Published online May 28, 2021. doi: 10.3748/wjg.v27.i20.2586
Peer-review started: November 18, 2020
First decision: January 23, 2021
Revised: February 10, 2021
Accepted: April 2, 2021
Article in press: April 2, 2021
Published online: May 28, 2021
Processing time: 182 Days and 23.3 Hours
Hepatocellular carcinoma (HCC) is a malignancy found globally. Accumulating studies have shown that long noncoding RNAs (lncRNAs) play critical roles in HCC. However, the function of lncRNA in HCC remains poorly understood.
To understand the effect of lncRNA W42 on HCC and dissect the underlying molecular mechanisms.
We measured the expression of lncRNA W42 in HCC tissues and cells (Huh7 and SMMC-7721) by quantitative reverse transcriptase polymerase chain reaction. Receiver operating characteristic curves were used to assess the sensitivity and specificity of lncRNA W42 expression. HCC cells were transfected with pcDNA3.1-lncRNA W42 or shRNA-lncRNA W42. Cell functions were detected by cell counting Kit-8 (CCK-8), colony formation, flow cytometry and Transwell assays. The interaction of lncRNA W42 and DBN1 was confirmed by RNA immunoprecipitation and RNA pull down assays. An HCC xenograft model was used to assess the role of lncRNA W42 on tumor growth in vivo. The Kaplan-Meier curve was used to evaluate the overall survival and recurrence-free survival after surgery in patients with HCC.
In this study, we identified a novel lncRNA (lncRNA W42), and investigated its biological functions and clinical significance in HCC. LncRNA W42 expression was upregulated in HCC tissues and cells. Overexpression of lncRNA W42 notably promoted the proliferative and invasion of HCC, and inhibited cell apoptosis. LncRNA W42 directly bound to DBN1 and activated the downstream pathway. LncRNA W42 knockdown suppressed HCC xenograft tumor growth in vivo. The clinical investigation revealed that HCC patients with high lncRNA W42 expression exhibited shorter survival times.
In vitro and in vivo results suggested that the novel lncRNA W42, which is upregulated in HCC, may serve as a potential candidate prognostic biomarker and therapeutic target in HCC patients.
Core Tip: The results of this study demonstrated, for the first time, that long noncoding RNA (lncRNA) W42 expression was significantly higher in hepatocellular carcinoma (HCC) tissues than in normal tissues and that dysregulation of lncRNA W42 was positively correlated with tumor number, liver cirrhosis and tumor recurrence in patients with HCC. Moreover, increased lncRNA W42 expression was associated with a poor prognosis in patients with HCC. These findings suggest that lncRNA W42 is an important marker for indicting prognosis and may have potential as a diagnostic and therapeutic target for HCC.
- Citation: Lei GL, Niu Y, Cheng SJ, Li YY, Bai ZF, Yu LX, Hong ZX, Liu H, Liu HH, Yan J, Gao Y, Zhang SG, Chen Z, Li RS, Yang PH. Upregulation of long noncoding RNA W42 promotes tumor development by binding with DBN1 in hepatocellular carcinoma. World J Gastroenterol 2021; 27(20): 2586-2602
- URL: https://www.wjgnet.com/1007-9327/full/v27/i20/2586.htm
- DOI: https://dx.doi.org/10.3748/wjg.v27.i20.2586
Liver cancer is a major health problem[1] and is currently the third leading cause of cancer-related mortality in the world[2]. Among all primary liver cancers, hepatocellular carcinoma (HCC) accounts for approximately 90% of cases[3]. Several complex pathways with frequent mutations in HCC, including the telomere maintenance, cell cycle, WNT-β-catenin, epigenetic and chromatin remodeling pathways, have been well defined[4-8]. Although advances in the diagnosis and treatment of HCC have been made, its prognosis remains poorly understood. Even with radical treatment of HCC, the median 5-year survival rate remains lower than 50%[9-11].
Long noncoding RNAs (lncRNAs) are noncoding transcripts that exceed 200 nt in length; they have been defined as the largest subclass in the noncoding transcriptome in humans[12-14]. The majority of lncRNAs with limited protein coding potential are transcribed by RNA polymerase II and polyadenylated at their 5′ and 3′ ends, respectively[15]. Recent reports have shown that a growing number of cancer transcriptomes have indeed revealed that thousands of aberrant lncRNAs are closely related to a variety of cancer types[16-18], including HCC. In particular, previous studies have revealed that a number of lncRNAs are dysregulated and exert essential roles in HCC progression; these lncRNAs include BZRAP1-AS1[19], RAB5IF[20], SNHG15[21], MAGI2-AS3[22], ATB[23], etc. We also previously demonstrated that the decreased expression of long intergenic noncoding RNA P7 predicts unfavorable prognosis and promotes tumor proliferation via the modulation of the STAT1-MAPK pathway in HCC[24], and lncRNA W5 inhibits progression and predicts favorable prognosis in HCC[25].
In this report, for the first time, we identified a novel lncRNA, W42, and investigated its expression levels in HCC tumor tissues and its association with the clinical-pathological parameters of HCC patients. The expression, biological function and underlying molecular mechanisms of lncRNA W42 in HCC were also studied in vivo and in vitro.
Data were obtained from 92 patients with HCC (72 males and 20 females) who underwent surgery for HCC at the Department of Hepatobiliary Surgery of the Fifth Medical Center, Chinese PLA General Hospital between October 2013 and July 2019. After obtaining written informed consent from all patients, tumor tissues and paired adjacent noncancerous tissue samples were collected from patients with HCC in accordance with the institutional guidelines of the hospital’s Ethics Committee. Resected tissues were immediately snap-frozen in liquid nitrogen within 30 min and preserved in an HCC Tissue Biobank until use. This study was approved by the Ethics Committee of the Fifth Medical Center of Chinese PLA General Hospital, and carried out according to the Declaration of Helsinki.
The human HCC cell lines HepG2 and Huh7 were purchased from the American Type Culture Collection (ATCC, Manassas, VA, United States). The normal liver cell line LO2 and human HCC cell line SMMC-7721 were obtained from Cell Bank, Shanghai Institutes for Biological Sciences, Chinese Academy of Science (Shanghai, China). SMMC-7721 cells were established from a 50-year-old Chinese male HCC patient with negative HBsAg and positive α-fetoprotein (AFP) by the scholars of Second Military Medical University (Shanghai, China) in 1977[26]. The use of the cell lines was approved by the Ethics Committee of the Fifth Medical Center of Chinese PLA General Hospital. All of the cell lines used in this study were maintained in DMEM (Thermo Fisher, Beijing, China) supplemented with 10% fetal bovine serum (Gibco, Beijing, China) in 5% CO2 at 37°C.
Total RNA from frozen fresh HCC tissues and paired adjacent noncancerous tissues was extracted using TRIzol reagent (Thermo Fisher) according to the manufacturer's instructions. RNA concentration and purity were detected using a NanoDrop ND-1000 (Thermo Fisher, Beijing, China). LncRNA W42 expression levels were examined by quantitative reverse transcriptase polymerase chain reaction (qRT-PCR) performed with Maxima SYBR Green on an ABI 7500 instrument (Applied Biosystems, CA, United States). All reactions were run at least three times using lncRNA W42-specific primers. GAPDH (glyceraldehyde-3-phosphate dehydrogenase) expression was monitored as the endogenous control, and all samples were normalized to human GAPDH[24,27,28]. The primer sequences of lncRNA W42 were as follows: forward: 5’-TGTGAACCAGGTTTGCTGGA-3’, reverse: 5’-CTCAACCATGCCGACGAGAA-3’; GAPDH forward: 5’-CAGCCTCAAGATCATCAGCA-3’ reverse: 5’-TGTGGTCATGAGTCCTTCCA-3’. The amplification procedure was 95°C for 5 min, followed by 40 cycles of denaturation at 95°C for 15 s and annealing at 60°C for 30 s, extension at 72°C for 30 s. The median of triplicate reactions was used to calculate relative lncRNA expression (ΔCt = Ct median lncRNA - Ct median GAPDH). Expression changes were calculated using the 2-ΔΔCt method.
