Published online Sep 28, 2020. doi: 10.3748/wjg.v26.i36.5420
Peer-review started: April 1, 2020
First decision: June 4, 2020
Revised: June 11, 2020
Accepted: August 25, 2020
Article in press: August 25, 2020
Published online: September 28, 2020
Processing time: 175 Days and 23.1 Hours
Hepatocellular carcinoma (HCC) is the third leading cause of cancer mortality worldwide. The gut microbiota can help maintain healthy metabolism and immunity. Granulocyte-macrophage colony-stimulating factor (GM-CSF) is a critical factor in promoting health and homeostasis; it promotes intestinal immunity, stimulates bone marrow precursors to generate macrophage colonies, and enhances the antibacterial and antitumor activity of circulating monocytes. As such, GM-CSF may protect against HCC development by regulating immunity as well as intestinal microecology.
To investigate the impact of GM-CSF on the gut microbiome and metabolic characteristics of HCC.
Thirty-six male BALB/c nude mice were divided into three groups: Control (n = 10), HCC (n = 13), and HCC + GM-CSF (GM-CSF overexpression, n = 13). We utilized HCC cells to establish orthotopic transplantation tumor models of HCC with normal and over-expressing GM-CSF. Liver injury, immune inflammatory function and intestinal barrier function were evaluated. The fecal microbiome and metabolome were studied using 16S rRNA absolute quantification sequencing and gas chromatography-mass spectrometry.
GM-CSF overexpression significantly affected the gut microbiome of mice with HCC and resulted in a high abundance of organisms of the genera Roseburia, Blautia and Butyricimonass, along with a significant reduction in Prevotella, Parabacteroides, Anaerotruncus, Streptococcus, Clostridium, and Mucispirillum. Likewise, GM-CSF overexpression resulted in a substantial increase in fecal biotin and oleic acid levels, along with a prominent decrease in the fecal succinic acid, adenosine, fumaric acid, lipoic acid, and maleic acid levels. Correlation analysis revealed that the intestinal microbiota and fecal metabolites induced by GM-CSF were primarily involved in pathways related to reducing the inflammatory response, biotin metabolism, and intestinal barrier dysfunction.
GM-CSF can protect against HCC development by regulating immunity and modulating the abundance of specific intestinal microorganisms and their metabolites. This study provides new insights into the therapeutic approaches for HCC.
Core Tip: Granulocyte-macrophage colony-stimulating factor (GM-CSF) is a critical factor in promoting intestinal immunity homeostasis. Whether GM-CSF plays a protective role against hepatocellular carcinoma (HCC) by regulating immunity and intestinal microecology is not completely understood. Orthotopic transplantation tumor models of HCC were established in mice by transfecting human GM-CSF sequences into a human HCC cell line using lentivirus. We measureed the absolute abundance of specific fecal bacterial communities using the absolute quantificative 16S rRNA amplicon sequencing technology. This study provides new insights into the therapeutic approaches for HCC.
- Citation: Wu YN, Zhang L, Chen T, Li X, He LH, Liu GX. Granulocyte-macrophage colony-stimulating factor protects mice against hepatocellular carcinoma by ameliorating intestinal dysbiosis and attenuating inflammation. World J Gastroenterol 2020; 26(36): 5420-5436
- URL: https://www.wjgnet.com/1007-9327/full/v26/i36/5420.htm
- DOI: https://dx.doi.org/10.3748/wjg.v26.i36.5420
Hepatocellular carcinoma (HCC) is currently the third leading cause of cancer mortality[1]. There are approximately 466000 new cases of HCC in China each year, accounting for approximately 55% of the total cases worldwide. HCC has become an extremely serious public health problem in China due to its high incidence, poor prognosis, and high rate of postsurgical recurrence[2,3]. Unfortunately, lack of effective treatment for liver cancer further increases the overall burden on the patients. There is currently substantial research on the characterization of the immunology and biochemistry of the gut, which scarcely involve metabolic organs. Recent interest has been focused on important physiological functions of the gut, such as maintaining energy homeostasis[4].
In recent decades, researchers have focused on the important contributions of the gut microbiota to the key aspects of human health, notably metabolism and immunity[5,6]. The liver is the largest solid organ in our body; it processes 80% of the blood derived from the gastrointestinal tract via outflow from the portal vein[7]. A large number of resident immune cells within the liver modulate immune responses, promote liver repair and detoxification, and resist invasion of pathogens[8,9]. The gut microbiome is a critical factor that regulates liver immunity and maintains liver homeostasis via a variety of mechanisms, including those involving endotoxin, Toll-like receptors (TLR), and bile acid metabolism[10-15]. Of particular interest, extensive studies have confirmed specific differences in the intestinal flora between patients with HCC and healthy control subjects. However, the specific mechanisms underlying these findings remain unclear[16,17].
Granulocyte–macrophage colony-stimulating factor (GM-CSF) is a host cytokine that stimulates bone marrow precursors to generate monocytes and macrophages. Furthermore, it enhances monocyte antibacterial and antitumor activities[18]. Microbiota can promote the release of GM-CSF; this process is of great importance in maintaining intestinal immune homeostasis. Interestingly, macrophage-mediated sensing of microbial signals controls GM-CSF levels[19]. Serum and tissue levels of GM-CSF were decreased in patients with HCC. Conversely, we found that GM-CSF overexpression inhibited the proliferation, invasion, and migration of liver cancer cells and promoted their apoptosis (unpublished data). We hypothesize that GM-CSF plays a protective role against HCC by regulating intestinal microecology. In this study, we examined the effects of GM-CSF overexpression on the intestinal microecology and metabolism of HCC. Our findings demonstrated that GM-CSF overexpression protects mice against HCC by ameliorating intestinal dysbiosis, stabilizing the intestinal barrier, attenuating inflammation, and reducing serum endotoxin levels. These findings may have profound implications for the development of therapies for HCC.
This study was approved by the Ethics Committee of the First Hospital of Lanzhou University (No. LLYYLL-2017-18) and was performed in accordance with the Guidelines for Experimental Animals of the Ministry of Science and Technology (Beijing, China).
Seven-week-old male BALB/c nude mice were housed in a specific pathogen-free environment with temperature (23 ± 2°C) and humidity controls (approximately 50%) and a daily cycle of 12-h light/12-h dark together with ad libitum access to food and drinking water. After 1 wk of acclimatization, the mice were randomly divided into three groups: Control (n = 10), HCC (n = 13), and HCC + GM-CSF (GM-CSF overexpression, n = 13) groups. In the HCC and HCC + GM-CSF groups, sub-cutaneous tumors were implanted in the early stage. Mouse orthotopic transplantation tumor models of HCC were established after subcutaneous tumors were successfully implanted. The specific operation method is as follows. The human GM-CSF sequence was transfected using lentivirus into the human HCC cell line HCCLM3. The cells growing in the log phase were injected into the armpit of nude mice under sterile conditions (1 × 107/mL). Then, the tumors were subcutaneously inoculated with cells for approximately 4 wks (approximately 1 cm in diameter, no rupture on the surface). The tumors were trimmed into a tumor mass of 1.5 mm × 2 mm × 1 mm, the abdominal wall was cut, and the right lobe of the liver was exposed and incised. The aforementioned spare tumor mass was implanted, followed by suturing and closing of the abdomen. After the effects of anesthesia wore off, the mice were returned to the cage for feeding. They were kept in this environment for 4 wk. The construction method of the HCC orthotopic tumor model was as described above.
Fecal samples were collected from all mice and stored in a -80°C freezer. Blood samples were collected and centrifuged at 3000 r/min for 15 min; all serum aliquots were stored at -80°C. Liver and colon samples were divided into halves; tissues from one part were fixed in 4% neutral buffered formaldehyde for paraffin embedding, whereas the other part was frozen in liquid nitrogen and stored at -80°C for subsequent RNA extraction.
Paraffin-embedded liver and colon tissue sections were stained with hematoxylin and eosin (H&E) to observe the morphology of these tissues and to detect liver injury. Images were scanned using a NanoZoomer digital pathology system (Hamamatsu Photonics, K.K., Japan), which digitally scans the sections into a specific image format for further evaluation.
Alanine aminotransferase (ALT) and aspartate aminotransferase (AST) levels were measured using the ROCHE CABAS C311 analyzer (Roche, Germany). Lipo-polysaccharide (LPS) concentrations as an indirect measure of endotoxin levels were measured with ELISA according to the manufacturer’s instructions (Guduo, Shanghai, China). Immune inflammatory factors were measured using commercial ELISA kits (Abbkine Scientific Co, United States).
Liver and colon tissue sections were stained using an immunohistochemistry kit (DAKO, Denmark) as previously described[20]. The NanoZoomer digital pathology scanner was used to scan the stained sections into a format suitable for analysis. Each section was randomly selected at 200× and 400× magnifications for analysis. Image-Pro Plus software was used to measure the average optical density of tight junction protein-1 (ZO-1).
