Published online Dec 28, 2021. doi: 10.3748/wjg.v27.i48.8302
Peer-review started: April 27, 2021
First decision: June 13, 2021
Revised: June 22, 2021
Accepted: November 11, 2021
Article in press: December 11, 2021
Published online: December 28, 2021
Processing time: 240 Days and 22.7 Hours
Hepatocellular carcinoma (HCC) is one of the most common malignant tumors. The association of hepatitis B virus (HBV) infection with HCC is hitherto documented. Exosomal miRNAs contribute to cancer progression and chemoresistance. HBV X protein has been known to modulate miRNAs that facilitate cell proliferation and the process of hepatocarcinogenesis. However, there has been no report on hepatitis B core antigen (HBc) regulating exosomal miRNAs to induce drug resistance of HCC cells.
To elucidate the mechanism by which HBc promotes Doxorubicin hydrochloride (Dox) resistance in HCC.
Exosomes were isolated by ultracentrifugation. The morphology and size of exosomes were evaluated by Dynamic Light Scattering (DLS) and transmission electron microscopy (TEM). The miRNAs differentially expressed in HCC were identified using The Cancer Genome Atlas (TCGA) database. The level of miR-135a-5p in patient tissue samples was detected by quantitative polymerase chain reaction. TargetScan and luciferase assay were used to predict and prove the target gene of miR-135a-5p. Finally, we identified the effects of miR-135a-5p on anti-apoptosis and the proliferation of HCC in the presence or absence of Dox using flow cytometry, Cell counting kit 8 (CCK-8) assay and western blot.
We found that HBc increased the expression of exosomal miR-135a-5p. Integrated analysis of bioinformatics and patient samples found that miR-135a-5p was increased in HCC tissues in comparison with paracancerous tissues. Bioinformatic analysis and in vitro validation identified vesicle-associated membrane protein 2 (VAMP2) as a novel target gene of miR-135a-5p. Functional assays showed that exosomal miR-135a-5p induced apoptosis protection, cell proliferation, and chemotherapy resistance in HCC. In addition, the rescue experiment demonstrated that VAMP2 reversed apoptosis protection, cell growth, and drug resistance by miR-135a-5p. Finally, HBc promoted HCC anti-apoptosis, proliferation, and drug resistance and prevented Dox-induced apoptosis via the miR-135a-5p/VAMP2 axis.
These data suggested that HBc upregulated the expression of exosomal miR-135a-5p and promoted anti-apoptosis, cell proliferation, and chemical resistance through miR-135a-5p/VAMP2. Thus, our work indicated an essential role of the miR-135a-5p/VAMP2 regulatory axis in chemotherapy resistance of HCC and a potential molecular therapeutic target for HCC.
Core Tip: Hepatitis B virus infection is the most common cause of hepatocellular carcinoma (HCC). Drug resistance is the primary reason for the high mortality of HCC patients. We demonstrated that hepatitis B core antigen (HBc) increased exosomal miR-135a-5p. Tissue samples showed that the level of miR-135a-5p was significantly elevated in HCC tissues. Vesicle-associated membrane protein 2 (VAMP2) was demonstrated to be a target gene of miR-135a-5p. Further investigation recommended that HBc enhanced the anti-apoptosis, cell proliferation, and chemotherapy resistance of HCC cells through exosomal miR-135a-5p by targeting VAMP2. Our findings reveal that HBc can cause anti-cancer drug resistance in HCC and provide us with a novel mechanism underlying drug resistance in cancer chemotherapy.
- Citation: Wei XC, Xia YR, Zhou P, Xue X, Ding S, Liu LJ, Zhu F. Hepatitis B core antigen modulates exosomal miR-135a to target vesicle-associated membrane protein 2 promoting chemoresistance in hepatocellular carcinoma. World J Gastroenterol 2021; 27(48): 8302-8322
- URL: https://www.wjgnet.com/1007-9327/full/v27/i48/8302.htm
- DOI: https://dx.doi.org/10.3748/wjg.v27.i48.8302
Hepatocellular carcinoma (HCC) is the fourth leading cause of cancer-related death worldwide, accounting for 90% of primary liver cancer[1]. Approximately 383000 individuals die from liver cancer every year in China, accounting for 51% of liver cancer deaths worldwide[2]. Surgical resection is the cornerstone of treatment for HCC patients with early stages. However, most patients with HCC are diagnosed at an advanced stage, which prevents surgical management. Chemotherapy is the primary treatment for patients with advanced HCC. Nevertheless, drug resistance has become more and more prominent in HCC[3]. Therefore, it is essential to understand the mechanism of pathology and drug resistance in HCC.
Hepatitis B virus (HBV) is one of the major causes of HCC development in Asia, including China[4]. Studies have shown that exosomes are critical mediators of cell-to-cell communication in HBV infection[5]. Exosomes are a class of lipid bilayer vesicles 30-150 nm in size and are secreted from cells into the extracellular environment[6]. Almost all cells can secrete exosomes. The number of circulating exosomes is elevated in various diseases, including cancers. However, exosomes from different cells contain several marker proteins (CD9, CD63, and CD81)[7]. Additionally, exosomes carry some signaling molecules, such as proteins, lipids, nucleic acids, and non-coding RNAs, to the recipient cell to perform their functions[8]. Among these cargos carried by exosomes, miRNAs receive sufficient attention due to their high conservation across species and extensive regulatory roles in gene expression[9].
MicroRNAs (miRNAs) belong to small non-coding RNAs, about 19-25 nucleotides in length. MiRNAs regulate posttranscriptional gene expression by binding to the 3’ untranslated regions (3’ UTRs) of messenger RNA to induce gene silencing or degradation[10]. In cancer, exosomal miRNAs play an essential role in cell apoptosis, proliferation, and chemical resistance[11,12]. Studies have shown that abnormal expression of miRNA is closely related to HBV-associated HCC[13]. The abnormal expression of miRNAs can affect the apoptosis, proliferation, and drug resistance in HCC[14,15]. In recent years, miR-135a has emerged as a critical miRNA in several cancers[16]. Several data suggest a markedly downregulated expression of miR-135a in some diseases and cancers[17,18]. Nonetheless, a high level of miR-135a-5p is associated with postoperative recurrence of HCC[19]. Hepatitis C virus (HCV) can drive the occurrence of HCV-associated hepatocarcinogenesis by upregulating miR-135a-5p[20]. Nevertheless, there is no existing literature on the roles and molecular mechanisms of miR-135a-5p in HCC chemotherapy resistance and the relationship between miR-135a-5p and HBV.
In this study, we discovered that Hepatitis B core antigen (HBc) changed the exosomes release and enhanced the expression of exosomal miR-135a-5p. Tissues and bioinformatics analysis revealed that the level of miR-135a-5p in HCC was higher than that in normal tissues. Vesicle-associated membrane protein 2 (VAMP2) was identified as the target gene of miR-135a-5p via the online prediction website TargetScan (http://www.targetscan.org) and luciferase assay. In vitro studies indicated that miR-135a-5p promoted anti-apoptosis, proliferation, and chemoresistance in HCC by targeting VAMP2. Additional experiments revealed that HBc enhanced anti-apoptosis, cell proliferation, and chemotherapy resistance in HCC via miR-135a-5p/VAMP2. In general, this study revealed a novel mechanism of HBV which counteracted apoptosis, enhanced cell proliferation, and developed chemotherapy resistance in HCC. Our findings also suggested that miR-135a-5p might be a potential therapeutic target in the treatment of HCC chemoresistance.
The Cancer Genome Atlas (TCGA) database (http://cancergenome.nih.gov/) was used to analyze the differentially expressed miRNAs in HCC. We analyzed the data obtained from TCGA through the R package (ggplot2, rjson, ggpubr, dplyr, limma, stringr) and determined the expression of miR-135a in HCC tissues and normal tissues.
Eighteen paired HCC and adjacent tissues were collected during surgical procedures at Ren-Min Hospital of Wuhan University in China. Samples were obtained under a consensus agreement approved by the Institutional Review Committee of the School of Medicine of Wuhan University. The samples were stored at -80℃ until experiments were carried out. Table 1 shows the patients’ information.
| Characteristics | Total | miR-135a-5p | P value | |
| Negative | Positive | |||
| Age (yr) | ||||
| < 55 | 12 | 3 | 9 | 0.82 |
| ≥ 55 | 6 | 2 | 4 | |
| Gender | ||||
| Male | 8 | 1 | 7 | 0.291 |
| Female | 10 | 3 | 7 | |
| Hepatitis B s antigen | ||||
| Negative | - | |||
| Positive | 18 | 5 | 13 | - |
The HepG2 cell line was purchased from American Type Culture Collection (Manassas, VA, United States). The HBV-transfected HepG2.2.15 cell line was obtained from the Japanese Collection of Research Bioresources Cell Bank (JCRB, Osaka, Japan). The cells were incubated at 37℃ in a humidified atmosphere with 5% CO2 in Dulbecco’s modified Eagle’s medium (DMEM; Gibco, United States) with fetal bovine serum (10%, Biological Industries, China), streptomycin (0.1 mg/mL, Gibco, United States) and penicillin (100 units/mL, Gibco, United States).
Doxorubicin hydrochloride (Dox) for injection was from Shenzhen Main Luck Pharmaceutical Company (10 mg, China). Cells were treated with Dox at a concentration of 1.2 μmol/L.
The HBV (strain ayw) genome (NC_003977.2; c1903-2454) was amplified using pUC18-HBV1.3 according to sequences in NCBI and cloned into the pcDNA3.1 (-) vector. Human VAMP2 (NM_001330125.1) gene was amplified from HepG2 cDNA and cloned into the pcDNA3.1 (-) vector. Wild-type (WT) VAMP2 3’UTR (NM_001330125.1) and mutant (MUT) VAMP2 3’UTR (70-76: AGCCATA to ACGTGCA) luciferase reporter vectors were constructed and subcloned into the pmiRGLO dual-luciferase miRNA target expression vector (Promega, Wisconsin, United States). All synthesized plasmids were sequenced at Sangon Biotech, Shanghai, China, and the sequences are completely consistent.