The lncRNA W42 overexpression vector was constructed and cloned into the BamHI and EcorI sites of the pcDNA3.1 (+) vector, yielding pcDNA3.1-lncRNA W42. The primers used in this experiment were as follows: forward: 5’-AGCTGAGGGAGCCGGCT-3’, reverse: 5’-ATTAATGAGAGAATTATGGTAT-3’. In addition, gene knockdown used RNA interference experiments. LncRNA W42 was knocked down by shRNA lentiviruses. shRNA-lncRNA W42-1 and shRNA-lncRNA W42-2 were designed and synthesized by RIBOBIO Co., Ltd. (Guangzhou, China). Their sequences were as follows: W42-1: 3’-dTdT GGCUCAGUCUUUGAUCAAA-5’; W42-2: 3’-dTdT CCUGUAGUAAACUACUCAA-5’; the shRNA negative control sequence number was siN05815122147 of RIBOBIO. SMMC7721 cells (1 × 104) were stably transfected with lentiviral vector containing lncRNA W42 with the help of Hanbio Biotechnology Co., Ltd. (Shanghai, China). After infection for 72 h, the cells were collected for RNA quantitation of lncRNA W42 and targeted genes using qRT-PCR.
A nucleocytoplasmic separation experiment was conducted with the PARISTM Protein and RNA Isolation System (Ambion, Life Technologies Lithuania). The experiment followed the manufacturer’s instructions.
Cell proliferation was assessed by the Cell Counting Kit-8 (CCK-8, Dojindo Laboratories, Kumamoto, Japan) assay according to the manufacturer’s protocols. Huh7 or SMMC7721 cells were plated in 12-well plates (5 × 105 cells/well) and then transfected with lncRNA W42 overexpression vector, and the numbers of cells per well were examined and counted by the absorbance (450 nm) of reduced WST-8 [2-(2-methoxy-4-nitrophenyl)-3-(4-nitrophenyl)-5-(2,4-isulfophenyl)-2H-tetrazolium, monosodium salt] at the indicated time.
Huh7 or SMMC7721 cells were plated in 6-well plates and incubated in DMEM with 10% fetal bovine serum (FBS) with 5% CO2 at 37°C. Six days later, the cells were fixed and stained with 0.1% crystal violet. The number of colonies, defined as > 50 cells/colony, was recorded and photographed[24].
Cell migration and invasion were determined using Transwell chambers (Sigma). The upper chamber of the Transwell was treated with Matrigel (without Matrigel coating for the migration assay). Briefly, SMMC7721 or Huh7 cells (50000 cells/well) were resuspended in serum-free DMEM (Gibco). Then, the medium containing the cells was seeded into the upper chamber. In addition, the lower chamber was supplemented with medium containing 10% FBS (Gibco) as a chemoattractant. After incubation for 48 h, the cells that did not migrate or invade the membrane were scraped away, while the migrated and invaded cells in the lower chamber of the Transwell were fixed with methanol and stained with 0.1% crystal violet (Sigma). The number of migrated and invaded cells was counted using a microscope.
An Annexin V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) apoptosis kit (BestBio, Shanghai, China) was used for the apoptosis assay and analyzed by flow cytometry. After treatment, SMMC7721 and Huh7 were washed three times with phosphate buffered saline (PBS) and stained with Annexin V-FITC/PI. After filtration, the suspension of each group was analyzed within 1 h using a BD FACSCalibur flow cytometer (BD Biosciences, Franklin Lakes, NJ, United States). The cells were transfected with lncRNA W42 overexpression vector or control and cultured for 48 h. Then the cells were washed twice with PBS and fixed in cold 70% ethanol for 1 h followed by incubation with 20 µg/mL PI (BestBio, Shanghai, China) and 200 µg/mL RNase A (Sigma) for 30 min. Finally, the cells were analyzed utilizing a flow cytometer (BD Biosciences, Franklin Lakes, NJ, United States).
RNA pull-down assays were performed as previously described[29,30]. Briefly, in vitro biotin-labeled RNAs (lncRNA W42 and its antisense RNA) were transcribed by biotin RNA labeling mix (Roche) and T7 RNA polymerase (Roche), treated with RNase-free DNase I (Promega) and purified using the RNeasy Mini Kit (QIAGEN). Biotinylated RNA was incubated with cell lysate, and precipitated proteins were separated via sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and subjected to mass spectrometric (MS) analysis. RNA pull-down of AVAN-associated proteins was performed using biotinylated lncRNA W42 or antisense probes (immunoglobulin G, IgG). Isolated proteins were resolved by SDS-PAGE followed by silver staining. Silver staining was performed with the SilverQuest™ Silver Staining Kit (Invitrogen) following the manufacturer’s recommendations.
The RNA immunoprecipitation (RIP) assay was performed as previously described[31,32]. With IgG antibodies as the isotype-matched control, nuclear extracts were immunoprecipitated with 2.5 mg hnRNP U (Abcam, clone 4D11, ab6106) overnight. RNA-protein-antibody complexes were captured using Dynabead Protein A/G (Thermo Fisher Scientific). RNA was eluted by adding TRIzol directly to magnetic beads and isolated according to the manufacturer’s instructions. cDNA was synthesized using TransScript First-Strand cDNA Synthesis SuperMix (TransGen Biotech) and analyzed by qPCR. The results were normalized to input RNA and are shown as fold-enrichment over control IgG RIP.
A total of 16 female 4-6-wk-old nude BALB/C mice were purchased from Charles River Laboratories (Beijing, China) and were maintained in a SPF (specific pathogen-free) facility under pathogen-free conditions at 22°C and 40%-50% humidity with a 12/12 h light/dark cycle, and had ad libitum access to food and water. Each mouse was randomly assigned to one of the following two groups: LncRNA W42 knockdown and control group (n = 8 per group). SMMC7721 cells (2 × 106) stably lncRNA W42 knockdown or empty vector were subcutaneously injected into the flanks of nude mice. Tumor growth was monitored by tumor volume and weight. After 4 d injection, tumor size was recorded every 3 d, and tumor volumes were calculated with the formula: Tumor volume (mm3) = length × width2 × 0.5. After 36 d, the mice were euthanized by i.p. injection of sodium pentobarbital (150 mg/kg), and tumor tissues were removed, weighed and fixed in polyformaldehyde after imaging of the tumors for further examination. All animal procedures were performed in strict accordance with the recommendations of the National Institutes of Health Laboratory Animal Care and Use Guidelines. This study was approved by the Ethics Committee of the Fifth Medical Center of Chinese PLA General Hospital (2020-002).
All data were analyzed using the SPSS 20.0 software package (Chicago, IL, United States). All statistical results are expressed as the mean ± SD. Kaplan-Meier analyses were used to represent the correlations between lncRNA W42 expression level and overall survival (OS) rate or recurrence-free survival (RFS). The data from the two groups were assessed using the Student t-test, the differences between various groups were analyzed by one-way analysis of variance (ANOVA), followed by Tukey’s post hoc test. All experiments were carried out at least three times. P values < 0.05 were considered significant.
