Published online Apr 28, 2021. doi: 10.3748/wjg.v27.i16.1664
Peer-review started: January 15, 2021
First decision: February 9, 2021
Revised: February 11, 2021
Accepted: March 17, 2021
Article in press: March 17, 2021
Published online: April 28, 2021
Processing time: 95 Days and 13 Hours
Originally proposed by John McCarthy in 1955, artificial intelligence (AI) has achieved a breakthrough and revolutionized the processing methods of clinical medicine with the increasing workloads of medical records and digital images. Doctors are paying attention to AI technologies for various diseases in the fields of gastroenterology and hepatology. This review will illustrate AI technology procedures for medical image analysis, including data processing, model establishment, and model validation. Furthermore, we will summarize AI applications in endoscopy, radiology, and pathology, such as detecting and evaluating lesions, facilitating treatment, and predicting treatment response and prognosis with excellent model performance. The current challenges for AI in clinical application include potential inherent bias in retrospective studies that requires larger samples for validation, ethics and legal concerns, and the incomprehensibility of the output results. Therefore, doctors and researchers should cooperate to address the current challenges and carry out further investigations to develop more accurate AI tools for improved clinical applications.
Core Tip: Artificial intelligence (AI) technologies are widely used for medical image analysis in the gastroenterology and hepatology fields. Several AI models have been developed for accurate diagnosis, treatment, and prognosis based on images of endoscopy, radiology, pathology, achieving high performance comparable to experts. However, we should be aware of the certain constraints that limit the acceptance and utilization of AI tools in clinical practice. To use AI wisely, doctors and researchers should work together to address the current challenges and develop more accurate AI tools to improve patient care.
- Citation: Cao JS, Lu ZY, Chen MY, Zhang B, Juengpanich S, Hu JH, Li SJ, Topatana W, Zhou XY, Feng X, Shen JL, Liu Y, Cai XJ. Artificial intelligence in gastroenterology and hepatology: Status and challenges. World J Gastroenterol 2021; 27(16): 1664-1690
- URL: https://www.wjgnet.com/1007-9327/full/v27/i16/1664.htm
- DOI: https://dx.doi.org/10.3748/wjg.v27.i16.1664
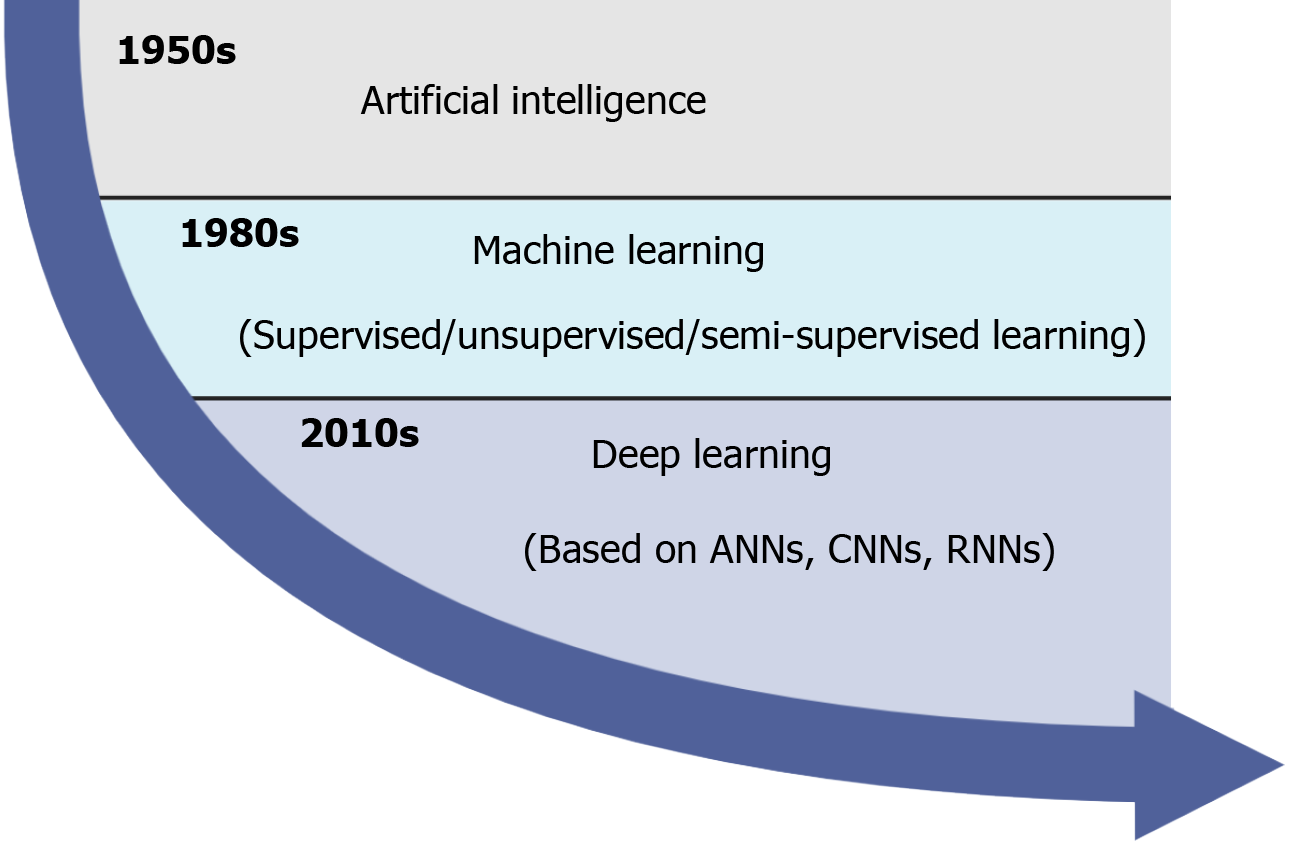
Originally proposed by John McCarthy in 1955, artificial intelligence (AI) which involves machine learning (ML) and problem solving, has achieved a breakthrough and revolutionized the processing methods of clinical medicine with the increasing workloads of medical records and digital images. In clinical practice, AI consists of several overlapping technologies such as ML, artificial neural networks (ANNs), deep learning (DL), convolutional neural networks (CNNs), and recurrent neural networks[1,2] (Figure 1). Since the 1980s, ML has been performed to construct a mathematical model and predict outcomes based on input data, and it is roughly divided into supervised (labeled data), unsupervised (unlabeled data), and semi-supervised (both labeled and unlabeled data) learning techniques[3]. Recently, as a subset of ML, ANNs have received increased interest because they can identify and learn input data by themselves instead of being labeled by experts[4]. In the last decade, DL, a new model of ML, holds great promise in clinical medicine. DL is particularly suitable for enormous complex or highly dimensional medical image analysis and predictive modeling tasks using the multilayers of ANNs, including CNNs and recurrent neural networks[5,6]. Notably, given that convolutional and pooling layers can extract distinct features and fully connected layers can make a final classification, CNNs have demonstrated excellent performance in image recognition such as endoscopy, radiology, and pathology[7,8].
In the fields of gastroenterology and hepatology, doctors are paying attention to AI technologies for the diagnosis, treatment, and prognosis of various diseases due to the heterogeneous expertise levels of doctors (majoring in endoscopy, radiology, and pathology), time-consuming procedures, and increasing workloads. Specifically, doctors usually assess medical images visually to detect and diagnose diseases based on personal expertise and experience. As the maturity of digitalization increases, a quantitative assessment of imaging information has become the reality instead of relatively inaccurate qualitative reasoning[9,10]. Although a lot of time is necessary to review and check image analysis traditionally, little information can be obtained. For example, using AI technologies to process pathology images can assess the histopathological classification and predict gene mutations in liver cancer[11], while only the mass nature can be identified by conventional pathology assessment. As a country with a high population, China has produced rapidly increasing medical records, which result in the high workloads[12]. Despite the progression of AI, gastroenterologists and hepatologists should always be aware of its limitations such as the retrospective manner of included studies and the utilization of not particularly suitable databases. In addition, it demands that doctors prepare for the effects and changes of AI on clinical practice in the real world.
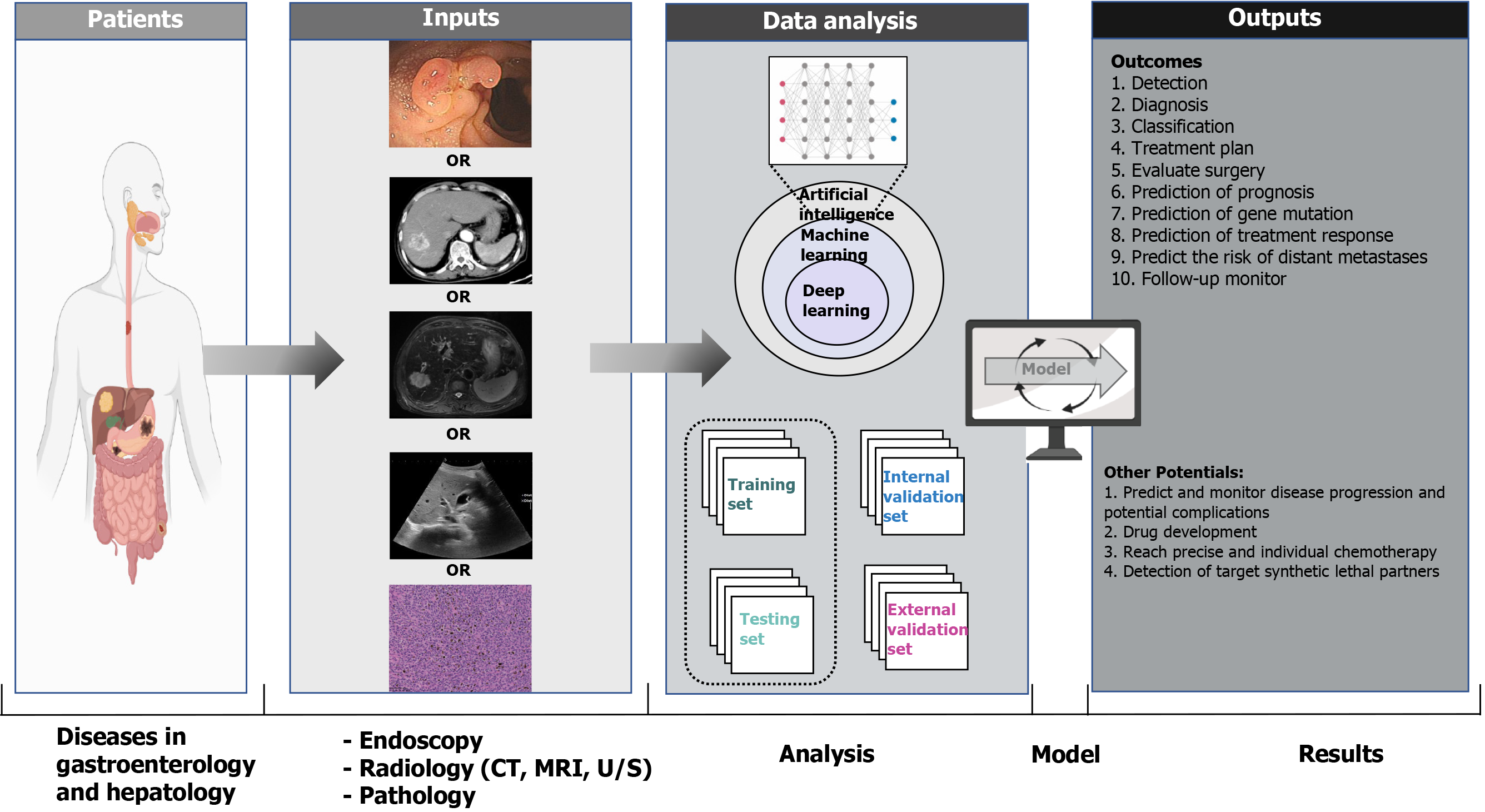
In this review, we aim to (1) introduce how AI technologies process input data, learn from input data, validate the established model; (2) summarize the AI applications in endoscopy, radiology, pathology for accurate diagnosis, treatment, and prognosis; and (3) discuss the current limitations and future considerations of AI applications in the fields of gastroenterology and hepatology (Figure 2).
As the most suitable approach for medical image analysis, the DL approach does not require shaped regions of interest on images to complete feature selection and extraction based on a neural network structure[13,14]. After data collection and processing, the correct neural network is chosen to establish a model, followed by model validation to assess its true generalizability.
Raw data are collected and analyzed, and corrupt data are identified and cleaned in the processing phase. Data selection methods are provided in Scikit-Learn[15], a Python machine learning library, which consist of univariate selection, feature importance, correlation matrix, and recursive feature elimination or addition. Other programming languages such as R Studio (http://www.r-project.org) or MATLAB software (University of New Mexico, New Mexico, United States) also offer a successful environment for AI, and they provide similar approaches to address specific tasks. Useful data and relevant variables from multiple data sources, which are applied to predict outcomes, are selected and divided into an initial training set and a testing set that allow training and internal validation of the model. Data in the training set should be different and nonredundant from that in the testing set. Notably, for small datasets, a higher proportion of data should be included in the testing set to measure the performance of the trained model accurately through cross-validation or in a bootstrapping procedure.
After transforming the data into an appropriate format, different tools are developed for implementing ML. Although several programming tools such as Python, R Studio, and MATLAB vary among themselves, they provide similar options and algorithms to adjust the parameters based on specific tasks. The major classification algorithms for testing are Naive Bayes, Decision Trees, Support Vector Machine, K-Nearest Neighbor, and Ensemble Classifiers. Oversampling or undersampling of the unbalanced training data can be utilized to improve the representation of classes and prevent model bias during the modeling stage. Currently, as the calculation workload of the batch learning process is heavy, minibatch learning is more popularized with repeating epochs, which usually decreases errors for the training and testing phases. However, an early stopping technique would be adopted to address the overfitting problem if repeating epochs cannot ensure error reduction. Based on the evaluations of model performance, developers conduct feature engineering again to manipulate the features and approve the predictive values of the model. After the optimization phase, selection for the model is primarily based on trial-and-error and the best performance for specific problem-solving. Finally, model optimization is performed with adjusted parameters by testing different configurations.
To evaluate the AI approaches, one of the most significant requirements is external validation, which is called the blind test. Any model developed within one dataset will merely reflect its idiosyncrasies, and will have poor performance in analyzing new settings. In addition, models can also be validated by internal data testing (e.g., k-fold cross-validation). In k-fold cross-validation, the dataset is separated into k subsets, including one subset for testing and the remaining (k-1) subsets for training a model. With all data used in both training and testing sets, the cross-validation process is repeated k times. The model performance is finally calculated as the average value of all k iterations. The k varies depending on the size of the dataset. For example, leave-one-out validation may be used in a small training set (< 200 data points), which means that k is equivalent to the dataset size. The appropriate and robust predictive model should have consistent performance between training and testing sets, preventing overfitting discrepancies.
