Published online Dec 7, 2021. doi: 10.3748/wjg.v27.i45.7792
Peer-review started: April 20, 2021
First decision: July 27, 2021
Revised: August 5, 2021
Accepted: November 20, 2021
Article in press: November 20, 2021
Published online: December 7, 2021
Processing time: 226 Days and 16.7 Hours
Allogeneic hematopoietic stem cell transplantation (aHSCT) is a standard validated therapy for patients suffering from malignant and nonmalignant hematological diseases. However, aHSCT procedures are limited by potentially life-threatening complications, and one of the most serious complications is acute graft-versus-host disease (GVHD). During the last decades, DNA sequencing technologies were used to investigate relationship between composition or function of the gut microbiome and disease states. Even if it remains unclear whether these microbiome alterations are causative or secondary to the presence of the disease, they may be useful for diagnosis, prevention and therapy in aHSCT recipients. Here, we summarized the most recent findings of the association between human gut microbiome changes and acute GVHD in patients receiving aHSCT.
Core Tip: This review reports the compositional and functional changes in gut microbiome of allogeneic hematopoietic stem cell transplantation recipients associated with acute graft-versus-host disease that could serve a biomarker for diagnosis and prevention in patients receiving allogeneic hematopoietic stem cell transplantation.
- Citation: Le Bastard Q, Chevallier P, Montassier E. Gut microbiome in allogeneic hematopoietic stem cell transplantation and specific changes associated with acute graft vs host disease. World J Gastroenterol 2021; 27(45): 7792-7800
- URL: https://www.wjgnet.com/1007-9327/full/v27/i45/7792.htm
- DOI: https://dx.doi.org/10.3748/wjg.v27.i45.7792
In human health, DNA sequencing technologies, including 16S rRNA gene-based amplicon sequence analysis and whole genome shotgun metagenomic analysis were used to discover how exogenous and intrinsic host factors influence gut microbiome composition[1,2]. In large cohorts, scientists investigated the impact of factors such as lifestyle, dietary information, anthropometrics and drugs on the gut microbiome communities[3,4]. They found that age, gender, dietary factors and intrinsic para
In addition, DNA sequencing technologies were used to investigate associations between composition or function of the gut microbiome and various disease states. Studies performed cross sectional comparisons between subjects with and without disease, and reported changes in the gut microbiome of patients with inflammatory bowel diseases, colorectal cancer, diabetes, obesity and metabolic disease, or auto
Allogeneic hematopoietic stem cell transplantation (aHSCT) is a standard validated therapy, which every year allows an increasing number of patients suffering from malignant and nonmalignant hematological diseases, to benefit from an HSC transplant. The donor can be related (called geno-identical if HLA matched or haploidentical if sharing only one haplotype) or unrelated (called pheno-identical if HLA matched and including cord blood). However, aHSCT procedures are limited by potentially life-threatening complications, and one of the most serious complications is acute graft-versus-host disease (GVHD). GVHD occurs when immune competent T cells in the donated tissue recognize the recipient as foreign[9], that induces tissue damages in target organs of the host[10], including skin, liver and the gut, and typically occurs 3 to 6 wk after transplantation in nearly 60% of related donor transplants and 80% of unrelated donor transplants[11]. Hahn et al[12] assessed acute GVHD risk factors in 1960 adults after HLA-identical sibling myeloablative transplant in 226 centers worldwide. They found that cyclophosphamide and total-body irra
Based on preclinical data demonstrating reduced acute GVHD severity in germ-free or antibiotic-treated mice (i.e., gut microbiome with reduced diversity and richness), the gut microbiome was suggested to play a pivotal role in the pathogenesis of acute GVHD following aHSCT[13]. These findings are in contradiction with the common paradigm that loss of microbiome diversity is associated with diseases[14]. Most recent studies, based on microbiome sequencing, however, reported that higher diversity of intestinal microbiota at the time of neutrophil engraftment was associated with lower mortality in patients undergoing allogeneic hematopoietic-cell transplantation[15].
Here, we summarized the most recent findings of the association between human gut microbiome changes and graft vs host disease of the intestinal tract in patients receiving aHSCT.
Several studies reported the profound alteration of the gut microbiota during the aHSCT procedure, as summarized in Table 1. Following various conditioning regimens, both myeloablative or of reduced-intensity, a recent 16S rRNA gene-based amplicon sequence analysis that profiled 8767 fecal samples from 1362 patients undergoing aHSCT at four different centers observed a significant gut microbiota disruption characterized by loss of diversity and domination by single taxa[15], defined as occupation of at least 30% of the gut microbiota by a single predominating bacterial taxon. They also identified an association between lower intestinal diversity and higher risks of transplantation-related death. Moreover, samples collected prior transplantation also showed evidence of microbiome disruption, and lower diversity before transplantation was associated with poor survival. In another study, mortality outcomes were significantly worse in patients with lower intestinal diversity, with an overall survival at 3 years of 36% in low gut microbiome diversity patients compared to 67% in high diversity groups. Overall, low diversity showed a strong effect on mortality after multivariate adjustment for other clinical predictors. Furthermore, in subjects with lower diversity, the gut microbiota was generally dominated by a single bacterial genus, including Enterococcus, Streptococcus, Enterobacteriaceae (Escherichia and Kluyvera), and Lactobacillus[16].