Based on our previous bioinformatics analysis, we chose another 10 candidate lncRNAs and re-validated their expression in the normal liver cell line LO2 and the human HCC cell line Huh7 (Supplementary Figure 1A). Subsequently, we re-validated the expression of lncRNA W42 in 25 pairs of HCC tissues and paired adjacent noncancerous tissues. The results showed that lncRNA W42 was the most differentially expressed among these lncRNAs. Hence, we chose lncRNA W42 as a candidate lncRNA for HCC in the following study. To define the role of lncRNA W42 in human HCC, we first measured lncRNA W42 expression in 92 pairs of HCC tissues and adjacent noncancerous tissues by qRT-PCR. As shown in Figure 1, we observed that lncRNA W42 was significantly increased in HCC tissues compared with paired adjacent noncancerous tissues (P < 0.001, Figure 1A). Next, qRT-PCR analysis also showed significantly higher expression of lncRNA W42 in three human HCC cell lines (SMMC-7721, HepG2 and Huh7) than in the normal liver cell line LO2 (P < 0.05, Figure 1B). Moreover, we used receiver operating characteristic (ROC) curves to assess the sensitivity and specificity of lncRNA W42 expression to discriminate between tumor and noncancerous samples, the sensitivity was 74.76% and the specificity was 72.82%. It was noticeable that lncRNA W42 had predictive value, with an area under the curve (AUC, denotes discrimination accuracy) of 0.808 (Figure 1C), revealing that lncRNA W42 has adequate sensitivity and specificity to discriminate between HCC tissues and paired adjacent noncancerous tissues. In addition, the nucleocytoplasmic separation experiment showed that lncRNA W42 was mainly located in the nucleus (Figure 1D). In addition, we sequenced full-length lncRNA W42 and found that its length was 1598 nt and that it was located on chromosome 7q: 94423057-94429430 and shares a transcript with the COL1A2 gene (Supplementary Figure 1B and C). Thus, these results suggest that lncRNA W42 may provide imperative clinical guidance in HCC diagnosis and serve a critical role in HCC progression.
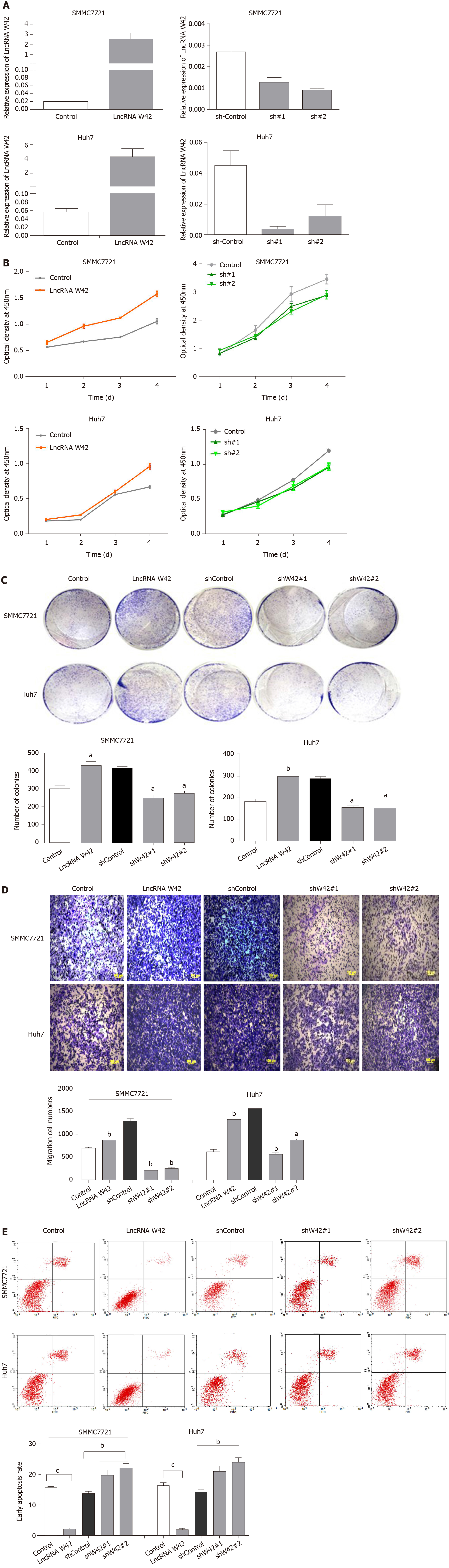
To further investigate the biological functions of lncRNA W42 in HCC, we constructed a lentiviral vector for stably overexpressing lncRNA W42 in HCC cell lines, i.e., SMMC7721 and Huh7, and designed two shRNAs for W42 (Figure 2A). CCK-8 assays revealed that overexpression of lncRNA W42 in these HCC cell lines significantly enhanced the cell proliferation ability (Figure 2B). Colony formation assays also indicated that the lncRNA W42-transfected HCC cells formed significantly more colonies than the control cells (Figure 2C). Conversely, we inhibited lncRNA W42 expression by transfecting lncRNA W42-specific shRNA into HCC cells. As indicated by CCK-8 assays, the repression of lncRNA W42 significantly decreased cell proliferation in HCC cells. Similarly, colony formation assays showed that compared to the control, knockdown of lncRNA W42 inhibited HCC cell colony formation. Moreover, overexpression of cellular lncRNA W42 not only increased HCC cell proliferation but also promoted the cell migration and invasion activity (Figure 2D) of HCC cells compared with that in the control group. In addition, we also examined apoptosis when over-expressing lncRNA W42 on HCC cells, the results showed that there was a significant decrease in the early apoptosis of HCC cells (Figure 2E). In addition, no significant difference was observed in the cell cycle analysis between the lncRNA W42 and control group (Supplementary Figure 2). Collectively, these data imply that lncRNA W42 exerts an important role in regulating HCC cell proliferation and invasion in vitro.
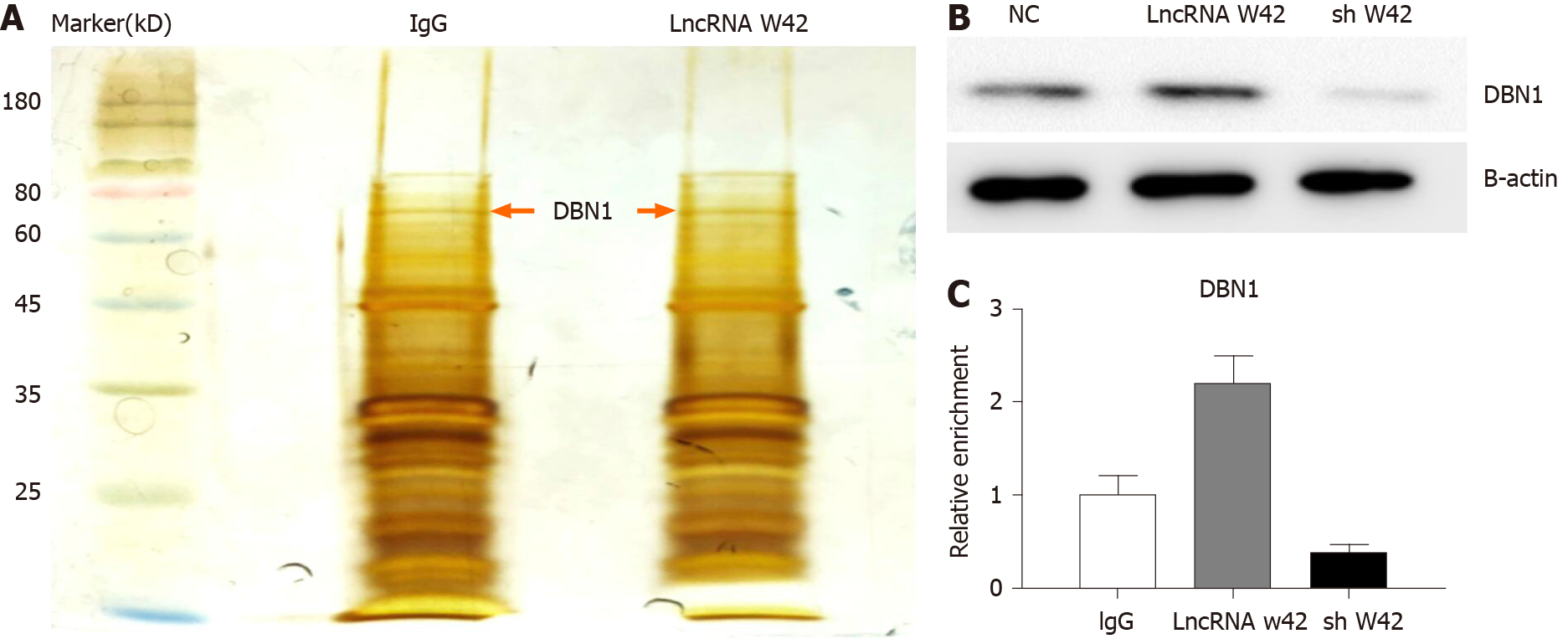
Given that lncRNA W42 is mainly located in the nucleus, we further investigated the molecular mechanism of lncRNA W42. RNA pull-down assays and MS analysis were performed in SMMC7721 cells. The results showed that DBN1, which is a key player in the advancement of several cancers, bound lncRNA W42 in SMMC7721 cells more readily than in lncRNA W42 antisense control cells (Figure 3A). This result was further confirmed by a lncRNA W42 RNA pull-down western blot assay (Figure 3B). To validate the interaction between lncRNA W42 and DBN1, we immunoprecipitated DBN1 from SMMC7721 cells and quantified the protein-bound lncRNA W42. Significantly higher expression of lncRNA W42 was detected with endogenous (Figure 3C). Together, these results indicate that lncRNA W42 RNA physically interacts with DBN1.