With the advent and continuous improvement of fiberoptics, endoscopy has been playing a significant role in the diagnosis and treatment of gastrointestinal diseases. However, gastrointestinal diseases remain an enormous economic burden and lead to high mortality worldwide. AI is applicable in the gastroenterology fields within endoscopy[16-18], such as identification of esophageal and gastric neoplasia in esophagogastroduodenoscopy (EGD), detection of gastrointestinal bleeding in wireless capsule endoscopy (WCE), and polyp detection and characterization in colonoscopy, etc[19-63] (Table 1).
| Ref. | Country | Disease studied | Design of study | Application | Number of cases | Type of machine learning algorithm | Outcomes (%) | |
| Accuracy | Sensitivity/Specificity | |||||||
| Esophagogastroduodenoscopy | ||||||||
| Takiyama et al[19], 2018 | Japan | Anatomical location of upper gastrointestinal tract | Retrospective | Recognition of the anatomical location of upper gastrointestinal tract | Training: 27335 images: 663 larynx, 3252 esophagus, 5479 upper stomach, 7184 middle stomach, 7539 lower stomach, and 3218 duodenum; Testing: 17081 images: 363 larynx, 2142 esophagus, 3532 upper stomach, 6379 middle stomach, 3137 lower stomach, and 1528 duodenum | CNNs | Larynx: 100; Esopha us: 100; Stomach: 99; Duodenum: 99 | Larynx: 93.9/100; Esophagus: 95.8/99.7; Stomach: 98.9/93; Duodenum: 87/99.2 |
| Wu et al[20], 2019 | China | Diseases of upper gastrointestinal tract | Prospective | Monitor blind spots of upper gastrointestinal tract | Training: 1.28 million images from 1000 object classes; Testing: 3000 images for DCNN1, and 2160 images for DCNN2 | CNNs | 90.4 | 87.57/95.02 |
| van der Sommen et al[21], 2016 | Netherlands | EN-BE | Retrospective | Detection of EN in BE | 21 patients with EN-BE (60 images), 23 patients without EN-BE (40 images) | SVM | NA | 86/87 |
| Swager et al[22], 2017 | Netherlands | EN-BE | Retrospective | Detection of EN in BE | 60 images: 40 with EN-BE and 30 without EN-BE | SVM | 95 | 90/93 |
| Hashimoto et al[23], 2020 | United States | EN-BE | Retrospective | Detection of EN in BE | Training: 916 images with EN-BE; Testing: 458 images: 225 dysplasia and 233 non-dysplasia | CNNs | 95.4 | 96.4/94.2 |
| Ebigbo et al[24], 2020 | Germany | EAC-BE | Retrospective | Detection of EAC in BE | Training: 129 images; Testing: 62 images: 36 EAC and 26 normal BE | CNNs | 89.9 | 83.7/100 |
| Horie et al[25], 2019 | Japan | EAC and ESCC | Retrospective | Detection of EAC and ESCC | Training: 384 patients with 32 EAC and 397 ESCC (8428 images); Testing: 47 patients with 8 EAC and 41 ESCC (1118 images) | CNNs | 98 | 98/79 |
| Kumagai et al[26], 2019 | Japan | ESCC | Retrospective | Detection of ESCC | Training: 240 patients (4715 images: 1141 ESCC and 3574 benign lesions); Testing: 55 patients (1520 images: 467 ESCC and 1053 benign) | CNNs | 90.9 | 92.6/89.3 |
| Zhao et al[27], 2019 | China | ESCC | Retrospective | Detection of ESCC | 165 patients with ESCC and 54 patients without ESCC (1383 images) | CNNs | 89.2 | 87.0/84.1 |
| Cai et al[28], 2019 | China | ESCC | Retrospective | Detection of ESCC | Training: 746 patients (2438 images: 1332 abnormal and 1096 normal); Testing: 52 patients (187 images) | CNNs | 91.4 | 97.8/85.4 |
| Nakagawa et al[29], 2019 | Japan | ESCC | Retrospective | Determination of invasion depth | Training: 804 patients with ESCC (14338 images: 8660 non-ME and 5678 ME); Testing: 155 patients with ESCC (914 images: 405 non-ME and 509 ME) | CNNs | SM1/SM2, 3: 91.0; Invasion depth: 89.6 | SM1/SM2, 3: 90.1/95.8; Invasion depth: 89.8/88.3 |
| Tokai et al[30], 2020 | Japan | ESCC | Retrospective | Determination of invasion depth | Training: 1751 images with ESCC; Testing: 42 patients with ESCC (293 images) | CNNs | 80.9 | 84.1/80.9 |
| Ali et al[31], 2018 | Pakistan | EGC | Retrospective | Detection of EGC | 56 patients with EGC, 120 patients without EGC | SVM | 87 | 91.0/82.0 |
| Sakai et al[32], 2018 | Japan | EGC | Retrospective | Detection of EGC | Training: 58 patients (348943 images: 172555 EGC and 176388 normal); Testing: 58 patients (9650 images: 4653 EGC and 4997 normal) | CNNs | 87.6 | 80.0/94.8 |
| Kanesaka et al[33], 2018 | Japan | EGC | Retrospective | Detection of EGC | Training: 126 images: 66 EGC and 60 normal; Testing: 81 images: 61 EGC and 20 normal | SVM | 96.3 | 96.7/95.0 |
| Wu et al[34], 2019 | China | EGC | Retrospective | Detection of EGC | Training: 9691 images: 3710 EGC and 5981 normal; Testing: 100 patients: 50 EGC and 50 normal | CNNs | 92.5 | 94.0/91.0 |
| Horiuchi et al[35], 2020 | Japan | EGC | Retrospective | Detection of EGC | Training: 2570 images: 1492 EGC and 1078 gastritis; Testing: 285 images: 151 EGC and 107 gastritis | CNNs | 85.3 | 95.4/71.0 |
| Zhu et al[36], 2019 | China | Invasive GC | Retrospective | Determination of invasion depth | Training: 245 patients with GC and 545 patients without GC (5056 images); Testing: 203 images: 68 GC and 135 normal | CNNs | 89.2 | 76.5/95.6 |
| Luo et al[37], 2019 | China | EAC, ESCC, and GC | Prospective | Detection of upper gastrointestinal cancers | Training: 15040 individuals (125898 images: 31633 cancer and 94265 control); Testing: 1886 individuals (15637 images: 3931 cancer and 11706 control) | CNNs | 91.5-97.7 | 94.2/85.8 |
| Nagao et al[38], 2020 | Japan | GC | Retrospective | Determination of invasion depth | 1084 patients with GC (16557 images); Training: Testing = 4:1 | CNNs | 94.5 | 84.4/99.4 |
| Wireless capsule endoscopy | ||||||||
| Ayaru et al[39], 2015 | United Kingdom | Small bowel bleeding | Retrospective | Prediction of outcomes | Training: 170 patients with small bowel bleeding; Testing: 130 patients with small bowel bleeding | ANNs | Recurrent bleeding 88; Therapeutic intervention: 88; Severe bleeding: 78 | Recurrent bleeding: 67/91; Therapeutic intervention: 80/89; Severe bleeding: 73/80 |
| Xiao et al[40], 2016 | China | Small bowel bleeding | Retrospective | Detection of bleeding in GI tract | Training: 8200 images: 2050 bleeding and 6150 non-bleeding; Testing: 1800 images: 800 bleeding and 1000 non-bleeding | CNNs | 99.6 | 99.2/99.9 |
| Usman et al[41], 2016 | South Korea | Small bowel bleeding | Retrospective | Detection of bleeding in GI tract | Training: 75000 pixels: 25000 bleeding and 50000 non-bleeding; Testing: 8000 pixels: 3000 bleeding and 5000 non-bleeding | SVM | 91.8 | 93.7/90.7 |
| Sengupta et al[42], 2017 | United States | Small bowel bleeding | Retrospective | Prediction of 30-d mortality | Training: 4044 patients with small bowel bleeding; Testing: 2060 patients with small bowel bleeding | ANNs | 81 | 87.8/90/9 |
| Leenhardt et al[43], 2019 | France | Small bowel bleeding | Retrospective | Detection of GIA | Training: 600 images: 300 hemorrhagic GIA and 300 non-hemorrhagic GIA; Testing: 600 images: 300 hemorrhagic GIA and 300 non-hemorrhagic GIA | CNNs | 98 | 100.0/96.0 |
| Aoki et al[44], 2020 | Japan | Small bowel bleeding | Retrospective | Detection of small bowel bleeding | Training: 41 patients (27847 images: 6503 bleeding and 21344 normal); Testing: 25 patients (10208 images: 208 bleeding and 10000 non-bleeding) | CNNs | 99.89 | 96.63/99.96 |
| Yang et al[45], 2020 | China | Small bowel polyps | Retrospective | Detection of small bowel polyps | 1000 images: 500 polyps and 500 non-polyps | SVM | 96.00 | 95.80/96.20 |
| Vieira et al[46], 2020 | Portugal | Small bowel tumors | Retrospective | Detection of small bowel tumors | 39 patients (3936 images: 936 tumors and 3000 normal) | SVM | 97.6 | 96.1/98.3 |
| Colonoscopy | ||||||||
| Fernández-Esparrach et al[47], 2016 | Spain | Colorectal polyps | Retrospective | Detection of polyps | 24 videos containing 31 different polyps | Energy maps | 79 | 70.4/72.4 |
| Komeda et al[48], 2017 | Japan | Colorectal polyps | Retrospective | Detection of polyps | Training: 1800 images: 1200 adenoma and 600 non-adenoma; Testing: 10 cases | CNNs | 70.0 | 83.3/50.0 |
| Misawa et al[49], 2017 | Japan | Colorectal polyps | Retrospective | Detection of polyps | Training: 1661 images: 1213 neoplasm and 448 non-neoplasm; Testing: 173 images: 124 neoplasm and 49 non-neoplasm | SVM | 87.8 | 94.3/71.4 |
| Misawa et al[50], 2018 | Japan | Colorectal polyps | Retrospective | Detection of polyps | 196631 frames: 63135 polyps and 133496 non-polyps | CNNs | 76.5 | 90.0/63.3 |
| Chen et al[51], 2018 | China | Colorectal polyps | Retrospective | Detection of diminutive colorectal polyps | Training: 2157 images: 681 hyperplastic and 1476 adenomas; Testing: 284 images: 96 hyperplastic and 188 adenomas | DNNs | 90.1 | 96.3/78.1 |
| Urban et al[52], 2018 | United States | Colorectal polyps | Retrospective | Detection of polyps | Training: 8561 images: 4008 polyps and 4553 non-polyps; Testing: 1330 images: 672 polyps and 658 non-polyps | CNNs | 96.4 | 96.9/95.0 |
| Renner et al[53], 2018 | Germany | Colorectal polyps | Retrospective | Differentiation of neoplastic from non-neoplastic polyps | Training: 788 images: 602 adenomas and 186 non-adenomatous polyps; Testing: 186 images: 52 adenomas and 48 hyperplastic lesions | DNNs | 78.0 | 92.3/62.5 |
| Wang et al[54], 2018 | United States | Colorectal polyps | Retrospective | Detection of polyps | Training: 5545 images: 3634 polyps and 1911 non-polyps; Testing: 27113 images: 5541 polyps and 21572 non-polyps | CNNs | 98 | 94.4/95.9 |
| Mori et al[55], 2018 | Japan | Colorectal polyps | Prospective | A diagnose-and-leave strategy for diminutive, non-neoplastic rectosigmoid polyps | Training: 61925 images; Testing: 466 cases (287 neoplastic polyps, 175 nonneoplastic polyps, and 4 missing specimens) | SVM | 96.5 | 93.8/91.0 |
| Byrne et al[56], 2019 | Canada | Colorectal polyps | Retrospective | Detection and classification of polyps | Training: 60089 frames of 223 videos (29% NICE type 1, 53% NICE type 2 and 18% of normal mucosa with no polyp); Testing: 125 videos: 51 hyperplastic polyps and 74 adenoma | CNNs | 94.0 | 98.0/83.0 |
| Blanes-Vidal et al[57], 2019 | Denmark | Colorectal polyps | Retrospective | Detection of polyps | 131 patients with polyps and 124 patients without polyps | CNNs | 96.4 | 97.1/93.3 |
| Lee et al[58], 2020 | South Korea | Colorectal polyps | Retrospective | Detection of polyps | Training: 306 patients (8593 images: 8495 polyp and 98 normal); Testing: 15 patients (15 polyps videos) | CNNs | 93.4 | 89.9/93.7 |
| Gohari et al[59], 2011 | Iran | CRC | Retrospective | Determination of prognostic factors of CRC | 1219 patients with CRC | ANNs | Colon cancer: 89; Rectum cancer: 82.7 | NA/NA |
| Biglarian et al[60], 2012 | Iran | CRC | Retrospective | Prediction of distant metastasis in CRC | 1219 patients with CRC | ANNs | 82 | NA/NA |
| Takeda et al[61], 2017 | Japan | CRC | Retrospective | Diagnosis of invasive CRC | Training: 5543 images: 2506 non-neoplasms, 2667 adenomas, and 370 invasive cancers; Testing: 200 images: 100 adenomas and 100 invasive cancers | SVM | 94.1 | 89.4/98.9 |
| Ito et al[62], 2019 | Japan | CRC | Retrospective | Diagnosis of cT1b CRC | Training: 9942 images: 5124 cTis + cT1a, 4818 cT1b, and 2604 cTis + cT1a; Testing: 5022 images: 2604 cTis + cT1a, and 2418 cT1b | CNNs | 81.2 | 67.5/89.0 |
| Zhou et al[63], 2020 | China | CRC | Retrospective | Diagnosis of CRC | Training: 3176 patients with CRC and 9003 patients without CRC (464105 images: 28071 CRC and 436034 non-CRC); Testing: 307 patients with CRC and 1956 patients without CRC (84615 images: 11675 CRC and 72940 non-CRC) | CNNs | 96.3 | 91.4/98.0 |
Inadequate examination of the upper gastrointestinal tract is one of the reasons for misdiagnosing several EGD diseases. Based on AI-assisted EGD, the upper gastrointestinal tract can be divided into the pharynx, esophagus, stomach (upper, middle, lower), and duodenum with high values of the area under the curve (AUC)[19]. Furthermore, several AI technologies have classified the images of the stomach within EGD to significantly monitor the blind spots, and their accuracy has reached the ability of experienced endoscopists[20,34].
Endoscopic surveillance for Barrett’s esophagus (BE) is the potential risk factor for esophageal adenocarcinoma (EAC), of which the prognosis is related to disease staging[64,65]. However, accurate detection of esophageal neoplasia and early EAC remains difficult for experienced endoscopists[66]. An AI system developed by Ebigbo et al[24] enabled early detection of EAC with high sensitivity and specificity, they subsequently designed a real-time system for neoplasia classification in magnification. Both accurate detection of early EAC is important in BE images and the novel system of high invasion accuracy also deserves clinical attention[23]. In esophageal squamous cell carcinoma (ESCC), these tumors are often diagnosed at advanced stages, while early ESCC seems to be detected based on endoscopists’ experience because they are almost impossible to visualize with white light endoscopy. Fortunately, with AI technologies, small esophageal lesions (< 10 mm) are recognized, and there is an AI system showing diagnostic accuracy of 91.4%, which is higher than that of high-level (with experience of > 15 years, 88.8%), mid-level (with experience of 5-15 years, 81.6%), and junior-level (with experience of < 5 years, 77.2%) endoscopists[28]. In addition, the prognosis of ESCC can be proved by differentiating tumor invasion depth[29,30].
The prognosis of gastric cancer (GC) mainly depends on the early detection and invasion depth of the disease. It is extremely difficult for endoscopists to recognize early gastric cancer (EGC), which is often accompanied by gastric mucosal inflammation, and the false-negative rate of EGC in EGD has reached nearly 25.0%[67,68]. AI-assisted EGD has the potential to address tough tissues. However, the first reported CNNs-based AI system for detection of EGC had a low positive predictive value of 30.6%, leading to misdiagnosis of gastritis and misinterpretation of the gastric angle as GC[69]. In 2019, Wu et al[34] examined the detection of GC by AI and validated 200 endoscopic images, with increased accuracy, sensitivity, and specificity values (92.5%, 94%, and 91%, respectively). Furthermore, the AI system, named GRAIDS, has achieved diagnostic sensitivity close to that of expert endoscopists (94.2% vs 94.5%), and it demonstrated a robust performance showing high diagnostic accuracy in a multicenter study[37]. Besides detection, one of the most important criteria for curative resection is the invasion depth. The invasion depth prediction of GC by AI was first developed by Kubota et al[70], and the model showed the accuracy of T-stages (T1 = 77%, T2 = 49%, T3 = 51%, T4 = 55%, respectively). Considering that endoscopic mucosal resection is appropriate for intramucosal cancers (M) and submucosal cancers (invasion < 500 μm) (SM1), a more detailed classification is urgently needed. Therefore, an AI system was developed to differentiate the depths of M or SM1 and SM2 (submucosal invasion ≥ 500 μm) for GC with significantly higher sensitivity, specificity, and accuracy than those of skilled endoscopists[38].
