Published online Jun 7, 2021. doi: 10.3748/wjg.v27.i21.2910
Peer-review started: February 13, 2021
First decision: March 14, 2021
Revised: April 1, 2021
Accepted: April 20, 2021
Article in press: April 20, 2021
Published online: June 7, 2021
Processing time: 102 Days and 18.8 Hours
Persistent liver inflammatory damage is the main risk factor for developing liver fibrosis, cirrhosis, and even hepatocellular carcinoma in chronic hepatitis B (CHB) patients. Thus, accurate prediction of the degree of liver inflammation is a high priority and a growing medical need.
To build an effective and robust non-invasive model for predicting hepatitis B-related hepatic inflammation.
A total of 650 treatment-naïve CHB (402 HBeAg-positive and 248 HBeAg-negative) patients who underwent liver biopsy were enrolled in this study. Histological inflammation grading was assessed by the Ishak scoring system. Serum quantitative hepatitis B core antibody (qAnti-HBc) levels and 21 immune-related inflammatory factors were measured quantitatively using a chemiluminescent microparticle immunoassay. A backward feature elimination (BFE) algorithm utilizing random forest (RF) was used to select optional features and construct a combined model. The diagnostic abilities of the model or variables were evaluated based on the estimated area under the receiver operating characteristics curve (AUROC) and compared using the DeLong test.
Four features were selected to predict moderate-to-severe inflammation in CHB patients using the RF-BFE method. These predictive features included qAnti-HBc, ALT, AST, and CXCL11. Spearman’s correlation analysis indicated that serum qAnti-HBc, ALT, AST, and CXCL11 levels were positively correlated with the histology activity index (HAI) score. These selected features were incorporated into the model to establish a novel model named I-3A index. The AUROC [0.822; 95% confidence interval (CI): 0.790-0.851] of the I-3A index was significantly increased compared with qAnti-HBc alone (0.760, 95%CI: 0.724-0.792, P < 0.0001) in all CHB patients. The use of an I-3A index cutoff value of 0.41 produced a sensitivity of 69.17%, specificity of 81.44%, and accuracy of 73.8%. Additionally, the I-3A index showed significantly improved diagnostic performance for predicting moderate-to-severe inflammation in HBeAg-positive and HBeAg-negative CHB patients (0.829, 95%CI: 0.789-0.865 and 0.810, 95%CI: 0.755-0.857, respectively).
The selected features of the I-3A index constructed using the RF-BFE algorithm can effectively predict moderate-to-severe liver inflammation in CHB patients.
Core Tip: We aimed to propose an effective backward feature elimination algorithm utilizing random forest to select optimal features and construct a novel non-invasive model for predicting hepatitis B-related hepatic inflammation based on a large, multicenter cohort. The results indicated that the I-3A index constructed based on the selected features significantly improved the diagnostic efficiency of quantitative hepatitis B core antibody alone for predicting moderate-to-severe inflammation. Additionally, the I-3A index showed high diagnostic accuracy for moderate-to-severe inflammation in both HBeAg-positive and HBeAg-negative chronic hepatitis B patients.
- Citation: Zhou JY, Song LW, Yuan R, Lu XP, Wang GQ. Prediction of hepatic inflammation in chronic hepatitis B patients with a random forest-backward feature elimination algorithm. World J Gastroenterol 2021; 27(21): 2910-2920
- URL: https://www.wjgnet.com/1007-9327/full/v27/i21/2910.htm
- DOI: https://dx.doi.org/10.3748/wjg.v27.i21.2910
It is estimated that greater than 257 million people are chronically infected with hepatitis B virus (HBV), and over 880000 deaths are caused by HBV infection annually worldwide due to complications of the disease, such as liver fibrosis, cirrhosis, and hepatocellular carcinoma (HCC)[1]. In general, hepatocellular lesions are not caused by HBV directly but are the result of the induction of the body's autoimmune response to inflammatory damage in hepatocytes. The burden of continuous inflammatory injury of hepatocytes not only hinders the body to clear HBV but is also the main risk factor for the development of liver fibrosis, cirrhosis, and even HCC[2]. Thus, it is essential to accurately evaluate the degree of hepatic inflammation and effectively reverse disease progression in chronic hepatitis B (CHB) patients.
Hepatitis B core antibody (anti-HBc) is a specific, nonprotective antibody against hepatitis B core antigen (HBcAg) that can be detected in almost all patients who have been exposed to HBV[3]. Recent studies have identified serum quantitative hepatitis B core antibody (qAnti-HBc) levels as an important immunological indicator that reflects immune activation status[4-7]. Furthermore, we previously reported that serum qAnti-HBc levels increase significantly with increasing histology activity index (HAI) score and have potential clinical value in assessing the degree of hepatitis B-related hepatic inflammation in CHB patients[8]. However, the optimal diagnostic efficacy may not be obtained by using qAnti-HBc alone, and its combination with other biomarkers potentially offers great clinical application value.
In this study, we aimed to assess the correlation between inflammatory immune-related factors and the degree of liver inflammation in CHB patients. Furthermore, we aimed to establish a novel model of serum qAnti-HBc combined with other immune inflammatory factors using a machine learning strategy to improve the diagnostic efficiency of qAnti-HBc in predicting the degree of hepatitis B-related hepatic inflammation.
Patients with chronic HBV infection were enrolled in this study from 24 hospitals between October 2013 and December 2015. Patients recruited in the cohort study met the following criteria: (1) Age 18-75 years; (2) Hepatitis B surface antigen (HBsAg) seropositive status beyond 6 mo; (3) Being treatment naïve; (4) Serum negative for anti-hepatitis Avirus (HAV) IgM, anti-HCV, anti-hepatitis E virus (HEV) IgM/IgG, anti-Epstein–Barr virus (EBV) IgM, and anti-cytomegalovirus (CMV) IgM; and (5) Withdrawal from potential transaminase-lowering agents such as bicyclol for at least 2 wk prior to blood biochemistry analysis.
The exclusion criteria for this study included overlapping etiologies for hepatitis, including HCV, HIV, alcoholism, autoimmune disease, genetic disease, drug-induced causes, and nonalcoholic fatty liver. Patients with decompensated cirrhosis or HCC were also excluded. All patents provided written informed consent for the scientific use of their clinical data and samples. The complete protocol for the clinical trial has been registered at clinicaltrials.gov (NCT01962155) and chictr.org (ChiCTR-DDT-13003724).