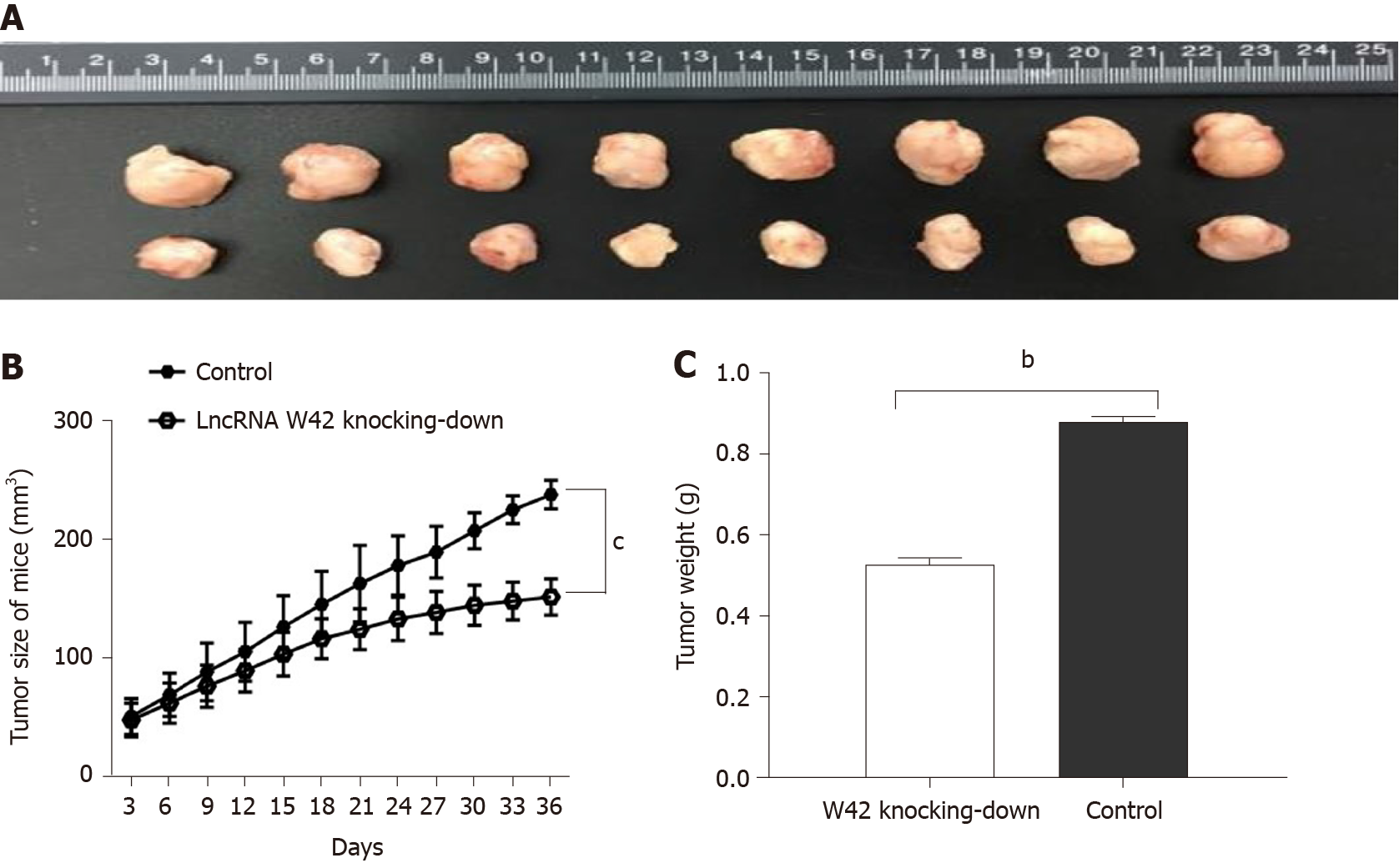
To elucidate the in vivo roles of lncRNA W42 in tumorigenesis, we determined whether decreased expression of lncRNA W42 could suppress tumor growth in the HCC xenograft tumor model. SMMC7721 cells with stable knockdown of lncRNA W42 or empty vector, generated by lentiviral vector transduction, were inoculated into nude mice. Thirty-six days after injection, the tumors were dissected from the mice (Supplementary Figure 3). Representative images of dissected tumors are shown in Figure 4A. As illustrated in Figure 4B, we observed that the growth rate of tumors in the lncRNA W42 knockdown group was lower than that in the control group. Six weeks after injection, the weight of tumors dissected from mice in the lncRNA W42 knockdown group was significantly lower than that in their control groups (Figure 4C). Consistent with the in vitro results, the tumor volumes and weights were markedly lower in the lncRNA W42 knockdown group than in the empty vector group, indicating that lncRNA W42 promotes HCC xenograft tumor growth in vivo.
