Published online Apr 21, 2020. doi: 10.3748/wjg.v26.i15.1775
Peer-review started: December 11, 2019
First decision: January 13, 2020
Revised: January 23, 2020
Accepted: March 19, 2020
Article in press: March 19, 2020
Published online: April 21, 2020
Processing time: 132 Days and 0.1 Hours
Esophageal cancer is a common digestive tract tumor that is generally treated with radiotherapy. Poor responses to radiotherapy in most patients generally result in local radiotherapy failure, so it is essential to find new radiosensitizers that can enhance the response of cancer cells to radiotherapy and improve the survival of esophageal cancer patients with radiation resistance. The long non-coding RNA (lncRNA) Rpph1 is highly expressed in human gastric cancer tissues, and represses breast cancer cell proliferation and tumorigenesis. However, the expression of lncRNA Rpph1 in esophageal cancer and its relationship with radio-sensitivity has not been studied.
To explore the value of lncRNA Rpph1 in esophageal cancer and its effect on cancer cell sensitivity to radiotherapy.
Eighty-three patients with esophageal cancer admitted to Qilu Hospital of Shandong University and 90 healthy participants who received physical examinations were collected as research participants. The expression of Rpph1 was determined by qRT-PCR. siRNA-NC and siRNA-Rpph1 were transfected into esophageal cancer cell lines, and cells without transfection were designated as the blank control group. Cell survival was tested by colony formation assays, and the levels of proteins related to apoptosis and epithelial-mesenchymal transitions were determined by Western blot assays. Cell proliferation was assessed by MTT assays, cell apoptosis by flow cytometry, and cell migration by wound-healing assays. Changes in cell cycle distribution were monitored.
Rpph1 was highly expressed in esophageal carcinoma, making it a promising marker for the diagnosis of esophageal cancer. Rpph1 could also be used to distinguish different short-term responses, T stages, N stages, and clinical stages of esophageal cancer patients. The results of 3-year overall survival favored patients with lower Rpph1 expression over patients with higher Rpph1 expression (P < 0.05). In vitro and in vivo experiments showed that silencing Rpph1 expression led to higher sensitivity of esophageal cancer cells to radiotherapy, stronger apoptosis in esophageal cancer cells induced by radiotherapy, higher expression of Bax and caspase-3, and lower expression of Bcl-2 (Bax, caspase-3, and Bcl-2 are apoptosis-related proteins). Additionally, silencing Rpph1 attenuated radiation-induced G2/M phase arrest, and significantly inhibited the expression of proteins involved in cell proliferation, migration, and epithelial-mesenchymal transition regulation in esophageal cancer cells.
Rpph1 is highly expressed in esophageal cancer. Silencing Rpph1 expression can promote cell apoptosis, inhibit cell proliferation and migration, and increase radio-sensitivity.
Core tip: This study investigated the clinical value of the long non-coding RNA Rpph1 in esophageal cancer, and analyzed the relationship between Rpph1 and the pathological parameters and prognosis of esophageal cancer. We also studied the effect of Rpph1 on the sensitivity of esophageal cancer to radiotherapy, on cell apoptosis, proliferation, migration, and cell cycle, as well as its effect on epithelial-mesenchymal transition-related proteins.
- Citation: Li ZY, Li HF, Zhang YY, Zhang XL, Wang B, Liu JT. Value of long non-coding RNA Rpph1 in esophageal cancer and its effect on cancer cell sensitivity to radiotherapy. World J Gastroenterol 2020; 26(15): 1775-1791
- URL: https://www.wjgnet.com/1007-9327/full/v26/i15/1775.htm
- DOI: https://dx.doi.org/10.3748/wjg.v26.i15.1775
Esophageal cancer is the eighth most prevalent cancer worldwide, as well as one of the main causes of death due to cancer in human[1,2]. The annual mortality of esophageal cancer is approximately 100/100000 people, with the highest incidence in Asia, northern France, eastern and southern Africa[3]. Esophageal squamous cell carcinoma and esophageal adenocarcinoma are the two major histological types of esophageal cancer[4]. Due to the lack of specific early symptoms, obvious signs of onset, and early diagnostic markers, most patients with esophageal cancer are already at advanced cancer stages at the time of treatment, demonstrating poor treatment response following surgical resection[5]. Radiotherapy is crucial for the treatment of patients not qualified for operations and those with locally advanced esophageal cancer[6]. Local radiotherapy failure may occur due to radiotherapy resistance or tumor heterogeneity. Persistent or recurrent disease is reported in 40%-60% of esophageal cancer patients receiving radiotherapy, and the prognosis of esophageal cancer is awful[7,8]. Therefore, the search for new radiosensitizers that can enhance radio-sensitivity in tumor cells and improve the survival of esophageal cancer patients with radiation-resistance is clinically urgent[9,10].
