Published online Dec 28, 2021. doi: 10.3748/wjg.v27.i48.8201
Peer-review started: March 19, 2021
First decision: May 1, 2021
Revised: May 8, 2021
Accepted: December 2, 2021
Article in press: December 2, 2021
Published online: December 28, 2021
Processing time: 279 Days and 22.1 Hours
S-palmitoylation is one of the most common post-translational modifications in nature; however, its importance has been overlooked for decades. Crohn’s disease (CD), a subtype of inflammatory bowel disease (IBD), is an autoimmune disease characterized by chronic inflammation involving the entire gastrointestinal tract. Bowel damage and subsequent disabilities caused by CD are a growing global health issue. Well-acknowledged risk factors for CD include genetic susceptibility, environmental factors, such as a westernized lifestyle, and altered gut microbiota. However, the pathophysiological mechanisms of this disorder are not yet comprehensively understood. With the rapidly increasing global prevalence of CD and the evident role of S-palmitoylation in CD, as recently reported, there is a need to investigate the relationship between CD and S-palmitoylation. In this review, we summarize the concept, detection, and function of S-palmitoylation as well as its potential effects on CD, and provide novel insights into the pathogenesis and treatment of CD.
Core Tip: S-palmitoylation is one of the most common post-translational modifications in nature; however, its importance has been overlooked for decades. Crohn’s disease (CD) is an autoimmune disease characterized by chronic inflammation of the entire gastrointestinal tract, whose underlying mechanisms of action remain poorly understood. Recent studies have revealed a key role of S-palmitoylation in CD; therefore, there is a need to elucidate the relationship between CD and S-palmitoylation. This review summarizes the basic facts of S-palmitoylation and its potential effect on CD to provide novel insights into the pathogenesis and treatment of CD.
- Citation: Cheng WX, Ren Y, Lu MM, Xu LL, Gao JG, Chen D, Kalyani FS, Lv ZY, Chen CX, Ji F, Lin HN, Jin X. Palmitoylation in Crohn’s disease: Current status and future directions. World J Gastroenterol 2021; 27(48): 8201-8215
- URL: https://www.wjgnet.com/1007-9327/full/v27/i48/8201.htm
- DOI: https://dx.doi.org/10.3748/wjg.v27.i48.8201
Cysteine palmitoylation or S-palmitoylation is the process of adding a 16-carbon saturated fatty acyl chain to the sulfhydryl group of cysteine residues of proteins via a labile thioester bond[1,2]. Initial reports on the modification of proteins by palmitate using 14C-labeled palmitic acid date back to the 1970s. Evidence supporting the modification of cysteine residues emerged in the 1980s[3]. Since then, accumulating evidence has shown that over 2000 proteins are S-palmitoylated in mammals, as documented in SwissPalm, an S-palmitoylation database (https://swisspalm.org/). Nevertheless, although S-palmitoylation widely occurs in nature, similar to phosphorylation, acetylation, and ubiquitination, its importance in human health and disease has been overlooked over the years. In fact, there are currently no approved drugs known to target S-palmitoylation. Crohn’s disease (CD), a subtype of inflammatory bowel disease (IBD), is characterized by chronic inflammation of the gastrointestinal tract with or without systemic symptoms, leading to bowel damage and disability[4]. Currently, genetic susceptibility, environmental factors, such as a western lifestyle, and an altered gut microbiota are well-known risk factors for CD[5]. However, the detailed mechanism underlying this disorder has yet to be elucidated. With the rapidly increasing global prevalence of CD[6] and recent reports on the effect of S-palmitoylation on CD[7], evaluating the relationship between CD and S-palmitoylation is a meaningful effort to gain insights into pathogenesis and treatment of CD. In this review, we summarize the concept, measurement, and function of S-palmitoylation, as well as its potential effect on CD, with the aim of providing insights into the pathogenesis and treatment of CD.
The addition of S-palmitoylation is catalyzed by palmitoyltransferases. Known palmitoyltransferases belong to the zinc finger aspartate-histidine-histidine-cysteine (ZDHHC) family[2]. There are 23 ZDHHC proteins in humans and mice, using palmitoyl-CoA as the major palmitoyl donor to acylate substrate proteins[8]. It should be noted that even though proteins prefer palmitoyl-CoA, they are also able to utilize other similar acyl-CoA molecules as substrates; therefore, some researchers prefer to use S-acylation over S-palmitoylation as a more general term to reflect the use of several different long-chain fatty acyl groups. Here, the term S-palmitoylation is used to represent all similar long-chain acylations on cysteine catalyzed by ZDHHCs. In most cases, these acylations are likely to have similar functions; thus, there is no need to specifically differentiate them for the purpose of this review.
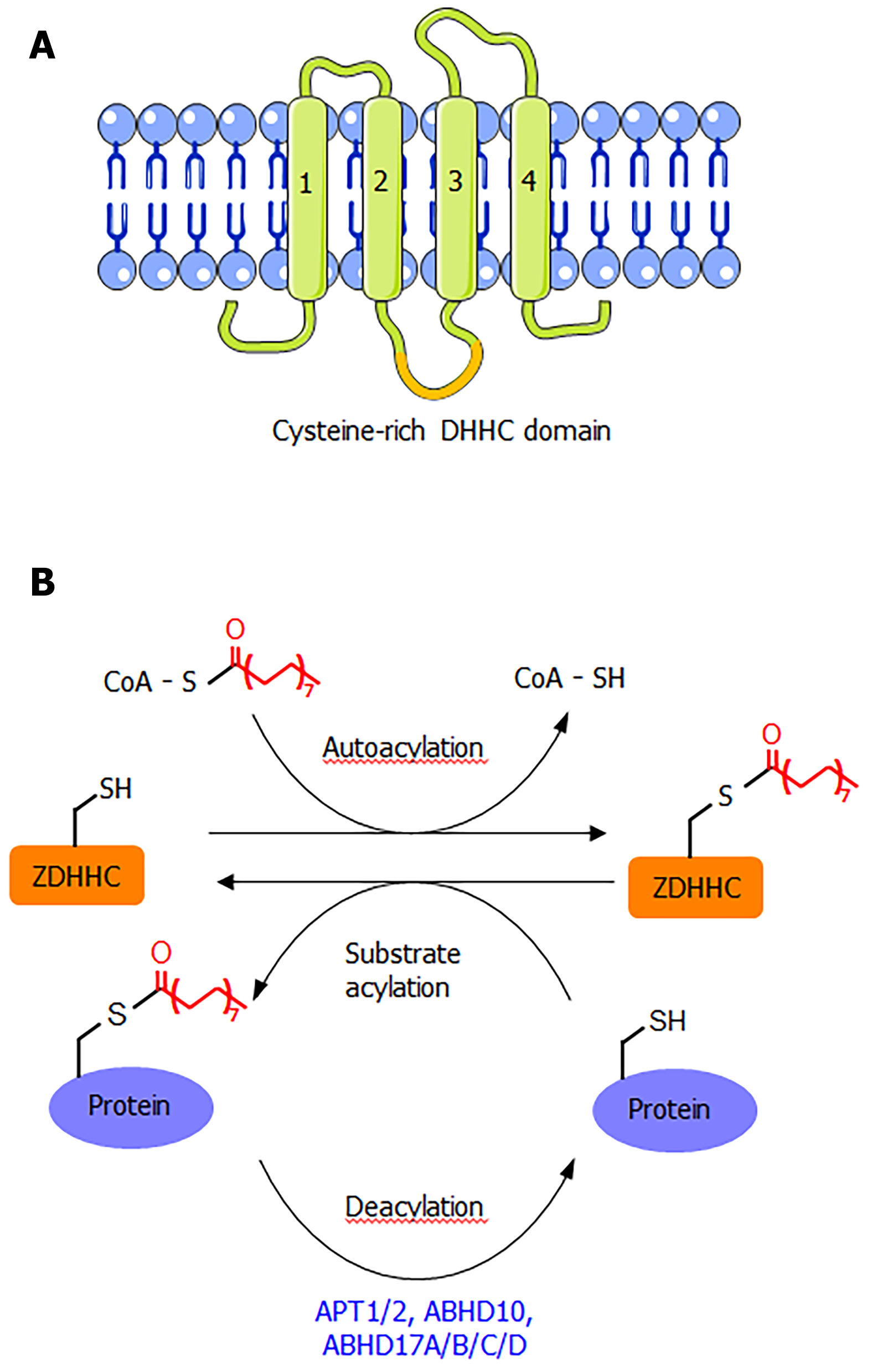
ZDHHC proteins are integral membrane proteins with at least four transmembrane helices (Figure 1A). The conserved DHHC cysteine-rich domain is present in the intracellular loop between transmembrane domains 2 and 3[1]. The cysteine residue in the conserved DHHC motif is known to serve as a catalytic nucleophile that reacts with the thioester bond in palmitoyl-CoA, forming a palmitoyl-enzyme intermediate, which then relays the palmitoyl group to the cysteine residues in the substrate proteins[1,2,8]. The crystal structure of DHHC20 has been reported[9], providing a structural basis for understanding this class of enzymes. Although S-palmitoylation is not a very stable modification due to the chemically labile nature of the thioester bond, the removal of S-palmitoylation is known to be catalyzed by several depalmitoylases (Figure 1B), including acyl protein thioesterase (APT1 and APT2), α/β-hydrolase domain 17 (ABHD17A/B/C/D), and α/β-hydrolase domain 10 (ABHD10)[1,10]. These enzymes belong to the alpha-beta hydrolase family, with a catalytic serine residue in the active site.