Total RNA was extracted from liver and colon tissues using an RNA kit (Qiagen, Germantown, MD, United States) according to the manufacturer’s protocol. Real-time PCR was performed with the Applied Biosystems 7500 system using the one-step SYBR PrimeScript plus RT-PCR kit (Takara, Japan). The PCR primer sequences used in this study are listed in Supplementary Table S1. The expression data of the samples were normalized to those of the internal control glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and were calculated according to the ΔΔCt method.
The QIAamp Rapid DNA Kit (Qiagen) was used to extract DNA from mouse stool samples. The DNA concentration and integrity were measured using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Hudson, NH, United States) and agarose gel electrophoresis, respectively. Suitable DNA samples were transported to Genesky Biotechnologies Inc., Shanghai, China, for absolute quantification and 16S rRNA gene amplicon sequencing. The spike-in sequences contained conserved regions identical to those of natural 16S rRNA genes and artificial variable regions and were distinct from those found in nucleotide sequences in public databases. These served as internal standards and facilitated absolute quantification across samples. Appropriate mixtures with known copy numbers of spike-in sequences were added to the sample DNAs. The V3-V4 regions of the 16S rRNA genes and spike-in sequences were PCR-amplified and sequenced using Illumina HiSeq.
Metabolomics analysis was performed using an Agilent 7890 gas chromatography time-of-flight mass spectrometry (GC-TOF-MS). ChromaTOF software was used to analyze the mass spectrum data for peak extraction, baseline correction, deconvolution, peak integration, and peak alignment[21]. For specific identification of biomolecules, the LECO-Fiehn Rtx5 database was used to match mass spectra and the retention time index. Finally, the peaks with a detection rate below 50% or relative standard deviation (RSD) > 30% in the quality control (QC) samples were removed[22]. Metabolites were identified from the US National Institutes of Standards and Technology (NIST) and Fiehn databases. Orthogonal partial least squares-discriminant analysis (OPLS-DA) was performed to visualize metabolic differences between experimental groups. Differential detection of metabolites was based on the statistical significance (P value) of the two-tailed Student’s t teston the normalized peak areas, and metabolites withvariable importance in the projection values > 1 and P values < 0.05 were included.
The data are presented as the means ± SE. For most data, one-way ANOVA with Tukey’s post hoc test was used to determine statistically significant differences between the groups. The Wilcoxon rank-sum test was performed to evaluate alpha diversity and principal coordinates between the different cohorts in absolute quantificative 16S rRNA amplicon sequencing analysis. PERMANOVA (Adonis) was used to test microbial community clustering using both weighted and unweighted UniFrac and Bray-Curtis distance matrices. Correlations between the variables were determined using Spearman’s rank correlation; P values < 0.05 were considered significant. The data were analyzed using SPSS version 19.0 for Windows (SPSS Inc., United States).
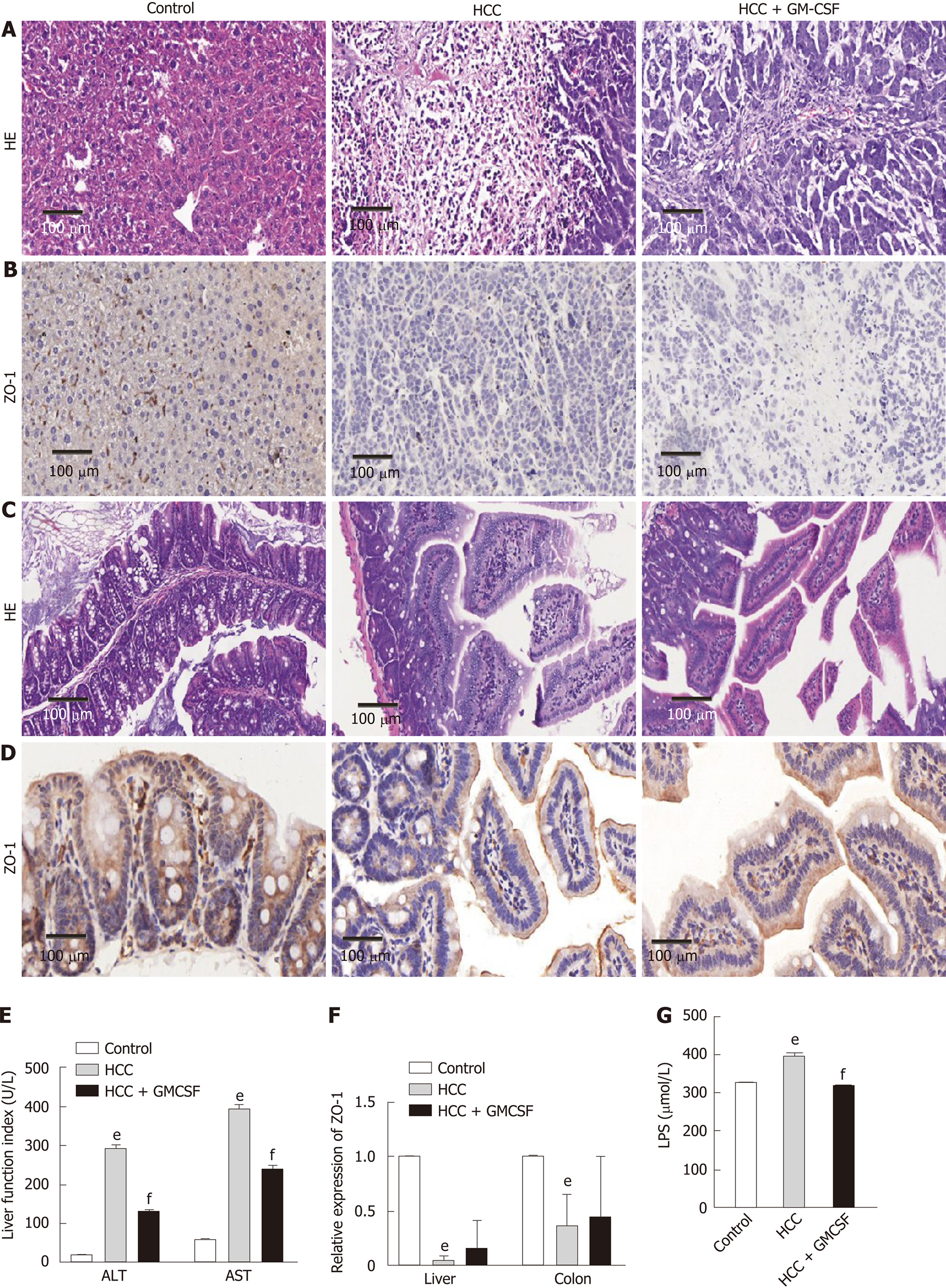
To assess the effects of GM-CSF on hepatic injury, liver function, intestinal barrier function, and serum endotoxin levels, we established an orthotopic transplantation tumor model of HCC by transecting GM-CSF-encoded lentivirus into an HCC cell line. The morphology of the livers in the HCC + GM-CSF group improved compared with that in the mice with HCC alone; tumor tissue infiltration was also reduced in the HCC + GM-CSF group (Figure 1A). The normal structure of the liver of the mice in the HCC group was extensively destroyed by tumor tissue infiltration, and there was considerable accumulation of vesicular fat, which is related to mixed inflammatory infiltration. In contrast, GM-CSF showed no impact on colon morphology (Figure 1C).
The mice in the HCC group had significantly higher serum ALT and AST levels than the controls. However, GM-CSF overexpression in HCC cells suppressed serum ALT and AST levels (Figure 1E). Zhu et al[23] reported that damage to intestinal tight junctions was associated with loss of intestinal barrier function. To explore this possibility as well as to examine the distribution of ZO-1 in the liver and colon[24], we first confirmed the increase in the levels of ZO-1 protein in the HCC + GM-CSF group via immunohistochemistry staining. These findings suggest that GM-CSF overexpression reversed intestinal barrier dysfunction observed in response to HCC (Figure 1B and D). In parallel, the expression of ZO-1 transcripts in the liver and colon was upregulated among mice in the HCC + GM-CSF group (Figure 1F).
Endotoxin activates specific Toll-like receptors[25]; serum LPS levels were significantly reduced in the mice in the HCC + GM-CSF group compared with those in the HCC alone group (Figure 1G). Taken together, these results suggest that GM-CSF overexpression alleviated hepatic injury, normalized liver function tests, stabilized the intestinal barrier, and reduced serum endotoxin levels observed in response to HCC.