The identification of molecular markers may improve the staging of esophageal cancer patients and develop new targeted therapies[11]. Long non-coding RNAs (lncRNAs), over 200 bp in length, have become the focus of public attention[12]. lncRNAs can interact with multiple RNA molecules or proteins both transcriptionally and post-transcriptionally to control the expression of target genes[13-15]. Related studies reported that lncRNA is crucial for dose compensation, epigenetic regulation, and regulation of cell cycle and cell differentiation[16,17]. The lncRNA Rpph1 is abnormally expressed in the neocortical tissue of patients with gastric cancer[18] and epilepsy[19]. A study revealed that lncRNA Rpph1 could inhibit breast cancer cell proliferation and tumor occurrence[20]. The study by Cheng et al[11] showed that XRCC 3 could increase the radio-sensitivity of esophageal cancer, so it is a latent target for treating esophageal cancer. So far, no study has been reported on lncRNA Rpph1 expression in esophageal cancer and its effect on the sensitivity to radiotherapy.
This study measured lncRNA Rpph1 expression in esophageal cancer, aiming to investigate the clinical value of lncRNA Rpph1 in esophageal cancer and its effect on cancer cell sensitivity to radiotherapy.
Eighty-three patients with esophageal cancer admitted to Qilu Hospital of Shandong University were enrolled, including 48 males and 35 females whose average age was (63.28 ± 8.83) years. Meanwhile, 90 healthy people who underwent physical examinations in Qilu Hospital of Shandong University during the same period were also enrolled, including 47 males and females whose average age was (62.65 ± 8.31) years. The general information of the two groups was not markedly different. With detailed descriptions of the experimental content, this study obtained approval from the Ethics Committee of Qilu Hospital of Shandong University. All participants signed a written informed consent. Inclusion criteria: Patients diagnosed with esophageal cancer by cytological and histopathological examination; patients with complete clinical data and good compliance; patients without mental illness; patients with an expected survival of longer than 1 mo; patients with no other malignant tumors. Exclusion criteria: Those with severe organic failure; those undergoing other anti-tumor treatments within 1 mo prior to the treatment; those with autoimmune system defects.
All patients received a concurrent radiotherapy regimen with a total dose of 60-70 Gy, 1.8-2.0 Gy each time, completed within 6-7 wk. All patients finished the complete course of treatment. Regular reexamination and follow-up were conducted. The follow-up was performed every 3 mo for 3 years.
RPMI-1640 medium and fetal bovine serum were provided by GIBCO, Inc. (item numbers: 31800002, 10099141). TRIzol reagent was from Invitrogen (Carlsbad, CA, United States), 0.25% trypsin from GIBCO, PBS buffer from Nanjing SenBeiJia Biological Technology Co., Ltd. Dimethyl sulfoxide (DMSO) was provided by Sigma, PVDF membrane by Millipore, DR5000 UV-Vis spectrophotometer by BioRad (Hercules, CA, United States), real-time PCR instrument by ABI (item number: 7500; Foster City, CA, United States). SYBR Premix Ex TaqTM was manufactured by Takara (Shiga, Japan). RIPA buffer was from Shanghai Haling Biological Technology Co. Ltd. (Shanghai, China). BCA Protein Assay Kit was provided by Shanghai Yubo Biological Technology Co. Ltd.. Flow cytometry was from Beckman (Brea, CA, United States), Annexin V-FITC Apoptosis Kit from BestBio. Primary antibodies including casepase-3, Bax, Bcl-2, and β-action were provided by Santa Cruz Biotechnology (Dallas, TX, United States). Vimentin, E-cadherin, N-cadherin, and horseradish peroxidase (HRP)-labeled goat anti-mouse secondary antibody were manufactured by Abcam (Cambridge, United Kingdom). MTT kit was provided by Beijing Baiao Laibo Technology Co. Ltd. ECL luminescence kit and MultiskanTM GO full-wavelength microplate reader by Thermo Fisher Scientific China Co. Ltd. (Shanghai, China).
We collected cancer tissues and adjacent tissues from esophageal cancer patients and stored them in liquid nitrogen. For each patient and healthy participant, 5 mL of fasting elbow vein blood was taken and centrifuged at 3000 × g for 10 min, and then the serum was collected.