The most common function of S-palmitoylation is to promote the membrane localization of proteins. This can be easily appreciated from a recent review that listed many S-palmitoylated proteins and the function of S-palmitoylation[1]. This function is consistent with the hydrophobic nature of the palmitoyl group, which is especially true for peripheral membrane proteins (proteins without integral transmembrane domains). One well-known example is the small GTPases of the Ras subfamily, H-Ras, N-Ras, and K-Ras4a[1,11]. These proteins are soluble cytosolic proteins, but function at the plasma membrane or intracellular membranes. Their targeting to the plasma membrane requires prenylation and palmitoylation at the C-terminal sequence. Interestingly, it has been shown that the palmitoylation-depalmitoylation cycle helps to actively promote the trafficking of Ras to the plasma membrane. Several non-receptor tyrosine kinases, such as Fyn and Lyn, also require palmitoylation to target the plasma membrane.
Many integral membrane proteins are also palmitoylated. Integral membrane proteins contain transmembrane domains; thus, in principle, they should not require palmitoylation for membrane targeting. Instead, many reports indicate that palmitoylation promotes the targeting of these proteins to lipid rafts, which are specific membrane microdomains. This phenomenon requires further exploration in future studies. Other functional effects of S-palmitoylation have also been reported, including the regulation of protein stability and the aggregation of proteins[1,8]. However, the exact mechanism of these effects is unclear and may be indirectly caused by the membrane-targeting effect of S-palmitoylation.
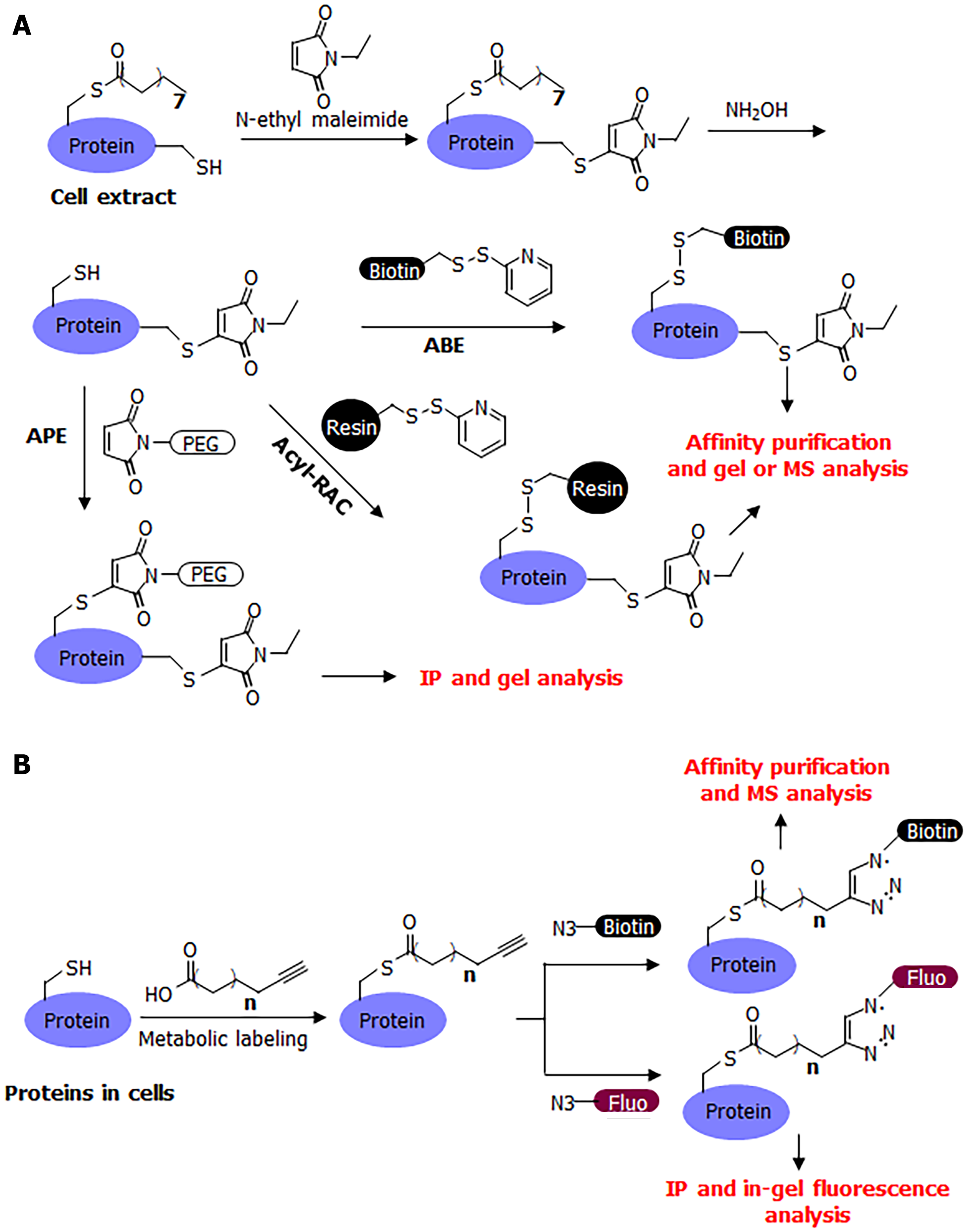
Many convenient tools have been developed for the study of S-palmitoylation, making it relatively easy to study compared to other post-translational modifications. The chemically labile nature of S-palmitoylation has enabled the development of several methods for its detection, including acyl-biotin exchange (ABE)[12,13], acyl-resin-assisted capture (Acyl-Rac)[14], and acyl-PEG exchange (APE)[1,15] (Figure 2A). A common procedure for these methods is to first cap free cysteine residues using a cysteine alkylation reagent, such as iodoacetamide or N-ethyl maleimide. Next, hydroxylamine is used to break down palmitoyl cysteine and release it as a free cysteine. The newly released free cysteine is then captured using a thiol-reactive group (HPDP-biotin in ABE, thiol-reactive resin in acyl-RAC, and thiol-reactive PEG in APE). In ABE, the biotinylation of palmitoylated proteins allows for affinity pulldown using streptavidin beads, and the palmitoylated proteins can then be detected after protein electrophoresis and western blotting, or analyzed by mass spectrometry (MS) in proteomic studies. In acyl-RAC, the palmitoylated proteins are pulled down using a resin and then analyzed using MS in proteomic studies. In a typical procedure for MS detection, the modified peptide is usually not detected by mass spectrometers because it is modified with a large biotin molecule or retained on the resin. However, certain modifications to this procedure can facilitate the detection of palmitoylated peptide. In APE, a large PEG molecule is attached to the palmitoylated protein of interest, which can change the protein size, which in turn can determine the number of palmitoyl cysteine modifications on the protein.
The ABE and acyl-RAC methods have the advantage of being able to detect S-palmitoylation in animal tissues as they reflect the endogenous palmitoylation level of endogenous proteins. A disadvantage of these methods is that there is no information on the identity of the acyl group on the cysteine residues as the hydroxylamine treatment removes all acyl modifications on cysteine residues. Theoretically, a short-chain acyl group modification could mistakenly be identified as palmitoylation; however, we are not aware of any such report for any protein. Another potential disadvantage is that a certain protein’s S-palmitoylation may be hydroxylamine-resistant and, therefore, may affect the outcome in acyl exchange assays[16].