MiR-135a-5p mimic, miR-135a-5p inhibitor, and the negative controls were synthesized at Sangon Biotech, Shanghai, China (the specific sequence is listed in Table 2). Cells with 80%-90% confluency were transfected using Lipofectamine 2000 Reagent (Invitrogen, Carlsbad, CA, United States) according to the manufacturer’s instructions.
| Category | Sequence (5’-3’) |
| miR-135a-5p mimic | F-UAUGGCUUUUUAUUCCUAUGUGA |
| R-UCACAUAGGAAUAAAAAGCCAUA | |
| miR-135a-5p inhibitor | UCACAUAGGAAUAAAAAGCCAUA |
| VAMP2 (NM_001330125.1) | F-CTAGCTAGCATGGACAGGTCTGCTAC |
| R-CGCGGATCCTTAAGTGCTGAAGT | |
| VAMP2 3’-UTR-WT | F-CTAGCTAGCATCCCCGAGGAGTCT |
| R-ACGCGTCGACAGAGAGGGGTGAAG | |
| VAMP2 3’-UTR-MUT | F-GTTCCTCCACCTCTCACGTGCATCTTTCAGCC CC |
| R-GGGGCTGAAAGATGCACGTGAGAGGTGGAGGAAC | |
| Hepatitis B virus-1903/2454 | F-CTAGCTAGCGCCACCATGGACATCGACCCTT |
| R-CCGCTCGAGCTAACATTGAGATTCCCGAGAT |
Exosomes were separated from the supernatant of cell cultures via ultracentrifugation, slightly modified, as reported[21]. Ultracentrifugation was performed using a fixed angle 70 Ti rotor (Beckman optimal L-100XP, CA, United States) with a speed of 110000 × g at 4°C for 70 min. The precipitate was refrigerated at -80℃ until it was used in the experiment.
For transmission electron microscopy (TEM), 10 μL of exosome suspension was absorbed onto carbon-coated copper grids (200 mesh) for 5 min. Samples were stained with 2% uranyl acetate for 2 min. After air drying, the sample was visualized under a microscope at 80 kV in TEM (HT7700, Tokyo, Japan).
Particle size distribution of purified exosomes was evaluated using dynamic light scattering (DLS). Briefly, about 200 μL of exosome sample was diluted in 1.5 mL PBS. DLS measurement was conducted using a Zetasizer Nano ZSP (Malvern Instruments Ltd., United Kingdom).
Exosomes isolated from HepG2 cells transfected with miR-135a-5p mimic were stained with PKH67 membrane dye (UR52303, Umibio, Shanghai, China) according to the manufacturer’s instructions. HepG2 cells were cultured in confocal Petri dishes 20 mm in diameter (801001, Nest Scientific USA Inc.). When confluency of 70%-80% was reached, 2.5 μg of PKH67-labeled exosomes was added to each well. After incubation for 4 h, the cells were washed with PBS and then stained with 0.5 μg of 4’, 6-diamidino-2-phenylindole (DAPI, Solarbio, Beijing, China) at 37°C. Cellular uptake of PKH67-labeled exosomes was visualized using confocal laser scanning microscopy LCS SP8 (Leica, Wetzlar, Germany).
Total RNA was extracted from HCC tissues, HCC cell lines and exosomes using TRIzol reagent (Invitrogen, Carlsbad, CA, United States) and complementary DNA (cDNA) was synthesized using ReverTra Ace quantitative real-time polymerase chain reaction (qPCR) RT Master Mix with gDNA Remover (TOYOBO, Osaka, Japan). qPCR was carried out using SYBR Green I dye master mix (Invitrogen, Carlsbad, CA, United States). The primer sequences are listed in Table 2. The mRNA expression levels of genes were normalized by glyceraldehyde-3-phosphate dehydrogenase (GAPDH) or U6.
Primer Premier 5.0 software (Premier, Delaware, Canada) was used to design the primers (primers for vector construction are listed in Table 2; qPCR Primers are listed in Table 3).
| Primer | Product size | Sequence (5’-3’) |
| VAMP2 (NM_001330125.1) | 182 bp | F-GGTCTCTCCTGCGTTCCC |
| R-TCGACCCGAAAAGACAGGC | ||
| GAPDH (NM_002046.7) | 197 bp | F-GGAGCGAGATCCCTCCAAAAT |
| R-GGCTGTTGTCATACTTCTCATGG | ||
| U6 (NR_004394.1) | 94 bp | F-CTCGCTTCGGCAGCACA |
| R-AACGCTTCACGAATTTGCGT | ||
| miR135a (NR_029677.1) | 68 bp | F-ACACTCCAGCTGGGTATGGCTTTTTATTCCT |
| R-TGGTGTCGTGGAGTCG |
Cells were collected for protein extraction using M-PER reagents (Pierce Chemical, Rockford, IL, United States) after 48 h transfection. Total protein content was quantified using the BCA Protein Quantification kit (Thermo Fisher Scientific, Waltham, MA, United States). Protein samples were separated on 12% SDS-polyacrylamide gel and transferred to polyvinylidene fluoride membranes (Millipore, United States). After blocking, the membranes were incubated with primary antibodies overnight at 4℃ and then with secondary antibodies for 1 h at room temperature. Using ECL chemiluminescence solution (Biosharp, Hefei, China), the band signal was visualized in an automatic chemiluminescence system (Tanon5200, Shanghai, China). The antibodies used in this article were all purchased from ABclonal (Wuhan, China), including anti-GAPDH (AC002), anti-VAMP2 (A1249), anti-CD63 (A5271), anti-CD9 (A1703), anti- Calnexin (CANX, A15631), anti-proliferating cell nuclear antigen (PCNA, A0264), anti-mini-chromosome maintenance protein-2 (MCM2, A1056), anti-B-cell lymphoma-2 (Bcl-2, A0208).
The luciferase reporter vectors containing the 3'UTR-WT or 3'UTR-MUT of VAMP2, along with miR-135a-5p mimics or negative control (NC), respectively, were co-transfected into HepG2 cells. Luciferase activities were assessed using the Dual Glo Luciferase Assay System (Promega, Madison, WI, United States) according to the manufacturer’s instructions.
After washing and collecting, cells were treated with the Annexin V-FITC/PI Apoptosis Assay Kit (Zomanbio, Beijing, China) according to the manufacturer's instructions. The apoptosis rate of cells was analyzed by flow cytometry (FACS Aria III, BD, United States) with FlowJo v10 software (Leonard Herzenberg, United States).
Cell counting kit 8 (CCK-8) (Zomanbio, Beijing, China) was used to assess cell proliferation according to the manufacturer’s instructions. Generally, 5 × 10³ cells were allowed to grow in 96-well plates. After incubation with Dox or tumor-derived exosomes for 0 h, 24 h, and 48 h, 10 μL CCK-8 solution was added to each sample and incubated for a further 30 min. The absorbance value was measured at 450 nm using the micro-plate reader.
Each experiment was carried out using at least three replicates. Clinical data analysis was performed using SPSS25.0. R software for bioinformatics analysis. Other data analysis was carried out with GraphPad Prism 5 software (GraphPad Software Inc., San Diego, CA, United States), and data were mentioned as mean ± standard error of the mean (SEM). The t-test was implemented to compare the data between 2 groups. P < 0.05 was considered to represent a statistically significant difference (aP < 0.05, bP < 0.01, cP < 0.001).
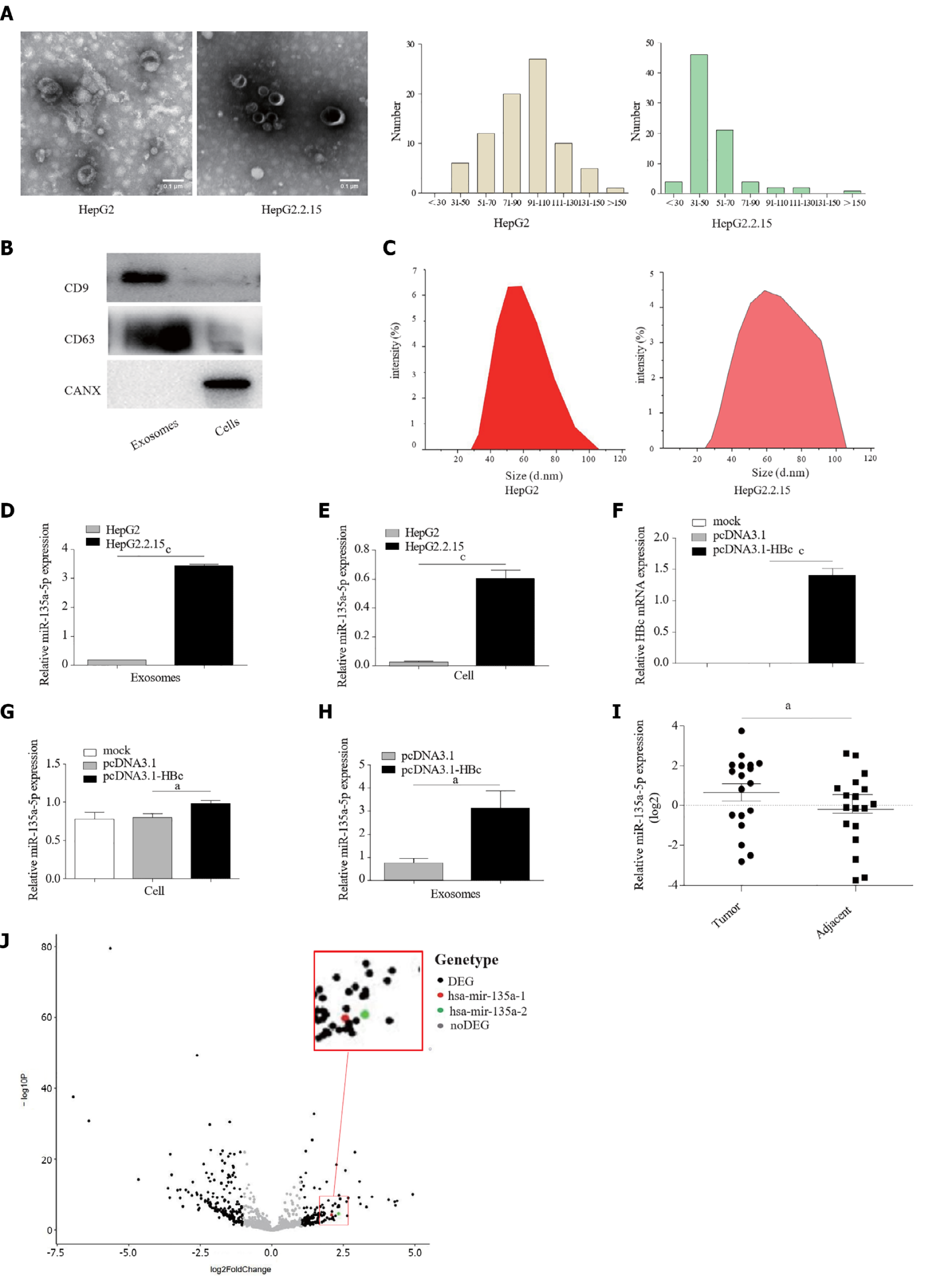
HBV infection changes the release of extracellular vesicles (EVs) from hepatocytes[22]. In this study, we extracted EVs from HepG2 cells and HepG2.2.15 cells. The TEM image showed that the EVs had a classic “cup” or “dish” morphology[23] (Figure 1A). EVs secreted from HepG2.2.15 cells with HBV replication contained exosomes, subviral particles, and virions[24]. Therefore, western blotting was utilized to verify the marker proteins of exosomes. The results revealed that CD63 and CD9, which commonly serve as specific marker proteins of exosomes, were present in purified EVs (Figure 1B). The negative control Calnexin was detected only in the cell lysate. Furthermore, DLS results demonstrated that the distribution of isolated EVs ranged from 30 nm to 150 nm (Figure 1C). These results suggested that we successfully isolated exosomes from HepG2 cells and HepG2.2.15 cells.