Esophageal cancer TE-1 and Kyse150 cell lines were provided by iCell Bioscience, Inc, Shanghai (item numbers: HDCL-040, HDCL-050). TE-1 and Kyse150 cells were cultured in RPMI1640 medium comprised of 10% fetal bovine serum, 100 U/mL penicillin, and 100 µg/mL streptomycin. After cell culture at 37 °C in an incubator with 5% CO2 and saturated humidity, cell passage was performed and cells were cryopreserved. Cells in logarithmic growth phase were harvested.
Grouping of transfection: Blank control group (not transfected), empty vector negative control group (siRNA-NC), Rpph1-silenced group (siRNA-Rpph1), empty vector combined with radiotherapy group (siRNA-NC + IR), Rpph1-silenced combined with radiotherapy group (siRNA-Rpph1 + IR). When the adherent growth of esophageal cancer cells from the TE-1 and Kyse150 lines reached 80%-90%, the transfection was carried out according to the manual of the LipofectamineTM 2000 transfection kit (Invitrogen). After 6 h of transfection, cell culture was performed in a new medium containing 10% fetal bovine serum. Cell transfection efficiency was measured by qRT-PCR. After 24 h of transfection, X-ray irradiation treatment was performed before subsequent experimentation.
TRIzol extraction kit was used to extract total RNA from serum, tissues, and cells. The purity, concentration, and integrity of total RNA were measured by UV spectrophotometer and agarose gel electrophoresis. The RNA was reverse transcribed into cDNA based on the instructions from the reverse transcription kit and was then stored at -20 °C. PCR was conducted with the SYBR Premix Ex TaqTM kit on a real-time PCR instrument, with GAPDH as the internal reference. Forward primer of Rpph1: 5'-CAGACTGGGCAGGAGAAGCC -3', reverse primer of Rpph1: 5'-TCACCTCAGCCATTGAACTCG-3'.Forward primer of GAPDH: 5'-AAGGGTGGAGCCAAAAGGG-3', reverse primer of GAPDH: 5' -TGGGGGT AGGAACACGGAA-3'. PCR amplification conditions: 40 cycles of 95 °C for 30 s, 95 °C for 5 s, 60 °C for 30 s. The experiment was repeated 3 times to obtain the final data, which were calculated using 2-ΔCT.
The esophageal cancer TE-1 and Kyse150 cells in the logarithmic growth phase were digested with trypsin and diluted to a suitable concentration. Cell counting was conducted under a microscope. There were six exposure groups at different doses (0, 2, 4, 6, 8 Gy, respectively). Cell culture was conducted in an incubator at 37 °C, with 5% CO2 and 2 mL of culture solution. After 24 h of adherence, cells received irradiation at various doses. Cells were kept in the incubator after the end of irradiation. When macroscopic clones appeared, cell culture was stopped and PBS washing was conducted twice, followed by fixation with formaldehyde and staining with crystal violet. The number of clones with 50 or more cell was counted under a microscope and the colony formation rate was calculated. Calculation formula: Plating efficiency (PE) = number of clones per well in the blank control group/ number of cells inoculated per well; surviving fraction (SF) = number of clones in an experimental group/(number of cells inoculated × PE). The experiment was done in triplicate to obtain the mean value and a colony formation curve was generated.
RIPA lysis buffer was used to extract the total protein that was then separated by 10% SDS-PAGE. The protein density was determined by BCA method. The separated protein was transferred to a polyvinylidene fluoride membrane, which was performed at constant voltage (100 V) for 100 min and blocked at 37 °C for 60 min. The membrane was then blocked in 5% skim milk, followed by an immune reaction: the membrane was incubated with primary antibody (1:1000) overnight at 4 °C and then incubated with secondary antibody (1:1000) at 37 °C for 1 h. The whole system was rinsed three times with TBST, 5 min each time. After the completion of the immune reaction, color development and fixation were performed using ECL luminescence reagent. Images were acquired using the Quantity One infrared imaging system. The relative expression level of protein = band gray value/internal reference gray value.
After digestion with trypsin, two round of PBS washing followed. After being mixed with 500 μL binding buffer, cells were resuspended and transferred to a flow tube, and then the system was added to 5 μL Annexin V-FITC and 5 μL PI and incubated at room temperature for 5 min away from light. Cell apoptosis was measured on the Kurt CytoFLEX LX flow cytometry system (Beckman). The procedures were conducted in triplicate to obtain the average value.