A complementary method that could address the limitations of these acyl-exchange methods is metabolic labeling with labeled fatty acid analogs (Figure 2B). Although 14C-labeled palmitic acid was commonly used in early studies, a more convenient and sensitive method that has become commonly used in recent decades is that of clickable fatty acid analogs. This method typically uses an alkyne-tagged fatty acid, such as Alk14, which has 16 carbons similar to palmitic acid, but ends with a C-C triple bond at the end[17,18]. The structure of Alk14 is very similar to that of palmitic acid and can be efficiently utilized by cellular machinery to convert to the corresponding acyl-CoA and acylate proteins. The Alk14-modified protein can then be conjugated to an azide-containing fluorescent or biotin tag using a highly efficient copper-catalyzed cyclo-addition reaction. The conjugation of a fluorescent dye allows for the in-gel fluorescent detection of the Alk14 modification, while the conjugation of biotin allows for affinity purification and MS identification in proteomic studies.
The Alk14 Labeling method is in many ways comparable to ABE, as both allow for the gel-based detection of the S-palmitoylation of proteins of interest and proteomic studies. Alk14 Labeling does not require S-palmitoylation to be sensitive to hydroxylamine and can readily label proteins with dynamic palmitoylation. In contrast to ABE, Alk14 Labeling reflects the ability of a protein to be palmitoylated, but it is technically not the endogenous palmitoylation and is rarely used in animal studies. Generally, Alk14 Labeling and ABE are highly complementary to each other, with Alk14 demonstrating the palmitoylation of target proteins and ABE able to determine endogenous modifications on endogenous proteins. These two methods are often used simultaneously to confirm the S-palmitoylation of a protein of interest.
In comparison to other modifications, such as lysine acetylation, these detection methods make S-palmitoylation relatively straightforward to study. For lysine acetylation, a pan-acetyl-lysine antibody is typically used for affinity pull-down modified proteins, which are then subjected to MS analysis[19]. For a given protein of interest, it can be pulled down using immunoprecipitation, and then acetyl-lysine modification can be detected using western blotting with pan-acetyl-lysine antibodies. These studies rely heavily on the pan-acetyl-lysine antibody, which is expensive and may not work for all acetyl-lysine peptides. For S-palmitoylation, there is no antibody currently available, but acyl exchange methods and metabolic labeling methods have been found to work extremely well.
To investigate the function of palmitoylation on a particular protein, the most common method involves the identification of the site of palmitoylation, followed by the evaluation of the effect of cysteine to serine or alanine mutations on protein function. Typically, the occurrence of cysteine residues in proteins is less frequent compared to other modified residues, such as lysine, making the task of mutating all cysteine residues in a protein of interest much more practical than mutating all lysine residues. If a cysteine residue of a protein is the major palmitoylation site, then mutating it to Ser/Ala would markedly decrease the S-palmitoylation of the protein (detected by ABE or Alk14 Labeling). Subsequently, the same mutant can be used to observe whether the mutation affects protein localization, stability, and interaction with other proteins, as well as other biochemical activities.
The mutagenesis method, although powerful, has limitations. The mutated cysteine residue may have other functions (structural function or other modifications), which in turn can affect the palmitoylation of the protein; therefore, complementary methods to further confirm its function should be used. These complementary methods include identifying the ZDHHC enzyme that is responsible for the S-palmitoylation of the protein and determining whether the knockdown or knockout of ZDHHC produces the same effect as mutating the modified Cys residues. Similarly, identifying and disrupting the depalmitoylase also produces results consistent with the mutation of palmitoylated Cys. Recently, a method using amber suppression technology and click chemistry to insert a palmitoyl cysteine mimic on proteins in live HEK293T cells has been reported[20]. This method may allow for the gain-of-function analysis of S-palmitoylation. However, how closely the palmitoyl cysteine mimic can replicate the functional effect of S-palmitoylation remains unclear and will need to be tested in more proteins in future studies.
It is widely acknowledged that the pathophysiology of CD involves multiple factors, including genetic, environmental, microbial, immunologic, epithelial, and gut mucosal factors[21-23]. These factors are explored in detail in this section.
Genetic factors: Genome-wide association studies (GWAS) have identified over 240 risk variants that affect the recognition of microbial products by intracellular pathways [such as nucleotide oligomerization domain (NOD)–like receptors 2 (NOD2)], autophagy pathways that promote intracellular organelle circulation and the clearance of intracellular microorganisms [such as autophagy-related protein 16 Like 1 (ATG16L1) and immunity-related GTPase M (IRGM)], genes that regulate epithelial barrier function [such as extracellular matrix protein 1 (ECM1)], and pathways that regulate innate and adaptive immunity [such as interleukin (IL)-23R and IL-10][21,22]. Interestingly, known associations between CD and NOD2 gene variants are mainly found in patients of European or Jewish origin, but not in patients of Japanese or Chinese origin[22,24,25]. Another GWAS study supports this, additionally revealing that the tumor necrosis factor superfamily member 15 (TNFSF15) variant is dominant in East Asian populations[26]. These results conclusively indicate that different genetic factors contribute to CD through different inflammatory pathways.
Environmental factors: A series of environmental factors have been reported to affect the incidence of CD, including breastfeeding, living on farms, childhood contact with animals, smoking, antibiotic exposure, and dietary pattern[4,27-29]. Although inconsistent, breastfeeding, living on farms, and childhood contact with animals are believed to represent protective factors for CD[4]. Smoking is one of the most consistently reported risk factors for CD and is associated with a two-fold increase in the risk of developing CD (OR = 1.76, 95%CI: 1.40–2.22)[4,27,28]. A meta-analysis revealed that exposure to antibiotics also markedly increased the risk of CD, especially in children (OR = 2.75, 95%CI: 1.724.38)[29]. Low dietary fiber and an increased intake of saturated fats are also associated with an increased risk of developing CD[4].
Microbial factors: Although the gut host-microbial relationship is symbiotic, close contact between a rich bacterial community and intestinal tissue poses a great risk to health. In humans, in excess of 1012/cm3 of bacteria over a span of approximately 200 m2 are separated from the intestinal tissues by a mere 10-μm epithelial layer[30]. Therefore, it is crucial to maintain homeostasis between the microbiota and mucosal immunity in the gut. Mucus, defensins, IgA, and RegIIIγ are products of the epithelial and immune cells that control the gut microbiota. Certain microbes are beneficial to the growth of various T cell subsets, promoting the induction of type 17 T helper (Th17), regulatory T (Treg), and type 1 T helper (Th1) cells, and regulate mucosal immunity[22,30]. In addition, gut microbes can produce essential components, such as vitamin K, an important coagulation cofactor, and short-chain fatty acids, which are energy sources for colon epithelial cells[21,31]. Numerous studies have shown that changes in the microbial community result in a dysregulation of homeostasis[31,32]. In these studies, CD was associated with a decrease in the total number, diversity, and richness of microbial species.
Immune factors: CD arises as a result of chronic gastrointestinal inflammation and is associated with tissue destruction via the aberrant expression of pro-and anti-inflammatory molecules in response to innate and adaptive immune systems[33,34]. Amongst the numerous immune cells involved, Th17 cells regulated by IL-23 play an important role in immune regulation in the progression of CD[34-36]. IL-23 not only acts on members of the innate immune system but also promotes the proliferation and maintenance of Th17 cells. It is generally acknowledged that Th17 cells promote tissue inflammation, while Treg cells suppress autoimmunity, which suggests that the balance of Th17/Treg cells is crucial in the pathogenesis of CD[37,38]. With the development of GWAS, evidence is increasingly supporting the role of the innate immune response in the pathological process of CD, which includes epithelial barrier integrity, innate microbial sensing, autophagy, and unfolded protein response[34]. Other factors, such as injuries of epithelial and mesenchymal cells, changes in intestinal permeability, and obesity, also contribute to the pathophysiology of CD.
