Published online Feb 28, 2021. doi: 10.3748/wjg.v27.i8.692
Peer-review started: September 9, 2020
First decision: December 4, 2020
Revised: December 16, 2020
Accepted: January 21, 2021
Article in press: January 21, 2021
Published online: February 28, 2021
Processing time: 170 Days and 3 Hours
Gallbladder cancer (GBC) is an aggressive type of biliary tract cancer that lacks effective therapeutic targets. Fork head box M1 (FoxM1) is an emerging molecular target associated with tumor progression in GBC, and accumulating evidence suggests that vascular endothelial growth factor (VEGF) promotes various tumors by inducing neoangiogenesis.
To investigate the role of FoxM1 and the angiogenesis effects of VEGF-A in primary GBC.
Using immunohistochemistry, we investigated FoxM1 and VEGF-A expression in GBC tissues, paracarcinoma tissues and cholecystitis tissues. Soft agar, cell invasion, migration and apoptosis assays were used to analyze the malignant phenotype influenced by FoxM1 in GBC. Kaplan-Meier survival analysis was performed to evaluate the impact of FoxM1 and VEGF-A expression in GBC patients. We investigated the relationship between FoxM1 and VEGF-A by regulating the level of FoxM1. Next, we performed MTT assays and Transwell invasion assays by knocking out or overexpressing VEGF-A to evaluate its function in GBC cells. The luciferase assay was used to reveal the relationship between FoxM1 and VEGF-A. BALB/c nude mice were used to establish the xenograft tumor model.
FoxM1 expression was higher in GBC tissues than in paracarcinoma tissues. Furthermore, the high expression of Foxm1 in GBC was significantly correlated with a malignant phenotype and worse overall survival. Meanwhile, high expression of FoxM1 influenced angiogenesis; high expression of FoxM1 combined with high expression of VEGF-A was related to poor prognosis. Attenuated FoxM1 significantly suppressed cell proliferation, transfer and invasion in vitro. Knockdown of FoxM1 in GBC cells reduced the expression of VEGF-A. Luciferase assay showed that FoxM1 was the transcription factor of VEGF-A, and knockdown VEGF-A in FoxM1 overexpressed cells could partly reverse the malignancy phenotype of GBC cells. In this study, we found that FoxM1 was involved in regulation of VEGF-A expression.
FoxM1 and VEGF-A overexpression were associated with the prognosis of GBC patients. FoxM1 regulated VEGF-A expression, which played an important role in the progression of GBC.
Core Tip: Fork head box M1 (FoxM1) is an emerging molecular target associated with early metastasis, drug resistance and poor patient survival in gallbladder cancer. Accumulating evidence suggests that vascular endothelial growth factor (VEGF) plays important roles in many kinds of tumors by inducing neoangiogenesis. In the present study, we found that VEGF-A overexpression was correlated with proliferation, invasion and poor prognosis. The results of Kaplan-Meier analysis showed that both FoxM1 and VEGF-A were associated with survival in gallbladder cancer. VEGF-A expression was regulated by FoxM1, which was the mechanism of FoxM1 enhancing the angiogenesis of gallbladder cancer.
- Citation: Wang RT, Miao RC, Zhang X, Yang GH, Mu YP, Zhang ZY, Qu K, Liu C. Fork head box M1 regulates vascular endothelial growth factor-A expression to promote the angiogenesis and tumor cell growth of gallbladder cancer. World J Gastroenterol 2021; 27(8): 692-707
- URL: https://www.wjgnet.com/1007-9327/full/v27/i8/692.htm
- DOI: https://dx.doi.org/10.3748/wjg.v27.i8.692
Gallbladder cancer (GBC), a frequent and aggressive type of biliary tract cancer, is now the sixth most common gastrointestinal cancer. The vast majority (65% to 90%) of GBCs are adenocarcinomas followed by squamous cell or adenosquamous cell carcinomas (5% to 10%) and undifferentiated carcinomas (5%)[1]. The incidence rate is 2.2 per 100000 per year worldwide[1]. Surgery combined with adjuvant chemotherapy and radiotherapy is the general therapy for patients with GBC. However, GBC is known to have an occult onset, rapid progression and abysmal prognosis, with a mean 6-mo survival time and a 5-yr survival rate of 5%[2]. Furthermore, a low diagnosis rate of GBC has been demonstrated in Japan, with only a 0.011% detection rate from mass screening[3]. Thus, accurate and specific diagnosis of GBC at the early stage is still urgently needed to improve therapeutic efficacy.
Cancer cells achieve invasion and metastasis through extracellular proteolysis, which affects cell proliferation, adhesion, migration and angiogenesis. In addition, Fork head box M1 (FoxM1) is characterized as a critical proliferation-associated transcription factor in cell cycle progression[4]. Inhibiting FoxM1 expression has been reported to induce the loss of the mitotic spindle, resulting in failed cell division. FoxM1 is widely expressed in normal proliferating cells and is also overexpressed in cancer cells, such as ovarian cancer cells, renal cell carcinoma cells, pancreatic cancer cells, breast cancer cells, laryngeal squamous carcinoma cells, glioma cells, prostate carcinoma cells and gastric cancer cells[5]. Moreover, the overexpression of FoxM1 has an impact on poor patient prognosis and can be regarded as an independent predictor of low survival[4]. However, in patients with GBC, the clinical correlation of FoxM1 expression remains unclear.
In recent research, investigators have found that the downregulation of FoxM1 inhibits the activity of vascular endothelial growth factor (VEGF), matrix metalloproteinase (MMP)-2, MMP-9, Cyclin B1, CDC25, p21 and p27 proteins, affecting cancer cell proliferation, migration and invasion[6]. Moreover, FoxM1 was proven to be upregulated at both the mRNA and protein levels[7]. In glioma, with the increase in FoxM1, cancer cell invasiveness and tumorigenicity are also promoted[8]. These investigations indicate that FoxM1 plays an important role in the progression of carcinoma and could have the same function in GBC cells. Two investigations also found a correlation between lymph node metastasis and FoxM1. However, the correlation between elevated levels of FoxM1 and clinical specimens has been controversial.
We found that FoxM1 overexpression promoted tumor cell migration and invasion, while FoxM1 downregulation led to an increase in cell apoptosis rates, indicating that FoxM1 might play a key role in disease progression of GBC. Moreover, we found that FoxM1 could be directly involved in the regulation of VEGF-A expression by combining with the VEGF-A promoter and activating VEGF-A transcription. Therefore, we indicated that FoxM1 could promote GBC progression via VEGF-A.
After surgery, 48 cases of fresh frozen GBC tumor tissue were collected from the First Affiliated Hospital of Xi’an Jiaotong University, together with paired tumor-free liver tissue (at least 2 cm from the tumor) from September 2015 to February 2017. GBC was confirmed by histopathological examination, and the study was approved by the Ethics Staff of the First Affiliated Hospital of Xi’an Jiaotong University.
The human GBC cell line SGC-996 was purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai branch) and was cultured in RPMI 1640 medium containing 10% fetal bovine serum (Gibco, Grand Island, NY, United States) at 37 °C with 5% CO2.
At 24 h after cell seeding in the culture dish, the recombinant adenovirus vector containing specific shRNA was transfected into SGC-996 cells with Lipofectamine 2000 (Invitrogen, United States) at different multiplicities of infection. Recombinant lentiviruses containing FoxM1 shRNA and VEGF-A shRNA were purchased from GenePharma (Shanghai, China). A negative control carrying green fluorescent protein, which expresses a scrambled RNA, was constructed as a control. The virus containing the construct was isolated using plaque screening, purification and amplification. The protocol for lentivirus infection was according to the GenePharma Recombinant Lentivirus Operation Manual (http://www.genepharma.com).