| Ref. | Sequencing technology | Change in diversity | Changes in composition | Change in functions |
| Peled et al[15] | 16S rRNA gene-based amplicon sequence | Loss of diversity | Gut microbiota dominate by single taxa (occupation of at least 30% of the gut microbiota by a single predominating bacterial taxon), including Enterococcus, Streptococcus, Enterobacteriaceae (Escherichia and Kluyvera), and Lactobacillus | |
| Taur et al[16] | 16S rRNA gene-based amplicon sequence | Decrease of the gut microbiome diversity index | Gut microbiota composition frequently dominated by a single bacterial taxon, including Enterococcus, Streptococcus or Proteobacteria | |
| Ilett et al[10] | Metagenomic shotgun | Loss of gut microbiota diversity | Post-aHSCT samples enriched in Staphylococcus, Eggerthella, Streptococcus, Enterococcus and Lactobacillus compared to pre-aHSCT samples, &t species level enrichment in Enterococcus faecium, Lactobacillus delbrieckii, Staphylococcus epidermidis, and Streptococcus thermophilus | |
| Montassier et al[19] | 16S rRNA gene-based amplicon sequence | Loss of gut microbiota diversity | Decreases in abundances of Firmicutes and Actinobacteria, and significant increases in abundances of Proteobacteria | Reduced capacity for nucleotide metabolism, energy metabolism, metabolism of cofactors and vitamins, and increased capacity for glycan metabolism, signal transduction and xenobiotics biodegradation |
Taur et al[16] analyzed a total of 439 fecal specimens obtained from 94 patients during their transplant hospitalization and reported that over the course of aHSCT, gut microbiome diversity index decreased and remained low until the end of the observation period (Day 35 post-transplant). Patients also demonstrated significant changes in gut microbiome composition, and in most patients, the microbial composition became dominated by a single bacterial taxon: Enterococcus (40% of the patients), Streptococcus (37%) and the phylum Proteobacteria (13%). Interestingly, they demonstrated that patients with enterococcal domination in the gut had a 9-fold increased risk of Vancomycin Resistant Enterococcus bacteremia, and intestinal domination by Proteobacteria increased the risk of bacteremia with aerobic gram-negative bacilli 5-fold[17]. In a recent study using metagenomic shotgun, able to achieve bacterial identification to species and strain level, Ilett et al[10] reported that, in addition to a significant loss of gut microbiota diversity, post-aHSCT samples were enriched in Staphylococcus, Eggerthella, Streptococcus, Enterococcus and Lactobacillus compared to pre-aHSCT samples, and several samples were dominated by a single micro-organism [Enterococcus (n = 41 of 112) or Streptococcus (n = 10 of 112)]. At species level, they observed an enrichment of Enterococcus faecium, Lactobacillus delbrieckii, Staphylococcus epidermidis, and Streptococcus thermophilus in the post-aHSCT samples compared to pre-aHSCT samples[10].