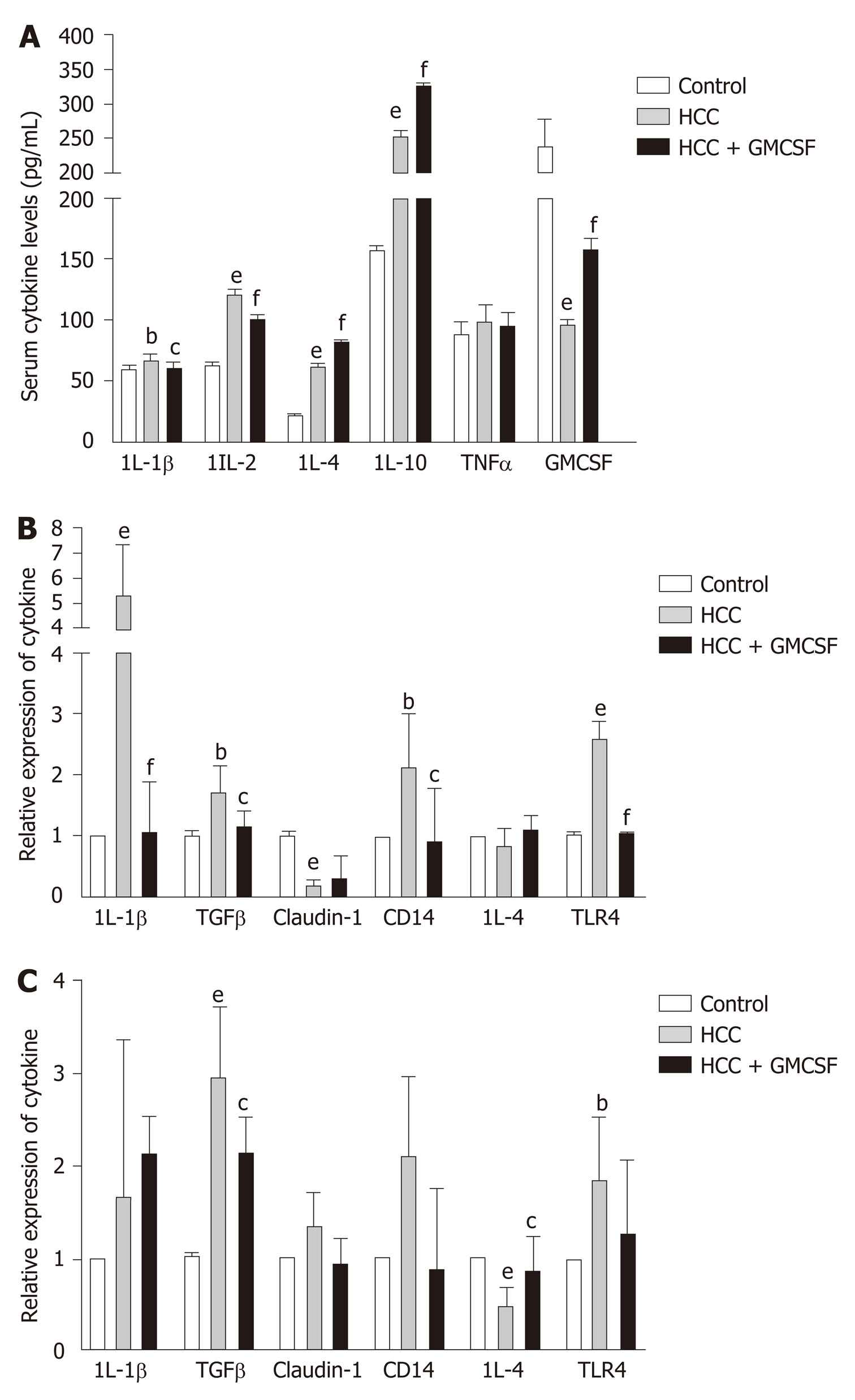
Current research indicates that cytokines play an important role in the progression of HCC[26]. GM-CSF promoted significant downregulation of proinflammatory cytokines [interleukin (IL)-1β and IL-2] and increased the levels of anti-inflammatory cytokines IL-4 and IL-10, with no effects on tumor necrosis factor levels. To explore and extend these findings with respect to our mouse model, we measured the expression of cytokine transcripts in the liver tissue. GM-CSF overexpression resulted in a significant decrease in the expression of IL-1β and TGFβ; however, no changes were observed with respect to IL-4 expression (Figure 2B). Furthermore, we found that GM-CSF overexpression resulted in increased IL-4 expression in the colon, whereas there were no changes in IL-1β expression (Figure 2C). Toll-like receptor (TLR)-4 and its coreceptor CD14 are central to LPS recognition[27]. Endotoxin-associated increases in both TLR4 and CD14 expression in the liver and colon were reversed by GM-CSF. These results suggest the direct effects of GM-CSF on modulating the expression of both TLR4 and CD14.
To obtain a greater understanding of the protective effects of GM-CSF, we examined the intestinal microbiome of the three study groups using the absolute quantificative 16S rRNA amplicon sequencing technology with an artificially synthesized DNA internal standard; use of this standard permits us to simultaneously measure the absolute and relative abundance of specific fecal bacterial communities. While previous studies have reported the relative abundance of microorganisms in specific fecal samples, increasing concern has been focused on determining the absolute abundance of targeted micro-organisms in different samples.
Alpha diversities were calculated using diversity indexes. Our results showed that the Chao index and number of operational taxonomic units were significantly increased in the fecal samples of HCC + GM-CSF mice; however, there were no significant differences in the Shannon index (Supplementary Figure 1A). Βeta diversity values were calculated by weighted UniFrac principal coordinates analysis to assess the phylogenetic relationship between microbial communities. The results from the control, HCC, and HCC + GM-CSF groups can be clearly separated into different clusters (Supplementary Figure 1C). Similarly, the structure of the intestinal flora of the HCC and HCC + GM-CSF groups was clearly divided into distinct clusters.
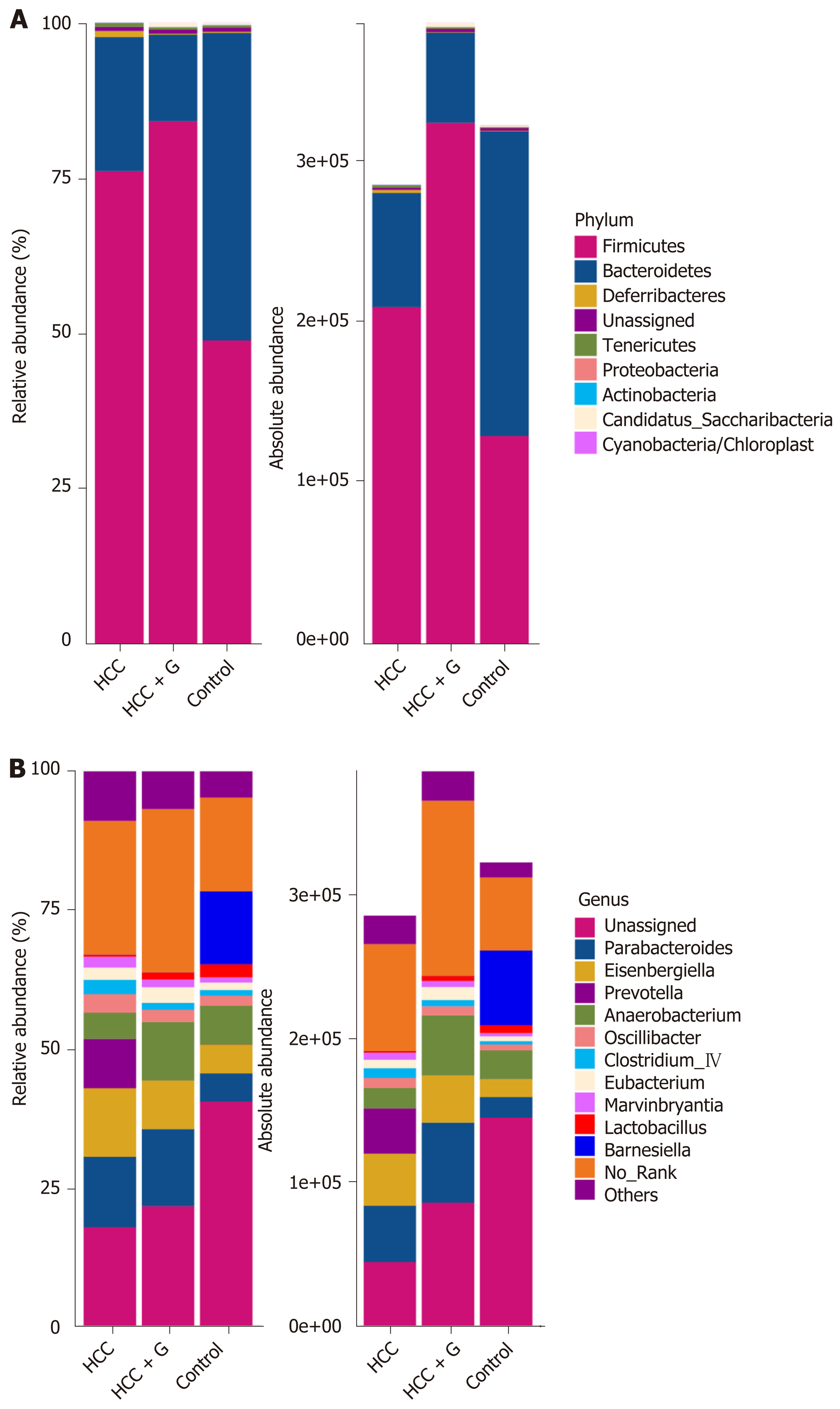
The relative abundance and absolute copy number of each bacterial group at the phylum level are shown in Figure 3A. The microbial composition of the HCC + GM-CSF group was significantly different from that of the HCC alone group. Of note, the absolute abundance of the micro-organisms in the HCC group was reduced by 12% compared with the control group. Furthermore, the absolute abundance of intestinal microorganisms among the fecal samples from the HCC + GM-CSF group was 1.35 times higher than among those in the HCC group; these findings suggest that GM-CSF limits HCC-induced intestinal dysbiosis. The intestinal microbiota identified in all three groups primarily included bacteria within the phyla Firmicutes, Bacteroidetes, Deferribacteres, Tenericutes, Proteobacteria, Actinobacteria, and Cyanobacteria (Figure 3A). Among them, the abundance of Tenericutes, which is related to hepatopancreatic necrosis, was 2.45 times higher in the samples from the HCC groups than those from the control. GM-CSF overexpression resulted in a 63% decrease in the absolute abundance of Tenericutes compared with that observed in samples from mice with HCC. At the genus level (Figure 3B), the intestinal flora mainly includes Parabacteroides, Prevotella, Anaerobacterium, Eubacterium, Clostridium_lV, Anaerotruncus, Mucispirillum, Rosecusburia, and Butyricicoccus. We analyzed the absolute abundance at the genus level as shown in Figure 4.