The miRNA content in exosomes is likely responsible for cancer progression, including anti-apoptosis, cell proliferation, and chemoresistance[12]. Notably, we detected the expression of several miRNAs in exosomes purified from HepG2 cells and HepG2.2.15 cells. The qPCR results indicated that the expression level of miR-135a-5p in exosomes isolated from HepG2.2.15 cells was significantly higher than that of HepG2 cells (Figure 1D). The same results were derived in cells (Figure 1E).
HCV promotes the expression of miR-135a-5p in HCC[25]. To our knowledge, there is no report on the effect of HBV on miR-135a-5p. Here, we found that high expression of HBc (Figure 1F) could significantly upregulate the level of miR-135a-5p (Figure 1G) in HCC cells and exosomes (Figure 1H). Moreover, patient tissue samples showed increased expression of miR-135a-5p in HCC tissues compared to paracancerous tissues (Figure 1I). TCGA data analysis identified high expression of miR-135a-5p in HCC tissues (Figure 1J). These results indicated that HBc might upregulate the expression of miR-135a-5p in HCC cell-derived exosomes.
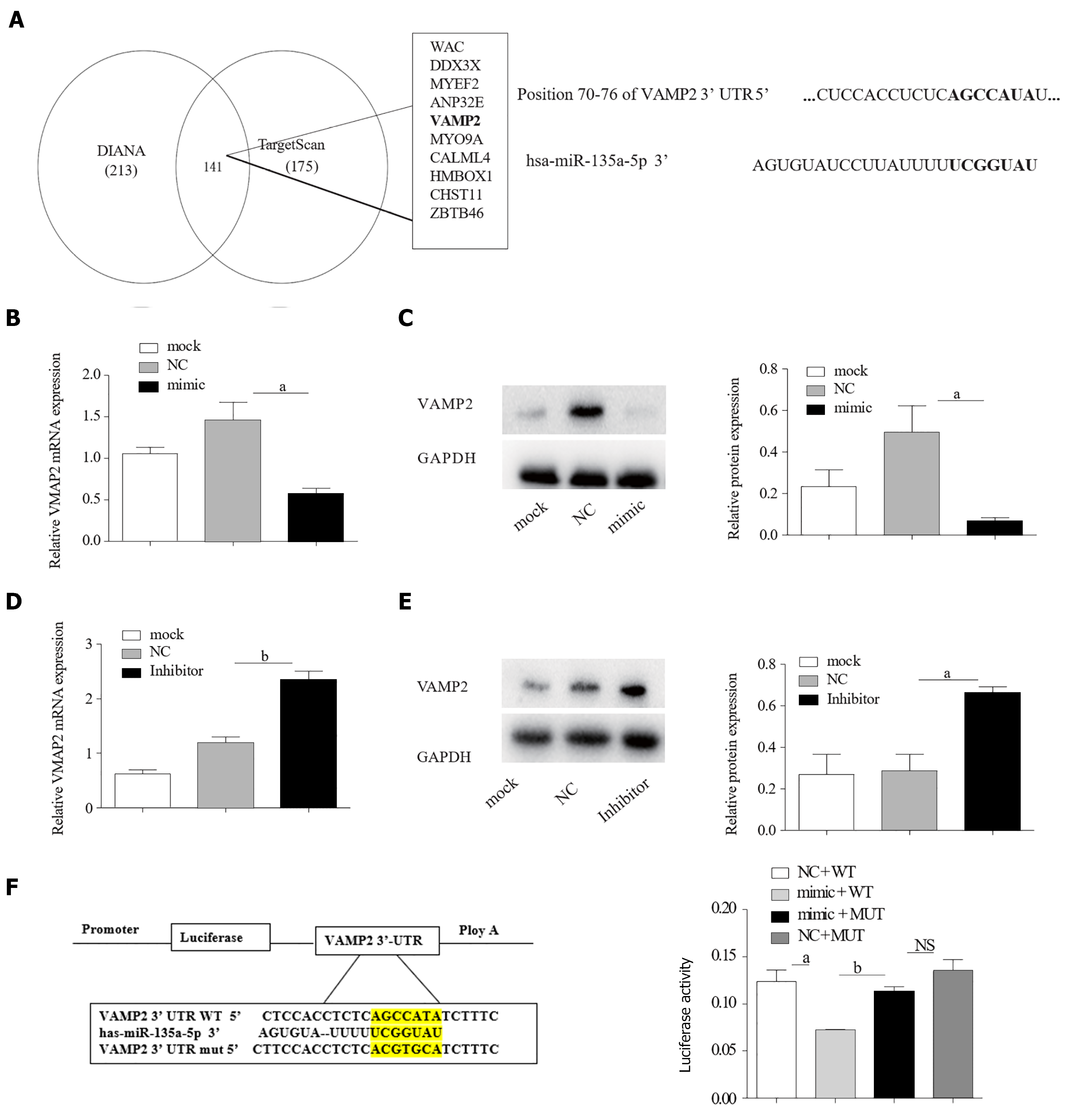
Generally, miRNAs exert their functions by inhibiting downstream target genes[26]. Thus, it is important to identify the biological targets of miR-135a-5p. Subsequently, TargetScan[27] and DIANA[28] predicted a potential binding site of miR-135a-5p on the 3’-UTR of VAMP2 (Figure 2A). To validate this bioinformatic prediction, HepG2 cells were transfected with miR-135a-5p mimics (mimic-HepG2). A high level of miR-135a-5p was found in HepG2 cells by qPCR assays (Figure 2B) and a down-regulation was seen when miR-135a-5p inhibitor was involved. The results of qPCR and western blot indicated miR-135a-5p inhibited the expression of VAMP2 (Figure 2C and D) and elevated VAMP2 mRNA and protein were observed (Figure 2F and G) when miR-135a-5p was knocked down (Figure 2E). Moreover, the fluorescence intensity in the cells co-transfected with miR-135a-5p and EGFP-VAMP2-3′UTR was significantly decreased as compared with that in the controls (Figure 2F), indicating that miR-135a-5p interacted with VAMP2. These results suggested that VAMP2 might be a target gene of miR-135a-5p.
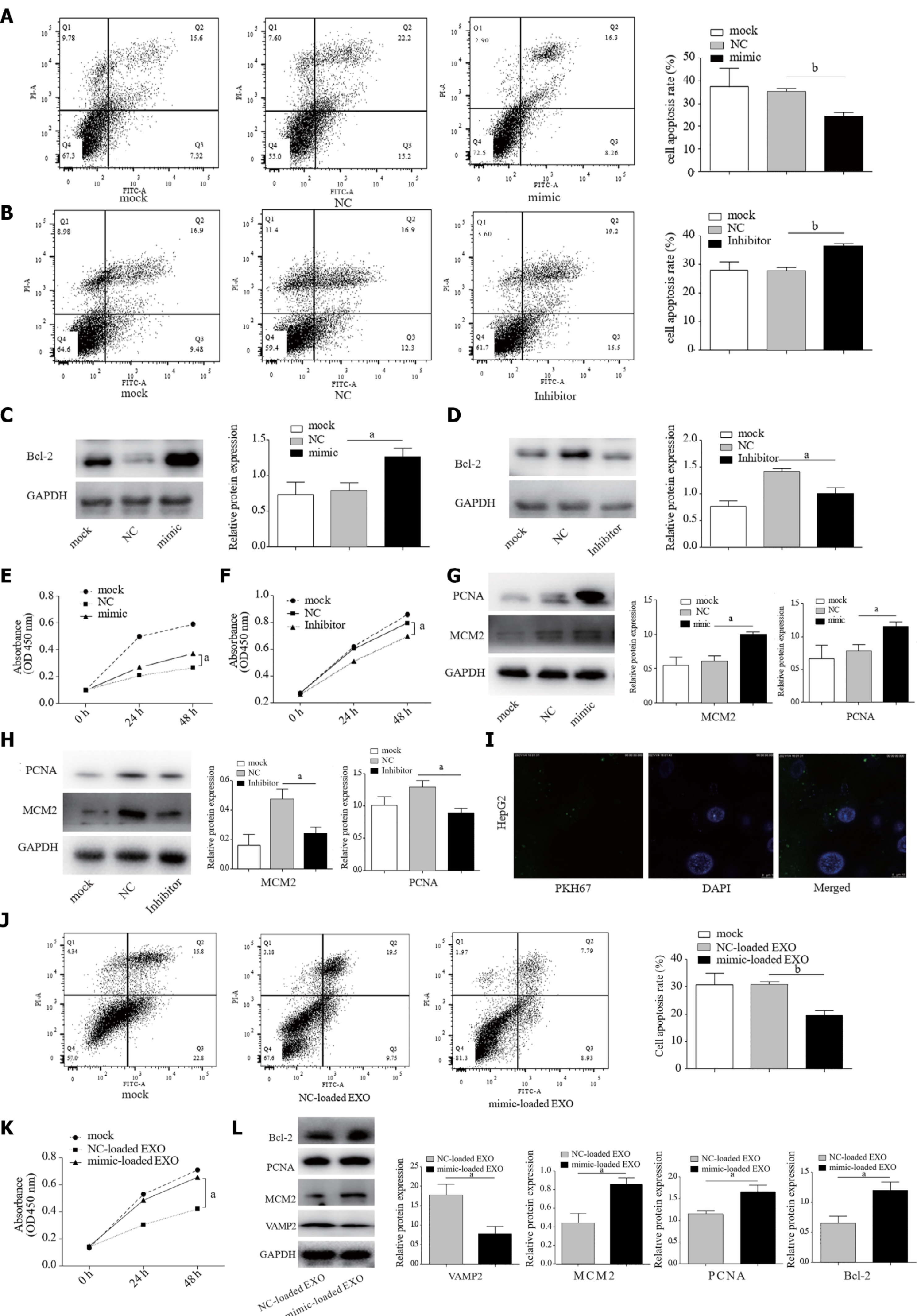
Our molecular analysis of patient tissue samples found that miR-135a-5p increased in HCC. Apoptosis can eliminate cancer cells. Apoptosis resistance commonly occurs in HCC[29]. Our experiment demonstrated reduced apoptosis in mimic-HepG2 cells when compared to the control group (Figure 3A). Moreover, miR-135a-5p inhibitor effectively increased apoptosis compared to control (Figure 3B). Western blot showed that miR-135a-5p enhanced the expression of Bcl-2 protein, one of the most common anti-apoptotic proteins[30] (Figure 3C), while the level of Bcl-2 protein was decreased in HepG2 cells transfected with miR-135a-5p inhibitor (inhibitor-HepG2) (Figure 3D).