In patients receiving intensive chemotherapy regimen used as myeloablative conditioning treatment to prepare patients for aHSCT, gut microbiota after che
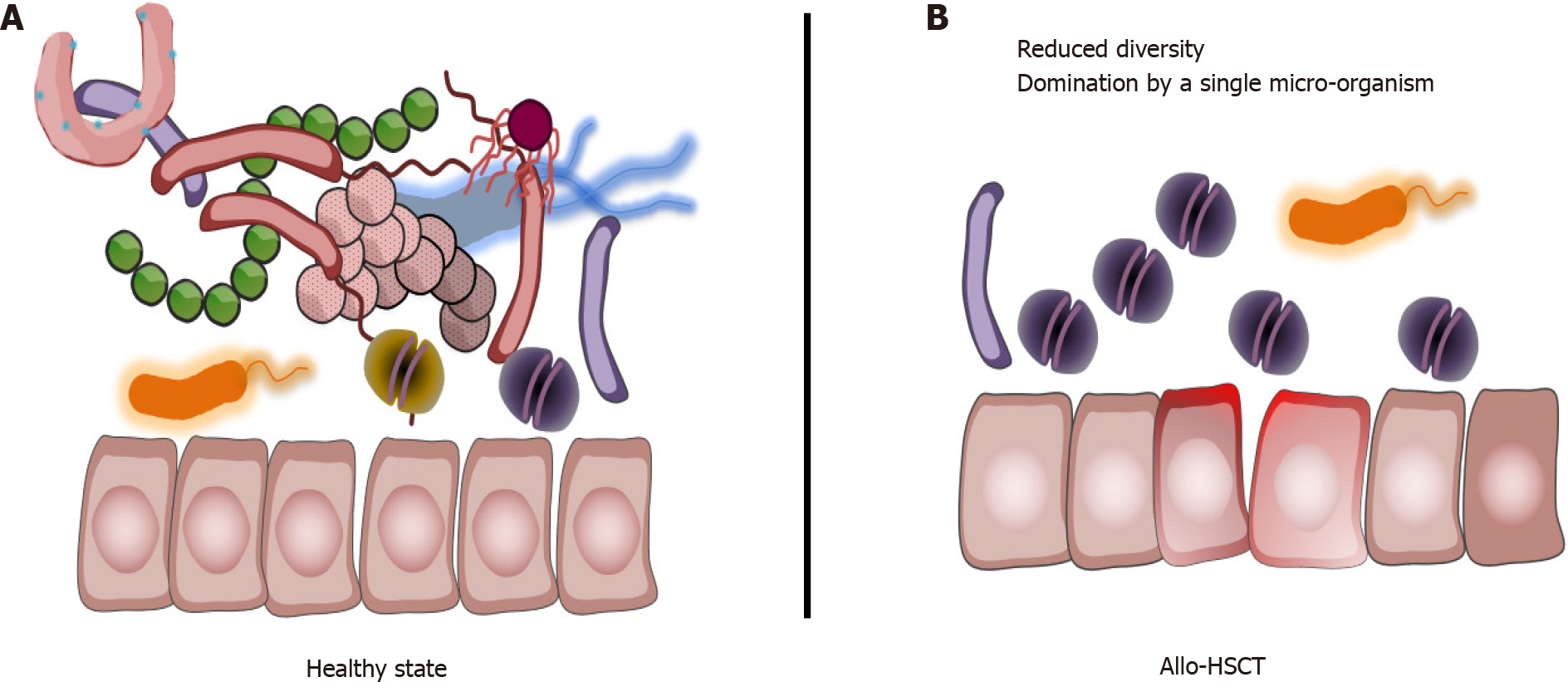
Thus, overall, aHSCT procedure is associated with a loss of gut microbiota diversity, a decrease in micro-organisms associated with health-promoting effects[20], and with a significant increase or domination by potentially pathobionts (Figure 1).
Studies reported that decreased gut microbiota diversity and richness was associated with the onset of acute intestinal GVHD. Jenq et al[21], in 2015, reported in a cohort of 115 patients receiving aHSCT, that increased bacterial diversity was associated with reduced GVHD-related mortality[21]. Moreover, in their recent 16S rRNA gene-based amplicon sequence analysis, Peled et al[15] found that higher intestinal diversity was associated with decreased risk of deaths attributable to GVHD (17 GVHD-related deaths among 244 patients in the higher-diversity group vs 26 such deaths among 184 patients in the lower-diversity group; hazard ratio, 0.49; 95%CI: 0.26-0.90)[15].
In a cohort of 44 patients, in which 16 (36%) experienced acute intestinal GVHD (median time to diagnosis: 53 d), Galloway-Peña et al[22] found that lower Shannon diversity index, popular diversity index in the ecological literature, of fecal samples collected at the time of engraftment was significantly associated with increased incidence of acute GVHD[22]. Golob et al[23] also found that diversity was statistically significantly lower in patients with acute GVHD when compared to those with no acute GVHD[23]. In another cohort of 57 patients, Liu et al[24] reported that decreased gut microbiota recipients’ diversity was not associated with decreased risk of aGVHD. However, they found that high gut microbiota donor diversity was associated with decreased risk of acute GVHD[24]. In a cohort of 70 patients, Payen et al[25] reported that patients with severe aGVHD had reduced gut microbiota diversity at disease onset, whereas patients with mild aGVHD had gut microbiota diversity more similar to those of controls[25].
Ilett et al[10] applied shotgun metagenomic sequencing to study a large cohort of adults (n = 150) undergoing aHSCT. Among them, 36 developed acute GVHD (median time to development: 34 d (interquartile range, 26-50 d post-aHSCT). They did not find significant association between diversity measures and acute GVHD in samples collected from the pre-aHSCT period. However, in samples collected in the early post-aHSCT period, patients who later developed acute GVHD had a significantly lower gene richness compared with those who did not develop acute GVHD[10].
Altogether, these findings clearly associate decreased diversity and richness of the gut microbiota, known to be associated with enhanced inflammation and impaired immunity[26], to onset of acute GVHD (Table 2).