We further determined whether lncRNA W42 expression was correlated with the clinicopathological characteristics and outcome of HCC patients. HCC patients (n = 92) were divided into a high lncRNA W42 expression group (above the average expression, n = 46) and a low lncRNA W42 expression group (below the average expression, n = 46). As illustrated in Table 1, no obvious differences were observed in age, tumor size, or AFP levels. However, lncRNA W42 expression was closely associated with tumor number, liver cirrhosis and tumor recurrence (P < 0.05).
| Parameters | Group | Total | W42 expression | P value | |
| Low | High | ||||
| Gender | Male | 72 | 35 | 37 | 0.810 |
| Female | 20 | 11 | 9 | ||
| Age (yr) | < 60 | 27 | 15 | 12 | 0.681 |
| ≥ 60 | 65 | 31 | 34 | ||
| Tumor size (cm) | < 5 | 52 | 25 | 27 | 0.846 |
| ≥ 5 | 40 | 21 | 19 | ||
| AFP | < 20 | 27 | 11 | 16 | 0.411 |
| ≥ 20 | 65 | 35 | 30 | ||
| Histological grade | Well/moderately | 14 | 8 | 6 | 0.776 |
| Poorly | 78 | 38 | 40 | ||
| Clinical stage | I and II | 69 | 31 | 38 | 0.182 |
| III | 23 | 15 | 8 | ||
| Tumor number | Solitary | 69 | 49 | 20 | 0.009b |
| Multiple | 23 | 6 | 17 | ||
| Drinking status | Yes | 34 | 19 | 23 | 0.999 |
| No | 58 | 31 | 27 | ||
| Smoking status | Yes | 48 | 22 | 26 | 0.563 |
| No | 44 | 24 | 20 | ||
| HBV | Yes | 66 | 30 | 36 | 0.419 |
| No | 26 | 14 | 12 | ||
| Recurrence | Yes | 29 | 9 | 20 | 0.016a |
| No | 79 | 49 | 30 | ||
| PVTT | Yes | 42 | 22 | 20 | 0.847 |
| No | 50 | 24 | 26 | ||
| Microvascular invasion | Yes | 73 | 37 | 35 | 0.999 |
| No | 19 | 9 | 10 | ||
| Liver cirrhosis | Absence | 45 | 16 | 29 | 0.030a |
| Presence | 47 | 30 | 17 | ||
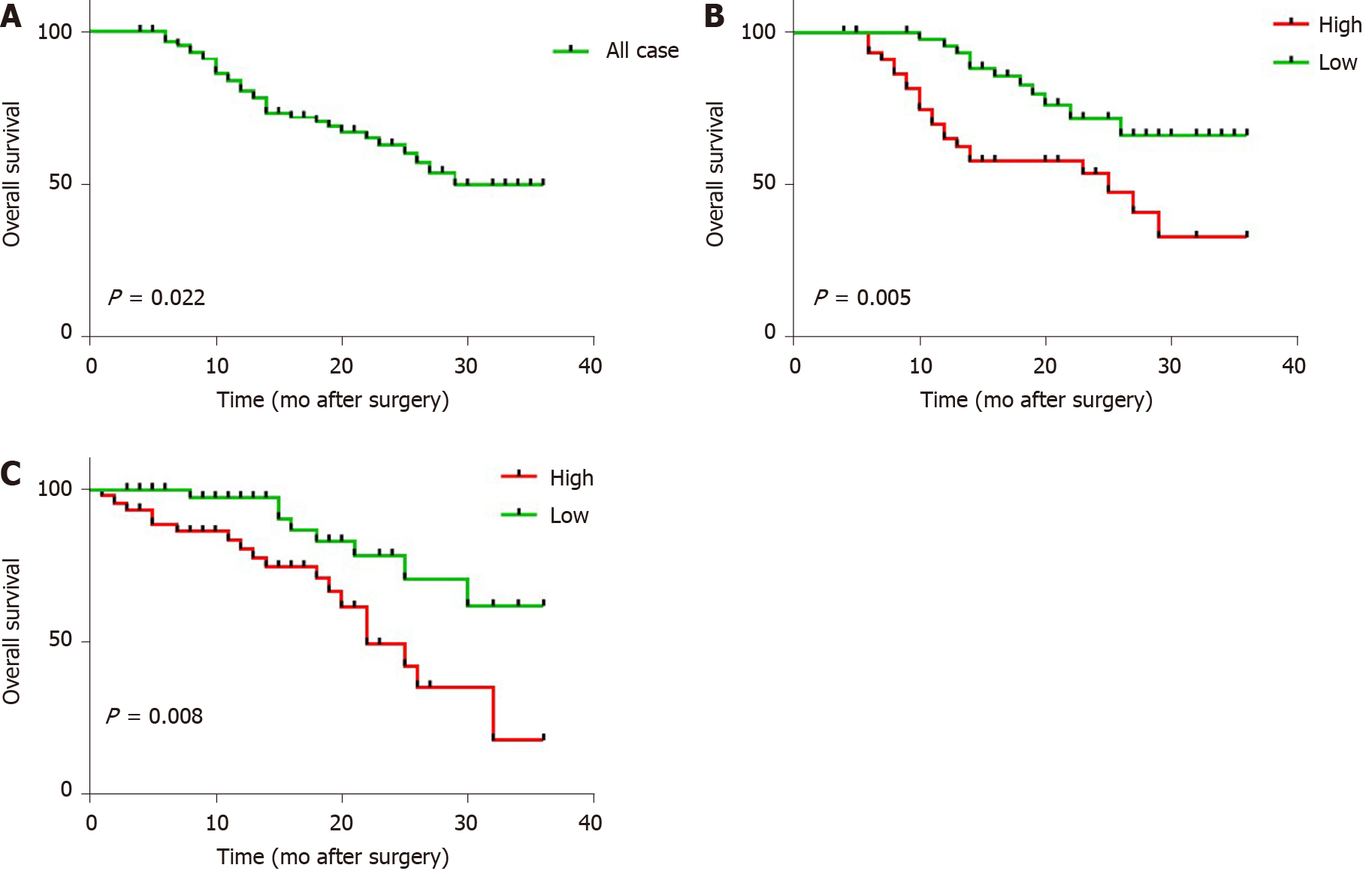
Subsequently, we determined whether lncRNA W42 expression was associated with the prognosis of HCC patients. All 92 HCC patients included in this experiment were classified into two cohorts with high or low lncRNA W42 expression according to median lncRNA W42 expression values. Kaplan-Meier analysis in the 92 patients with HCC showed that the 3-year OS rate after surgery was 64.13% (P = 0.022; Figure 5A). Specifically, a higher lncRNA W42 expression level in HCC tissues was significantly associated with a reduction in OS (P = 0.005; Figure 5B) and RFS (P = 0.008; Figure 5C) during the 3 year follow up period, supporting the critical roles of lncRNA W42 in the pathogenesis of HCC.
With the advances of next-generation sequencing technology, lncRNAs have been reported to be aberrantly expressed in multiple types of cancers[33-35]. A number of lncRNAs have been validated to be critical regulators of biological processes in cancer[36]. In particular, several HCC hot lncRNAs, such as HULC[37], P53[38], MCM3AP-AS1[39], ATB[40], HOTAIR[14] and lnc00624[41], have been reported, and increasingly evidence showed that further investigation on the relationship between lncRNA and HCC might provide potential targets for targeted therapy of HCC. Herein, we revealed differential expression of lncRNA W42 using qRT-PCR analysis of HCC and paired adjacent noncancerous tissues. Our results showed that lncRNA W42 was upregulated in HCC tissues compared to their normal counterparts. The ROC AUC of lncRNA W42 was 0.808, suggesting its specificity and sensitivity in the diagnosis of HCC. We also identified the function of lncRNA W42 in HCC cells by applying gain-of-function and loss-of-function approaches. Overexpression of lncRNA W42 promoted the proliferation and invasion and suppressed early apoptosis of HCC cells. However, this phenomenon was reversed when lncRNA W42 was downregulated.
DBN1 is an actin-binding protein that was initially found in embryonic chicken brains, and it was identified to be a neuron-specific protein[42]. However, several studies have shown the presence of DBN1 in various nonneuronal tissues, even in tumor tissues, such as recurrent non-small-cell lung cancer[43] and lymphoblastic leukemia[44] tissues. Herein, we showed that the overexpression of lncRNA W42 and DBN1 may be involved in the progression of HCC. Our results also showed that lncRNA W42 RNA physically interacts with DBN1 and that lncRNA W42 might directly bind to DBN1 and enhance the DBN1-mediated tumor-promoting effects. In addition, the mechanism by which lncRNA W42 regulates DBN1 warrants further investigation in subsequent studies.