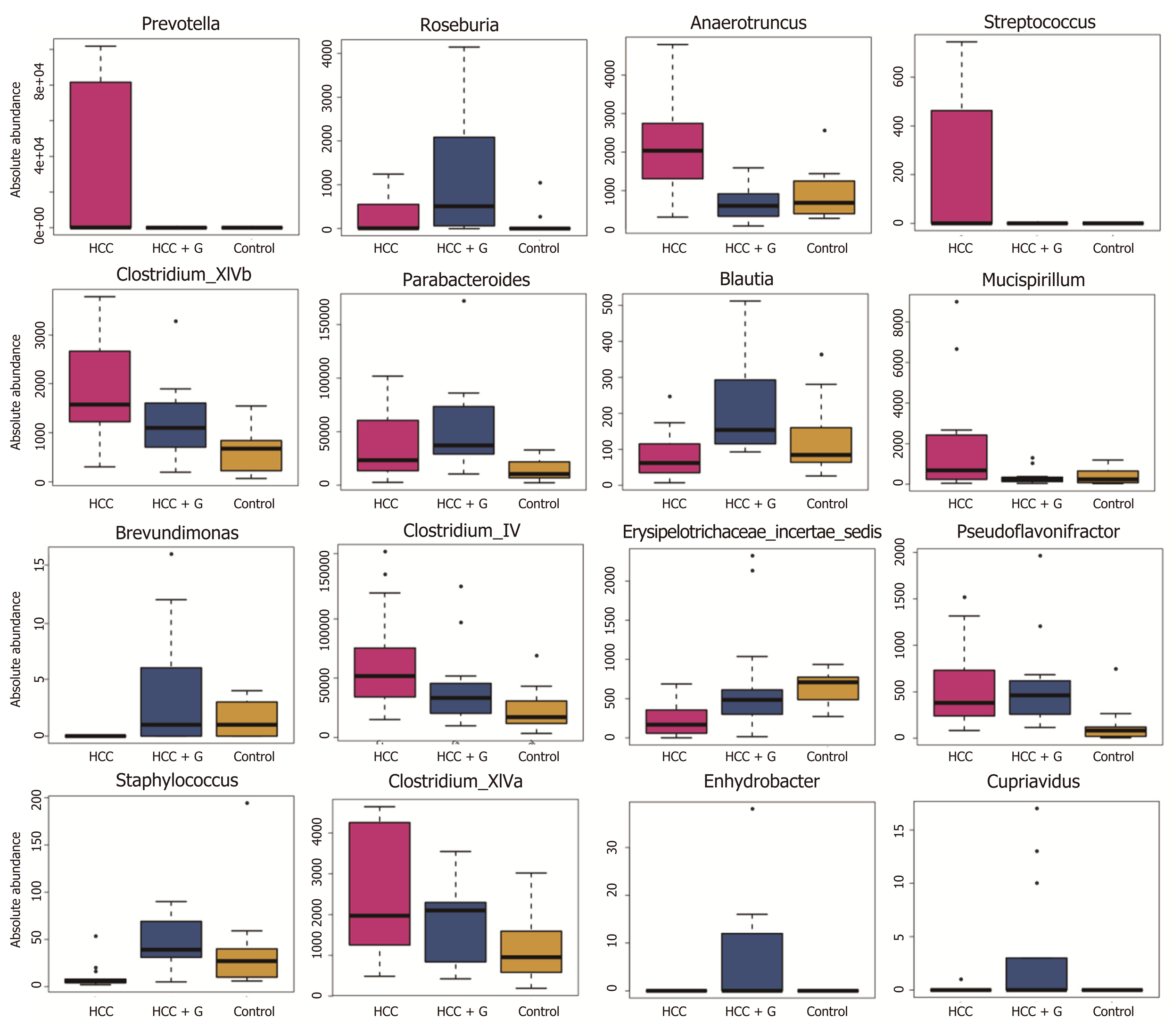
To evaluate the differences in the intestinal microbiome at the bacterial genus level, we performed statistical evaluation focusing on the most prominent differences among the three groups. GM-CSF overexpression contributed to the high abundance of protective genera (e.g., Roseburia, Blautia, and Butyricimonass), along with a significant reduction in Prevotella, Parabacteroides, Anaerotruncu, Streptococcus, Clostridium, and Mucispirillum. Particularly notable was the genus Prevotella, which was reduced 10 000-fold in response to GM-CSF overexpression. To identify additional differences in the intestinal flora among the different groups, we performed the linear discriminant analysis effect size (LEfse) analysis based on the ribosomal database project classification data (Supplementary Figures 2C and D) and identified the most important differences between the HCC and HCC + GM-CSF groups, including those identified among the Prevotella, Clostridium, and Anaerotruncus genera.