A FoxM1 expression plasmid was purchased from Sino Biological Inc. (Beijing, China). SGC-996 cells were transfected with the FoxM1 expression vector or an empty control vector (400 ng/well) using HiPerFect. After 48 h of transfection, the cells were collected, and protein levels were analyzed by Western blotting and quantitative reverse transcription (qRT)-PCR. The overexpression of VEGF-A was performed as described above.
Western blot analysis was performed as previously reported. Briefly, the membranes were probed with the following primary antibodies: FoxM1 (1:1000, Proteintech, Wuhan, China), VEGF-A (1:1000, Proteintech, Wuhan, China) and β-actin (1:3000, Abways, Shanghai, China). After being washed with TBS-T, the membranes were incubated with horseradish peroxidase-conjugated anti-rabbit or anti-mouse secondary antibody (1:10000, Proteintech, Wuhan, China). Mouse anti-β-actin (1:1000, Proteintech, Wuhan, China) with goat anti-mouse (1:10000, Proteintech, Wuhan, China) antibodies were used as a loading control. Chemiluminescence detection was performed with ChemiGlow detection reagents (Bio-Rad Western ECL Substrate). The blots were visualized with a Bio-Rad ChemiDoc MP and quantified with a densitometer using the imager application program (Alpha Innotech, San Leandro, CA, United States).
RNA isolation and RT-PCR analysis were performed as previously described. The cDNA for FoxM1, VEGF-A and β-actin were amplified using the Platinum Taq DNA Polymerase kit (Life Technologies) with specific primers. β-actin was used as an internal control. Primer sequences were as follows: FoxM1 Forward, 5’-CAC CCC AGT GCC AAC CGC TAC TTG-3’; FoxM1 Reverse, 5’-AAA GAG GAG CTA TCC CCT CCT CAG-3’; VEGF-A Forward, 5′- CAG ATT ATG CGG ATC AAA CCT CA -3′; VEGF-A Reverse, 5’-CAA GGC CCA CAG GGA CTG CAA -3’.
Tissue specimens were fixed in neutral buffered formalin (10% vol/vol formalin in water, pH 7.4) and embedded in paraffin wax. Serial sections of 5 mm thickness were cut and mounted on charged glass slides. Conditions for FoxM1 and VEGF-A were optimized and evaluated by two independent pathologists. The goat monoclonal antibodies against FoxM1 (sc-26688; Santa Cruz Biotechnology, Santa Cruz, CA, United States) and VEGF-A (sc-152; Santa Cruz Biotechnology, Santa Cruz, CA, United States) were used at dilutions of 1:200. The streptavidin–peroxidase kit (SP-9000 Golden Bridge Int., Beijing, China) was used in accordance with the manufacturer’s instructions. An irrelevant goat antiserum served as a negative control. Sections were counterstained with Mayer’s hematoxylin. The immunohistochemical score and analysis were described as previously reported.
Experiments with soft agar were carried out on 6-well plates. Agar was used for the bottom (0.8%) and top (0.4%) containing cells and fed every 4 d in a complete DMEM medium. After 20 d, colonies with diameters greater than 150 μm were scored under a 10-fold fluorescence microscope.
The SGC-996 cells were inoculated into a 6-well plate with a density of 1 x 106 cells per well. Cells at logarithmic growth stage were digested by 0.25% trypsin, and the supernatant was discarded after 800 rpm centrifugation for 5 min. The mixture was resuspended with PBS, centrifuged at 800 rpm for 5 min, and washed twice with PBS. Annexin V (10 μL) was added at a concentration of 50 μg/mL and left standing for 40min (Bender Medsystems, Australia). Flow cytometry was used to detect apoptosis (Beckman, United States).
SGC-996 cells were seeded at 5 × 103 cells per well in 96-well flat-bottom plates before transfection. For the assay, 20 μL of MTT solution (5 g/L) was added to each well, and the cells were incubated for 4 h. Then, supernatants were removed, and formazan crystals were dissolved in 200 mL dimethyl sulfoxide. After the insoluble crystals were completely dissolved, the absorbance values at 490 nm were measured using a microplate reader (Bio-Rad, Hercules, CA, United States).
Cell invasion and migration assays were performed using Transwell permeable supports with 8 μm pore size (Costar, Cambridge, MA, United States). Cells were suspended in serum-free medium and seeded into Transwell inserts either uncoated (for the migration assay) or coated (for the invasion assay) with growth factor-reduced Matrigel (BD Biosciences, Bedford, MA, United States). Bottom wells were filled with complete medium. After 24 h, the invaded cells were fixed with methanol and stained with a crystal violet solution. The number of cells that penetrated the membrane was determined by counting the mean cell number in five randomly selected high-power fields.
For the clonogenic assay, cells were plated in the 6-well plate with 1000 cells per well. The samples were fixed with 4% paraformaldehyde for 30 min and stained with 0.05% crystal violet for 20 min. Representative photos were collected and more than 40 colonies were counted.
The pre-cooled 96 well plates were coated with matrix gel (35 μL/well) and then placed in the humidified CO2 incubator at 37 degrees Celsius for 1 h to solidify the coating. HUVEC cells were starved overnight under 1% bovine serum albumin. Then, the cells were digested with trypsin and collected and inoculated on a 96 well plate with a density of 2.5 × 104 cells per well. SGC-996 cells were cocultured with HUVEC for 4 h. The tube formation of HUVECs was observed at 4 h by a microscope.
A pmirGLO Dual luciferase miRNA Target Expression Vector (E1330; Promega Corporation) contained the fragment of FOXM1 3’UTR and VEGF 3’UTR was used to establish luciferase reporter plasmids: WT-FOXM1; MUT-FOXM1; WT-VEGF; and MUT-VEGF. HEK293T cells were cultured in 24-well plates. When the cells reached 70% confluence, cells were transfected with 2 µg WT-FOXM1 or MUT-FOXM1 and 2 µg WT-VEGF or MUT-VEGF using Lipofectamine® 3000 (Invitrogen, Thermo Fisher Scientific, Inc). After 48 h, luciferase activity was detected by the dual-enzyme reporter assay kit (Promega).
Animal experiments were approved by the Animal Ethics Committee of the Medical College of Xi’an Jiaotong University. BALB/c nude mice aged 4-6 wk were purchased from Medical Laboratory Animal Center of Xi’an Jiaotong University. Mice weighing 16 to 20 g were randomly divided into three groups: (1) Lenti-FoxM1-shRNA; (2) cDNA-FoxM1; or (3) control (1 x 107 SGC-996 cells were injected into the back of mice to establish xenograft tumor model). After 1 mo, the mice were killed, and the tumors were collected.
All data are expressed as the mean ± SD. To compare the means of normally distributed variables, analysis of variance or Student’s t test was applied. P value less than 0.05 was considered significant in all tests. All analyses were performed using SPSS software version 19.0 (SPSS Inc., Chicago, IL, United States).
