Published online Jul 7, 2021. doi: 10.3748/wjg.v27.i25.3913
Peer-review started: February 9, 2021
First decision: March 28, 2021
Revised: April 10, 2021
Accepted: May 27, 2021
Article in press: May 27, 2021
Published online: July 7, 2021
Processing time: 146 Days and 20.9 Hours
Helicobacter pylori (H. pylori) is an important pathogen that can cause a variety of diseases. Yet, full eradication of H. pylori remains a significant challenge in clinical practice. H. pylori and other microbial communities have complex interactions in the unique gastric microecological environment. However, it is not clear whether the interactions have any effect on the therapeutic effect of H. pylori.
The aim was to investigate the characteristics of the gastric microbiota with H. pylori infection and the influence on the H. pylori eradication treatment.
Patients with H. pylori infection underwent gastroscopy and received treatment for eradication. The prescription included esomeprazole 20 mg bid, Livzon Dele 220 mg bid, amoxicillin 1000 mg bid, and clarithromycin 500 mg bid for 14 d. Patients who did not respond to treatment and failed eradication were compared with those who achieved eradication by 1:2 propensity matching. High-throughput sequencing of the gastric mucosal microbiota was performed, and the results were evaluated by alpha diversity analysis, beta diversity analysis, species correlation analysis, and metabolic pathway correlation analysis.
The eradication rate of all the patients was 95.5% (171/179). Twenty-four patients were enrolled in the study after propensity-matched scoring. There were eight cases in the failure group (patients who did not respond well to therapy) and 16 cases in the success group. The majority phyla in the two groups were the same, and included Proteobacteria, Bacteroides, Firmicutes, Actinomycetes, and Fusobacteria. The microbial diversity in the failure group had a decreasing trend (P = 0.092) and the species abundance was significantly lower (P = 0.031) compared with the success group. The high rate of H. pylori eradication was associated with Rhodococcus, Lactobacillus, and Sphingomonas, as they were significantly enriched in the successful group (P < 0.05). Veronococcus and Cilium were enriched in the mucosa of chronic atrophic gastritis patients compared with chronic superficial gastritis patients (P = 0.0466 and 0.0122, respectively). In both study groups, H. pylori was negatively correlated with other bacterial genera. More bacterial genera were directly related to H. pylori in the successful group compared with the failure group.
The effectiveness of quadruple H. pylori eradication therapy containing bismuth depended on gastric microbiota, and the high rate of H. pylori eradication was associated with the presence of Rhodococcus, Lactobacillus, and Sphingomonas.
Core Tip: Helicobacter pylori (H. pylori) is an important pathogen that can cause a variety of diseases. Its eradication can be affected by many factors. In this study, we explored the effect of the gastric microbiota on quadruple H. pylori eradication therapy containing bismuth. The results indicated that quadruple H. pylori eradication therapy containing bismuth was affected by the gastric microbiota. A high rate of H. pylori eradication was associated with the presence of Rhodococcus, Lactobacillus, and Sphingomonas. Our findings may provide the basis for clinical treatment.
- Citation: Niu ZY, Li SZ, Shi YY, Xue Y. Effect of gastric microbiota on quadruple Helicobacter pylori eradication therapy containing bismuth. World J Gastroenterol 2021; 27(25): 3913-3924
- URL: https://www.wjgnet.com/1007-9327/full/v27/i25/3913.htm
- DOI: https://dx.doi.org/10.3748/wjg.v27.i25.3913
The gastric mucosal environment was long thought to be sterile. Yet, the discovery of Helicobacter pylori (H. pylori) put an end to the traditional view of the sterile stomach[1]. Furthermore, the development of modern technology led to a deeper understan
H. pylori is an important pathogen associated with a variety of diseases[5-7] including gastric cancer[8]. About 50% of the global population is infected with H. pylori[9]. H. pylori infection is an infectious disease and a "screening-treatment" strategy has been recommended for the treatment of H. pylori. However, the eradi
Patients diagnosed with H. pylori infection in the Gastroenterology Department of Peking University Third Hospital between were enrolled between July 2018 and July 2019. Patients who were (1) 18-70 years of age; (2) with H. pylori infection confirmed by gastroscopy and histopathology were eligible. Patients (1) with previous H. pylori eradication therapy; (2) using proton pump inhibitors, H2 receptor blockers, bismuth, antibiotics, or other drugs that might affect the study results within 4 wk of inclusion; (3) with gastrointestinal tumors; (4) with a history of gastric or esophageal surgery; (5) with Zollinger-Ellison syndrome; (6) with abnormal liver or kidney function; (7) with severe cardiovascular, respiratory, blood, endocrine, neurological, or mental disease; (8) with allergy to a study drug; (9) were pregnant or lactating; and (10) or with histories of alcohol abuse or clinical conditions that might increase the risk of side effects were excluded.
Before inclusion, all patients provided informed consent for clinical sample collection. A biopsy was taken from the antrum before H. pylori eradication treatment. A rapid urease test (RUT) was performed during the gastroscopy, and if the result was positive, gastric mucosa biopsies from the lesser curvature of antrum and corpus were collected, placed in a cryovial, and stored −80 ℃. At the same time, mucosa specimens were collected from the gastric antrum and corpus were collected for histopathological examination and Warthin-Starry (WS) staining.
Patients diagnosed with H. pylori infection by positive RUT results and WS staining were treated with esomeprazole 20 mg bid, amoxicillin 1000 mg bid, clarithromycin 500 mg bid, Livzon Dele bismuth potassium citrate 220 mg bid for14 d. A 13C urease breath test (13C-UBT) was performed 8 wk after treatment, which was considered successful if the 13C-UBT was negative.