Moreover, in vivo experiments revealed that lncRNA W42 knockdown inhibited tumor growth in nude mice, as indicated by tumor volume and weight. Subsequently, we will conduct in vivo experiments to analyze the effects of lncRNA W42 overexpression on liver tumors in nude mice. A clinical investigation showed that lncRNA W42 upregulation was closely correlated with tumor number, liver cirrhosis and tumor recurrence. More importantly, we observed that upregulated lncRNA W42 expression was closely associated with decreased OS rates and RFS rates. Based on these data, we propose that lncRNA W42 may play an important role in the development and progression of HCC.
Several previous studies have shown that liver cirrhosis is a predictor of and an independent risk factor for HCC recurrence and that patients with liver cirrhosis tend to express different levels lncRNAs[45-47]. Patients with liver cirrhosis usually have a higher probability of tumor development once a virus infection occurs because they are resistant to direct-acting antiviral agents. HCC develops frequently in the advanced fibrosis stage, and eradicating HBV or HCV infection has been a promising prophylactic therapy for preventing the occurrence of liver fibrosis and HCC. To date, several lncRNAs have been validated to be involved in HCC progression, but the role of lncRNAs in cirrhosis has not been well explained. On the other hand, lncRNA HULC is upregulated in the plasma samples of patients with HBV-related cirrhosis[48]. In line with these findings, we also noted that high expression of lncRNA W42 in patients was associated with cirrhosis, suggesting that the lncRNA profiles may be changed in HCC patients with cirrhosis. Furthermore, factors different from carcinogenesis or genetic and epigenetic factors are related to the flow from chronic hepatitis to liver cirrhosis or from focal HCC carcinogenesis in the liver to multi-organ metastasis. As shown in Table 1, we observed that lncRNA W42 expression was closely associated with liver cirrhosis and tumor recurrence, but no obvious differences were observed in age, gender, tumor size, HBV and AFP levels. Of course, the relationship between lncRNA W42 and the etiology of HCC (HCV, HBV, NAFLD, alcohol, etc.) needs to be validated in a much larger cohort. Moreover, further study will explore the levels of lncRNA W42 in serum and the AUC values of AFP in tumor tissue to ascertain if lncRNA W42 provides a higher diagnostic capacity compared to AFP in HCC patients.
Several studies have shown that microRNA (miRNA) and circulating tumor DNA detected in blood and body fluids such as urine, are useful for screening the presence of various cancers[49,50]. Similarly, the abundant mRNA, circular RNA, and lncRNA in human blood could be utilized as potential biomarkers for the diagnosis of various cancers, which were revealed by extracellular vesicles long RNA sequencing[51]. However, whether lncRNA W42 could be detected in peripheral blood needs to be investigated in subsequent studies.
These results further support that lncRNA W42 might participate in the proliferation and apoptosis regulation of HCC cells. Of course, the proliferation results need to be confirmed by flow cytometry (e.g., CFSE assay) in subsequent studies. In addition, the effect and underlying molecular mechanism of lncRNA W42 in HCC progression and potential lncRNA W42 targets, for example, the modulation of lncRNA W42 impacts on the response to chemo-or-targeted therapies, remain to be investigated in the future.
In conclusion, our results demonstrated, for the first time, that lncRNA W42 expression was significantly higher in HCC tissues than in normal tissues and that dysregulation of lncRNA W42 was positively correlated with tumor number, liver cirrhosis and tumor recurrence in patients with HCC. Moreover, increased lncRNA W42 expression was associated with a poor prognosis in patients with HCC. These findings suggest that lncRNA W42 is an important marker for indicting prognosis and may have potential as a diagnostic and therapeutic target for HCC.
Hepatocellular carcinoma (HCC) is a malignancy found globally.
The function of long noncoding RNAs (lncRNAs) in HCC requires further investigation.
We aimed to understand the effect of lncRNA W42 on HCC and dissect the underlying molecular mechanisms.
LncRNA W42 expression in HCC tissues and cells (Huh7 and SMMC-7721) was detected by quantitative reverse transcriptase polymerase chain reaction. After transfection with pcDNA3.1-lncRNA W42 or shRNA-lncRNA W42, cell functions were assessed by cell counting Kit-8 (CCK-8), colony formation, flow cytometry and Transwell assays. An HCC xenograft model was used to assess the role of lncRNA W42 on tumor growth in vivo. Receiver operating characteristic curves and Kaplan-Meier curves were used for clinical investigation.
LncRNA W42 expression was upregulated in HCC tissues and cells. LncRNA W42 directly bound to DBN1 and activated the downstream pathway to promote cell proliferation, and invasion of HCC. LncRNA W42 knockdown suppressed HCC xenograft tumor growth in vivo. HCC patients with high lncRNA W42 expression exhibited shorter survival times.
Upregulation of lncRNA W42 promotes tumor development by binding with DBN1 in HCC.
LncRNA W42, which is upregulated in HCC, may serve as a potential candidate prognostic biomarker and therapeutic target in HCC.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Li J, Watanabe T S-Editor: Gao CC L-Editor: Webster JR P-Editor: Liu JH
| 1. | Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87-108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18694] [Cited by in RCA: 21367] [Article Influence: 2136.7] [Reference Citation Analysis (3)] |
| 2. | Wang H, Huo X, Yang XR, He J, Cheng L, Wang N, Deng X, Jin H, Wang C, Zhao F, Fang J, Yao M, Fan J, Qin W. STAT3-mediated upregulation of lncRNA HOXD-AS1 as a ceRNA facilitates liver cancer metastasis by regulating SOX4. Mol Cancer. 2017;16:136. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 397] [Cited by in RCA: 444] [Article Influence: 55.5] [Reference Citation Analysis (0)] |
| 3. | Sartorius K, Sartorius B, Aldous C, Govender PS, Madiba TE. Global and country underestimation of hepatocellular carcinoma (HCC) in 2012 and its implications. Cancer Epidemiol. 2015;39:284-290. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 104] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 4. | Nault JC, Datta S, Imbeaud S, Franconi A, Mallet M, Couchy G, Letouzé E, Pilati C, Verret B, Blanc JF, Balabaud C, Calderaro J, Laurent A, Letexier M, Bioulac-Sage P, Calvo F, Zucman-Rossi J. Recurrent AAV2-related insertional mutagenesis in human hepatocellular carcinomas. Nat Genet. 2015;47:1187-1193. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 315] [Cited by in RCA: 376] [Article Influence: 37.6] [Reference Citation Analysis (0)] |
| 5. | Rogacki K, Kasprzak A, Stępiński A. Alterations of Wnt/β-catenin signaling pathway in hepatocellular carcinomas associated with hepatitis C virus. Pol J Pathol. 2015;66:9-21. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 17] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 6. | Zucman-Rossi J, Villanueva A, Nault JC, Llovet JM. Genetic Landscape and Biomarkers of Hepatocellular Carcinoma. Gastroenterology 2015; 149: 1226-1239. e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 744] [Cited by in RCA: 951] [Article Influence: 95.1] [Reference Citation Analysis (0)] |