| Ref. | Sequencing technology | Change in diversity | Changes in composition | Change in functions |
| Ilett et al[10] (150 patients) | Shotgun metagenomic | Decrease in gene richness during the early post-aHSCT period in patients with aGVHD | Decrease in Akkermansia muciniphila, Blautia obeum, Blautia hydrogenotropica, and Blautia hansenii during the early post-aHSCT period in patients with aGVHD | Increase in toxin named PetZ, that triggers bacterial autolysis in pathological bacteria during the pre-aHSCT and the early post-aHSCT period in patients with aGVHD |
| Holler et al[27] (31 patients) | 16S rRNA V3 sequencing | Increase in enterococci in patients who subsequently developed acute GVHD | ||
| Galloway-Peña et al[22] (44 patients) | 16S rRNA V4 sequencing | Lower Shannon diversity index in fecal samples collected at the time of engraftment in patients with subsequent aGVHD | Coriobacteriia negatively correlated with the incidence of acute GVHD | Fecal metabolites (Fecal indole and butyrate levels determined using liquid chromatography tandem mass spectrometry) associated with acute GVHD |
| Liu et al[24] (57 patients) | 16S rRNA V4 sequencing | High gut microbiota donor diversity associated with decreased risk of aGVHD in recipient | ||
| Doki et al[29] (107 patients) | 16S rRNA V4 sequencing | Higher abundance of phylum Firmicutes in samples collected before aHSCT in patients with acute GVHD | ||
| Golob et al[23] (66 patients) | 16S rRNA V3-V4 sequencing | Diversity significantly lower in patients with GVHD | Butyricicoccus, Bacteroides luti, Bacteroides thetaiotaomicron, Bacteroides ovatus, and Bacteroides caccae negatively correlated with subsequent acute GVHD, Rothia mucilaginosa, Solobacterium moorei, Veillonella parvula, and Bacteroides dorei positively correlated with subsequent onset of GVHD | |
| Michonneau et al[31] (99 patients) | Metabolomics | Significant decrease in tryptophan metabolites, including microbiota-produced compounds, such as 3-indoxyl sulfate, indoleacetate, indoleacetylglutamine, and indolepropionate in patients with aGVHD | ||
| Payen et al[25] (70 patients) | 16S rRNA V3-V4 sequencing | Decreased diversity in acute GVHD | Lachnoclostridium, Blautia, Sellimonas, Anaerostipes, Faecalibacterium, Flavonifractor, Erysipelatoclostridium and Lactococcus negatively associated with subsequent acute GVHD |
Using a 16S rRNA-based sequencing analysis, Jenq et al[21] reported that genus Blautia were most significantly associated with reduced GVHD-related mortality, whereas genus Veillonella, was associated with increased GVHD-related mortality[21].
Holler et al[27] reported that post-transplant samples were increased in enterococci, and that the increase was more pronounced in patients developing acute GVHD compared to patients who did not develop aGVDH. They confirmed this trend using enterococcal polymerase chain reaction (PCR) and observed a predominance of Enterococcus faecium and Enterococcus faecalis in samples collected in patients who developed acute GVHD. Moreover, post-transplant samples collected in patients with acute GVHD were significantly depleted in Clostridia and Eubacterium rectale[27]. Stein-Thoeringer et al[28] also reported that fecal domination by Enterococcus (i.e., relative genus abundance ≥ 30% in samples collected during early post-transplant period) was associated with increased risk of acute GVHD, and increased GVHD-related mortality[28]. Importantly, in a in gnotobiotic model, the authors demonstrated that Enterococcus growth is dependent on disaccharide lactose, and that dietary lactose depletion attenuates Enterococcus outgrowth and reduces the severity of GVHD[28].
In their cohort of 44 patients, Galloway-Peña et al[22] reported that only one taxon at the time of engraftment, Coriobacteriia, a class of Gram-positive bacteria within the Actinobacteria phylum, was negatively correlated with the incidence of aGVHD[22]. In another cohort of 107 patients, Doki et al[29] reported patients who developed acute GVHD exhibited a significantly higher abundance of phylum Firmicutes than patients who did not develop aGVHD[29].
Golob et al[23] found in a cohort of 66 patients, that in samples collected at neutrophil recovery post-HCT, the presence of Actinobacteria and Firmicutes was positively correlated with subsequent acute GVHD, whereas Lachnospiraceae were negatively correlated. In detail, Butyricicoccus, Bacteroides luti, Bacteroides thetaiotaomicron, Bacteroides ovatus, and Bacteroides caccae were negatively correlated with subsequent acute severe GVHD, while Rothia mucilaginosa, Solobacterium moorei, Veillonella parvula, and Bacteroides dorei were positively correlated[23]. Payen et al[25] reported that Lachnoclostridium, Blautia, Sellimonas, Anaerostipes, Faecalibacterium, Flavonifractor, Erysipelatoclostridium and Lactococcus were negatively associated with subsequent acute severe GVHD, whereas Prevotella and Stenotrophomonas were considered positive biomarkers of severe aGVHD. Moreover, using qPCR, they observed a significant depletion of the Blautia coccoides group (cluster XIVa) in patients with aGVHD compared with controls and patients with no aGVHD[25].
Based on a shotgun metagenomic sequencing analysis in 150 patients, Ilett et al[10] found that no bacteria were associated with acute GVHD in samples collected during the pre-aHSCT period. In samples collected during the early post-aHSCT period, they found that Blautia, Akkermansia, and Campylobacter, as well as the specific species Akkermansia muciniphila, Blautia obeum, Blautia hydrogenotropica, and Blautia hansenii were all significantly associated with reduced risk of a GVHD[10]. Still using shotgun metagenomic sequencing analysis, Turner assessed samples collected in nine patients with aGVHD and treated with standard-of-care high-dose steroids. Three of these patients were steroid-refractory, whereas six had a response. They showed that Dorea longicatena was associated with response to high-dose steroids treatment whereas Akkermansia muciniphila was associated with refractoriness. They also reported that maintenance of a stable Dorea/Akkermansia ratio predicted steroid response, whereas a decline in this ratio preceded refractory disease. Importantly, Shono et al[30] pre
Further studies are needed to confirm or not the controversial role of Akkermansia in acute GVHD. Moreover, some species of the genus Blautia should be investigated as a potential biomarker: High relative abundance at the time of engraftment being protective against GVHD, while low relative abundance could be considered a risk factor for secondary development of GVHD.