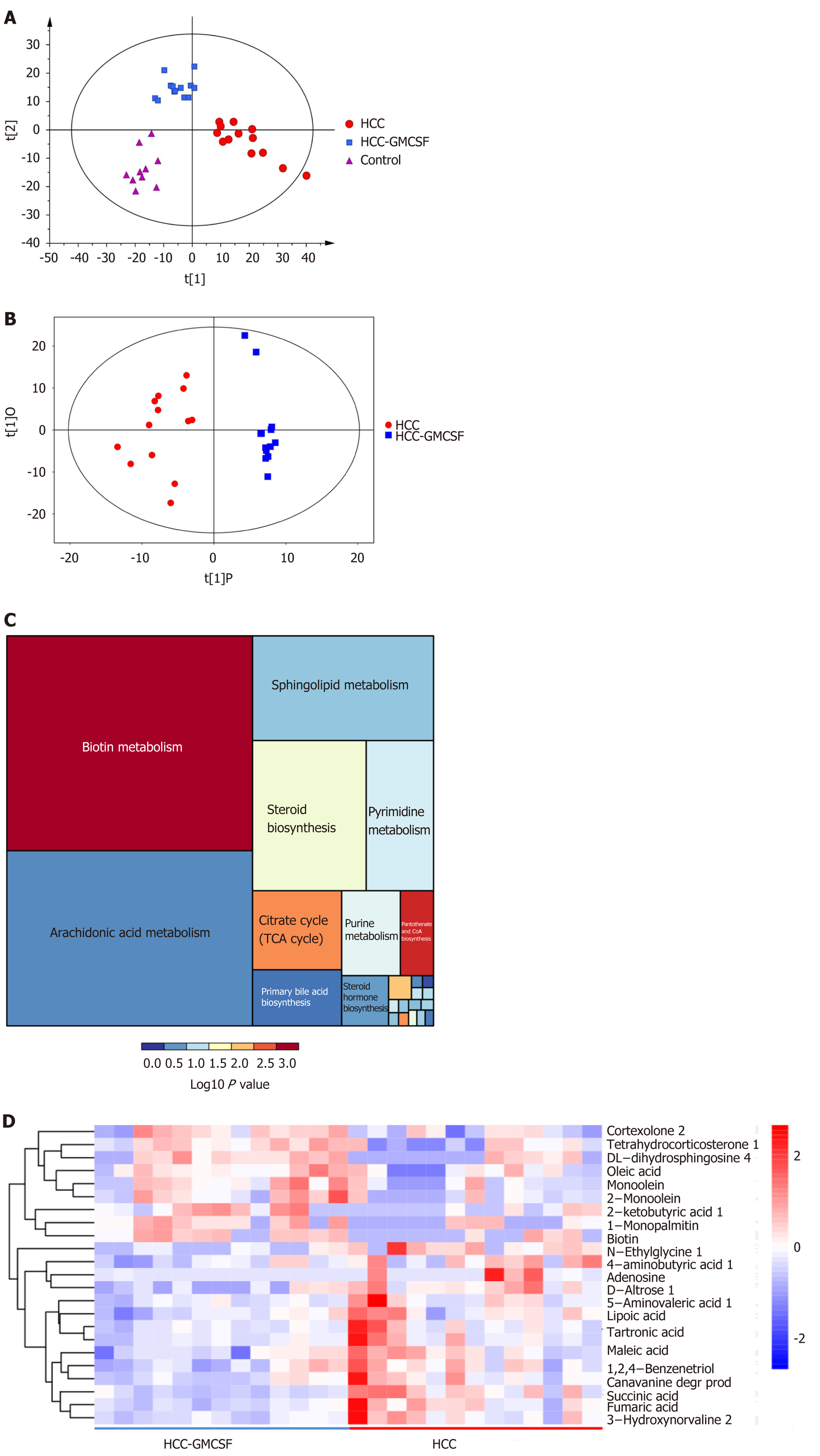
We further elucidated the intestinal dysfunction secondary to HCC through a nontargeted metabolomics study. A total of 305 metabolites included in the following Kyoto Encyclopedia of Genes and Genomes (KEGG) metabolic pathways were identified: Biotin (vitamin) anabolic, sphingolipid, steroid anabolic, pyrimidine, citric acid cycle (TCA cycle), purine, and primary bile acid metabolism (Figure 5C). A corresponding OPLS-DA analysis was performed. As shown in Figure 5A, the score graph reveals that the metabolomic profiles from each group are clearly clustered. We further established the OPLS-DA model corresponding to the HCC and HCC + GM-CSF groups; the results demonstrated that the HCC and HCC + GM-CSF groups could be clearly divided (Figure 5B). A total of 69 metabolites were screened; we identified significantly higher concentrations of biotin and oleic acid in the HCC + GM-CSF group. These two metabolites were involved in biotin and lipid metabolism, respectively. Likewise, succinic acid, fumaric acid, adenosine, and maleic acid were detected at significantly reduced concentrations in samples from the HCC + GM-CSF group (Figure 5D).
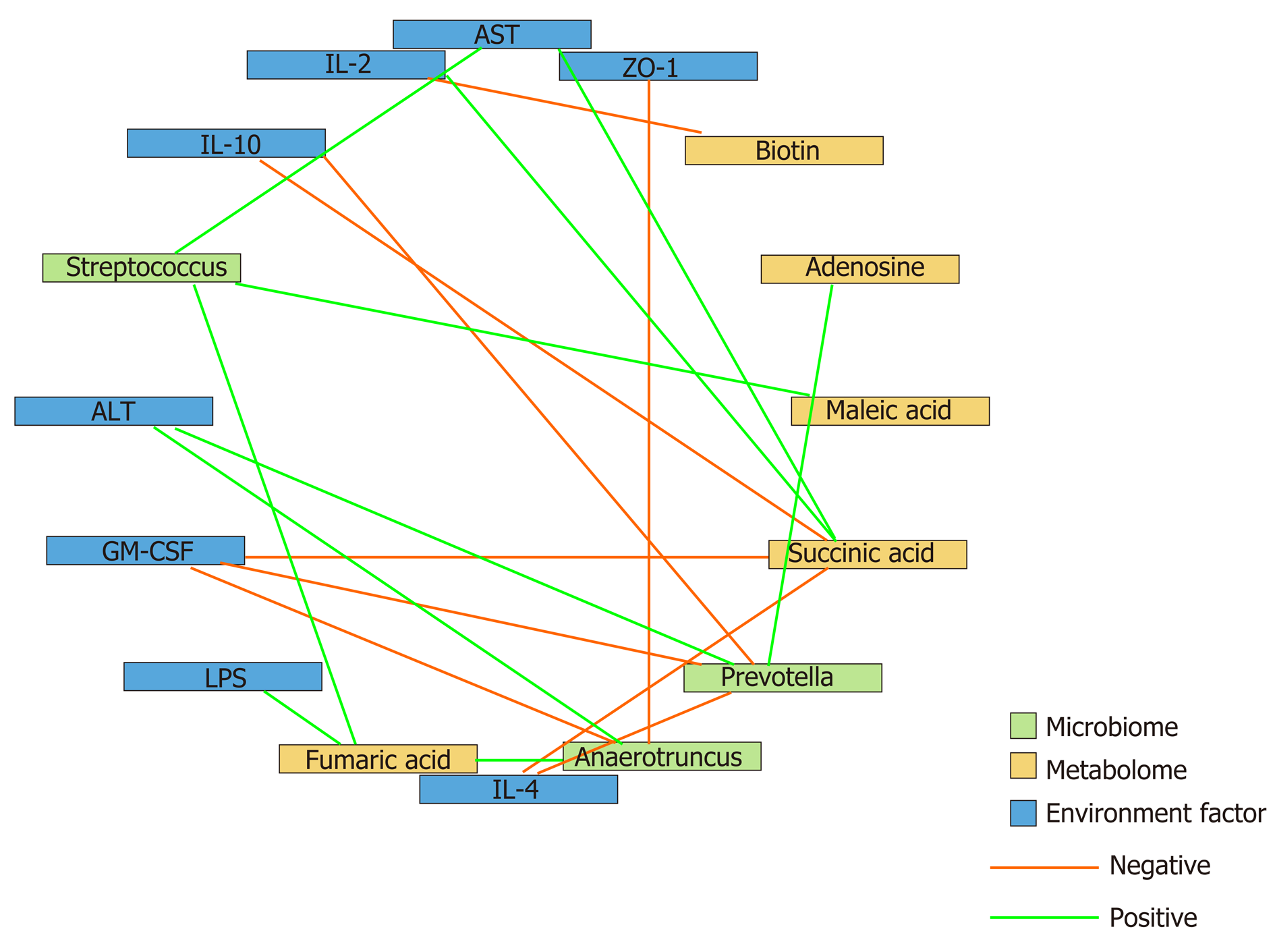
To determine the beneficial impact of GM-CSF on intestinal microecology, we conducted a correlation analysis of the interaction matrix. As part of the analysis, we defined the following: Representative microbial genera with significant alterations in abundance in the HCC and HCC + GM-CSF groups, metabolites detected at significantly different concentrations in the aforementioned two groups, and other representative parameters related to liver and intestine function (Figure 6). GM-CSF overexpression can promote remodeling of the intestinal microecology and function resulting from the development of HCC. A significant reduction in the abundance of Prevotella in the HCC + GM-CSF group positively correlated with serum ALT and LPS levels and inversely correlated with serum IL-10 and GM-CSF levels. Additionally, the abundance of bacteria of the Prevotella genus positively correlated with succinic acid levels, with the latter being significantly higher in fecal samples from the HCC group. Similarly, the abundance of the bacterial genus Anaerotruncus, which may be involved in the inflammatory response, was strongly positively correlated with serum ALT levels and inversely correlated with ZO-1 levels and IL-4 expression. Biotin levels, which were high in samples in response to GM-CSF overexpression, inversely correlated with IL-2 expression. Likewise, succinic acid, which was detected at relatively low levels in the fecal samples of mice in the HCC + GM-CSF group, positively correlated with ALT, AST, and LPS levels and inversely correlated with IL-4 and IL-10 expression. Taken together, these data provide evidence that GM-CSF plays a dominant role in remodeling the aberrant intestinal microecology and function that develop in response to HCC.
This study demonstrated that GM-CSF overexpression can repair intestinal dysbiosis, liver damage, inflammation, and damage to liver function induced by HCC. Specifically, several complementary GM-CSF-mediated mechanisms have been revealed, including a role in (1) protecting the intestinal barrier; (2) regulating biotin and lipid metabolism; (3) attenuating inflammation; and (4) reducing endotoxin levels.