AI-assisted WCE enables endoscopists to highlight suspicious regions on examination of the digestive tract noninvasively, including detection of small bowel bleeding, ulcers, and polyps, celiac disease, etc. Based on specific AI classifiers and validation techniques (mainly k-fold cross-validation), these models utilized still frames, pixels, or real-time videos to identify patients with small bowel bleeding with accuracy above 90% for most studies[40,41,43,44]. A CNNs-based algorithm, established in a retrospective analysis of 10000 WCE images (8200 and 1800 in the training and testing set, respectively) and validated by 10-fold cross-validation, was proposed for automatic detection of small bowel bleeding. The model was performed with a high F-1 score of 99.6% and precision of 99.9% for both active and inactive bleeding frames[40]. Besides detection, several emerging AI tools have been developed to stratify patients for the possibility of recurrent bleeding, treatment requirement, and mortality estimate to prevent repeated endoscopies in a significant proportion of patients with potential recurrent upper or lower gastrointestinal bleeding[39,42].
Colorectal polyp detection and appropriate polypectomy during colonoscopy is the standard way to prevent colorectal cancer (CRC). Since missed colorectal polyps can potentially progress into CRC, AI-assisted colonoscopy has been developed for polyp detection and characterization, and predicting the prognosis of CRC. In terms of polyp detection, an automated AI system using an energy map was developed in 2016, and it showed barely satisfactory performance[47]. Urban et al[52] used 8641 labeled images and 20 colonoscopy videos as the training and testing set to establish a CNNs model to identify colonic polyps, and the model had an accuracy of 96.4%[52]. Notably, the models should be validated to improve accuracy. After validating the model developed by Wang et al[54] with 27113 newly collected images from 1138 patients, the model showed acceptable performance (sensitivity = 94.38%, specificity = 95.2%, and AUC = 0.984). In addition, polyp characterization with magnifying endoscopic images is useful for identifying pit or vascular patterns to improve performance. AI tools with narrow-band imaging[51] or endoscopic videos[56] can be used to differentiate diminutive hyperplastic polyps and adenomas with high accuracy. Specifically, diminutive polyps (≤ 5 mm) may also be identified during colonoscopy[55]. In addition, AI may assist doctors in predicting the prognosis of CRC. An ANNs model, which was developed from a dataset of 1219 CRC patients, may predict patient survival and influential factors more accurately than a Cox regression model[59], and it also enables doctors to predict the risk of distant metastases[60].
There is a disproportionate growing rate between radiological imaging data and the number of trained radiologists, and it has forced radiologists to compensate by increasing productivity[71]. The emergence of AI technologies has eased the current dilemma and dramatically advanced radiology image analysis, including ultrasound, computed tomography (CT), and magnetic resonance imaging (MRI) in the fields of gastroenterology and hepatology. In addition, the rise of radiomics, which is a new technology in radiology and cancer, can extract abundant quantifiable objective data to evaluate surgical resection and predict treatment response[72-112] (Table 2).
| Ref. | Country | Disease studied | Design of study | Application | Number of cases | Type of machine learning algorithm | Outcomes (%) | |
| Accuracy | Sensitivity/Specificity | |||||||
| Ultrasound-based medical image recognition | ||||||||
| Gatos et al[72], 2016 | United States | Hepatic fibrosis | Retrospective | Classification of CLD | 85 images: 54 healthy and 31 CLD | SVM | 87 | 83.3/89.1 |
| Gatos et al[73], 2017 | United States | Hepatic fibrosis | Retrospective | Classification of CLD | 124 images: 54 healthy and 70 CLD | SVM | 87.3 | 93.5/81.2 |
| Chen et al[74], 2017 | China | Hepatic fibrosis | Retrospective | Classification of the stages of hepatic fibrosis in HBV patients | 513 HBV patients with different hepatic fibrosis (119 S0, 164 S1, 88 S2, 72 S3, and 70 S4) | SVM, Naive Bayes, RF, KNN | 82.87 | 92.97/82.50 |
| Li et al[75], 2019 | China | Hepatic fibrosis | Prospective | Classification of the stages of hepatic fibrosis in HBV patients | 144 HBV patients | Adaptive boosting, decision tree, RF, SVM | 85 | 93.8/76.9 |
| Gatos et al[76], 2019 | United States | Hepatic fibrosis | Retrospective | Classification of CLD | 88 healthy individuals (88 F0 fibrosis stage images) and 112 CLD patients (112 images: 46 F1, 16 F2, 22 F3, and 28 F4) | CNNs | 82.5 | NA/NA |
| Wang et al[77], 2019 | China | Hepatic fibrosis | Prospective | Classification of the stages of hepatic fibrosis in HBV patients | Training: 266 HBV patients (1330 images); Testing: 132 HBV patients (660 images) | CNNs | F4: 100; ≥ F3: 99; ≥ F2: 99 | F4: 100.0/100.0; ≥ F3: 97.4/95.7; ≥ F2: 100.0/97.7 |
| Kuppili et al[78], 2017 | United States | MAFLD | Retrospective | Detection and characterization of FLD | 63 patients: 27 healthy and 36 MAFLD | ELM, SVM | ELM: 96.75; SVM: 89.01 | NA/NA |
| Byra et al[79], 2018 | Poland | MAFLD | Retrospective | Diagnosis of the amount of fat in the liver | 55 severely obese patients | CNNs, SVM | 96.3 | 100/88.2 |
| Biswas et al[80], 2018 | United States | MAFLD | Retrospective | Detection and risk stratification of FLD | 63 patients: 27 healthy and 36 MAFLD | CNNs, SVM, ELM | CNNs: 100; SVM: 82; ELM: 92 | NA/NA |
| Cao et al[81], 2020 | China | MAFLD | Retrospective | Detection and classification of MAFLD | 240 patients: 106 healthy, 57 mild MAFLD, 67 moderate MAFLD, and 10 severe MAFLD | CNNs | 95.8 | NA/NA |
| Guo et al[82], 2018 | China | Liver tumors | Retrospective | Diagnosis of liver tumors | 93 patients with liver tumors: 47 malignant lesions (22 HCC, 5 CC, and 10 RCLM), and 46 benign lesions | DNNs | 90.41 | 93.56/86.89 |
| Schmauch et al[83], 2019 | France | FLL | Retrospective | Detection and characterization of FLL | Training: 367 patients (367 images); Testing: 177 patients | CNNs | Detection: 93.5; Characterization: 91.6 | NA/NA |
| Yang et al[84], 2020 | China | FLL | Retrospective | Detection of FLL | Training: 1815 patients with FLL (18000 images); Testing: 328 patients with FLL (3718 images) | CNNs | 84.7 | 86.5/85.5 |
| CT/MRI-based medical image recognition | ||||||||
| Choi et al[85], 2018 | South Korea | Hepatic fibrosis | Retrospective | Staging liver fibrosis by using CT images | Training: 7461 patients: 3357 F0, 113 F1, 284 F2, 460 F3, 3247 F4; Testing: 891 patients: 118 F0, 109 F1, 161 F2, 173 F3, 330 F4 | CNNs | 92.1–95.0 | 84.6–95.5/89.9–96.6 |
| He et al[86], 2019 | United States | Hepatic fibrosis | Retrospective | Staging liver fibrosis by using MRI images | Training: 225 CLD patients; Testing: 84 patients | SVM | 81.8 | 72.2/87.0 |
| Ahmed et al[87], 2020 | Egypt | Hepatic fibrosis | Retrospective | Detection and staging of liver fibrosis by using MRI images | 37 patients: 15 healthy and 22 CLD | SVM | 83.7 | 81.8/86.6 |
| Hectors et al[88], 2020 | United States | Liver fibrosis | Retrospective | Staging liver fibrosis by using MRI images | Training: 178 patients with liver fibrosis; Testing: 54 patients with liver fibrosis | CNNs | F1-F4: 85; F2-F4: 89; F3-F4: 91; F4: 83 | F1-F4: 84/90; F2-F4: 87/93; F3-F4: 97/83; F4: 68/94 |
| Vivanti et al[89], 2017 | Israel | Liver tumors | Retrospective | Detection and segmentation of new tumors in follow-up by using CT images | 246 liver tumors (97 new tumors) | CNNs | 86 | 70/NA |
| Yasaka et al[90], 2018 | Japan | Liver masses | Retrospective | Detection and differentiation of liver masses by using CT images | Training: 460 patients with liver masses (1068 images: 240 Category A, 121 Category B, 320 Category C, 207 Category D, 180 Category E); Testing: 100 images with liver masses: 21 Category A, 9 Category B, 35 Category C, 20 Category D, 15 Category E | CNNs | 84 | Category A: 71/NA; Category B: 33/NA; Category C: 94/NA; Category D: 90/NA; Category E: 100/NA |
| Ibragimov et al[91], 2018 | United States | Liver diseases requiring SBRT | Retrospective | Prediction of hepatotoxicity after liver SBRT by using CT images | 125 patients undergone liver SBRT: 58 liver metastases, 36 HCC, 27 cholangiocarcinoma, and 4 other histopathologies | CNNs | 85 | NA/NA |
| Abajian et al[92], 2018 | United States | HCC | Retrospective | Prediction of HCC response to TACE by using MRI images | 36 HCC patients treated with TACE | RF | 78 | 62.5/82.1 |
| Zhang et al[93], 2018 | United States | HCC | Retrospective | Classification of HCC by using MRI images | 20 patients with HCC | CNNs | 80 | NA/NA |
| Morshid et al[94], 2019 | United States | HCC | Retrospective | Prediction of HCC response to TACE by using CT images | 105 HCC patients received first-line treatment with TACE | CNNs | 74.2 | NA/NA |
| Nayak et al[95], 2019 | India | Cirrhosis; HCC | Retrospective | Detection of cirrhosis and HCC by using CT images | 40 patients: 14 healthy, 12 cirrhosis, 14 cirrhosis with HCC | SVM | 86.9 | 100/95 |
| Hamm et al[96], 2019 | United States | Common hepatic lesions | Retrospective | Classification of common hepatic lesions by using MRI images | Training: 434 patients with common hepatic lesions; Testing: 60 patients with common hepatic lesions | CNNs | 92 | 92/98 |
| Wang et al[97], 2019 | United States | Common hepatic lesions | Retrospective | Demonstration of a proof-of-concept interpretable DL system by using MRI images | 60 common hepatic lesions patients | CNNs | NA | 82.9/NA |
| Jansen et al[98], 2019 | Netherlands | FLL | Retrospective | Classification of FLL by using MRI images | 95 patients with FLL (125 benign lesions: 40 adenomas, 29 cysts, and 56 hemangiomas; and 88 malignant lesions: 30 HCC and 58 metastases) | RF | 77 | Adenoma: 80/78; Cyst: 93/93; Hemangioma: 84/82; HCC: 73/56; Metastasis: 62/77 |
| Mokrane et al[99], 2020 | France | HCC | Retrospective | Diagnosis of HCC in patients with cirrhosis by using CT images | Training: 106 patients: 85 HCC and 21 non-HCC; Testing: 36 patients: 23 HCC and 13 non-HCC | SVM, KNN, RF | 70 | 70/54 |
| Shi et al[100], 2020 | China | HCC | Retrospective | Detection of HCC from FLL by using CT images | Training: 359 lesions: 155 HCC and 204 non-HCC; Testing: 90 lesions: 39 HCC and 51 non-HCC | CNNs | 85.6 | 74.4/94.1 |
| Alirr et al[101], 2020 | Kuwait | Liver tumors | Retrospective | Segmentation of liver tumors | Training: 100 images with liver tumors;Testing: 31 images with liver tumors | CNNs | 95.2 | NA/NA |
| Zheng et al[102], 2020 | China | Pancreatic cancer | Retrospective | Pancreas segmentation by using MRI images | 20 patients with PDAC | CNNs | 99.86 | NA/NA |
| Radiomics | ||||||||
| Liang et al[103], 2014 | China | HCC | Retrospective | Prediction of recurrence for HCC patients who received RFA | 83 patients with HCC receiving RFA as first treatment (18 recurrence and 65 non-recurrence) | SVM | 82 | 67/86 |
| Zhou et al[104], 2017 | China | HCC | Retrospective | Characterization of HCC | 46 patients with HCC: 21 low-grade (Edmondson grades I and II) and 25 high-grade (Edmondson grades III and IV) | Free-form curve-fitting | 86.95 | 76.00/100.00 |
| Abajian et al[105], 2018 | United States | HCC | Retrospective | Prediction of response to intra-arterial treatment | 36 patients undergone trans-arterial treatment | RF | 78 | 62.5/82.1 |
| Ibragimov et al[91], 2018 | United States | Liver tumors | Retrospective | Prediction of hepatobiliary toxicity of SBRT | 125 patients undergone liver SBRT: 58 metapatients, 36 HCC, 27 cholangiocarcinoma, and 4 other primary liver tumor histopathologies | CNNs | 85 | NA/NA |
| Morshid et al[94], 2019 | United States | HCC | Retrospective | Prediction of HCC response to TACE | 105 patients with HCC: 11 BCLC stage A, 24 BCLC stage B, 67 BCLC stage C, and 3 BCLC stage D | CNNs | 74.2 | NA/NA |
| Ma et al[106], 2019 | China | HCC | Retrospective | Prediction of MVI in HCC | Training: 110 patients with HCC: 37 with MVI and 73 without MVI; Testing: 47 patients with HCC: 18 with MVI and 29 without MVI | SVM | 76.6 | 65.6/94.4 |
| Dong et al[107], 2020 | China | HCC | Retrospective | Prediction and differentiation of MVI in HCC | Prediction: 322 patients with HCC: 144 with MVI and 178 without MVI; Differentiation: 144 patients with HCC and MVI | RF, mRMR | Prediction: 63.4; Differentiation: 73.0 | Prediction: 89.2/48.4; Differentiation: 33.3/80.0 |
| He et al[108], 2020 | China | HCC | Prospective | Prediction of MVI in HCC | Training: 101 patients with HCC; Testing: 18 patients with HCC | LASSO | 84.4 | NA/NA |
| Schoenberg et al[109], 2020 | Germany | HCC | Prospective | Prediction of disease-free survival after HCC resection | Training: 127 patients with HCC; Testing: 53 patients with HCC | RF | 78.8 | NA/NA |
| Zhao et al[110], 2020 | China | HCC | Retrospective | Prediction of ER of HCC after partial hepatectomy | Training: 78 patients with HCC: 40 with ER and 38 without ER; Testing: 35 patients with HCC: 18 with ER and 17 without ER | LASSO | 80.8 | 80.0/81.6 |
| Liu et al[111], 2020 | China | HCC | Retrospective | Prediction of progression-free survival of HCC patients after RFA and SR | RFA: Training: 149 HCC patients undergone RFA Testing: 65 HCC patients undergone RFA; SR: Training: 144 HCC patients undergone SR Testing: 61 HCC patients undergone SR | Cox-CNNs | RFA: 82.0; SR: 86.3 | NA/NA |
| Chen et al[112], 2021 | China | HCC | Retrospective | Prediction of HCC response to first TACE by using CT images | Training: 355 patients with HCC; Testing: 118 patients with HCC | LASSO | 81 | 85.2/77.2 |
AI technologies have been applied to abdominal ultrasound-based medical images for the assessment of liver diseases, such as hepatic fibrosis and mass lesions. A support vector machine-derived approach was developed by Gatos et al[72] to detect and classify chronic liver disease (CLD) based on abdominal ultrasound. After quantifying 85 ultrasound images (54 healthy and 31 with CLD), the proposed model showed superior results (accuracy = 87.0%, sensitivity = 83.3%, and specificity = 89.1%), which greatly improved the diagnostic and classification accuracy of CLD. Furthermore, CNNs are employed to identify and isolate regions of different stiffness temporal stability under ultrasound to explore the impact on CLD diagnosis. The updated detection algorithm has augmented the accuracy to 95.5% after excluding unreliable areas and reducing interobserver variability[76]. Detecting and classifying hepatic mass lesions as benign or malignant is equally important. Schmauch et al[83] performed supervised training (367 ultrasonic images together with the radiological reports) to build a DL model, and the resulting algorithm had high receiver operating characteristic curves of 0.93 and 0.916 in lesion detection and characterization, respectively. Although the model could increase the diagnostic accuracy and detect potential malignant mass lesions, it should be further validated. In addition, combining AI technologies with contrast-enhanced ultrasound may improve the performance to identify and characterize liver cancer. For example, after AI-assisted contrast-enhanced ultrasound was applied to detect liver lesions in the arterial, portal, and late phases, the accuracy, sensitivity and specificity of the examination were markedly increased[82]. Due to the misty demonstration of gastroenterology within ultrasound, an AI-assisted ultrasound tool was limited.