All patients underwent ultrasonographic-guided liver biopsy according to a standardized protocol. Pathological interpretations were conducted at the Department of Pathology of Youan Hospital affiliated with Capital Medical University. Each section was blindly and independently assessed by two pathologists. When discrepancies occurred, the samples were reviewed by experienced pathologists who were also responsible for reassessing in 10% of the samples selected randomly. The modified HAI was used to grade disease inflammatory activity. HAI ≥ 5 represented moderate-to-severe inflammation, and fibrosis score ≥ 3 was considered significant fibrosis[9,10].
At the time of liver biopsy, biochemical, blood cell, and coagulation tests were further performed using routine automated analyzers. Serum HBsAg and HBeAg levels were quantified by commercially available enzyme immunoassays (Roche Diagnostics, Penzberg, Germany). Serum HBV DNA levels (range 2.0 × 101-1.7 × 108 IU/mL) were measured via a COBAS AmpliPrep/COBAS TaqMan method. Serum qAnti-HBc levels were measured using double-sandwich enzyme-linked immunosorbent assays (Wantai, Xiamen, China).
Six cytokines (CXCL9, CXCL10, CXCL11, IL-2R, IL-33, and IL-34) were measured with a Luminex screening system (LXSAHM-6, R&D, Minneapolis, MN, United States). Additionally, 15 cytokines (IL-1β, IL-2, IL-4, IL-6, IL-8, IL-10, IL-13, IL-17A, CCL2, CCL3, IL-12 p70, IFN-γ, TNF-α, TGF-α, and granulocyte-monocyte colony stimulating factor) were measured with human cytokines panel I (Cat. No. HCYTOMAG-60K, EMD Millipore, Billerica, MA, United States). The results were analyzed with a Luminex 200 system (EMD Millipore, Billerica, MA, United States) according to the manufacturer’s instructions. The coefficient of variation between the duplicate wells was controlled within 10%, and R2 of the standard curve was at least 0.999.
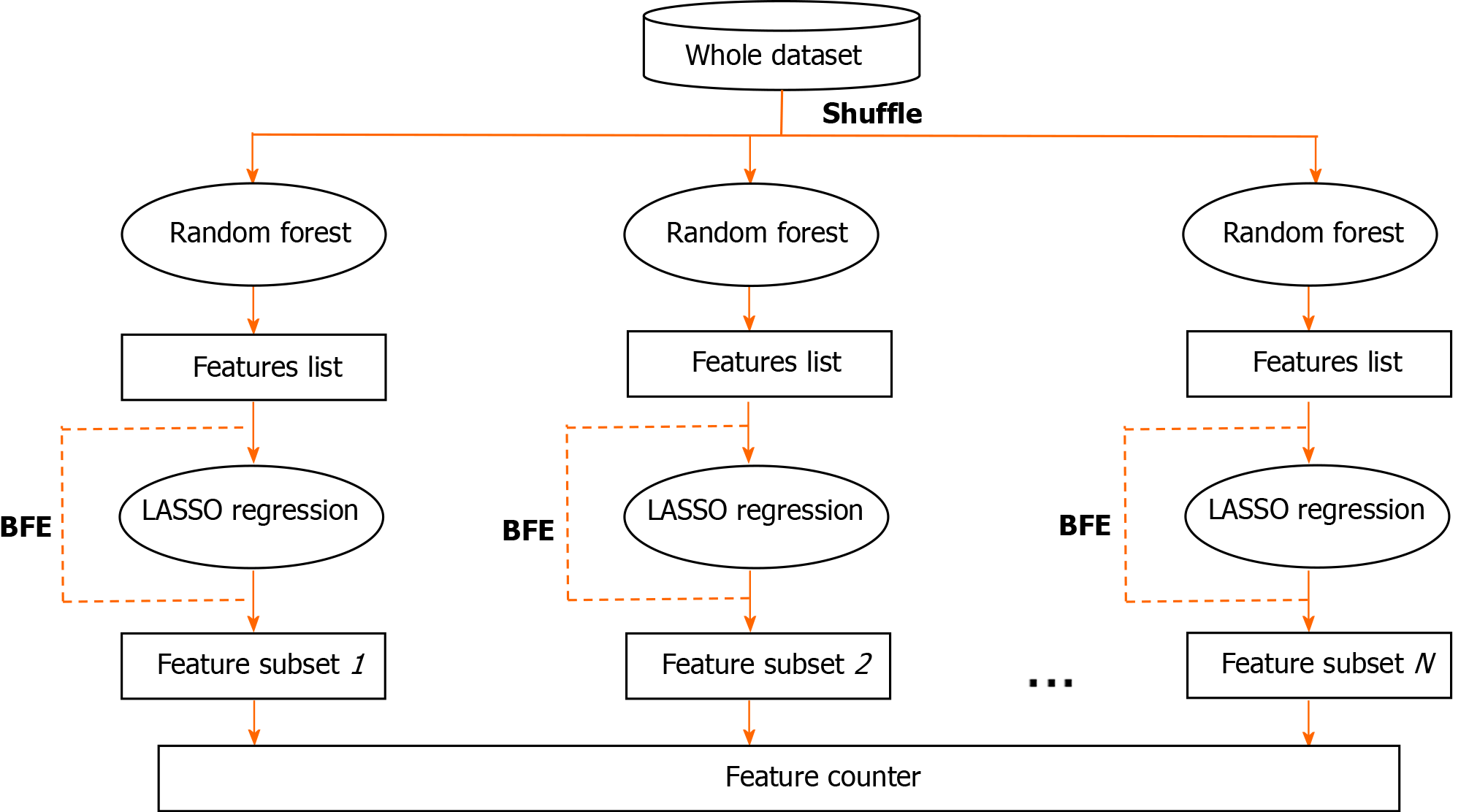
In this study, a random forest (RF)-backward feature elimination (BFE) algorithm was used to automatically select the optimal feature subset, and the main procedure of the RF-BFE algorithm is illustrated in Figure 1. First, the entire dataset was shuffled and the features were ranked by RF importance measures. Then, least absolute shrinkage and selection operator (LASSO) regression was iteratively used to perform both regularization and calculation of accuracy in the BFE. LASSO constructs a linear model, which penalizes the regression coefficients with an L1 penalty, shrinking many of them to zero. It performs both variable selection and regularization in order to enhance the prediction accuracy and interpretability of the resulting statistical model. The iterations ended when further removals did not result in an improvement, and produced a series of feature subsets. Finally, all of the feature subsets were gathered in a feature counter, and the final feature subset was determined by hard voting with equal weight.