Patients were divided into failure and the success groups after their treatment was completed. Patients who did not respond well to therapy were included in the failure group. The success group was evaluated by nearest-neighbor matching, which is a type of propensity score matching and paired with patients in the failure group who had a similar propensity index. The propensity index was estimated by the model so as to equalize the covariates between the two groups. Gender, age, body-mass index (BMI kg/m2), gastroscopy diagnosis, and background gastric mucosa were the covariables used to calculate the propensity values. Taking the sample size and matching quality, the allowable error was set to 0.1. The failure and success groups were matched at a ratio of 1:2. The gastric mucosa microbiota were assayed and compared according to the results of propensity score matching.
The total DNA of the microbiota was extracted following the instructions with E.Z.N.A.® soil DNA kits (Omega Bio-Tek, Norcross, GA, United States) following the manufacturer’s instructions, and the quality of DNA extraction was assayed by 1% agarose gel electrophoresis. DNA concentration and purity were determined by a NanoDrop 2000 spectrophotometer, and 338 F (5'-ACTCCTACGGGAGGCAGCAG-3') and 806R (5'-GGACTACHVGGGTWTCTAAT-3') primers were used for PCR amplification of the V3-V4 region of the 16S rRNA gene. The PCR products were recovered on 2% agarose gels after combining the PCR products of the same sample. Recovered products were purified with AxyPrep DNA gel extraction kits (Axygen Biosciences, Union City, CA, United States) and assayed with a Quantus™ Fluorometer (Promega, United States). The library was built with NEXTFLEX Rapid DNA-Seq Kits and sequenced with a Miseq PE300 platform (Illumina).
Fastp software was used for quality control of the original sequence, and Flash software was used for stitching. Usearch software (version7.0, http://drive5.com/uparse/) was used to for operational taxonomic unit (OTU) clustering of sequences based on 97% similarity and elimination of chimeras. RDP classifier (http://rdp.cme.msu.edu/) was used to compare each sequence with the sequences included in Silva database (SSU132). The threshold was set to 70%, and the results of species classification annotations were obtained. The sample species composition was analyzed based on the annotation results.
The alpha diversity analysis (i.e. within-sample diversity) reflected species diversity, including Shannon, ace, and coverage indices. Beta diversity analysis (i.e. diversity between samples) or between-group differences in species composition was by principal coordinates analysis (PCoA). The Wilcoxon rank-sum test was used to compare the two groups. A P value of < 0.05 was considered significant. Correlation analysis of species was carried out by correlation heatmaps, network analysis, and metabolic pathway analysis. The relationship between the microbiota and the result of H. pylori eradication was preliminarily explained.
SPSS 23.0 was used for data analysis. measurement data were reported as means ± SD and compared by t-tests. Categorical data were compared by χ2 tests P values of < 0.05 were considered statistically significant.
Of the 179 enrolled patients, 171 responded well to therapy with successful H. pylori eradication. Eight patients failed treatment. The eradication therapy success rate was 95.5%. Propensity scoring resulted in matching eight failed cases with 16 successful cases. The results of gastroscopy revealed chronic gastritis in all patients; and after matching, there were no significant differences in the baseline characteristics between the two groups (Table 1).
| Failure group | Success group | P value | ||
| Sex | Male | 7 | 14 | 1.000 |
| Female | 1 | 2 | ||
| Age in yr, mean ± SD | 40.13 ± 14.55 | 37.44 ± 6.42 | 0.532 | |
| BMI in kg/m2, mean ± SD | 25.01 ± 5.32 | 25.20 ± 2.84 | 0.913 | |
| Background mucosa | Chronic superficial gastritis | 6 | 13 | 1.000 |
| Chronic atrophic gastritis | 2 | 3 | ||
A total of 1 204 878 reads and 1028 OTUs were obtained from 24 samples. The samples contained a mean of 50 203 reads and 191 OTUs. The reads in the failed eradication group (58 487) were significantly higher (P = 0.013) than those in the successful eradication group (46 061); the difference between the OTUs in the two groups was not significant (166 and 203, respectively, P = 0.719). The samples were randomly flattened according to the minimum number of sample sequences to avoid analysis deviation. A total of 980 OTUs were obtained, and each sample contained 30 043 reads. All samples were dominated by H. pylori at the genus level; H. pylori infection was pathologically confirmed.
The proportions of bacterial species in the failure and the success groups were evaluated by Good's species coverage index, which found that the difference between the two groups was not significant (P = 0.125). The coverage index in both groups was higher than 0.99 and confirmed that the test results covered most bacterial species in the gastric mucosa. Analysis of community composition showed that the gastric mucosa microbiota mainly contained Proteobacteria, Bacteroidetes, Firmicutes, Actinomycetes, and Fusobacteria, regardless of the study group. The abundance of Proteobacteria was higher in the failure group than in the success group, and that of Actinobacteria was lower (Table 2).