Suppression of apoptosis can lead to cell proliferation[31], one of the prerequisites for cancer progression or carcinogenesis[32]. We found that miR-135a-5p promoted HCC cell proliferation as compared with the control group (Figure 3E). Subsequently, miR-135a-5p inhibitor suppressed cell proliferation in HepG2 cells (Figure 3F). PCNA[33] and MCM2[34] are the traditional proliferating protein molecules. MiR-135a-5p upregulated the expression levels of PCNA and MCM2 (Figure 3G), while miR-135a-5p inhibitor downregulated the levels of these two genes in HepG2 cells (Figure 3H).
Our previous study suggested increased miR-135a-5p in exosomes from HepG2.2.15 cells. Here, we found that these purified exosomes from HepG2 cells transfected with miR-135a-5p mimic (mimic-loaded EXO) could be absorbed by HepG2 cells (Figure 3I). QPCR showed an increased level of miR-135a-5p and a decreased expression of VAMP2 in the recipient cells (Supplementary Figure 1). It is worth mentioning that after absorbing exosomes, the recipient cells exerted anti-apoptotic (Figure 3J) and proliferative effects (Figure 3K). Interestingly, the protein expression levels of Bcl-2, PCNA, and MCM2 increased in the recipient cells, while target gene VAMP2 decreased (Figure 3L).
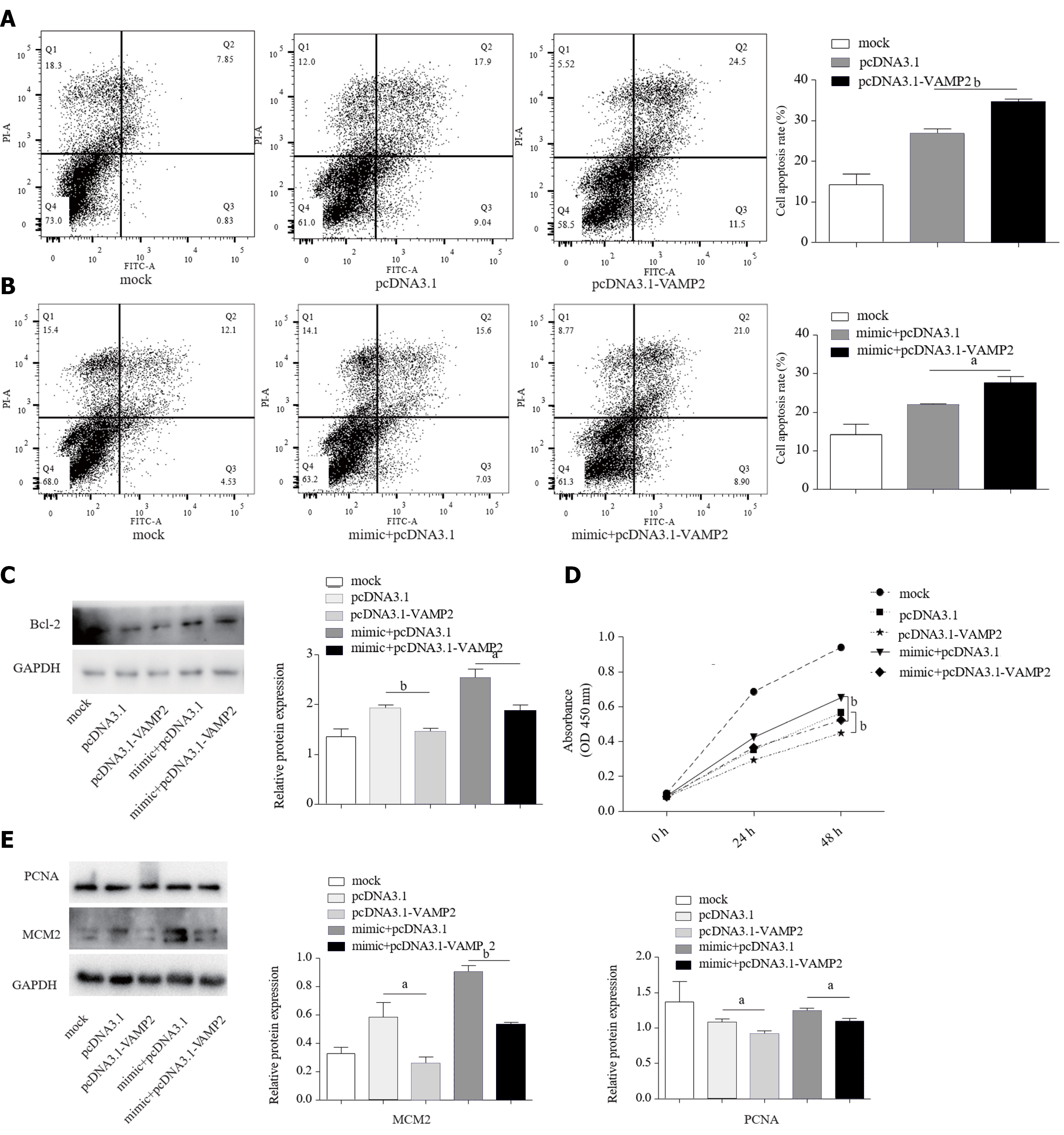
As the target gene of miR-135a-5p, increased VAMP2 (Supplementary Figure 2) induced the apoptosis in HepG2 cells (Figure 4A). As miR-135a-5p induced anti-apoptosis, we also measured the apoptosis rates in mimic-HepG2 cells co-transfected with pcDNA3.1-VAMP2 or pcDNA3.1 and found that VAMP2 led to excessive apoptosis (Figure 4B). Western blot demonstrated that VAMP2 markedly down-regulated the Bcl-2 protein (Figure 4C). The CCK-8 assay further demonstrated that VAMP2 restrained the proliferation of HCC cells (Figure 4D). In addition, VAMP2 suppressed the protein levels of PCNA and MCM2 in HepG2 cells (Figure 4E).
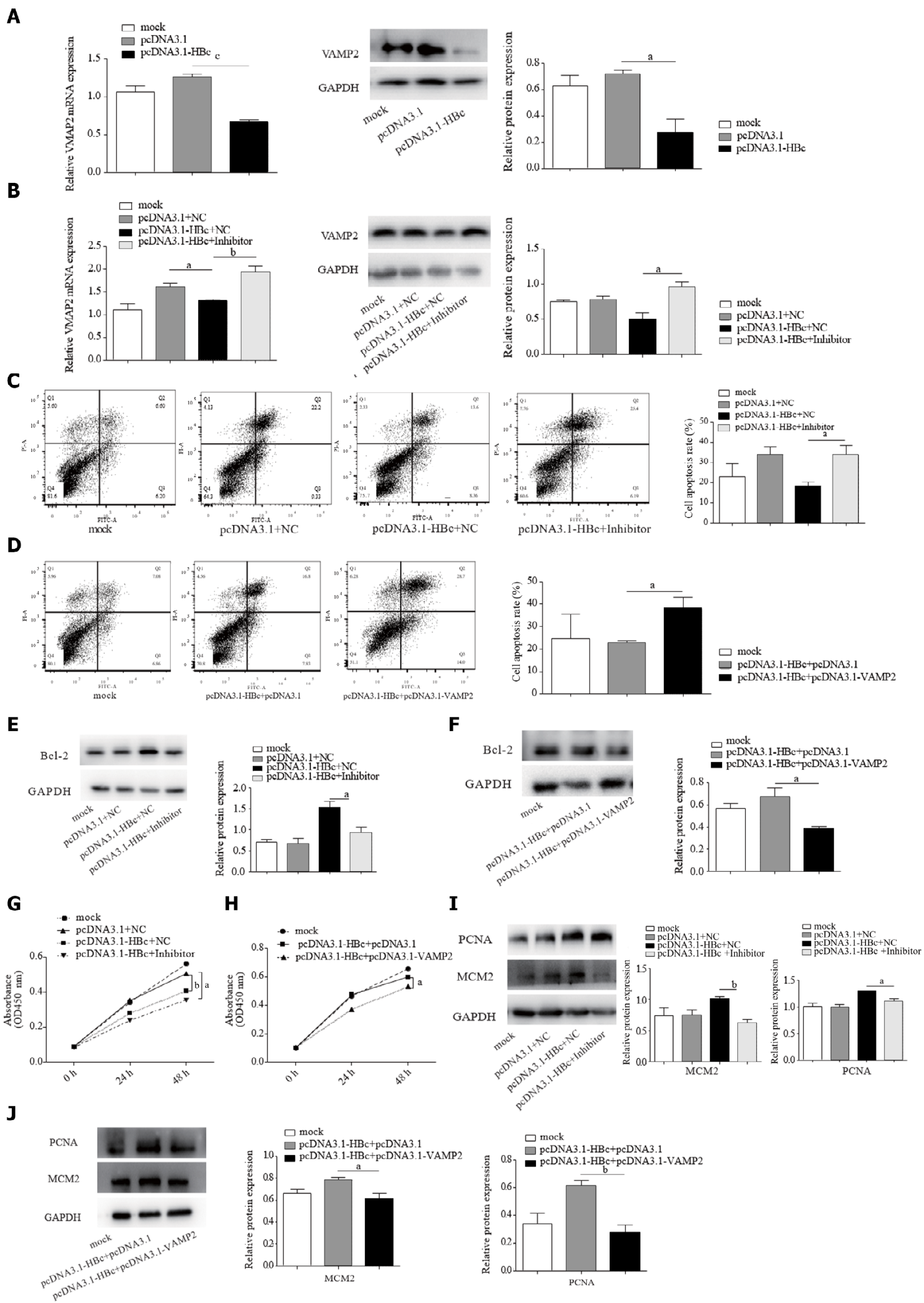
HBc protects HCC cells against apoptosis and promotes proliferation by miR-135a-5p/VAMP2
HBc has been reported to inhibit apoptosis[35] and promote HCC proliferation[36]. Our data also confirmed this (Supplementary Figure 3). Combined with our data that suggested that HBc upregulated miR-135a-5p, we attempted to determine the functions of miR-135a-5p and its target VAMP2 in the process of anti-apoptosis and proliferation induced by HBc. HBc restrained the expression of VAMP2 in HCC (Figure 5A). Noticeably, we found that miR-135a-5p inhibitors recovered the level of VAMP2 (Figure 5B). To further investigate the role of the miR-135a-5p/VAMP2 axis in the effect of HBc on anti-apoptosis, we co-transfected HBc and miR-135a-5p inhibitors or VAMP2 into HepG2 cells. The data showed that the expression of miR-135a-5p was decreased (Supplementary Figure 4A), and VAMP2 was upregulated (Supplementary Figure 5). As expected, both miR-135a-5p inhibitors (Figure 5C) and VAMP2 (Figure 5D) reversed the effect of HBc against apoptosis. Western blotting showed that anti-apoptotic protein decreased (Figure 5E and F). Subsequently, both miR-135a-5p inhibitors (Figure 5G) and VAMP2 (Figure 5H) impaired the enhancement of HCC cell proliferation by HBc. In addition, MCM2 and PCNA decreased (Figure 5I and J). These results suggested that HBc protects HCC cells against apoptosis and promotes proliferation by miR-135a-5p/VAMP2.