In recent decades, complex molecular pathways have been reported to be involved in the pathogenesis of CD. The identification of the main pathways and key factors may provide novel therapeutic targets. Current clinical therapies for IBD include anti-tumor necrosis factor (TNF) antibodies (such as infliximab, adalimumab, and certolizumab pegol), anti-IL-12/23 antibodies (such as ustekinumab), anti-sense oligonucleotides inhibiting SMAD7 transcription (such as mongersen), Janus kinase (JAK) inhibitors (such as tofacitinib and filgotinib), and anti-adhesion molecules (such as vedolizumab, etrolizumab, and anti-MAdCAM1 antibody)[4,39,40]. The main pathways and key factors are discussed in detail in this section (Table 1).
| Signaling pathway | Relative function | Targeted factor | Drug application |
| NF-κB | Maintenance of epithelial integrity and intestinal immune homeostasis | TNF-α | infliximab, adalimumab, and certolizumab |
| IL-12/23 | ustekinumab | ||
| IκBa | corticosteroids | ||
| TGF-β/SMAD3 | Immunosuppression and fibrosis | SMAD7 | mongerson |
| JAK/STAT | Immunoregulation, anti-inflammation and epithelial barrier function | JAK | tofacitinib and filgotinib |
| Chemokines/integrins | Leukocytes trafficking to targeted location | α4β7 integrin | vedolizumab |
| α4β7 and αEβ7 integrins | etrolizumab | ||
| MAdCAM1 | PF-00547659 | ||
| Wnt | Regulation of epithelial proliferation and gut mucosal homeostasis | NA | NA |
Nuclear factor kappa B signaling pathway: The targeting of TNF-α is a first-line treatment for CD, as well as for several other autoimmune diseases[40]. TNF-α is a pro-inflammatory mediator that plays a crucial role in the immune response to CD. It can induce T cell activation, inflammatory cell recruitment to local inflammatory sites, edema, coagulation, and granuloma formation[41]. Nuclear factor kappa B (NF-κB) signaling is considered the key pathway in lieu of TNF-α. Previous studies have shown that CD patients with high NF-κB activation have specific clinical manifestations, such as a higher frequency of ileocolonic involvement and higher histologic scores, compared to patients with low NF-κB activation[42]. The NF-κB signaling pathway also regulates the expression of IL-1, IL-6, IL-12, and IL-23[43-45] which are involved in mucosal damage within the inflammatory parts of the intestine. Furthermore, the differentiation of Th1 influenced by IL-12, IL-23, and TNF-α is actively involved in CD[45-47]. Corticosteroids, another first-line drug for the treatment of CD, have immunosuppressive effects and can induce the increased expression of IκBα, a key factor in the NF-κB pathway. These findings indicate that the NF-κB pathway plays a central role in the pathogenesis of CD.
Transforming growth factor-β/SMAD signaling pathway: Transforming growth factor-β (TGF-β) is an immunosuppressive cytokine produced by a variety of cells and activated by integrins. The role of TGF-β in intestinal immunity has been intensively investigated in previous studies[48]. Tregs have been suggested to produce anti-inflammatory cytokines, such as IL-10 and TGF-β. IL-10 promotes Treg cell proliferation by activating the signal transducer and activator of transcription (STAT)3, while TGF-β inhibits the proinflammatory responses of macrophages and effector T cells by activating SMAD3 and SMAD4[22]. Therefore, the upregulation of the Treg cell population and the reduction of effector T cells in CD indicate that the TGF-β/SMAD pathway plays a crucial role. In addition, SMAD7 is a downstream target of the TGF-β pathway, inhibiting the TGF-β pathway through negative feedback. In CD, the expression of SMAD7 is increased, leading to a reduction in SMAD3 phosphorylation and the suppression of TGF-β signaling, which may contribute to CD pathogenesis[48].
JAK/STAT signaling pathway: Although clinical trials using tofacitinib for the treatment of CD were canceled due to poor results, the efficacy of filgotinib, a selective JAK1 inhibitor, was confirmed in a randomized, double-blind, placebo-controlled phase II trial[49,50]. JAK tyrosine kinases and STAT DNA-binding proteins mediate signal transduction and downstream biological effects in response to cytokine receptor binding, some of which are associated with CD pathology. The cytokines mentioned above, which play essential roles in immunoregulation and the maintenance of epithelial barrier function (such as IL-6, IL-10, IL-12, and IL-23) are all dependent on JAK/STAT signaling[51]. STAT3 has also been reported to be crucial for the differentiation of Th17 cells and Th17 cell-dependent colitis, such as CD[37,52]. Interestingly, there is crosstalk between TNF and the JAK/STAT signaling pathway: TNF can amplify JAK-dependent receptor signal transduction by upregulating the expression of STAT[53]. Therefore, the role of JAK/STAT in the pathology of CD should be emphasized.
Wingless/Int1 signaling pathway: The Wingless/Int1 (Wnt) pathway is a key regulator of epithelial proliferation and gut mucosal homeostasis[54,55]. Wnt signaling is crucial for maintaining the stability of epithelial homeostasis, where the inhibition of this pathway leads to crypt loss and tissue degradation[56]. This pathway stimulates the differentiation and maturation of Paneth cells and regulates the expression of the α-defensins HD5 and HD6, in addition to mediating the stabilization of β-catenin[57]. Recently, Courth et al[58] found that the relationship between Paneth cells and bone marrow-derived monocytes participates in the mechanism of CD, which is characterized by the reduction of Wnt ligand expression in peripheral blood mononuclear cells (PBMCs) to attenuate intestinal barrier function. Furthermore, Wnt signaling is involved in various inflammatory signaling pathways, including NF-κB, mitogen-activated protein kinase (MAPK), protein kinase B (PKB/AKT), and STAT signaling. This complex network of signaling pathways may explain the contribution of Wnt to inflammatory injury repair[54].
Chemokines and integrins: In CD, chemokines induce the recruitment of immune cells to inflamed and epithelial-damaged sites. A highly effective and sequential adhesion system is involved in this process, in which integrins are activated by chemokines and interact with the addressins on the endothelium. For example, the antibody blockade of CCL25/CCR9 has been found to reduce early chronic ileitis in mice[59]. In addition, the ligation of CCR9 by CCL25 can result in a conformational change in α4β7 integrin, subsequently leading to the stable adhesion of MAdCAM-1[60]. Collectively, these results indicate that anti-adhesion molecules can be used clinically for CD therapy.
Many of the molecular pathways associated with CD have been reported to be modulated by S-palmitoylation. For example, a high frequency of mutations in NOD1/2 are found in IBD patients, and ZDHHC5-mediated NOD1/2 palmitoylation is responsible for normal gut functions. However, most reported CD-associated pathways in which palmitoylation occurs don’t specifically connect CD to palmitoylated factors such as Myd88. Myd88 is a component of TLR signaling that has been reported to be palmitoylated by ZDHHC6, but its palmitoylation hasn’t been associated with a gut phenotype. In CD, Myd88 participates in the recognition of extracellular and/or vacuolar intracellular pathogen-associated molecular patterns (PAMPs), which mediate sensing of microbial antigens[34]. The effects of palmitoylation on function of CD-associated factors need further exploration. Whether the effects of palmitoylation on CD symptoms are positive or negative might depend on a varied array of factors. Present opinion suggests that the functional effects of palmitoylation predominantly act to retain normal gut structures and functions. However, it is too early to conclude that all instances of palmitoylation exert positive effects. For instance, palmitoylation-mediated NF-kB activation probably results in negative consequences for CD patients. In this section, we summarize the S-palmitoylation events that have been reported to be associated with signaling pathways implicated in CD.
In the presence of damaged DNA, cyclic GMP-AMP synthase (cGAS) is activated and catalyzes the synthesis of cyclic GMP–AMP (cGAMP), which binds and activates its receptor stimulator of interferon genes (STING). STING is a membrane protein typically associated with endoplasmic reticulum (ER) stress. Upon activation, it translocates to the Golgi apparatus, where it is palmitoylated on Cys88 and Cys91, most likely by ZDHHC3, ZDHHC7, or ZDHHC15. Cysteine palmitoylation is important for its ability to activate TANK-binding kinase 1 (TBK1), which in turn phosphorylates interferon regulatory factor 3 (IRF3), which subsequently activates the transcription of immune response genes. STING palmitoylation has been proposed to promote the localization of STING to lipid rafts in the Golgi apparatus, which recruits both TBK1 and IRF3 to allow for downstream signal propagation[61]. Small molecules that can covalently label the palmitoylated Cys residues of STING have been identified and shown to suppress inflammation[62]. Interestingly, 9- or 10-nitro-oleic acid, which can be produced endogenously under inflammation, can also covalently modify the Cys residue of STING and inhibit its palmitoylation. This is likely to be a negative feedback regulation that inhibits STING signaling[62].