| Phyla | Failure group, % | Success group, % | P value |
| Proteobacteria | 98.26 | 94.60 | 0.0346 |
| Bacteroidetes | 0.60 | 1.23 | 0.4623 |
| Firmicutes | 0.52 | 1.23 | 0.2839 |
| Actinomycetes | 0.21 | 0.96 | 0.0016 |
| Fusobacteria | 0.11 | 0.27 | 0.6025 |
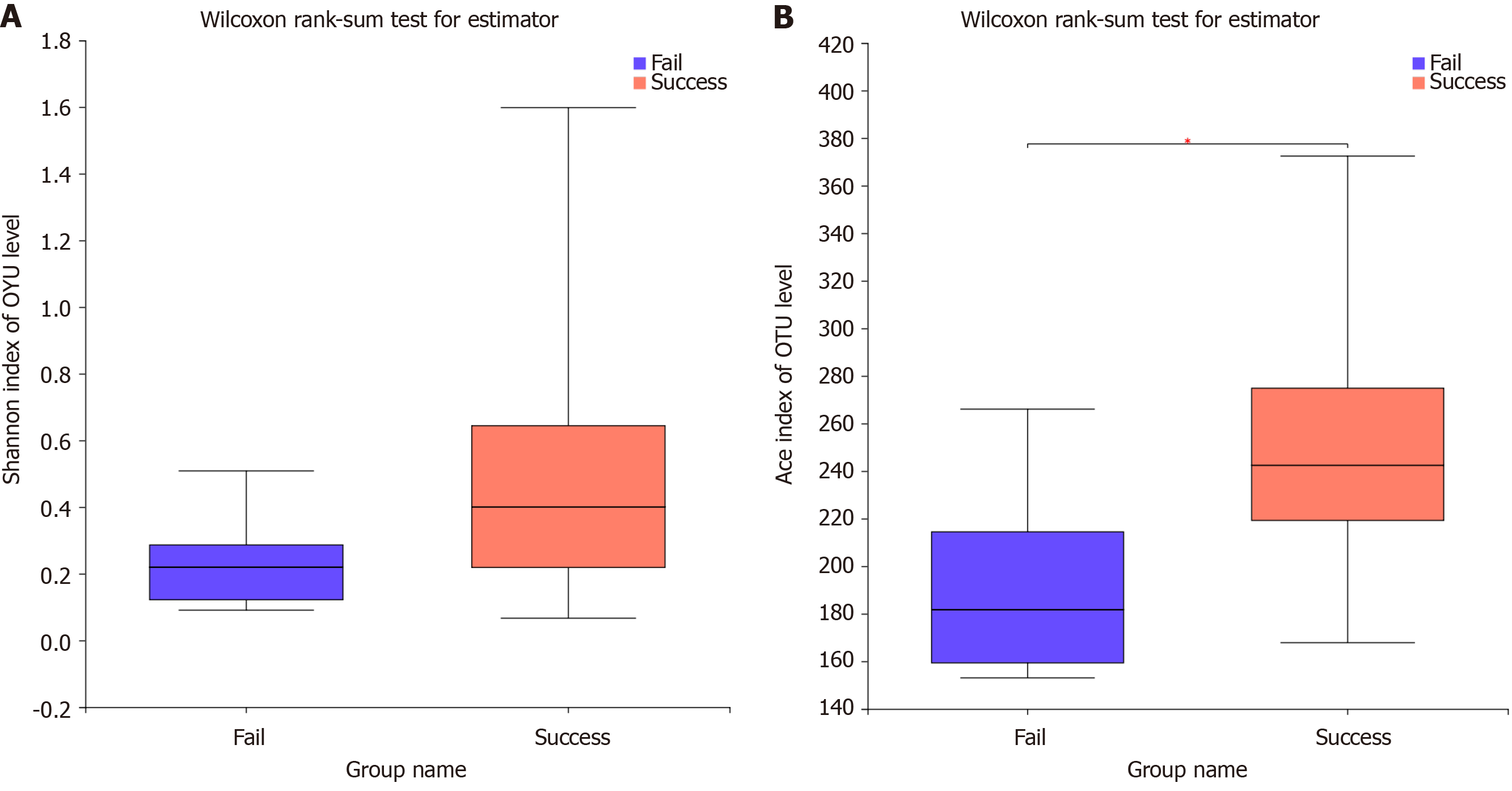
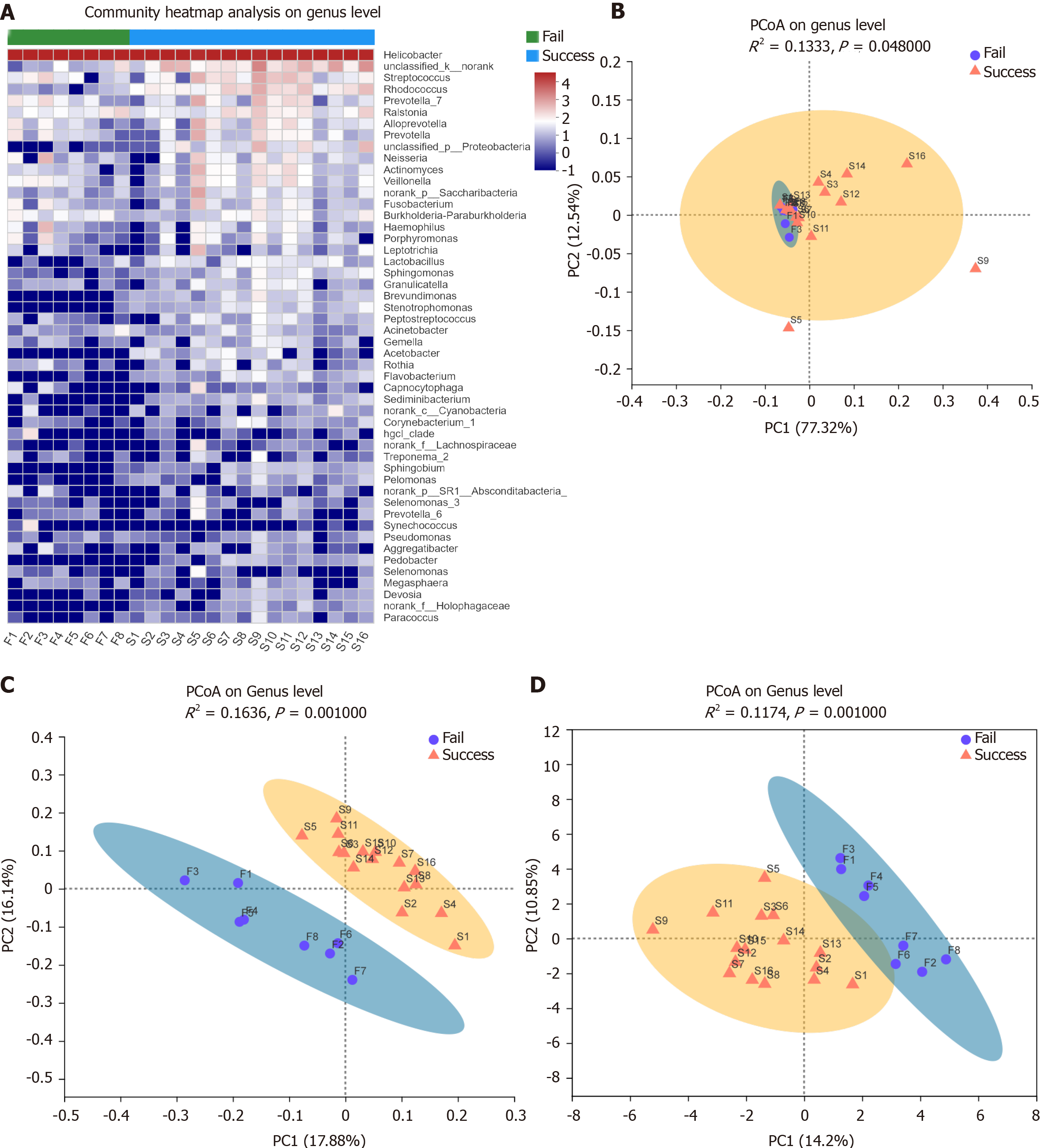
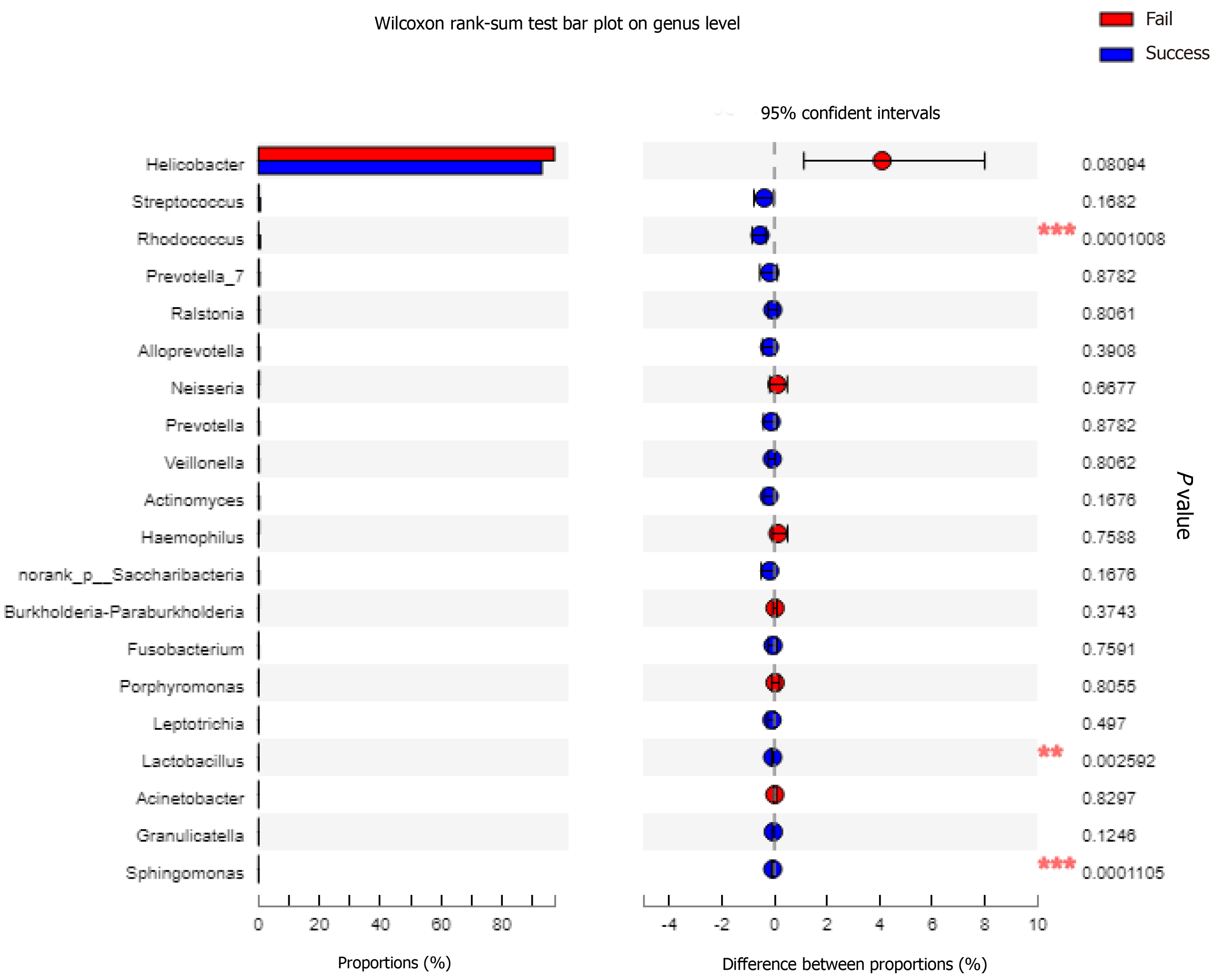
Alpha and beta diversity reflect differences in the microbial composition. The results of the Shannon index, which is one an indexes of alpha diversity, showed that the diversity in the failure group was reduced compared with the success group, but the difference was not significant (P = 0.092; Figure 1A). The Ace index showed that the species abundance was significantly lower in the failure group than in the success group (P = 0.031; Figure 1B). The dominant species in both study samples was H. pylori, but the microbiota composition differed at the genus level (Figure 2A). PCoA analysis of beta diversity resulted in a weighted UniFrac showing that the total diversity of the first two principal coordinates was 89.96% and the difference between the two groups was significant (ANOSIM, P = 0.048; Figure 2B). According to the unweighted UniFrac, the difference between the two groups was significant (ANOSIM, P = 0.001; Figure 2C). Binary Euclidean analysis, which was used to assess the difference in species composition, showed a significant difference between the two groups (ANOSIM, P = 0.001; Figure 2D). As shown in Figure 3, H. pylori was more abundance in the failure than in the success group, but the difference was not significant (Wilcoxon rank-sum test, P = 0.0809). Rhodococcus, Lactobacillus, and Sphingomonas were significantly enriched in the success group, (Wilcoxon rank-sum test, P < 0.05).
