate the need for reliable and individualized diagnostic, prognostic and treatment selection tools. Recent years have seen AI technologies tested by researchers in all phases of the CRLM natural history, aiming at overcoming the current difficulties and limitations faced by the multidisciplinary team responsible of the patients’ care (Figure 1). The possibility of identifying the subgroup of patients at higher risk of CRLM development before the occurrence of the disease from the radiomics baseline CT scan analysis with high accuracy (AUC ≥ 0.75) and in less than 5 min could give such patients the best chances of an early diagnosis, more effective treatment, and therefore, a better outcome thanks to a personalized approach[23-25]. Radiomics has also demonstrated a great potential in assisting the radiologists in diagnosing CRLM from CT and MRI scans also by optimizing the identification of the optimal phases for lesions recognition and characterizing small nodules of uncertain nature[27-31,34,38]. A more efficient diagnostic process would help reduce timings and costs, resulting in a potential benefit for both patients and healthcare systems. AI application in order to rapidly and accurately identify CRLM tissue and its different histopathological growth patterns[41,47] could give a significant contribution towards a rapid oncological individualized approach and treatments. AI technologies have also shown potential as a prognostic and outcome tool, predicting with good accuracy response to chemotherapy[54,55,57,58], early local tumor progression after ablation treatment[59], and patient survival after surgery or chemotherapy[60,64-66].
The possibility of reducing human factors and error, increase accuracy and contain timings and costs while adopting a personalized medicine approach is undoubtedly fascinating and appealing, but despite showing promising results, the role of AI in CRLM patients has not yet been fully elucidated. The implementation of AI resources supports the contemporary paradigm shift that sees healthcare focus moving from a generalized, disease-oriented to an individual, patient-centered, precision medicine approach. The effectiveness of ML models lie on a rigid framework in which a well-defined problem and ground truth along with quantitative objective measures to train and validate the algorithm are needed, making the process efficient but rigid. There is also a balance to be struck between the accuracy and artificial logic and the risk of AI becoming less intelligible and explainable. On the other hand, AI medical technologies could represent a way to enable patients to take ownership of their own care, increa
AI will likely affect the immediate future of medicine and patients’ management, but rather than replacing the human roles, it will probably be aimed to assist and facilitate physicians in their practice, while being supervised to ensure maximum safety. This could be in the context of diagnostic uncertainty or to assist in planning optimal treatment strategies. A possible future development would be to improve diagnosis and management through the AI analysis and integration of clinical information, radiomic and genetic data thanks to the recent developments in gene sequencing and liquid biopsies, that have showed great potential in gastrointestinal tumors including CRLM[69-72]. A personalized holistic approach providing reliable data for the diagnosis, management and outcome estimation of cancer patients would assist clinicians in the prevention as well as selecting the most appropriate individualized treatment that would grant the patient the best outcome as well as helping patients to make fully informed decisions.
In order to continue to pursue the ambitious goal of improving patients’ care through AI healthcare technologies, further larger, prospective, randomized controlled and rigorous studies are needed.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Italy
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Liu Y S-Editor: Wang JJ L-Editor: Kerr C P-Editor: Wang JJ
| 1. | Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75126] [Cited by in RCA: 64104] [Article Influence: 16026.0] [Reference Citation Analysis (174)] |