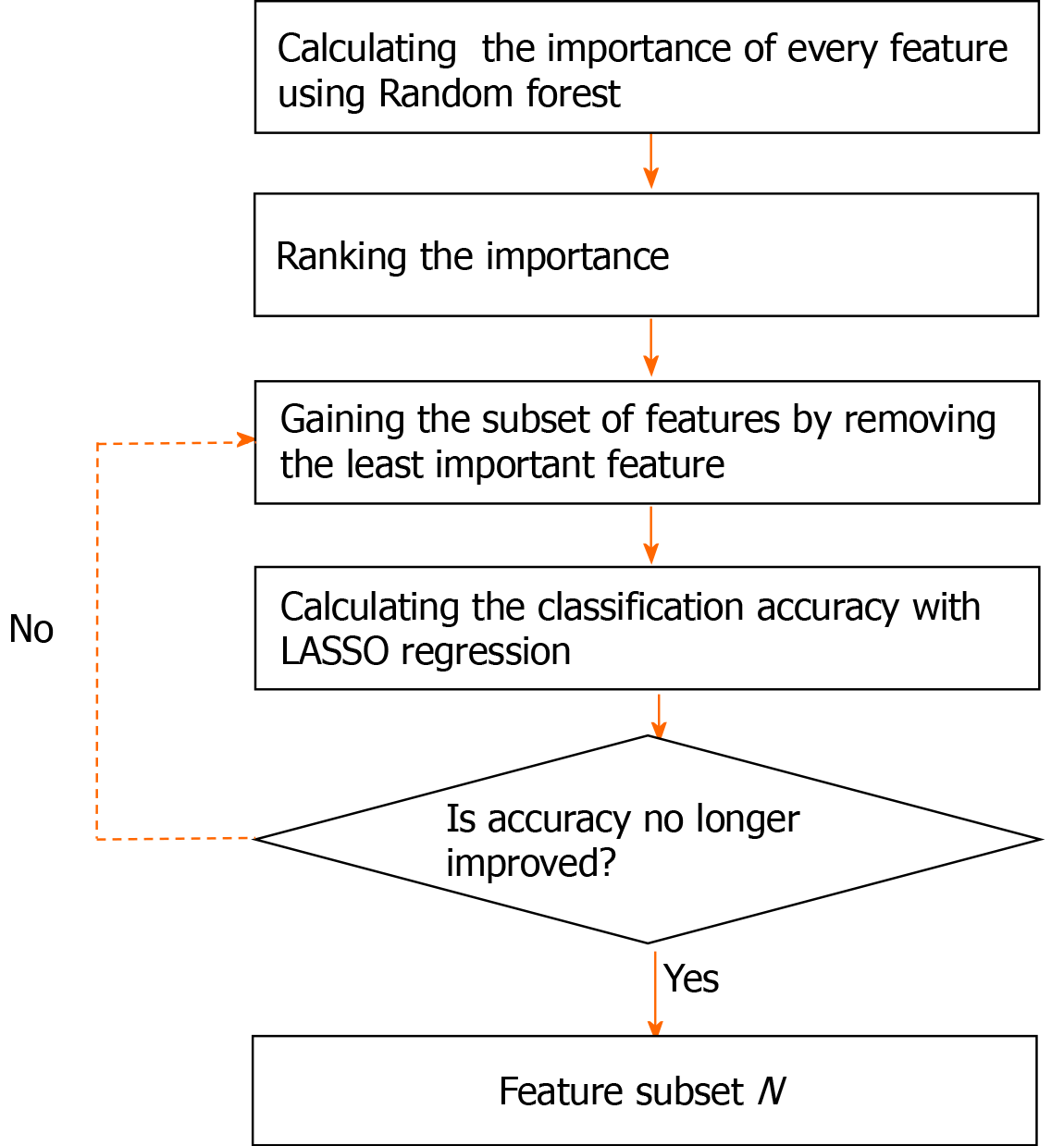
To provide more robust results, the RF importance measure of each feature was calculated in every iteration. The detailed feature selection scheme in an iteration is illustrated in Figure 2. The least important feature was eliminated, and then the updated features were used to retrain the LASSO regression. The BFE process ended when the accuracy no longer improved. The accuracy was calculated as follows:
Sensitivity = TP/(TP + FN)
Specificity = TN/(TN + FP)
Accuracy = (Sensitivity + Specificity)/2 (1)
where TP, TN, FP, and FN represent true positive, true negative, false positive, and false negative, respectively.
In our study, 100 iterations were implemented. Therefore, we used a hard vote strategy to determine the final features after the 100 subsets of the features were obtained. All of the selected features were gathered in a feature counter. The final feature subset Fs with k features was determined as follows:
Fs =Fsub :{f1, f2, ..., fk}|vf > v0
vf ,v0∈(0,N) (2)
where Fsubis the set of features addressed in all subsets, vfis the vote of the feature, and v0is the threshold. Given that we used 100 iterations, vfand v0ranged from 0 to 100.
Statistical analyses were conducted with SPSS (version16.0) and Python (version 3.5). Quantitative variables are expressed as the mean ± SD. Student’s t test was used to analyze single specific differences of biological interest, and the chi-square test was used to analyze relationships between categorical variables. Spearman’s rank tests were used to analyze associations between features and the HAI score. The LASSO logistic regression was performed using the “glmnet” package. The AUROC of models for diagnosing moderate-to-severe inflammation was analyzed using MedCalc version 15.6 (Ostend, Belgium). The diagnostic abilities of the different models were compared using the DeLong test. Additionally, the sensitivity, specificity, positive predictive value, (PPV) and negative predictive value (NPV) for the cut-off values were calculated. The level of statistical significance was defined as P < 0.05 (two-tailed).