The functional alterations of the intestinal microbiome in patients receiving aHSCT are currently poorly described because the majority of studies have used 16S ribosomal RNA gene-based technics, which boil down to providing bacterial taxonomy only at the genus level, but are ineffective in obtaining functional information. In a cohort of 44 patients, Galloway-Peña et al[22] reported that fecal metabolites (fecal indole and butyrate levels) were not associated with aGVHD[22]. In another study, Michonneau et al[31] reported that aGVHD was characterized by specific metabolomics changes in two cohorts of patients (n = 99). They found that bile acids, plasmalogens, tryptophan, and arginine metabolites were the main contributors involved, especially the aryl hydrocarbon receptor ligand 3-indoxyl sulfate. In addition to host-derived metabolites, they also identified significant variation in microbiota-derived indole compounds, especially in aryl hydrocarbon receptor ligands. The authors suggested that allogeneic immune response during aGVHD might be influenced by bile acids and by the decreased production of aryl hydrocarbon receptor ligands by gut microbiome that could limit indoleamine 2,3-dioxygenase induction and influence allogeneic T cell reactivity[31]. To assess if altered composition of the gut microbiota may result in an altered metabolome, which potentially disrupts functionalities at the onset of aGVHD, Payen et al[25] quantified the Short Chain Fatty Acids (SCFA) content in fecal samples with measurement of total SCFAs and acetate, propionate, and butyrate. They found that the fecal amount of all SCFAs was drastically diminished at aGVHD onset. In detail, they observed that total SCFAs, acetate and butyrate respectively decreased by 80%, 75% and 95% in severe aGVHD patients as compared with controls[25]. Importantly, in a mouse model, Mathewson et al[32] demonstrated that butyrate restoration improved intestinal epithelial cells junctional integrity and mitigated aGVHD[32].
Based on a shotgun metagenomic sequencing analysis, Ilett et al[10] found that a total of 1267 and 1289 genes were present in significantly different amounts among those who developed aGVHD vs those who did not develop aGVHD during pre-HSCT and early post-aHSCT, respectively. Of these genes, 24 overlapped between the 2 time periods, all being significantly higher in abundance among those who did not develop aGVHD. They pointed out that 1 gene (O2.CD1-0-PT_GL0039283) had a 3-log fold-change in both periods and is known to function as a toxin named Petz, that triggers bacterial autolysis in pathological bacteria[10].
The investigations described in this mini review provide an understanding of the role of the gut microbiome in the pathophysiology of aGVHD in patients receiving aHSCT. Observational studies have shown that a decrease in diversity of the gut microbiome and specific species or metabolic pathways were associated with aGVHD. These specific changes could serve as biomarkers for diagnosis and prevention in patients receiving aHSCT. Moreover, functional alterations of the gut microbiome during aHSCT should be more investigated so that modulations of the gut microbiome could be tested to prevent this potentially life-threatening complication.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: France
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Hegazy MAE, Sameer S S-Editor: Fan JR L-Editor: A P-Editor: Fan JR
| 1. | Kurilshikov A, Medina-Gomez C, Bacigalupe R, Radjabzadeh D, Wang J, Demirkan A, Le Roy CI, Raygoza Garay JA, Finnicum CT, Liu X, Zhernakova DV, Bonder MJ, Hansen TH, Frost F, Rühlemann MC, Turpin W, Moon JY, Kim HN, Lüll K, Barkan E, Shah SA, Fornage M, Szopinska-Tokov J, Wallen ZD, Borisevich D, Agreus L, Andreasson A, Bang C, Bedrani L, Bell JT, Bisgaard H, Boehnke M, Boomsma DI, Burk RD, Claringbould A, Croitoru K, Davies GE, van Duijn CM, Duijts L, Falony G, Fu J, van der Graaf A, Hansen T, Homuth G, Hughes DA, Ijzerman RG, Jackson MA, Jaddoe VWV, Joossens M, Jørgensen T, Keszthelyi D, Knight R, Laakso M, Laudes M, Launer LJ, Lieb W, Lusis AJ, Masclee AAM, Moll HA, Mujagic Z, Qibin Q, Rothschild D, Shin H, Sørensen SJ, Steves CJ, Thorsen J, Timpson NJ, Tito RY, Vieira-Silva S, Völker U, Völzke H, Võsa U, Wade KH, Walter S, Watanabe K, Weiss S, Weiss FU, Weissbrod O, Westra HJ, Willemsen G, Payami H, Jonkers DMAE, Arias Vasquez A, de Geus EJC, Meyer KA, Stokholm J, Segal E, Org E, Wijmenga C, Kim HL, Kaplan RC, Spector TD, Uitterlinden AG, Rivadeneira F, Franke A, Lerch MM, Franke L, Sanna S, D'Amato M, Pedersen O, Paterson AD, Kraaij R, Raes J, Zhernakova A. Large-scale association analyses identify host factors influencing human gut microbiome composition. Nat Genet. 2021;53:156-165. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 590] [Cited by in RCA: 1051] [Article Influence: 262.8] [Reference Citation Analysis (0)] |
