Published online Sep 14, 2021. doi: 10.3748/wjg.v27.i34.5666
Peer-review started: January 27, 2021
First decision: May 2, 2021
Revised: May 14, 2021
Accepted: August 3, 2021
Article in press: August 3, 2021
Published online: September 14, 2021
Processing time: 225 Days and 3 Hours
Gastrointestinal (GI) cancers are among the most common cancer types and leading causes of cancer-related deaths worldwide. There is a tremendous clinical need for effective early diagnosis for better healthcare of GI cancer patients. In this article, we provide a short overview of the recent advances in GI cancer diagnosis. In the first part, we discuss the applications of blood-based biomarkers, such as plasma circulating cell-free DNA, circulating tumor cells, extracellular vesicles, and circulating cell-free RNA, for cancer liquid biopsies. In the second part, we review the current trends of artificial intelligence (AI) for pathology image and tissue biopsy analysis for GI cancer, as well as deep learning-based approaches for purity assessment of tissue biopsies. We further provide our opinions on the future directions in blood-based and AI-enhanced approaches for GI cancer diagnosis, and we think that these fields will have more intensive integrations with clinical needs in the near future.
Core Tip: Recent studies have discovered a variety of blood-based biomarkers with great potential in improving the diagnosis and surveillance of gastrointestinal (GI) cancers. In this article, we review the latest advances in the diagnosis of various GI cancers, focusing on emerging blood-based liquid biopsy assays and artificial intelligence-enhanced approaches. We also discuss purity assessment approaches for tissue biopsies, which is an important issue in cancer studies, especially those applicable in metastatic GI cancers.
- Citation: Li LS, Guo XY, Sun K. Recent advances in blood-based and artificial intelligence-enhanced approaches for gastrointestinal cancer diagnosis. World J Gastroenterol 2021; 27(34): 5666-5681
- URL: https://www.wjgnet.com/1007-9327/full/v27/i34/5666.htm
- DOI: https://dx.doi.org/10.3748/wjg.v27.i34.5666
Gastrointestinal (GI) cancers, which include tumors from the colon, rectum, stomach, pancreas, esophagus, anus, gallbladder, liver, and bile duct, are among the most common cancer types and leading causes of cancer-related deaths worldwide[1]. Colorectal cancer (CRC), in particular, is the third most common cancer in men (9%) and women (8%) in the United States[1-3]. Most patients with GI cancers are in the advanced stage upon diagnosis and have relatively poor prognosis outcome. For example, the overall 5-year survival rate for patients with hepatocellular carcinoma (HCC) is around 20%[3], while the survival rate for early stage patients is as high as 70%[1,3]. This is because various efficient treatment strategies, such as surgical resection and organ transplantation, are feasible for early-stage patients only. Hence, early diagnosis is of high clinical significance for better healthcare of cancer patients.
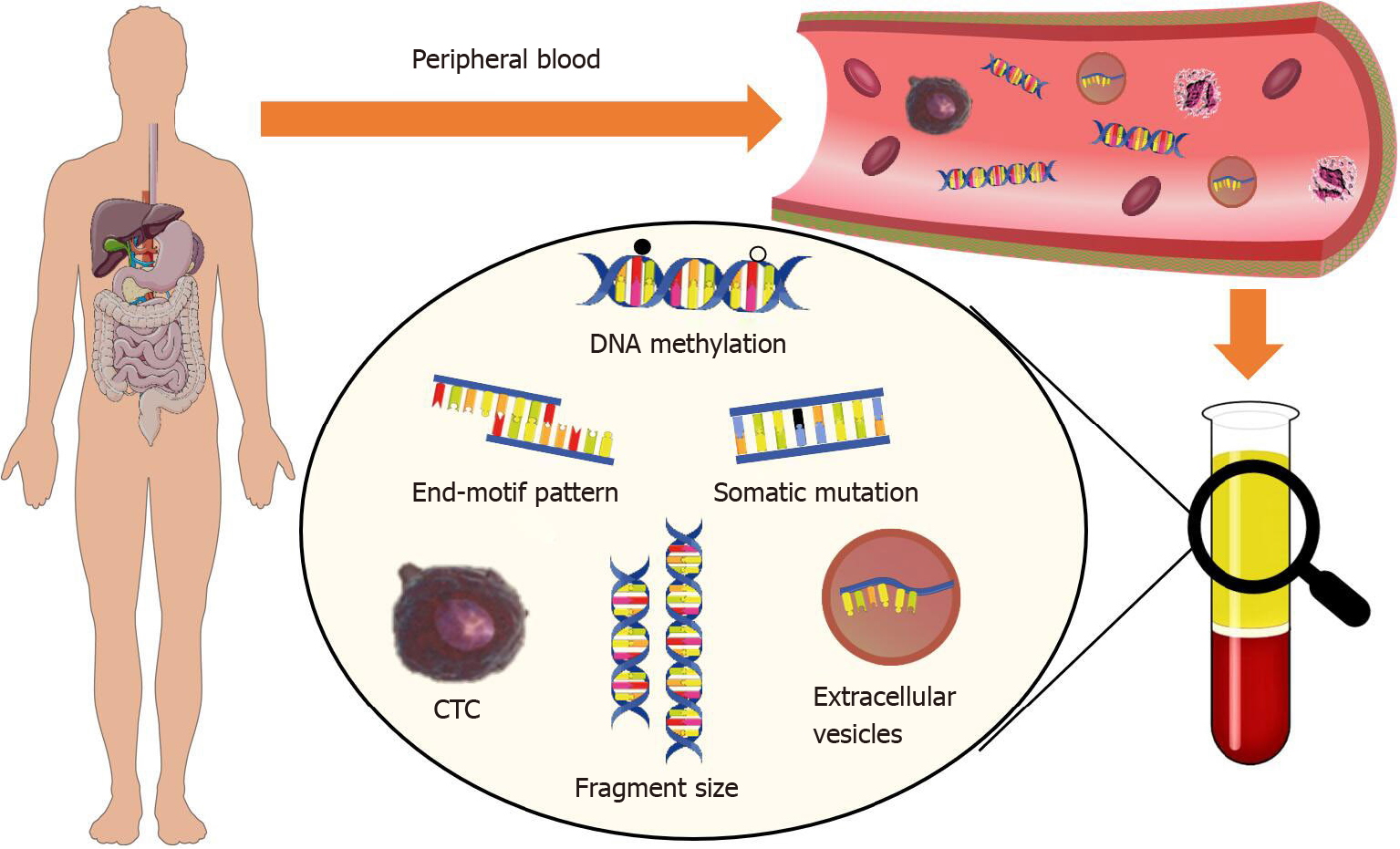
Pathological analysis of the tumor tissue is currently the “gold standard” for clinical diagnosis of cancers[4,5]. In particular, image-based computed tomography (CT) colonography, optical colonoscopy, and endoscopic biopsy are mostly widely used in the diagnosis of GI cancers. Recent studies have demonstrated that blood-based approaches, also known as liquid biopsy, offer substantial advantages over conventional tissue biopsy-based diagnostic methods, including minimally invasive nature, cost-efficiency, clinical convenience for real-time monitoring, and the potential to promote higher patient compliance. Major analytes in current liquid biopsy studies include circulating cell-free DNA (cfDNA), circulating tumor cells (CTCs), as well as extracellular vesicles (EVs) that transport the biomolecules from tumor cells, such as lipids, proteins, DNA fragment, and RNAs[6-9]. Moreover, advances and integration of artificial intelligence (AI) have brought revolutionary changes to the clinical and translational cancer studies[10-12], especially in cancer diagnosis. The identification and utilization of disease-specific biomarkers, and development of exquisite diagnostic algorithms are both highly valuable for early diagnosis of cancers.
In this article, we provide an overview of current approaches for GI cancer diagnosis, especially in CRC and HCC. We focus on blood-based biomarkers, as well as AI algorithms for image-based tissue biopsies. We review the identification of biomarkers and development of diagnostic methods, and provide our opinions on future directions in early diagnosis of GI cancers.