Cells were seeded in six-well plates and then exposed to irradiation at 0 Gy or 6 Gy according to the grouping of siRNA-NC, siRNA-Rpph1, siRNA-NC + IR, and siRNA-Rpph1 + IR. After 24 h of cell culture, PBS washing was performed twice and cells were mixed with 200 μL PBS to obtain a cell suspension which was fixed in 70% ethanol at 4 °C for 2 h. After centrifugation at 1000 r/min for 5 min, cells were washed twice with PBS and mixed with PI stain and then incubated for 30 min away from light. Finally, the changes in esophageal cancer cell cycle in each transfection group were determined by flow cytometry and Single Histogram Statistics.
Logarithmic growth cells were harvested and trypsinized before centrifugation. Cells were then inoculated into 96-well plates at a concentration of 4000 cells/well. Cells were cultured at 37 °C under 5% CO2 for 24, 48, 72, and 96 h. The upper original medium was discarded, and 20 μL MTT solution and 150 μL dimethyl sulfoxide were added into each well. The plate was shaken for 5-10 min until all purple crystals disappeared. The optical density (OD) value of each well was measured at 450 nm by a microplate reader.
After cell counting, logarithmic growth phase cells were inoculated in plates and cultured in a medium with 2.5% fetal bovine serum. A scratch was made on cells with a sterilized 100 μL disposable pipette tip perpendicular to the horizontal plane and then cells were carefully washed three times with PBS before being transferred to serum-free medium. The scratches of each group were observed under a microscope to evaluate the cell migration at 0 h and 24 h after dissection.
The expression of epithelial-mesenchymal transition (EMT)-related proteins: cytoskeletal protein (Vimentin), epithelial cadherin (E-cadherin), and neuro-cadherin (N-cadherin) levels were determined by Western blot assay following the same steps as described above.
Statistical analysis was performed sing SPSS19.0 (Asia Analytics Formerly SPSS China). The measurement data were expressed with the mean ± SD and compared between any two groups using independent sample t-tests, and between multiple groups using one-way analysis of variance. LSD-t test was used as the post hoc test. The diagnostic value was assessed by the receiver operating characteristic (referred to as ROC) curve. Correlation analysis was performed using the Pearson correlation coefficient. The survival of patients was displayed in a Kaplan-Meier survival curve and compared by Log-rank test. A statistical difference was determined for P < 0.05.
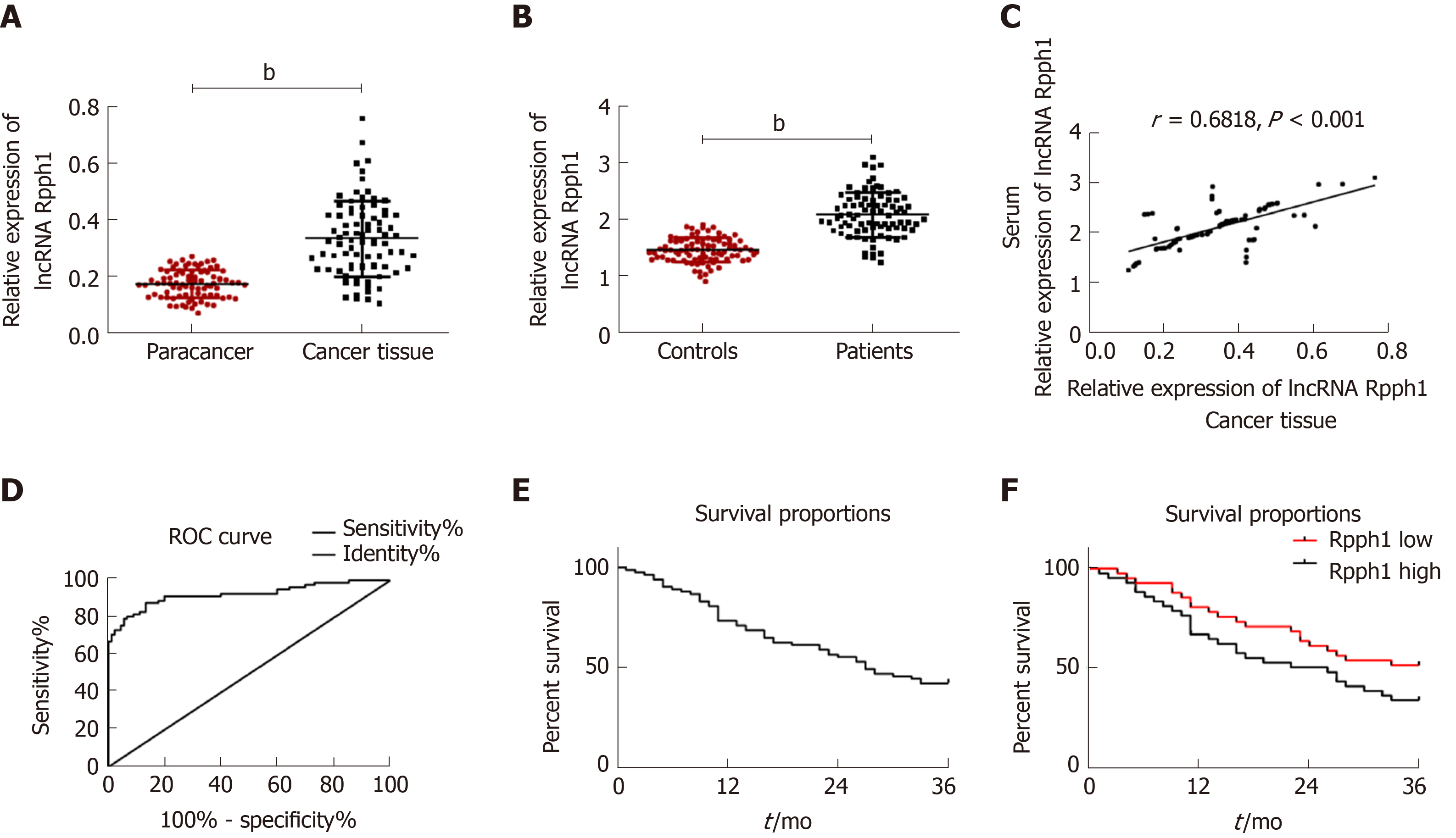