| 2. | Global Burden of Disease Cancer Collaboration, Fitzmaurice C, Abate D, Abbasi N, Abbastabar H, Abd-Allah F, Abdel-Rahman O, Abdelalim A, Abdoli A, Abdollahpour I, Abdulle ASM, Abebe ND, Abraha HN, Abu-Raddad LJ, Abualhasan A, Adedeji IA, Advani SM, Afarideh M, Afshari M, Aghaali M, Agius D, Agrawal S, Ahmadi A, Ahmadian E, Ahmadpour E, Ahmed MB, Akbari ME, Akinyemiju T, Al-Aly Z, AlAbdulKader AM, Alahdab F, Alam T, Alamene GM, Alemnew BTT, Alene KA, Alinia C, Alipour V, Aljunid SM, Bakeshei FA, Almadi MAH, Almasi-Hashiani A, Alsharif U, Alsowaidi S, Alvis-Guzman N, Amini E, Amini S, Amoako YA, Anbari Z, Anber NH, Andrei CL, Anjomshoa M, Ansari F, Ansariadi A, Appiah SCY, Arab-Zozani M, Arabloo J, Arefi Z, Aremu O, Areri HA, Artaman A, Asayesh H, Asfaw ET, Ashagre AF, Assadi R, Ataeinia B, Atalay HT, Ataro Z, Atique S, Ausloos M, Avila-Burgos L, Avokpaho EFGA, Awasthi A, Awoke N, Ayala Quintanilla BP, Ayanore MA, Ayele HT, Babaee E, Bacha U, Badawi A, Bagherzadeh M, Bagli E, Balakrishnan S, Balouchi A, Bärnighausen TW, Battista RJ, Behzadifar M, Bekele BB, Belay YB, Belayneh YM, Berfield KKS, Berhane A, Bernabe E, Beuran M, Bhakta N, Bhattacharyya K, Biadgo B, Bijani A, Bin Sayeed MS, Birungi C, Bisignano C, Bitew H, Bjørge T, Bleyer A, Bogale KA, Bojia HA, Borzì AM, Bosetti C, Bou-Orm IR, Brenner H, Brewer JD, Briko AN, Briko NI, Bustamante-Teixeira MT, Butt ZA, Carreras G, Carrero JJ, Carvalho F, Castro C, Castro F, Catalá-López F, Cerin E, Chaiah Y, Chanie WF, Chattu VK, Chaturvedi P, Chauhan NS, Chehrazi M, Chiang PP, Chichiabellu TY, Chido-Amajuoyi OG, Chimed-Ochir O, Choi JJ, Christopher DJ, Chu DT, Constantin MM, Costa VM, Crocetti E, Crowe CS, Curado MP, Dahlawi SMA, Damiani G, Darwish AH, Daryani A, das Neves J, Demeke FM, Demis AB, Demissie BW, Demoz GT, Denova-Gutiérrez E, Derakhshani A, Deribe KS, Desai R, Desalegn BB, Desta M, Dey S, Dharmaratne SD, Dhimal M, Diaz D, Dinberu MTT, Djalalinia S, Doku DT, Drake TM, Dubey M, Dubljanin E, Duken EE, Ebrahimi H, Effiong A, Eftekhari A, El Sayed I, Zaki MES, El-Jaafary SI, El-Khatib Z, Elemineh DA, Elkout H, Ellenbogen RG, Elsharkawy A, Emamian MH, Endalew DA, Endries AY, Eshrati B, Fadhil I, Fallah Omrani V, Faramarzi M, Farhangi MA, Farioli A, Farzadfar F, Fentahun N, Fernandes E, Feyissa GT, Filip I, Fischer F, Fisher JL, Force LM, Foroutan M, Freitas M, Fukumoto T, Futran ND, Gallus S, Gankpe FG, Gayesa RT, Gebrehiwot TT, Gebremeskel GG, Gedefaw GA, Gelaw BK, Geta B, Getachew S, Gezae KE, Ghafourifard M, Ghajar A, Ghashghaee A, Gholamian A, Gill PS, Ginindza TTG, Girmay A, Gizaw M, Gomez RS, Gopalani SV, Gorini G, Goulart BNG, Grada A, Ribeiro Guerra M, Guimaraes ALS, Gupta PC, Gupta R, Hadkhale K, Haj-Mirzaian A, Hamadeh RR, Hamidi S, Hanfore LK, Haro JM, Hasankhani M, Hasanzadeh A, Hassen HY, Hay RJ, Hay SI, Henok A, Henry NJ, Herteliu C, Hidru HD, Hoang CL, Hole MK, Hoogar P, Horita N, Hosgood HD, Hosseini M, Hosseinzadeh M, Hostiuc M, Hostiuc S, Househ M, Hussen MM, Ileanu B, Ilic MD, Innos K, Irvani SSN, Iseh KR, Islam SMS, Islami F, Jafari Balalami N, Jafarinia M, Jahangiry L, Jahani MA, Jahanmehr N, Jakovljevic M, James SL, Javanbakht M, Jayaraman S, Jee SH, Jenabi E, Jha RP, Jonas JB, Jonnagaddala J, Joo T, Jungari SB, Jürisson M, Kabir A, Kamangar F, Karch A, Karimi N, Karimian A, Kasaeian A, Kasahun GG, Kassa B, Kassa TD, Kassaw MW, Kaul A, Keiyoro PN, Kelbore AG, Kerbo AA, Khader YS, Khalilarjmandi M, Khan EA, Khan G, Khang YH, Khatab K, Khater A, Khayamzadeh M, Khazaee-Pool M, Khazaei S, Khoja AT, Khosravi MH, Khubchandani J, Kianipour N, Kim D, Kim YJ, Kisa A, Kisa S, Kissimova-Skarbek K, Komaki H, Koyanagi A, Krohn KJ, Bicer BK, Kugbey N, Kumar V, Kuupiel D, La Vecchia C, Lad DP, Lake EA, Lakew AM, Lal DK, Lami FH, Lan Q, Lasrado S, Lauriola P, Lazarus JV, Leigh J, Leshargie CT, Liao Y, Limenih MA, Listl S, Lopez AD, Lopukhov PD, Lunevicius R, Madadin M, Magdeldin S, El Razek HMA, Majeed A, Maleki A, Malekzadeh R, Manafi A, Manafi N, Manamo WA, Mansourian M, Mansournia MA, Mantovani LG, Maroufizadeh S, Martini SMS, Mashamba-Thompson TP, Massenburg BB, Maswabi MT, Mathur MR, McAlinden C, McKee M, Meheretu HAA, Mehrotra R, Mehta V, Meier T, Melaku YA, Meles GG, Meles HG, Melese A, Melku M, Memiah PTN, Mendoza W, Menezes RG, Merat S, Meretoja TJ, Mestrovic T, Miazgowski B, Miazgowski T, Mihretie KMM, Miller TR, Mills EJ, Mir SM, Mirzaei H, Mirzaei HR, Mishra R, Moazen B, Mohammad DK, Mohammad KA, Mohammad Y, Darwesh AM, Mohammadbeigi A, Mohammadi H, Mohammadi M, Mohammadian M, Mohammadian-Hafshejani A, Mohammadoo-Khorasani M, Mohammadpourhodki R, Mohammed AS, Mohammed JA, Mohammed S, Mohebi F, Mokdad AH, Monasta L, Moodley Y, Moosazadeh M, Moossavi M, Moradi G, Moradi-Joo M, Moradi-Lakeh M, Moradpour F, Morawska L, Morgado-da-Costa J, Morisaki N, Morrison SD, Mosapour A, Mousavi SM, Muche AA, Muhammed OSS, Musa J, Nabhan AF, Naderi M, Nagarajan AJ, Nagel G, Nahvijou A, Naik G, Najafi F, Naldi L, Nam HS, Nasiri N, Nazari J, Negoi I, Neupane S, Newcomb PA, Nggada HA, Ngunjiri JW, Nguyen CT, Nikniaz L, Ningrum DNA, Nirayo