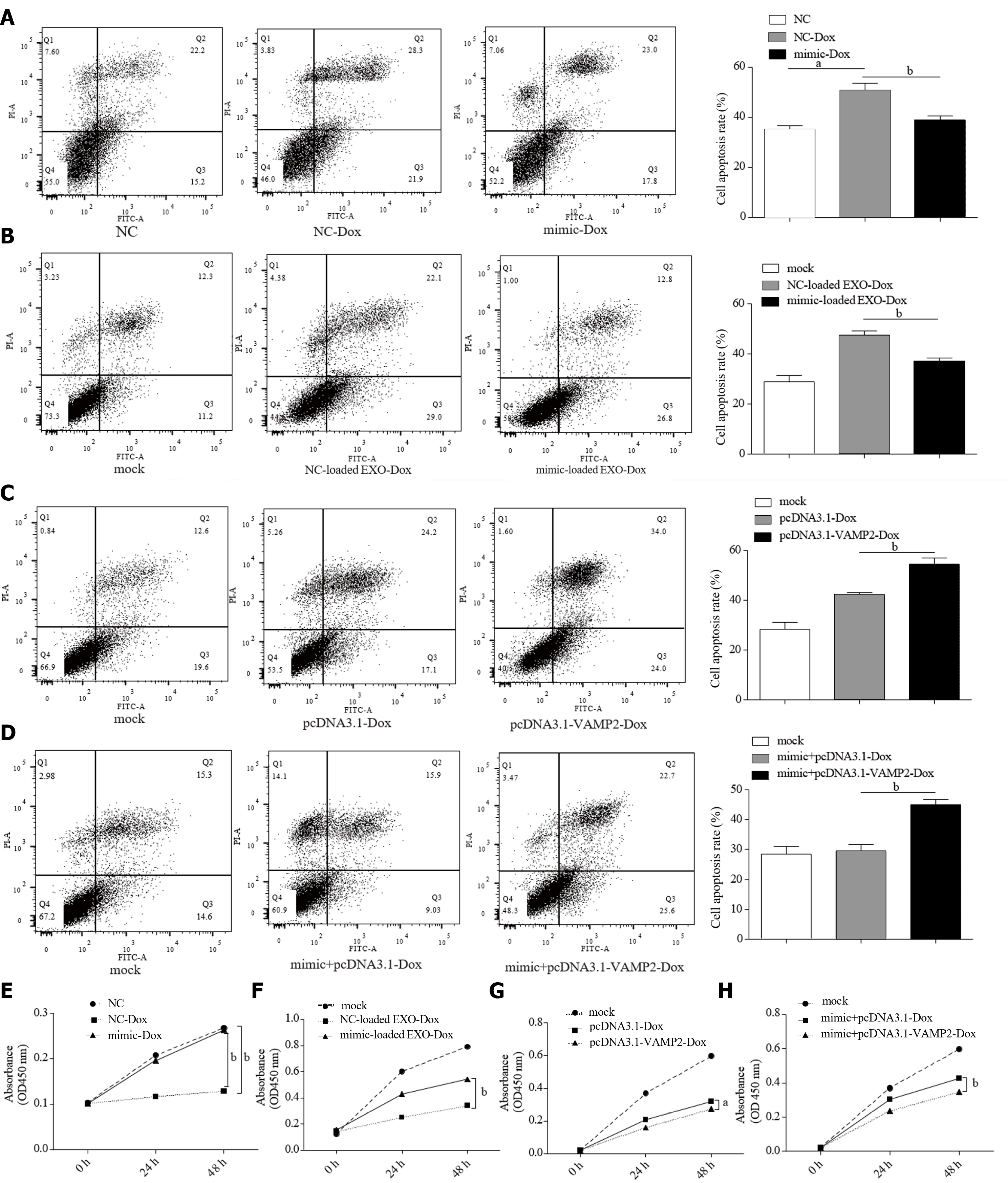
Cell survival and proliferation usually counter the chemotherapy drug effect[37]. Herein, we tried to demonstrate whether miR-135a-5p/VAMP2 is involved in the resistance to anti-cancer drugs. Intriguingly, miR-135a-5p reversed the apoptosis caused by Dox (Figure 6A). Mimic-loaded EXO confirmed this result (Figure 6B). On the contrary, VAMP2 enhanced the effect of Dox-induced apoptosis in HepG2 cells (Figure 6C). The results from co-transfected miR-135a-5p mimics and pcDNA3.1-VAMP2 suggested that VAMP2 reversed Dox resistance induced by miR-135a-5p (Figure 6D).
Similarly, miR-135a-5p recovered cell proliferation in HepG2 cells treated with Dox (Figure 6E and F). Moreover, VAMP2 played a critical role in the Dox resistance triggered by miR-135a-5p (Figure 6G and H). Taken together, these results suggest that miR-135a-5p could be transported to other cells by exosomes and lead to Dox resistance of recipient cells by down-regulating VAMP2.
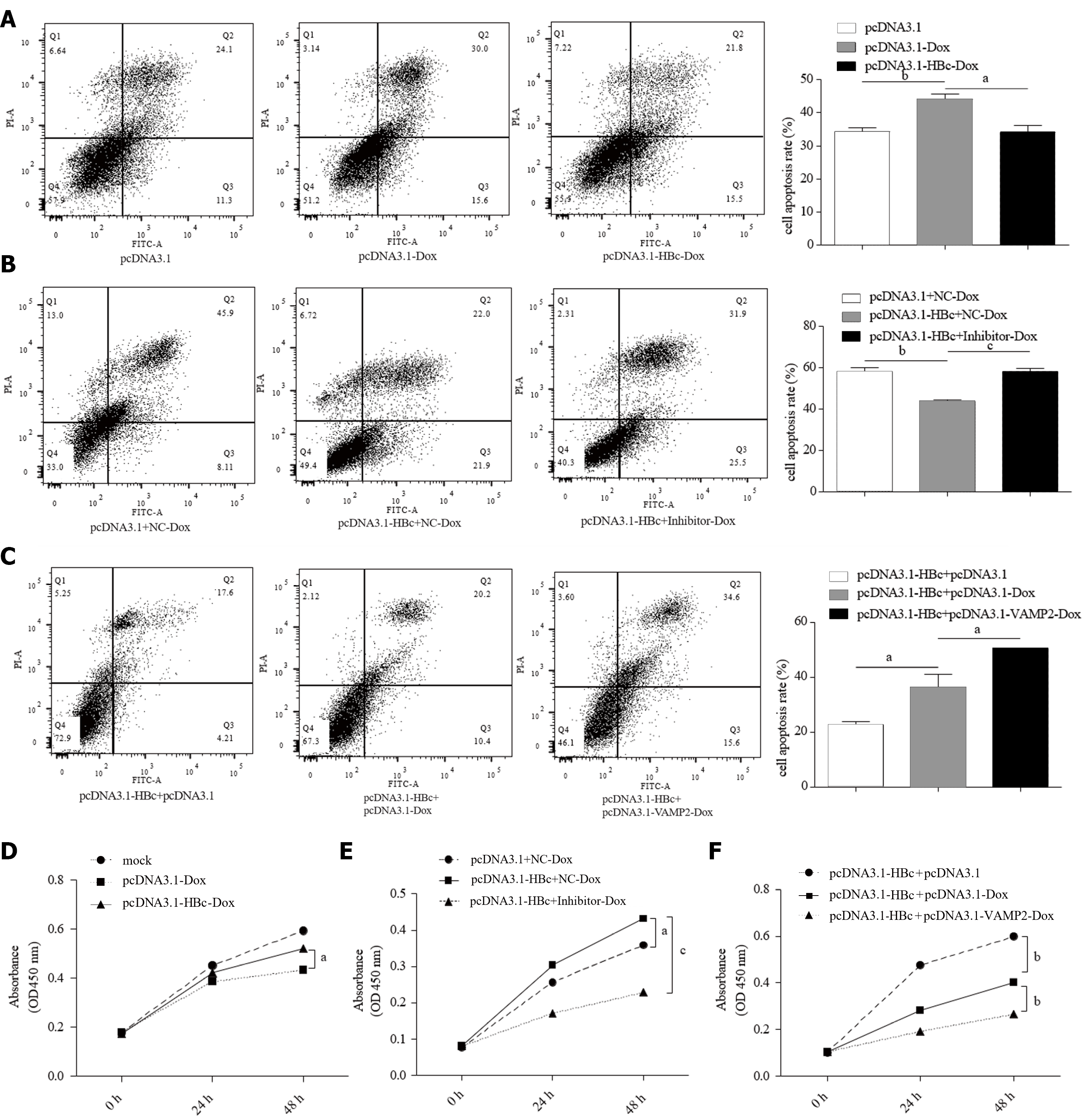
Dox can directly promote HBV replication[38]. However, there are no publicly available data on the effect of HBV or HBV proteins on the chemotherapy resistance of HCC. We noted that HBc protects HCC cells against apoptosis in the Dox treatment groups (Figure 7A). Since HBc increased miR-135a-5p and decreased VAMP2, we co-transfected HBc and miR-135a-5p inhibitors or VAMP2 in HepG2 cells. Flow cytometry revealed that the apoptosis rate was higher in HepG2 cells co-transfected with pcDNA3.1-HBc plasmid and miR-135a-5p inhibitors than in the control after treatment with Dox (Figure 7B). Similarly, VAMP2 also recovered the apoptosis rate (Figure 7C), suggesting that miR-135a-5p/VAMP2 participated in the HBc-mediated chemotherapy resistance of HCC.
The cell proliferation assay further demonstrated that HBc mediated resistance of HCC cells to Dox (Figure 7D) and miR-135a-5p/VAMP2 played an essential role in this (Figure 7E and F). In summary, HBc mediated Dox resistance in HCC cells via miR-135a-5p/VAMP2.