With the advantages of being highly efficient, minimally invasive, and cost-effective, liquid biopsy has become an emerging technique for early diagnosis of cancers. Tumor-derived cell-free DNA in peripheral blood provide a surrogate for resear
| Cancer type | Biomarker type | Biomarkers | No. of patients /controls | Accuracy | Ref. |
| GC | cfDNA copy number | HER2 gene | 60/30 | Sn = 73.3%, Sp = 93.3% | [19] |
| GC | cfDNA methylation | BARHL2 | 128/30 | Sn = 90%, Sp = 100% | [104] |
| GC | CTC | CTC level | 116/31 | Sn = 85.3%, Sp = 90.3% | [49] |
| GC | Exosomal lncRNA | lncRNA-UEGC1 | 10/5 | AUC = 0.876 | [105] |
| GC | Exosomal lncRNA | lncRNA-pcsk2-2:1 | 63/29 | Sn = 84%, Sp = 86.5% | [77] |
| GC | Exosomal lncRNA | lncRNA-GNAQ6:1 | 43/27 | AUC = 0.732 | [71] |
| GC | Exosomal lncRNA | lncRNA-HOTTIP | 126/120 | AUC = 0.827 | [106] |
| GC | Exosomal miRNA | miR-1246 | 117/82 | AUC = 0.843 | [107] |
| GC | Serum cfRNA | miR-10b-5p, miR-195-5p, miR-185-5p, | 441/233 | AUC = 0.702 | [68] |
| GC | Serum cfRNA | miR-21 | 50/50 | Sn = 88.4%, Sp = 79.6% | [81] |
| GC | Serum cfRNA | miR-30a-5p, miR-659-3p and miR-3917 | 354 | AUC = 0.82 | [82] |
| GC | Serum cfRNA | B3GALT5-AS1 | 107/87 | AUC = 0.816 | [108] |
| CRC | CTC mRNA | ECT2 gene | 90/151 | AUC = 0.821 | [61] |
| CRC | Serum cfRNA | miR-21, miR-29a, miR-125b | 160/77 | AUC = 0.827 | [86] |
| CRC | Serum cfRNA | miR-30a-5p | 138/60 | Sn = 77.5%, Sp = 78.3% | [109] |
| CRC | cfDNA methylation | SFRP2 | 62/55 | Sn = 69.4%, Sp = 87.3% | [21] |
| CRC | CTC protein | CD133+CD54+CD44+ Protein | 10/10 | Sn = 88.2%, Sp = 92.4% | [62] |
| CRC | Exosomal miRNA | miR-27a, miR-130a | 40/40 | AUC = 0.773 | [110] |
| CRC | Exosomal miRNA | miR-125a-3p | 50/50 | AUC = 0.685 | [111] |
| CRC | Exosomal protein | CPNE3 | 92/32 | Sn = 67.5%, Sp = 84.4% | [112] |
| PC | CTC protein | Vimentin | 100/30 | AUC = 0.968 | [58] |
| PC | cfDNA methylation | BNC1, ADAMTS1 | 39/95 | Sn = 94.8%, Sp = 91.6% | [23] |
| PC | cfDNA methylation | CDO1 | 160 | Sn = 95% | [24] |
| PC | CTC | CTC level | 126 | Sn = 100%, Sp = 88.6% | [113] |
| PC | Exosomal lncRNA | CLDN1, TIMP1, MAL2, MARCH2, ITIH2, | 284/117 | AUC = 0.931 | [79] |
| PC | Exosomal miRNA | miR-21 | 27 | AUC = 0.9 | [76] |
| PC | Exosomal miRNA | miR-155 | 27 | AUC = 0.89 | [76] |
| PC | Exosomal miRNA | miR-451a | 56 | Sn = 69.2%, Sp = 70.8% | [114] |
| PC | Exosomal miRNA | miR-133a | 110/64 | Sn = 90.6%, Sp = 87.2%, AUC = 0.893 | [115] |
| HCC | cfDNA methylation | cfDNA 5hmC | 1204/958 | AUC = 0.846 | [30] |
| HCC | Exosomal miRNA | miR-10b-5p | 90/28 | AUC = 0.934 | [75] |
| HCC | Exosomal mRNA | hnRNPH1 | 88/68 | Sn = 85.2%, Sp = 76.5% | [80] |
| HCC | Plasma cfRNA | hsa_circ_0000976, hsa_circ_0007750 and | 158/53 | AUC = 0.863 | [85] |
| HCC | Serum cfRNA | miR-143 | 131/122 | Sn = 80.3%, Sp = 82.4% | [87] |
| HCC | Serum cfRNA | miR-132, miR-212` | 80/42 | Sn = 93.75%, Sp = 63.44% | [116] |
cfDNA molecules in plasma originate from various tissues in human body. In cancer patients, the dying tumor cells release their DNA, which contains the tumor-specific signatures, including somatic mutations, copy number aberrations (CNAs), and altered DNA methylation profiles, into the circulation (which is known as ctDNA). However, tumor-derived cfDNA usually account for a small fraction of the total cfDNA, and the concentration is even lower in early-stage cancer patients, which largely limits the accuracy of most cfDNA-based liquid biopsy assays. To this end, in recent studies, researcher have explored the application of extra-deep sequencing and discovered a broad range of tumor-related signals in cfDNA to further improve their performance in bother diagnosis and prognosis monitoring.
Conventional genomic and epigenomic alterations: Somatic mutations are one of the most widely studied tumor-associated genomic alterations. In a proof-of-concept study, through extra-high sequencing of cfDNA in HCC patients, researchers are able to non-invasively resolve the mutation landscape of the tumor with high accuracy[16], which would be informative in guiding the therapeutic strategies for cancer patients. The somatic mutations in cfDNA are widely utilized used as biomarkers for early diagnosis and prognosis monitoring. For example, in pancreatic cancer (PC), the overall survival of patients with detectable KRAS mutation in cfDNA was significantly shorter than that of patients without such mutations[17]. Notably, the somatic mutations detected in cfDNA may not solely come from the tumors. In fact, recent studies have proven that clonal hematopoiesis also introduces somatic mutations which are detectable in cfDNA. Hence, it is highly recommended to analyze the blood cells in parallel to cfDNA in somatic mutation-oriented studies.
CNA is another widely used biomarker in cancer diagnosis and prognosis monitoring. For example, Shoda et al[18] found that the copy number status of HER2 gene in cfDNA is a potential biomarker for predicting the risk of recurrence and monitoring the effects of treatment for patients with gastric cancer (GC); Chen et al[19] showed that in GC, patients with stable chromosomes were sensitive to drug treatment while patients with detectable chromosomal instability in cfDNA were resistant to treatment, demonstrating the potential of cfDNA in predicting treatment responses.
DNA modifications, mostly 5-methylcytosine (5mC) and 5-hydroxymethylcytosine (5hmC), are abundant epigenetic regulators in the human genome. These modifications are known to affect gene expression, and suffer from gross abnormalities in almost all kinds of tumors. In cancer patients, such modifications are detectable in cfDNA and could be used to point to the tissue origin of the tumor. For example, Chan et al[20] analyzed the CNA and 5mC profiles of cfDNA simultaneously using whole-genome bisulfite sequencing technology, demonstrating that flexible combination of these two biomarkers would benefit in cancer diagnosis in various clinical scenarios; Li et al[21] showed that methylated SFRP2 in cfDNA has the potential to serve as a non-invasive biomarker for CRC screening; Gai et al[22] reported the application of DNA methylation biomarkers in differentiating CRC patients with and without liver metastases. Eissa et al[23] reported that the methylation of BNC1 and ADAMTS1 gene promoters could be used for PC diagnosis and screening with an area under the curve (AUC) of 0.95. The hypermethylation of cysteine dioxygenase 1 gene (CDO1) could distinguish pancreatic ductal adenocarcinoma (PDAC) patients from healthy controls with a sensitivity of 95%[24]. Besides diagnosis, Kim et al[25] showed that low methylation of ST-alpha and L1 of cfDNA are associated with a shorter overall survival in GC patients; Harada et al[26] suggested that GC patients with a high methylation level in the promoter of CDO1 gene in cfDNA had a significantly worse prognosis. Similarly, Zhang et al[27] found that, in PDAC patients, the methylation of PCDH10 gene was associated with a shorter disease-free survival. On the other hand, recent studies have suggested that 5hmC-based biomarkers in cfDNA are highly predictive of colorectal, gastric, and liver cancers with superior performance over conventional protein-based biomarkers[28-30]. Through genome-wide profiling of 5hmC in cfDNA as well as genomic DNA (gDNA) of paired tumor and adjacent normal tissues from patients with colorectal, gastric, liver, and other cancers, Li et al[21] discovered robust cancer-associated 5hmC signatures in cfDNA for cancer diagnosis. In another study, Cai et al[29] utilized the highly sensitive 5hmC-Seal assay to profile the genome-wide 5hmC landscape in cfDNA samples from 2554 HCC patients and found that liver-related 5hmC biomarkers show a remarkable capacity for distinguishing early HCC from high-risk non-cancerous subjects with chronic hepatitis B virus infection or liver cirrhosis. These studies demonstrated that DNA methylation-based biomarkers have favorable clinical value in cancer diagnosis, molecular characterization of the tumor, and prognosis monitoring of the patients, especially in GI cancers.
Emerging cfDNA fragmentomic features: Plasma cell-free DNA is known as naturally fragmented, small molecules. Recent studies have revealed that the tumor-derived DNA possess a remarkable difference in fragmentation characteristics compared to the background DNA (which mostly comes from the hematopoietic system)[31], and demonstrated the clinical significance of these features in the diagnosis of various GI cancers.
CfDNA molecules show a dominant size peak at 166 bp and most of them are shorter than 200 bp, and this phenomenon is believed to correlate with the nucleosome structure[32]. In cancer patients, tumor-derived cfDNA is even shorter than the background DNA[33]. Hence, it is doable to enrich the tumor-derived cfDNA through selection of shorter cfDNA molecules in principle. Indeed, Mouliere et al[34] demonstrated the enrichment of tumor-derived cfDNA in fragment sizes between 90 and 150 bp in a cohort of 344 plasma samples from various cancer patients. They also showed that using an integrated analysis of cfDNA at different size ranges, they could classify CRC patients from healthy individuals with a high specificity of 94%[34].
Besides size pattern, recent studies further showed that the relationship between the fragmentation pattern and nucleosome structure could point to the tissue origin of cfDNA[35,36]. In particular, cfDNA of different origins tends to have different preferences on fragment ends. Jiang et al[16] compared the genome-wide fragment end patterns of cfDNA in one HCC patient and one non-cancerous control; they reported that many genomic loci show significantly higher chances to serve as fragment ends (which they called “preferred-ends”) in the HCC patient, and the usage of such genomic loci could differentiate HCC patients from non-cancerous controls with a favorable accuracy. Moreover, the same team further showed that the nucleotide frequencies (or motifs) at the cfDNA end, which are believed to correlate with nuclease cutting preference, also change in various cancers[37,38]. In fact, they found a nuclease, DNAS1L3, which plays key roles in the apoptotic fragmentation process and shows strong sequence preferences when cutting the DNA[39]. Interestingly, this gene is significantly down-regulated in many cancers and thus affects the end motif pattern of the tumor-derived DNA[37]. The discovery of these novel biomarkers largely extends our knowledgebase on both the biology of cfDNA and cancer biomarkers; these novel biomarkers also promise the development of less complex and cheaper assays than those based on the conventional genomic/epigenomic biomarkers. However, most current studies are still in proof-of-principle stage, thus large-scale validation studies are essential to assure the performances before clinical investigations.
CTCs are released into the bloodstream of patients through the blood vessels and lymphatic vessels. When the primary cancer cells acquire an increased propensity for migration, invasiveness, and resistance, these tumor cells will invade through the tissue surrounding their site of origin and keep invading until they enter the blood or the lymphatic system, and become the CTCs[40,41]. Although most of CTCs will die within hours, only a small part of them can potentially form a new cancerous lesion at a distant site and grow into a new metastasis[42,43].