YL, Nixon MR, Nnaji CA, Nojomi M, Nosratnejad S, Shiadeh MN, Obsa MS, Ofori-Asenso R, Ogbo FA, Oh IH, Olagunju AT, Olagunju TO, Oluwasanu MM, Omonisi AE, Onwujekwe OE, Oommen AM, Oren E, Ortega-Altamirano DDV, Ota E, Otstavnov SS, Owolabi MO, P A M, Padubidri JR, Pakhale S, Pakpour AH, Pana A, Park EK, Parsian H, Pashaei T, Patel S, Patil ST, Pennini A, Pereira DM, Piccinelli C, Pillay JD, Pirestani M, Pishgar F, Postma MJ, Pourjafar H, Pourmalek F, Pourshams A, Prakash S, Prasad N, Qorbani M, Rabiee M, Rabiee N, Radfar A, Rafiei A, Rahim F, Rahimi M, Rahman MA, Rajati F, Rana SM, Raoofi S, Rath GK, Rawaf DL, Rawaf S, Reiner RC, Renzaho AMN, Rezaei N, Rezapour A, Ribeiro AI, Ribeiro D, Ronfani L, Roro EM, Roshandel G, Rostami A, Saad RS, Sabbagh P, Sabour S, Saddik B, Safiri S, Sahebkar A, Salahshoor MR, Salehi F, Salem H, Salem MR, Salimzadeh H, Salomon JA, Samy AM, Sanabria J, Santric Milicevic MM, Sartorius B, Sarveazad A, Sathian B, Satpathy M, Savic M, Sawhney M, Sayyah M, Schneider IJC, Schöttker B, Sekerija M, Sepanlou SG, Sepehrimanesh M, Seyedmousavi S, Shaahmadi F, Shabaninejad H, Shahbaz M, Shaikh MA, Shamshirian A, Shamsizadeh M, Sharafi H, Sharafi Z, Sharif M, Sharifi A, Sharifi H, Sharma R, Sheikh A, Shirkoohi R, Shukla SR, Si S, Siabani S, Silva DAS, Silveira DGA, Singh A, Singh JA, Sisay S, Sitas F, Sobngwi E, Soofi M, Soriano JB, Stathopoulou V, Sufiyan MB, Tabarés-Seisdedos R, Tabuchi T, Takahashi K, Tamtaji OR, Tarawneh MR, Tassew SG, Taymoori P, Tehrani-Banihashemi A, Temsah MH, Temsah O, Tesfay BE, Tesfay FH, Teshale MY, Tessema GA, Thapa S, Tlaye KG, Topor-Madry R, Tovani-Palone MR, Traini E, Tran BX, Tran KB, Tsadik AG, Ullah I, Uthman OA, Vacante M, Vaezi M, Varona Pérez P, Veisani Y, Vidale S, Violante FS, Vlassov V, Vollset SE, Vos T, Vosoughi K, Vu GT, Vujcic IS, Wabinga H, Wachamo TM, Wagnew FS, Waheed Y, Weldegebreal F, Weldesamuel GT, Wijeratne T, Wondafrash DZ, Wonde TE, Wondmieneh AB, Workie HM, Yadav R, Yadegar A, Yadollahpour A, Yaseri M, Yazdi-Feyzabadi V, Yeshaneh A, Yimam MA, Yimer EM, Yisma E, Yonemoto N, Younis MZ, Yousefi B, Yousefifard M, Yu C, Zabeh E, Zadnik V, Moghadam TZ, Zaidi Z, Zamani M, Zandian H, Zangeneh A, Zaki L, Zendehdel K, Zenebe ZM, Zewale TA, Ziapour A, Zodpey S, Murray CJL. Global, Regional, and National Cancer Incidence, Mortality, Years of Life Lost, Years Lived With Disability, and Disability-Adjusted Life-Years for 29 Cancer Groups, 1990 to 2017: A Systematic Analysis for the Global Burden of Disease Study. JAMA Oncol. 2019;5:1749-1768. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1723] [Cited by in RCA: 1742] [Article Influence: 290.3] [Reference Citation Analysis (0)] |
| 3. | Chan AT, Giovannucci EL. Primary prevention of colorectal cancer. Gastroenterology. 2010;138:2029-2043.e10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 475] [Cited by in RCA: 438] [Article Influence: 29.2] [Reference Citation Analysis (0)] |
| 4. | Kastrinos F, Syngal S. Inherited colorectal cancer syndromes. Cancer J. 2011;17:405-415. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 80] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 5. | van der Pool AE, Damhuis RA, Ijzermans JN, de Wilt JH, Eggermont AM, Kranse R, Verhoef C. Trends in incidence, treatment and survival of patients with stage IV colorectal cancer: a population-based series. Colorectal Dis. 2012;14:56-61. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 148] [Cited by in RCA: 176] [Article Influence: 13.5] [Reference Citation Analysis (2)] |
| 6. | Miller KD, Nogueira L, Mariotto AB, Rowland JH, Yabroff KR, Alfano CM, Jemal A, Kramer JL, Siegel RL. Cancer treatment and survivorship statistics, 2019. CA Cancer J Clin. 2019;69:363-385. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2417] [Cited by in RCA: 3067] [Article Influence: 511.2] [Reference Citation Analysis (0)] |
| 7. | Sheth KR, Clary BM. Management of hepatic metastases from colorectal cancer. Clin Colon Rectal Surg. 2005;18:215-223. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 69] [Article Influence: 3.5] [Reference Citation Analysis (1)] |
| 8. | Chow FC, Chok KS. Colorectal liver metastases: An update on multidisciplinary approach. World J Hepatol. 2019;11:150-172. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 173] [Cited by in RCA: 150] [Article Influence: 25.0] [Reference Citation Analysis (3)] |