In this study, a total of 650 patients were included in the statistical analysis, including 402 HBeAg-positive CHB patients and 248 HBeAg-negative CHB patients. Among the HBeAg-positive patients, 248 (61.69%) had at least moderate inflammation, and 249 (61.49%) had significant fibrosis. Among the HBeAg-negative patients, 138 (55.64%) had at least moderate inflammation, and 101 (40.73%) had significant fibrosis. The specific demographic characteristics are shown in Table 1. Serum levels of 4 (IL-2, IL-4, IL-12 p70, and CXCL9) of 21 cytokines were below the limits of detection in CHB patients. The remaining 17 cytokines were detected.
| Parameter | HBeAg-positive patients (n = 402) | HBeAg-negative patients (n = 248) | P value |
| Gender, male | 79.4.4% (319/402) | 76.2% (189/248) | 0.38 |
| Age (yr) | 36.35 ± 10.36 | 41.67 ± 9.65 | < 0.001 |
| BMI (kg/m2) | 23.12 ± 3.31 | 23.62 ± 2.84 | 0.01 |
| HBsAg (log10 IU/mL) | 3.83 ± 0.81 | 3.16 ± 0.73 | < 0.001 |
| HBV DNA (log10IU/mL) | 6.93 ± 1.66 | 4.82 ± 1.64 | < 0.001 |
| ALT (U/L) | 106.74 ± 139.40 | 80.58 ± 105.61 | < 0.001 |
| AST (U/L) | 65.48 ± 78.67 | 56.69 ± 70.06 | 0.029 |
| TC (mmol/L) | 4.61 ± 3.05 | 4.44 ± 0.98 | 0.691 |
| TBil (μmol/L) | 16.54 ± 15.48 | 18.49 ± 27.32 | 0.009 |
| DBil (μmol/L) | 5.61 ± 7.54 | 5.77 ± 6.59 | 0.527 |
| Albumin (g/L) | 43.72 ± 5.14 | 44.91 ± 6.07 | 0.011 |
| PT (s) | 12.81 ± 1.49 | 12.55 ± 1.49 | 0.035 |
| Cr (μmol/L) | 70.21 ± 25.41 | 69.19 ± 15.93 | 0.818 |
| BUN (mmol/L) | 5.28 ± 5.57 | 4.90 ± 1.33 | 0.681 |
| Platelet counts (× 109/L) | 178.6 ± 56.18 | 158.5 ± 55.28 | < 0.001 |
| Histology (n, %) | |||
| HAI 0-4 | 38.31% (154/402) | 44.35% (110/248) | 0.14 |
| HAI ≥ 5 | 61.69% (248/402) | 55.64% (138/248) | |
| Fibrosis score 0-2 | 38.06% (153/402) | 59.27% (147/248) | < 0.001 |
| Fibrosis score ≥ 3 | 61.49% (249/402) | 40.73% (101/248) |
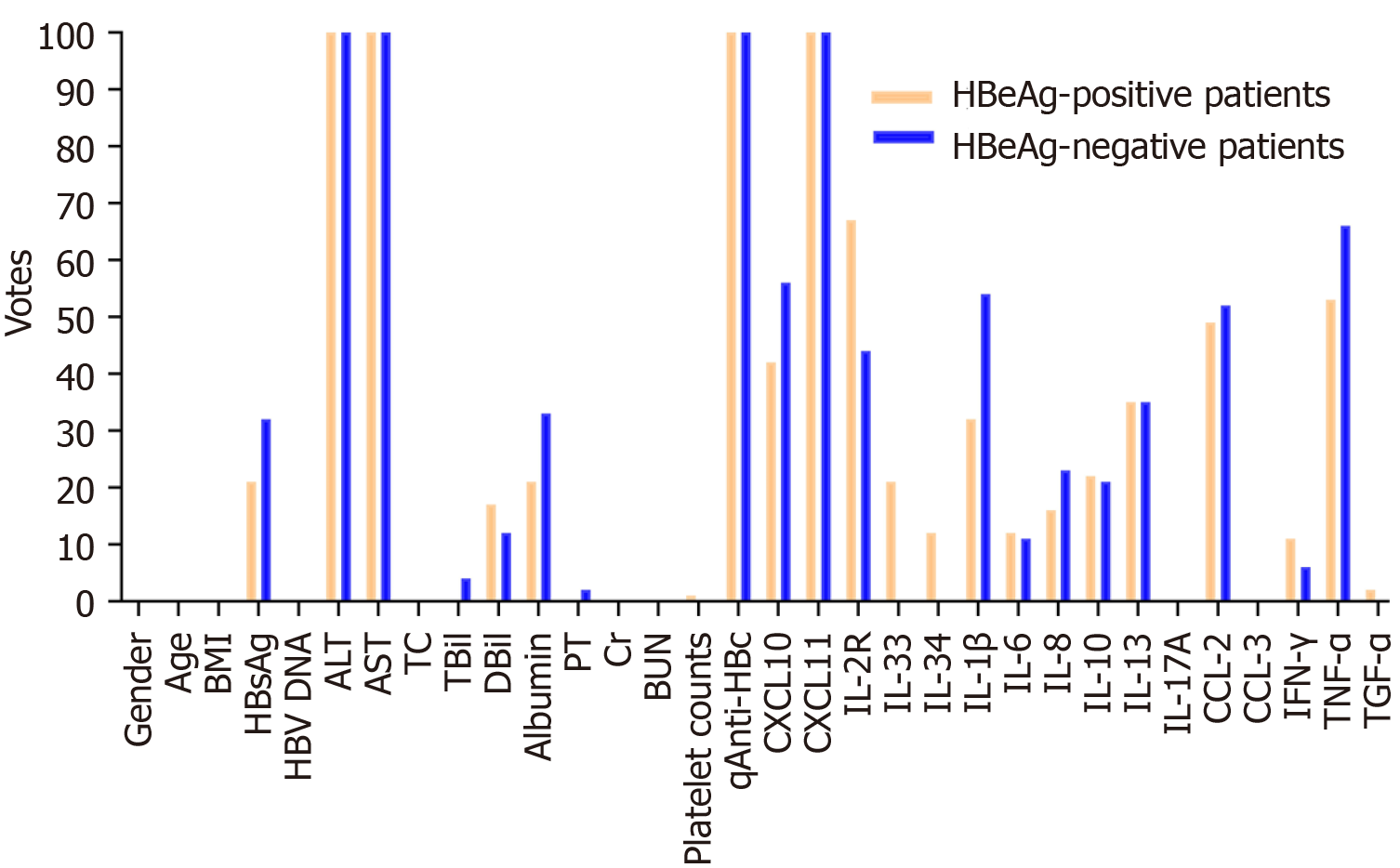
In this study, we set the threshold v0 to 100, and four features (qAnti-HBc, ALT, AST, and CXCL11) were selected using the RF-BFE method (Figure 3 and Table 2). To validate the correlation between the selected features and HAI score, Spearman’s rank tests were further performed. In HBeAg-positive patients, serum qAnti-HBc, ALT, AST, and CXCL11 levels were positively correlated with the HAI score (r = 0.55, 0.45, 0.51, and 0.34, respectively, P < 0.001). In HBeAg-negative patients, serum qAnti-HBc, ALT, AST, and CXCL11 Levels were also positively correlated with the HAI score (r = 