The flora composition of gastric mucosa that were histologically different was also analyzed. The abundance of H. pylori was higher in the mucosa of chronic superficial gastritis but the difference was not significant (Wilcoxon rank-sum test, P = 0.1179). Veronococcus and Cilium were more enriched in the mucosa of chronic atrophic gastritis (Wilcoxon rank-sum test, P = 0.0466 and 0.0122).
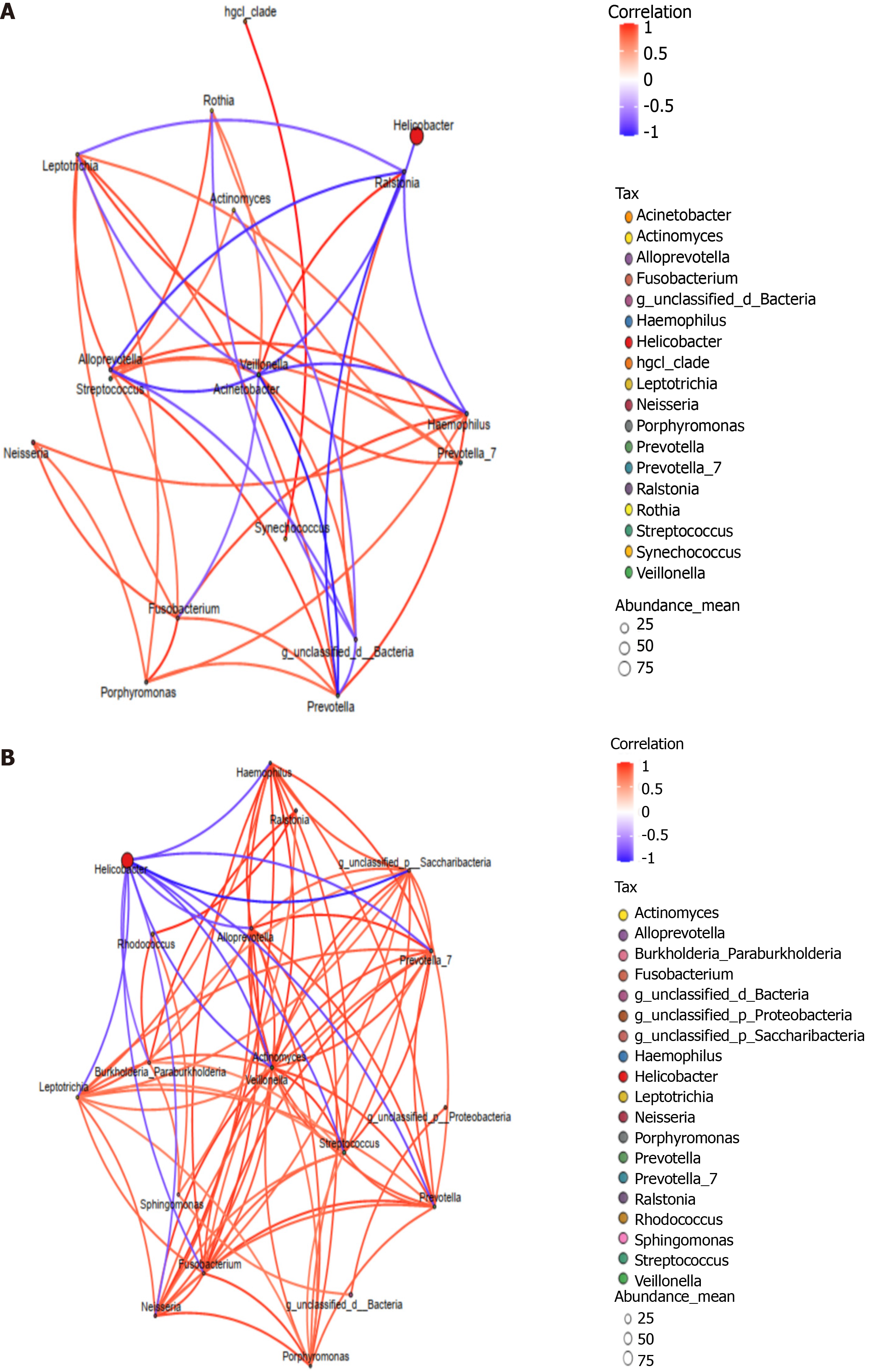
Heatmap results of the species correlation analysis found that H. pylori was negatively correlated with other bacterial genera (Figure 4), H. pylori was negatively correlated with Ralstonia in the failure group (genus level) and negatively correlated with Haemophilus, Prevotella, Streptococcus, Actinomycetes, Veillonella, Neisseria, Fusobacterium, and Leptotrichia in the success group (genus level). The PICRUSt metabolic pathway function prediction showed that the level two metabolic pathways in the two study groups were basically the same, mainly including carbohydrates, amino acids, energy, coenzyme factors, and vitamins.