| 9. | Lillemoe HA, Vauthey JN. Surgical approach to synchronous colorectal liver metastases: staged, combined, or reverse strategy. Hepatobiliary Surg Nutr. 2020;9:25-34. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 27] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 10. | Arshad U, Sutton PA, Ashford MB, Treacher KE, Liptrott NJ, Rannard SP, Goldring CE, Owen A. Critical considerations for targeting colorectal liver metastases with nanotechnology. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2020;12:e1588. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 11] [Article Influence: 2.2] [Reference Citation Analysis (1)] |
| 11. | Ismaili N. Treatment of colorectal liver metastases. World J Surg Oncol. 2011;9:154. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 59] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 12. | Weledji EP. Centralization of Liver Cancer Surgery and Impact on Multidisciplinary Teams Working on Stage IV Colorectal Cancer. Oncol Rev. 2017;11:331. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 15] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 13. | Topol EJ. High-performance medicine: the convergence of human and artificial intelligence. Nat Med. 2019;25:44-56. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2376] [Cited by in RCA: 2784] [Article Influence: 464.0] [Reference Citation Analysis (0)] |
| 14. | Moloi T, Marwala, T. Artificial Intelligence in Economics and Finance Theories. In: Introduction to Artificial Intelligence in Economics and Finance Theories. 2020: 1-12. [RCA] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 15. | Matheny ME, Whicher D, Thadaney Israni S. Artificial Intelligence in Health Care: A Report From the National Academy of Medicine. JAMA. 2020;323:509-510. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 175] [Cited by in RCA: 239] [Article Influence: 47.8] [Reference Citation Analysis (0)] |
| 16. | Xi Q, Yang Q, Wang M, Huang B, Zhang B, Li Z, Liu S, Yang L, Zhu L, Jin L. Individualized embryo selection strategy developed by stacking machine learning model for better in vitro fertilization outcomes: an application study. Reprod Biol Endocrinol. 2021;19:53. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 8] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 17. | Ulloa Cerna AE, Jing L, Good CW, vanMaanen DP, Raghunath S, Suever JD, Nevius CD, Wehner GJ, Hartzel DN, Leader JB, Alsaid A, Patel AA, Kirchner HL, Pfeifer JM, Carry BJ, Pattichis MS, Haggerty CM, Fornwalt BK. Deep-learning-assisted analysis of echocardiographic videos improves predictions of all-cause mortality. Nat Biomed Eng. 2021;5:546-554. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 29] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 18. | Muehlematter UJ, Daniore P, Vokinger KN. Approval of artificial intelligence and machine learning-based medical devices in the USA and Europe (2015-20): a comparative analysis. Lancet Digit Health. 2021;3:e195-e203. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 278] [Article Influence: 69.5] [Reference Citation Analysis (0)] |
| 19. | Benjamens S, Dhunnoo P, Meskó B. The state of artificial intelligence-based FDA-approved medical devices and algorithms: an online database. NPJ Digit Med. 2020;3:118. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 418] [Cited by in RCA: 487] [Article Influence: 97.4] [Reference Citation Analysis (0)] |
| 20. | Gillies RJ, Kinahan PE, Hricak H. Radiomics: Images Are More than Pictures, They Are Data. Radiology. 2016;278:563-577. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4541] [Cited by in RCA: 5501] [Article Influence: 611.2] [Reference Citation Analysis (3)] |
| 21. | Hackl C, Neumann P, Gerken M, Loss M, Klinkhammer-Schalke M, Schlitt HJ. Treatment of colorectal liver metastases in Germany: a ten-year population-based analysis of 5772 cases of primary colorectal adenocarcinoma. BMC Cancer. 2014;14:810. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 141] [Cited by in RCA: 245] [Article Influence: 22.3] [Reference Citation Analysis (0)] |
| 22. | Engstrand J, Nilsson H, Strömberg C, Jonas E, Freedman J. Colorectal cancer liver metastases — a population-based study on incidence, management and survival. BMC Cancer. 2018;18:78. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 299] [Cited by in RCA: 590] [Article Influence: 84.3] [Reference Citation Analysis (1)] |