0.49, 0.44, 0.49, and 0.33, respectively, all P < 0.001) (Table 2). Therefore, these selected features were incorporated into model construction to predict moderate-to-severe inflammation. Furthermore, the selected features were normalized using LASSO regression according to the following formula: ALT’ = (ALT-100.1)/130.25; AST’ = (AST-64.28)/78.33; Log(qAnti-HBc)’ = [log(qAnti-HBc) - 4.408]/0.7835; CXCL11’ = (CXCL11-55.02)/68.80. The established model, namely, the I-3A score, was calculated as follows:
| Feature name | HBeAg-positive patients | HBeAg-negative patients | ||||
| Votes | Spearman’r (95%CI)1 | P value1 | Votes | Spearman r (95%CI)1 | P value1 | |
| ALT | 100 | 0.45 (0.36-0.53) | < 0.001 | 100 | 0.44 (0.33-0.53) | < 0.001 |
| AST | 100 | 0.51 (0.43-0.58) | < 0.001 | 100 | 0.49 (0.38-0.58) | < 0.001 |
| qAnti-HBc | 100 | 0.55 (0.47-0.62) | < 0.001 | 100 | 0.49 (0.39-0.58) | < 0.001 |
| CXCL10 | 46 | 0.34 (0.25-0.43) | < 0.001 | 51 | 0.35 (0.23-0.46) | < 0.001 |
| CXCL11 | 100 | 0.34 (0.25-0.43) | < 0.001 | 100 | 0.33 (0.21-0.44) | < 0.001 |
| IL-2R | 71 | 0.30 (0.20-0.39) | < 0.001 | 44 | 0.27 (0.14-0.38) | < 0.001 |
| CCL2 | 42 | -0.01 (-0.09-0.07) | 0.79 | 52 | 0.03 (-0.10-0.16) | 0.65 |
| TNF-α | 56 | 0.01 (-0.17-0.09) | 0.78 | 62 | 0.01 (-0.17-0.09) | 0.78 |
| IL-1β | 27 | -0.10 (-0.23-0.03) | 0.12 | 54 | 0.15 (0.03-0.28) | 0.02 |
Logit P = 1/[1+ exp(0.4986 × ALT’ + 1.190 × AST’ + 0.9154 × log(qAnti-HBc)’ + 0.5333 × CXCL11’ + 0.8099)]. (3)
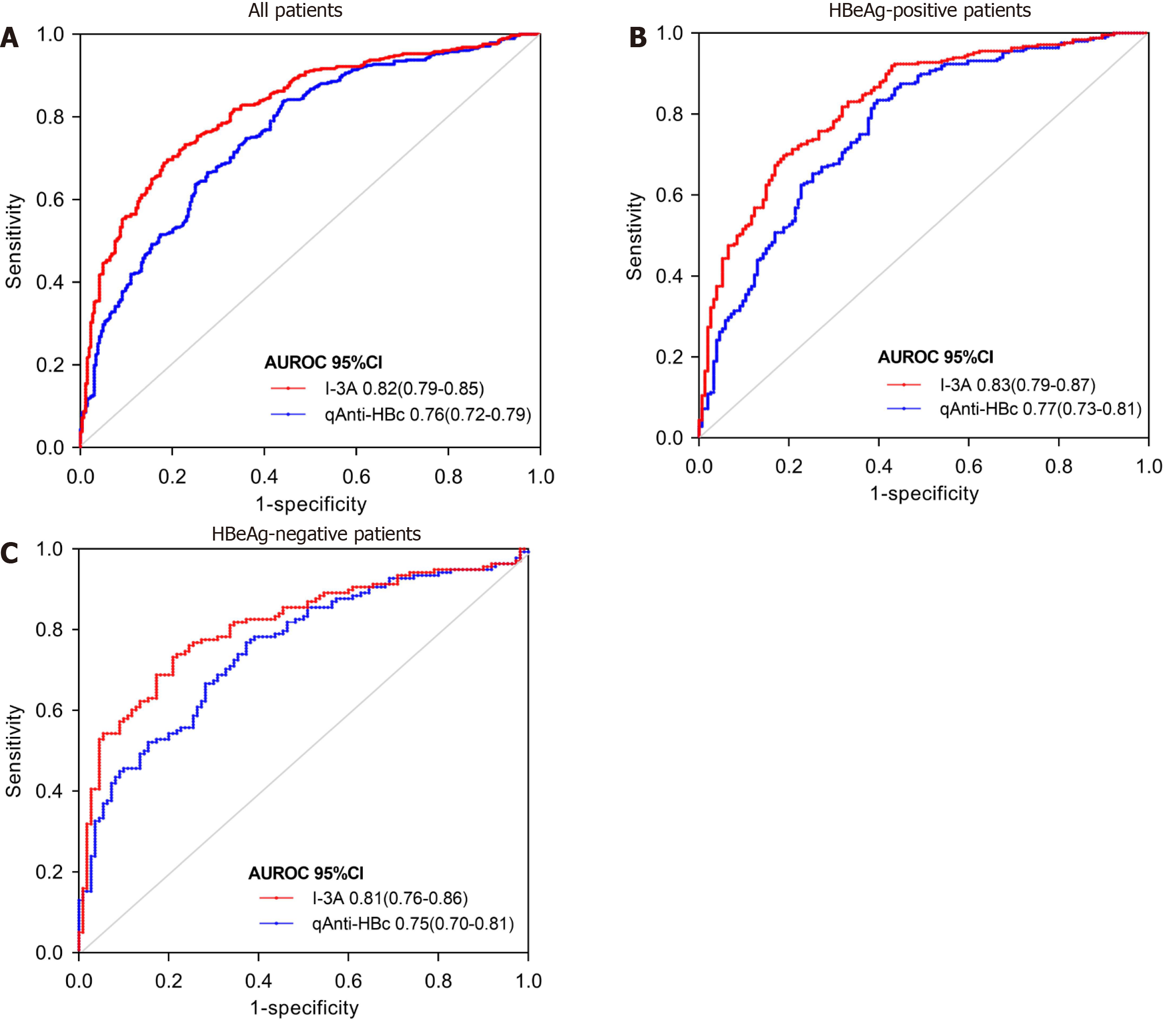
The ROC curves for predicting moderate-to-severe inflammation are shown in Figure 4 and Table 3. In all patients, the AUROC of the I-3A index to differentiate patients with moderate-to-severe inflammation was 0.822 [95% confidence interval (CI): 0.790-0.851], showing a significant increase compared with the value of 0.760 (95%CI: 0.724-0.792) for qAnti-HBc alone (P < 0.0001). The use of an I-3A index cutoff value of 0.41 (< 0.41) for predicting moderate-to-severe liver inflammation produced a sensitivity of 69.17%, specificity of 81.44%, and accuracy of 73.8%. The use of a qAnti-HBc cutoff value of 4.20 Log10IU/mL produced a sensitivity of 83.94%, specificity of 55.68%, and accuracy of 72.13% (Table 3). Similar results were obtained in diagnostic efficiency analyses of both in HBeAg-positive and HBeAg-negative CHB patients. Thus, these results indicate that the I-3A index has better diagnostic efficiency for predicting moderate-to-severe inflammation.