Esophageal cancer tissues had higher lncRNA Rpph1 expression than tumor-adjacent tissues (P < 0.001). LncRNA Rpph1 expression was higher in the serum of esophageal cancer patients than in the serum of healthy participants (P < 0.001). Pearson analysis revealed a positive correlation between lncRNA Rpph1 expression in esophageal cancer tissues and in the serum of esophageal cancer patients (r = 0.6818, P < 0.001). ROC curves demonstrated that serum lncRNA Rpph1 for diagnosing esophageal cancer had an area under the curve (AUC) value of 0.9248, a sensitivity of 87.95%, and a specificity of 86.67%. More details are shown in Figure 1. The follow-up lasted for 3 years and revealed that 3 years of survival was achieved in 35 patients (42.17%). With the median value of serum lncRNA Rpph1 expression (2.060) as the division line, patients with a lncRNA Rpph1 expression higher than 2.060 were assigned into the high expression group (n = 41), and those with an expression equal to or lower than 2.060 were assigned into the low expression group (n = 42). The results of 3-year overall survival favored the low expression group over the high expression group (51.22% vs 33.33%, P = 0.01).
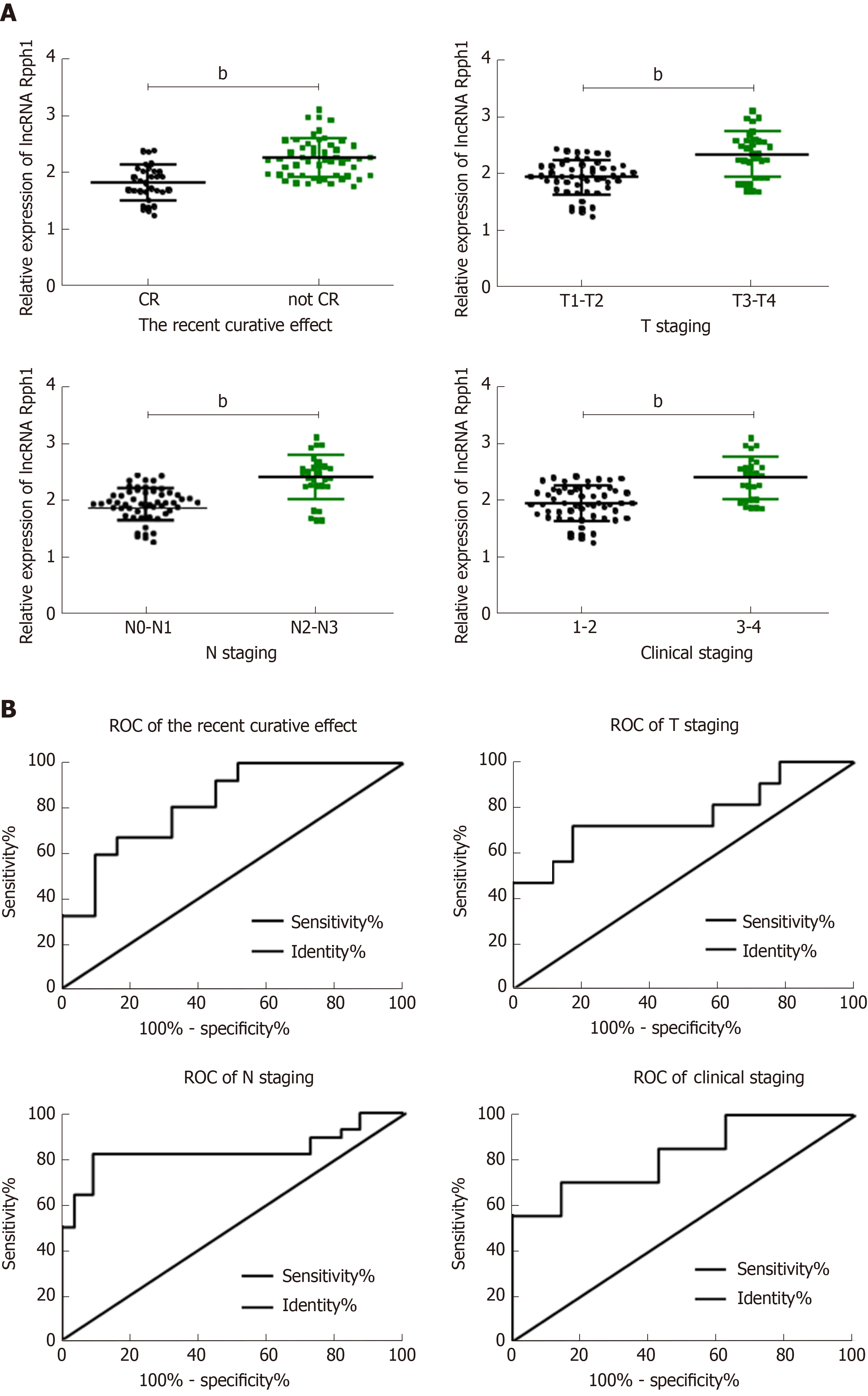