| 23. | Li M, Li X, Guo Y, Miao Z, Liu X, Guo S, Zhang H. Development and assessment of an individualized nomogram to predict colorectal cancer liver metastases. Quant Imaging Med Surg. 2020;10:397-414. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 41] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 24. | Taghavi M, Trebeschi S, Simões R, Meek DB, Beckers RCJ, Lambregts DMJ, Verhoef C, Houwers JB, van der Heide UA, Beets-Tan RGH, Maas M. Machine learning-based analysis of CT radiomics model for prediction of colorectal metachronous liver metastases. Abdom Radiol (NY). 2021;46:249-256. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 59] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 25. | Lee S, Choe EK, Kim SY, Kim HS, Park KJ, Kim D. Liver imaging features by convolutional neural network to predict the metachronous liver metastasis in stage I-III colorectal cancer patients based on preoperative abdominal CT scan. BMC Bioinformatics. 2020;21:382. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 19] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 26. | Vorontsov E, Tang A, Roy D, Pal CJ, Kadoury S. Metastatic liver tumour segmentation with a neural network-guided 3D deformable model. Med Biol Eng Comput. 2017;55:127-139. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 14] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 27. | Zheng Q, Yang L, Zeng B, Li J, Guo K, Liang Y, Liao G. Artificial intelligence performance in detecting tumor metastasis from medical radiology imaging: A systematic review and meta-analysis. EclinicalMedicine. 2021;31:100669. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 48] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 28. | Vorontsov E, Cerny M, Régnier P, Di Jorio L, Pal CJ, Lapointe R, Vandenbroucke-Menu F, Turcotte S, Kadoury S, Tang A. Deep Learning for Automated Segmentation of Liver Lesions at CT in Patients with Colorectal Cancer Liver Metastases. Radiol Artif Intell. 2019;1:180014. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 64] [Article Influence: 10.7] [Reference Citation Analysis (1)] |
| 29. | Schima W, Kulinna C, Langenberger H, Ba-Ssalamah A. Liver metastases of colorectal cancer: US, CT or MR? Cancer Imaging. 2005;5 Spec No A:S149-S156. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 62] [Cited by in RCA: 65] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 30. | Ma J, Dercle L, Lichtenstein P, Wang D, Chen A, Zhu J, Piessevaux H, Zhao J, Schwartz LH, Lu L, Zhao B. Automated Identification of Optimal Portal Venous Phase Timing with Convolutional Neural Networks. Acad Radiol. 2020;27:e10-e18. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 31. | Kim K, Kim S, Han K, Bae H, Shin J, Lim JS. Diagnostic Performance of Deep Learning-Based Lesion Detection Algorithm in CT for Detecting Hepatic Metastasis from Colorectal Cancer. Korean J Radiol. 2021;22:912-921. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 25] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 32. | Jang HJ, Lim HK, Lee WJ, Lee SJ, Yun JY, Choi D. Small hypoattenuating lesions in the liver on single-phase helical CT in preoperative patients with gastric and colorectal cancer: prevalence, significance, and differentiating features. J Comput Assist Tomogr. 2002;26:718-724. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 36] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 33. | Lim GH, Koh DC, Cheong WK, Wong KS, Tsang CB. Natural history of small, “indeterminate” hepatic lesions in patients with colorectal cancer. Dis Colon Rectum. 2009;52:1487-1491. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 23] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 34. | Khalili K, Lawlor RL, Pourafkari M, Lu H, Tyrrell P, Kim TK, Jang HJ, Johnson SA, Martel AL. Convolutional neural networks versus radiologists in characterization of small hypoattenuating hepatic nodules on CT: a critical diagnostic challenge in staging of colorectal carcinoma. Sci Rep. 2020;10:15248. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 35. | Xu LH, Cai SJ, Cai GX, Peng WJ. Imaging diagnosis of colorectal liver metastases. World J Gastroenterol. 2011;17:4654-4659. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 27] [Cited by in RCA: 27] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 36. | Böttcher J, Hansch A, Pfeil A, Schmidt P, Malich A, Schneeweiss A, Maurer MH, Streitparth F, Teichgräber UK, Renz DM. Detection and classification of different liver lesions: comparison of Gd-EOB-DTPA-enhanced MRI versus multiphasic spiral CT in a clinical single centre investigation. Eur J Radiol. 2013;82:1860-1869. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 39] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 37. | Mao Y, Chen B, Wang H, Zhang Y, Yi X, Liao W, Zhao L. Diagnostic performance of magnetic resonance imaging for colorectal liver metastasis: A systematic review and meta-analysis. Sci Rep. 2020;10:1969. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 34] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 38. | Jansen MJA, Kuijf HJ, Niekel M, Veldhuis WB, Wessels FJ, Viergever MA, Pluim JPW. Liver segmentation and metastases detection in MR images using convolutional neural networks. J Med Imaging (Bellingham). 