| 2. | Rothschild D, Weissbrod O, Barkan E, Kurilshikov A, Korem T, Zeevi D, Costea PI, Godneva A, Kalka IN, Bar N, Shilo S, Lador D, Vila AV, Zmora N, Pevsner-Fischer M, Israeli D, Kosower N, Malka G, Wolf BC, Avnit-Sagi T, Lotan-Pompan M, Weinberger A, Halpern Z, Carmi S, Fu J, Wijmenga C, Zhernakova A, Elinav E, Segal E. Environment dominates over host genetics in shaping human gut microbiota. Nature. 2018;555:210-215. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1342] [Cited by in RCA: 1842] [Article Influence: 263.1] [Reference Citation Analysis (0)] |
| 3. | Zhernakova A, Kurilshikov A, Bonder MJ, Tigchelaar EF, Schirmer M, Vatanen T, Mujagic Z, Vila AV, Falony G, Vieira-Silva S, Wang J, Imhann F, Brandsma E, Jankipersadsing SA, Joossens M, Cenit MC, Deelen P, Swertz MA; LifeLines cohort study, Weersma RK, Feskens EJ, Netea MG, Gevers D, Jonkers D, Franke L, Aulchenko YS, Huttenhower C, Raes J, Hofker MH, Xavier RJ, Wijmenga C, Fu J. Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science. 2016;352:565-569. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1029] [Cited by in RCA: 1304] [Article Influence: 144.9] [Reference Citation Analysis (0)] |
| 4. | Vandeputte D, Falony G, Vieira-Silva S, Tito RY, Joossens M, Raes J. Stool consistency is strongly associated with gut microbiota richness and composition, enterotypes and bacterial growth rates. Gut. 2016;65:57-62. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 575] [Cited by in RCA: 708] [Article Influence: 78.7] [Reference Citation Analysis (0)] |
| 5. | Haiser HJ, Gootenberg DB, Chatman K, Sirasani G, Balskus EP, Turnbaugh PJ. Predicting and manipulating cardiac drug inactivation by the human gut bacterium Eggerthella lenta. Science. 2013;341:295-298. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 410] [Cited by in RCA: 499] [Article Influence: 41.6] [Reference Citation Analysis (0)] |
| 6. | ElRakaiby M, Dutilh BE, Rizkallah MR, Boleij A, Cole JN, Aziz RK. Pharmacomicrobiomics: the impact of human microbiome variations on systems pharmacology and personalized therapeutics. OMICS. 2014;18:402-414. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 112] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 7. | Young VB. The role of the microbiome in human health and disease: an introduction for clinicians. BMJ. 2017;356:j831. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 287] [Cited by in RCA: 311] [Article Influence: 38.9] [Reference Citation Analysis (0)] |
| 8. | Yang J, McDowell A, Kim EK, Seo H, Lee WH, Moon CM, Kym SM, Lee DH, Park YS, Jee YK, Kim YK. Development of a colorectal cancer diagnostic model and dietary risk assessment through gut microbiome analysis. Exp Mol Med. 2019;51:1-15. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 77] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 9. | Zeiser R, Blazar BR. Acute Graft-versus-Host Disease - Biologic Process, Prevention, and Therapy. N Engl J Med. 2017;377:2167-2179. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 630] [Cited by in RCA: 915] [Article Influence: 114.4] [Reference Citation Analysis (0)] |
| 10. | Ilett EE, Jørgensen M, Noguera-Julian M, Nørgaard JC, Daugaard G, Helleberg M, Paredes R, Murray DD, Lundgren J, MacPherson C, Reekie J, Sengeløv H. Associations of the gut microbiome and clinical factors with acute GVHD in allogeneic HSCT recipients. Blood Adv. 2020;4:5797-5809. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 49] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 11. | Vargas-Díez E, García-Díez A, Marín A, Fernández-Herrera J. Life-threatening graft-vs-host disease. Clin Dermatol. 2005;23:285-300. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 27] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 12. | Hahn T, McCarthy PL Jr, Zhang MJ, Wang D, Arora M, Frangoul H, Gale RP, Hale GA, Horan J, Isola L, Maziarz RT, van Rood JJ, Gupta V, Halter J, Reddy V, Tiberghien P, Litzow M, Anasetti C, Pavletic S, Ringdén O. Risk factors for acute graft-versus-host disease after human leukocyte antigen-identical sibling transplants for adults with leukemia. J Clin Oncol. 2008;26:5728-5734. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 145] [Cited by in RCA: 151] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 13. | van Bekkum DW, Roodenburg J, Heidt PJ, van der Waaij D. Mitigation of secondary disease of allogeneic mouse radiation chimeras by modification of the intestinal microflora. J Natl Cancer Inst. 1974;52:401-404. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 239] [Cited by in RCA: 236] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 14. | Mosca A, Leclerc M, Hugot JP. Gut Microbiota Diversity and Human Diseases: Should We Reintroduce Key Predators in Our Ecosystem? Front Microbiol. 2016;7:455. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 289] [Cited by in RCA: 389] [Article Influence: 43.2] [Reference Citation Analysis (0)] |