NOD1/2 are receptors for pathogen-associated molecular patterns, sensing bacterial peptidoglycans and initiating immune signaling, mainly by activating NF-κB. They are cytosolic proteins associated with bacteria-containing phagosomes upon bacterial infection. The cysteine palmitoylation of NOD1/2 is important for the phagosome translocation of NOD1/2. Palmitoylation occurs on multiple cysteine residues and is catalyzed by ZDHHC5[63]. NOD1/2 mutations are also associated with IBD. Interestingly, several of these mutations decrease the palmitoylation of NOD1/2 and inhibit NF-κB activation[63]. Therefore, methods to modulate the palmitoylation and signaling NOD1/2 hold potential for use in the treatment of CD.
Intriguingly, both the ligand TNF-α and its receptor TNFR1 are known to be regulated by cysteine palmitoylation. TNF-α is palmitoylated on Cys47, however, the enzymes regulating palmitoylation have not been reported[64]. Palmitoylation promotes the targeting of membrane TNF-α (before cleavage and secretion) to lipid rafts. TNF-α palmitoylation does not affect the secretion of soluble TNF-α, but affects the stability of the N-terminal intracellular domain[65]. A recent report showed that TNFR1 is palmitoylated and that palmitoylation is regulated by APT2 and TNF-α[66]. However, the site of modification, the exact ZDHHC responsible for palmitoylation, and the effect of palmitoylation on TNF signaling requires further exploration. Given the importance of anti-TNF therapy in CD, the palmitoylation of TNF and TNFR1 deserves further investigation in future studies.
Toll-like receptors (TLRs) and transmembrane proteins initiate immune signaling by sensing PAMPs. A proteomic study identified several TLRs as palmitoylated proteins. TLR2 palmitoylation was found to occur on a membrane-proximal cysteine residue, Cys609. Palmitoylation is important for TLR2 and NF-κB activation[67]. TLR signaling requires an adaptor protein, Myd88, which is palmitoylated on Cys113 and Cys274 by ZDHHC6. The palmitoylation of Cys113 is important for the recruitment of interleukin-1 receptor-associated kinase 4 (IRAK4) and NF-κB activation. The palmitoylation of Myd88 is also affected by the fatty acid synthase (FASN). Small molecule inhibitors of FASN reduce Myd88 palmitoylation and NF-κB activation[68]. Though no report indicates that Myd88 palmitoylation influences CD, it may exert effects related to sensing of microbial antigens, which is mediated by Myd88. However, as NF-kB activation displayed a high correlation to clinical CD manifestations, impaired palmitoylation resulting in NF-kB inhibition could be beneficial for CD patients.
STAT3-mediated Th17 differentiation is important for IBD. STAT3 is a transcription factor that, when phosphorylated by JAK in response to cytokines, such as IL-6, activates the transcription of genes that promote Th17 cell differentiation. Recently, STAT3 was reported to be regulated by S-palmitoylation of Cys108[7]. Palmitoylation is regulated by ZDHHC7 and depalmitoylated by APT2. Interestingly, the palmitoylation-depalmitoylation cycle has been found to be important for the activation of STAT3. Palmitoylation promotes STAT3 membrane localization and phosphorylation by JAK2. However, to translocate to the nucleus, phosphorylated STAT3 needs to be depalmitoylated. Therefore, APT2 is required for STAT3 activation. Interestingly, APT2 seems to prefer phosphorylated STAT3 over unphosphorylated STAT3, which ensures that the palmitoylation-depalmitoylation cycle moves in one direction, that which promotes STAT3 activation. Accordingly, the deletion or inhibition of either ZDHHC7 or APT2 decreases STAT3 activation, Th17 differentiation, and colitis in a mouse model. Furthermore, APT2 and ZDHHC7 are upregulated in human IBD patients, and the levels of IL-17 are closely correlated with the levels of APT2. This study provides strong evidence that the palmitoylation of STAT3 is a promising target for the treatment of IBD.
STAT3 activation occurs downstream of IL-6 receptor activation. Interestingly, one subunit of the IL-6 receptor, IL6ST (also called Gp130), is also regulated by palmitoylation. In neurons, ZDHHC5 and ZDHHC8 can palmitoylate IL6ST and promote JAK-STAT3 signaling[69]. Thus, it is possible that other proteins in the IL-6 signaling pathway, in addition to IL6ST and STAT3, could be regulated by cysteine palmitoylation. Future studies in this direction could identify additional targets, which would prove useful for advances in the treatment of IBD. Currently, there are no reports regarding the palmitoylation of the SMAD signaling pathway. However, SMAD2 has been reported to work with STAT3 to affect Th17 differentiation[70]. Therefore, SMAD signaling may be indirectly affected by STAT3 palmitoylation.
Inflammation involves the migration of various immune cells to the site of infection or inflammation. Thus, the inhibition of immune cell migration is an effective strategy to inhibit inflammation and autoimmune responses. Immune cell migration is typically mediated by chemotactic chemokine signaling. Multiple components of the chemokine signaling pathway can be regulated by cysteine palmitoylation. Chemotactic signaling is initiated by the binding of chemotactic ligands to the cell surface G protein-coupled receptors (GPCRs). Sphingosine 1-phosphate (S1P) receptor 1 (S1PR1), which binds to S1P, is important for the migration of mature T cells from the thymus into the blood stream and peripheral lymphoid organs. S1PR1 is palmitoylated by ZDHHC5 on multiple Cys residues at the C-terminus, and palmitoylation is important for its downstream signaling, which is mediated by trimeric G proteins[71,72]. Similarly, another chemotactic receptor, CCR5, is also regulated by cysteine palmitoylation[73]. Despite this, there are currently no reports on the palmitoylation of CCR9, which has been implicated in CD.
Chemotactic GPCR signaling requires coupling with the downstream trimeric G proteins. Interestingly, many trimeric G proteins are known to be regulated by palmitoylation[74,75]. Similarly, RGS proteins, which are regulators of G protein signaling, are also reported to be regulated by palmitoylation[75-77]. However, most of these examples have been reported in neuronal systems, and their role in the regulation of the immune system has yet to be studied extensively. The Rac1 small GTPase is important for cytoskeletal reorganization, which is required for immune cell adhesion and migration. Rac1 is palmitoylated on Cys176, which promotes its targeting to lipid rafts and inhibits its oligomerization, and is required for its signaling function. Palmitoylation-deficient Rac1 mutant cells are defective in cell spreading and migration[78]. However, the enzymes responsible for regulating Rac1 palmitoylation have yet to be identified. Targeting Rac1 palmitoyltransferases may potentially inhibit immune cell migration, thus representing a potential strategy for the treatment of autoimmune diseases.
Accumulating evidence has recently provided novel insights into the role of palmitoylation in the pathological mechanism of CD, highlighting potential drug targets for the control of palmitoylation. Since STING signaling is associated with palmitoylation, it is reasonable to assume that STING-associated autoimmune diseases, such as systemic lupus erythematosus (SLE) and Aicardi–Goutières syndrome (AGS), are related to the process of palmitoylation[79]. However, the contribution of STING to CD requires further study. If this relationship is confirmed, a novel promising drug target for the treatment of CD could be identified based on STING-related factors. Other factors related to CD have also been found to undergo palmitoylation during normal functional processes. These findings support the potential of palmitoylation as drug targets in CD, and we hope this area will attract more intensive research in the future.
S-palmitoylation is one of the most common post-translational modifications in nature which has been overlooked for decades. With the rapidly increasing global prevalence of CD and recent reports on the effect of S-palmitoylation on CD, elucidating the relationship between CD and S-palmitoylation becomes an urgent task. The basic facts of S-palmitoylation and its potential effect on CD summarized by this review will provide novel insights into the pathogenesis and treatment of CD.