Liver diseases often present indeterminate behaviors on abdominal CT, and a biopsy is recommended according to the European Association for the Study of the Liver guidelines[113]. Based on a large dataset of CT images (7461 patients diagnosed with liver fibrosis), a CNNs model was developed and it outperformed the radiologists’ interpretation[85]. Furthermore, depending on ANNs-based contrast-enhanced CT images from 460 patients, Yasaka et al[90] conducted a retrospective study to classify liver masses into five categories with high accuracy, including (1) primary hepatocellular carcinoma (HCC); (2) malignant tumors apart from HCC; (3) early HCC, indeterminate masses, or dysplastic nodules; (4) hemangiomas; and (5) cysts. For patients diagnosed with liver tumors or pancreatic cancer, it is crucial to complete the liver or pancreas segmentation to assess the lesions and make the ideal treatment plan. Instead of conventional manual segmentation, a CNNs model was proposed to segment liver tumors based on CT images, with an accuracy of more than 80.0%, favoring suitable decision-making[101]. Additionally, a CNNs model was also developed for pancreas localization and segmentation using CT images[102]. Furthermore, monitoring tumor recurrence plays an important role in follow-up CT. Vivanti et al[89] collected and integrated the initial appearance of tumors, CT behaviors, and quantification of the tumor loads throughout the disease course, and then they designed an automated detection model of tumor recurrence with an accuracy of 86%.
Besides depending on CT images, a DL approach for pancreas segmentation can also be designed from MRI images. Several AI-assisted studies have shown promising results in classifying MRI liver lesions with/without risk factors and patients’ clinical data, improving the accuracy and yields of reference models[93,96,98,102].
Currently, radiomics has received great interest from doctors because this AI-assisted technology can extract indiscoverable quantifiable objective data of the radiological images and reveal the association with potential biological processes[114,115]. Preoperative stratification of patients at different risk of recurrence and prediction of survival after resection is fundamental to improve prognosis. As an independent risk factor of recurrence, microvascular invasion (MVI) cannot be provided in conventional radiological techniques[116]. Several studies have managed to use radiomic algorithms based on ultrasound, CT, or MRI to elaborate radiomic signatures for preoperative prediction of MVI[106-108]. Besides the prediction of recurrence, radiomics may also be utilized to predict survival after surgical resection. However, compared to the excellent AI models based on pathologic images, radiomics-based predictive models merely attain a low value of 0.78[109].
Beyond recurrence and survival prediction purposes, radiomics can also be utilized for prediction of patients’ response to transarterial chemoembolization (TACE) and radiofrequency ablation (RFA), and post-radiotherapy hepatotoxicity. A CNNs model developed from 105 HCC patients’ CT images had higher accuracy in predicting response to TACE than the Barcelona Clinic Liver Cancer stages[94]. In addition, Chen et al[112] designed an excellent clinical-radiomic model to predict objective response to first TACE based on 595 HCC patients’ CT images, which could assist the selection of HCC patients for TACE. Another study used radiomics of MRI images with clinical data to perform prediction of TACE response[105]. For HCC in the early stages, RFA is a recommended option. Based on radiomics, Liang et al[103] designed a model to predict the RFA response and HCC recurrence after RFA, obtaining high AUC, sensitivity, and specificity. Additionally, post-radiotherapy hepatotoxicity should be monitored to adjust the position and dose of radiotherapy. A CNNs model not only identified that irradiation of the proximal portal vein was associated with poor prognosis, it also predicted post-radiotherapy hepatotoxicity with an AUC of 0.85[91]. Ibragimov et al[91] applied a CNNs model to determine the consistent patterns in toxicity-related dose plans, and the AUC of the model for dose planned analysis was increased from 0.79 to 0.85 after the combination with some pre-treatment clinical features, showing that the combined framework can indicate the accurate position and dose of radiotherapy.
Pathological analysis is considered the gold standard for the diagnosis of diseases in the fields of gastroenterology and hepatology. Currently, there is a shortage of pathologists around the world, which has become an obstruction for maintaining the accuracy of pathological analysis[117]. With the development of the whole-slide imaging (WSI) scanner and AI technologies, a combination of both technologies can ease the medical burden, improve the diagnosis accuracy, and even predict gene mutations and prognosis[118-147] (Table 3).
| Ref. | Country | Disease studied | Design of study | Application | Number of cases | Type of machine learning algorithm | Outcomes (%) | |
| Accuracy | Sensitivity/Specificity | |||||||
| Basic AI-based pathology: diagnosis | ||||||||
| Tomita et al[118], 2019 | United States | BE and EAC | Retrospective | Detection and classification of cancerous and precancerous esophagus tissue | Training: 379 images with 4 classes: normal, BE-no-dysplasia, BE-with-dysplasia, and adenocarcinoma; Testing: 123 images with 4 classes: normal, BE-no-dysplasia, BE-with-dysplasia, and adenocarcinoma | CNNs | Mean: 83; BE-no-dysplasia: 85; BE-with-dysplasia: 89; Adenocarcinoma: 88 | Normal: 69/71 BE-no-dysplasia: 77/88; BE-with-dysplasia: 21/97; Adenocarcinoma: 71/91 |
| Sharma et al[119], 2017 | Germany | GC | Retrospective | Classification and necrosis detection of GC | 454 patients (6810 WSIs: 4994 for cancer classification and 1816 for necrosis detection) (HER2 immunohistochemical stain and HE stained) | CNNs | Cancer classification: 69.90; Necrosis detection: 81.44 | NA/NA |
| Li et al[120], 2018 | China | GC | Retrospective | Detection of GC | 700 images: 560 GC and 140 normal (HE stained) | CNNs | 100 | NA/NA |
| Leon et al[121], 2019 | Colombia | GC | Retrospective | Detection of GC | 40 images: 20 benign and 20 malignant | CNNs | 89.72 | NA/NA |
| Sun et al[122], 2019 | China | GC | Retrospective | Diagnosis of GC | 500 WSIs of gastric areas with typical cancerous regions | DNNs | 91.6 | NA/NA |
| Ma et al[123], 2020 | China | GC | Retrospective | Classification of lesions in the gastric mucosa | Training: 534 WSIs (1616713 images: 544925 normal, 544624 chronic gastritis, and 527164 cancer) (HE stained) Testing: 153 WSIs (399240 images: 135446 normal, 125783 chronic gastritis, and 138011 cancer) (HE stained) | CNNs, RF | Benign and cancer: 98.4; Normal, chronic gastritis, and GC: 94.5 | Benign and cancer: 98.0/98.9; Normal, chronic gastritis, and GC: NA/NA |
| Yoshida et al[124], 2018 | Japan | Gastric lesions | Retrospective | Classification of gastric biopsy specimens | 3062 gastric biopsy specimens (HE stained) | CNNs | 55.6 | 89.5/50.7 |
| Qu et al[125], 2018 | Japan | Gastric lesions | Retrospective | Classification of gastric pathology images | Training: 1080 patches: 540 benign and 540 malignant; Testing: 5400 patches: 2700 benign and 2700 malignant | CNNs | 96.5 | NA/NA |
| Iizuka et al[126], 2020 | Japan | Gastric and colonic epithelial tumors | Retrospective | Classification of gastric and colonic epithelial tumors | 4128 cases of human gastric epithelial lesions and 4036 of colonic epithelial lesions (HE stained) | CNNs, RNNs | Gastric adenocarcinoma: 97; Gastric adenoma: 99; Colonic adenocarcinoma: 96; Colonic adenoma: 99 | NA/NA |
| Korbar et al[127], 2017 | United States | Colorectal polyps | Retrospective | Classification of different types of colorectal polyps on WSIs | Training: 458 WSIs; Testing: 239 WSIs | A modified version of a residual network | 93 | 88.3/NA |
| Wei et al[128], 2020 | United States | Colorectal polyps | Retrospective | Classification of colorectal polyps on WSIs | Training: 326 slides with colorectal polyps: 37 tubular, 30 tubulovillous or villous, 111 hyperplastic, 140 sessile serrated, and 8 normal; Testing: 238 slides with colorectal polyps: 95 tubular, 78 tubulovillous or villous, 41 hyperplastic, and 24 sessile serrated | CNNs | Tubular: 84.5; Tubulovillous or villous: 89.5; Hyperplastic: 85.3; Sessile serrated: 88.7 | Tubular: 73.7/91.6; Tubulovillous or villous: 97.6/87.8; Hyperplastic: 60.3/97.5; Sessile serrated: 79.2/89.7 |
| Shapcott et al[129], 2018 | UnitedKingdom | CRC | Retrospective | Diagnosis of CRC | 853 hand-marked images | CNNs | 84 | NA/NA |
| Geessink et al[130], 2019 | Netherlands | CRC | Retrospective | Quantification of intratumoral stroma in CRC | 129 patients with CRC | CNNs | 94.6 | 91.1/99.4 |
| Song et al[131], 2020 | China | CRC | Retrospective | Diagnosis of CRC | Training: 177 slides: 156 adenoma and 21 non-neoplasm; Testing: 362 slides: 167 adenoma and 195 non-neoplasm | CNNs | 90.4 | 89.3/79.0 |
| Wang et al[132], 2015 | China | Hepatic fibrosis | Retrospective | Assessment of HBV-related liver fibrosis and detection of liver cirrhosis | Training: 105 HBV patients; Testing: 70 HBV patients | SVM | 82 | NA/NA |
| Forlano et al[133], 2020 | UnitedKingdom | MAFLD | Retrospective | Detection and quantification of histological features of MAFLD | Training: 100 MAFLD patients; Testing: 146 MAFLD patients | K-means | Steatosis: 97; Inflammation: 96; Ballooning: 94; Fibrosis: 92 | NA/NA |
| Li et al[134], 2017 | China | HCC | Retrospective | Nuclei grading of HCC | 4017 HCC nuclei patches | CNNs | 96.7 | G1: 94.3/97.5; G2: 96.0/97.0;G3: 97.1/96.6; G4: 99.5/95.8 |
| Kiani et al[135], 2020 | United States | Liver cancer (HCC and CC) | Retrospective | Histopathologic classification of liver cancer | Training: 70 WSIs: 35 HCC and 35 CC Testing: 80 WSIs: 40 HCC and 40 CC | SVM | 84.2 | 72/95 |
| Advanced AI-based pathology: prediction of gene mutations and prognosis | ||||||||
| Steinbuss et al[136], 2020 | Germany | Gastritis | Retrospective | Identification of gastritis subtypes | Training: 92 patients (825 images: 398 low inflammation, 305 severe inflammation, and 122 A gastritis) (HE stained) Testing: 22 patients (209 images: 122 low inflammation, 38 severe inflammation, and 49 A gastritis) (HE stained) | CNNs | 84 | A gastritis: 88/89; B gastritis: 100/93; C gastritis: 83/100 |
| Liu et al[137], 2020 | China | Gastrointestinal neuroendocrine tumor | Retrospective | Prediction of Ki-67 positive cells | 12 patients (18762 images: 5900 positive cells, 6086 positive cells, and 6776 background from ROIs) (HE and IHC stained) | CNNs | 97.8 | 97.8/NA |
| Kather et al[138], 2019 | Germany | GC and CRC | Retrospective | Prediction of MSI in GC and CRC | Training: 360 patients (93408 tiles); Testing: 378 patients (896530 tiles) | CNNs | 84 | NA/NA |
| Bychkov et al[139], 2018 | Finland | CRC | Retrospective | Prediction of CRC outcome | 420 CRC tumor tissue microarray samples | CNNs, RNNs | 69 | NA/NA |
| Kather et al[140], 2019 | Germany | CRC | Retrospective | Prediction of survival from CRC histology slides | Training: 86 CRC tissue slides (> 100000 HE image patches); Testing: 25 CRC patients (7180 images) | CNNs | 98.7 | NA/NA |
| Echle et al[141], 2020 | Germany | CRC | Retrospective | Detection of dMMR or MSI in CRC | Training: 5500 patients; Testing: 906 patients | A modified shufflenet DL system | 92 | 98/52 |
| Skrede et al[142], 2020 | 3R23 Song 2020 | CRC | Retrospective | Prediction of CRC outcome after resection | Training: 828 patients (> 12000000 image tiles); Testing: 920 patients | CNNs | 76 | 52/78 |
| Sirinukunwattana et al[143], 2020 | UnitedKingdom | CRC | Retrospective | Identification of consensus molecular subtypes of CRC | Training: 278 patients with CRC; Testing: 574 patients with CRC: 144 biopsies and 430 TCGA | Neural networks with domain-adversarial learning | Biopsies: 85; TCGA: 84 | NA/NA |
| Jang et al[144], 2020 | South Korea | CRC | Retrospective | Prediction of gene mutations in CRC | Training: 629 WSIs with CRC (HE stained) Testing: 142 WSIs with CRC (HE stained) | CNNs | 64.8-88.0 | NA/NA |
| Chaudhary et al[145], 2018 | United States | HCC | Retrospective | Identification of survival subgroups of HCC | Training: 360 HCC patients’ data using RNA-seq, miRNA-seq and methylation data from TCGA; Testing: 684 HCC patients’ data (LIRI-JP cohort: 230; NCI cohort: 221; Chinese cohort: 166, E-TABM-36 cohort: 40, and Hawaiian cohort: 27) | DL | LIRI-JP cohort: 75; NCI cohort: 67; Chinese cohort: 69; E-TABM-36 cohort: 77; Hawaiian cohort: 82 | NA/NA |
| Saillard et al[146], 2020 | France | HCC | Retrospective | Prediction of the survival of HCC patients treated by surgical resection | Training: 206 HCC (390 WSIs); Testing: 328 HCC (342 WSIs) | CNNs (SCHMOWDER and CHOWDER) | SCHMOWDER: 78; CHOWDER: 75 | NA/NA |
| Chen et al[11], 2020 | China | HCC | Retrospective | Classification and gene mutation prediction of HCC | Training: 472 WSIs: 383 HCC and 89 normal liver tissue; Testing: 101 WSIs: 67 HCC and 34 normal liver tissue | CNNs | Classification: 96.0; Tumor differentiation: 89.6; Gene mutation: 71-89 | NA/NA |
| Fu et al[147], 2020 | UnitedKingdom | EAC, GC, CRC, and liver cancers | Retrospective | Prediction of mutations, tumor composition and prognosis | 17335 HE-stained images of 28 cancer types | CNNs | Variable across tumors/gene alterations | NA/NA |
The basic role of pathology is disease diagnosis. In the fields of gastroenterology, there is an increasing need for automatic pathological analysis and diagnosis of GC. Based on the virtual version of pathological slices, several studies were performed to identify and classify GC automatically with high AUCs[120-122,126]. For example, a CNNs model was developed to distinguish gastric mass lesions including gastric adenocarcinoma, adenoma and non-neoplastic lesions, and it has achieved the highest AUC of 0.97 for the identification of gastric adenocarcinoma[126]. With regard to colorectal lesions, Wei et al[128] trained an AI-assisted model to classify colorectal polyps on WSIs, and notably, the performance of the model was similar to that of local pathologists whether in a single institution or other institutions. Besides diagnosis, a model based on more than 400 WSIs was developed to differentiate five common subtypes of colorectal polyps with accuracy of 93%[127]. In CRC, Shapcott et al[129] performed a retrospective study to develop a CNNs model for diagnosis based on 853 hand-marked images with an accuracy of 84%.