There are many commercial kits for CTC isolation and enrichments. CTC isolation and enrichment are mostly based on their physical and immunological properties. The physical features of CTCs, such as the size, density, and electric charge, are utilized to enrich CTCs from blood. For example, ISET (isolation by size of epithelial tumor cells) system and a microfluidic chip developed by Haber’ lab filter CTCs from blood by the size of CTCs[44-46], CAP (centrifugal affinity plate) system isolates CTCs by the density gradient centrifugation[47], and ApoStream system enriches CTCs by dielectrophoretic field-flow fractionation[48,49]. Enriching CTCs based on the immunological properties of CTCs depends on the antigens expressed on the surface of epithelial cells, including epithelial cell adhesion molecule (EpCAM) and cytokeratin (CK). CellSearch is the first and only FDA-approved test for capturing CTCs that express EpCAM by using immunomagnetic beads[50,51]. There also are some methods that combine epithelial marker immunofluorescence staining with automated microscopy, such as NYONE[52]. At the protein level, fluorescence in situ hybridization (FISH) has been utilized for identifying chromosomal rearrangements in CTCs[53]. Immunocytochemistry (ICC) is the major technique that is used by several commercial kits for isolating CTCs, including ApoStream and OcoQuick. Flow cytometry is another protein analysis method employed for CTC isolation. The analysis of genetic information of CTCs is mainly based on PCR and next-generation sequencing (NGS), such as RNA-based digital PCR and single-cell sequencing[54,55]. Due to the low concentration and fragile property of CTCs and limitations of current methods, combination and further optimization may achieve better enrichment and detection for CTCs.
The clinical application of CTCs depends on the number of CTCs and the information about protein expression and gene mutation in CTCs. Kang et al[56] showed that for individuals with two or more CTCs per 7.5 mL of blood, 97.1% of them were GC patients in their cohort. The increased number of CTCs after treatment was also correlated with early recurrence[27]. Zhang et al[27] showed that in GC patients, the patients with five or more CTCs per 7.5 mL in postoperative blood had a significantly shorter overall survival and disease-free survival compared to those with less CTCs. The protein expression patterns in CTCs are also an important biomarker for cancer diagnosis. Around 50% GC patients with CK+CD44+ CTCs were diagnosed with distant metastases[57]. Wei et al[58] found that, in 76% of PDAC patients, vimentin was upregulated in CTCs, and a high level of vimentin in CTCs was associated with a shorter recurrence-free survival. Similarly, PDAC patients with CTCs that expressed high levels of mucin 1 (MUC-1) and cancer stem cell markers, such as ALDH, CD133, and CD44, were associated with a worse overall survival and tumor recurrence[59,60]. Chen et al[61] found up-regulated expression of ECT2 in CTCs from patients with CRC. Fang et al[62] found that CRC patients with liver metastasis showed a higher percentage of +CD133+CD54+ CD44+ cell subpopulations. Moreover, the upregulated genes in CTCs, such as SHH, SMO, POU5F1B, and ALCAM, were demonstrated as potential markers for therapeutic response and prognosis prediction in PC patients[63]. In HCC patients, Zhou et al[64] discovered that the presence of EpCAM mRNA+ CTCs was associated with early recurrence when combined with Treg/CD4+ T cell ratio.
Exosomes are membrane-bound extracellular nanovesicles (EVs) released by many types of cells, having a size ranging from 30 to 150 nm. Exosomes have been found in various bodily fluids, including plasma, urine, and malignant ascites[65]. Exosomes are involved in mechanisms of cell-cell communication in physiological and pathological tissues through transporting biomolecules, such as lipids, proteins, mRNA, microRNAs, and DNA fragments, and the intercellular transfer of signal components[66,67]. Tumor-associated exosomes are larger than those derived from healthy cells and are enriched with different types of mediators of tumorigenesis, including proteins, growth factors, immunomodulatory molecules, and nucleic acids that arise from the cytoplasm of donor cancer cells[68].
In the last decade, a significant number of studies have focused their attention on the exosome-mediated cross-talk which can promote the activation of signaling pathways and reprogram the functions of recipient cells through their cargo transfer that occurs between cancer and normal cells, especially in the tumor microenvironment[65,69-71].
Several examples of exosome biomarkers, such as proteins, microRNAs, and long non-coding RNAs (lncRNAs), have been shown to have the potential for the diagnosis and prognosis of specific types of cancer. For example, GPC-1 was identified as a diagnostic exosome marker for PC and CRC. CD9 and CD147 were found to be upregulated in exosomes derived from serum of CRC patients[72]. Previously, seven miRNAs including let-7a, miR-21, miR-23a, miR-155, miR-223, miR-1246, and miR-1229 were reported to be highly expressed in the serum exosomes of CRC patients and were significantly decreased after surgical resection[73]. MiR-103 was reported to be delivered from hepatoma cell-derived exosomes into endothelial cells, and induced metastasis[74]. In HCC patients, serum exosomal miR-10b-5p was fond to upregulated and could be used to distinguish HCC patients from healthy controls with an AUC of 0.934[75]. Nakamura et al[76] found that exosomal miR-21 and miR-155 were significantly upregulated in PDAC patients compared to chronic pancreatitis patients. In GC patients, exosomal lncRNA pcsk2-2:1 and lncRNA GNAQ-6:1 were significantly downregulated and could be served as biomarkers for early diagnosis for GC[77,78]. Yu et al[79] developed a panel of eight exosomal RNAs, which could distinguish PDAC patients from chronic pancreatitis patients with an AUC of 0.931. The exosomal mRNA hnRNPH1 was found to have a high expression level in HCC patients and could be used to distinguish HCC patients from chronic hepatitis B patients with an AUC of 0.865[80]. Furthermore, lncRNAs, such as LncRNA-ARSR, Lnc-sox2ot, and LncRNA-h19, were showed to be associated with tumor progression[72,81].
Circulating cell-free RNAs (cfRNAs) are also present in the blood circulation and have the potential to serve as cancer biomarkers for cancer diagnosis. CfRNAs may be released into the blood through mechanisms of cell death or exosome-mediated signaling by living cells.
Historically, the study of cfRNA has focused on a small group of known cancer-related mRNA, miRNAs, and lncRNAs[76-78,82]. For instance, miR-21 showed an increased expression level in GC patients, which could serve as a classification biomarker with an 88.4% sensitivity and 79.6% specificity[83]. Huang et al[84] designed a panel of six miRNAs that were upregulated in the serum of GC patients, which showed an AUC of 0.702 in distinguishing GC patients from healthy controls. Shimura et al[85] found that three miRNAs (miR-30a-5p, miR-659-3p, and miR-3917) were overexpressed in GC patients who had peritoneal metastasis and could be used to distinguish patients with or without peritoneal metastasis with an AUC of 0.82. Yamada et al[86] showed that the expression of three serum miRNAs, including miR-21, miR-29a, and miR-125b, could discriminate CRC patients from healthy ones with an AUC of 0.827. In HCC patients, serum miR-143 was downregulated and could distinguish HCC patients from healthy controls with an AUC of 0.831[87]. Another study showed that a panel of three plasma circRNAs were found to distinguish HCC patients from healthy ones with an AUC of 0.863[88].
The early diagnosis and prognosis of a cancer type have become a necessity in cancer research. Image-based and tissue biopsy-based pathology analyses are considered to be the “gold standard” for their accuracy and effectiveness for cancer screening.
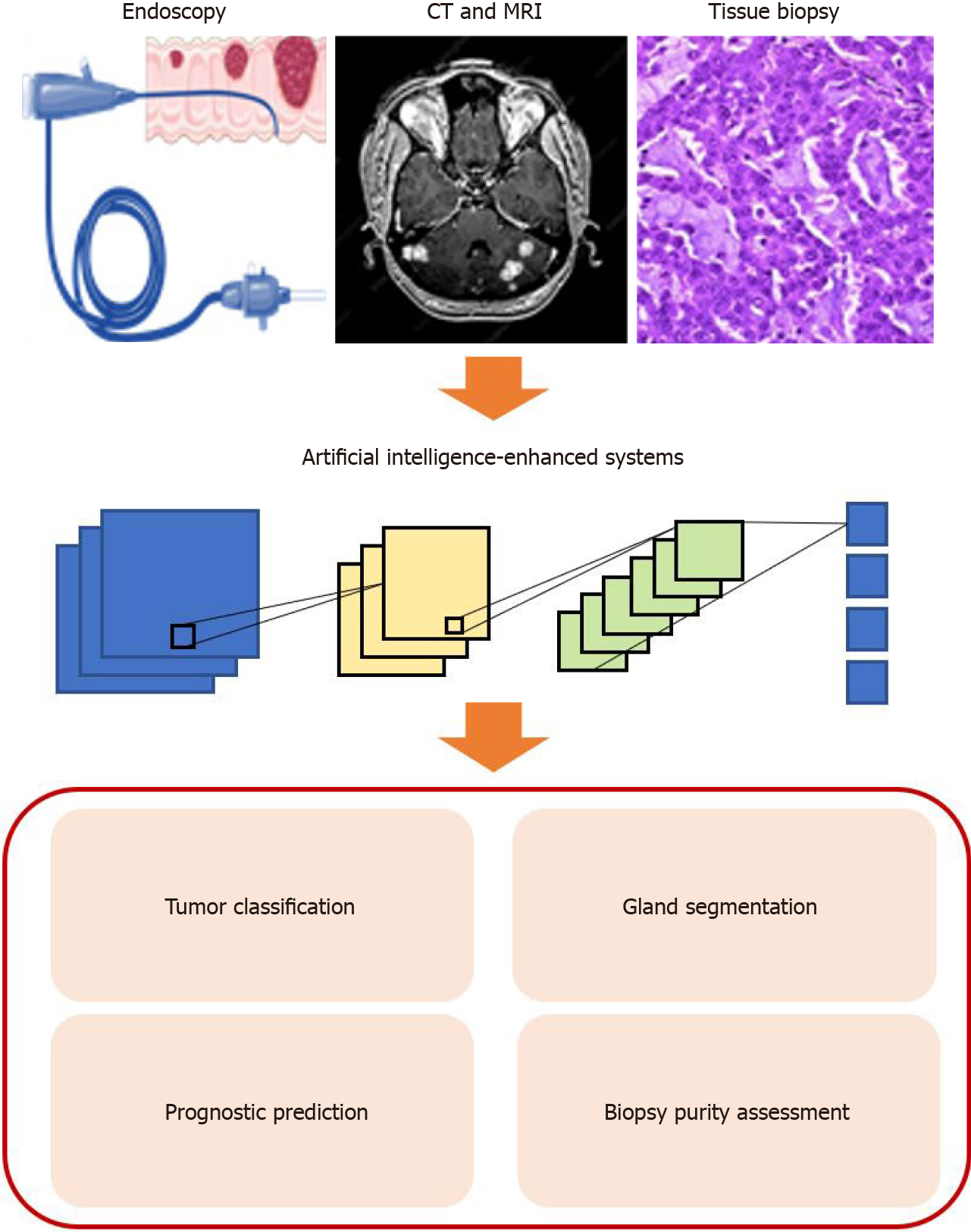
In recent years, AI has become an emerging field with the advances of computational power and massive amounts of learning data. Over the last two decades, different machine learning (ML) algorithms, such as support vector machines (SVM), random forest (RF), and artificial neural networks (ANNs), have been widely used in the field of medicine[89-91]. More recently, convolutional neural networks (CNN), a novel algorithm, show its great potential in processing and interpreting radiological and pathological images (Figure 2)[92,93]. Previous studies demonstrated that a multi-layer CNN with pre-trained GoogleNet Inception v3 architecture on skin cancer level prediction could achieve comparable results to dermatologists[94]. Similarly, another image-based deep learning model proposed by Kermany et al[10] in analyzing optical coherence tomography (OCT) images of the retina is on par with the ophthalmologist, with an accuracy of 96.6%, sensitivity of 97.8%, and specificity of 97.4% on the four-class classification problem. Most importantly, both studies utilized transfer learning when training the model to save the training time, which is also a widely used technique in training an ML model in medical images due to the lack of training data in most cases[95].