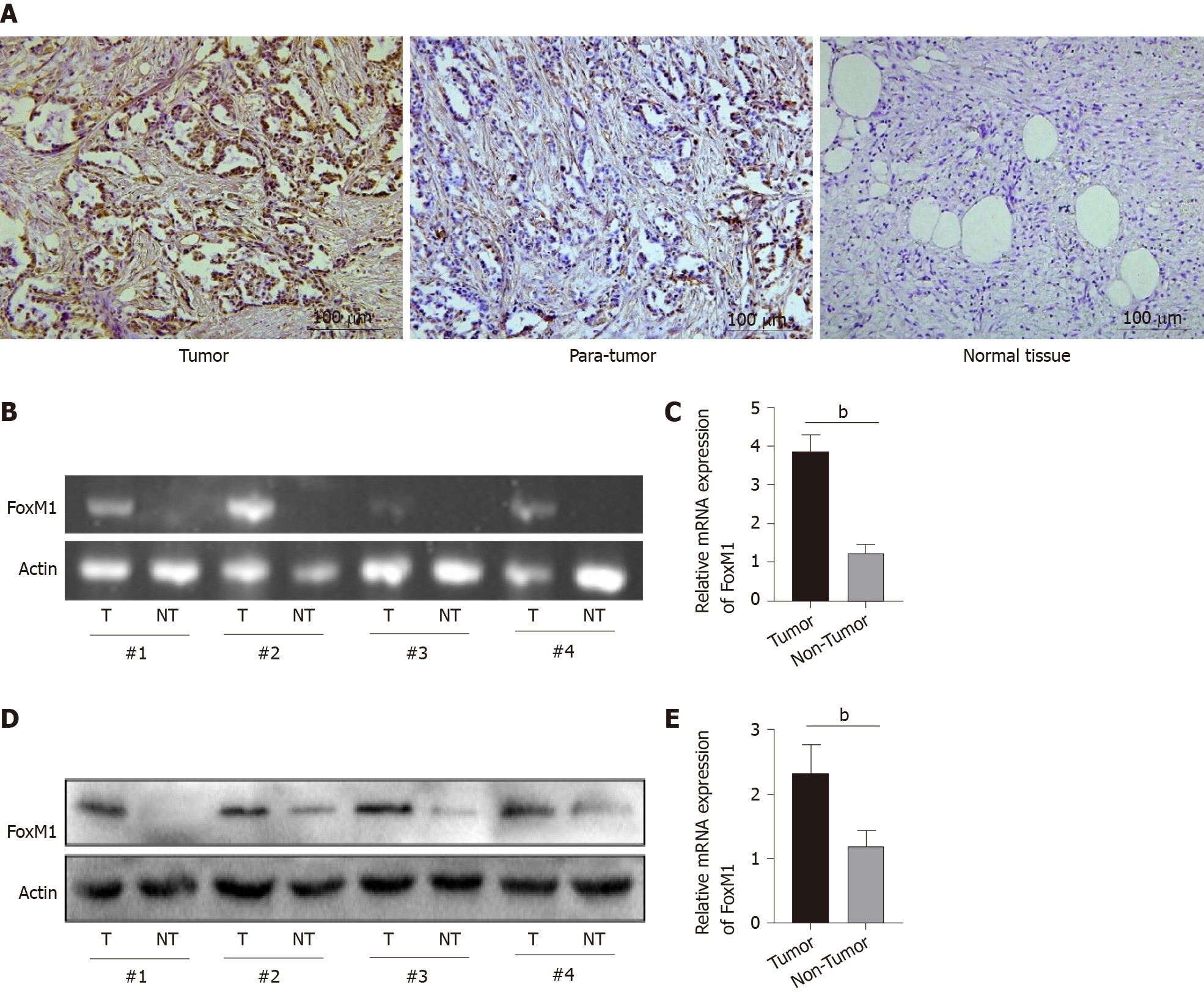
First, the expression of FoxM1 was higher than that in paracarcinoma tissues or cholecystitis tissues (Figure 1A). Moreover, we compared the levels of FoxM1 expression in four paired GBC clinical samples by qRT-PCR and Western blot analysis. The results revealed that the expression of FoxM1 in GBC was significantly higher than that in paracarcinoma tissue (Figure 1B and 1C). Western blot results showed that the protein level of FoxM1 in GBC tissues was higher than the expression of FoxM1 in paracarcinoma tissues (Figure 1D and 1E). This suggested that FoxM1 expression was closely related to the development of GBC.
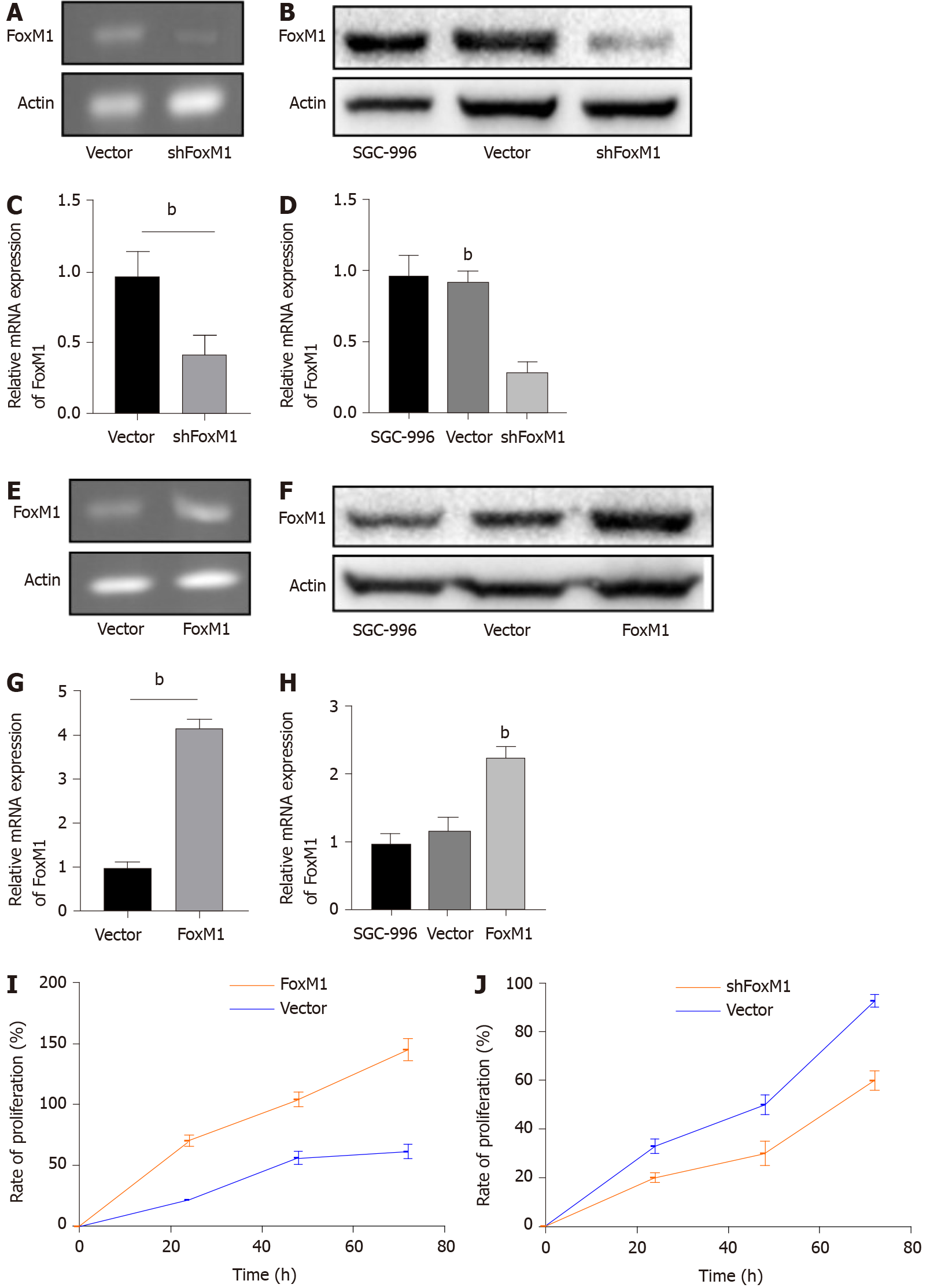
To investigate the function of FoxM1 in GBC, the expression of FoxM1 was suppressed by lenti-FoxM1 shRNA and was elevated by lenti- FoxM1 in the SGC-996 cell line. The protein and mRNA levels of FoxM1 were significantly decreased after lenti-FoxM1 shRNA transfection (Figure 2A-D). The protein and mRNA levels of FoxM1 were significantly increased after lenti- FoxM1 transfection (Figure 2E-H).