2019;6:044003. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 39. | Steenhuis EGM, Schoenaker IJH, de Groot JWB, Fiebrich HB, de Graaf JC, Brohet RM, van Dijk JD, van Westreenen HL, Siersema PD, de Vos Tot Nederveen Cappel WH. Feasibility of volatile organic compound in breath analysis in the follow-up of colorectal cancer: A pilot study. Eur J Surg Oncol. 2020;46:2068-2073. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 20] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 40. | Miller-Atkins G, Acevedo-Moreno LA, Grove D, Dweik RA, Tonelli AR, Brown JM, Allende DS, Aucejo F, Rotroff DM. Breath Metabolomics Provides an Accurate and Noninvasive Approach for Screening Cirrhosis, Primary, and Secondary Liver Tumors. Hepatol Commun. 2020;4:1041-1055. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 35] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 41. | Kiritani S, Yoshimura K, Arita J, Kokudo T, Hakoda H, Tanimoto M, Ishizawa T, Akamatsu N, Kaneko J, Takeda S, Hasegawa K. A new rapid diagnostic system with ambient mass spectrometry and machine learning for colorectal liver metastasis. BMC Cancer. 2021;21:262. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 10] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 42. | Vermeulen PB, Colpaert C, Salgado R, Royers R, Hellemans H, Van Den Heuvel E, Goovaerts G, Dirix LY, Van Marck E. Liver metastases from colorectal adenocarcinomas grow in three patterns with different angiogenesis and desmoplasia. J Pathol. 2001;195:336-342. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 210] [Cited by in RCA: 219] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 43. | Nielsen K, Rolff HC, Eefsen RL, Vainer B. The morphological growth patterns of colorectal liver metastases are prognostic for overall survival. Mod Pathol. 2014;27:1641-1648. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 77] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 44. | van Dam PJ, van der Stok EP, Teuwen LA, Van den Eynden GG, Illemann M, Frentzas S, Majeed AW, Eefsen RL, Coebergh van den Braak RRJ, Lazaris A, Fernandez MC, Galjart B, Laerum OD, Rayes R, Grünhagen DJ, Van de Paer M, Sucaet Y, Mudhar HS, Schvimer M, Nyström H, Kockx M, Bird NC, Vidal-Vanaclocha F, Metrakos P, Simoneau E, Verhoef C, Dirix LY, Van Laere S, Gao ZH, Brodt P, Reynolds AR, Vermeulen PB. International consensus guidelines for scoring the histopathological growth patterns of liver metastasis. Br J Cancer. 2017;117:1427-1441. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 190] [Cited by in RCA: 173] [Article Influence: 21.6] [Reference Citation Analysis (0)] |
| 45. | Stremitzer S, Vermeulen P, Graver S, Kockx M, Dirix L, Yang D, Zhang W, Stift J, Wrba F, Gruenberger T, Lenz HJ, Scherer SJ. Immune phenotype and histopathological growth pattern in patients with colorectal liver metastases. Br J Cancer. 2020;122:1518-1524. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 29] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 46. | Frentzas S, Simoneau E, Bridgeman VL, Vermeulen PB, Foo S, Kostaras E, Nathan M, Wotherspoon A, Gao ZH, Shi Y, Van den Eynden G, Daley F, Peckitt C, Tan X, Salman A, Lazaris A, Gazinska P, Berg TJ, Eltahir Z, Ritsma L, Van Rheenen J, Khashper A, Brown G, Nystrom H, Sund M, Van Laere S, Loyer E, Dirix L, Cunningham D, Metrakos P, Reynolds AR. Vessel co-option mediates resistance to anti-angiogenic therapy in liver metastases. Nat Med. 2016;22:1294-1302. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 362] [Cited by in RCA: 325] [Article Influence: 36.1] [Reference Citation Analysis (0)] |
| 47. | Han Y, Chai F, Wei J, Yue Y, Cheng J, Gu D, Zhang Y, Tong T, Sheng W, Hong N, Ye Y, Wang Y, Tian J. Identification of Predominant Histopathological Growth Patterns of Colorectal Liver Metastasis by Multi-Habitat and Multi-Sequence Based Radiomics Analysis. Front Oncol. 2020;10:1363. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 37] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 48. | Abdalla EK, Vauthey JN, Ellis LM, Ellis V, Pollock R, Broglio KR, Hess K, Curley SA. Recurrence and outcomes following hepatic resection, radiofrequency ablation, and combined resection/ablation for colorectal liver metastases. Ann Surg. 2004;239:818-825; discussion 825-827. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1364] [Cited by in RCA: 1286] [Article Influence: 61.2] [Reference Citation Analysis (0)] |