Chronic HBV infection is still a significant risk factor for HCC. Various studies have underlined the usefulness of exosomal miRNAs as potential biomarkers to detect early stages of HBV-related HCC[39]. Hepatitis B virus X protein (HBx) has been reported to modulate several exosomal miRNAs that facilitate the process of hepatocarcinogenesis[22]. A recent finding revealed that HBc promotes liver cancer metastasis through the miR-382-5p/DLC-1 axis[40]. However, it is less clear on the effect of HBc on drug resistance in HCC. Here, we reported that HBc reduced apoptosis, induced cell proliferation, and mediated resistance of HCC to chemotherapeutic drugs by increasing and modulating exosomal miR-135a-5p to target VAMP2.
Viral infections can induce exosomal cargos, including miRNAs, to change them profoundly[41]. This study successfully isolated exosomes from HepG2 cells and HepG2.2.15 cells and found that HBc could induce the overexpression of miR-135a-5p in exosomes. HBV-associated miRNAs can distinguish HBV-related HCC from healthy controls[39]. Our clinical data revealed that miR-135a was upregulated in liver cancer tissues, consistent with other studies.
As a small non-coding RNA, miRNA mainly inhibits the expression of downstream target genes. Most miRNAs may regulate more than one target gene[42]. Forkhead box O1 (FOXO1)[43], protein tyrosine phosphatase receptor delta (PTPRD)[20], Kruppel-like factor-4 (KLF4)[44], signal transducer and activator of transcription 6 (STAT6)[45], ELK1 and ELK3[46] have been proven to be direct target genes of miR-135a-5p. We tried to identify a novel target gene of miR-135a-5p in HCC. Both TargetScan and DIANA predicted VAMP2 as a candidate target gene of miR-135a-5p. The present study verified the prediction and added VAMP2 as one more target gene of miR-135a-5p.
Exosomal miRNAs have a significant function in the regulation of tumor progression[47]. Numerous studies have suggested that miR-135a has shown protective effects under some conditions[46,48,49]. Zhou and his collaborators showed that apoptosis was induced by miR-135a through the Janus kinase 2 (JAK2)/signal transducer and activator of transcription 3 (STAT3) signaling pathway in human renal cancer cells[50]. Moreover, miR-135a-5p also induces the apoptosis of glioma[48] and cardiomyocyte cells[51], whereas miR-135a-5p inhibitor significantly protects nerve cells against epilepsy-induced apoptosis[52]. However, our findings suggested an opposite role of miR-135a-5p in mediating cell apoptosis, indicating that miR-135a-5p might serve a dual role as a regulator of cancer progression. In gastric cancer, miR-135a has been reported to have an anti-apoptotic effect consistent with our results[53].
Abnormal cell apoptosis is one of the causes of excessive proliferation and oncogenesis[31]. It is interesting to note that miR-135a-5p also exerts different functions in cell proliferation. It is clear that miR-135a-5p acts as a tumor suppressor miRNA in some cancers, including prostate cancer[45], renal carcinoma cells[50], nasopharyngeal carcinoma[54], and as an oncogenic miRNA in bladder cancer[55] and HCC[19,44]. Our experimental results also demonstrated that miR-135a-5p acts as an onco-miRNA to promote HCC proliferation via inhibition of VAMP2. Many recent studies showed that the same individual miRNA has different purposes in different diseases[56]. This study also showed that miR-135a has a distinct purpose in HCC, implying that miR-135a might also play diverse roles in different cancers. Therefore, the effects of miR-135a on diseases depend on its target genes.
There are two different conclusions regarding the effect of HBc on apoptosis in HCC[57,58]. Several studies report that HBc, involved in HBV self-regulation, can inhibit apoptosis or enhance anti-apoptosis in HCC[35,57]. Liu and his partners reported that HBc inhibits Fas-mediated hepatocyte apoptosis[35]. Du et al[57] found that HBc enhances anti-apoptosis of hepatocytes by blocking death receptor 5 (DR5) expression. On the contrary, researchers in the Institut Pasteur of Shanghai revealed that HBc increases tumor necrosis factor alpha (TNF-α) -induced apoptosis in HCC cells[58]. Our experimental results showed that HBc prevented cell apoptosis and promoted cell proliferation through the miR-135a-5p/VAMP2 axis in HCC cells, which is similar to the report that HBc fosters the proliferation of HCC by upregulating the expression of c-Ets2[36].
Chemotherapy is the primary treatment for patients with advanced cancer. Exosomes secreted by drug-resistant cell lines can deliver miRNAs to sensitive cells and induce drug-resistant characteristics[59]. A few articles describe that miR-135a increases chemical resistance in some cancers[60-62]. Upregulation of miR-135a contributes to paclitaxel resistance in human non-small cell lung cancer cells[60]. High levels of miR-135b-5p promote resistance to cisplatin treatment in endometrial cancer cells[62] and gastric cancer cells[63]. MiR-135a also seems to have different effects on drug resistance, as well as cell apoptosis. A report from Nanjing Medical University shows that enforced miR-135a/b expression sensitizes A549/Cisplatin (CDDP) cells to CDDP-induced apoptosis[64]. Our results suggested that miR-135a-5p could resist Dox-induced apoptosis by targeting VAMP2 in HCC.
Our research group and other groups have published several articles on HBx protein promoted chemotherapeutic resistance in HCC[65-67]. A recently published paper concludes that HBx protein leads to resistance to the chemotherapy drug 5-Fluorouracil in HCC by downregulating SHIP2 through SKP2[65]. We also reported that HBx protein can promote Dox chemoresistance in HCC through overexpression of Variant 1 of KIAA0101[66] and Transcript variant 2 of the chemokine-like factor (CKLF1)[67]. However, there is no relevant study to assess the effect of HBc on HCC drug resistance. Herein, we found that HBc protected HCC from Dox-induced apoptosis through the miR-135a-5p/VAMP2 axis.
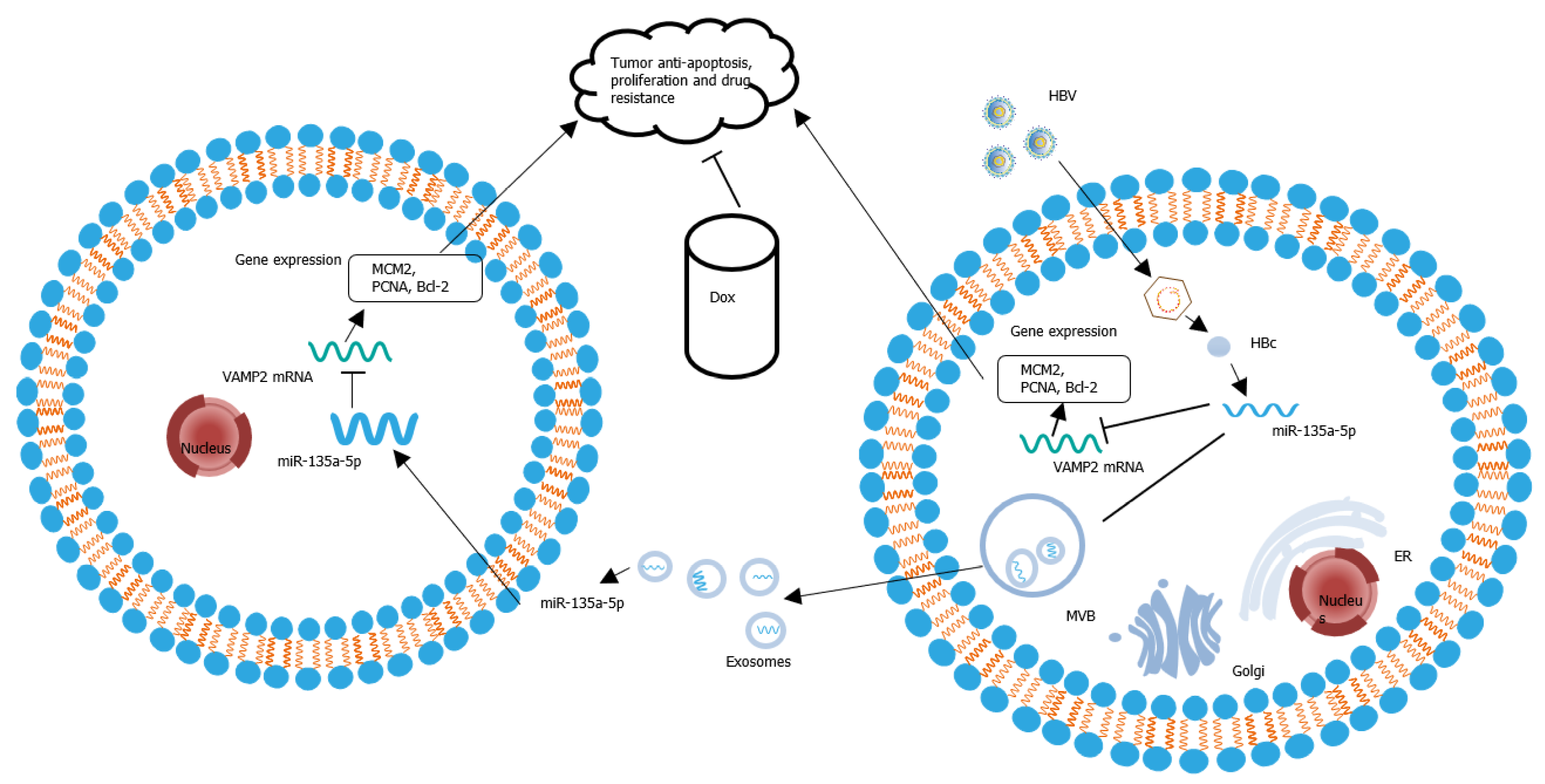
HBc could upregulate the expression of miR-135a-5p in HBV-infected hepatocytes. Then, miR-135a-5p was packaged into exosomes. After adjacent or distant recipient cells absorbed these exosomes, miR-135a-5p was delivered into recipient cells and led to a decrease in VAMP2 transcription, a novel target gene. The decreased VAMP2 facilitated tumor anti-apoptosis, cell proliferation, and drug resistance in HCC (Figure 8).
Hepatocellular carcinoma (HCC) is a frequently diagnosed malignant tumor caused by its main risk factor, hepatitis B virus (HBV) infection. HBV infection alters the level of miRNA in cells, which can be delivered to surrounding cells by exosomes to affect disease progression.
HCC is a common malignant tumor with relatively insipid early symptoms, rapid disease progression, burdensome treatment, and poor prognosis. Since HBV infection is still one of the major causes of HCC in China, the mechanism of HBV in HCC resistance remains unclear.
To explore the role of hepatitis B core antigen (HBc) on Dox-induced HCC resistance and the underlying mechanism.