In the fields of hepatology, AI-assisted pathology is applied in patients with hepatitis B virus (HBV), metabolic associated fatty liver disease, HCC, etc. An automated, stain-free AI system can quantify the amount of fibrillar collagen to evaluate the degree of HBV-related fibrosis with the AUC > 0.82[132]. For patients with metabolic associated fatty liver disease, AI-assisted pathology tools were used to identify and quantify pathological changes including steatosis, macrosteatosis, lobular inflammation, ballooning, and fibrosis[133], and the algorithm output scores for quantitative comparison with experienced pathologists achieved good agreement. However, limited AI-assisted pathology tools have been built for HCC diagnosis. Notably, the MFC-CNN-ELM program was designed for nuclei grading of biopsy specimens from HCC patients, which revealed high performance in classifying tumor cells of different differentiation stages[134].
Apart from AI-assisted pathology tools in diagnosis, it is no surprise that many tools have been developed for the prediction of gene mutations and prognosis in the fields of gastroenterology and hepatology. In CRC, AI tools have shown great effectiveness in predicting prognosis across all tumor stages based on WSIs[139,140], and several prospective multicenter studies have further validated the high prognosis performance[142]. Notably, a subset of genetic defects occurring in gastroenterology is related to some morphological features detected on WSIs. Among screened genetic defects, microsatellite instability and mismatch-repair deficiency are associated with the survival of gastrointestinal and colorectal cancer patients receiving immunotherapy. Therefore, an AI tool was designed to predict microsatellite instability and mismatch-repair deficiency directly from pathology, and it finally showed reasonably good performance in assisting immunotherapy[138,141]. Notably, Kather et al[140] further validated the above model’s performance in predicting overall survival from CRC pathology slides with a hazard ratio of 2.29 in CRC-specific overall survival (OS) and an hazard ratio of 1.63 in OS, respectively. However, besides the above-mentioned studies that have focused on tumor detection of CRC, few studies were designed to predict gene mutations and prognosis due to the more complicated and heterogeneous histomorphology in gastric diseases than that in the colon[136,137].
In the fields of hepatology, AI tools are mainly used to predict gene mutations and prognosis in HCC. For example, a model has higher accuracy in predicting survival postoperatively than using a composite score of clinical and pathological factors in HCC. In addition, the model may generalize well after validating the performance in an external dataset with different staining and scanning methods[146]. Chen et al[11] investigated a CNN (Inception V3) for automatic classification (benign/malignant classification with 96.0% accuracy, and differentiation degree with 89.6% accuracy) and gene mutation prediction from WSIs after resection of HCC. It was found that CTNNB1, FMN2, TP53, and ZFX4 could be predicted from WSIs with external AUCs from 0.71 to 0.89. Currently, after integrating clinical data, biological data, genetic data, and pathological data, the novel model may also be a promising approach. The first multi-omics model combined ribonucleic acid (RNA) sequencing, miRNA sequencing and methylation data from The Cancer Genome Atlas, and then employed AI technologies to predict and differentiate survival of HCC patients[145]. Other attempts have been made to develop models that can predict gene mutations directly based on WSIs of HCC. Using AI-assisted pathology, some approaches can predict gene expression and RNA sequencing, which may have the potential for clinical translation[147]. Interestingly, some gene expression such as PD-1 and PD-L1 expression, inflammatory gene signatures, and biomarkers of inflammation did trend with improved survival and response in HCC patients[148].
This review retrospectively summarized some key and representative articles with the possibility of missing some publications in AI-related journals. Although various studies have shown promising results in the fields of AI-assisted gastroenterology and hepatology, there are still several limitations to be discussed and resolved. One of the major criticisms is the lack of high-quality training, testing, and validation datasets for the development and validation of AI models. Due to the retrospective manner of most studies, selection bias must be considered at the training stage, meanwhile, overfitting and spectrum bias may result in overestimation of the model accuracy and generalization. According to the rigorous “six-steps” translation pipelines[149], doctors and AI researchers should join the calls that advocate for developing interconnected networks of collecting raw acquisition data which was shifted from processed medical images over the world and training AI on a large scale to obtain robust and generalizable models. Furthermore, the black-box nature of AI technologies has become a barrier to clinical practice, because developers and users do not know the details about how computers output the conclusion. Explainable AI for reliable healthcare is worth investigating to reach clinical interpretability and transparency. In addition, from the perspective of ethics and legal liabilities, AI models may potentially cause errors and challenge the patient-doctor relationship despite the fact that they improve the clinical workflow with enhanced precision. Especially in the fields of gastroenterology and hepatology, cancer discrimination may mean a completely different treatment. If misdiagnosis occurs during AI application, who should take responsibility- the doctor, the programmer, the company providing the system, or the patient? Issues such as ethics and legal liabilities should be demonstrated in the early phase to maintain the balance between minimal error rates and maximal patient benefits[150,151].
There have been an increasing number of studies applying AI to gastroenterology and hepatology over the past decade. In the future, the trend will continue and larger studies will be carried out to compare the performance of medical professionals with AI vs professionals without AI to highlight the importance of AI assistance. AI technologies will be utilized to develop more accurate models to predict and monitor disease progression and potential complications, and these models may ameliorate the insufficiency of medical resources in the remote underserved or developing regions. Besides, AI-assisted personalized imaging protocols and immediate three-dimensional reconstruction may further improve the diagnostic efficiency and accuracy. Researchers will be able to realize the mechanism of disease progression and treatment response through the combination of multi-modality images or multi-omics data. In addition, there is an emerging trend applying AI to drug development, such as prediction of compound toxicity, physical properties, and biological activities, which may assist chemotherapy for digestive system malignancy. Furthermore, AI could be used to process the data generated from the tissue-on-a-chip platform which could better summarize the tumor microenvironment, thus reach precise and individual chemotherapy in gastroenterology and hepatology. As synthetic lethality becomes a promising genetically targeted cancer therapy[152,153], AI could also be used for the detection of target synthetic lethal partners of overexpressed or mutated genes in tumor cells to kill cancers. Finally, AI tools could not replace endoscopists, radiologists, and pathologists in the near and even distant future. Computers would make predictions and doctors would make the final decision, in other words, they would always work together to benefit patients.
AI is rapidly developing and becoming a promising tool in medical image analysis of endoscopy, radiology, and pathology to improve disease diagnosis and treatment in the fields of gastroenterology and hepatology. Nevertheless, we should be aware of the constraints that limit the acceptance and utilization of AI tools in clinical practice. To use AI wisely, doctors and researchers should cooperate to address the current challenges and develop more accurate AI tools to improve patient care.
We thank Yun Cai for polishing our manuscript. We are grateful to our colleagues for their assistance in checking the data of the studies.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): 0
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Santos-García G S-Editor: Zhang L L-Editor: Webster JR P-Editor: Wang LL
| 1. | Laskaris R. Artificial Intelligence: A Modern Approach, 3rd edition. Library Journal, 2015; 140: 45-45. . |
| 2. | Colom R, Karama S, Jung RE, Haier RJ. Human intelligence and brain networks. Dialogues Clin Neurosci. 2010;12:489-501. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 154] [Cited by in RCA: 156] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 3. | Darcy AM, Louie AK, Roberts LW. Machine Learning and the Profession of Medicine. JAMA. 2016;315:551-552. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 244] [Cited by in RCA: 232] [Article Influence: 25.8] [Reference Citation Analysis (0)] |
| 4. | Esteva A, Robicquet A, Ramsundar B, Kuleshov V, DePristo M, Chou K, Cui C, Corrado G, Thrun S, Dean J. A guide to deep learning in healthcare. Nat Med. 2019;25:24-29. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1123] [Cited by in RCA: 1509] [Article Influence: 251.5] [Reference Citation Analysis (0)] |
| 5. | Yang YJ, Bang CS. Application of artificial intelligence in gastroenterology. World J Gastroenterol. 2019;25:1666-1683. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 211] [Cited by in RCA: 160] [Article Influence: 26.7] [Reference Citation Analysis (5)] |
| 6. | Le Berre C, Sandborn WJ, Aridhi S, Devignes MD, Fournier L, Smaïl-Tabbone M, Danese S, Peyrin-Biroulet L. Application of Artificial Intelligence to Gastroenterology and Hepatology. Gastroenterology 2020; 158: 76-94. e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 230] [Cited by in RCA: 325] [Article Influence: 65.0] [Reference Citation Analysis (1)] |
| 7. | Bengio Y, Courville A, Vincent P. Representation learning: a review and new perspectives. IEEE Trans Pattern Anal Mach Intell. 2013;35:1798-1828. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6542] [Cited by in RCA: 2586] [Article Influence: 215.5] [Reference Citation Analysis (0)] |
| 8. | Kumar A, Kim J, Lyndon D, Fulham M, Feng D. An Ensemble of Fine-Tuned Convolutional Neural Networks for Medical Image Classification. IEEE J Biomed Health Inform. 2017;21:31-40. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 264] [Cited by in RCA: 167] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 9. | Ambinder EP. A history of the shift toward full computerization of medicine. J Oncol Pract. 2005;1:54-56. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 26] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 10. | Chen H, Sung JJY. Potentials of AI in medical image analysis in Gastroenterology and Hepatology. J Gastroenterol Hepatol. 2021;36:31-38. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 24] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 11. | Chen M, Zhang B, Topatana W, Cao J, Zhu H, Juengpanich S, Mao Q, Yu H, Cai X. Classification and mutation prediction based on histopathology H&E images in liver cancer using deep learning. NPJ Precis Oncol. 2020;4:14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 132] [Article Influence: 26.4] [Reference Citation Analysis (0)] |
| 12. | Fu Y, Schwebel DC, Hu G. Physicians' Workloads in China: 1998⁻2016. Int J Environ Res Public Health. 2018;15. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 34] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 13. | Miotto R, Wang F, Wang S, Jiang X, Dudley JT. Deep learning for healthcare: review, opportunities and challenges. Brief Bioinform. 2018;19:1236-1246. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1189] [Cited by in RCA: 915] [Article Influence: 130.7] [Reference Citation Analysis (0)] |
| 14. | Shen D, Wu G, Suk HI. Deep Learning in Medical Image Analysis. Annu Rev Biomed Eng. 2017;19:221-248. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2581] [Cited by in RCA: 1955] [Article Influence: 244.4] [Reference Citation Analysis (0)] |
| 15. | Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, Vanderplas J, Passos A, Cournapeau D, Brucher M, Perrot M, Duchesnay E. Scikit-learn: Machine Learning in Python. J Mach Learn Res. 2011;12:2825-2830. |
| 16. | Yu H, Singh R, Shin SH, Ho KY. Artificial intelligence in upper GI endoscopy - current status, challenges and future promise. J Gastroenterol Hepatol. 2021;36:20-24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 7] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 17. | Mori Y, Neumann H, Misawa M, Kudo SE, Bretthauer M. Artificial intelligence in colonoscopy - Now on the market. What's next? J Gastroenterol Hepatol. 2021;36:7-11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 51] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 18. | Wu J, Chen J, Cai J. Application of Artificial Intelligence in Gastrointestinal Endoscopy. J Clin Gastroenterol. 2021;55:110-120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 7] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 19. | Takiyama H, Ozawa T, Ishihara S, Fujishiro M, Shichijo S, Nomura S, Miura M, Tada T. Automatic anatomical classification of esophagogastroduodenoscopy images using deep convolutional neural networks. Sci Rep. 2018;8:7497. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 119] [Cited by in RCA: 88] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 20. | Wu L, Zhang J, Zhou W, An P, Shen L, Liu J, Jiang X, Huang X, Mu G, Wan X, Lv X, Gao J, Cui N, Hu S, Chen Y, Hu X, Li J, Chen D, Gong D, He X, Ding Q, Zhu X, Li S, Wei X, Li X, Wang X, Zhou J, Zhang M, Yu HG. Randomised controlled trial of WISENSE, a real-time quality improving system for monitoring blind spots during esophagogastroduodenoscopy. Gut. 2019;68:2161-2169. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 244] [Cited by in RCA: 214] [Article Influence: 35.7] [Reference Citation Analysis (0)] |
| 21. | van der Sommen F, Zinger S, Curvers WL, Bisschops R, Pech O, Weusten BL, Bergman JJ, de With PH, Schoon EJ. Computer-aided detection of early neoplastic lesions in Barrett's esophagus. Endoscopy. 2016;48:617-624. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 127] [Article Influence: 14.1] [Reference Citation Analysis (2)] |
| 22. | Swager AF, van der Sommen F, Klomp SR, Zinger S, Meijer SL, Schoon EJ, Bergman JJGHM, de With PH, Curvers WL. Computer-aided detection of early Barrett's neoplasia using volumetric laser endomicroscopy. Gastrointest Endosc. 2017;86:839-846. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 102] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 23. | Hashimoto R, Requa J, Dao T, Ninh A, Tran E, Mai D, Lugo M, El-Hage Chehade N, Chang KJ, Karnes WE, Samarasena JB. Artificial intelligence using convolutional neural networks for real-time detection of early esophageal neoplasia in Barrett's esophagus (with video). Gastrointest Endosc 2020; 91: 1264-1271. e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 172] [Cited by in RCA: 144] [Article Influence: 28.8] [Reference Citation Analysis (0)] |
| 24. | Ebigbo A, Mendel R, Probst A, Manzeneder J, Prinz F, de Souza LA Jr, Papa J, Palm C, Messmann H. Real-time use of artificial intelligence in the evaluation of cancer in Barrett's oesophagus. Gut. 2020;69:615-616. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 84] [Cited by in RCA: 124] [Article Influence: 24.8] [Reference Citation Analysis (0)] |