Enterohepatic recycling depends heavily on the nature and function of the intestinal micro-organisms; the intestinal flora also plays a critical role in protecting against infections[28]. TLR signaling triggers the initial inflammatory response by releasing cytokines, which may promote intestinal dysbiosis along with secondary TLR activation and signaling[29]. In this study, we found that GM-CSF overexpression resulted in significant downregulation of TLR4 expression in both liver and colon tissues. These findings suggest that GM-CSF provides protection from HCC-mediated destruction of the intestinal barrier and limits the inflammatory response secondary to LPS-mediated TLR4 signaling. The gut microbiota and metabolic products are translocated to the liver via the portal circulation, consequently activating TLR4 in the immune cells localized in the hepatic tissue. Emerging studies have shown that intestinal mucosal damage and epithelial barrier disruption result in bacterial translocation and overgrowth; as such, endotoxins can accumulate in the liver. As the liver loses its toxin clearing ability, endotoxins will find their way into the systemic circulation[30]. This scenario has been reported in patients with alcoholic fatty liver in which significant elevations in systemic LPS have been detected[13]. In this study, we found that LPS levels were significantly elevated in the serum of mice with HCC; these levels were markedly reduced in mice with HCC and GM-CSF overexpression. Recent studies have shown that elevated LPS levels in patients with liver injury are mainly due to impaired intestinal barrier function and intestinal dysbiosis[31]. LPS is a pathogen-associated molecular pattern molecule that can trigger severe inflammatory responses both directly and indirectly[14]. Taken together, our data suggest that GM-CSF may limit LPS-mediated activation of the TLR4 signaling pathway, thereby protecting against the inflammatory mic-roenvironment generated in response to HCC.
Previous studies have revealed that Bacillus subtilis promotes the expression of the tight junction protein ZO-1, alters intestinal immune activity, and affects the integrity of the intestinal barrier[32,33]. In this study, we detected ZO-1 expression via immu-nohistochemistry and gene transcription analysis, and the results revealed that mice with HCC experienced intestinal barrier dysfunction, as indicated by a significant decrease in ZO-1 expression. Conversely, GM-CSF resulted in an increase in ZO-1 expression. Overall, our results indicate that GM-CSF overexpression protects against HCC-mediated disruption of intestinal barrier function.
Tumor growth typically takes place in a complex environment with cytokines, chemokines, and extracellular matrix components, which are collectively considered to be the “tumor microenvironment”[34]. HCC is a typical inflammation-related tumor; its initiation and development are closely associated with a specific tumor mic-roenvironment[35]. Pollard et al[36] reported that macrophages in the tumor mic-roenvironment are polarized into M1 and M2 phenotypes in response to GM-CSF. Macrophages can be recruited to tumor tissues where they can modulate and regulate the local immune response[37]. In this study, we found that liver and systemic inflammatory responses were ameliorated in response to GM-CSF overexpression; these observations may be directly related to the impact of GM-CSF on macrophage activation. In addition, we found that serum levels of the proinflammatory cytokines IL-1β and IL-2 and the liver enzymes ALT and AST were significantly downregulated in response to GM-CSF overexpression, whereas the expression of the anti-inflammatory cytokines IL-4 and IL-10 was increased. These findings suggest that liver function was protected by GM-CSF overexpression and that liver cell damage and the associated inflammation were reduced.
A previous study has shown that translocation of intestinal flora results in chronic inflammation in the liver, thereby exacerbating HCC[38]. To generate an accurate evaluation of the structural characteristics of the intestinal flora, we determined the absolute copy number of the 16S rRNA genes in fecal samples from control mice and mice with HCC with or without GM-CSF overexpression. We identified an increase in the overall abundance of Tenericutes associated with hepatopancreatic necrosis disease in mice with HCC. In contrast, GM-CSF overexpression in association with HCC led to a 63% reduction in the overall abundance of bacteria of this phylum. It is also worth noting that GM-CSF overexpression in association with HCC resulted in a reduction in the overall abundance of Parabacteroide, Prevotella, Streptococcus, and Clostridium spp. and increased the overall abundance of anti-inflammatory bacterial species, including Blautia, Butyricimonass, and Roseburia[39-41].
Recent research findings have suggested that patients with type 2 diabetes mellitus, inflammatory bowel diseases, colorectal adenocarcinoma, and liver cancer present with moderate dysbiosis, including a net decrease in the anti-inflammatory bacterial genera and an increase in various potential pathogens[42-45]. These results parallel our findings on the beneficial effects of GM-CSF overexpression in association with HCC.
In our previous research, we found an increase in the abundance of Prevotella in the feces of patients with HCC compared with healthy volunteers[17]. Prevotella is a conditional pathogen that was originally identified in the oral cavity and vagina; Prevotella spp. is associated with periodontal disease. The identification of a large number of these bacteria in the feces of patients with HCC suggests translocation of oral bacteria to the gastrointestinal tract. The bacterial genomes of Prevotella spp. encode superoxide reductase, which facilitates resistance to host-derived reactive oxygen species and promotes intestinal inflammation by generating thioredoxin[46]. In this study, we found that Prevotella positively correlated with ALT, IL-2, and LPS levels and inversely correlated with IL-10 and GM-CSF; this finding is consistent with the conclusions from the aforementioned previous studies. Absolute numbers of bacteria from the phylum Anaerotruncus were also present in significantly reduced numbers in the fecal samples of mice overexpressing GM-CSF; these bacteria may promote both intestinal and hepatic inflammatory responses. Anaerotruncus levels strongly positively correlated with serum ALT levels and inversely correlated with the expression of the intestinal barrier function protein ZO-1 and the anti-inflammatory factor IL-4. Overall, these results imply that bacterial populations responding significantly to GM-CSF overexpression may be those regulating inflammation, immunity, and barrier functions.
Microbes are connected to each other via complex metabolic chains. Studying microbial metabolism in the intestines may help in determining the interactions among the members of the microbial community. Therefore, to achieve a better understanding of the protective effects of GM-CSF against HCC, we integrated metabolomics into this study. We found that GM-CSF overexpression was associated with a significant increase in biotin and oleic acid levels and a concomitant decrease in succinic acid, adenosine, fumaric acid, lipoic acid, and maleic acid levels. As a group, these metabolites are closely related to biotin and lipid metabolism pathways. Biotin is a vitamin synthesized by intestinal microorganisms[47] that enhanced expression of glucokinase in rat liver[48]. When biotin levels are low, protein synthesis in the liver and intestinal mucosa is inhibited; protein synthesis is restored after biotin sup-plementation[49]. Likewise, succinic acid levels were diminished in the fecal samples of GM-CSF-overexpressing mice. Earlier studies have reported that succinic acid is a toxic factor directly associated with inflammation and disease[50]. However, no changes in bile acids were detected in our study. This may be because our research used a GC-MS detection platform to collect data and the Fiehn and National Institute of Standards and Technology (NIST) databases to determine the nature of the substances. Because the database of the GC platform itself contains relatively few bile acids, there are no qualitative bile acids available. In the future, the LC platform will be used to detect related bile acid substances, and relevant research will be conducted. We found that succinic acid levels positively correlated with serum ALT, AST, and LPS levels and inversely correlated with IL-4 and IL-10 levels; these findings are consistent with previous results. Interestingly, Prevotella are succinic acid-producing bacteria, explaining the positive correlation between succinic acid and Prevotella. GM-CSF overexpression resulted in the reduced abundance of Prevotella[51], possibly constituting an effective feedback loop in the intestinal microecosystem, serving to magnify the protective effects of GM-CSF overexpression.
In summary, we have demonstrated that GM-CSF overexpression can ameliorate intestinal dysbiosis, attenuate inflammation, and reduce endotoxin levels associated with HCC pathogenesis. In light of our findings, a more comprehensive understanding of the impact of GM-CSF in this setting might lead to new insights into the treatment for HCC.
Granulocyte-macrophage colony-stimulating factor (GM-CSF) plays a contributing role in the pathogenesis of hepatocellular carcinoma (HCC) progression, and GM-CSF is a critical factor in promoting intestinal immunity.
GM-CSF may protect against the development of HCC by regulating immunity as well as the intestinal microecology.
To investigate the impact of GM-CSF on the gut microbiome and metabolic characteristics of HCC.
We utilized established mouse models of HCC and overexpressed GM-CSF in HCC. Liver injury, intestinal barrier function, immune inflammation and the fecal microbiome and metabolome were studied.
Overexpression of GM-CSF had a significant impact on the gut microbiome of mice with HCC and resulted in a high abundance of organisms from the genera Roseburia, Blautia and Butyricimonass, along with a significant reduction in Prevotella, Parabacteroides, Anaerotruncus, Streptococcus, Clostridium, and Mucispirillum. GM-CSF overexpression resulted in a substantial increase in fecal biotin and oleic acid, along with a prominent decrease in the fecal levels of succinic acid, adenosine, fumaric acid, lipoic acid, and maleic acid. The intestinal microbiota and fecal metabolites induced by GM-CSF are primarily involved in the pathways related to the reduction in the inflammatory response, biotin metabolism and intestinal barrier dysfunction.
GM-CSF protects mice against HCC by ameliorating intestinal dysbiosis and attenuating inflammation.