| Cutoff value | Se (%) | Spe (%) | PPV (%) | NPV (%) | Accuracy (%) | AUROC (95%CI) | P value | |
| All patients | ||||||||
| I-3A | < 0.41 | 69.17 | 81.44 | 84.49 | 64.37 | 73.8 | 0.82 (0.79-0.85) | < 0.0001 |
| qAnti-HBc | > 4.20 log10 IU/mL | 83.94 | 55.68 | 73.47 | 70.33 | 72.13 | 0.76 (0.72-0.79) | |
| HBeAg-positive patients | ||||||||
| I-3A | < 0.41 | 70.16 | 80.52 | 85.29 | 62.63 | 74.13 | 0.83 (0.79-0.87) | < 0.0001 |
| qAnti-HBc | > 4.20 log10 IU/ml | 83.06 | 60.39 | 77.15 | 68.89 | 74.38 | 0.77 (0.73-0.81) | |
| HBeAg-negative patients | ||||||||
| I-3A | < 0.44 | 73.19 | 78.18 | 80.8 | 69.92 | 75.4 | 0.81 (0.76-0.86) | 0.002 |
| qAnti-HBc | > 4.40 log10 IU/mL | 76.81 | 61.13 | 71.62 | 68 | 70.16 | 0.75 (0.70-0.81) | |
The BFE algorithm is widely used to reduce the dimensionality of feature space and is commonly employed using the support vector machine (SVM). RF is an importance measure that reveals the impact of each variable and has several advantages compared with SVM, as RF is an unbiased estimator, easy to parallel, and effectively and efficiently employed[11]. In this study, we proposed an effective BFE algorithm utilizing RF to select optional features and constructed a novel non-invasive model for predicting the degree of hepatitis B-related hepatic inflammation based on serum qAnti-HBc and other immune inflammatory factors. The results indicated that the I-3A index constructed based on the selected features significantly improved the diagnostic efficiency of qAnti-HBc alone for predicting moderate-to-severe inflammation. Additionally, the I-3A index showed high diagnostic accuracy for moderate-to-severe inflammation in both HBeAg-positive and HBeAg-negative CHB patients (AUROC = 0.829 and 0.810, respectively).
Histological analysis based on liver biopsy is the gold standard for evaluating the severity of hepatic inflammation. However, the practical use of this diagnostic method is limited by the traumatic nature of the procedures, which has inherent risks such as bleeding and sampling error[12,13]. ALT is the most widely used parameter of liver inflammation in clinical practice[14]. However, the diagnostic efficiency of ALT is limited due to its poor performance in CHB patients[15,16]. Therefore, non-invasive biomarkers that can accurately assess hepatic inflammation are still urgently needed. Our previous study of large, multicenter cohorts revealed that the serum qAnti-HBc level was remarkably correlated with the degree of liver inflammation in CHB patients[8]. Further analysis indicated that qAnti-HBc was an independent risk factor for moderate-to-severe inflammation in CHB patients and exhibited high diagnostic accuracy for hepatic inflammation in patients with normal ALT levels[8]. A recent study by Chen et al[17] reported that serum qAnti-HBc levels could reflect the degree of hepatic inflammation in children infected with HBV who have normal ALT levels. Together, these findings suggest that serum qAnti-HBc is a promising biomarker of hepatic inflammation. In this study, the AUROC value of serum qAnti-HBc levels for predicting CHB patients with moderate-to-severe inflammation was 0.760, similar to the value of 0.768 reported by Li et al[18]. To improve the diagnostic efficiency of qAnti-HBc in the CHB population, a combined model was constructed. Li et al[18] reported a novel model (AC index) combing qAnti-HBc and ALT that exhibited improved diagnostic efficiency for predicting patients with moderate-to-severe inflammation with an AUROC of 0.810. In this study, the RF-BFE algorithm was used to evaluate immune inflammatory factors that may be related to the degree of CHB liver inflammation, and the importance of each factor was evaluated. The results showed that in addition to qAnti-HBc, CXCL11, ALT, and AST exhibited relatively high significance in measuring liver inflammation. The AUROC value of the combined I-3A model based on LASSO regression was as high as 0.822, showing a significant increase compared with the diagnostic value of qAnti-HBc alone (LeLong P < 0.0001). Furthermore, the AUROC value of the I-3A index was significantly higher than that of the AC index reported by Li et al[18] (0.822 vs 0.810, DeLong P =0.041).