| 49. | Di Martino M, Rompianesi G, Mora-Guzmán I, Martín-Pérez E, Montalti R, Troisi RI. Systematic review and meta-analysis of local ablative therapies for resectable colorectal liver metastases. Eur J Surg Oncol. 2020;46:772-781. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 52] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 50. | Sorbye H. Recurrence patterns after resection of liver metastases from colorectal cancer. Recent Results Cancer Res. 2014;203:243-252. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 16] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 51. | Bredt LC, Rachid AF. Predictors of recurrence after a first hepatectomy for colorectal cancer liver metastases: a retrospective analysis. World J Surg Oncol. 2014;12:391. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 28] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 52. | Folprecht G, Gruenberger T, Bechstein W, Raab HR, Weitz J, Lordick F, Hartmann JT, Stoehlmacher-Williams J, Lang H, Trarbach T, Liersch T, Ockert D, Jaeger D, Steger U, Suedhoff T, Rentsch A, Köhne CH. Survival of patients with initially unresectable colorectal liver metastases treated with FOLFOX/cetuximab or FOLFIRI/cetuximab in a multidisciplinary concept (CELIM study). Ann Oncol. 2014;25:1018-1025. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 169] [Cited by in RCA: 176] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 53. | Bonanni L, de’Liguori Carino N, Deshpande R, Ammori BJ, Sherlock DJ, Valle JW, Tam E, O’Reilly DA. A comparison of diagnostic imaging modalities for colorectal liver metastases. Eur J Surg Oncol. 2014;40:545-550. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 33] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 54. | Maaref A, Romero FP, Montagnon E, Cerny M, Nguyen B, Vandenbroucke F, Soucy G, Turcotte S, Tang A, Kadoury S. Predicting the Response to FOLFOX-Based Chemotherapy Regimen from Untreated Liver Metastases on Baseline CT: a Deep Neural Network Approach. J Digit Imaging. 2020;33:937-945. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 8] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 55. | Wei J, Cheng J, Gu D, Chai F, Hong N, Wang Y, Tian J. Deep learning-based radiomics predicts response to chemotherapy in colorectal liver metastases. Med Phys. 2021;48:513-522. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 14] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 56. | Meric-Bernstam F, Hurwitz H, Raghav KPS, McWilliams RR, Fakih M, VanderWalde A, Swanton C, Kurzrock R, Burris H, Sweeney C, Bose R, Spigel DR, Beattie MS, Blotner S, Stone A, Schulze K, Cuchelkar V, Hainsworth J. Pertuzumab plus trastuzumab for HER2-amplified metastatic colorectal cancer (MyPathway): an updated report from a multicentre, open-label, phase 2a, multiple basket study. Lancet Oncol. 2019;20:518-530. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 311] [Cited by in RCA: 392] [Article Influence: 65.3] [Reference Citation Analysis (0)] |
| 57. | Giannini V, Rosati S, Defeudis A, Balestra G, Vassallo L, Cappello G, Mazzetti S, De Mattia C, Rizzetto F, Torresin A, Sartore-Bianchi A, Siena S, Vanzulli A, Leone F, Zagonel V, Marsoni S, Regge D. Radiomics predicts response of individual HER2-amplified colorectal cancer liver metastases in patients treated with HER2-targeted therapy. Int J Cancer. 2020;147:3215-3223. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 22] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 58. | Nakanishi R, Oki E, Hasuda H, Sano E, Miyashita Y, Sakai A, Koga N, Kuriyama N, Nonaka K, Fujimoto Y, Jogo T, Hokonohara K, Hu Q, Hisamatsu Y, Ando K, Kimura Y, Yoshizumi T, Mori M. Radiomics Texture Analysis for the Identification of Colorectal Liver Metastases Sensitive to First-Line Oxaliplatin-Based Chemotherapy. Ann Surg Oncol. 2021;28:2975-2985. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 20] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 59. | Taghavi M, Staal F, Gomez Munoz F, Imani F, Meek DB, Simões R, Klompenhouwer LG, van der Heide UA, Beets-Tan RGH, Maas M. CT-Based Radiomics Analysis Before Thermal Ablation to Predict Local Tumor Progression for Colorectal Liver Metastases. Cardiovasc Intervent Radiol. 2021;44:913-920. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 22] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 60. | Mühlberg A, Holch JW, Heinemann V, Huber T, Moltz J, Maurus S, Jäger N, Liu L, Froelich MF, Katzmann A, Gresser E, Taubmann O, Sühling M, Nörenberg D. The relevance of CT-based geometric and radiomics analysis of whole liver tumor burden to predict survival of patients with metastatic colorectal cancer. Eur Radiol. 2021;31:834-846. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 27] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 61. | Sasaki K, Margonis GA, Andreatos N, Zhang XF, Buettner S, Wang J, Deshwar A, He J, Wolfgang CL, Weiss M, Pawlik TM. The prognostic utility of the “Tumor Burden Score” based on preoperative radiographic features of colorectal liver metastases. J Surg Oncol. 2017;116:515-523. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 52] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 62. | Hao X, Luo H, Krawczyk M, Wei W, Wang W, Wang J, Flagg K, Hou J, Zhang H, Yi S, Jafari M, Lin D, Chung C, Caughey BA, Li G, Dhar D, Shi W, Zheng L, Hou R, Zhu J, Zhao L, Fu X, Zhang E, Zhang C, Zhu JK, Karin M, Xu RH, Zhang K. DNA methylation markers for diagnosis and prognosis of common cancers. Proc Natl Acad Sci U S A. 2017;114:7414-7419. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 251] [Cited by in RCA: 335] [Article Influence: 41.9] [Reference Citation Analysis (0)] |
| 63. | Martinelli E, Ciardiello D, Martini G, Troiani T, Cardone C, Vitiello PP, Normanno N, Rachiglio AM, Maiello E, Latiano T, De Vita F, Ciardiello F. Implementing anti-epidermal growth factor receptor (EGFR) therapy in metastatic colorectal cancer: challenges and future perspectives. Ann Oncol. 2020;31:30-40. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 124] [Article Influence: 31.0] [Reference Citation Analysis (0)] |
| 64. | Dercle L, Lu L, Schwartz LH, Qian M, Tejpar S, Eggleton P, Zhao B, Piessevaux H. Radiomics Response Signature for Identification of Metastatic Colorectal Cancer Sensitive to Therapies Targeting EGFR Pathway. J Natl Cancer Inst. 2020;112:902-912. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 105] [Article Influence: 26.3] [Reference Citation Analysis (0)] |
| 65. | Spelt L, Nilsson J, Andersson R, Andersson B. Artificial neural networks—a method for prediction of survival following liver resection for colorectal cancer metastases. Eur J Surg Oncol. 2013;39:648-654. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 37] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 66. | Paredes AZ, Hyer JM, Tsilimigras DI, Moro A, Bagante F, Guglielmi A, Ruzzenente A, Alexandrescu S, Makris EA, Poultsides GA, Sasaki K, Aucejo FN, Pawlik TM. A Novel Machine-Learning Approach to Predict Recurrence After Resection of Colorectal Liver Metastases. Ann Surg Oncol. 2020;27:5139-5147. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 25] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 67. | Fong Y, Fortner J, Sun RL, Brennan MF, Blumgart LH. Clinical score for predicting recurrence after hepatic resection for metastatic colorectal cancer: analysis of 1001 consecutive cases. Ann Surg. 1999;230:309-318; discussion 318-321. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2693] [Cited by in RCA: 2793] [Article Influence: 107.4] [Reference Citation Analysis (1)] |
| 68. | Brudvik KW, Jones RP, Giuliante F, Shindoh J, Passot G, Chung MH, Song J, Li L, Dagenborg VJ, Fretland ÅA, Røsok B, De Rose AM, Ardito F, Edwin B, Panettieri E, Larocca LM, Yamashita S, Conrad C, Aloia TA, Poston GJ, Bjørnbeth BA, Vauthey JN. RAS Mutation Clinical Risk Score to Predict Survival After Resection of Colorectal Liver Metastases. Ann Surg. 2019;269:120-126. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 175] [Cited by in RCA: 175] [Article Influence: 29.2] [Reference Citation Analysis (0)] |
| 69. | Qi ZH, Xu HX, Zhang SR, Xu JZ, Li S, Gao HL, Jin W, Wang WQ, Wu CT, Ni QX, Yu XJ, Liu L. The Significance of Liquid Biopsy in Pancreatic Cancer. J Cancer. 2018;9:3417-3426. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 40] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 70. | Mason MC, Tzeng CD, Tran Cao HS, Aloia TA, Newhook TE, Overman MJ, Kopetz SE, Vauthey JN, Chun YS. Preliminary Analysis of Liquid Biopsy after Hepatectomy for Colorectal Liver Metastases. J Am Coll Surg. 2021;233:82-89.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 21] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 71. | Saini A, Pershad Y, Albadawi H, Kuo M, Alzubaidi S, Naidu S, Knuttinen MG, Oklu R. Liquid Biopsy in Gastrointestinal Cancers. Diagnostics (Basel). 2018;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 14] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 72. | Rompianesi G, Di Martino M, Gordon-Weeks A, Montalti R, Troisi R. Liquid biopsy in cholangiocarcinoma: Current status and future perspectives. World J Gastrointest Oncol. 2021;13:332-350. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 41] [Cited by in RCA: 25] [Article Influence: 6.3] [Reference Citation Analysis (0)] |