Our findings indicate that GM-CSF can protect against the development of HCC by regulating immunity as well as by modulating the abundance of specific intestinal micro-organisms and their metabolites. This study provides new insights into the treatment of HCC.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): A, A
Grade B (Very good): 0
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Mukherjee S S-Editor: Gong ZM L-Editor: MedE-Ma JY P-Editor: Zhang YL
| 1. | Villanueva A. Hepatocellular Carcinoma. N Engl J Med. 2019;380:1450-1462. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2066] [Cited by in RCA: 3176] [Article Influence: 529.3] [Reference Citation Analysis (37)] |
| 2. | Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67:7-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11065] [Cited by in RCA: 12187] [Article Influence: 1523.4] [Reference Citation Analysis (3)] |
| 3. | Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115-132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11444] [Cited by in RCA: 13214] [Article Influence: 1468.2] [Reference Citation Analysis (3)] |
| 4. | Yu LX, Schwabe RF. The gut microbiome and liver cancer: mechanisms and clinical translation. Nat Rev Gastroenterol Hepatol. 2017;14:527-539. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 334] [Cited by in RCA: 414] [Article Influence: 51.8] [Reference Citation Analysis (0)] |
| 5. | Rooks MG, Garrett WS. Gut microbiota, metabolites and host immunity. Nat Rev Immunol. 2016;16:341-352. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1428] [Cited by in RCA: 2249] [Article Influence: 281.1] [Reference Citation Analysis (1)] |
| 6. | Tremaroli V, Bäckhed F. Functional interactions between the gut microbiota and host metabolism. Nature. 2012;489:242-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2779] [Cited by in RCA: 3085] [Article Influence: 237.3] [Reference Citation Analysis (0)] |
| 7. | Wisse E, Braet F, Luo D, De Zanger R, Jans D, Crabbé E, Vermoesen A. Structure and function of sinusoidal lining cells in the liver. Toxicol Pathol. 1996;24:100-111. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 204] [Cited by in RCA: 175] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 8. | Mikulak J, Bruni E, Oriolo F, Di Vito C, Mavilio D. Hepatic Natural Killer Cells: Organ-Specific Sentinels of Liver Immune Homeostasis and Physiopathology. Front Immunol. 2019;10:946. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 63] [Cited by in RCA: 105] [Article Influence: 17.5] [Reference Citation Analysis (0)] |
| 9. | Robinson MW, Harmon C, O'Farrelly C. Liver immunology and its role in inflammation and homeostasis. Cell Mol Immunol. 2016;13:267-276. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 682] [Cited by in RCA: 775] [Article Influence: 86.1] [Reference Citation Analysis (0)] |
| 10. | Dorrestein PC, Mazmanian SK, Knight R. Finding the missing links among metabolites, microbes, and the host. Immunity. 2014;40:824-832. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 206] [Cited by in RCA: 233] [Article Influence: 21.2] [Reference Citation Analysis (0)] |
| 11. | Abu-Shanab A, Quigley EM. The role of the gut microbiota in nonalcoholic fatty liver disease. Nat Rev Gastroenterol Hepatol. 2010;7:691-701. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 332] [Cited by in RCA: 354] [Article Influence: 23.6] [Reference Citation Analysis (0)] |
| 12. | Spruss A, Kanuri G, Wagnerberger S, Haub S, Bischoff SC, Bergheim I. Toll-like receptor 4 is involved in the development of fructose-induced hepatic steatosis in mice. Hepatology. 2009;50:1094-1104. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 403] [Cited by in RCA: 438] [Article Influence: 27.4] [Reference Citation Analysis (0)] |
| 13. | Uhrig A, Banafsche R, Kremer M, Hegenbarth S, Hamann A, Neurath M, Gerken G, Limmer A, Knolle PA. Development and functional consequences of LPS tolerance in sinusoidal endothelial cells of the liver. J Leukoc Biol. 2005;77:626-633. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 130] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 14. | Atarashi K, Nishimura J, Shima T, Umesaki Y, Yamamoto M, Onoue M, Yagita H, Ishii N, Evans R, Honda K, Takeda K. ATP drives lamina propria T(H)17 cell differentiation. Nature. 2008;455:808-812. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 815] [Cited by in RCA: 866] [Article Influence: 50.9] [Reference Citation Analysis (0)] |
| 15. | Ma C, Han M, Heinrich B, Fu Q, Zhang Q, Sandhu M, Agdashian D, Terabe M, Berzofsky JA, Fako V, Ritz T, Longerich T, Theriot CM, McCulloch JA, Roy S, Yuan W, Thovarai V, Sen SK, Ruchirawat M, Korangy F, Wang XW, Trinchieri G, Greten TF. Gut microbiome-mediated bile acid metabolism regulates liver cancer via NKT cells. Science. 2018;360. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 549] [Cited by in RCA: 1038] [Article Influence: 148.3] [Reference Citation Analysis (0)] |
| 16. | Ren Z, Li A, Jiang J, Zhou L, Yu Z, Lu H, Xie H, Chen X, Shao L, Zhang R, Xu S, Zhang H, Cui G, Chen X, Sun R, Wen H, Lerut JP, Kan Q, Li L, Zheng S. Gut microbiome analysis as a tool towards targeted non-invasive biomarkers for early hepatocellular carcinoma. Gut. 2019;68:1014-1023. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 542] [Cited by in RCA: 509] [Article Influence: 84.8] [Reference Citation Analysis (0)] |
| 17. | Zhang L, Wu YN, Chen T, Ren CH, Li X, Liu GX. Relationship between intestinal microbial dysbiosis and primary liver cancer. Hepatobiliary Pancreat Dis Int. 2019;18:149-157. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 56] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 18. | Egea L, McAllister CS, Lakhdari O, Minev I, Shenouda S, Kagnoff MF. GM-CSF produced by nonhematopoietic cells is required for early epithelial cell proliferation and repair of injured colonic mucosa. J Immunol. 2013;190:1702-1713. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 58] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 19. | Zhan Y, Xu Y, Lew AM. The regulation of the development and function of dendritic cell subsets by GM-CSF: more than a hematopoietic growth factor. Mol Immunol. 2012;52:30-37. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 66] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 20. | Keren-Shaul H, Spinrad A, Weiner A, Matcovitch-Natan O, Dvir-Szternfeld R, Ulland TK, David E, Baruch K, Lara-Astaiso D, Toth B, Itzkovitz S, Colonna M, Schwartz M, Amit I. A Unique Microglia Type Associated with Restricting Development of Alzheimer's Disease. Cell. 2017;169:1276-1290.e17. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2095] [Cited by in RCA: 3447] [Article Influence: 430.9] [Reference Citation Analysis (1)] |
| 21. | Kind T, Wohlgemuth G, Lee DY, Lu Y, Palazoglu M, Shahbaz S, Fiehn O. FiehnLib: mass spectral and retention index libraries for metabolomics based on quadrupole and time-of-flight gas chromatography/mass spectrometry. Anal Chem. 2009;81:10038-10048. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 964] [Cited by in RCA: 1033] [Article Influence: 68.9] [Reference Citation Analysis (0)] |
| 22. | Dunn WB, Broadhurst D, Begley P, Zelena E, Francis-McIntyre S, Anderson N, Brown M, Knowles JD, Halsall A, Haselden JN, Nicholls AW, Wilson ID, Kell DB, Goodacre R; Human Serum Metabolome (HUSERMET) Consortium. Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat Protoc. 2011;6:1060-1083. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2495] [Cited by in RCA: 2108] [Article Influence: 150.6] [Reference Citation Analysis (0)] |
| 23. | Zhu L, Baker SS, Gill C, Liu W, Alkhouri R, Baker RD, Gill SR. Characterization of gut microbiomes in nonalcoholic steatohepatitis (NASH) patients: a connection between endogenous alcohol and NASH. Hepatology. 2013;57:601-609. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1015] [Cited by in RCA: 1283] [Article Influence: 106.9] [Reference Citation Analysis (1)] |
| 24. | Zhu L, Baker RD, Zhu R, Baker SS. Sequencing the gut metagenome as a noninvasive diagnosis for advanced nonalcoholic steatohepatitis. Hepatology. 2017;66:2080-2083. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 5] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 25. | Tremellen K, McPhee N, Pearce K. Metabolic endotoxaemia related inflammation is associated with hypogonadism in overweight men. Basic Clin Androl. 2017;27:5. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 31] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 26. | Tilg H, Diehl AM. Cytokines in alcoholic and nonalcoholic steatohepatitis. N Engl J Med. 2000;343:1467-1476. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 688] [Cited by in RCA: 685] [Article Influence: 27.4] [Reference Citation Analysis (0)] |