Traditionally, the most effective and direct method of screening colorectal carcinoma is endoscopy, particularly colonoscopy[96]. Abdominal computerized tomography (CT) or abdominal magnetic resonance imaging (MRI) are employed when more precise and additional information is needed about the lesion to help assess cancer and look for any signs of spread. However, these methods are usually labor-intensive and time-consuming and require professional knowledge. In the era of big data, the number of colonoscopy examinations and radiology screening is enormously increasing[96]. To this end, the deep learning-based radiology and digital pathology workflow could assist the radiologists and pathologists to diagnose diseases in various aspects, such as tumor classification, gland segment, tumor microenvironment analysis, and prognostic survival prediction, to ease their ever-growing work
Apart from AI application in colonoscopy, CT, and MRI images, the digital pathology also drew much attention from researchers, in particular adopting deep learning on the whole slide images (WSIs)[92,98-100]. However, the WSIs are multi-gigabyte images, present high morphological variance, and contain various types of artifacts. Such circumstances prevent CNN directly from applying to the high-resolution images. On the other hand, typically small labeled datasets in pathological images impede the generality of the deep learning model[101]. Kermany et al[102] developed a multiple-instance learning-based deep learning system that only used the reported annotated datasets to tackle the limited number of labeled images and the AUCs were all above 0.98 when testing on multiple cancer types. Joseph et al[96] introduced a novel machine learning-based system on prediction of CRC outcome for whole digitized haematoxylin and eosin (HE) stained histopathology slides in learning heterogeneous and discriminant contents. They demonstrated the effectiveness of the method and presented a detailed analysis of its different elements which corroborate its ability to extract and learn salient, discriminative, and clinically meaningful content.
At present, pathological analysis of tissue biopsies is still the most widely approach in cancer diagnosis. Recent studies have shown that combination of molecular data and deep learning approach could predict the cancerous status and tissue origin of pan-cancer biopsies with high accuracy. For instance, Sun et al[103] developed GeneCT, which showed an overall accuracy of 98.2% in differentiating tumor samples from adjacent normal samples, and an overall accuracy of 98.6% in predicting the tissue origins of the biopsies. In addition, as tissue biopsy is one of the most widely materials in cancer studies, and the purity of the biopsies is crucial for appropriate project design and correct interpretation of the data. To this end, Fan et al[12] showed that the low prediction accuracy of GeneCT in some datasets suggested impurity of the samples. The performance of GeneCT was further evaluated using metastatic tumor samples, which is a common scenario in GI cancers. As a result, GeneCT shows potential as an accurate classification tool for tissue biopsy-based diagnosis, as well as an easy-to-use quality-control tool for cancer studies.
GI malignancies, especially CRC and HCC, are common cancers that lead to death of patients with cancers worldwide. Liquid biopsy plays an important role in early-stage GI cancer diagnosis as a noninvasive tumor detection strategy. Biomarkers including cfDNA, CTCs, and exosomes of liquid biopsy are widely used for the detection of tumor-associated molecules. The advantages of cfDNA, such as high amount, being relatively stable and easy for extraction and preservation, mostly representing the information from the major tumor clone, and the significant features including fragment sizes, the proportion of fragment, the end-motif, and genomic alterations, make it the preferential and accurate biomarker for the cancer diagnosis. CTCs are also important liquid biomarkers because they not only contain direct molecular information from the tumor clone but also can be cultured in vitro. However, CTCs are very rare and require fresh blood for isolation.
The potential of liquid biopsy in clinical practice is enormous in the future. However, there are several challenges that need to be resolved for the utility of liquid biopsy in clinical diagnosis. First, standardization should be established for the sample collection and storage conditions, and biomarker molecules and the suitable detection and analysis strategy depend on sample type. Second, tumor-associated molecule detection techniques require high sensitivity for early tumor detection and high specificity for early-stage tumor screening. At the same time, the low content of tumor biomarkers and the varied interference factors in the circulation make the high requirements for the high sensitivity and specificity of detection strategies. Reducing purification steps could shorten the time of sample treatment and reduce the cost, and optimization of the throughput platform could help to increase both the sensitivity and specificity of the biomarkers.
AI based on machine learning and deep learning is widely used in all walks of life. Recently, rapid developments have been made in the utility of AI in medicine areas and showed promising results in terms of accuracy for cancer diagnosis. However, the scale and quality of the training and validation datasets of most of the studies were relatively limited to apply this technique in clinical practice. Moreover, it also requires the external cross-validation, especially in tumor classification. Therefore, future studies with a larger number of datasets with high-quality annotations and external cross-validation are required for routine practice-level validation.
We would like to thank Ms. Qi Wang from Shenzhen Bay Laboratory for her technical assistance.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Park WS, Vymetalkova V S-Editor: Ma YJ L-Editor: Wang TQ P-Editor: Liu JH
| 1. | Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12667] [Cited by in RCA: 15314] [Article Influence: 3062.8] [Reference Citation Analysis (4)] |
| 2. | Siegel RL, Miller KD, Goding Sauer A, Fedewa SA, Butterly LF, Anderson JC, Cercek A, Smith RA, Jemal A. Colorectal cancer statistics, 2020. CA Cancer J Clin. 2020;70:145-164. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2268] [Cited by in RCA: 3270] [Article Influence: 654.0] [Reference Citation Analysis (2)] |
| 3. | Society AC. Cancer Facts & Figures 2021: Atlanta: American Cancer Society, 2021. |
| 4. | Moore H, Dodd N. Computed tomographic colonography (CTC); colorectal cancer diagnosis with CTC in an Auckland population. J Med Imaging Radiat Oncol. 2013;57:572-575. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 5. | Greenhill C. Colorectal cancer: CTC for diagnosing symptomatic colorectal cancer. Nat Rev Gastroenterol Hepatol. 2013;10:198. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 6. | Fici P. Cell-Free DNA in the Liquid Biopsy Context: Role and Differences Between ctDNA and CTC Marker in Cancer Management. Methods Mol Biol. 2019;1909:47-73. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 22] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 7. | Keup C, Storbeck M, Hauch S, Hahn P, Sprenger-Haussels M, Tewes M, Mach P, Hoffmann O, Kimmig R, Kasimir-Bauer S. Cell-Free DNA Variant Sequencing Using CTC-Depleted Blood for Comprehensive Liquid Biopsy Testing in Metastatic Breast Cancer. Cancers (Basel). 2019;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 23] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 8. | Nanduri LK, Hissa B, Weitz J, Schölch S, Bork U. The prognostic role of circulating tumor cells in colorectal cancer. Expert Rev Anticancer Ther. 2019;19:1077-1088. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 32] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 9. | Maltoni R, Palleschi M, Ravaioli S, Tumedei MM, Rocca A, Melegari E, Altini M, Puccetti M, Manunta S, Bravaccini S. Cell-Free DNA Variant Sequencing Using CTC-Depleted Blood for Comprehensive Liquid Biopsy Testing in Metastatic Breast Cancer. Cell Transplant. 2020;29:963689720925057. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 10. | Kermany DS, Goldbaum M, Cai W, Valentim CCS, Liang H, Baxter SL, McKeown A, Yang G, Wu X, Yan F, Dong J, Prasadha MK, Pei J, Ting MYL, Zhu J, Li C, Hewett S, Ziyar I, Shi A, Zhang R, Zheng L, Hou R, Shi W, Fu X, Duan Y, Huu VAN, Wen C, Zhang ED, Zhang CL, Li O, Wang X, Singer MA, Sun X, Xu J, Tafreshi A, Lewis MA, Xia H, Zhang K. Identifying Medical Diagnoses and Treatable Diseases by Image-Based Deep Learning. Cell. 2018;172:1122-1131.e9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2132] [Cited by in RCA: 1702] [Article Influence: 283.7] [Reference Citation Analysis (0)] |