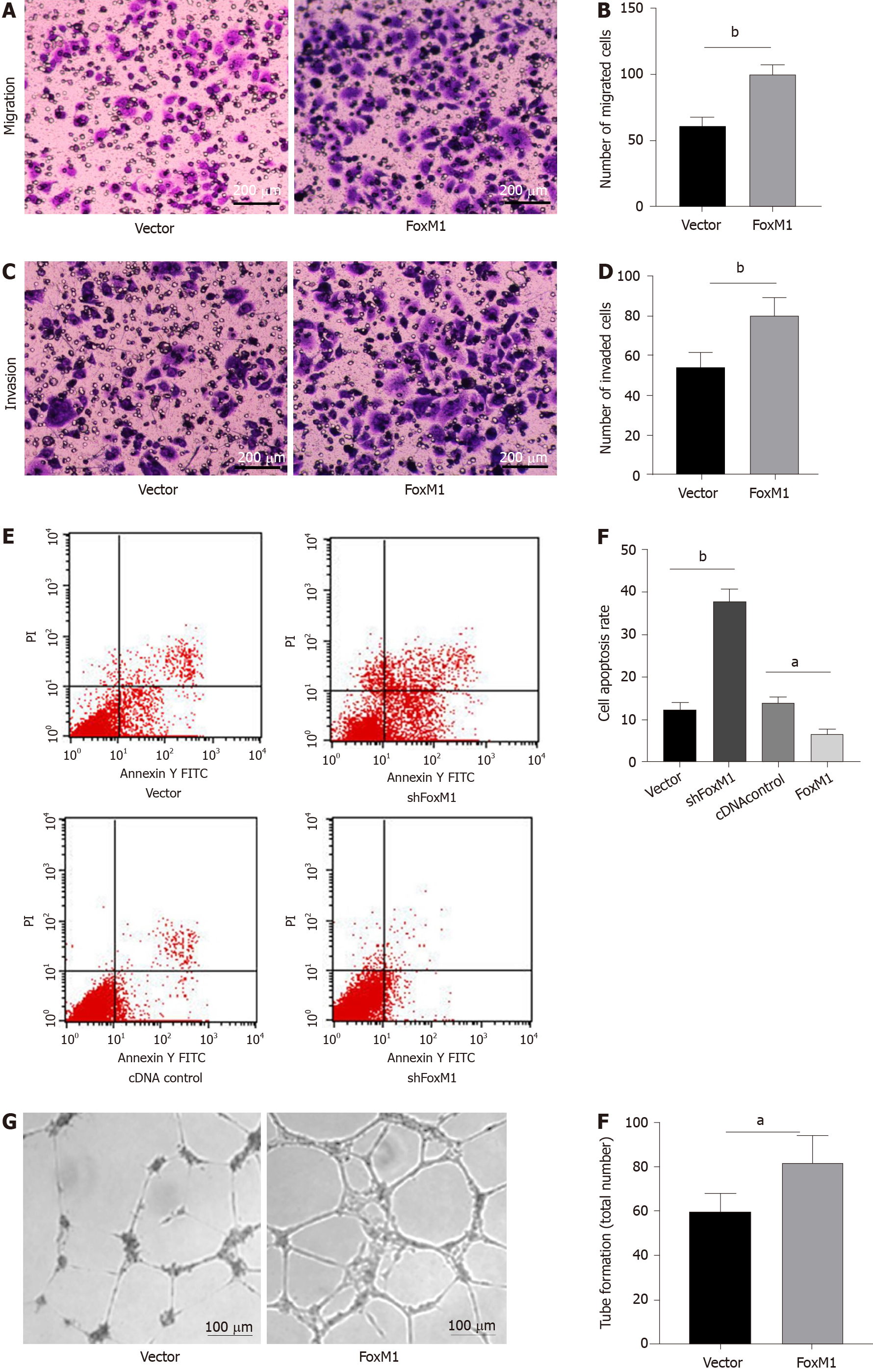
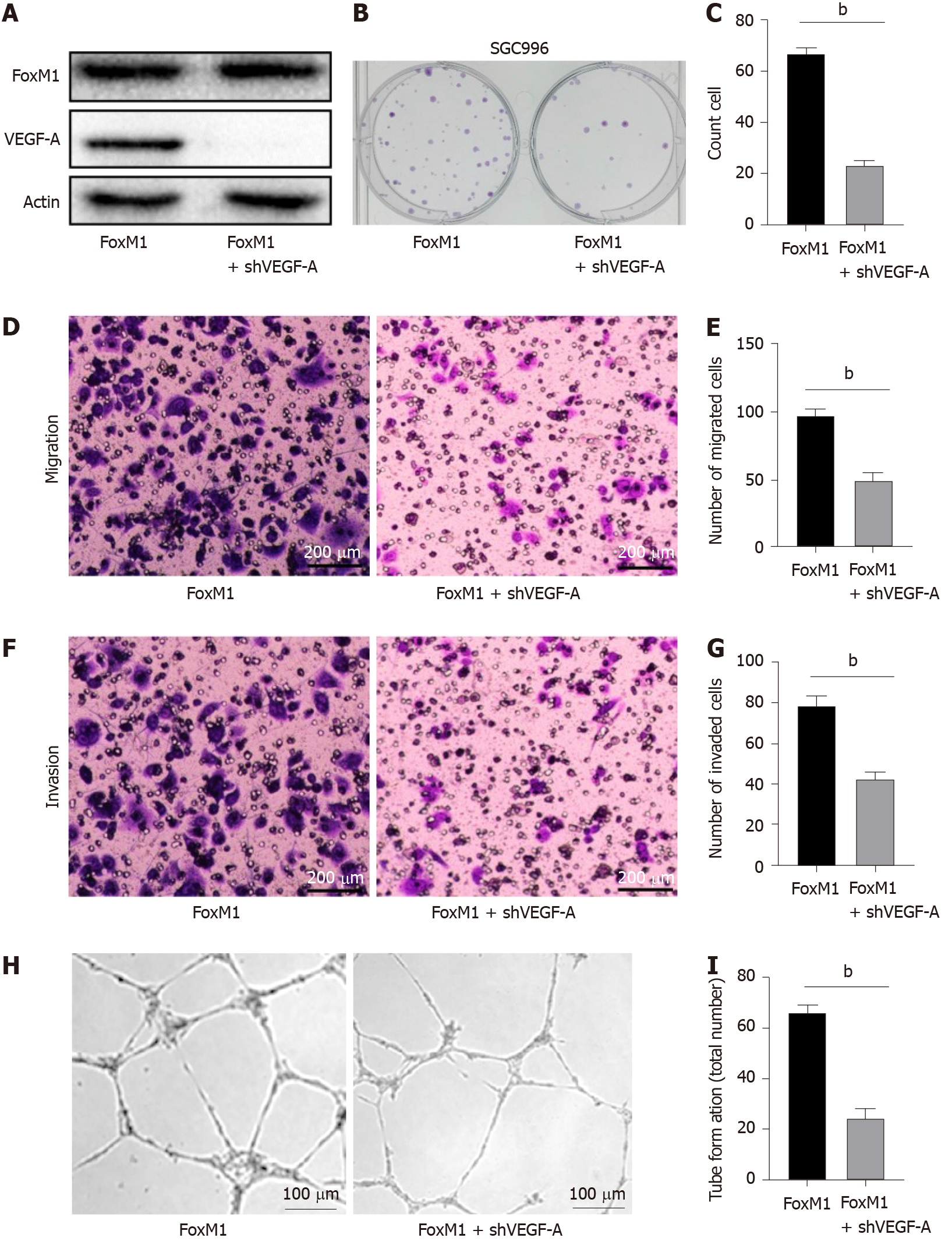
As demonstrated by the soft agar cloning assay, lenti-FoxM1 shRNA significantly decreased proliferation in SGC-996 cells. Conversely, lenti-FoxM1 significantly increased SGC-996 cell proliferation (Figure 2I and 2J). To determine the potential metastatic-promoting effect of FoxM1 in GBC cells, we investigated cell migration and invasion abilities. Compared with that in SGC-996 cells, less cell penetration was observed in lenti-FoxM1 shRNA-transfected cells. However, lenti-FoxM1-transfected cells had stronger invasion abilities in the presence of Matrigel (Figure 3A-D). As shown in the apoptosis assay, knockdown-FoxM1 cells increased apoptotic levels (Figure 3E and 3F). Tube formation also showed that in FoxM1 up-regulated cells vessel generation ability was significantly enhanced (Figure 3G and 3H). These data suggested that FoxM1 plays important roles in GBC progression.