We thank Dr. Zhou XX from the Department of Gastroenterology, The First Affiliated Hospital, Zhejiang University School of Medicine for her constructive suggestions for the section on Crohn’s disease. We also thank Dr. Zhang MM for his elegant work revealing the effect of the STAT3 palmitoylation circle on CD under the supervision of Dr. Lin H, which constitutes one of the major bases for this paper.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Flemming S, Liakina V S-Editor: Ma YJ L-Editor: A P-Editor: Ma YJ
| 1. | Jiang H, Zhang X, Chen X, Aramsangtienchai P, Tong Z, Lin H. Protein Lipidation: Occurrence, Mechanisms, Biological Functions, and Enabling Technologies. Chem Rev. 2018;118:919-988. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 195] [Cited by in RCA: 340] [Article Influence: 48.6] [Reference Citation Analysis (0)] |
| 2. | Linder ME, Jennings BC. Mechanism and function of DHHC S-acyltransferases. Biochem Soc Trans. 2013;41:29-34. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 52] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 3. | Chen ZQ, Ulsh LS, DuBois G, Shih TY. Posttranslational processing of p21 ras proteins involves palmitylation of the C-terminal tetrapeptide containing cysteine-186. J Virol. 1985;56:607-612. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 102] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 4. | Torres J, Mehandru S, Colombel JF, Peyrin-Biroulet L. Crohn's disease. Lancet. 2017;389:1741-1755. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1121] [Cited by in RCA: 1804] [Article Influence: 225.5] [Reference Citation Analysis (111)] |
| 5. | Li N, Shi RH. Updated review on immune factors in pathogenesis of Crohn's disease. World J Gastroenterol. 2018;24:15-22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 52] [Cited by in RCA: 45] [Article Influence: 6.4] [Reference Citation Analysis (6)] |
| 6. | Ng SC, Shi HY, Hamidi N, Underwood FE, Tang W, Benchimol EI, Panaccione R, Ghosh S, Wu JCY, Chan FKL, Sung JJY, Kaplan GG. Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: a systematic review of population-based studies. Lancet. 2017;390:2769-2778. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2677] [Cited by in RCA: 4105] [Article Influence: 513.1] [Reference Citation Analysis (110)] |
| 7. | Zhang M, Zhou L, Xu Y, Yang M, Komaniecki GP, Kosciuk T, Chen X, Lu X, Zou X, Linder ME, Lin H. A STAT3 palmitoylation cycle promotes TH17 differentiation and colitis. Nature. 2020;586:434-439. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 223] [Article Influence: 44.6] [Reference Citation Analysis (0)] |
| 8. | Linder ME, Deschenes RJ. Palmitoylation: policing protein stability and traffic. Nat Rev Mol Cell Biol. 2007;8:74-84. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 906] [Cited by in RCA: 891] [Article Influence: 49.5] [Reference Citation Analysis (0)] |
| 9. | Rana MS, Kumar P, Lee CJ, Verardi R, Rajashankar KR, Banerjee A. Fatty acyl recognition and transfer by an integral membrane S-acyltransferase. Science. 2018;359. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 178] [Cited by in RCA: 181] [Article Influence: 25.9] [Reference Citation Analysis (0)] |
| 10. | Dekker FJ, Rocks O, Vartak N, Menninger S, Hedberg C, Balamurugan R, Wetzel S, Renner S, Gerauer M, Schölermann B, Rusch M, Kramer JW, Rauh D, Coates GW, Brunsveld L, Bastiaens PI, Waldmann H. Small-molecule inhibition of APT1 affects Ras localization and signaling. Nat Chem Biol. 2010;6:449-456. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 292] [Cited by in RCA: 325] [Article Influence: 21.7] [Reference Citation Analysis (0)] |
| 11. | Ko PJ, Dixon SJ. Protein palmitoylation and cancer. EMBO Rep. 2018;19. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 265] [Article Influence: 37.9] [Reference Citation Analysis (0)] |
| 12. | Drisdel RC, Green WN. Labeling and quantifying sites of protein palmitoylation. Biotechniques. 2004;36:276-285. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 251] [Cited by in RCA: 272] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 13. | Wan J, Roth AF, Bailey AO, Davis NG. Palmitoylated proteins: purification and identification. Nat Protoc. 2007;2:1573-1584. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 305] [Cited by in RCA: 349] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 14. | Forrester MT, Thompson JW, Foster MW, Nogueira L, Moseley MA, Stamler JS. Proteomic analysis of S-nitrosylation and denitrosylation by resin-assisted capture. Nat Biotechnol. 2009;27:557-559. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 330] [Cited by in RCA: 323] [Article Influence: 20.2] [Reference Citation Analysis (0)] |
| 15. | Percher A, Ramakrishnan S, Thinon E, Yuan X, Yount JS, Hang HC. Mass-tag labeling reveals site-specific and endogenous levels of protein S-fatty acylation. Proc Natl Acad Sci U S A. 2016;113:4302-4307. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 134] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 16. | Aramsangtienchai P, Spiegelman NA, Cao J, Lin H. S-Palmitoylation of Junctional Adhesion Molecule C Regulates Its Tight Junction Localization and Cell Migration. J Biol Chem. 2017;292:5325-5334. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 41] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 17. | Charron G, Zhang MM, Yount JS, Wilson J, Raghavan AS, Shamir E, Hang HC. Robust fluorescent detection of protein fatty-acylation with chemical reporters. J Am Chem Soc. 2009;131:4967-4975. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 249] [Cited by in RCA: 263] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 18. | Martin BR, Cravatt BF. Large-scale profiling of protein palmitoylation in mammalian cells. Nat Methods. 2009;6:135-138. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 443] [Cited by in RCA: 421] [Article Influence: 26.3] [Reference Citation Analysis (0)] |
| 19. | Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, Cheng T, Kho Y, Xiao H, Xiao L, Grishin NV, White M, Yang XJ, Zhao Y. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell. 2006;23:607-618. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1168] [Cited by in RCA: 1219] [Article Influence: 64.2] [Reference Citation Analysis (0)] |
| 20. | Li Y, Wang S, Chen Y, Li M, Dong X, Hang HC, Peng T. Site-specific chemical fatty-acylation for gain-of-function analysis of protein S-palmitoylation in live cells. Chem Commun (Camb). 2020;56:13880-13883. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 27] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 21. | Chang JT. Pathophysiology of Inflammatory Bowel Diseases. N Engl J Med. 2020;383:2652-2664. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 274] [Cited by in RCA: 771] [Article Influence: 154.2] [Reference Citation Analysis (0)] |
| 22. | de Souza HS, Fiocchi C. Immunopathogenesis of IBD: current state of the art. Nat Rev Gastroenterol Hepatol. 2016;13:13-27. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1185] [Cited by in RCA: 1120] [Article Influence: 124.4] [Reference Citation Analysis (1)] |
| 23. | Coufal S, Galanova N, Bajer L, Gajdarova Z, Schierova D, Jiraskova Zakostelska Z, Kostovcikova K, Jackova Z, Stehlikova Z, Drastich P, Tlaskalova-Hogenova H, Kverka M. Inflammatory Bowel Disease Types Differ in Markers of Inflammation, Gut Barrier and in Specific Anti-Bacterial Response. Cells. 2019;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 32] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 24. | Inoue N, Tamura K, Kinouchi Y, Fukuda Y, Takahashi S, Ogura Y, Inohara N, Núñez G, Kishi Y, Koike Y, Shimosegawa T, Shimoyama T, Hibi T. Lack of common NOD2 variants in Japanese patients with Crohn's disease. Gastroenterology. 2002;123:86-91. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 326] [Cited by in RCA: 327] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 25. | Leong RW, Armuzzi A, Ahmad T, Wong ML, Tse P, Jewell DP, Sung JJ. NOD2/CARD15 gene polymorphisms and Crohn's disease in the Chinese population. Aliment Pharmacol Ther. 2003;17:1465-1470. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 166] [Cited by in RCA: 172] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 26. | Liu JZ, van Sommeren S, Huang H, Ng SC, Alberts R, Takahashi A, Ripke S, Lee JC, Jostins L, Shah T, Abedian S, Cheon JH, Cho J, Dayani NE, Franke L, Fuyuno Y, Hart A, Juyal RC, Juyal G, Kim WH, Morris AP, Poustchi H, Newman WG, Midha V, Orchard TR, Vahedi H, Sood A, Sung JY, Malekzadeh R, Westra HJ, Yamazaki K, Yang SK; International Multiple Sclerosis Genetics Consortium; International IBD Genetics Consortium, Barrett JC, Alizadeh BZ, Parkes M, Bk T, Daly MJ, Kubo M, Anderson CA, Weersma RK. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet. 2015;47:979-986. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1898] [Cited by in RCA: 1866] [Article Influence: 186.6] [Reference Citation Analysis (0)] |