As displayed in Table 1, the expression of lncRNA Rpph1 varied in patients with different short-term responses, T stages, N stages, and other clinical stages (P < 0.05). ROC curves were employed to analyze the diagnostic value of lncRNA Rpph1 for different pathological parameters of esophageal cancer and revealed that lncRNA Rpph1 could be used to distinguish different short-term responses, T stages, N stages, and clinical stages. More details are shown in Figure 2 and Tables 1 and 2.
| n | lncRNA Rpph11 | t | P value | |
| Sex | 0.226 | 0.822 | ||
| Male | 48 | 2.10 ± 0.43 | ||
| Female | 35 | 2.08 ± 0.35 | ||
| Age | 0.685 | 0.460 | ||
| < 60 | 36 | 2.06 ± 0.39 | ||
| ≥ 60 | 47 | 2.12 ± 0.40 | ||
| Smoking | 0.396 | 0.693 | ||
| Yes | 29 | 2.12 ± 0.48 | ||
| No | 54 | 2.08 ± 0.35 | ||
| Short-term response | 5.828 | < 0.001 | ||
| CR | 31 | 1.82 ± 0.32 | ||
| Not CR | 52 | 2.26 ± 0.34 | ||
| T stage | 5.190 | < 0.001 | ||
| T1-T2 | 51 | 1.94 ± 0.30 | ||
| T3-T4 | 32 | 2.34 ± 0.40 | ||
| N stage | 6.121 | < 0.001 | ||
| N0-N1 | 55 | 1.94 ± 0.29 | ||
| N2-N3 | 28 | 2.41 ± 0.40 | ||
| Clinical stage | 5.749 | < 0.001 | ||
| I-II | 56 | 1.95 ± 0.31 | ||
| III-IV | 27 | 2.40 ± 0.38 |
| Pathological parameters | AUC | 95%CI | Sensitivity | Specificity | Cut-off |
| Short-term response | 0.826 | 0.735-0.918 | 67.31% | 83.87% | > 2.093 |