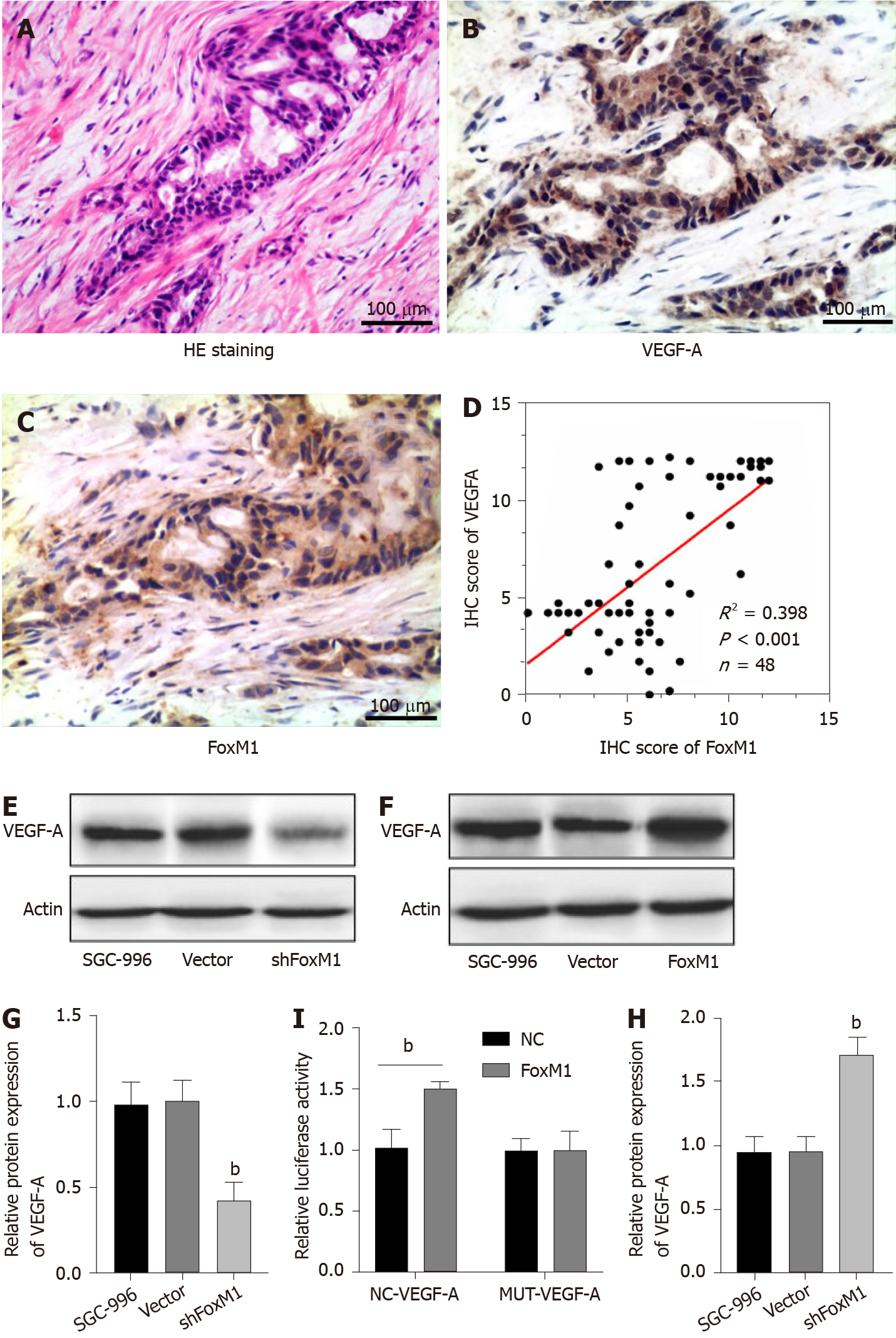
As shown in Figure 3G and 3H, the high expression level of FoxM1 influenced vessel generation in GBC. Meanwhile, research revealed that VEGF-A was associated with endothelial cell proliferation, division, migration and lumen formation. Therefore, we examined the correlation between FoxM1 and VEGF-A. As shown in Figure 3, FoxM1 and VEGF-A were both associated with GBC, and the expression of VEGF-A was obviously decreased in FoxM1 siRNA-transfected SGC-996 cells. We detected the expression of FoxM1 and VEGF-A by consecutive section immunohistochemistry in 48 GBC tissues. A positive correlation between FoxM1 and VEGF-A was observed (Figure 4A-D). Thus, we hypothesized that VEGF-A is regulated by FoxM1 to promote GBC cell migration and invasion.
First, we investigated whether FoxM1 regulated the protein and mRNA levels of VEGF-A in GBC cells. Therefore, we determined the VEGF-A protein and mRNA expression levels after silencing FoxM1 by Western blot and RT-PCR analyses, respectively. We found that FoxM1 knockdown led to significant downregulation of VEGF-A protein and mRNA expression levels in SGC-996 cells (Figure 4E and 4G). To further prove the link between FoxM1 and VEGF-A expression, we overexpressed FoxM1 by transfecting lenti-FoxM1 in SGC-996 cells and found that VEGF-A expression was significantly increased at the transcriptional and translational levels (Figure 4F and 4H). Moreover, the luciferase assay showed that FoxM1 was the transcription factor of VEGF-A (Figure 4I). Nevertheless, knockdown VEGF-A in FoxM1 overexpressed cells could partly reverse the malignant phenotype of GBC cells (Figure 5A-I). These results indicated that FoxM1 was involved in the regulation of VEGF-A expression.