Of note, serum CXCL11 levels increased significantly as the HAI score increased in CHB patients. CXCL11, which is also known as human interferon-inducible T cell alpha chemokine (I-TAC), is a member of the chemokine family and is involved in various inflammatory and angiogenic processes[19]. In liver disease, hepatocytes and sinusoidal cells release CXCL11, selectively recruit activated T cells to sites of inflammation, and are involved in the progression of hepatic damage[20]. In our study, CXCL11 was selected using the RF-BFE method and represents a potential biomarker for predicting moderate-to-severe inflammation in CHB patients. Serum CXCL10 levels were also positively correlated with the severity of histological inflammation, consistent with the findings of previous studies[21,22]. However, in our study, CXCL10 was not selected and incorporated into the model. CXCR3 (CD183) is a surface marker of B lymphocyte activation, and its ligands include CXCL10 and CXCL11. Increased CXCR3 expression is observed in chronic viral hepatitis and correlates with increases in CXCL10 and CXCL11 expression[21,23]. In our study, serum CXCL10 and CXCL11 levels were positively correlated not only with the degree of liver inflammation but also with the level of serum qAnti-HBc in CHB patients (data not shown). This finding suggests that a large proportion of activated B lymphocytes may be present in CHB patients with hepatic inflammatory injury and that the specific level of qAnti-HBc secreted by B lymphocytes in circulating blood may indirectly reflect the degree of immune inflammatory injury of the liver. However, the specific mechanisms of B lymphocytes, Anti-HBc, and liver immune injury must be further explored.
There are several limitations to our study. First, this study examined only serological inflammatory factors, but imaging techniques or transient elastography may improve performance. Radiomics is a promising tool that extracts high-throughput quantitative features from medical images and can provide valuable information for the diagnosis of liver damage. Second, it was a cross-sectional study and longitudinal data were not obtained to verify our results. Finally, our previous studies reported that qAnti-HBc alone has high diagnostic accuracy for predicting liver inflammation in CHB patients with normal ALT levels, and the I-3A index does not remarkably improve the diagnostic value of qAnti-HBc. Therefore, future prospective studies are needed to achieve increased diagnostic accuracy.
In conclusion, the selected features of the I-3A index constructed using the RF-BFE method can effectively predict liver inflammation in CHB patients.
The burden of continuous inflammatory injury of hepatocytes is the main risk factor for the development of liver fibrosis, cirrhosis, and even hepatocellular carcinoma. Thus, it is essential to accurately evaluate the degree of hepatic inflammation and effectively reverse disease progression in chronic hepatitis B (CHB) patients.
Recent studies have identified that serum quantitative hepatitis B core antibody (qAnti-HBc) levels have potential clinical value in assessing the degree of hepatitis B-related hepatic inflammation in CHB patients. However, the optimal diagnostic efficacy may not be obtained by using qAnti-HBc alone, and its combination with other biomarkers potentially offers great clinical application value.
The objective of this study was to build an effective and robust noninvasive model for predicting hepatitis B-related hepatic inflammation.
Serum qAnti-HBc levels and 21 immune-related inflammatory factors were measured quantitatively in 650 treatment-naïve CHB patients who underwent liver biopsy. A backward feature elimination (BFE) algorithm utilizing Random Forest (RF) was used to select optional features and construct a combined model. The diagnostic abilities of the model or variables were evaluated based on estimated area under the receiver operating characteristics curve (AUROC) and compared using the DeLong test.
Four features (qAnti-HBc, ALT, AST, and CXCL11) were selected and incorporated into the model to establish a novel I-3A index. The AUROC of the I-3A index to predict moderate-to-severe liver inflammation was significantly increased compared with qAnti-HBc alone in all CHB patients. The I-3A index showed significantly improved diagnostic performance for predicting moderate-to-severe inflammation in HBeAg-positive and HBeAg-negative CHB patients.
The selected features of the I-3A index constructed using the RF-BFE algorithm can effectively predict moderate-to-severe liver inflammation in CHB patients.
The novel I-3A index is a promising non-invasive tool to predict liver inflammation. A longitudinal study is needed to verify our results and more emerging strategies such as radiomics are needed to further achieve increased diagnostic efficiency.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Sujatha R S-Editor: Gong ZM L-Editor: Wang TQ P-Editor: Wang LL
| 1. | Polaris Observatory Collaborators. Global prevalence, treatment, and prevention of hepatitis B virus infection in 2016: a modelling study. Lancet Gastroenterol Hepatol. 2018;3:383-403. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1260] [Cited by in RCA: 1213] [Article Influence: 173.3] [Reference Citation Analysis (2)] |
| 2. | Berasain C, Castillo J, Perugorria MJ, Latasa MU, Prieto J, Avila MA. Inflammation and liver cancer: new molecular links . Ann N Y Acad Sci. 2009;1155:206-221. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 286] [Cited by in RCA: 294] [Article Influence: 18.4] [Reference Citation Analysis (0)] |
| 3. | Wu T, Kwok RM, Tran TT. Isolated anti-HBc: The Relevance of Hepatitis B Core Antibody-A Review of New Issues. Am J Gastroenterol. 2017;112:1780-1788. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 33] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 4. | Fan R, Sun J, Yuan Q, Xie Q, Bai X, Ning Q, Cheng J, Yu Y, Niu J, Shi G, Wang H, Tan D, Wan M, Chen S, Xu M, Chen X, Tang H, Sheng J, Lu F, Jia J, Zhuang H, Xia N, Hou J; Chronic Hepatitis B Study Consortium. Baseline quantitative hepatitis B core antibody titre alone strongly predicts HBeAg seroconversion across chronic hepatitis B patients treated with peginterferon or nucleos(t)ide analogues. Gut. 2016;65:313-320. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 96] [Cited by in RCA: 110] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 5. | Song LW, Liu PG, Liu CJ, Zhang TY, Cheng XD, Wu HL, Yang HC, Hao XK, Yuan Q, Zhang J, Kao JH, Chen DS, Chen PJ, Xia NS. Quantitative hepatitis B core antibody levels in the natural history of hepatitis B virus infection. Clin Microbiol Infect. 2015;21:197-203. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 97] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 6. | Yang HC, Tsou HH, Pei SN, Chang CS, Chen JH, Yao M, Lin SJ, Lin J, Yuan Q, Xia N, Liu TW, Chen PJ, Cheng AL, Hsu C; Taiwan Cooperative Oncology Group. Quantification of HBV core antibodies may help predict HBV reactivation in patients with lymphoma and resolved HBV infection. J Hepatol. 2018;69:286-292. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 75] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 7. | Chi H, Li Z, Hansen BE, Yu T, Zhang X, Sun J, Hou J, Janssen HLA, Peng J. Serum Level of Antibodies Against Hepatitis B Core Protein Is Associated With Clinical Relapse After Discontinuation of Nucleos(t)ide Analogue Therapy. Clin Gastroenterol Hepatol 2019; 17: 182-191. e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 62] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 8. | Zhou J, Song L, Zhao H, Yan L, Ma A, Xie S, Zhang X, Zhang D, Xie Q, Zhang G, Shang J, Cheng J, Zhao W, Zou Z, Zhang M, Xia N, Wang G. Serum hepatitis B core antibody as a biomarker of hepatic inflammation in chronic hepatitis B patients with normal alanine aminotransferase. Sci Rep. 2017;7:2747. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 45] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 9. | Wai CT, Greenson JK, Fontana RJ, Kalbfleisch JD, Marrero JA, Conjeevaram HS, Lok AS. A simple noninvasive index can predict both significant fibrosis and cirrhosis in patients with chronic hepatitis C. Hepatology. 2003;38:518-526. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2762] [Cited by in RCA: 3242] [Article Influence: 147.4] [Reference Citation Analysis (0)] |
| 10. | Hui AY, Chan HL, Wong VW, Liew CT, Chim AM, Chan FK, Sung JJ. Identification of chronic hepatitis B patients without significant liver fibrosis by a simple noninvasive predictive model. Am J Gastroenterol. 2005;100:616-623. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 151] [Cited by in RCA: 156] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 11. | Breiman L. Random Forests. Mach Learn. 2001;45:5-32. [RCA] [DOI] [Full Text] [Cited by in Crossref: 56052] [Cited by in RCA: 34052] [Article Influence: 2837.7] [Reference Citation Analysis (0)] |
| 12. | Cadranel JF, Rufat P, Degos F. Practices of liver biopsy in France: results of a prospective nationwide survey. For the Group of Epidemiology of the French Association for the Study of the Liver (AFEF). Hepatology. 2000;32:477-481. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 723] [Cited by in RCA: 731] [Article Influence: 29.2] [Reference Citation Analysis (0)] |
| 13. | Regev A, Berho M, Jeffers LJ, Milikowski C, Molina EG, Pyrsopoulos NT, Feng ZZ, Reddy KR, Schiff ER. Sampling error and intraobserver variation in liver biopsy in patients with chronic HCV infection. Am J Gastroenterol. 2002;97:2614-2618. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1504] [Cited by in RCA: 1569] [Article Influence: 68.2] [Reference Citation Analysis (0)] |
| 14. | Kim WR, Flamm SL, Di Bisceglie AM, Bodenheimer HC; Public Policy Committee of the American Association for the Study of Liver Disease. Serum activity of alanine aminotransferase (ALT) as an indicator of health and disease. Hepatology. 2008;47:1363-1370. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 521] [Cited by in RCA: 585] [Article Influence: 34.4] [Reference Citation Analysis (0)] |
| 15. | Chao DT, Lim JK, Ayoub WS, Nguyen LH, Nguyen MH. Systematic review with meta-analysis: the proportion of chronic hepatitis B patients with normal alanine transaminase ≤ 40 IU/L and significant hepatic fibrosis. Aliment Pharmacol Ther. 2014;39:349-358. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 86] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 16. | Sarin SK, Kumar M, Lau GK, Abbas Z, Chan HL, Chen CJ, Chen DS, Chen HL, Chen PJ, Chien RN, Dokmeci AK, Gane E, Hou JL, Jafri W, Jia J, Kim JH, Lai CL, Lee HC, Lim SG, Liu CJ, Locarnini S, Al Mahtab M, Mohamed R, Omata M, Park J, Piratvisuth T, Sharma BC, Sollano J, Wang FS, Wei L, Yuen MF, Zheng SS, Kao JH. Asian-Pacific clinical practice guidelines on the management of hepatitis B: a 2015 update. Hepatol Int. 2016;10:1-98. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1985] [Cited by in RCA: 1955] [Article Influence: 217.2] [Reference Citation Analysis (0)] |
| 17. | Chen HS, Wu JF, Su TH, Chen HL, Hsu HY, Xia NS, Chen PJ, Chang MH. Baseline Level of Hepatitis B Core Antibody Predicts Spontaneous Hepatitis B e Antigen (HBeAg) Seroconversion in HBeAg-Positive Children With a Normal Alanine Aminotransferase Level. Hepatology. 2019;70:1903-1912. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 10] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 18. | Li J, Zhang TY, Song LW, Qi X, Yu XP, Li FH, Zhou P, Qin YL, Yang L, Zhao JH, Mao RC, Zhang YM, Wang JY, Yang FF, Zhu HX, Yang SS, Huang YX, Yuan Q, Zhang J, Zhang JM, Xia NS. Role of quantitative hepatitis B core antibody levels in predicting significant liver inflammation in chronic hepatitis B patients with normal or near-normal alanine aminotransferase levels. Hepatol Res. 2018;48:E133-E145. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 35] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 19. | Lacotte S, Brun S, Muller S, Dumortier H. CXCR3, inflammation, and autoimmune diseases. Ann N Y Acad Sci. 2009;1173:310-317. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 233] [Cited by in RCA: 261] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 20. | Helbig KJ, Ruszkiewicz A, Semendric L, Harley HA, McColl SR, Beard MR. Expression of the CXCR3 ligand I-TAC by hepatocytes in chronic hepatitis C and its correlation with hepatic inflammation. Hepatology. 2004;39:1220-1229. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 98] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 21. | Zeremski M, Petrovic LM, Chiriboga L, Brown QB, Yee HT, Kinkhabwala M, Jacobson IM, Dimova R, Markatou M, Talal AH. Intrahepatic levels of CXCR3-associated chemokines correlate with liver inflammation and fibrosis in chronic hepatitis C. Hepatology. 2008;48:1440-1450. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 176] [Cited by in RCA: 180] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 22. | Chalin A, Lefevre B, Devisme C, Barget N, Amiot L, Samson M. Circulating levels of CXCL11 and CXCL12 are biomarkers of cirrhosis in patients with chronic hepatitis C infection. Cytokine. 2019;117:72-78. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 13] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 23. | Butera D, Marukian S, Iwamaye AE, Hembrador E, Chambers TJ, Di Bisceglie AM, Charles ED, Talal AH, Jacobson IM, Rice CM, Dustin LB. Plasma chemokine levels correlate with the outcome of antiviral therapy in patients with hepatitis C. Blood. 2005;106:1175-1182. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 135] [Cited by in RCA: 140] [Article Influence: 7.0] [Reference Citation Analysis (0)] |