| 25. | Horie Y, Yoshio T, Aoyama K, Yoshimizu S, Horiuchi Y, Ishiyama A, Hirasawa T, Tsuchida T, Ozawa T, Ishihara S, Kumagai Y, Fujishiro M, Maetani I, Fujisaki J, Tada T. Diagnostic outcomes of esophageal cancer by artificial intelligence using convolutional neural networks. Gastrointest Endosc. 2019;89:25-32. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 240] [Cited by in RCA: 272] [Article Influence: 45.3] [Reference Citation Analysis (0)] |
| 26. | Kumagai Y, Takubo K, Kawada K, Aoyama K, Endo Y, Ozawa T, Hirasawa T, Yoshio T, Ishihara S, Fujishiro M, Tamaru JI, Mochiki E, Ishida H, Tada T. Diagnosis using deep-learning artificial intelligence based on the endocytoscopic observation of the esophagus. Esophagus. 2019;16:180-187. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 65] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 27. | Zhao YY, Xue DX, Wang YL, Zhang R, Sun B, Cai YP, Feng H, Cai Y, Xu JM. Computer-assisted diagnosis of early esophageal squamous cell carcinoma using narrow-band imaging magnifying endoscopy. Endoscopy. 2019;51:333-341. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 86] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 28. | Cai SL, Li B, Tan WM, Niu XJ, Yu HH, Yao LQ, Zhou PH, Yan B, Zhong YS. Using a deep learning system in endoscopy for screening of early esophageal squamous cell carcinoma (with video). Gastrointest Endosc 2019; 90: 745-753. e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 110] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 29. | Nakagawa K, Ishihara R, Aoyama K, Ohmori M, Nakahira H, Matsuura N, Shichijo S, Nishida T, Yamada T, Yamaguchi S, Ogiyama H, Egawa S, Kishida O, Tada T. Classification for invasion depth of esophageal squamous cell carcinoma using a deep neural network compared with experienced endoscopists. Gastrointest Endosc. 2019;90:407-414. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 132] [Cited by in RCA: 110] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 30. | Tokai Y, Yoshio T, Aoyama K, Horie Y, Yoshimizu S, Horiuchi Y, Ishiyama A, Tsuchida T, Hirasawa T, Sakakibara Y, Yamada T, Yamaguchi S, Fujisaki J, Tada T. Application of artificial intelligence using convolutional neural networks in determining the invasion depth of esophageal squamous cell carcinoma. Esophagus. 2020;17:250-256. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 83] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 31. | Ali H, Yasmin M, Sharif M, Rehmani MH. Computer assisted gastric abnormalities detection using hybrid texture descriptors for chromoendoscopy images. Comput Methods Programs Biomed. 2018;157:39-47. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 32] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 32. | Sakai Y, Takemoto S, Hori K, Nishimura M, Ikematsu H, Yano T, Yokota H. Automatic detection of early gastric cancer in endoscopic images using a transferring convolutional neural network. Annu Int Conf IEEE Eng Med Biol Soc. 2018;2018:4138-4141. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 48] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 33. | Kanesaka T, Lee TC, Uedo N, Lin KP, Chen HZ, Lee JY, Wang HP, Chang HT. Computer-aided diagnosis for identifying and delineating early gastric cancers in magnifying narrow-band imaging. Gastrointest Endosc. 2018;87:1339-1344. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 129] [Article Influence: 18.4] [Reference Citation Analysis (0)] |
| 34. | Wu L, Zhou W, Wan X, Zhang J, Shen L, Hu S, Ding Q, Mu G, Yin A, Huang X, Liu J, Jiang X, Wang Z, Deng Y, Liu M, Lin R, Ling T, Li P, Wu Q, Jin P, Chen J, Yu H. A deep neural network improves endoscopic detection of early gastric cancer without blind spots. Endoscopy. 2019;51:522-531. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 194] [Cited by in RCA: 156] [Article Influence: 26.0] [Reference Citation Analysis (0)] |
| 35. | Horiuchi Y, Aoyama K, Tokai Y, Hirasawa T, Yoshimizu S, Ishiyama A, Yoshio T, Tsuchida T, Fujisaki J, Tada T. Convolutional Neural Network for Differentiating Gastric Cancer from Gastritis Using Magnified Endoscopy with Narrow Band Imaging. Dig Dis Sci. 2020;65:1355-1363. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 102] [Article Influence: 20.4] [Reference Citation Analysis (1)] |
| 36. | Zhu Y, Wang QC, Xu MD, Zhang Z, Cheng J, Zhong YS, Zhang YQ, Chen WF, Yao LQ, Zhou PH, Li QL. Application of convolutional neural network in the diagnosis of the invasion depth of gastric cancer based on conventional endoscopy. Gastrointest Endosc 2019; 89: 806-815. e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 201] [Cited by in RCA: 231] [Article Influence: 38.5] [Reference Citation Analysis (0)] |
| 37. | Luo H, Xu G, Li C, He L, Luo L, Wang Z, Jing B, Deng Y, Jin Y, Li Y, Li B, Tan W, He C, Seeruttun SR, Wu Q, Huang J, Huang DW, Chen B, Lin SB, Chen QM, Yuan CM, Chen HX, Pu HY, Zhou F, He Y, Xu RH. Real-time artificial intelligence for detection of upper gastrointestinal cancer by endoscopy: a multicentre, case-control, diagnostic study. Lancet Oncol. 2019;20:1645-1654. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 155] [Cited by in RCA: 253] [Article Influence: 42.2] [Reference Citation Analysis (0)] |
| 38. | Nagao S, Tsuji Y, Sakaguchi Y, Takahashi Y, Minatsuki C, Niimi K, Yamashita H, Yamamichi N, Seto Y, Tada T, Koike K. Highly accurate artificial intelligence systems to predict the invasion depth of gastric cancer: efficacy of conventional white-light imaging, nonmagnifying narrow-band imaging, and indigo-carmine dye contrast imaging. Gastrointest Endosc 2020; 92: 866-873. e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 70] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 39. | Ayaru L, Ypsilantis PP, Nanapragasam A, Choi RC, Thillanathan A, Min-Ho L, Montana G. Prediction of Outcome in Acute Lower Gastrointestinal Bleeding Using Gradient Boosting. PLoS One. 2015;10:e0132485. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 47] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 40. | Xiao Jia, Meng MQ. A deep convolutional neural network for bleeding detection in Wireless Capsule Endoscopy images. Annu Int Conf IEEE Eng Med Biol Soc. 2016;2016:639-642. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 67] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 41. | Usman MA, Satrya GB, Usman MR, Shin SY. Detection of small colon bleeding in wireless capsule endoscopy videos. Comput Med Imaging Graph. 2016;54:16-26. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 23] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 42. | Sengupta N, Tapper EB. Derivation and Internal Validation of a Clinical Prediction Tool for 30-Day Mortality in Lower Gastrointestinal Bleeding. Am J Med 2017; 130: 601.e1-601. e8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 41] [Article Influence: 5.1] [Reference Citation Analysis (1)] |
| 43. | Leenhardt R, Vasseur P, Li C, Saurin JC, Rahmi G, Cholet F, Becq A, Marteau P, Histace A, Dray X; CAD-CAP Database Working Group. A neural network algorithm for detection of GI angiectasia during small-bowel capsule endoscopy. Gastrointest Endosc. 2019;89:189-194. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 148] [Article Influence: 24.7] [Reference Citation Analysis (1)] |
| 44. | Aoki T, Yamada A, Kato Y, Saito H, Tsuboi A, Nakada A, Niikura R, Fujishiro M, Oka S, Ishihara S, Matsuda T, Nakahori M, Tanaka S, Koike K, Tada T. Automatic detection of blood content in capsule endoscopy images based on a deep convolutional neural network. J Gastroenterol Hepatol. 2020;35:1196-1200. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 72] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 45. | Yang J, Chang L, Li S, He X, Zhu T. WCE polyp detection based on novel feature descriptor with normalized variance locality-constrained linear coding. Int J Comput Assist Radiol Surg. 2020;15:1291-1302. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 46. | Vieira PM, Freitas NR, Valente J, Vaz IF, Rolanda C, Lima CS. Automatic detection of small bowel tumors in wireless capsule endoscopy images using ensemble learning. Med Phys. 2020;47:52-63. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 13] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 47. | Fernández-Esparrach G, Bernal J, López-Cerón M, Córdova H, Sánchez-Montes C, Rodríguez de Miguel C, Sánchez FJ. Exploring the clinical potential of an automatic colonic polyp detection method based on the creation of energy maps. Endoscopy. 2016;48:837-842. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 87] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 48. | Komeda Y, Handa H, Watanabe T, Nomura T, Kitahashi M, Sakurai T, Okamoto A, Minami T, Kono M, Arizumi T, Takenaka M, Hagiwara S, Matsui S, Nishida N, Kashida H, Kudo M. Computer-Aided Diagnosis Based on Convolutional Neural Network System for Colorectal Polyp Classification: Preliminary Experience. Oncology. 2017;93 Suppl 1:30-34. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 174] [Cited by in RCA: 129] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 49. | Misawa M, Kudo SE, Mori Y, Takeda K, Maeda Y, Kataoka S, Nakamura H, Kudo T, Wakamura K, Hayashi T, Katagiri A, Baba T, Ishida F, Inoue H, Nimura Y, Oda M, Mori K. Accuracy of computer-aided diagnosis based on narrow-band imaging endocytoscopy for diagnosing colorectal lesions: comparison with experts. Int J Comput Assist Radiol Surg. 2017;12:757-766. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 49] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 50. | Misawa M, Kudo SE, Mori Y, Cho T, Kataoka S, Yamauchi A, Ogawa Y, Maeda Y, Takeda K, Ichimasa K, Nakamura H, Yagawa Y, Toyoshima N, Ogata N, Kudo T, Hisayuki T, Hayashi T, Wakamura K, Baba T, Ishida F, Itoh H, Roth H, Oda M, Mori K. Artificial Intelligence-Assisted Polyp Detection for Colonoscopy: Initial Experience. Gastroenterology 2018; 154: 2027-2029. e3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 317] [Cited by in RCA: 269] [Article Influence: 38.4] [Reference Citation Analysis (0)] |
| 51. | Chen PJ, Lin MC, Lai MJ, Lin JC, Lu HH, Tseng VS. Accurate Classification of Diminutive Colorectal Polyps Using Computer-Aided Analysis. Gastroenterology. 2018;154:568-575. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 250] [Cited by in RCA: 276] [Article Influence: 39.4] [Reference Citation Analysis (0)] |
| 52. | Urban G, Tripathi P, Alkayali T, Mittal M, Jalali F, Karnes W, Baldi P. Deep Learning Localizes and Identifies Polyps in Real Time With 96% Accuracy in Screening Colonoscopy. Gastroenterology 2018; 155: 1069-1078. e8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 398] [Cited by in RCA: 433] [Article Influence: 61.9] [Reference Citation Analysis (1)] |
| 53. | Renner J, Phlipsen H, Haller B, Navarro-Avila F, Saint-Hill-Febles Y, Mateus D, Ponchon T, Poszler A, Abdelhafez M, Schmid RM, von Delius S, Klare P. Optical classification of neoplastic colorectal polyps - a computer-assisted approach (the COACH study). Scand J Gastroenterol. 2018;53:1100-1106. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 35] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 54. | Wang P, Xiao X, Glissen Brown JR, Berzin TM, Tu M, Xiong F, Hu X, Liu P, Song Y, Zhang D, Yang X, Li L, He J, Yi X, Liu J, Liu X. Development and validation of a deep-learning algorithm for the detection of polyps during colonoscopy. Nat Biomed Eng. 2018;2:741-748. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 248] [Cited by in RCA: 270] [Article Influence: 38.6] [Reference Citation Analysis (0)] |
| 55. | Mori Y, Kudo SE, Misawa M, Saito Y, Ikematsu H, Hotta K, Ohtsuka K, Urushibara F, Kataoka S, Ogawa Y, Maeda Y, Takeda K, Nakamura H, Ichimasa K, Kudo T, Hayashi T, Wakamura K, Ishida F, Inoue H, Itoh H, Oda M, Mori K. Real-Time Use of Artificial Intelligence in Identification of Diminutive Polyps During Colonoscopy: A Prospective Study. Ann Intern Med. 2018;169:357-366. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 412] [Cited by in RCA: 355] [Article Influence: 50.7] [Reference Citation Analysis (1)] |
| 56. | Byrne MF, Chapados N, Soudan F, Oertel C, Linares Pérez M, Kelly R, Iqbal N, Chandelier F, Rex DK. Real-time differentiation of adenomatous and hyperplastic diminutive colorectal polyps during analysis of unaltered videos of standard colonoscopy using a deep learning model. Gut. 2019;68:94-100. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 363] [Cited by in RCA: 412] [Article Influence: 68.7] [Reference Citation Analysis (0)] |
| 57. | Blanes-Vidal V, Baatrup G, Nadimi ES. Addressing priority challenges in the detection and assessment of colorectal polyps from capsule endoscopy and colonoscopy in colorectal cancer screening using machine learning. Acta Oncol. 2019;58:S29-S36. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 62] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 58. | Lee JY, Jeong J, Song EM, Ha C, Lee HJ, Koo JE, Yang DH, Kim N, Byeon JS. Real-time detection of colon polyps during colonoscopy using deep learning: systematic validation with four independent datasets. Sci Rep. 2020;10:8379. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 48] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 59. | Gohari MR, Biglarian A, Bakhshi E, Pourhoseingholi MA. Use of an artificial neural network to determine prognostic factors in colorectal cancer patients. Asian Pac J Cancer Prev. 2011;12:1469-1472. [PubMed] |
| 60. | Biglarian A, Bakhshi E, Gohari MR, Khodabakhshi R. Artificial neural network for prediction of distant metastasis in colorectal cancer. Asian Pac J Cancer Prev. 2012;13:927-930. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 24] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 61. | Takeda K, Kudo SE, Mori Y, Misawa M, Kudo T, Wakamura K, Katagiri A, Baba T, Hidaka E, Ishida F, Inoue H, Oda M, Mori K. Accuracy of diagnosing invasive colorectal cancer using computer-aided endocytoscopy. Endoscopy. 2017;49:798-802. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 101] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 62. | Ito N, Kawahira H, Nakashima H, Uesato M, Miyauchi H, Matsubara H. Endoscopic Diagnostic Support System for cT1b Colorectal Cancer Using Deep Learning. Oncology. 2019;96:44-50. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 60] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 63. | Zhou D, Tian F, Tian X, Sun L, Huang X, Zhao F, Zhou N, Chen Z, Zhang Q, Yang M, Yang Y, Guo X, Li Z, Liu J, Wang J, Wang B, Zhang G, Sun B, Zhang W, Kong D, Chen K, Li X. Diagnostic evaluation of a deep learning model for optical diagnosis of colorectal cancer. Nat Commun. 2020;11:2961. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 55] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 64. | Shaheen NJ, Falk GW, Iyer PG, Gerson LB; American College of Gastroenterology. ACG Clinical Guideline: Diagnosis and Management of Barrett's Esophagus. Am J Gastroenterol. 2016;111:30-50; quiz 51. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 990] [Cited by in RCA: 1060] [Article Influence: 117.8] [Reference Citation Analysis (0)] |
| 65. | Fitzgerald RC, di Pietro M, Ragunath K, Ang Y, Kang JY, Watson P, Trudgill N, Patel P, Kaye PV, Sanders S, O'Donovan M, Bird-Lieberman E, Bhandari P, Jankowski JA, Attwood S, Parsons SL, Loft D, Lagergren J, Moayyedi P, Lyratzopoulos G, de Caestecker J; British Society of Gastroenterology. British Society of Gastroenterology guidelines on the diagnosis and management of Barrett's oesophagus. Gut. 2014;63:7-42. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1016] [Cited by in RCA: 878] [Article Influence: 79.8] [Reference Citation Analysis (0)] |