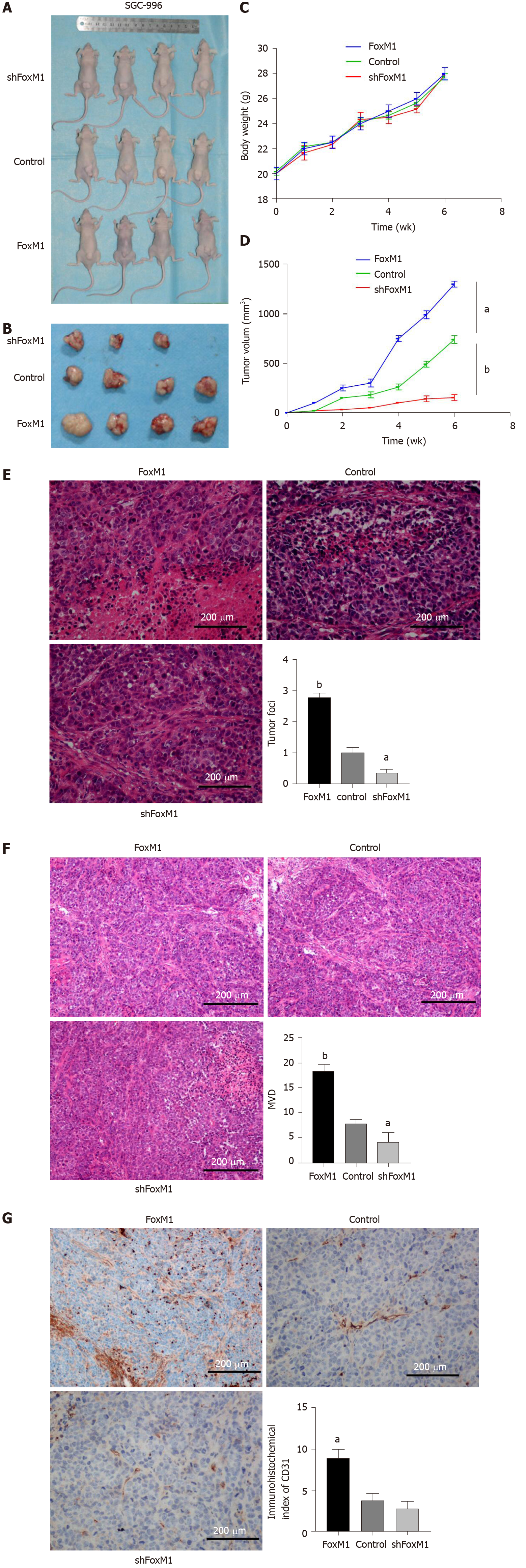
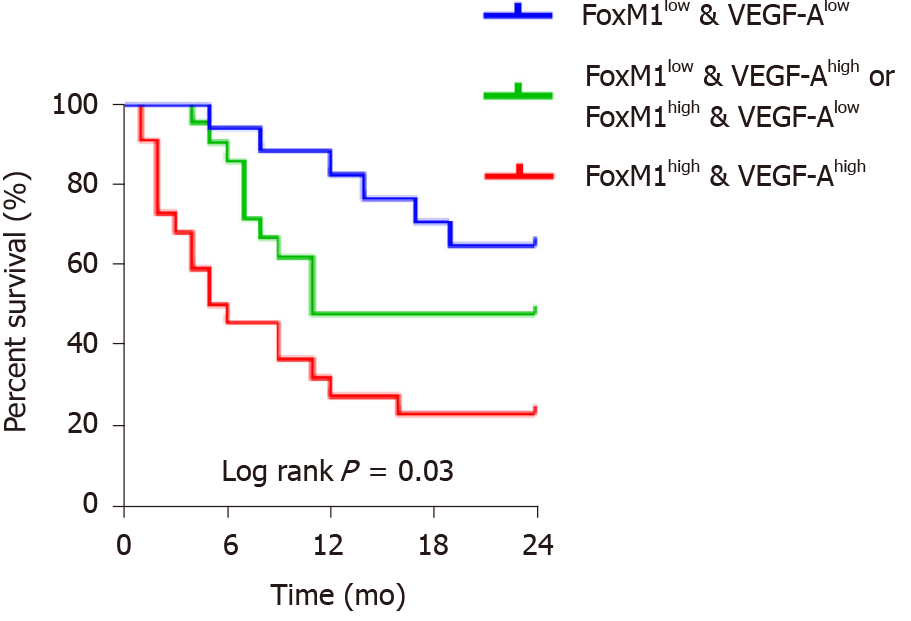
Because the in vitro study showed that VEGF-A, a potent vascular active molecule, promoted proliferation, invasion and metastasis regulated by FoxM1, we further investigated the function of the FoxM1-VEGF-A axis on angiogenesis in GBC. As a result, the tumor volume and weight of mice transfected with lenti-FoxM1-shRNA or lenti-FoxM1 were determined (Figure 6A-D). The tumors of mice transfected with lenti-FoxM1 were larger than the control group, and the tumors of mice transfected with lenti-FoxM1-shRNA were smaller than the control group. Next, analysis of hematoxylin-eosin staining was performed to detect the number of tumor foci (Figure 6E). The most tumor foci were in tumor tissues of mice overexpressing FoxM1. Additionally, microvascular density, CD31 and VEGF-A were measured to investigate the angiogenic ability (Figure 6F and 6G). The above results showed that FoxM1 can improve the angiogenesis of GBC by regulating the expression of VEGF-A. Kaplan-Meier analysis was used to assess the impact of FoxM1 and VEGF-A on survival. The results showed that the group with high expression of FoxM1 or VEGF-A showed poor survival compared with the patients with low expression of FoxM1 and VEGF-A (Figure 7).
GBC is still a frequent malignancy with a dismal prognosis. Because of the rapid progression and low diagnosis rate of GBC, it is necessary to explore the inherent mechanisms of tumorigenesis. The essential interrelation between FoxM1 and VEGF-A provided us with a broader approach to malignancy therapy. In this investigation, we found that FoxM1 played an essential role in the expression of VEGF. Previous studies have indicated that high expression levels of FoxM1 are relevant to adverse prognosis in multiple malignancies[9]. We proved that the upregulation of FoxM1 in GBC cells influenced the expression of VEGF-A in vivo and in vitro. In GBC patients, VEGF-A expression was closely related to the overexpression of FoxM1 and was highly expressed to promote vascularization. FoxM1 could directly regulate the activity of the VEGF gene promoter to induce its expression. We detected the opposite results in GBC cells with inhibited FoxM1. Our conclusions could provide a novel strategy for treating GBC.
The contribution of FoxM1 on numerous kinds of malignancies has been demonstrated by previous investigations[8,10-13]. As an important factor of poor prognosis, the expression level of FoxM1 has a negative correlation with survival duration[14-16]. In our previous study, we found that FoxM1 was upregulated in GBC tissues compared with normal tissues and as a prognostic biomarker had greater relevance to prognosis and survival[17]. Likewise, in this study, the role of VEGF-A in the progression of GBC was investigated. The same tumor-promoting function through enhanced proliferation, invasion and metastasis was observed in GBC cells. The function of VEGF-A in promoting cancer was also shown in other investigations[18,19]. A significant decrease in the expression of VEGF-A was detected after knocking down the expression of FoxM1.