| 27. | Ananthakrishnan AN. Epidemiology and risk factors for IBD. Nat Rev Gastroenterol Hepatol. 2015;12:205-217. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 930] [Cited by in RCA: 1239] [Article Influence: 123.9] [Reference Citation Analysis (0)] |
| 28. | Mahid SS, Minor KS, Soto RE, Hornung CA, Galandiuk S. Smoking and inflammatory bowel disease: a meta-analysis. Mayo Clin Proc. 2006;81:1462-1471. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 479] [Cited by in RCA: 522] [Article Influence: 27.5] [Reference Citation Analysis (0)] |
| 29. | Ungaro R, Bernstein CN, Gearry R, Hviid A, Kolho KL, Kronman MP, Shaw S, Van Kruiningen H, Colombel JF, Atreja A. Antibiotics associated with increased risk of new-onset Crohn's disease but not ulcerative colitis: a meta-analysis. Am J Gastroenterol. 2014;109:1728-1738. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 229] [Cited by in RCA: 263] [Article Influence: 23.9] [Reference Citation Analysis (1)] |
| 30. | Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336:1268-1273. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3003] [Cited by in RCA: 2988] [Article Influence: 229.8] [Reference Citation Analysis (0)] |
| 31. | Schirmer M, Garner A, Vlamakis H, Xavier RJ. Microbial genes and pathways in inflammatory bowel disease. Nat Rev Microbiol. 2019;17:497-511. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 249] [Cited by in RCA: 594] [Article Influence: 118.8] [Reference Citation Analysis (0)] |
| 32. | Marchesi JR, Adams DH, Fava F, Hermes GD, Hirschfield GM, Hold G, Quraishi MN, Kinross J, Smidt H, Tuohy KM, Thomas LV, Zoetendal EG, Hart A. The gut microbiota and host health: a new clinical frontier. Gut. 2016;65:330-339. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1341] [Cited by in RCA: 1564] [Article Influence: 173.8] [Reference Citation Analysis (0)] |
| 33. | Park JH, Peyrin-Biroulet L, Eisenhut M, Shin JI. IBD immunopathogenesis: A comprehensive review of inflammatory molecules. Autoimmun Rev. 2017;16:416-426. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 166] [Cited by in RCA: 215] [Article Influence: 26.9] [Reference Citation Analysis (0)] |
| 34. | Geremia A, Biancheri P, Allan P, Corazza GR, Di Sabatino A. Innate and adaptive immunity in inflammatory bowel disease. Autoimmun Rev. 2014;13:3-10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 484] [Cited by in RCA: 706] [Article Influence: 58.8] [Reference Citation Analysis (1)] |
| 35. | Cătană CS, Berindan Neagoe I, Cozma V, Magdaş C, Tăbăran F, Dumitraşcu DL. Contribution of the IL-17/IL-23 axis to the pathogenesis of inflammatory bowel disease. World J Gastroenterol. 2015;21:5823-5830. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 141] [Cited by in RCA: 144] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 36. | Geremia A, Jewell DP. The IL-23/IL-17 pathway in inflammatory bowel disease. Expert Rev Gastroenterol Hepatol. 2012;6:223-237. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 78] [Article Influence: 6.0] [Reference Citation Analysis (1)] |
| 37. | Yan JB, Luo MM, Chen ZY, He BH. The Function and Role of the Th17/Treg Cell Balance in Inflammatory Bowel Disease. J Immunol Res. 2020;2020:8813558. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 205] [Article Influence: 41.0] [Reference Citation Analysis (0)] |
| 38. | Noack M, Miossec P. Th17 and regulatory T cell balance in autoimmune and inflammatory diseases. Autoimmun Rev. 2014;13:668-677. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 566] [Cited by in RCA: 692] [Article Influence: 62.9] [Reference Citation Analysis (0)] |
| 39. | Sandborn WJ, Vermeire S, Tyrrell H, Hassanali A, Lacey S, Tole S, Tatro AR; Etrolizumab Global Steering Committee. Etrolizumab for the Treatment of Ulcerative Colitis and Crohn's Disease: An Overview of the Phase 3 Clinical Program. Adv Ther. 2020;37:3417-3431. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 65] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 40. | Cushing K, Higgins PDR. Management of Crohn Disease: A Review. JAMA. 2021;325:69-80. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 211] [Article Influence: 52.8] [Reference Citation Analysis (0)] |
| 41. | Berns M, Hommes DW. Anti-TNF-α therapies for the treatment of Crohn's disease: the past, present and future. Expert Opin Investig Drugs. 2016;25:129-143. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 53] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 42. | Han YM, Koh J, Kim JW, Lee C, Koh SJ, Kim B, Lee KL, Im JP, Kim JS. NF-kappa B activation correlates with disease phenotype in Crohn's disease. PLoS One. 2017;12:e0182071. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 43] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 43. | Becker C, Wirtz S, Blessing M, Pirhonen J, Strand D, Bechthold O, Frick J, Galle PR, Autenrieth I, Neurath MF. Constitutive p40 promoter activation and IL-23 production in the terminal ileum mediated by dendritic cells. J Clin Invest. 2003;112:693-706. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 117] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 44. | Becker C, Wirtz S, Ma X, Blessing M, Galle PR, Neurath MF. Regulation of IL-12 p40 promoter activity in primary human monocytes: roles of NF-kappaB, CCAAT/enhancer-binding protein beta, and PU.1 and identification of a novel repressor element (GA-12) that responds to IL-4 and prostaglandin E(2). J Immunol. 2001;167:2608-2618. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 79] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 45. | Atreya I, Atreya R, Neurath MF. NF-kappaB in inflammatory bowel disease. J Intern Med. 2008;263:591-596. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 536] [Cited by in RCA: 645] [Article Influence: 37.9] [Reference Citation Analysis (0)] |
| 46. | Oppmann B, Lesley R, Blom B, Timans JC, Xu Y, Hunte B, Vega F, Yu N, Wang J, Singh K, Zonin F, Vaisberg E, Churakova T, Liu M, Gorman D, Wagner J, Zurawski S, Liu Y, Abrams JS, Moore KW, Rennick D, de Waal-Malefyt R, Hannum C, Bazan JF, Kastelein RA. Novel p19 protein engages IL-12p40 to form a cytokine, IL-23, with biological activities similar as well as distinct from IL-12. Immunity. 2000;13:715-725. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2046] [Cited by in RCA: 2110] [Article Influence: 84.4] [Reference Citation Analysis (0)] |
| 47. | Neurath MF, Finotto S, Glimcher LH. The role of Th1/Th2 polarization in mucosal immunity. Nat Med. 2002;8:567-573. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 451] [Cited by in RCA: 451] [Article Influence: 19.6] [Reference Citation Analysis (0)] |
| 48. | Ihara S, Hirata Y, Koike K. TGF-β in inflammatory bowel disease: a key regulator of immune cells, epithelium, and the intestinal microbiota. J Gastroenterol. 2017;52:777-787. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 146] [Cited by in RCA: 213] [Article Influence: 26.6] [Reference Citation Analysis (0)] |
| 49. | Panés J, Sandborn WJ, Schreiber S, Sands BE, Vermeire S, D'Haens G, Panaccione R, Higgins PDR, Colombel JF, Feagan BG, Chan G, Moscariello M, Wang W, Niezychowski W, Marren A, Healey P, Maller E. Tofacitinib for induction and maintenance therapy of Crohn's disease: results of two phase IIb randomised placebo-controlled trials. Gut. 2017;66:1049-1059. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 222] [Cited by in RCA: 286] [Article Influence: 35.8] [Reference Citation Analysis (0)] |
| 50. | Vermeire S, Schreiber S, Petryka R, Kuehbacher T, Hebuterne X, Roblin X, Klopocka M, Goldis A, Wisniewska-Jarosinska M, Baranovsky A, Sike R, Stoyanova K, Tasset C, Van der Aa A, Harrison P. Clinical remission in patients with moderate-to-severe Crohn's disease treated with filgotinib (the FITZROY study): results from a phase 2, double-blind, randomised, placebo-controlled trial. Lancet. 2017;389:266-275. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 282] [Cited by in RCA: 337] [Article Influence: 42.1] [Reference Citation Analysis (0)] |
| 51. | Salas A, Hernandez-Rocha C, Duijvestein M, Faubion W, McGovern D, Vermeire S, Vetrano S, Vande Casteele N. JAK-STAT pathway targeting for the treatment of inflammatory bowel disease. Nat Rev Gastroenterol Hepatol. 2020;17:323-337. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 188] [Cited by in RCA: 425] [Article Influence: 85.0] [Reference Citation Analysis (0)] |