| 15. | Peled JU, Gomes ALC, Devlin SM, Littmann ER, Taur Y, Sung AD, Weber D, Hashimoto D, Slingerland AE, Slingerland JB, Maloy M, Clurman AG, Stein-Thoeringer CK, Markey KA, Docampo MD, Burgos da Silva M, Khan N, Gessner A, Messina JA, Romero K, Lew MV, Bush A, Bohannon L, Brereton DG, Fontana E, Amoretti LA, Wright RJ, Armijo GK, Shono Y, Sanchez-Escamilla M, Castillo Flores N, Alarcon Tomas A, Lin RJ, Yáñez San Segundo L, Shah GL, Cho C, Scordo M, Politikos I, Hayasaka K, Hasegawa Y, Gyurkocza B, Ponce DM, Barker JN, Perales MA, Giralt SA, Jenq RR, Teshima T, Chao NJ, Holler E, Xavier JB, Pamer EG, van den Brink MRM. Microbiota as Predictor of Mortality in Allogeneic Hematopoietic-Cell Transplantation. N Engl J Med. 2020;382:822-834. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 304] [Cited by in RCA: 493] [Article Influence: 98.6] [Reference Citation Analysis (1)] |
| 16. | Taur Y, Jenq RR, Perales MA, Littmann ER, Morjaria S, Ling L, No D, Gobourne A, Viale A, Dahi PB, Ponce DM, Barker JN, Giralt S, van den Brink M, Pamer EG. The effects of intestinal tract bacterial diversity on mortality following allogeneic hematopoietic stem cell transplantation. Blood. 2014;124:1174-1182. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 544] [Cited by in RCA: 674] [Article Influence: 61.3] [Reference Citation Analysis (2)] |
| 17. | Taur Y, Xavier JB, Lipuma L, Ubeda C, Goldberg J, Gobourne A, Lee YJ, Dubin KA, Socci ND, Viale A, Perales MA, Jenq RR, van den Brink MR, Pamer EG. Intestinal domination and the risk of bacteremia in patients undergoing allogeneic hematopoietic stem cell transplantation. Clin Infect Dis. 2012;55:905-914. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 609] [Cited by in RCA: 756] [Article Influence: 58.2] [Reference Citation Analysis (0)] |
| 18. | Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Vega Thurber RL, Knight R, Beiko RG, Huttenhower C. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 2013;31:814-821. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5694] [Cited by in RCA: 6303] [Article Influence: 525.3] [Reference Citation Analysis (0)] |
| 19. | Montassier E, Gastinne T, Vangay P, Al-Ghalith GA, Bruley des Varannes S, Massart S, Moreau P, Potel G, de La Cochetière MF, Batard E, Knights D. Chemotherapy-driven dysbiosis in the intestinal microbiome. Aliment Pharmacol Ther. 2015;42:515-528. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 238] [Cited by in RCA: 354] [Article Influence: 35.4] [Reference Citation Analysis (1)] |
| 20. | Eisenstein M. The hunt for a healthy microbiome. Nature. 2020;577:S6-S8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 48] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 21. | Jenq RR, Taur Y, Devlin SM, Ponce DM, Goldberg JD, Ahr KF, Littmann ER, Ling L, Gobourne AC, Miller LC, Docampo MD, Peled JU, Arpaia N, Cross JR, Peets TK, Lumish MA, Shono Y, Dudakov JA, Poeck H, Hanash AM, Barker JN, Perales MA, Giralt SA, Pamer EG, van den Brink MR. Intestinal Blautia Is Associated with Reduced Death from Graft-versus-Host Disease. Biol Blood Marrow Transplant. 2015;21:1373-1383. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 587] [Cited by in RCA: 572] [Article Influence: 57.2] [Reference Citation Analysis (0)] |
| 22. | Galloway-Peña JR, Peterson CB, Malik F, Sahasrabhojane PV, Shah DP, Brumlow CE, Carlin LG, Chemaly RF, Im JS, Rondon G, Felix E, Veillon L, Lorenzi PL, Alousi AM, Jenq RR, Kontoyiannis DP, Shpall EJ, Shelburne SA, Okhuysen PC. Fecal Microbiome, Metabolites, and Stem Cell Transplant Outcomes: A Single-Center Pilot Study. Open Forum Infect Dis. 2019;6:ofz173. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 27] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 23. | Golob JL, Pergam SA, Srinivasan S, Fiedler TL, Liu C, Garcia K, Mielcarek M, Ko D, Aker S, Marquis S, Loeffelholz T, Plantinga A, Wu MC, Celustka K, Morrison A, Woodfield M, Fredricks DN. Stool Microbiota at Neutrophil Recovery Is Predictive for Severe Acute Graft vs Host Disease After Hematopoietic Cell Transplantation. Clin Infect Dis. 2017;65:1984-1991. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 99] [Cited by in RCA: 152] [Article Influence: 21.7] [Reference Citation Analysis (0)] |