| 11. | He B, Lu Q, Lang J, Yu H, Peng C, Bing P, Li S, Zhou Q, Liang Y, Tian G. A New Method for CTC Images Recognition Based on Machine Learning. Front Bioeng Biotechnol. 2020;8:897. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 35] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 12. | Fan F, Chen D, Zhao Y, Wang H, Sun H, Sun K. Rapid preliminary purity evaluation of tumor biopsies using deep learning approach. Comput Struct Biotechnol J. 2020;18:1746-1753. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 3] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 13. | Yeh P, Hunter T, Sinha D, Ftouni S, Wallach E, Jiang D, Chan YC, Wong SQ, Silva MJ, Vedururu R, Doig K, Lam E, Arnau GM, Semple T, Wall M, Zivanovic A, Agarwal R, Petrone P, Jones K, Westerman D, Blombery P, Seymour JF, Papenfuss AT, Dawson MA, Tam CS, Dawson SJ. Circulating tumour DNA reflects treatment response and clonal evolution in chronic lymphocytic leukaemia. Nat Commun. 2017;8:14756. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 76] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 14. | Hao X, Luo H, Krawczyk M, Wei W, Wang W, Wang J, Flagg K, Hou J, Zhang H, Yi S, Jafari M, Lin D, Chung C, Caughey BA, Li G, Dhar D, Shi W, Zheng L, Hou R, Zhu J, Zhao L, Fu X, Zhang E, Zhang C, Zhu JK, Karin M, Xu RH, Zhang K. DNA methylation markers for diagnosis and prognosis of common cancers. Proc Natl Acad Sci USA. 2017;114:7414-7419. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 251] [Cited by in RCA: 335] [Article Influence: 41.9] [Reference Citation Analysis (0)] |
| 15. | Buechler C, Aslanidis C. Role of lipids in pathophysiology, diagnosis and therapy of hepatocellular carcinoma. Biochim Biophys Acta Mol Cell Biol Lipids. 2020;1865:158658. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 49] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 16. | Jiang P, Sun K, Tong YK, Cheng SH, Cheng THT, Heung MMS, Wong J, Wong VWS, Chan HLY, Chan KCA, Lo YMD, Chiu RWK. Preferred end coordinates and somatic variants as signatures of circulating tumor DNA associated with hepatocellular carcinoma. Proc Natl Acad Sci USA. 2018;115:E10925-E10933. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 91] [Cited by in RCA: 157] [Article Influence: 22.4] [Reference Citation Analysis (0)] |
| 17. | Zhang Q, Peng J, Ye M, Weng W, Tan C, Ni S, Huang D, Sheng W, Wang L. KRAS Mutation Predicted More Mirometastases and Closer Resection Margins in Patients with Colorectal Cancer Liver Metastases. Ann Surg Oncol. 2020;27:1164-1173. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 18] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 18. | Shoda K, Ichikawa D, Fujita Y, Masuda K, Hiramoto H, Hamada J, Arita T, Konishi H, Komatsu S, Shiozaki A, Kakihara N, Okamoto K, Taniguchi H, Imoto I, Otsuji E. Monitoring the HER2 copy number status in circulating tumor DNA by droplet digital PCR in patients with gastric cancer. Gastric Cancer. 2017;20:126-135. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 104] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 19. | Chen Z, Zhang C, Zhang M, Li B, Niu Y, Chen L, Yang J, Lu S, Gao J, Shen L. Chromosomal instability of circulating tumor DNA reflect therapeutic responses in advanced gastric cancer. Cell Death Dis. 2019;10:697. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 22] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 20. | Chan KC, Jiang P, Chan CW, Sun K, Wong J, Hui EP, Chan SL, Chan WC, Hui DS, Ng SS, Chan HL, Wong CS, Ma BB, Chan AT, Lai PB, Sun H, Chiu RW, Lo YM. Noninvasive detection of cancer-associated genome-wide hypomethylation and copy number aberrations by plasma DNA bisulfite sequencing. Proc Natl Acad Sci USA. 2013;110:18761-18768. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 276] [Cited by in RCA: 329] [Article Influence: 27.4] [Reference Citation Analysis (0)] |
| 21. | Li H, Wang Z, Zhao G, Ma Y, Chen Y, Xue Q, Zheng M, Fei S. Performance of a MethyLight assay for methylated SFRP2 DNA detection in colorectal cancer tissue and serum. Int J Biol Markers. 2019;34:54-59. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 24] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 22. | Gai W, Ji L, Lam WKJ, Sun K, Jiang P, Chan AWH, Wong J, Lai PBS, Ng SSM, Ma BBY, Wong GLH, Wong VWS, Chan HLY, Chiu RWK, Lo YMD, Chan KCA. Liver- and Colon-Specific DNA Methylation Markers in Plasma for Investigation of Colorectal Cancers with or without Liver Metastases. Clin Chem. 2018;64:1239-1249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 55] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 23. | Eissa MAL, Lerner L, Abdelfatah E, Shankar N, Canner JK, Hasan NM, Yaghoobi V, Huang B, Kerner Z, Takaesu F, Wolfgang C, Kwak R, Ruiz M, Tam M, Pisanic TR 2nd, Iacobuzio-Donahue CA, Hruban RH, He J, Wang TH, Wood LD, Sharma A, Ahuja N. Promoter methylation of ADAMTS1 and BNC1 as potential biomarkers for early detection of pancreatic cancer in blood. Clin Epigenetics. 2019;11:59. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 66] [Cited by in RCA: 115] [Article Influence: 19.2] [Reference Citation Analysis (0)] |
| 24. | Nishizawa N, Harada H, Kumamoto Y, Kaizu T, Katoh H, Tajima H, Ushiku H, Yokoi K, Igarashi K, Fujiyama Y, Okuwaki K, Iwai T, Watanabe M, Yamashita K. Diagnostic potential of hypermethylation of the cysteine dioxygenase 1 gene (CDO1) promoter DNA in pancreatic cancer. Cancer Sci. 2019;110:2846-2855. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 27] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 25. | Kim Y, Wen X, Jeong S, Cho NY, Kim WH, Kang GH. Combinatory low methylation statuses of SAT-α and L1 are associated with shortened survival time in patients with advanced gastric cancer. Gastric Cancer. 2019;22:37-47. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 15] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 26. | Harada H, Hosoda K, Moriya H, Mieno H, Ema A, Ushiku H, Washio M, Nishizawa N, Ishii S, Yokota K, Tanaka Y, Kaida T, Soeno T, Kosaka Y, Watanabe M, Yamashita K. Cancer-specific promoter DNA methylation of Cysteine dioxygenase type 1 (CDO1) gene as an important prognostic biomarker of gastric cancer. PLoS One. 2019;14:e0214872. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 19] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 27. | Zhang Q, Shan F, Li Z, Gao J, Li Y, Shen L, Ji J, Lu M. A prospective study on the changes and clinical significance of pre-operative and post-operative circulating tumor cells in resectable gastric cancer. J Transl Med. 2018;16:171. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 29] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 28. | Li W, Zhang X, Lu X, You L, Song Y, Luo Z, Zhang J, Nie J, Zheng W, Xu D, Wang Y, Dong Y, Yu S, Hong J, Shi J, Hao H, Luo F, Hua L, Wang P, Qian X, Yuan F, Wei L, Cui M, Zhang T, Liao Q, Dai M, Liu Z, Chen G, Meckel K, Adhikari S, Jia G, Bissonnette MB, Zhao Y, Zhang W, He C, Liu J. 5-Hydroxymethylcytosine signatures in circulating cell-free DNA as diagnostic biomarkers for human cancers. Cell Res. 2017;27:1243-1257. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 171] [Cited by in RCA: 265] [Article Influence: 33.1] [Reference Citation Analysis (0)] |
| 29. | Cai J, Chen L, Zhang Z, Zhang X, Lu X, Liu W, Shi G, Ge Y, Gao P, Yang Y, Ke A, Xiao L, Dong R, Zhu Y, Yang X, Wang J, Zhu T, Yang D, Huang X, Sui C, Qiu S, Shen F, Sun H, Zhou W, Zhou J, Nie J, Zeng C, Stroup EK, Chiu BC, Lau WY, He C, Wang H, Zhang W, Fan J. Genome-wide mapping of 5-hydroxymethylcytosines in circulating cell-free DNA as a non-invasive approach for early detection of hepatocellular carcinoma. Gut. 2019;68:2195-2205. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 206] [Cited by in RCA: 200] [Article Influence: 33.3] [Reference Citation Analysis (0)] |
| 30. | Xiao Z, Wu W, Wu C, Li M, Sun F, Zheng L, Liu G, Li X, Yun Z, Tang J, Yu Y, Luo S, Sun W, Feng X, Cheng Q, Tao X, Wu S, Tao J. 5-Hydroxymethylcytosine signature in circulating cell-free DNA as a potential diagnostic factor for early-stage colorectal cancer and precancerous adenoma. Mol Oncol. 2021;15:138-150. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 21] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 31. | Sun K, Jiang P, Chan KC, Wong J, Cheng YK, Liang RH, Chan WK, Ma ES, Chan SL, Cheng SH, Chan RW, Tong YK, Ng SS, Wong RS, Hui DS, Leung TN, Leung TY, Lai PB, Chiu RW, Lo YM. Plasma DNA tissue mapping by genome-wide methylation sequencing for noninvasive prenatal, cancer, and transplantation assessments. Proc Natl Acad Sci USA. 2015;112:E5503-E5512. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 416] [Cited by in RCA: 539] [Article Influence: 53.9] [Reference Citation Analysis (0)] |
| 32. | Sun K, Jiang P, Wong AIC, Cheng YKY, Cheng SH, Zhang H, Chan KCA, Leung TY, Chiu RWK, Lo YMD. Size-tagged preferred ends in maternal plasma DNA shed light on the production mechanism and show utility in noninvasive prenatal testing. Proc Natl Acad Sci USA. 2018;115:E5106-E5114. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 75] [Cited by in RCA: 97] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 33. | Underhill HR, Kitzman JO, Hellwig S, Welker NC, Daza R, Baker DN, Gligorich KM, Rostomily RC, Bronner MP, Shendure J. Fragment Length of Circulating Tumor DNA. PLoS Genet. 2016;12:e1006162. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 374] [Cited by in RCA: 500] [Article Influence: 55.6] [Reference Citation Analysis (0)] |
| 34. | Mouliere F, Chandrananda D, Piskorz AM, Moore EK, Morris J, Ahlborn LB, Mair R, Goranova T, Marass F, Heider K, Wan JCM, Supernat A, Hudecova I, Gounaris I, Ros S, Jimenez-Linan M, Garcia-Corbacho J, Patel K, Østrup O, Murphy S, Eldridge MD, Gale D, Stewart GD, Burge J, Cooper WN, van der Heijden MS, Massie CE, Watts C, Corrie P, Pacey S, Brindle KM, Baird RD, Mau-Sørensen M, Parkinson CA, Smith CG, Brenton JD, Rosenfeld N. Enhanced detection of circulating tumor DNA by fragment size analysis. Sci Transl Med. 2018;10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 622] [Cited by in RCA: 706] [Article Influence: 100.9] [Reference Citation Analysis (0)] |
| 35. | Snyder MW, Kircher M, Hill AJ, Daza RM, Shendure J. Cell-free DNA Comprises an In Vivo Nucleosome Footprint that Informs Its Tissues-Of-Origin. Cell. 2016;164:57-68. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 758] [Cited by in RCA: 1027] [Article Influence: 114.1] [Reference Citation Analysis (0)] |