| 66. | Schölvinck DW, van der Meulen K, Bergman JJGHM, Weusten BLAM. Detection of lesions in dysplastic Barrett's esophagus by community and expert endoscopists. Endoscopy. 2017;49:113-120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 23] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 67. | Menon S, Trudgill N. How commonly is upper gastrointestinal cancer missed at endoscopy? Endosc Int Open. 2014;2:E46-E50. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 176] [Cited by in RCA: 230] [Article Influence: 20.9] [Reference Citation Analysis (0)] |
| 68. | Hosokawa O, Hattori M, Douden K, Hayashi H, Ohta K, Kaizaki Y. Difference in accuracy between gastroscopy and colonoscopy for detection of cancer. Hepatogastroenterology. 2007;54:442-444. [PubMed] |
| 69. | Hirasawa T, Aoyama K, Tanimoto T, Ishihara S, Shichijo S, Ozawa T, Ohnishi T, Fujishiro M, Matsuo K, Fujisaki J, Tada T. Application of artificial intelligence using a convolutional neural network for detecting gastric cancer in endoscopic images. Gastric Cancer. 2018;21:653-660. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 568] [Cited by in RCA: 427] [Article Influence: 61.0] [Reference Citation Analysis (0)] |
| 70. | Kubota K, Kuroda J, Yoshida M, Ohta K, Kitajima M. Medical image analysis: computer-aided diagnosis of gastric cancer invasion on endoscopic images. Surg Endosc. 2012;26:1485-1489. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 58] [Article Influence: 4.1] [Reference Citation Analysis (1)] |
| 71. | Boland GW, Guimaraes AS, Mueller PR. The radiologist's conundrum: benefits and costs of increasing CT capacity and utilization. Eur Radiol. 2009;19:9-11; discussion 12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 37] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 72. | Gatos I, Tsantis S, Spiliopoulos S, Karnabatidis D, Theotokas I, Zoumpoulis P, Loupas T, Hazle JD, Kagadis GC. A new computer aided diagnosis system for evaluation of chronic liver disease with ultrasound shear wave elastography imaging. Med Phys. 2016;43:1428-1436. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 20] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 73. | Gatos I, Tsantis S, Spiliopoulos S, Karnabatidis D, Theotokas I, Zoumpoulis P, Loupas T, Hazle JD, Kagadis GC. A Machine-Learning Algorithm Toward Color Analysis for Chronic Liver Disease Classification, Employing Ultrasound Shear Wave Elastography. Ultrasound Med Biol. 2017;43:1797-1810. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 73] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 74. | Chen Y, Luo Y, Huang W, Hu D, Zheng RQ, Cong SZ, Meng FK, Yang H, Lin HJ, Sun Y, Wang XY, Wu T, Ren J, Pei SF, Zheng Y, He Y, Hu Y, Yang N, Yan H. Machine-learning-based classification of real-time tissue elastography for hepatic fibrosis in patients with chronic hepatitis B. Comput Biol Med. 2017;89:18-23. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 60] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 75. | Li W, Huang Y, Zhuang BW, Liu GJ, Hu HT, Li X, Liang JY, Wang Z, Huang XW, Zhang CQ, Ruan SM, Xie XY, Kuang M, Lu MD, Chen LD, Wang W. Multiparametric ultrasomics of significant liver fibrosis: A machine learning-based analysis. Eur Radiol. 2019;29:1496-1506. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 62] [Cited by in RCA: 96] [Article Influence: 13.7] [Reference Citation Analysis (1)] |
| 76. | Gatos I, Tsantis S, Spiliopoulos S, Karnabatidis D, Theotokas I, Zoumpoulis P, Loupas T, Hazle JD, Kagadis GC. Temporal stability assessment in shear wave elasticity images validated by deep learning neural network for chronic liver disease fibrosis stage assessment. Med Phys. 2019;46:2298-2309. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 42] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 77. | Wang K, Lu X, Zhou H, Gao Y, Zheng J, Tong M, Wu C, Liu C, Huang L, Jiang T, Meng F, Lu Y, Ai H, Xie XY, Yin LP, Liang P, Tian J, Zheng R. Deep learning Radiomics of shear wave elastography significantly improved diagnostic performance for assessing liver fibrosis in chronic hepatitis B: a prospective multicentre study. Gut. 2019;68:729-741. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 226] [Cited by in RCA: 354] [Article Influence: 59.0] [Reference Citation Analysis (1)] |
| 78. | Kuppili V, Biswas M, Sreekumar A, Suri HS, Saba L, Edla DR, Marinho RT, Sanches JM, Suri JS. Extreme Learning Machine Framework for Risk Stratification of Fatty Liver Disease Using Ultrasound Tissue Characterization. J Med Syst. 2017;41:152. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 76] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 79. | Byra M, Styczynski G, Szmigielski C, Kalinowski P, Michałowski Ł, Paluszkiewicz R, Ziarkiewicz-Wróblewska B, Zieniewicz K, Sobieraj P, Nowicki A. Transfer learning with deep convolutional neural network for liver steatosis assessment in ultrasound images. Int J Comput Assist Radiol Surg. 2018;13:1895-1903. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 206] [Cited by in RCA: 154] [Article Influence: 22.0] [Reference Citation Analysis (0)] |
| 80. | Biswas M, Kuppili V, Edla DR, Suri HS, Saba L, Marinhoe RT, Sanches JM, Suri JS. Symtosis: A liver ultrasound tissue characterization and risk stratification in optimized deep learning paradigm. Comput Methods Programs Biomed. 2018;155:165-177. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 119] [Article Influence: 17.0] [Reference Citation Analysis (1)] |
| 81. | Cao W, An X, Cong L, Lyu C, Zhou Q, Guo R. Application of Deep Learning in Quantitative Analysis of 2-Dimensional Ultrasound Imaging of Nonalcoholic Fatty Liver Disease. J Ultrasound Med. 2020;39:51-59. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 50] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 82. | Guo LH, Wang D, Qian YY, Zheng X, Zhao CK, Li XL, Bo XW, Yue WW, Zhang Q, Shi J, Xu HX. A two-stage multi-view learning framework based computer-aided diagnosis of liver tumors with contrast enhanced ultrasound images. Clin Hemorheol Microcirc. 2018;69:343-354. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 72] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 83. | Schmauch B, Herent P, Jehanno P, Dehaene O, Saillard C, Aubé C, Luciani A, Lassau N, Jégou S. Diagnosis of focal liver lesions from ultrasound using deep learning. Diagn Interv Imaging. 2019;100:227-233. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 104] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 84. | Yang Q, Wei J, Hao X, Kong D, Yu X, Jiang T, Xi J, Cai W, Luo Y, Jing X, Yang Y, Cheng Z, Wu J, Zhang H, Liao J, Zhou P, Song Y, Zhang Y, Han Z, Cheng W, Tang L, Liu F, Dou J, Zheng R, Yu J, Tian J, Liang P. Improving B-mode ultrasound diagnostic performance for focal liver lesions using deep learning: A multicentre study. EBioMedicine. 2020;56:102777. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 71] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 85. | Choi KJ, Jang JK, Lee SS, Sung YS, Shim WH, Kim HS, Yun J, Choi JY, Lee Y, Kang BK, Kim JH, Kim SY, Yu ES. Development and Validation of a Deep Learning System for Staging Liver Fibrosis by Using Contrast Agent-enhanced CT Images in the Liver. Radiology. 2018;289:688-697. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 151] [Article Influence: 21.6] [Reference Citation Analysis (0)] |
| 86. | He L, Li H, Dudley JA, Maloney TC, Brady SL, Somasundaram E, Trout AT, Dillman JR. Machine Learning Prediction of Liver Stiffness Using Clinical and T2-Weighted MRI Radiomic Data. AJR Am J Roentgenol. 2019;213:592-601. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 42] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 87. | Ahmed Y, Hussein RS, Basha TA, Khalifa AM, Ibrahim AS, Abdelmoaty AS, Abdella HM, Fahmy AS. Detecting liver fibrosis using a machine learning-based approach to the quantification of the heart-induced deformation in tagged MR images. NMR Biomed. 2020;33:e4215. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 16] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 88. | Hectors SJ, Kennedy P, Huang KH, Stocker D, Carbonell G, Greenspan H, Friedman S, Taouli B. Fully automated prediction of liver fibrosis using deep learning analysis of gadoxetic acid-enhanced MRI. Eur Radiol. 2020;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 47] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 89. | Vivanti R, Szeskin A, Lev-Cohain N, Sosna J, Joskowicz L. Automatic detection of new tumors and tumor burden evaluation in longitudinal liver CT scan studies. Int J Comput Assist Radiol Surg. 2017;12:1945-1957. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 50] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 90. | Yasaka K, Akai H, Abe O, Kiryu S. Deep Learning with Convolutional Neural Network for Differentiation of Liver Masses at Dynamic Contrast-enhanced CT: A Preliminary Study. Radiology. 2018;286:887-896. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 293] [Cited by in RCA: 394] [Article Influence: 49.3] [Reference Citation Analysis (0)] |
| 91. | Ibragimov B, Toesca D, Chang D, Yuan Y, Koong A, Xing L. Development of deep neural network for individualized hepatobiliary toxicity prediction after liver SBRT. Med Phys. 2018;45:4763-4774. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 83] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 92. | Abajian A, Murali N, Savic LJ, Laage-Gaupp FM, Nezami N, Duncan JS, Schlachter T, Lin M, Geschwind JF, Chapiro J. Predicting Treatment Response to Intra-arterial Therapies for Hepatocellular Carcinoma with the Use of Supervised Machine Learning-An Artificial Intelligence Concept. J Vasc Interv Radiol 2018; 29: 850-857. e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 126] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 93. | Zhang F, Yang J, Nezami N, Laage-Gaupp F, Chapiro J, De Lin M, Duncan J. Liver Tissue Classification Using an Auto-context-based Deep Neural Network with a Multi-phase Training Framework. Patch Based Tech Med Imaging (2018). 2018;11075:59-66. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 17] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 94. | Morshid A, Elsayes KM, Khalaf AM, Elmohr MM, Yu J, Kaseb AO, Hassan M, Mahvash A, Wang Z, Hazle JD, Fuentes D. A machine learning model to predict hepatocellular carcinoma response to transcatheter arterial chemoembolization. Radiol Artif Intell. 2019;1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 83] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 95. | Nayak A, Baidya Kayal E, Arya M, Culli J, Krishan S, Agarwal S, Mehndiratta A. Computer-aided diagnosis of cirrhosis and hepatocellular carcinoma using multi-phase abdomen CT. Int J Comput Assist Radiol Surg. 2019;14:1341-1352. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 42] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 96. | Hamm CA, Wang CJ, Savic LJ, Ferrante M, Schobert I, Schlachter T, Lin M, Duncan JS, Weinreb JC, Chapiro J, Letzen B. Deep learning for liver tumor diagnosis part I: development of a convolutional neural network classifier for multi-phasic MRI. Eur Radiol. 2019;29:3338-3347. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117] [Cited by in RCA: 207] [Article Influence: 34.5] [Reference Citation Analysis (0)] |
| 97. | Wang CJ, Hamm CA, Savic LJ, Ferrante M, Schobert I, Schlachter T, Lin M, Weinreb JC, Duncan JS, Chapiro J, Letzen B. Deep learning for liver tumor diagnosis part II: convolutional neural network interpretation using radiologic imaging features. Eur Radiol. 2019;29:3348-3357. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 103] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 98. | Jansen MJA, Kuijf HJ, Veldhuis WB, Wessels FJ, Viergever MA, Pluim JPW. Automatic classification of focal liver lesions based on MRI and risk factors. PLoS One. 2019;14:e0217053. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 47] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 99. | Mokrane FZ, Lu L, Vavasseur A, Otal P, Peron JM, Luk L, Yang H, Ammari S, Saenger Y, Rousseau H, Zhao B, Schwartz LH, Dercle L. Radiomics machine-learning signature for diagnosis of hepatocellular carcinoma in cirrhotic patients with indeterminate liver nodules. Eur Radiol. 2020;30:558-570. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 125] [Article Influence: 25.0] [Reference Citation Analysis (0)] |
| 100. | Shi W, Kuang S, Cao S, Hu B, Xie S, Chen S, Chen Y, Gao D, Zhu Y, Zhang H, Liu H, Ye M, Sirlin CB, Wang J. Deep learning assisted differentiation of hepatocellular carcinoma from focal liver lesions: choice of four-phase and three-phase CT imaging protocol. Abdom Radiol (NY). 2020;45:2688-2697. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 50] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 101. | Alirr OI. Deep learning and level set approach for liver and tumor segmentation from CT scans. J Appl Clin Med Phys. 2020;21:200-209. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 29] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 102. | Zheng H, Chen Y, Yue X, Ma C, Liu X, Yang P, Lu J. Deep pancreas segmentation with uncertain regions of shadowed sets. Magn Reson Imaging. 2020;68:45-52. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 32] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 103. | Liang JD, Ping XO, Tseng YJ, Huang GT, Lai F, Yang PM. Recurrence predictive models for patients with hepatocellular carcinoma after radiofrequency ablation using support vector machines with feature selection methods. Comput Methods Programs Biomed. 2014;117:425-434. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 31] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 104. | Zhou W, Zhang L, Wang K, Chen S, Wang G, Liu Z, Liang C. Malignancy characterization of hepatocellular carcinomas based on texture analysis of contrast-enhanced MR images. J Magn Reson Imaging. 2017;45:1476-1484. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 88] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 105. | Abajian A, Murali N, Savic LJ, Laage-Gaupp FM, Nezami N, Duncan JS, Schlachter T, Lin M, Geschwind JF, Chapiro J. Predicting Treatment Response to Image-Guided Therapies Using Machine Learning: An Example for Trans-Arterial Treatment of Hepatocellular Carcinoma. J Vis Exp. 2018;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 7] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 106. | Ma X, Wei J, Gu D, Zhu Y, Feng B, Liang M, Wang S, Zhao X, Tian J. Preoperative radiomics nomogram for microvascular invasion prediction in hepatocellular carcinoma using contrast-enhanced CT. Eur Radiol. 2019;29:3595-3605. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 163] [Article Influence: 27.2] [Reference Citation Analysis (1)] |
| 107. | Dong Y, Zhou L, Xia W, Zhao XY, Zhang Q, Jian JM, Gao X, Wang WP. Preoperative Prediction of Microvascular Invasion in Hepatocellular Carcinoma: Initial Application of a Radiomic Algorithm Based on Grayscale Ultrasound Images. Front Oncol. 2020;10:353. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 49] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 108. | He M, Zhang P, Ma X, He B, Fang C, Jia F. Radiomic Feature-Based Predictive Model for Microvascular Invasion in Patients With Hepatocellular Carcinoma. Front Oncol. 2020;10:574228. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 27] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 109. | Schoenberg MB, Bucher JN, Koch D, Börner N, Hesse S, De Toni EN, Seidensticker M, Angele MK, Klein C, Bazhin AV, Werner J, Guba MO. A novel machine learning algorithm to predict disease free survival after resection of hepatocellular carcinoma. Ann Transl Med. 2020;8:434. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 19] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 110. | Zhao Y, Wu J, Zhang Q, Hua Z, Qi W, Wang N, Lin T, Sheng L, Cui D, Liu J, Song Q, Li X, Wu T, Guo Y, Cui J, Liu A. Radiomics Analysis Based on Multiparametric MRI for Predicting Early Recurrence in Hepatocellular Carcinoma After Partial Hepatectomy. J Magn Reson Imaging. 2020;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 55] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 111. | Liu F, Liu D, Wang K, Xie X, Su L, Kuang M, Huang G, Peng B, Wang Y, Lin M, Tian J. Deep Learning Radiomics Based on Contrast-Enhanced Ultrasound Might Optimize Curative Treatments for Very-Early or Early-Stage Hepatocellular Carcinoma Patients. Liver Cancer. 2020;9:397-413. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 103] [Article Influence: 20.6] [Reference Citation Analysis (0)] |