| 7. | Ding SL, Yang ZW, Wang J, Zhang XL, Chen XM, Lu FM. Integrative analysis of aberrant Wnt signaling in hepatitis B virus-related hepatocellular carcinoma. World J Gastroenterol. 2015;21:6317-6328. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 27] [Cited by in RCA: 36] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 8. | Yao S, Johnson C, Hu Q, Yan L, Liu B, Ambrosone CB, Wang J, Liu S. Differences in somatic mutation landscape of hepatocellular carcinoma in Asian American and European American populations. Oncotarget. 2016;7:40491-40499. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 33] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 9. | Li X, Li C, Zhang L, Wu M, Cao K, Jiang F, Chen D, Li N, Li W. The significance of exosomes in the development and treatment of hepatocellular carcinoma. Mol Cancer. 2020;19:1. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 162] [Cited by in RCA: 394] [Article Influence: 78.8] [Reference Citation Analysis (0)] |
| 10. | Chen B, Garmire L, Calvisi DF, Chua MS, Kelley RK, Chen X. Harnessing big 'omics' data and AI for drug discovery in hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol. 2020;17:238-251. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 91] [Article Influence: 18.2] [Reference Citation Analysis (0)] |
| 11. | Vogel A, Saborowski A. Current strategies for the treatment of intermediate and advanced hepatocellular carcinoma. Cancer Treat Rev. 2020;82:101946. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 97] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 12. | Wei L, Wang X, Lv L, Liu J, Xing H, Song Y, Xie M, Lei T, Zhang N, Yang M. The emerging role of microRNAs and long noncoding RNAs in drug resistance of hepatocellular carcinoma. Mol Cancer. 2019;18:147. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 159] [Cited by in RCA: 252] [Article Influence: 42.0] [Reference Citation Analysis (0)] |
| 13. | Lim LJ, Wong SYS, Huang F, Lim S, Chong SS, Ooi LL, Kon OL, Lee CG. Roles and Regulation of Long Noncoding RNAs in Hepatocellular Carcinoma. Cancer Res. 2019;79:5131-5139. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 155] [Article Influence: 25.8] [Reference Citation Analysis (0)] |
| 14. | Wong CM, Tsang FH, Ng IO. Non-coding RNAs in hepatocellular carcinoma: molecular functions and pathological implications. Nat Rev Gastroenterol Hepatol. 2018;15:137-151. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 247] [Cited by in RCA: 341] [Article Influence: 48.7] [Reference Citation Analysis (0)] |
| 15. | Dhanoa JK, Sethi RS, Verma R, Arora JS, Mukhopadhyay CS. Long non-coding RNA: its evolutionary relics and biological implications in mammals: a review. J Anim Sci Technol. 2018;60:25. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 113] [Cited by in RCA: 172] [Article Influence: 24.6] [Reference Citation Analysis (0)] |
| 16. | Chen X, Wang S. LncRNA-ZFAS1 contributes to colon cancer progression through the miR-150-5p/VEGFA axis. Ann Oncol. 2018;29:v52-v53. [RCA] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 17. | Kirchhoff T, Simpson D, Hekal T, Ferguson R, Kazlow E, Moran U, Lee Y, Izsak A, Wilson MA, Shapiro R, Pavlick A, Osman I. Discovery of novel germline genetic biomarkers of melanoma recurrence impacting exonic and long non-coding RNA (lncRNA) transcripts. Ann Oncol. 2018;29 (suppl_8):viii442-viii466. [RCA] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 42] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 18. | Zhang J, Han X, Jiang L, Han Z, Wang Z. LncRNA CERNA2 is an independent predictor for clinical prognosis and is related to tumor development in gastric cancer. Int J Clin Exp Pathol. 2018;11:5783-5791. [PubMed] |
| 19. | Wang W, Chen G, Wang B, Yuan Z, Liu G, Niu B, Chen Y, Zhou S, He J, Xue H. Long non-coding RNA BZRAP1-AS1 silencing suppresses tumor angiogenesis in hepatocellular carcinoma by mediating THBS1 methylation. J Transl Med. 2019;17:421. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 38] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 20. | Koo JI, Lee HJ, Jung JH, Im E, Kim JH, Shin N, Sim DY, Hwang J, Kim SH. The Pivotal Role of Long Noncoding RNA RAB5IF in the Proliferation of Hepatocellular Carcinoma Via LGR5 Mediated β-Catenin and c-Myc Signaling. Biomolecules. 2019;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 13] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 21. | Dai W, Dai JL, Tang MH, Ye MS, Fang S. lncRNA-SNHG15 accelerates the development of hepatocellular carcinoma by targeting miR-490-3p/ histone deacetylase 2 axis. World J Gastroenterol. 2019;25:5789-5799. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 25] [Cited by in RCA: 26] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 22. | Pu J, Wang J, Wei H, Lu T, Wu X, Wu Y, Shao Z, Luo C, Lu Y. lncRNA MAGI2-AS3 Prevents the Development of HCC via Recruiting KDM1A and Promoting H3K4me2 Demethylation of the RACGAP1 Promoter. Mol Ther Nucleic Acids. 2019;18:351-362. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 40] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 23. | Wang CZ, Yan GX, Dong DS, Xin H, Liu ZY. LncRNA-ATB promotes autophagy by activating Yes-associated protein and inducing autophagy-related protein 5 expression in hepatocellular carcinoma. World J Gastroenterol. 2019;25:5310-5322. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 47] [Cited by in RCA: 59] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 24. | Cheng S, Li T, Wang C, Wang K, Lai C, Yan J, Fan H, Sun F, Wang Z, Zhang P, Yu L, Hong Z, Lei G, Sun B, Gao Y, Xiao Z, Ji X, Wang R, Wu J, Wang X, Zhang S, Yang P. Decreased long intergenic noncoding RNA P7 predicts unfavorable prognosis and promotes tumor proliferation via the modulation of the STAT1-MAPK pathway in hepatocellular carcinoma. Oncotarget. 2018;9:36057-36066. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 9] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 25. | Lei GL, Fan HX, Wang C, Niu Y, Li TL, Yu LX, Hong ZX, Yan J, Wang XL, Zhang SG, Ren MJ, Yang PH. Long non-coding ribonucleic acid W5 inhibits progression and predicts favorable prognosis in hepatocellular carcinoma. World J Gastroenterol. 2021;27:55-68. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 2] [Cited by in RCA: 3] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 26. | Dong RC. Establishment of SMMC-7721 human liver cancer cell line and preliminary observation of its biological characteristics. J Sec Milit Med Univ. 1980;1:5-9. |
| 27. | Wang Y, Fu J, Jiang M, Zhang X, Cheng L, Xu X, Fan Z, Zhang J, Ye Q, Song H. MiR-410 is overexpressed in liver and colorectal tumors and enhances tumor cell growth by silencing FHL1 via a direct/indirect mechanism. PLoS One. 2014;9:e108708. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 46] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 28. | Fu J, Cheng L, Wang Y, Yuan P, Xu X, Ding L, Zhang H, Jiang K, Song H, Chen Z, Ye Q. The RNA-binding protein RBPMS1 represses AP-1 signaling and regulates breast cancer cell proliferation and migration. Biochim Biophys Acta. 2015;1853:1-13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 33] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 29. | Xiu B, Chi Y, Liu L, Chi W, Zhang Q, Chen J, Guo R, Si J, Li L, Xue J, Shao ZM, Wu ZH, Huang S, Wu J. LINC02273 drives breast cancer metastasis by epigenetically increasing AGR2 transcription. Mol Cancer. 2019;18:187. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 75] [Cited by in RCA: 147] [Article Influence: 24.5] [Reference Citation Analysis (0)] |