Exosomes were isolated by ultracentrifugation. The miRNAs differentially expressed in HCC were identified using the Cancer Genome Atlas (TCGA) database. The level of miR-135a-5p in patient tissues and exosomes was detected by quantitative polymerase chain reaction. After transfection with the indicated plasmids, cell functions affected by the HBV-regulated miR-135a/vesicle-associated membrane protein 2 (VAMP2) axis were assessed by flow cytometry and cell counting kit 8 assay.
miR-135a-5p expression was upregulated in HCC tissues and cells. HBc increased the expression of exosomal miR-135a-5p. VAMP2 is one of the potential target genes of miR-135a-5p, and functional assays showed that HBc mediated the miR-135a/VAMP2 axis to induce apoptosis protection, cell proliferation, and chemotherapy resistance in HCC.
HBc elevated the expression of exosomal miR-135a-5p and promoted anti-apoptosis, cell proliferation, and chemical resistance through miR-135a-5p/VAMP2 in HCC.
The role of the miR-135a-5p/VAMP2 regulatory axis in chemotherapy resistance of HCC may serve as a potential molecular therapeutic target for HCC.
We are very grateful to Wang Y for her skillful statistical analysis guidance. Many thanks to Medical Research Center for Structural biology of Wuhan University for providing Ultracentrifuge- Beckman optimal L-100XP; Transmission electron microscopy- HT7700; Confocal microscopy- Leica-LCS-SP8-STED; Flow cytometry- FACS AriaIII and Wuhan University Testing Center for providing Dynamic Light Scatterer-Zetasizer Nano ZSP. We appreciate the contributors of TCGA databases for providing open access resources for exploring cancer genomics data.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Sahin TT, Yu JS S-Editor: Wu YXJ L-Editor: Webster JR P-Editor: Wu YXJ
| 1. | Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53206] [Cited by in RCA: 55820] [Article Influence: 7974.3] [Reference Citation Analysis (132)] |
| 2. | Wang FS, Fan JG, Zhang Z, Gao B, Wang HY. The global burden of liver disease: the major impact of China. Hepatology. 2014;60:2099-2108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 985] [Cited by in RCA: 943] [Article Influence: 85.7] [Reference Citation Analysis (4)] |
| 3. | Llovet JM, Montal R, Sia D, Finn RS. Molecular therapies and precision medicine for hepatocellular carcinoma. Nat Rev Clin Oncol. 2018;15:599-616. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1458] [Cited by in RCA: 1382] [Article Influence: 197.4] [Reference Citation Analysis (0)] |
| 4. | El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557-2576. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3846] [Cited by in RCA: 4265] [Article Influence: 236.9] [Reference Citation Analysis (2)] |
| 5. | Yang Y, Han Q, Hou Z, Zhang C, Tian Z, Zhang J. Exosomes mediate hepatitis B virus (HBV) transmission and NK-cell dysfunction. Cell Mol Immunol. 2017;14:465-475. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 115] [Cited by in RCA: 172] [Article Influence: 19.1] [Reference Citation Analysis (0)] |
| 6. | Kalluri R. The biology and function of exosomes in cancer. J Clin Invest. 2016;126:1208-1215. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 944] [Cited by in RCA: 1418] [Article Influence: 157.6] [Reference Citation Analysis (0)] |
| 7. | Kowal J, Arras G, Colombo M, Jouve M, Morath JP, Primdal-Bengtson B, Dingli F, Loew D, Tkach M, Théry C. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc Natl Acad Sci U S A. 2016;113:E968-E977. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2360] [Cited by in RCA: 2571] [Article Influence: 285.7] [Reference Citation Analysis (0)] |
| 8. | Wu Q, Zhou L, Lv D, Zhu X, Tang H. Exosome-mediated communication in the tumor microenvironment contributes to hepatocellular carcinoma development and progression. J Hematol Oncol. 2019;12:53. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 113] [Cited by in RCA: 212] [Article Influence: 35.3] [Reference Citation Analysis (0)] |
| 9. | Zhang J, Li S, Li L, Li M, Guo C, Yao J, Mi S. Exosome and exosomal microRNA: trafficking, sorting, and function. Genomics Proteomics Bioinformatics. 2015;13:17-24. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1443] [Cited by in RCA: 1558] [Article Influence: 155.8] [Reference Citation Analysis (0)] |
| 10. | Bracken CP, Scott HS, Goodall GJ. A network-biology perspective of microRNA function and dysfunction in cancer. Nat Rev Genet. 2016;17:719-732. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 437] [Cited by in RCA: 532] [Article Influence: 59.1] [Reference Citation Analysis (0)] |
| 11. | Zheng P, Chen L, Yuan X, Luo Q, Liu Y, Xie G, Ma Y, Shen L. Exosomal transfer of tumor-associated macrophage-derived miR-21 confers cisplatin resistance in gastric cancer cells. J Exp Clin Cancer Res. 2017;36:53. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 326] [Cited by in RCA: 489] [Article Influence: 61.1] [Reference Citation Analysis (0)] |
| 12. | Sun Z, Shi K, Yang S, Liu J, Zhou Q, Wang G, Song J, Li Z, Zhang Z, Yuan W. Effect of exosomal miRNA on cancer biology and clinical applications. Mol Cancer. 2018;17:147. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 311] [Cited by in RCA: 588] [Article Influence: 84.0] [Reference Citation Analysis (0)] |
| 13. | Sartorius K, Makarova J, Sartorius B, An P, Winkler C, Chuturgoon A, Kramvis A. The Regulatory Role of MicroRNA in Hepatitis-B Virus-Associated Hepatocellular Carcinoma (HBV-HCC) Pathogenesis. Cells. 2019;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 63] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 14. | Lu C, Jia S, Zhao S, Shao X. MiR-342 regulates cell proliferation and apoptosis in hepatocellular carcinoma through Wnt/β-catenin signaling pathway. Cancer Biomark. 2019;25:115-126. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 24] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 15. | Zhou Y, Chen E, Tang Y, Mao J, Shen J, Zheng X, Xie S, Zhang S, Wu Y, Liu H, Zhi X, Ma T, Ni H, Chen J, Chai K, Chen W. miR-223 overexpression inhibits doxorubicin-induced autophagy by targeting FOXO3a and reverses chemoresistance in hepatocellular carcinoma cells. Cell Death Dis. 2019;10:843. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 80] [Cited by in RCA: 91] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 16. | Cao Z, Qiu J, Yang G, Liu Y, Luo W, You L, Zheng L, Zhang T. MiR-135a biogenesis and regulation in malignancy: a new hope for cancer research and therapy. Cancer Biol Med. 2020;17:569-582. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 36] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 17. | Yang C, Zheng X, Ye K, Sun Y, Lu Y, Fan Q, Ge H. miR-135a Inhibits the Invasion and Migration of Esophageal Cancer Stem Cells through the Hedgehog Signaling Pathway by Targeting Smo. Mol Ther Nucleic Acids. 2020;19:841-852. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 13] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 18. | Xie Y, Li F, Li Z, Shi Z. miR-135a suppresses migration of gastric cancer cells by targeting TRAF5-mediated NF-κB activation. Onco Targets Ther. 2019;12:975-984. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 28] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 19. | von Felden J, Heim D, Schulze K, Krech T, Ewald F, Nashan B, Lohse AW, Wege H. High expression of micro RNA-135A in hepatocellular carcinoma is associated with recurrence within 12 months after resection. BMC Cancer. 2017;17:60. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 22] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 20. | Van Renne N, Roca Suarez AA, Duong FHT, Gondeau C, Calabrese D, Fontaine N, Ababsa A, Bandiera S, Croonenborghs T, Pochet N, De Blasi V, Pessaux P, Piardi T, Sommacale D, Ono A, Chayama K, Fujita M, Nakagawa H, Hoshida Y, Zeisel MB, Heim MH, Baumert TF, Lupberger J. miR-135a-5p-mediated downregulation of protein tyrosine phosphatase receptor delta is a candidate driver of HCV-associated hepatocarcinogenesis. Gut. 2018;67:953-962. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 57] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 21. | Théry C, Amigorena S, Raposo G, Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol. 2006;Chapter 3:Unit 3.22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2443] [Cited by in RCA: 3726] [Article Influence: 196.1] [Reference Citation Analysis (0)] |
| 22. | Kouwaki T, Fukushima Y, Daito T, Sanada T, Yamamoto N, Mifsud EJ, Leong CR, Tsukiyama-Kohara K, Kohara M, Matsumoto M, Seya T, Oshiumi H. Extracellular Vesicles Including Exosomes Regulate Innate Immune Responses to Hepatitis B Virus Infection. Front Immunol. 2016;7:335. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 99] [Cited by in RCA: 149] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 23. | Zhao L, Liu W, Xiao J, Cao B. The role of exosomes and "exosomal shuttle microRNA" in tumorigenesis and drug resistance. Cancer Lett. 2015;356:339-346. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 115] [Cited by in RCA: 132] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 24. | Hu X, Jiang J, Ni C, Xu Q, Ye S, Wu J, Ge F, Han Y, Mo Y, Huang D, Yang L. HBV Integration-mediated Cell Apoptosis in HepG2.2.15. J Cancer. 2019;10:4142-4150. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 18] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 25. | Sodroski C, Lowey B, Hertz L, Jake Liang T, Li Q. MicroRNA-135a Modulates Hepatitis C Virus Genome Replication through Downregulation of Host Antiviral Factors. Virol Sin. 2019;34:197-210. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 20] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 26. | Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281-297. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25833] [Cited by in RCA: 27842] [Article Influence: 1325.8] [Reference Citation Analysis (0)] |
| 27. | Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4013] [Cited by in RCA: 5415] [Article Influence: 541.5] [Reference Citation Analysis (0)] |
| 28. | Karagkouni D, Paraskevopoulou MD, Chatzopoulos S, Vlachos IS, Tastsoglou S, Kanellos I, Papadimitriou D, Kavakiotis I, Maniou S, Skoufos G, Vergoulis T, Dalamagas T, Hatzigeorgiou AG. DIANA-TarBase v8: a decade-long collection of experimentally supported miRNA-gene interactions. Nucleic Acids Res. 2018;46:D239-D245. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 507] [Cited by in RCA: 811] [Article Influence: 135.2] [Reference Citation Analysis (0)] |
| 29. | Liu M, Jiang L, Guan XY. The genetic and epigenetic alterations in human hepatocellular carcinoma: a recent update. Protein Cell. 2014;5:673-691. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 116] [Cited by in RCA: 133] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 30. | Tsujimoto Y. Role of Bcl-2 family proteins in apoptosis: apoptosomes or mitochondria? Genes Cells. 1998;3:697-707. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 469] [Cited by in RCA: 553] [Article Influence: 20.5] [Reference Citation Analysis (0)] |