The composition of the gastric mucosa microbiota infected by H. pylori mainly included Proteobacteria, Bacteroidetes, Firmicutes, Actinobacteria, and Fusobacteria, which is consistent with the results reported by previous studies[14,15]. H. pylori infection can significantly reduce the diversity of gastric mucosal microbiota[16]. In this study, the Shannon index did not reveal a significant difference in the diversity of the microbiota between the success and the failure group. However, the microbiota diversity in the failure group was lower, which suggests that eradication of H. pylori may be associated with the diversity of gastric mucosal microbiota.
The abundance of Proteobacteria was higher in the failure group. The genus level data suggests that the higher abundance of H. pylori in the eradication failure group might have caused an increased abundance of Proteobacteria. Although the difference in H. pylori abundance between the two groups was not significant, previous studies have shown that a higher abundance of H. pylori may reduce the effectiveness of empirical eradication therapy[17]. It has been reported that the abundance of H. pylori impacted only the result of traditional triple therapy[18]. Nevertheless, our results implied that the abundance of H. pylori had an impact on the eradication effect of quadruple therapy containing bismuth. In addition, 16s RNA may be more accurate than the 13C/14C-UBT or histological evaluation for the determination of H. pylori abundance.
At the genus level, the gastric mucosa microbiota species diversity was similar in the two groups. However, PCoA analysis showed that there were differences in the species composition and abundance between the two groups. The Ace index confirmed the differences in abundance of the two groups, and statistical analysis confirmed that the success group was significantly enriched in Rhodococcus, Lactobacillus, and Sphingomonas. The reason for the increased abundance of Actinomycetes in the success group may be related to the enrichment of Rhodococcus, an opportunistic pathogen[19,20] that can be detected in the stool of healthy people. Thiamine is required for the growth of Rhodococcus[21] and is essential nutrient for the growth of H. pylori[22]. H. pylori is a thiamine auxotroph that lacks the gene that synthesizes thiamine[23]. Vitamin metabolism is one of the microbiota's main metabolic pathways predicted by PICRUSt. Therefore, Rhodococcus may inhibit the growth of H. pylori through the acquisition of thiamine. Lactobacilli, which are beneficial bacteria, effectively improve H. pylori eradication if added to the prescription, especially in 7-d and 14-d triple therapy[24]. Previous studies have shown that Lactobacillus is a strong antagonist of H. pylori by preventing colonization and growth[25,26] and inhibiting adhesion to and invasion of gastric epithelial cells. Lactobacillus even has an antagonistic effect on multidrug resistant H. pylori[27]. In this study, the abundance of
Analysis of the microbiota against different gastric mucosa backgrounds showed that the genera Veronococcus and Cilium were enriched in chronic atrophic gastritis. Previous studies have shown bacterial overgrowth in the gastric mucosa of pre-gastric cancer. Veronococcus and Cilium are enriched in gastric mucosa of gastric cancer[30]. Veronococcus can convert nitrate to nitrite, and the increased concentration of nitrite may promote gastric cancer[31]. In addition, Veronococcus is enriched in patients with oral cancer, colorectal cancer, and lung cancer[32-35]. A study conducted in Colombia showed that the gastric mucosal flora of a population at high risk of gastric cancer was significantly enriched in Veronococcus and Cilium but the rate of H. pylori infection was not increased. Therefore, the genus Veronococcus and Cilium may be factors promoting the occurrence of gastric cancer. Our results confirmed a trend in pre-gastric cancer.
A negative correlation was found between H. pylori and other microbiota, which was more significant in the success group than in the failure group. The negative correlation of H. pylori and other microbiota was associated with a positive correlation among the other bacterial communities. Co-inhibition of H. pylori by a variety of mutually-promoting microbiota may improve the effectiveness of H. pylori eradication. There was no difference in the abundance of mutually-promoting microbiota present in the two groups. The lack of correlation between mutually-promoting microbiota and H. pylori in the failure group may have been related to a difference in H. pylori strains and needs to be confirmed by further studies.
Differences in the gastric microbiota may have contributed to H. pylori eradication failure, because all samples were collected before H. pylori eradication. This study has some limitations. It had a relatively small sample size because of the high eradication rate. The result preliminarily showed an effect of the gastric microbiota on H. pylori eradication, but studies that have larger sample sizes are needed to confirm these findings. This study did not determine H. pylori resistance, but the results provide clinically significant guidance for empirical treatment.
The effect of quadruple H. pylori eradication therapy containing bismuth depends on the gastric microbiota. A high rate of H. pylori eradication was associated with the presence of Rhodococcus, Lactobacillus, and Sphingomonas. The study identified gastric microbiota beneficial to H. pylori eradication and laid a foundation for further research on how the gastric microbiota influences H. pylori eradication. In addition, the study results may help to improve the eradication rate of H. pylori in the future.
There are complex interactions between Helicobacter pylori (H. pylori) and other microbial communities in the gastric microecological environment. Yet, it remains unclear whether the interactions affect the eradication of H. pylori.
The motivation was to explore the interaction between gastric microbiota and H. pylori and to determine the influence of gastric microbiota on the eradication of H. pylori.
To investigate the characteristics of the gastric mucosa microbiota with H. pylori infection and the influence on H. pylori eradication treatment. This may help improve the eradication rate of H. pylori in the future.
Patients with H. pylori infection underwent gastroscopy and received treatment. Propensity matching analysis was conducted, including the number of patients who did not respond to treatment. The gastric microbiota was assayed by high-throughput sequencing and subsequent analysis of alpha diversity, beta diversity, species correlations, and predicted metabolic pathways.