| 30. | Li D, Liu X, Zhou J, Hu J, Zhang D, Liu J, Qiao Y, Zhan Q. Long noncoding RNA HULC modulates the phosphorylation of YB-1 through serving as a scaffold of extracellular signal-regulated kinase and YB-1 to enhance hepatocarcinogenesis. Hepatology. 2017;65:1612-1627. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 125] [Cited by in RCA: 155] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 31. | Yan X, Zhang D, Wu W, Wu S, Qian J, Hao Y, Yan F, Zhu P, Wu J, Huang G, Huang Y, Luo J, Liu X, Liu B, Chen X, Du Y, Chen R, Fan Z. Mesenchymal Stem Cells Promote Hepatocarcinogenesis via lncRNA-MUF Interaction with ANXA2 and miR-34a. Cancer Res. 2017;77:6704-6716. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 145] [Cited by in RCA: 195] [Article Influence: 24.4] [Reference Citation Analysis (0)] |
| 32. | Li O, Li Z, Tang Q, Li Y, Yuan S, Shen Y, Zhang Z, Li N, Chu K, Lei G. Long Stress Induced Non-Coding Transcripts 5 (LSINCT5) Promotes Hepatocellular Carcinoma Progression Through Interaction with High-Mobility Group AT-hook 2 and MiR-4516. Med Sci Monit. 2018;24:8510-8523. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 17] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 33. | An HX, Xu B, Wang Q, Li YS, Shen LF, Li SG. Up-regulation of long non-coding RNA SNHG6 predicts poor prognosis in renal cell carcinoma. Eur Rev Med Pharmacol Sci. 2018;22:8624-8629. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 34. | Wang D, Hu Y. Long Non-coding RNA PVT1 Competitively Binds MicroRNA-424-5p to Regulate CARM1 in Radiosensitivity of Non-Small-Cell Lung Cancer. Mol Ther Nucleic Acids. 2019;16:130-140. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 75] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 35. | Li L, Peng M, Xue W, Fan Z, Wang T, Lian J, Zhai Y, Lian W, Qin D, Zhao J. Integrated analysis of dysregulated long non-coding RNAs/microRNAs/mRNAs in metastasis of lung adenocarcinoma. J Transl Med. 2018;16:372. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 51] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 36. | Liu L, Zhou XY, Zhang JQ, Wang GG, He J, Chen YY, Huang C, Li L, Li SQ. LncRNA HULC promotes non-small cell lung cancer cell proliferation and inhibits the apoptosis by up-regulating sphingosine kinase 1 (SPHK1) and its downstream PI3K/Akt pathway. Eur Rev Med Pharmacol Sci. 2018;22:8722-8730. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 17] [Reference Citation Analysis (0)] |
| 37. | Xin X, Wu M, Meng Q, Wang C, Lu Y, Yang Y, Li X, Zheng Q, Pu H, Gui X, Li T, Li J, Jia S, Lu D. Long noncoding RNA HULC accelerates liver cancer by inhibiting PTEN via autophagy cooperation to miR15a. Mol Cancer. 2018;17:94. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 116] [Cited by in RCA: 166] [Article Influence: 23.7] [Reference Citation Analysis (0)] |
| 38. | Qin G, Tu X, Li H, Cao P, Chen X, Song J, Han H, Li Y, Guo B, Yang L, Yan P, Li P, Gao C, Zhang J, Yang Y, Zheng J, Ju HQ, Lu L, Wang X, Yu C, Sun Y, Xing B, Ji H, Lin D, He F, Zhou G. Long Noncoding RNA p53-Stabilizing and Activating RNA Promotes p53 Signaling by Inhibiting Heterogeneous Nuclear Ribonucleoprotein K deSUMOylation and Suppresses Hepatocellular Carcinoma. Hepatology. 2020;71:112-129. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 107] [Article Influence: 21.4] [Reference Citation Analysis (0)] |
| 39. | Wang Y, Yang L, Chen T, Liu X, Guo Y, Zhu Q, Tong X, Yang W, Xu Q, Huang D, Tu K. A novel lncRNA MCM3AP-AS1 promotes the growth of hepatocellular carcinoma by targeting miR-194-5p/FOXA1 axis. Mol Cancer. 2019;18:28. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 216] [Cited by in RCA: 315] [Article Influence: 52.5] [Reference Citation Analysis (0)] |
| 40. | Li W, Kang Y. A new Lnc in metastasis: long noncoding RNA mediates the prometastatic functions of TGF-β. Cancer Cell. 2014;25:557-559. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 74] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 41. | Li Z, Lu X, Liu Y, Zhao J, Ma S, Yin H, Huang S, Zhao Y, He X. Gain of LINC00624 Enhances Liver Cancer Progression by Disrupting the Histone Deacetylase 6/Tripartite Motif Containing 28/Zinc Finger Protein 354C Corepressor Complex. Hepatology. 2020;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 45] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 42. | Lin Q, Tan HT, Lim TK, Khoo A, Lim KH, Chung MC. iTRAQ analysis of colorectal cancer cell lines suggests Drebrin (DBN1) is overexpressed during liver metastasis. Proteomics. 2014;14:1434-1443. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 21] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 43. | Mitra R, Lee J, Jo J, Milani M, McClintick JN, Edenberg HJ, Kesler KA, Rieger KM, Badve S, Cummings OW, Mohiuddin A, Thomas DG, Luo X, Juliar BE, Li L, Mesaros C, Blair IA, Srirangam A, Kratzke RA, McDonald CJ, Kim J, Potter DA. Prediction of postoperative recurrence-free survival in non-small cell lung cancer by using an internationally validated gene expression model. Clin Cancer Res. 2011;17:2934-2946. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 37] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 44. | Vaskova M, Kovac M, Volna P, Angelisova P, Mejstrikova E, Zuna J, Brdicka T, Hrusak O. High expression of cytoskeletal protein drebrin in TEL/AML1pos B-cell precursor acute lymphoblastic leukemia identified by a novel monoclonal antibody. Leuk Res. 2011;35:1111-1113. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 13] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 45. | Zhang K, Shi Z, Zhang M, Dong X, Zheng L, Li G, Han X, Yao Z, Han T, Hong W. Silencing lncRNA Lfar1 alleviates the classical activation and pyoptosis of macrophage in hepatic fibrosis. Cell Death Dis. 2020;11:132. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 81] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 46. | Li LJ, Wu XY, Tan SW, Xie ZJ, Pan XM, Pan SW, Bai WR, Li HJ, Liu HL, Jiang J, Wu B. Lnc-TCL6 is a potential biomarker for early diagnosis and grade in liver-cirrhosis patients. Gastroenterol Rep (Oxf). 2019;7:434-443. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 47. | Zhang K, Zhang M, Yao Q, Han X, Zhao Y, Zheng L, Li G, Liu Q, Chang Y, Zhang P, Cui H, Shi Z, Chen T, Yao Z, Han T, Hong W. The hepatocyte-specifically expressed lnc-HSER alleviates hepatic fibrosis by inhibiting hepatocyte apoptosis and epithelial-mesenchymal transition. Theranostics. 2019;9:7566-7582. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 39] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 48. | Zhang Y, Li Z, Zhang Y, Zhong Q, Chen Q, Zhang L. Molecular mechanism of HEIH and HULC in the proliferation and invasion of hepatoma cells. Int J Clin Exp Med. 2015;8:12956-12962. [PubMed] |
| 49. | He Q, Fang Y, Lu F, Pan J, Wang L, Gong W, Fei F, Cui J, Zhong J, Hu R, Liang M, Fang L, Wang H, Yu M, Zhang ZF. Analysis of differential expression profile of miRNA in peripheral blood of patients with lung cancer. J Clin Lab Anal. 2019;33:e23003. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 37] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 50. | Patel H, Okamura R, Fanta P, Patel C, Lanman RB, Raymond VM, Kato S, Kurzrock R. Clinical correlates of blood-derived circulating tumor DNA in pancreatic cancer. J Hematol Oncol. 2019;12:130. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 72] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 51. | Li Y, Zhao J, Yu S, Wang Z, He X, Su Y, Guo T, Sheng H, Chen J, Zheng Q, Li Y, Guo W, Cai X, Shi G, Wu J, Wang L, Wang P, Huang S. Extracellular Vesicles Long RNA Sequencing Reveals Abundant mRNA, circRNA, and lncRNA in Human Blood as Potential Biomarkers for Cancer Diagnosis. Clin Chem. 2019;65:798-808. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 204] [Article Influence: 34.0] [Reference Citation Analysis (0)] |