| 31. | Goyal L. Cell death inhibition: keeping caspases in check. Cell. 2001;104:805-808. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 184] [Cited by in RCA: 177] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 32. | Sarig R, Tzahor E. The cancer paradigms of mammalian regeneration: can mammals regenerate as amphibians? Carcinogenesis. 2017;38:359-366. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 12] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 33. | Gramantieri L, Trerè D, Chieco P, Lacchini M, Giovannini C, Piscaglia F, Cavallari A, Bolondi L. In human hepatocellular carcinoma in cirrhosis proliferating cell nuclear antigen (PCNA) is involved in cell proliferation and cooperates with P21 in DNA repair. J Hepatol. 2003;39:997-1003. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 35] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 34. | Maiorano D, Lutzmann M, Méchali M. MCM proteins and DNA replication. Curr Opin Cell Biol. 2006;18:130-136. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 178] [Cited by in RCA: 199] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 35. | Liu W, Lin YT, Yan XL, Ding YL, Wu YL, Chen WN, Lin X. Hepatitis B virus core protein inhibits Fas-mediated apoptosis of hepatoma cells via regulation of mFas/FasL and sFas expression. FASEB J. 2015;29:1113-1123. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 38] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 36. | Gai X, Zhao P, Pan Y, Shan H, Yue X, Du J, Zhang Z, Liu P, Ma H, Guo M, Yang X, Sun W, Gao L, Ma C, Liang X. Hepatitis B virus core protein enhances human telomerase reverse transcriptase expression and hepatocellular carcinoma cell proliferation in a c-Ets2-dependent manner. Int J Biochem Cell Biol. 2013;45:1174-1185. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 19] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 37. | Jones VS, Huang RY, Chen LP, Chen ZS, Fu L, Huang RP. Cytokines in cancer drug resistance: Cues to new therapeutic strategies. Biochim Biophys Acta. 2016;1865:255-265. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 111] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 38. | Wang W, Peng H, Li J, Zhao X, Zhao F, Hu K. Controllable inhibition of hepatitis B virus replication by a DR1-targeting short hairpin RNA (shRNA) expressed from a DOX-inducible lentiviral vector. Virus Genes. 2013;46:393-403. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 39. | Bandopadhyay M, Bharadwaj M. Exosomal miRNAs in hepatitis B virus related liver disease: a new hope for biomarker. Gut Pathog. 2020;12:23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 32] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 40. | Du J, Bai F, Zhao P, Li X, Gao L, Ma C, Liang X. Hepatitis B core protein promotes liver cancer metastasis through miR-382-5p/DLC-1 axis. Biochim Biophys Acta Mol Cell Res. 2018;1865:1-11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 28] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 41. | Kouwaki T, Okamoto M, Tsukamoto H, Fukushima Y, Oshiumi H. Extracellular Vesicles Deliver Host and Virus RNA and Regulate Innate Immune Response. Int J Mol Sci. 2017;18. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 73] [Cited by in RCA: 87] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 42. | Lu TX, Rothenberg ME. MicroRNA. J Allergy Clin Immunol. 2018;141:1202-1207. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 853] [Cited by in RCA: 1821] [Article Influence: 227.6] [Reference Citation Analysis (0)] |
| 43. | Zeng YB, Liang XH, Zhang GX, Jiang N, Zhang T, Huang JY, Zhang L, Zeng XC. miRNA-135a promotes hepatocellular carcinoma cell migration and invasion by targeting forkhead box O1. Cancer Cell Int. 2016;16:63. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51] [Cited by in RCA: 68] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 44. | Yao S, Tian C, Ding Y, Ye Q, Gao Y, Yang N, Li Q. Down-regulation of Krüppel-like factor-4 by microRNA-135a-5p promotes proliferation and metastasis in hepatocellular carcinoma by transforming growth factor-β1. Oncotarget. 2016;7:42566-42578. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 34] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 45. | Xu B, Lu X, Zhao Y, Liu C, Huang X, Chen S, Zhu W, Zhang L, Chen M. MicroRNA-135a induces prostate cancer cell apoptosis via inhibition of STAT6. Oncol Lett. 2019;17:1889-1895. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 12] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 46. | Ahmad A, Zhang W, Wu M, Tan S, Zhu T. Tumor-suppressive miRNA-135a inhibits breast cancer cell proliferation by targeting ELK1 and ELK3 oncogenes. Genes Genomics. 2018;40:243-251. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 43] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 47. | Bach DH, Hong JY, Park HJ, Lee SK. The role of exosomes and miRNAs in drug-resistance of cancer cells. Int J Cancer. 2017;141:220-230. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 150] [Cited by in RCA: 213] [Article Influence: 26.6] [Reference Citation Analysis (0)] |
| 48. | Wu S, Lin Y, Xu D, Chen J, Shu M, Zhou Y, Zhu W, Su X, Qiu P, Yan G. MiR-135a functions as a selective killer of malignant glioma. Oncogene. 2012;31:3866-3874. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 75] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 49. | Zhang T, Shao Y, Chu TY, Huang HS, Liou YL, Li Q, Zhou H. MiR-135a and MRP1 play pivotal roles in the selective lethality of phenethyl isothiocyanate to malignant glioma cells. Am J Cancer Res. 2016;6:957-972. [PubMed] |
| 50. | Zhou W, Bi X, Gao G, Sun L. miRNA-133b and miRNA-135a induce apoptosis via the JAK2/STAT3 signaling pathway in human renal carcinoma cells. Biomed Pharmacother. 2016;84:722-729. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 47] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 51. | Liu N, Shi YF, Diao HY, Li YX, Cui Y, Song XJ, Tian X, Li TY, Liu B. MicroRNA-135a Regulates Apoptosis Induced by Hydrogen Peroxide in Rat Cardiomyoblast Cells. Int J Biol Sci. 2017;13:13-21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 18] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 52. | Wang Y, Yang Z, Zhang K, Wan Y, Zhou Y. miR-135a-5p inhibitor protects glial cells against apoptosis via targeting SIRT1 in epilepsy. Exp Ther Med. 2021;21:431. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 14] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 53. | Pan Y, Ren F, Zhang W, Liu G, Yang D, Hu J, Feng K, Feng Y. Regulation of BGC-823 cell sensitivity to adriamycin via miRNA-135a-5p. Oncol Rep. 2014;32:2549-2556. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 18] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 54. | Wang LX, Kang ZP, Yang ZC, Ma RX, Tan Y, Peng XB, Dai RZ, Li J, Yu Y, Xu M. MicroRNA-135a Inhibits Nasopharyngeal Carcinoma Cell Proliferation Through Targeting Interleukin-17. Cell Physiol Biochem. 2018;46:2232-2238. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 18] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 55. | Mao XP, Zhang LS, Huang B, Zhou SY, Liao J, Chen LW, Qiu SP, Chen JX. Mir-135a enhances cellular proliferation through post-transcriptionally regulating PHLPP2 and FOXO1 in human bladder cancer. J Transl Med. 2015;13:86. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 53] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 56. | Iacona JR, Lutz CS. miR-146a-5p: Expression, regulation, and functions in cancer. Wiley Interdiscip Rev RNA. 2019;10:e1533. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 126] [Article Influence: 21.0] [Reference Citation Analysis (0)] |
| 57. | Du J, Liang X, Liu Y, Qu Z, Gao L, Han L, Liu S, Cui M, Shi Y, Zhang Z, Yu L, Cao L, Ma C, Zhang L, Chen Y, Sun W. Hepatitis B virus core protein inhibits TRAIL-induced apoptosis of hepatocytes by blocking DR5 expression. Cell Death Differ. 2009;16:219-229. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 68] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 58. | Jia B, Guo M, Li G, Yu D, Zhang X, Lan K, Deng Q. Hepatitis B virus core protein sensitizes hepatocytes to tumor necrosis factor-induced apoptosis by suppression of the phosphorylation of mitogen-activated protein kinase kinase 7. J Virol. 2015;89:2041-2051. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 35] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 59. | Fu X, Liu M, Qu S, Ma J, Zhang Y, Shi T, Wen H, Yang Y, Wang S, Wang J, Nan K, Yao Y, Tian T. Exosomal microRNA-32-5p induces multidrug resistance in hepatocellular carcinoma via the PI3K/Akt pathway. J Exp Clin Cancer Res. 2018;37:52. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 129] [Cited by in RCA: 193] [Article Influence: 27.6] [Reference Citation Analysis (0)] |
| 60. | Holleman A, Chung I, Olsen RR, Kwak B, Mizokami A, Saijo N, Parissenti A, Duan Z, Voest EE, Zetter BR. miR-135a contributes to paclitaxel resistance in tumor cells both in vitro and in vivo. Oncogene. 2011;30:4386-4398. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 85] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 61. | Yan LH, Chen ZN, Li-Li, Chen J, Wei WE, Mo XW, Qin YZ, Lin Y, Chen JS. miR-135a promotes gastric cancer progression and resistance to oxaliplatin. Oncotarget. 2016;7:70699-70714. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 32] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 62. | Wang J, Zhang L, Jiang W, Zhang R, Zhang B, Silayiding A, Duan X. MicroRNA-135a promotes proliferation, migration, invasion and induces chemoresistance of endometrial cancer cells. Eur J Obstet Gynecol Reprod Biol X. 2020;5:100103. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 28] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 63. | Shao L, Chen Z, Soutto M, Zhu S, Lu H, Romero-Gallo J, Peek R, Zhang S, El-Rifai W. Helicobacter pylori-induced miR-135b-5p promotes cisplatin resistance in gastric cancer. FASEB J. 2019;33:264-274. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 59] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 64. | Zhou L, Qiu T, Xu J, Wang T, Wang J, Zhou X, Huang Z, Zhu W, Shu Y, Liu P. miR-135a/b modulate cisplatin resistance of human lung cancer cell line by targeting MCL1. Pathol Oncol Res. 2013;19:677-683. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 37] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 65. | Su KJ, Yu YL. Downregulation of SHIP2 by Hepatitis B Virus X Promotes the Metastasis and Chemoresistance of Hepatocellular Carcinoma through SKP2. Cancers (Basel). 2019;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 66. | Liu L, Chen X, Xie S, Zhang C, Qiu Z, Zhu F. Variant 1 of KIAA0101, overexpressed in hepatocellular carcinoma, prevents doxorubicin-induced apoptosis by inhibiting p53 activation. Hepatology. 2012;56:1760-1769. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 45] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 67. | Liu Y, Liu L, Zhou Y, Zhou P, Yan Q, Chen X, Ding S, Zhu F. CKLF1 Enhances Inflammation-Mediated Carcinogenesis and Prevents Doxorubicin-Induced Apoptosis via IL6/STAT3 Signaling in HCC. Clin Cancer Res. 2019;25:4141-4154. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 53] [Article Influence: 8.8] [Reference Citation Analysis (0)] |