The main phyla in the two groups were the same in the eight failure group patients who did not respond well to therapy and the 16 success group patients and included Proteobacteria, Bacteroides, Firmicutes, Actinomycetes, and Fusobacteria. The high rate of H. pylori eradication was associated with Rhodococcus, Lactobacillus, and Sphin
The effectiveness of quadruple H. pylori eradication therapy containing bismuth depended on the gastric microbiota. The high rate of H. pylori eradication was associated with Rhodococcus, Lactobacillus, and Sphingomonas.
This study laid a foundation for further research on the mechanism of the influence of the gastric microbiota on H. pylori eradication, which will help to improve the eradication rate of H. pylori in the future.
We thank all medical staff and technicians from the Department of Gastroenterology and Clinical Epidemiology Research Center who agreed to participate in this study.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Baryshnikova NV, Slomiany BL S-Editor: Ma YJ L-Editor: Filipodia P-Editor: Wang LL
| 1. | Marshall BJ, Warren JR. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet. 1984;1:1311-1315. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3302] [Cited by in RCA: 3263] [Article Influence: 79.6] [Reference Citation Analysis (1)] |
| 2. | Petrosino JF, Highlander S, Luna RA, Gibbs RA, Versalovic J. Metagenomic pyrosequencing and microbial identification. Clin Chem. 2009;55:856-866. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 389] [Cited by in RCA: 318] [Article Influence: 19.9] [Reference Citation Analysis (0)] |
| 3. | Mustapha P, Paris I, Garcia M, Tran CT, Cremniter J, Garnier M, Faure JP, Barthes T, Boneca IG, Morel F, Lecron JC, Burucoa C, Bodet C. Chemokines and antimicrobial peptides have a cag-dependent early response to Helicobacter pylori infection in primary human gastric epithelial cells. Infect Immun. 2014;82:2881-2889. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 27] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 4. | Yin YN, Wang CL, Liu XW, Cui Y, Xie N, Yu QF, Li FJ, Lu FG. Gastric and duodenum microflora analysis after long-term Helicobacter pylori infection in Mongolian Gerbils. Helicobacter. 2011;16:389-397. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 37] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 5. | Correa P, Houghton J. Carcinogenesis of Helicobacter pylori. Gastroenterology. 2007;133:659-672. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 467] [Cited by in RCA: 505] [Article Influence: 28.1] [Reference Citation Analysis (0)] |
| 6. | Wang AY, Peura DA. The prevalence and incidence of Helicobacter pylori-associated peptic ulcer disease and upper gastrointestinal bleeding throughout the world. Gastrointest Endosc Clin N Am. 2011;21:613-635. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 48] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 7. | Franceschi F, Zuccalà G, Roccarina D, Gasbarrini A. Clinical effects of Helicobacter pylori outside the stomach. Nat Rev Gastroenterol Hepatol. 2014;11:234-242. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 104] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 8. | Malfertheiner P, Megraud F, O'Morain CA, Gisbert JP, Kuipers EJ, Axon AT, Bazzoli F, Gasbarrini A, Atherton J, Graham DY, Hunt R, Moayyedi P, Rokkas T, Rugge M, Selgrad M, Suerbaum S, Sugano K, El-Omar EM; European Helicobacter and Microbiota Study Group and Consensus panel. Management of Helicobacter pylori infection-the Maastricht V/Florence Consensus Report. Gut. 2017;66:6-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2220] [Cited by in RCA: 1973] [Article Influence: 246.6] [Reference Citation Analysis (1)] |
| 9. | Khalifa MM, Sharaf RR, Aziz RK. Helicobacter pylori: a poor man's gut pathogen? Gut Pathog. 2010;2:2. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 106] [Cited by in RCA: 122] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 10. | Graham DY, Dore MP. Helicobacter pylori therapy: a paradigm shift. Expert Rev Anti Infect Ther. 2016;14:577-585. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 95] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 11. | Kotilea K, Mekhael J, Salame A, Mahler T, Miendje-Deyi VY, Cadranel S, Bontems P. Eradication rate of Helicobacter Pylori infection is directly influenced by adherence to therapy in children. Helicobacter. 2017;22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 39] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 12. | Liou JM, Fang YJ, Chen CC, Bair MJ, Chang CY, Lee YC, Chen MJ, Tseng CH, Hsu YC, Lee JY, Yang TH, Luo JC, Chang CC, Chen CY, Chen PY, Shun CT, Hsu WF, Hu WH, Chen YN, Sheu BS, Lin JT, Wu JY, El-Omar EM, Wu MS; Taiwan Gastrointestinal Disease and Helicobacter Consortium. Concomitant, bismuth quadruple, and 14-day triple therapy in the first-line treatment of Helicobacter pylori: a multicentre, open-label, randomised trial. Lancet. 2016;388:2355-2365. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 134] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 13. | Wang D, Li Q, Gong Y, Yuan Y. The association between vacA or cagA status and eradication outcome of Helicobacter pylori infection: A meta-analysis. PLoS One. 2017;12:e0177455. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 13] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 14. | Ferreira RM, Pereira-Marques J, Pinto-Ribeiro I, Costa JL, Carneiro F, Machado JC, Figueiredo C. Gastric microbial community profiling reveals a dysbiotic cancer-associated microbiota. Gut. 2018;67:226-236. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 341] [Cited by in RCA: 476] [Article Influence: 68.0] [Reference Citation Analysis (1)] |