| 36. | Ulz P, Thallinger GG, Auer M, Graf R, Kashofer K, Jahn SW, Abete L, Pristauz G, Petru E, Geigl JB, Heitzer E, Speicher MR. Inferring expressed genes by whole-genome sequencing of plasma DNA. Nat Genet. 2016;48:1273-1278. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 209] [Cited by in RCA: 298] [Article Influence: 33.1] [Reference Citation Analysis (0)] |
| 37. | Jiang P, Sun K, Peng W, Cheng SH, Ni M, Yeung PC, Heung MMS, Xie T, Shang H, Zhou Z, Chan RWY, Wong J, Wong VWS, Poon LC, Leung TY, Lam WKJ, Chan JYK, Chan HLY, Chan KCA, Chiu RWK, Lo YMD. Plasma DNA End-Motif Profiling as a Fragmentomic Marker in Cancer, Pregnancy, and Transplantation. Cancer Discov. 2020;10:664-673. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 192] [Article Influence: 38.4] [Reference Citation Analysis (0)] |
| 38. | Sun K, Jiang P, Cheng SH, Cheng THT, Wong J, Wong VWS, Ng SSM, Ma BBY, Leung TY, Chan SL, Mok TSK, Lai PBS, Chan HLY, Sun H, Chan KCA, Chiu RWK, Lo YMD. Orientation-aware plasma cell-free DNA fragmentation analysis in open chromatin regions informs tissue of origin. Genome Res. 2019;29:418-427. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 108] [Cited by in RCA: 176] [Article Influence: 29.3] [Reference Citation Analysis (0)] |
| 39. | Serpas L, Chan RWY, Jiang P, Ni M, Sun K, Rashidfarrokhi A, Soni C, Sisirak V, Lee WS, Cheng SH, Peng W, Chan KCA, Chiu RWK, Reizis B, Lo YMD. Dnase1 L3 deletion causes aberrations in length and end-motif frequencies in plasma DNA. Proc Natl Acad Sci USA. 2019;116:641-649. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 83] [Cited by in RCA: 146] [Article Influence: 20.9] [Reference Citation Analysis (0)] |
| 40. | Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646-674. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51728] [Cited by in RCA: 47135] [Article Influence: 3366.8] [Reference Citation Analysis (5)] |
| 41. | Plaks V, Koopman CD, Werb Z. Cancer. Circulating tumor cells. Science. 2013;341:1186-1188. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 444] [Cited by in RCA: 497] [Article Influence: 41.4] [Reference Citation Analysis (0)] |
| 42. | Heitzer E, Auer M, Gasch C, Pichler M, Ulz P, Hoffmann EM, Lax S, Waldispuehl-Geigl J, Mauermann O, Lackner C, Höfler G, Eisner F, Sill H, Samonigg H, Pantel K, Riethdorf S, Bauernhofer T, Geigl JB, Speicher MR. Complex tumor genomes inferred from single circulating tumor cells by array-CGH and next-generation sequencing. Cancer Res. 2013;73:2965-2975. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 406] [Cited by in RCA: 399] [Article Influence: 33.3] [Reference Citation Analysis (0)] |
| 43. | Vignot S, Frampton GM, Soria JC, Yelensky R, Commo F, Brambilla C, Palmer G, Moro-Sibilot D, Ross JS, Cronin MT, André F, Stephens PJ, Lazar V, Miller VA, Brambilla E. Next-generation sequencing reveals high concordance of recurrent somatic alterations between primary tumor and metastases from patients with non-small-cell lung cancer. J Clin Oncol. 2013;31:2167-2172. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 134] [Cited by in RCA: 145] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 44. | Hofman V, Ilie MI, Long E, Selva E, Bonnetaud C, Molina T, Vénissac N, Mouroux J, Vielh P, Hofman P. Detection of circulating tumor cells as a prognostic factor in patients undergoing radical surgery for non-small-cell lung carcinoma: comparison of the efficacy of the CellSearch Assay™ and the isolation by size of epithelial tumor cell method. Int J Cancer. 2011;129:1651-1660. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 192] [Cited by in RCA: 246] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 45. | Pailler E, Oulhen M, Billiot F, Galland A, Auger N, Faugeroux V, Laplace-Builhé C, Besse B, Loriot Y, Ngo-Camus M, Hemanda M, Lindsay CR, Soria JC, Vielh P, Farace F. Method for semi-automated microscopy of filtration-enriched circulating tumor cells. BMC Cancer. 2016;16:477. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 28] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 46. | Edd JF, Mishra A, Dubash TD, Herrera S, Mohammad R, Williams EK, Hong X, Mutlu BR, Walsh JR, Machado de Carvalho F, Aldikacti B, Nieman LT, Stott SL, Kapur R, Maheswaran S, Haber DA, Toner M. Microfluidic concentration and separation of circulating tumor cell clusters from large blood volumes. Lab Chip. 2020;20:558-567. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 47] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 47. | Lee SW, Hyun KA, Kim SI, Kang JY, Jung HI. Enrichment of circulating tumor cells using a centrifugal affinity plate system. J Chromatogr A. 2014;1373:25-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 48. | Balasubramanian P, Kinders RJ, Kummar S, Gupta V, Hasegawa D, Menachery A, Lawrence SM, Wang L, Ferry-Galow K, Davis D, Parchment RE, Tomaszewski JE, Doroshow JH. Antibody-independent capture of circulating tumor cells of non-epithelial origin with the ApoStream® system. PLoS One. 2017;12:e0175414. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 46] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 49. | Le Du F, Fujii T, Kida K, Davis DW, Park M, Liu DD, Wu W, Chavez-MacGregor M, Barcenas CH, Valero V, Tripathy D, Reuben JM, Ueno NT. EpCAM-independent isolation of circulating tumor cells with epithelial-to-mesenchymal transition and cancer stem cell phenotypes using ApoStream® in patients with breast cancer treated with primary systemic therapy. PLoS One. 2020;15:e0229903. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 28] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 50. | Swennenhuis JF, Reumers J, Thys K, Aerssens J, Terstappen LW. Efficiency of whole genome amplification of single circulating tumor cells enriched by CellSearch and sorted by FACS. Genome Med. 2013;5:106. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 68] [Cited by in RCA: 69] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 51. | Huebner H, Fasching PA, Gumbrecht W, Jud S, Rauh C, Matzas M, Paulicka P, Friedrich K, Lux MP, Volz B, Gass P, Häberle L, Meier-Stiegen F, Hartkopf A, Neubauer H, Almstedt K, Beckmann MW, Fehm TN, Ruebner M. Filtration based assessment of CTCs and CellSearch® based assessment are both powerful predictors of prognosis for metastatic breast cancer patients. BMC Cancer. 2018;18:204. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 30] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 52. | Hendricks A, Brandt B, Geisen R, Dall K, Röder C, Schafmayer C, Becker T, Hinz S, Sebens S. Isolation and Enumeration of CTC in Colorectal Cancer Patients: Introduction of a Novel Cell Imaging Approach and Comparison to Cellular and Molecular Detection Techniques. Cancers (Basel). 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 16] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 53. | Wang A, Li Z, Wang Q, Bai Y, Ji X, Fu T, Ji K, Xue Y, Han T, Wu X, Zhang J, Yang Y, Xu G, Bu Z, Ji J. Diagnostic value of negative enrichment and immune fluorescence in situ hybridization for intraperitoneal free cancer cells of gastric cancer. Chin J Cancer Res. 2019;31:945-954. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 7] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 54. | Miyamoto DT, Lee RJ, Kalinich M, LiCausi JA, Zheng Y, Chen T, Milner JD, Emmons E, Ho U, Broderick K, Silva E, Javaid S, Kwan TT, Hong X, Dahl DM, McGovern FJ, Efstathiou JA, Smith MR, Sequist LV, Kapur R, Wu CL, Stott SL, Ting DT, Giobbie-Hurder A, Toner M, Maheswaran S, Haber DA. An RNA-Based Digital Circulating Tumor Cell Signature Is Predictive of Drug Response and Early Dissemination in Prostate Cancer. Cancer Discov. 2018;8:288-303. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 111] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 55. | Winter M, Cai Z, Winkler K, Georgiou K, Inglis D, Lavranos T, Rezaei M, Warkiani M, Thierry B. Circulating tumour cell RNA characterisation from colorectal cancer patient blood after inertial microfluidic enrichment. MethodsX. 2019;6:1512-1520. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 8] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 56. | Kang HM, Kim GH, Jeon HK, Kim DH, Jeon TY, Park DY, Jeong H, Chun WJ, Kim MH, Park J, Lim M, Kim TH, Cho YK. Circulating tumor cells detected by lab-on-a-disc: Role in early diagnosis of gastric cancer. PLoS One. 2017;12:e0180251. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 66] [Cited by in RCA: 56] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 57. | Szczepanik A, Sierzega M, Drabik G, Pituch-Noworolska A, Kołodziejczyk P, Zembala M. CD44+ cytokeratin-positive tumor cells in blood and bone marrow are associated with poor prognosis of patients with gastric cancer. Gastric Cancer. 2019;22:264-272. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 29] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 58. | Wei T, Zhang X, Zhang Q, Yang J, Chen Q, Wang J, Li X, Chen J, Ma T, Li G, Gao S, Lou J, Que R, Wang Y, Dang X, Zheng L, Liang T, Bai X. Vimentin-positive circulating tumor cells as a biomarker for diagnosis and treatment monitoring in patients with pancreatic cancer. Cancer Lett. 2019;452:237-243. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 92] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 59. | Dotan E, Alpaugh RK, Ruth K, Negin BP, Denlinger CS, Hall MJ, Astsaturov I, McAleer C, Fittipaldi P, Thrash-Bingham C, Meropol NJ, Cohen SJ. Prognostic Significance of MUC-1 in Circulating Tumor Cells in Patients With Metastatic Pancreatic Adenocarcinoma. Pancreas. 2016;45:1131-1135. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 41] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 60. | Poruk KE, Blackford AL, Weiss MJ, Cameron JL, He J, Goggins M, Rasheed ZA, Wolfgang CL, Wood LD. Circulating Tumor Cells Expressing Markers of Tumor-Initiating Cells Predict Poor Survival and Cancer Recurrence in Patients with Pancreatic Ductal Adenocarcinoma. Clin Cancer Res. 2017;23:2681-2690. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 87] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 61. | Chen CJ, Sung WW, Chen HC, Chern YJ, Hsu HT, Lin YM, Lin SH, Peck K, Yeh KT. Early Assessment of Colorectal Cancer by Quantifying Circulating Tumor Cells in Peripheral Blood: ECT2 in Diagnosis of Colorectal Cancer. Int J Mol Sci. 2017;18. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 17] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 62. | Fang C, Fan C, Wang C, Huang Q, Meng W, Yu Y, Yang L, Peng Z, Hu J, Li Y, Mo X, Zhou Z. CD133+CD54+CD44+ circulating tumor cells as a biomarker of treatment selection and liver metastasis in patients with colorectal cancer. Oncotarget. 2016;7:77389-77403. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 37] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 63. | Amantini C, Morelli MB, Nabissi M, Piva F, Marinelli O, Maggi F, Bianchi F, Bittoni A, Berardi R, Giampieri R, Santoni G. Expression Profiling of Circulating Tumor Cells in Pancreatic Ductal Adenocarcinoma Patients: Biomarkers Predicting Overall Survival. Front Oncol. 2019;9:874. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 50] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 64. | Zhou Y, Wang B, Wu J, Zhang C, Zhou Y, Yang X, Zhou J, Guo W, Fan J. Association of preoperative EpCAM Circulating Tumor Cells and peripheral Treg cell levels with early recurrence of hepatocellular carcinoma following radical hepatic resection. BMC Cancer. 2016;16:506. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 69] [Cited by in RCA: 70] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 65. | Mittelbrunn M, Sánchez-Madrid F. Intercellular communication: diverse structures for exchange of genetic information. Nat Rev Mol Cell Biol. 2012;13:328-335. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 420] [Cited by in RCA: 539] [Article Influence: 41.5] [Reference Citation Analysis (0)] |
| 66. | Théry C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol. 2002;2:569-579. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4254] [Cited by in RCA: 4119] [Article Influence: 179.1] [Reference Citation Analysis (0)] |
| 67. | Scavo MP, Depalo N, Tutino V, De Nunzio V, Ingrosso C, Rizzi F, Notarnicola M, Curri ML, Giannelli G. Exosomes for Diagnosis and Therapy in Gastrointestinal Cancers. Int J Mol Sci. 2020;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 36] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 68. | Bebelman MP, Smit MJ, Pegtel DM, Baglio SR. Biogenesis and function of extracellular vesicles in cancer. Pharmacol Ther. 2018;188:1-11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 337] [Cited by in RCA: 593] [Article Influence: 84.7] [Reference Citation Analysis (1)] |
| 69. | Carpelan-Holmström MA, Haglund CH, Roberts PJ. Differences in serum tumor markers between colon and rectal cancer. Comparison of CA 242 and carcinoembryonic antigen. Dis Colon Rectum. 1996;39:799-805. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 15] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 70. | Brzozowski JS, Jankowski H, Bond DR, McCague SB, Munro BR, Predebon MJ, Scarlett CJ, Skelding KA, Weidenhofer J. Lipidomic profiling of extracellular vesicles derived from prostate and prostate cancer cell lines. Lipids Health Dis. 2018;17:211. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 94] [Cited by in RCA: 114] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 71. | Alzahrani FA, El-Magd MA, Abdelfattah-Hassan A, Saleh AA, Saadeldin IM, El-Shetry ES, Badawy AA, Alkarim S. Potential Effect of Exosomes Derived from Cancer Stem Cells and MSCs on Progression of DEN-Induced HCC in Rats. Stem Cells Int. 2018;2018:8058979. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 77] [Cited by in RCA: 116] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 72. | Xiao Y, Zhong J, Zhong B, Huang J, Jiang L, Jiang Y, Yuan J, Sun J, Dai L, Yang C, Li Z, Wang J, Zhong T. Exosomes as potential sources of biomarkers in colorectal cancer. Cancer Lett. 2020;476:13-22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 132] [Article Influence: 26.4] [Reference Citation Analysis (0)] |
| 73. | Lane RE, Korbie D, Hill MM, Trau M. Extracellular vesicles as circulating cancer biomarkers: opportunities and challenges. Clin Transl Med. 2018;7:14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 114] [Cited by in RCA: 184] [Article Influence: 26.3] [Reference Citation Analysis (0)] |
| 74. | Nedaeinia R, Manian M, Jazayeri MH, Ranjbar M, Salehi R, Sharifi M, Mohaghegh F, Goli M, Jahednia SH, Avan A, Ghayour-Mobarhan M. Circulating exosomes and exosomal microRNAs as biomarkers in gastrointestinal cancer. Cancer Gene Ther. 2017;24:48-56. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 172] [Cited by in RCA: 164] [Article Influence: 20.5] [Reference Citation Analysis (0)] |
| 75. | Cho HJ, Eun JW, Baek GO, Seo CW, Ahn HR, Kim SS, Cho SW, Cheong JY. Serum Exosomal MicroRNA, miR-10b-5p, as a Potential Diagnostic Biomarker for Early-Stage Hepatocellular Carcinoma. J Clin Med. 2020;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 97] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 76. | Nakamura S, Sadakari Y, Ohtsuka T, Okayama T, Nakashima Y, Gotoh Y, Saeki K, Mori Y, Nakata K, Miyasaka Y, Onishi H, Oda Y, Goggins M, Nakamura M. Pancreatic Juice Exosomal MicroRNAs as Biomarkers for Detection of Pancreatic Ductal Adenocarcinoma. Ann Surg Oncol. 2019;26:2104-2111. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 68] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 77. | Cai C, Zhang H, Zhu Y, Zheng P, Xu Y, Sun J, Zhang M, Lan T, Gu B, Li S, Ma P. Serum Exosomal Long Noncoding RNA pcsk2-2:1 As A Potential Novel Diagnostic Biomarker For Gastric Cancer. Onco Targets Ther. 2019;12:10035-10041. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 56] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 78. | Li S, Zhang M, Zhang H, Hu K, Cai C, Wang J, Shi L, Ma P, Xu Y, Zheng P. Exosomal long noncoding RNA lnc-GNAQ-6:1 may serve as a diagnostic marker for gastric cancer. Clin Chim Acta. 2020;501:252-257. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 49] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 79. | Yu S, Li Y, Liao Z, Wang Z, Qian L, Zhao J, Zong H, Kang B, Zou WB, Chen K, He X, Meng Z, Chen Z, Huang S, Wang P. Plasma extracellular vesicle long RNA profiling identifies a diagnostic signature for the detection of pancreatic ductal adenocarcinoma. Gut. 2020;69:540-550. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 137] [Article Influence: 27.4] [Reference Citation Analysis (0)] |
| 80. | Xu H, Dong X, Chen Y, Wang X. Serum exosomal hnRNPH1 mRNA as a novel marker for hepatocellular carcinoma. Clin Chem Lab Med. 2018;56:479-484. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 96] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 81. | Baassiri A, Nassar F, Mukherji D, Shamseddine A, Nasr R, Temraz S. Exosomal Non Coding RNA in LIQUID Biopsies as a Promising Biomarker for Colorectal Cancer. Int J Mol Sci. 2020;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 75] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 82. | Redis RS, Calin S, Yang Y, You MJ, Calin GA. Cell-to-cell miRNA transfer: from body homeostasis to therapy. Pharmacol Ther. 2012;136:169-174. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 128] [Cited by in RCA: 141] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 83. | Wu J, Li G, Wang Z, Yao Y, Chen R, Pu X, Wang J. Circulating MicroRNA-21 Is a Potential Diagnostic Biomarker in Gastric Cancer. Dis Markers. 2015;2015:435656. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 75] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 84. | Huang Z, Zhu D, Wu L, He M, Zhou X, Zhang L, Zhang H, Wang W, Zhu J, Cheng W, Chen Y, Fan Y, Qi L, Yin Y, Zhu W, Shu Y, Liu P. Six Serum-Based miRNAs as Potential Diagnostic Biomarkers for Gastric Cancer. Cancer Epidemiol Biomarkers Prev. 2017;26:188-196. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 110] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 85. | Shimura T, Toden S, Kandimalla R, Toiyama Y, Okugawa Y, Kanda M, Baba H, Kodera Y, Kusunoki M, Goel A. Genomewide Expression Profiling Identifies a Novel miRNA-Based Signature for the Detection of Peritoneal Metastasis in Patients With Gastric Cancer. Ann Surg. 2019;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 25] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 86. | Yamada A, Horimatsu T, Okugawa Y, Nishida N, Honjo H, Ida H, Kou T, Kusaka T, Sasaki Y, Yagi M, Higurashi T, Yukawa N, Amanuma Y, Kikuchi O, Muto M, Ueno Y, Nakajima A, Chiba T, Boland CR, Goel A. Serum miR-21, miR-29a, and miR-125b Are Promising Biomarkers for the Early Detection of Colorectal Neoplasia. Clin Cancer Res. 2015;21:4234-4242. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 125] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 87. | Zhang J, Lin H, Wang XY, Zhang DQ, Chen JX, Zhuang Y, Zheng XL. Predictive value of microRNA-143 in evaluating the prognosis of patients with hepatocellular carcinoma. Cancer Biomark. 2017;19:257-262. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 13] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 88. | Yu J, Ding WB, Wang MC, Guo XG, Xu J, Xu QG, Yang Y, Sun SH, Liu JF, Qin LX, Liu H, Yang F, Zhou WP. Plasma circular RNA panel to diagnose hepatitis B virus-related hepatocellular carcinoma: A large-scale, multicenter study. Int J Cancer. 2020;146:1754-1763. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 83] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 89. | Kourou K, Exarchos TP, Exarchos KP, Karamouzis MV, Fotiadis DI. Machine learning applications in cancer prognosis and prediction. Comput Struct Biotechnol J. 2015;13:8-17. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1362] [Cited by in RCA: 1289] [Article Influence: 117.2] [Reference Citation Analysis (0)] |