| 112. | Chen M, Cao J, Hu J, Topatana W, Li S, Juengpanich S, Lin J, Tong C, Shen J, Zhang B, Wu J, Pocha C, Kudo M, Amedei A, Trevisani F, Sung PS, Zaydfudim VM, Kanda T, Cai X. Clinical-Radiomic Analysis for Pretreatment Prediction of Objective Response to First Transarterial Chemoembolization in Hepatocellular Carcinoma. Liver Cancer. 2021;10:38-51. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 82] [Article Influence: 20.5] [Reference Citation Analysis (0)] |
| 113. | European Association for the Study of the Liver. EASL Clinical Practice Guidelines: Management of hepatocellular carcinoma. J Hepatol. 2018;69:182-236. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5593] [Cited by in RCA: 6064] [Article Influence: 866.3] [Reference Citation Analysis (3)] |
| 114. | Gillies RJ, Kinahan PE, Hricak H. Radiomics: Images Are More than Pictures, They Are Data. Radiology. 2016;278:563-577. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4541] [Cited by in RCA: 5556] [Article Influence: 617.3] [Reference Citation Analysis (3)] |
| 115. | Lambin P, Leijenaar RTH, Deist TM, Peerlings J, de Jong EEC, van Timmeren J, Sanduleanu S, Larue RTHM, Even AJG, Jochems A, van Wijk Y, Woodruff H, van Soest J, Lustberg T, Roelofs E, van Elmpt W, Dekker A, Mottaghy FM, Wildberger JE, Walsh S. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol. 2017;14:749-762. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1825] [Cited by in RCA: 3569] [Article Influence: 446.1] [Reference Citation Analysis (0)] |
| 116. | Erstad DJ, Tanabe KK. Prognostic and Therapeutic Implications of Microvascular Invasion in Hepatocellular Carcinoma. Ann Surg Oncol. 2019;26:1474-1493. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 139] [Cited by in RCA: 288] [Article Influence: 48.0] [Reference Citation Analysis (0)] |
| 117. | Metter DM, Colgan TJ, Leung ST, Timmons CF, Park JY. Trends in the US and Canadian Pathologist Workforces From 2007 to 2017. JAMA Netw Open. 2019;2:e194337. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 123] [Cited by in RCA: 183] [Article Influence: 30.5] [Reference Citation Analysis (0)] |
| 118. | Tomita N, Abdollahi B, Wei J, Ren B, Suriawinata A, Hassanpour S. Attention-Based Deep Neural Networks for Detection of Cancerous and Precancerous Esophagus Tissue on Histopathological Slides. JAMA Netw Open. 2019;2:e1914645. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 133] [Cited by in RCA: 101] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 119. | Sharma H, Zerbe N, Klempert I, Hellwich O, Hufnagl P. Deep convolutional neural networks for automatic classification of gastric carcinoma using whole slide images in digital histopathology. Comput Med Imaging Graph. 2017;61:2-13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 176] [Cited by in RCA: 172] [Article Influence: 21.5] [Reference Citation Analysis (0)] |
| 120. | Li Y, Li X, Xie X, Shen L. Deep learning based gastric cancer identification. Proceedings of the 2018 IEEE 15th International Symposium on Biomedical Imaging; 2018: 182-185. [DOI] [Full Text] |
| 121. | Leon F, Gelvez M, Jaimes Z, Gelvez T, Arguello H. Supervised Classification of Histopathological Images Using Convolutional Neuronal Networks for Gastric Cancer Detection. Proceedings of the 2019 XXII Symposium on Image, Signal Processing and Artificial Vision; 2019: 1-5. [DOI] [Full Text] |
| 122. | Sun M, Zhang G, Dang H, Qi X, Zhou X, Chang Q. Accurate Gastric Cancer Segmentation in Digital Pathology Images Using Deformable Convolution and Multi-Scale Embedding Networks. IEEE Access 2019; 7: 75530-75541. [DOI] [Full Text] |
| 123. | Ma B, Guo Y, Hu W, Yuan F, Zhu Z, Yu Y, Zou H. Artificial Intelligence-Based Multiclass Classification of Benign or Malignant Mucosal Lesions of the Stomach. Front Pharmacol. 2020;11:572372. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 17] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 124. | Yoshida H, Shimazu T, Kiyuna T, Marugame A, Yamashita Y, Cosatto E, Taniguchi H, Sekine S, Ochiai A. Automated histological classification of whole-slide images of gastric biopsy specimens. Gastric Cancer. 2018;21:249-257. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 72] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 125. | Qu J, Hiruta N, Terai K, Nosato H, Murakawa M, Sakanashi H. Gastric Pathology Image Classification Using Stepwise Fine-Tuning for Deep Neural Networks. J Healthc Eng. 2018;2018:8961781. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 36] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 126. | Iizuka O, Kanavati F, Kato K, Rambeau M, Arihiro K, Tsuneki M. Deep Learning Models for Histopathological Classification of Gastric and Colonic Epithelial Tumours. Sci Rep. 2020;10:1504. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 132] [Cited by in RCA: 215] [Article Influence: 43.0] [Reference Citation Analysis (0)] |
| 127. | Korbar B, Olofson AM, Miraflor AP, Nicka CM, Suriawinata MA, Torresani L, Suriawinata AA, Hassanpour S. Deep Learning for Classification of Colorectal Polyps on Whole-slide Images. J Pathol Inform. 2017;8:30. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 140] [Cited by in RCA: 168] [Article Influence: 21.0] [Reference Citation Analysis (0)] |
| 128. | Wei JW, Suriawinata AA, Vaickus LJ, Ren B, Liu X, Lisovsky M, Tomita N, Abdollahi B, Kim AS, Snover DC, Baron JA, Barry EL, Hassanpour S. Evaluation of a Deep Neural Network for Automated Classification of Colorectal Polyps on Histopathologic Slides. JAMA Netw Open. 2020;3:e203398. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 69] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 129. | Shapcott M, Hewitt KJ, Rajpoot N. Deep Learning With Sampling in Colon Cancer Histology. Front Bioeng Biotechnol. 2019;7:52. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 41] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 130. | Geessink OGF, Baidoshvili A, Klaase JM, Ehteshami Bejnordi B, Litjens GJS, van Pelt GW, Mesker WE, Nagtegaal ID, Ciompi F, van der Laak JAWM. Computer aided quantification of intratumoral stroma yields an independent prognosticator in rectal cancer. Cell Oncol (Dordr). 2019;42:331-341. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 83] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 131. | Song Z, Yu C, Zou S, Wang W, Huang Y, Ding X, Liu J, Shao L, Yuan J, Gou X, Jin W, Wang Z, Chen X, Chen H, Liu C, Xu G, Sun Z, Ku C, Zhang Y, Dong X, Wang S, Xu W, Lv N, Shi H. Automatic deep learning-based colorectal adenoma detection system and its similarities with pathologists. BMJ Open. 2020;10:e036423. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 28] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 132. | Wang TH, Chen TC, Teng X, Liang KH, Yeh CT. Automated biphasic morphological assessment of hepatitis B-related liver fibrosis using second harmonic generation microscopy. Sci Rep. 2015;5:12962. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 33] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 133. | Forlano R, Mullish BH, Giannakeas N, Maurice JB, Angkathunyakul N, Lloyd J, Tzallas AT, Tsipouras M, Yee M, Thursz MR, Goldin RD, Manousou P. High-Throughput, Machine Learning-Based Quantification of Steatosis, Inflammation, Ballooning, and Fibrosis in Biopsies From Patients With Nonalcoholic Fatty Liver Disease. Clin Gastroenterol Hepatol 2020; 18: 2081-2090. e9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 94] [Article Influence: 18.8] [Reference Citation Analysis (0)] |
| 134. | Li S, Jiang H, Pang W. Joint multiple fully connected convolutional neural network with extreme learning machine for hepatocellular carcinoma nuclei grading. Comput Biol Med. 2017;84:156-167. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 39] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 135. | Kiani A, Uyumazturk B, Rajpurkar P, Wang A, Gao R, Jones E, Yu Y, Langlotz CP, Ball RL, Montine TJ, Martin BA, Berry GJ, Ozawa MG, Hazard FK, Brown RA, Chen SB, Wood M, Allard LS, Ylagan L, Ng AY, Shen J. Impact of a deep learning assistant on the histopathologic classification of liver cancer. NPJ Digit Med. 2020;3:23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 181] [Cited by in RCA: 160] [Article Influence: 32.0] [Reference Citation Analysis (0)] |
| 136. | Steinbuss G, Kriegsmann K, Kriegsmann M. Identification of Gastritis Subtypes by Convolutional Neuronal Networks on Histological Images of Antrum and Corpus Biopsies. Int J Mol Sci. 2020;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 20] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 137. | Liu Y, Li X, Zheng A, Zhu X, Liu S, Hu M, Luo Q, Liao H, Liu M, He Y, Chen Y. Predict Ki-67 Positive Cells in H&E-Stained Images Using Deep Learning Independently From IHC-Stained Images. Front Mol Biosci. 2020;7:183. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 25] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 138. | Kather JN, Pearson AT, Halama N, Jäger D, Krause J, Loosen SH, Marx A, Boor P, Tacke F, Neumann UP, Grabsch HI, Yoshikawa T, Brenner H, Chang-Claude J, Hoffmeister M, Trautwein C, Luedde T. Deep learning can predict microsatellite instability directly from histology in gastrointestinal cancer. Nat Med. 2019;25:1054-1056. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 992] [Cited by in RCA: 761] [Article Influence: 126.8] [Reference Citation Analysis (0)] |
| 139. | Bychkov D, Linder N, Turkki R, Nordling S, Kovanen PE, Verrill C, Walliander M, Lundin M, Haglund C, Lundin J. Deep learning based tissue analysis predicts outcome in colorectal cancer. Sci Rep. 2018;8:3395. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 427] [Cited by in RCA: 365] [Article Influence: 52.1] [Reference Citation Analysis (0)] |
| 140. | Kather JN, Krisam J, Charoentong P, Luedde T, Herpel E, Weis CA, Gaiser T, Marx A, Valous NA, Ferber D, Jansen L, Reyes-Aldasoro CC, Zörnig I, Jäger D, Brenner H, Chang-Claude J, Hoffmeister M, Halama N. Predicting survival from colorectal cancer histology slides using deep learning: A retrospective multicenter study. PLoS Med. 2019;16:e1002730. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 555] [Cited by in RCA: 492] [Article Influence: 82.0] [Reference Citation Analysis (0)] |
| 141. | Echle A, Grabsch HI, Quirke P, van den Brandt PA, West NP, Hutchins GGA, Heij LR, Tan X, Richman SD, Krause J, Alwers E, Jenniskens J, Offermans K, Gray R, Brenner H, Chang-Claude J, Trautwein C, Pearson AT, Boor P, Luedde T, Gaisa NT, Hoffmeister M, Kather JN. Clinical-Grade Detection of Microsatellite Instability in Colorectal Tumors by Deep Learning. Gastroenterology 2020; 159: 1406-1416. e11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 210] [Cited by in RCA: 220] [Article Influence: 44.0] [Reference Citation Analysis (0)] |
| 142. | Skrede OJ, De Raedt S, Kleppe A, Hveem TS, Liestøl K, Maddison J, Askautrud HA, Pradhan M, Nesheim JA, Albregtsen F, Farstad IN, Domingo E, Church DN, Nesbakken A, Shepherd NA, Tomlinson I, Kerr R, Novelli M, Kerr DJ, Danielsen HE. Deep learning for prediction of colorectal cancer outcome: a discovery and validation study. Lancet. 2020;395:350-360. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 332] [Cited by in RCA: 364] [Article Influence: 72.8] [Reference Citation Analysis (0)] |
| 143. | Sirinukunwattana K, Domingo E, Richman SD, Redmond KL, Blake A, Verrill C, Leedham SJ, Chatzipli A, Hardy C, Whalley CM, Wu CH, Beggs AD, McDermott U, Dunne PD, Meade A, Walker SM, Murray GI, Samuel L, Seymour M, Tomlinson I, Quirke P, Maughan T, Rittscher J, Koelzer VH; S:CORT consortium. Image-based consensus molecular subtype (imCMS) classification of colorectal cancer using deep learning. Gut. 2021;70:544-554. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 150] [Cited by in RCA: 148] [Article Influence: 37.0] [Reference Citation Analysis (0)] |
| 144. | Jang HJ, Lee A, Kang J, Song IH, Lee SH. Prediction of clinically actionable genetic alterations from colorectal cancer histopathology images using deep learning. World J Gastroenterol. 2020;26:6207-6223. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 46] [Cited by in RCA: 48] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 145. | Chaudhary K, Poirion OB, Lu L, Garmire LX. Deep Learning-Based Multi-Omics Integration Robustly Predicts Survival in Liver Cancer. Clin Cancer Res. 2018;24:1248-1259. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 483] [Cited by in RCA: 589] [Article Influence: 84.1] [Reference Citation Analysis (0)] |
| 146. | Saillard C, Schmauch B, Laifa O, Moarii M, Toldo S, Zaslavskiy M, Pronier E, Laurent A, Amaddeo G, Regnault H, Sommacale D, Ziol M, Pawlotsky JM, Mulé S, Luciani A, Wainrib G, Clozel T, Courtiol P, Calderaro J. Predicting Survival After Hepatocellular Carcinoma Resection Using Deep Learning on Histological Slides. Hepatology. 2020;72:2000-2013. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 189] [Article Influence: 37.8] [Reference Citation Analysis (0)] |
| 147. | Fu Y, Jung AW, Torne RV, Gonzalez S, Vöhringer H, Shmatko A, Yates LR, Jimenez-Linan M, Moore L, Gerstung M. Pan-cancer computational histopathology reveals mutations, tumor composition and prognosis. Nature Cancer. 2020;1:800-810. [RCA] [DOI] [Full Text] [Cited by in Crossref: 168] [Cited by in RCA: 337] [Article Influence: 67.4] [Reference Citation Analysis (0)] |
| 148. | Sangro B, Melero I, Wadhawan S, Finn RS, Abou-Alfa GK, Cheng AL, Yau T, Furuse J, Park JW, Boyd Z, Tang HT, Shen Y, Tschaika M, Neely J, El-Khoueiry A. Association of inflammatory biomarkers with clinical outcomes in nivolumab-treated patients with advanced hepatocellular carcinoma. J Hepatol. 2020;73:1460-1469. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 155] [Cited by in RCA: 309] [Article Influence: 61.8] [Reference Citation Analysis (0)] |
| 149. | Stead WW. Clinical Implications and Challenges of Artificial Intelligence and Deep Learning. JAMA. 2018;320:1107-1108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 137] [Cited by in RCA: 137] [Article Influence: 19.6] [Reference Citation Analysis (4)] |
| 150. | Sung JJ, Poon NC. Artificial intelligence in gastroenterology: where are we heading? Front Med. 2020;14:511-517. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 151. | Poon NC, Sung JJ. Self-driving cars and AI-assisted endoscopy: Who should take the responsibility when things go wrong? J Gastroenterol Hepatol. 2019;34:625-626. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 5] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 152. | Li S, Topatana W, Juengpanich S, Cao J, Hu J, Zhang B, Ma D, Cai X, Chen M. Development of synthetic lethality in cancer: molecular and cellular classification. Signal Transduct Target Ther. 2020;5:241. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 68] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 153. | Topatana W, Juengpanich S, Li S, Cao J, Hu J, Lee J, Suliyanto K, Ma D, Zhang B, Chen M, Cai X. Advances in synthetic lethality for cancer therapy: cellular mechanism and clinical translation. J Hematol Oncol. 2020;13:118. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 113] [Article Influence: 22.6] [Reference Citation Analysis (0)] |