| 27. | Csak T, Velayudham A, Hritz I, Petrasek J, Levin I, Lippai D, Catalano D, Mandrekar P, Dolganiuc A, Kurt-Jones E, Szabo G. Deficiency in myeloid differentiation factor-2 and toll-like receptor 4 expression attenuates nonalcoholic steatohepatitis and fibrosis in mice. Am J Physiol Gastrointest Liver Physiol. 2011;300:G433-G441. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 170] [Cited by in RCA: 198] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 28. | Schubert K, Olde Damink SWM, von Bergen M, Schaap FG. Interactions between bile salts, gut microbiota, and hepatic innate immunity. Immunol Rev. 2017;279:23-35. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 70] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 29. | Szabo G, Velayudham A, Romics L, Mandrekar P. Modulation of non-alcoholic steatohepatitis by pattern recognition receptors in mice: the role of toll-like receptors 2 and 4. Alcohol Clin Exp Res. 2005;29:140S-145S. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 112] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 30. | Velloso LA, Folli F, Saad MJ. TLR4 at the Crossroads of Nutrients, Gut Microbiota, and Metabolic Inflammation. Endocr Rev. 2015;36:245-271. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 177] [Cited by in RCA: 207] [Article Influence: 20.7] [Reference Citation Analysis (0)] |
| 31. | Ye J, Lv L, Wu W, Li Y, Shi D, Fang D, Guo F, Jiang H, Yan R, Ye W, Li L. Butyrate Protects Mice Against Methionine-Choline-Deficient Diet-Induced Non-alcoholic Steatohepatitis by Improving Gut Barrier Function, Attenuating Inflammation and Reducing Endotoxin Levels. Front Microbiol. 2018;9:1967. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 96] [Cited by in RCA: 149] [Article Influence: 21.3] [Reference Citation Analysis (0)] |
| 32. | Derrien M, van Hylckama Vlieg JE. Fate, activity, and impact of ingested bacteria within the human gut microbiota. Trends Microbiol. 2015;23:354-366. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 343] [Cited by in RCA: 394] [Article Influence: 39.4] [Reference Citation Analysis (0)] |
| 33. | Gu JF, Su SL, Guo J, Zhu Y, Zhao M, Duan J. The aerial parts of Salvia miltiorrhiza Bge. strengthen intestinal barrier and modulate gut microbiota imbalance in streptozocin-induced diabetic mice. J Funct Foods. 2017;36:362-374. [RCA] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 32] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 34. | Sundaram S, Johnson AR, Makowski L. Obesity, metabolism and the microenvironment: Links to cancer. J Carcinog. 2013;12:19. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 76] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 35. | Shirabe K, Mano Y, Muto J, Matono R, Motomura T, Toshima T, Takeishi K, Uchiyama H, Yoshizumi T, Taketomi A, Morita M, Tsujitani S, Sakaguchi Y, Maehara Y. Role of tumor-associated macrophages in the progression of hepatocellular carcinoma. Surg Today. 2012;42:1-7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 136] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 36. | Pollard JW. Tumour-educated macrophages promote tumour progression and metastasis. Nat Rev Cancer. 2004;4:71-78. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2416] [Cited by in RCA: 2566] [Article Influence: 122.2] [Reference Citation Analysis (0)] |
| 37. | Shalapour S, Lin XJ, Bastian IN, Brain J, Burt AD, Aksenov AA, Vrbanac AF, Li W, Perkins A, Matsutani T, Zhong Z, Dhar D, Navas-Molina JA, Xu J, Loomba R, Downes M, Yu RT, Evans RM, Dorrestein PC, Knight R, Benner C, Anstee QM, Karin M. Inflammation-induced IgA+ cells dismantle anti-liver cancer immunity. Nature. 2017;551:340-345. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 259] [Cited by in RCA: 425] [Article Influence: 53.1] [Reference Citation Analysis (0)] |
| 38. | Ringelhan M, Pfister D, O'Connor T, Pikarsky E, Heikenwalder M. The immunology of hepatocellular carcinoma. Nat Immunol. 2018;19:222-232. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 440] [Cited by in RCA: 736] [Article Influence: 105.1] [Reference Citation Analysis (0)] |
| 39. | Schirmer M, Garner A, Vlamakis H, Xavier RJ. Microbial genes and pathways in inflammatory bowel disease. Nat Rev Microbiol. 2019;17:497-511. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 249] [Cited by in RCA: 594] [Article Influence: 118.8] [Reference Citation Analysis (0)] |
| 40. | García-Agudo L, Nilsen E. Butyricimonas virosa: A rare cause of bacteremia. Anaerobe. 2018;54:121-123. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 8] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 41. | Wang K, Liao M, Zhou N, Bao L, Ma K, Zheng Z, Wang Y, Liu C, Wang W, Wang J, Liu SJ, Liu H. Parabacteroides distasonis Alleviates Obesity and Metabolic Dysfunctions via Production of Succinate and Secondary Bile Acids. Cell Rep. 2019;26:222-235.e5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 373] [Cited by in RCA: 751] [Article Influence: 150.2] [Reference Citation Analysis (1)] |
| 42. | Zhong H, Ren H, Lu Y, Fang C, Hou G, Yang Z, Chen B, Yang F, Zhao Y, Shi Z, Zhou B, Wu J, Zou H, Zi J, Chen J, Bao X, Hu Y, Gao Y, Zhang J, Xu X, Hou Y, Yang H, Wang J, Liu S, Jia H, Madsen L, Brix S, Kristiansen K, Liu F, Li J. Distinct gut metagenomics and metaproteomics signatures in prediabetics and treatment-naïve type 2 diabetics. EBioMedicine. 2019;47:373-383. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 126] [Article Influence: 21.0] [Reference Citation Analysis (0)] |
| 43. | Vich Vila A, Imhann F, Collij V, Jankipersadsing SA, Gurry T, Mujagic Z, Kurilshikov A, Bonder MJ, Jiang X, Tigchelaar EF, Dekens J, Peters V, Voskuil MD, Visschedijk MC, van Dullemen HM, Keszthelyi D, Swertz MA, Franke L, Alberts R, Festen EAM, Dijkstra G, Masclee AAM, Hofker MH, Xavier RJ, Alm EJ, Fu J, Wijmenga C, Jonkers DMAE, Zhernakova A, Weersma RK. Gut microbiota composition and functional changes in inflammatory bowel disease and irritable bowel syndrome. Sci Transl Med. 2018;10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 208] [Cited by in RCA: 379] [Article Influence: 63.2] [Reference Citation Analysis (0)] |
| 44. | Zhang Y, Yu X, Yu E, Wang N, Cai Q, Shuai Q, Yan F, Jiang L, Wang H, Liu J, Chen Y, Li Z, Jiang Q. Changes in gut microbiota and plasma inflammatory factors across the stages of colorectal tumorigenesis: a case-control study. BMC Microbiol. 2018;18:92. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 56] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 45. | Ponziani FR, Bhoori S, Castelli C, Putignani L, Rivoltini L, Del Chierico F, Sanguinetti M, Morelli D, Paroni Sterbini F, Petito V, Reddel S, Calvani R, Camisaschi C, Picca A, Tuccitto A, Gasbarrini A, Pompili M, Mazzaferro V. Hepatocellular Carcinoma Is Associated With Gut Microbiota Profile and Inflammation in Nonalcoholic Fatty Liver Disease. Hepatology. 2019;69:107-120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 300] [Cited by in RCA: 475] [Article Influence: 79.2] [Reference Citation Analysis (1)] |
| 46. | Maeda Y, Takeda K. Role of Gut Microbiota in Rheumatoid Arthritis. J Clin Med. 2017;6. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 121] [Cited by in RCA: 157] [Article Influence: 19.6] [Reference Citation Analysis (0)] |
| 47. | Agrawal S, Agrawal A, Said HM. Biotin deficiency enhances the inflammatory response of human dendritic cells. Am J Physiol Cell Physiol. 2016;311:C386-C391. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 86] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 48. | Chauhan J, Dakshinamurti K. Transcriptional regulation of the glucokinase gene by biotin in starved rats. J Biol Chem. 1991;266:10035-10038. [PubMed] |
| 49. | Dakshinamurti K, Litvak S. Biotin and protein synthesis in rat liver. J Biol Chem. 1970;245:5600-5605. [PubMed] |
| 50. | Nadjsombati MS, McGinty JW, Lyons-Cohen MR, Jaffe JB, DiPeso L, Schneider C, Miller CN, Pollack JL, Nagana Gowda GA, Fontana MF, Erle DJ, Anderson MS, Locksley RM, Raftery D, von Moltke J. Detection of Succinate by Intestinal Tuft Cells Triggers a Type 2 Innate Immune Circuit. Immunity. 2018;49:33-41.e7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 241] [Cited by in RCA: 397] [Article Influence: 66.2] [Reference Citation Analysis (0)] |
| 51. | Serena C, Ceperuelo-Mallafré V, Keiran N, Queipo-Ortuño MI, Bernal R, Gomez-Huelgas R, Urpi-Sarda M, Sabater M, Pérez-Brocal V, Andrés-Lacueva C, Moya A, Tinahones FJ, Fernández-Real JM, Vendrell J, Fernández-Veledo S. Elevated circulating levels of succinate in human obesity are linked to specific gut microbiota. ISME J. 2018;12:1642-1657. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 291] [Cited by in RCA: 277] [Article Influence: 39.6] [Reference Citation Analysis (0)] |