| 90. | Jiménez Pérez M, Grande RG. Application of artificial intelligence in the diagnosis and treatment of hepatocellular carcinoma: A review. World J Gastroenterol. 2020;26:5617-5628. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 52] [Cited by in RCA: 32] [Article Influence: 6.4] [Reference Citation Analysis (2)] |
| 91. | Jin P, Ji X, Kang W, Li Y, Liu H, Ma F, Ma S, Hu H, Li W, Tian Y. Artificial intelligence in gastric cancer: a systematic review. J Cancer Res Clin Oncol. 2020;146:2339-2350. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 84] [Cited by in RCA: 60] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 92. | Thakur N, Yoon H, Chong Y. Current Trends of Artificial Intelligence for Colorectal Cancer Pathology Image Analysis: A Systematic Review. Cancers (Basel). 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 53] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 93. | Wang Y, He X, Nie H, Zhou J, Cao P, Ou C. Application of artificial intelligence to the diagnosis and therapy of colorectal cancer. Am J Cancer Res. 2020;10:3575-3598. [PubMed] |
| 94. | Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, Thrun S. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542:115-118. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5683] [Cited by in RCA: 5359] [Article Influence: 669.9] [Reference Citation Analysis (0)] |
| 95. | Cheplygina V, de Bruijne M, Pluim JPW. Not-so-supervised: A survey of semi-supervised, multi-instance, and transfer learning in medical image analysis. Med Image Anal. 2019;54:280-296. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 315] [Cited by in RCA: 381] [Article Influence: 63.5] [Reference Citation Analysis (0)] |
| 96. | Joseph DA, Meester RG, Zauber AG, Manninen DL, Winges L, Dong FB, Peaker B, van Ballegooijen M. Colorectal cancer screening: Estimated future colonoscopy need and current volume and capacity. Cancer. 2016;122:2479-2486. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 168] [Cited by in RCA: 176] [Article Influence: 19.6] [Reference Citation Analysis (0)] |
| 97. | Luo H, Xu G, Li C, He L, Luo L, Wang Z, Jing B, Deng Y, Jin Y, Li Y, Li B, Tan W, He C, Seeruttun SR, Wu Q, Huang J, Huang DW, Chen B, Lin SB, Chen QM, Yuan CM, Chen HX, Pu HY, Zhou F, He Y, Xu RH. Real-time artificial intelligence for detection of upper gastrointestinal cancer by endoscopy: a multicentre, case-control, diagnostic study. Lancet Oncol. 2019;20:1645-1654. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 155] [Cited by in RCA: 253] [Article Influence: 42.2] [Reference Citation Analysis (0)] |
| 98. | Zhen SH, Cheng M, Tao YB, Wang YF, Juengpanich S, Jiang ZY, Jiang YK, Yan YY, Lu W, Lue JM, Qian JH, Wu ZY, Sun JH, Lin H, Cai XJ. Deep Learning for Accurate Diagnosis of Liver Tumor Based on Magnetic Resonance Imaging and Clinical Data. Front Oncol. 2020;10:680. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 99] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 99. | Kiani A, Uyumazturk B, Rajpurkar P, Wang A, Gao R, Jones E, Yu Y, Langlotz CP, Ball RL, Montine TJ, Martin BA, Berry GJ, Ozawa MG, Hazard FK, Brown RA, Chen SB, Wood M, Allard LS, Ylagan L, Ng AY, Shen J. Impact of a deep learning assistant on the histopathologic classification of liver cancer. NPJ Digit Med. 2020;3:23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 181] [Cited by in RCA: 160] [Article Influence: 32.0] [Reference Citation Analysis (0)] |
| 100. | Chen M, Zhang B, Topatana W, Cao J, Zhu H, Juengpanich S, Mao Q, Yu H, Cai X. Classification and mutation prediction based on histopathology H&E images in liver cancer using deep learning. NPJ Precis Oncol. 2020;4:14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 131] [Article Influence: 26.2] [Reference Citation Analysis (0)] |
| 101. | Dimitriou N, Arandjelović O, Caie PD. Deep Learning for Whole Slide Image Analysis: An Overview. Front Med (Lausanne). 2019;6:264. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 91] [Cited by in RCA: 146] [Article Influence: 24.3] [Reference Citation Analysis (0)] |
| 102. | Campanella G, Hanna MG, Geneslaw L, Miraflor A, Werneck Krauss Silva V, Busam KJ, Brogi E, Reuter VE, Klimstra DS, Fuchs TJ. Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat Med. 2019;25:1301-1309. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 710] [Cited by in RCA: 1199] [Article Influence: 199.8] [Reference Citation Analysis (0)] |
| 103. | Sun K, Wang J, Wang H, Sun H. GeneCT: a generalizable cancerous status and tissue origin classifier for pan-cancer biopsies. Bioinformatics. 2018;34:4129-4130. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 104. | Yamamoto H, Watanabe Y, Oikawa R, Morita R, Yoshida Y, Maehata T, Yasuda H, Itoh F. BARHL2 Methylation Using Gastric Wash DNA or Gastric Juice Exosomal DNA is a Useful Marker For Early Detection of Gastric Cancer in an H. pylori-Independent Manner. Clin Transl Gastroenterol. 2016;7:e184. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 65] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 105. | Lin LY, Yang L, Zeng Q, Wang L, Chen ML, Zhao ZH, Ye GD, Luo QC, Lv PY, Guo QW, Li BA, Cai JC, Cai WY. Tumor-originated exosomal lncUEGC1 as a circulating biomarker for early-stage gastric cancer. Mol Cancer. 2018;17:84. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 109] [Cited by in RCA: 181] [Article Influence: 25.9] [Reference Citation Analysis (0)] |
| 106. | Oehme F, Krahl S, Gyorffy B, Muessle B, Rao V, Greif H, Ziegler N, Lin K, Thepkaysone ML, Polster H, Tonn T, Schneider M, Weitz J, Baenke F, Kahlert C. Low level of exosomal long non-coding RNA HOTTIP is a prognostic biomarker in colorectal cancer. RNA Biol. 2019;16:1339-1345. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 70] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 107. | Shi Y, Wang Z, Zhu X, Chen L, Ma Y, Wang J, Yang X, Liu Z. Exosomal miR-1246 in serum as a potential biomarker for early diagnosis of gastric cancer. Int J Clin Oncol. 2020;25:89-99. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 90] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 108. | Feng W, Zong W, Li Y, Shen X, Cui X, Ju S. Abnormally expressed long noncoding RNA B3GALT5-AS1 may serve as a biomarker for the diagnostic and prognostic of gastric cancer. J Cell Biochem. 2020;121:557-565. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 27] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 109. | Sun Y, Yang B, Lin M, Yu H, Chen H, Zhang Z. Identification of serum miR-30a-5p as a diagnostic and prognostic biomarker in colorectal cancer. Cancer Biomark. 2019;24:299-305. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 32] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 110. | Liu X, Pan B, Sun L, Chen X, Zeng K, Hu X, Xu T, Xu M, Wang S. Circulating Exosomal miR-27a and miR-130a Act as Novel Diagnostic and Prognostic Biomarkers of Colorectal Cancer. Cancer Epidemiol Biomarkers Prev. 2018;27:746-754. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 104] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 111. | Wang J, Yan F, Zhao Q, Zhan F, Wang R, Wang L, Zhang Y, Huang X. Circulating exosomal miR-125a-3p as a novel biomarker for early-stage colon cancer. Sci Rep. 2017;7:4150. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 100] [Cited by in RCA: 146] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 112. | Sun B, Li Y, Zhou Y, Ng TK, Zhao C, Gan Q, Gu X, Xiang J. Circulating exosomal CPNE3 as a diagnostic and prognostic biomarker for colorectal cancer. J Cell Physiol. 2019;234:1416-1425. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 91] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 113. | Court CM, Ankeny JS, Sho S, Winograd P, Hou S, Song M, Wainberg ZA, Girgis MD, Graeber TG, Agopian VG, Tseng HR, Tomlinson JS. Circulating Tumor Cells Predict Occult Metastatic Disease and Prognosis in Pancreatic Cancer. Ann Surg Oncol. 2018;25:1000-1008. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 79] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 114. | Takahasi K, Iinuma H, Wada K, Minezaki S, Kawamura S, Kainuma M, Ikeda Y, Shibuya M, Miura F, Sano K. Usefulness of exosome-encapsulated microRNA-451a as a minimally invasive biomarker for prediction of recurrence and prognosis in pancreatic ductal adenocarcinoma. J Hepatobiliary Pancreat Sci. 2018;25:155-161. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 109] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 115. | Wang Z. Diagnostic performance for declined microRNA-133a in pancreatic cancer. J Cell Biochem. 2019;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 116. | Wang F, Wang J, Ju L, Chen L, Cai W, Yang J. Diagnostic and prognostic potential of serum miR-132/212 cluster in patients with hepatocellular carcinoma. Ann Clin Biochem. 2018;55:576-582. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 13] [Article Influence: 1.9] [Reference Citation Analysis (0)] |