| 15. | Sohn SH, Kim N, Jo HJ, Kim J, Park JH, Nam RH, Seok YJ, Kim YR, Lee DH. Analysis of Gastric Body Microbiota by Pyrosequencing: Possible Role of Bacteria Other Than Helicobacter pylori in the Gastric Carcinogenesis. J Cancer Prev. 2017;22:115-125. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 49] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 16. | Sung J, Kim N, Kim J, Jo HJ, Park JH, Nam RH, Seok YJ, Kim YR, Lee DH, Jung HC. Comparison of Gastric Microbiota Between Gastric Juice and Mucosa by Next Generation Sequencing Method. J Cancer Prev. 2016;21:60-65. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 53] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 17. | Lai YC, Wang TH, Huang SH, Yang SS, Wu CH, Chen TK, Lee CL. Density of Helicobacter pylori may affect the efficacy of eradication therapy and ulcer healing in patients with active duodenal ulcers. World J Gastroenterol. 2003;9:1537-1540. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 23] [Cited by in RCA: 27] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 18. | Onal IK, Gokcan H, Benzer E, Bilir G, Oztas E. What is the impact of Helicobacter pylori density on the success of eradication therapy: a clinico-histopathological study. Clin Res Hepatol Gastroenterol. 2013;37:642-646. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 17] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 19. | Prescott JF. Rhodococcus equi: an animal and human pathogen. Clin Microbiol Rev. 1991;4:20-34. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 466] [Cited by in RCA: 460] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 20. | Meijer WG, Prescott JF. Rhodococcus equi. Vet Res. 2004;35:383-396. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 84] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 21. | Goodfellow M, Alderson G. The actinomycete-genus Rhodococcus: a home for the "rhodochrous" complex. J Gen Microbiol. 1977;100:99-122. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 191] [Cited by in RCA: 159] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 22. | Testerman TL, Conn PB, Mobley HL, McGee DJ. Nutritional requirements and antibiotic resistance patterns of Helicobacter species in chemically defined media. J Clin Microbiol. 2006;44:1650-1658. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 61] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 23. | Nosaka K, Uchiyama R, Tadano K, Endo Y, Hayashi M, Konno H, Mimuro H. Thiamin transport in Helicobacter pylori lacking the de novo synthesis of thiamin. Microbiology (Reading). 2019;165:224-232. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 24. | Wang F, Feng J, Chen P, Liu X, Ma M, Zhou R, Chang Y, Liu J, Li J, Zhao Q. Probiotics in Helicobacter pylori eradication therapy: Systematic review and network meta-analysis. Clin Res Hepatol Gastroenterol. 2017;41:466-475. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 77] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 25. | Khosravi Y, Dieye Y, Loke MF, Goh KL, Vadivelu J. Streptococcus mitis induces conversion of Helicobacter pylori to coccoid cells during co-culture in vitro. PLoS One. 2014;9:e112214. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 37] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 26. | Zaman C, Osaki T, Hanawa T, Yonezawa H, Kurata S, Kamiya S. Analysis of the microflora in the stomach of Mongolian gerbils infected with Helicobacter pylori. J Gastroenterol Hepatol. 2010;25 Suppl 1:S11-S14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 15] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 27. | Chen YH, Tsai WH, Wu HY, Chen CY, Yeh WL, Chen YH, Hsu HY, Chen WW, Chen YW, Chang WW, Lin TL, Lai HC, Lin YH, Lai CH. Probiotic Lactobacillus spp. act Against Helicobacter pylori-induced Inflammation. J Clin Med. 2019;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 68] [Article Influence: 11.3] [Reference Citation Analysis (2)] |
| 28. | Richard ML, Liguori G, Lamas B, Brandi G, da Costa G, Hoffmann TW, Pierluigi Di Simone M, Calabrese C, Poggioli G, Langella P, Campieri M, Sokol H. Mucosa-associated microbiota dysbiosis in colitis associated cancer. Gut Microbes. 2018;9:131-142. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 148] [Article Influence: 18.5] [Reference Citation Analysis (1)] |
| 29. | Qian Y, Yang X, Xu S, Wu C, Song Y, Qin N, Chen SD, Xiao Q. Alteration of the fecal microbiota in Chinese patients with Parkinson's disease. Brain Behav Immun. 2018;70:194-202. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 215] [Cited by in RCA: 297] [Article Influence: 42.4] [Reference Citation Analysis (0)] |
| 30. | Schulz C, Koch N, Schütte K, Pieper DH, Malfertheiner P. H. pylori and its modulation of gastrointestinal microbiota. J Dig Dis. 2015;16:109-117. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 31] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 31. | Castaño-Rodríguez N, Goh KL, Fock KM, Mitchell HM, Kaakoush NO. Dysbiosis of the microbiome in gastric carcinogenesis. Sci Rep. 2017;7:15957. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 109] [Cited by in RCA: 182] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 32. | Plottel CS, Blaser MJ. Microbiome and malignancy. Cell Host Microbe. 2011;10:324-335. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 319] [Cited by in RCA: 484] [Article Influence: 37.2] [Reference Citation Analysis (0)] |
| 33. | Geng J, Song Q, Tang X, Liang X, Fan H, Peng H, Guo Q, Zhang Z. Co-occurrence of driver and passenger bacteria in human colorectal cancer. Gut Pathog. 2014;6:26. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 73] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 34. | Yan X, Yang M, Liu J, Gao R, Hu J, Li J, Zhang L, Shi Y, Guo H, Cheng J, Razi M, Pang S, Yu X, Hu S. Discovery and validation of potential bacterial biomarkers for lung cancer. Am J Cancer Res. 2015;5:3111-3122. [PubMed] |
| 35. | Guerrero-Preston R, Godoy-Vitorino F, Jedlicka A, Rodríguez-Hilario A, González H, Bondy J, Lawson F, Folawiyo O, Michailidi C, Dziedzic A, Thangavel R, Hadar T, Noordhuis MG, Westra W, Koch W, Sidransky D. 16S rRNA amplicon sequencing identifies microbiota associated with oral cancer, human papilloma virus infection and surgical treatment. Oncotarget. 2016;7:51320-51334. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 160] [Cited by in RCA: 236] [Article Influence: 33.7] [Reference Citation Analysis (0)] |