Angiogenesis is indispensable to the proliferation of tumor cells, auxesis, migration and invasion. In addition, directly controlling angiogenesis could inhibit the progression of tumors. Angiogenesis is influenced by various factors, which has been shown in many studies[20]. Oxygen deficiency is one such factor. VEGF expression could be promoted by oxygen deficiency in a mouse model, especially in the adjacent areas of the necrotic regions[18-20]. For example, as glioma is a hypoxic tumor, the overexpression of VEGF in glioma is associated at least with hypoxia and has been observed in many studies[21-23]. Moreover, the following two observations were made in our research. The expression of VEGF in GBC cells was significantly higher than that in paracarcinoma tissue. The expression of VEGF in the adjacent areas of the necrotic regions was significantly related to the cells in other normal areas (P < 0.001).
In addition to VEGF, MMP-2, MMP-9 and uPAR are thought to participate in the migration and invasion of tumor cells[24]. It is generally accepted that VEGF is a predominant facilitator of angiogenesis and the angiogenic signaling cascade[25]. VEGF-A, a member of the VEGF family, is also considered to be critical for tumor growth, invasion and metastasis[26,27]. The direct interaction of FoxM1 with the VEGF-A promoter was discovered in our study, which could prove the direct regulation between FoxM1 and VEGF-A. First, the participation of FoxM1 in the regulation of the VEGF promoter was evidenced by our results, and the expression of FoxM1 could activate the VEGF promoter-mediated luciferase expression. Then, we selected two consensus binding sequences on the VEGF-A promoter to which FoxM1 could bind both in vivo and in vitro. After destroying them, the promoter of VEGF-A could not mediate its transcription. The activity of the FoxM1 binding areas could be evaluated.
It is also widely accepted that FoxM1 is a crucial mediator in the cell cycle[28]. FoxM1 is accumulated in the late G1 and S phases during the cell cycle. It could also be downregulated by many tumor suppressors, such as p53. FoxM1-siRNA has been used to decrease the proliferation and division of cells by suppressing mRNA and protein related to the cell cycle. As essential components of tumor progression, MMP-2, MMP9, uPA and uPAR were reduced by decreasing FoxM1, which has been proven in numerous studies[6].
Previous investigations have shown that as a consequence of downregulating FoxM1 with FoxM1-siRNA, the colonies of tumor cells decrease significantly[29]. Other studies also revealed that downregulation of FoxM1 could decrease the average number of cells crossing the Matrigel-coated membrane in one high power field and the migrating distance[5,30]. This evidence suggests that FoxM1 plays an essential role in tumor progression. In the present study, we also found that the downregulation of FoxM1 could suppress the viability, migration and invasion of tumor cells. Moreover, decreased FoxM1 has been shown to be relevant to prognosis and survival.
In conclusion, this study indicated that FoxM1 could be directly involved in the regulation of VEGF-A expression by activating and regulating the VEGF-A promoter. The overexpression of FoxM1 leading to excess VEGF-A and angiogenesis was proven to be enhanced in the adjacent areas of the necrotic regions. Therefore, it is conceivable that controlling the expression of FoxM1 could inhibit the angiogenesis and growth of GBC cells with the possibility of decreasing the invasion and metastasis of cancer cells, indicating that FoxM1 may play a key role in the pathogenesis and disease progression of GBC. This is the first work on the effects of FoxM1 and VEGF-A on GBC cells, and we hope the data can offer some reference and evidence to other studies or promote further discussion.
In conclusion, FoxM1 and VEGF-A overexpression were associated with prognosis of GBC patients. FoxM1 regulated VEGF-A expression, which played an important role in the progression of GBC.
Gallbladder cancer (GBC), with low detectable diagnosis and high metastasis, has a short survival time and a low 5-yr survival rate. Fork head box M1 (FoxM1) is relevant to poor prognosis and malignant behaviors, including proliferation, invasion and metastasis. Vascular endothelial growth factor-A (VEGF-A) plays an important role in angiogenesis. In GBC, it is meaningful to investigate the functional correlation between FoxM1 and VEGF-A.
FoxM1 is relevant to poor prognosis and malignant behaviors including proliferation, invasion and metastasis. VEGF-A plays an important role in angiogenesis. However, it is unclear whether FoxM1 could regulate VEGF-A.
This study aimed to investigate whether FoxM1 enhanced the angiogenesis of GBC via regulating VEGF-A.
Using immunohistochemistry, we investigated FoxM1 and VEGF-A expression in GBC tissues, paracarcinoma tissues and cholecystitis tissues. Soft agar, cell invasion, migration and apoptosis assays were used to analyze the malignant phenotype influenced by FoxM1 in GBC. Kaplan-Meier survival analysis was performed to evaluate the impact of FoxM1 and VEGF-A expression in GBC patients. We investigated the relationship between FoxM1 and VEGF-A by regulating the level of FoxM1. Next, we performed MTT assays and Transwell invasion assays by knocking out or overexpressing VEGF-A to evaluate its function in GBC cells. The luciferase assay was used to reveal the relationship between FoxM1 and VEGF-A. BALB/c nude mice were used to establish the xenograft tumor model.
FoxM1 expression was higher in GBC tissues than in paracarcinoma tissues. Furthermore, the high expression of FoxM1 in GBC was significantly correlated with the malignant phenotype and worse overall survival. Meanwhile, high expression of FoxM1 influenced angiogenesis. Attenuated FoxM1 significantly suppressed cell proliferation, transfer and invasion in vitro. Knockdown of FoxM1 in GBC cells reduced the expression of VEGF-A. The luciferase assay showed that FoxM1 was the transcription factor of VEGF-A. Knockdown VEGF-A in FoxM1 overexpressed cells could partly reverse the malignant phenotype of GBC cells. High expression of FoxM1 combined with high expression of VEGF-A was related to poor prognosis. In this study, we found that FoxM1 was involved in regulation of VEGF-A expression.
FoxM1 and VEGF-A overexpression were associated with prognosis of GBC patients. FoxM1 upregulated VEGF-A expression, which played an important role in the progression of GBC.
By comprehending the way FoxM1 induced angiogenesis through regulating VEGF-A in GBC, the present study showed a possible method for treatment strategy of GBC patients with metastasis and in late stage.