| 52. | Lu D, Liu L, Ji X, Gao Y, Chen X, Liu Y, Zhao X, Li Y, Jin Y, Zhang Y, McNutt MA, Yin Y. The phosphatase DUSP2 controls the activity of the transcription activator STAT3 and regulates TH17 differentiation. Nat Immunol. 2015;16:1263-1273. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 114] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 53. | Rogler G. Efficacy of JAK inhibitors in Crohn's Disease. J Crohns Colitis. 2020;14:S746-S754. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 48] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 54. | Moparthi L, Koch S. Wnt signaling in intestinal inflammation. Differentiation. 2019;108:24-32. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 119] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 55. | Armbruster NS, Stange EF, Wehkamp J. In the Wnt of Paneth Cells: Immune-Epithelial Crosstalk in Small Intestinal Crohn's Disease. Front Immunol. 2017;8:1204. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 23] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 56. | Kuhnert F, Davis CR, Wang HT, Chu P, Lee M, Yuan J, Nusse R, Kuo CJ. Essential requirement for Wnt signaling in proliferation of adult small intestine and colon revealed by adenoviral expression of Dickkopf-1. Proc Natl Acad Sci U S A. 2004;101:266-271. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 483] [Cited by in RCA: 506] [Article Influence: 23.0] [Reference Citation Analysis (0)] |
| 57. | van Es JH, Jay P, Gregorieff A, van Gijn ME, Jonkheer S, Hatzis P, Thiele A, van den Born M, Begthel H, Brabletz T, Taketo MM, Clevers H. Wnt signalling induces maturation of Paneth cells in intestinal crypts. Nat Cell Biol. 2005;7:381-386. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 472] [Cited by in RCA: 515] [Article Influence: 25.8] [Reference Citation Analysis (0)] |
| 58. | Courth LF, Ostaff MJ, Mailänder-Sánchez D, Malek NP, Stange EF, Wehkamp J. Crohn's disease-derived monocytes fail to induce Paneth cell defensins. Proc Natl Acad Sci U S A. 2015;112:14000-14005. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 58] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 59. | Rivera-Nieves J, Ho J, Bamias G, Ivashkina N, Ley K, Oppermann M, Cominelli F. Antibody blockade of CCL25/CCR9 ameliorates early but not late chronic murine ileitis. Gastroenterology. 2006;131:1518-1529. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 106] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 60. | Thomas S, Baumgart DC. Targeting leukocyte migration and adhesion in Crohn's disease and ulcerative colitis. Inflammopharmacology. 2012;20:1-18. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 139] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 61. | Mukai K, Konno H, Akiba T, Uemura T, Waguri S, Kobayashi T, Barber GN, Arai H, Taguchi T. Activation of STING requires palmitoylation at the Golgi. Nat Commun. 2016;7:11932. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 266] [Cited by in RCA: 508] [Article Influence: 56.4] [Reference Citation Analysis (0)] |
| 62. | Haag SM, Gulen MF, Reymond L, Gibelin A, Abrami L, Decout A, Heymann M, van der Goot FG, Turcatti G, Behrendt R, Ablasser A. Targeting STING with covalent small-molecule inhibitors. Nature. 2018;559:269-273. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 337] [Cited by in RCA: 787] [Article Influence: 112.4] [Reference Citation Analysis (0)] |
| 63. | Lu Y, Zheng Y, Coyaud É, Zhang C, Selvabaskaran A, Yu Y, Xu Z, Weng X, Chen JS, Meng Y, Warner N, Cheng X, Liu Y, Yao B, Hu H, Xia Z, Muise AM, Klip A, Brumell JH, Girardin SE, Ying S, Fairn GD, Raught B, Sun Q, Neculai D. Palmitoylation of NOD1 and NOD2 is required for bacterial sensing. Science. 2019;366:460-467. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 154] [Cited by in RCA: 152] [Article Influence: 25.3] [Reference Citation Analysis (0)] |
| 64. | Utsumi T, Takeshige T, Tanaka K, Takami K, Kira Y, Klostergaard J, Ishisaka R. Transmembrane TNF (pro-TNF) is palmitoylated. FEBS Lett. 2001;500:1-6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 35] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 65. | Poggi M, Kara I, Brunel JM, Landrier JF, Govers R, Bonardo B, Fluhrer R, Haass C, Alessi MC, Peiretti F. Palmitoylation of TNF alpha is involved in the regulation of TNF receptor 1 signalling. Biochim Biophys Acta. 2013;1833:602-612. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 37] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 66. | Zingler P, Särchen V, Glatter T, Caning L, Saggau C, Kathayat RS, Dickinson BC, Adam D, Schneider-Brachert W, Schütze S, Fritsch J. Palmitoylation is required for TNF-R1 signaling. Cell Commun Signal. 2019;17:90. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 37] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 67. | Chesarino NM, Hach JC, Chen JL, Zaro BW, Rajaram MV, Turner J, Schlesinger LS, Pratt MR, Hang HC, Yount JS. Chemoproteomics reveals Toll-like receptor fatty acylation. BMC Biol. 2014;12:91. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 60] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 68. | Kim YC, Lee SE, Kim SK, Jang HD, Hwang I, Jin S, Hong EB, Jang KS, Kim HS. Toll-like receptor mediated inflammation requires FASN-dependent MYD88 palmitoylation. Nat Chem Biol. 2019;15:907-916. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 122] [Article Influence: 20.3] [Reference Citation Analysis (0)] |
| 69. | Collura KM, Niu J, Sanders SS, Montersino A, Holland SM, Thomas GM. The palmitoyl acyltransferases ZDHHC5 and ZDHHC8 are uniquely present in DRG axons and control retrograde signaling via the Gp130/JAK/STAT3 pathway. J Biol Chem. 2020;295:15427-15437. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 24] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 70. | Yoon JH, Sudo K, Kuroda M, Kato M, Lee IK, Han JS, Nakae S, Imamura T, Kim J, Ju JH, Kim DK, Matsuzaki K, Weinstein M, Matsumoto I, Sumida T, Mamura M. Phosphorylation status determines the opposing functions of Smad2/Smad3 as STAT3 cofactors in TH17 differentiation. Nat Commun. 2015;6:7600. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 65] [Cited by in RCA: 85] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 71. | Ohno Y, Ito A, Ogata R, Hiraga Y, Igarashi Y, Kihara A. Palmitoylation of the sphingosine 1-phosphate receptor S1P is involved in its signaling functions and internalization. Genes Cells. 2009;14:911-923. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 16] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 72. | Badawy SMM, Okada T, Kajimoto T, Ijuin T, Nakamura SI. DHHC5-mediated palmitoylation of S1P receptor subtype 1 determines G-protein coupling. Sci Rep. 2017;7:16552. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 15] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 73. | Blanpain C, Wittamer V, Vanderwinden JM, Boom A, Renneboog B, Lee B, Le Poul E, El Asmar L, Govaerts C, Vassart G, Doms RW, Parmentier M. Palmitoylation of CCR5 is critical for receptor trafficking and efficient activation of intracellular signaling pathways. J Biol Chem. 2001;276:23795-23804. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 108] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 74. | Tsutsumi R, Fukata Y, Noritake J, Iwanaga T, Perez F, Fukata M. Identification of G protein alpha subunit-palmitoylating enzyme. Mol Cell Biol. 2009;29:435-447. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 107] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 75. | Lin H. Protein cysteine palmitoylation in immunity and inflammation. FEBS J. 2021;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 53] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 76. | Castro-Fernández C, Janovick JA, Brothers SP, Fisher RA, Ji TH, Conn PM. Regulation of RGS3 and RGS10 palmitoylation by GnRH. Endocrinology. 2002;143:1310-1317. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 24] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 77. | Wang J, Xie Y, Wolff DW, Abel PW, Tu Y. DHHC protein-dependent palmitoylation protects regulator of G-protein signaling 4 from proteasome degradation. FEBS Lett. 2010;584:4570-4574. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 24] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 78. | Navarro-Lérida I, Sánchez-Perales S, Calvo M, Rentero C, Zheng Y, Enrich C, Del Pozo MA. A palmitoylation switch mechanism regulates Rac1 function and membrane organization. EMBO J. 2012;31:534-551. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 143] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 79. | Lee-Kirsch MA, Gong M, Chowdhury D, Senenko L, Engel K, Lee YA, de Silva U, Bailey SL, Witte T, Vyse TJ, Kere J, Pfeiffer C, Harvey S, Wong A, Koskenmies S, Hummel O, Rohde K, Schmidt RE, Dominiczak AF, Gahr M, Hollis T, Perrino FW, Lieberman J, Hübner N. Mutations in the gene encoding the 3'-5' DNA exonuclease TREX1 are associated with systemic lupus erythematosus. Nat Genet. 2007;39:1065-1067. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 510] [Cited by in RCA: 547] [Article Influence: 30.4] [Reference Citation Analysis (0)] |