| 24. | Liu C, Frank DN, Horch M, Chau S, Ir D, Horch EA, Tretina K, van Besien K, Lozupone CA, Nguyen VH. Associations between acute gastrointestinal GvHD and the baseline gut microbiota of allogeneic hematopoietic stem cell transplant recipients and donors. Bone Marrow Transplant. 2017;52:1643-1650. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 59] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 25. | Payen M, Nicolis I, Robin M, Michonneau D, Delannoye J, Mayeur C, Kapel N, Berçot B, Butel MJ, Le Goff J, Socié G, Rousseau C. Functional and phylogenetic alterations in gut microbiome are linked to graft-versus-host disease severity. Blood Adv. 2020;4:1824-1832. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 65] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 26. | Simms-Waldrip TR, Sunkersett G, Coughlin LA, Savani MR, Arana C, Kim J, Kim M, Zhan X, Greenberg DE, Xie Y, Davies SM, Koh AY. Antibiotic-Induced Depletion of Anti-inflammatory Clostridia Is Associated with the Development of Graft-versus-Host Disease in Pediatric Stem Cell Transplantation Patients. Biol Blood Marrow Transplant. 2017;23:820-829. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 130] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 27. | Holler E, Butzhammer P, Schmid K, Hundsrucker C, Koestler J, Peter K, Zhu W, Sporrer D, Hehlgans T, Kreutz M, Holler B, Wolff D, Edinger M, Andreesen R, Levine JE, Ferrara JL, Gessner A, Spang R, Oefner PJ. Metagenomic analysis of the stool microbiome in patients receiving allogeneic stem cell transplantation: loss of diversity is associated with use of systemic antibiotics and more pronounced in gastrointestinal graft-versus-host disease. Biol Blood Marrow Transplant. 2014;20:640-645. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 354] [Cited by in RCA: 415] [Article Influence: 37.7] [Reference Citation Analysis (0)] |
| 28. | Stein-Thoeringer CK, Nichols KB, Lazrak A, Docampo MD, Slingerland AE, Slingerland JB, Clurman AG, Armijo G, Gomes ALC, Shono Y, Staffas A, Burgos da Silva M, Devlin SM, Markey KA, Bajic D, Pinedo R, Tsakmaklis A, Littmann ER, Pastore A, Taur Y, Monette S, Arcila ME, Pickard AJ, Maloy M, Wright RJ, Amoretti LA, Fontana E, Pham D, Jamal MA, Weber D, Sung AD, Hashimoto D, Scheid C, Xavier JB, Messina JA, Romero K, Lew M, Bush A, Bohannon L, Hayasaka K, Hasegawa Y, Vehreschild MJGT, Cross JR, Ponce DM, Perales MA, Giralt SA, Jenq RR, Teshima T, Holler E, Chao NJ, Pamer EG, Peled JU, van den Brink MRM. Lactose drives Enterococcus expansion to promote graft-versus-host disease. Science. 2019;366:1143-1149. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 212] [Cited by in RCA: 248] [Article Influence: 41.3] [Reference Citation Analysis (0)] |
| 29. | Doki N, Suyama M, Sasajima S, Ota J, Igarashi A, Mimura I, Morita H, Fujioka Y, Sugiyama D, Nishikawa H, Shimazu Y, Suda W, Takeshita K, Atarashi K, Hattori M, Sato E, Watakabe-Inamoto K, Yoshioka K, Najima Y, Kobayashi T, Kakihana K, Takahashi N, Sakamaki H, Honda K, Ohashi K. Clinical impact of pre-transplant gut microbial diversity on outcomes of allogeneic hematopoietic stem cell transplantation. Ann Hematol. 2017;96:1517-1523. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 46] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 30. | Shono Y, Docampo MD, Peled JU, Perobelli SM, Velardi E, Tsai JJ, Slingerland AE, Smith OM, Young LF, Gupta J, Lieberman SR, Jay HV, Ahr KF, Porosnicu Rodriguez KA, Xu K, Calarfiore M, Poeck H, Caballero S, Devlin SM, Rapaport F, Dudakov JA, Hanash AM, Gyurkocza B, Murphy GF, Gomes C, Liu C, Moss EL, Falconer SB, Bhatt AS, Taur Y, Pamer EG, van den Brink MRM, Jenq RR. Increased GVHD-related mortality with broad-spectrum antibiotic use after allogeneic hematopoietic stem cell transplantation in human patients and mice. Sci Transl Med. 2016;8:339ra71. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 391] [Cited by in RCA: 421] [Article Influence: 46.8] [Reference Citation Analysis (0)] |
| 31. | Michonneau D, Latis E, Curis E, Dubouchet L, Ramamoorthy S, Ingram B, de Latour RP, Robin M, de Fontbrune FS, Chevret S, Rogge L, Socié G. Metabolomics analysis of human acute graft-versus-host disease reveals changes in host and microbiota-derived metabolites. Nat Commun. 2019;10:5695. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 101] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 32. | Mathewson ND, Jenq R, Mathew AV, Koenigsknecht M, Hanash A, Toubai T, Oravecz-Wilson K, Wu SR, Sun Y, Rossi C, Fujiwara H, Byun J, Shono Y, Lindemans C, Calafiore M, Schmidt TM, Honda K, Young VB, Pennathur S, van den Brink M, Reddy P. Gut microbiome-derived metabolites modulate intestinal epithelial cell damage and mitigate graft-versus-host disease. Nat Immunol. 2016;17:505-513. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 506] [Cited by in RCA: 505] [Article Influence: 56.1] [Reference Citation Analysis (0)] |