We are indebted to the individuals who have participated in or have helped with this article.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Merigo F, Zhang L S-Editor: Gao CC L-Editor: Filipodia P-Editor: Liu JH
| 1. | Mehrotra R, Tulsyan S, Hussain S, Mittal B, Singh Saluja S, Singh S, Tanwar P, Khan A, Javle M, Hassan MM, Pant S, De Aretxabala X, Sirohi B, Rajaraman P, Kaur T, Rath GK. Genetic landscape of gallbladder cancer: Global overview. Mutat Res. 2018;778:61-71. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 37] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 2. | Montalvo-Jave EE, Rahnemai-Azar AA, Papaconstantinou D, Deloiza ME, Tsilimigras DI, Moris D, Mendoza-Barrera GE, Weber SM, Pawlik TM. Molecular pathways and potential biomarkers in gallbladder cancer: A comprehensive review. Surg Oncol. 2019;31:83-89. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 19] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 3. | Hickman L, Contreras C. Gallbladder Cancer: Diagnosis, Surgical Management, and Adjuvant Therapies. Surg Clin North Am. 2019;99:337-355. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 107] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 4. | Choi HJ, Jhe YL, Kim J, Lim JY, Lee JE, Shin MK, Cheong JH. FoxM1-dependent and fatty acid oxidation-mediated ROS modulation is a cell-intrinsic drug resistance mechanism in cancer stem-like cells. Redox Biol. 2020;36:101589. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 71] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 5. | Nandi D, Cheema PS, Jaiswal N, Nag A. FoxM1: Repurposing an oncogene as a biomarker. Semin Cancer Biol. 2018;52:74-84. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 102] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 6. | Liao GB, Li XZ, Zeng S, Liu C, Yang SM, Yang L, Hu CJ, Bai JY. Regulation of the master regulator FOXM1 in cancer. Cell Commun Signal. 2018;16:57. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 231] [Cited by in RCA: 294] [Article Influence: 42.0] [Reference Citation Analysis (0)] |
| 7. | Song X, Fiati Kenston SS, Zhao J, Yang D, Gu Y. Roles of FoxM1 in cell regulation and breast cancer targeting therapy. Med Oncol. 2017;34:41. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 33] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 8. | Kim H, Park KJ, Ryu BK, Park DH, Kong DS, Chong K, Chae YS, Chung YG, Park SI, Kang SH. Forkhead box M1 (FOXM1) transcription factor is a key oncogenic driver of aggressive human meningioma progression. Neuropathol Appl Neurobiol. 2020;46:125-141. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 27] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 9. | Liu L, Wu J, Guo Y, Xie W, Chen B, Zhang Y, Li S, Hua Y, Peng B, Shen S. Overexpression of FoxM1 predicts poor prognosis of intrahepatic cholangiocarcinoma. Aging (Albany NY). 2018;10:4120-4140. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 18] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 10. | Wang H, Li H, Hu L, Wang J, Liu Q, Wang D, Sun X. Overexpression of FoxM1 in Sinonasal Inverted Papilloma and Associated Squamous Cell Carcinoma. Am J Rhinol Allergy. 2019;33:706-715. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 11. | Zhou W, Yu X, Sun S, Zhang X, Yang W, Zhang J, Zhang X, Jiang Z. Increased expression of MMP-2 and MMP-9 indicates poor prognosis in glioma recurrence. Biomed Pharmacother. 2019;118:109369. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 82] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 12. | Hu G, Yan Z, Zhang C, Cheng M, Yan Y, Wang Y, Deng L, Lu Q, Luo S. FOXM1 promotes hepatocellular carcinoma progression by regulating KIF4A expression. J Exp Clin Cancer Res. 2019;38:188. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 122] [Article Influence: 20.3] [Reference Citation Analysis (0)] |
| 13. | Marchand B, Pitarresi JR, Reichert M, Suzuki K, Laczkó D, Rustgi AK. PRRX1 isoforms cooperate with FOXM1 to regulate the DNA damage response in pancreatic cancer cells. Oncogene. 2019;38:4325-4339. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 30] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 14. | Cai H, Chen J, He B, Li Q, Li Y, Gao Y. A FOXM1 related long non-coding RNA contributes to gastric cancer cell migration. Mol Cell Biochem. 2015;406:31-41. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 27] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 15. | Huang W, Chen Z, Zhang L, Tian D, Wang D, Fan D, Wu K, Xia L. Interleukin-8 Induces Expression of FOXC1 to Promote Transactivation of CXCR1 and CCL2 in Hepatocellular Carcinoma Cell Lines and Formation of Metastases in Mice. Gastroenterology 2015; 149: 1053-67. e14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 114] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 16. | Okada K, Fujiwara Y, Takahashi T, Nakamura Y, Takiguchi S, Nakajima K, Miyata H, Yamasaki M, Kurokawa Y, Mori M, Doki Y. Overexpression of forkhead box M1 transcription factor (FOXM1) is a potential prognostic marker and enhances chemoresistance for docetaxel in gastric cancer. Ann Surg Oncol. 2013;20:1035-1043. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 78] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 17. | Wang R, Song Y, Xu X, Wu Q, Liu C. The expression of Nek7, FoxM1, and Plk1 in gallbladder cancer and their relationships to clinicopathologic features and survival. Clin Transl Oncol. 2013;15:626-632. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 36] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 18. | Wang Y, Huang Q, Liu J, Wang Y, Zheng G, Lin L, Yu H, Tang W, Huang Z. Vascular endothelial growth factor A polymorphisms are associated with increased risk of coronary heart disease: a meta-analysis. Oncotarget. 2017;8:30539-30551. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 27] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 19. | Marisi G, Scarpi E, Passardi A, Nanni O, Ragazzini A, Valgiusti M, Casadei Gardini A, Neri LM, Frassineti GL, Amadori D, Ulivi P. Circulating VEGF and eNOS variations as predictors of outcome in metastatic colorectal cancer patients receiving bevacizumab. Sci Rep. 2017;7:1293. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 23] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 20. | Xie Y, Mansouri M, Rizk A, Berger P. Regulation of VEGFR2 trafficking and signaling by Rab GTPase-activating proteins. Sci Rep. 2019;9:13342. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 26] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 21. | Wei Y, Sun Q, Zhao L, Wu J, Chen X, Wang Y, Zang W, Zhao G. LncRNA UCA1-miR-507-FOXM1 axis is involved in cell proliferation, invasion and G0/G1 cell cycle arrest in melanoma. Med Oncol. 2016;33:88. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 75] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 22. | Engeland K. Cell cycle arrest through indirect transcriptional repression by p53: I have a DREAM. Cell Death Differ. 2018;25:114-132. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 309] [Cited by in RCA: 488] [Article Influence: 61.0] [Reference Citation Analysis (0)] |
| 23. | Jin H, Li XJ, Park MH, Kim SM. FOXM1-mediated downregulation of uPA and MMP9 by 3,3'-diindolylmethane inhibits migration and invasion of human colorectal cancer cells. Oncol Rep. 2015;33:3171-3177. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 17] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 24. | Elaimy AL, Mercurio AM. Convergence of VEGF and YAP/TAZ signaling: Implications for angiogenesis and cancer biology. Sci Signal. 2018;11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 71] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 25. | Ramjiawan RR, Griffioen AW, Duda DG. Anti-angiogenesis for cancer revisited: Is there a role for combinations with immunotherapy? Angiogenesis. 2017;20:185-204. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 426] [Cited by in RCA: 506] [Article Influence: 63.3] [Reference Citation Analysis (0)] |
| 26. | Frezzetti D, Gallo M, Maiello MR, D'Alessio A, Esposito C, Chicchinelli N, Normanno N, De Luca A. VEGF as a potential target in lung cancer. Expert Opin Ther Targets. 2017;21:959-966. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 158] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 27. | Liang L, Yue Z, Du W, Li Y, Tao H, Wang D, Wang R, Huang Z, He N, Xie X, Han Z, Liu N, Li Z. Molecular Imaging of Inducible VEGF Expression and Tumor Progression in a Breast Cancer Model. Cell Physiol Biochem. 2017;42:407-415. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 15] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 28. | Zhang J, Niu Y, Huang C. Role of FoxM1 in the Progression and Epithelial to Mesenchymal Transition of Gastrointestinal Cancer. Recent Pat Anticancer Drug Discov. 2017;12:247-259. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 22] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 29. | O'Regan RM, Nahta R. Targeting forkhead box M1 transcription factor in breast cancer. Biochem Pharmacol. 2018;154:407-413. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 31] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 30. | Laissue P. The forkhead-box family of transcription factors: key molecular players in colorectal cancer pathogenesis. Mol Cancer. 2019;18:5. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 118] [Article Influence: 19.7] [Reference Citation Analysis (0)] |