| T stage | 0.765 | 0.651-0.879 | 71.88% | 82.35% | > 2.210 |
| N stage | 0.835 | 0.720-0.950 | 82.14% | 90.91% | > 2.254 |
| Clinical stage | 0.823 | 0.723-0.923 | 70.37% | 85.71% | > 2.255 |
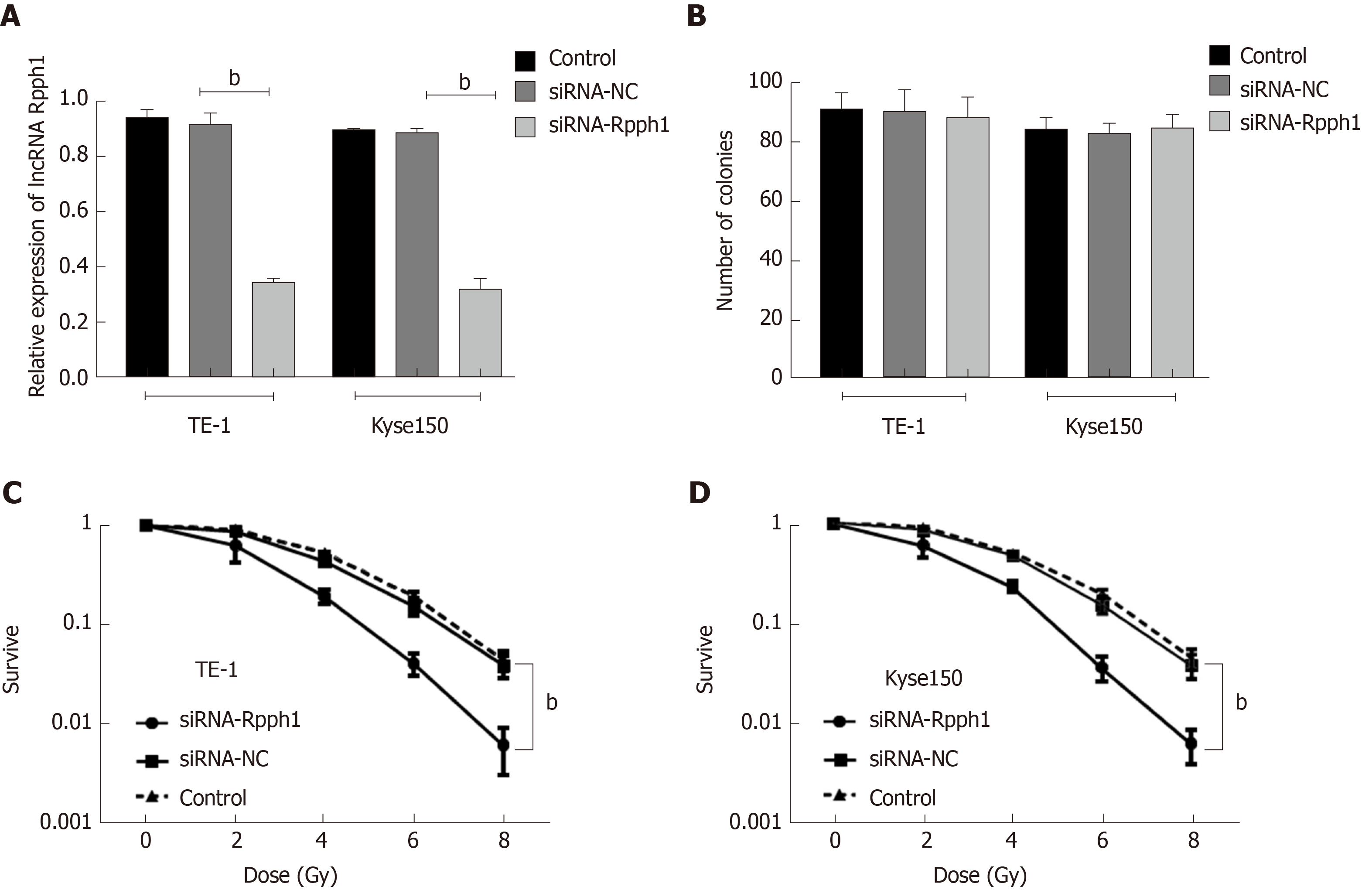
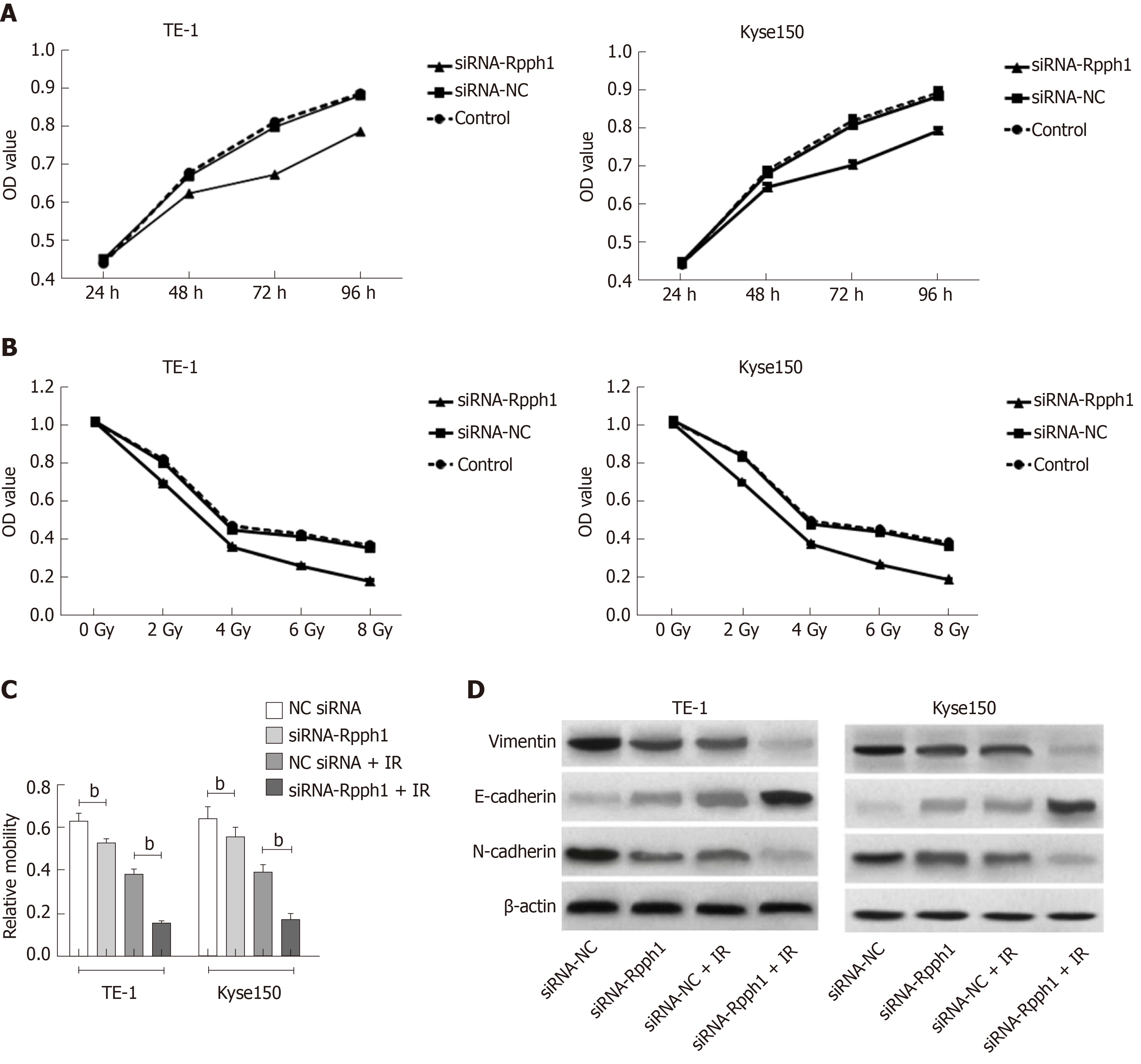
The blank control group, siRNA-NC, and siRNA-Rpph1 were designed according to the experimental requirements. The expression of Rpph1 in TE-1 and Kyse150 cells was detected by qRT-PCR 48 h after siRNA was transfected into TE-1 and Kyse150 cells. Comparison between the blank control group and the siRNA-NC group in Rpph1 expression showed no statistical difference, while the siRNA-Rpph1 group had significantly lower Rpph1 expression than the other two groups (P < 0.05). The colony formation assay demonstrated that the expression of siRNA-Rpph1 did not affect the colony sizes of TE-1 and Kyse150 cells. The surviving fractions of TE-1 and Kyse150 cells in the three groups before radiotherapy were not markedly different. The cell surviving fraction in each group decreased with increasing radiotherapy doses, with a greater decrease in the siRNA-Rpph1 group. The siRNA-Rpph1 group had a markedly smaller cell surviving fraction than the blank control group and the siRNA-NC group after radiotherapy (P < 0.05) (Figure 3).
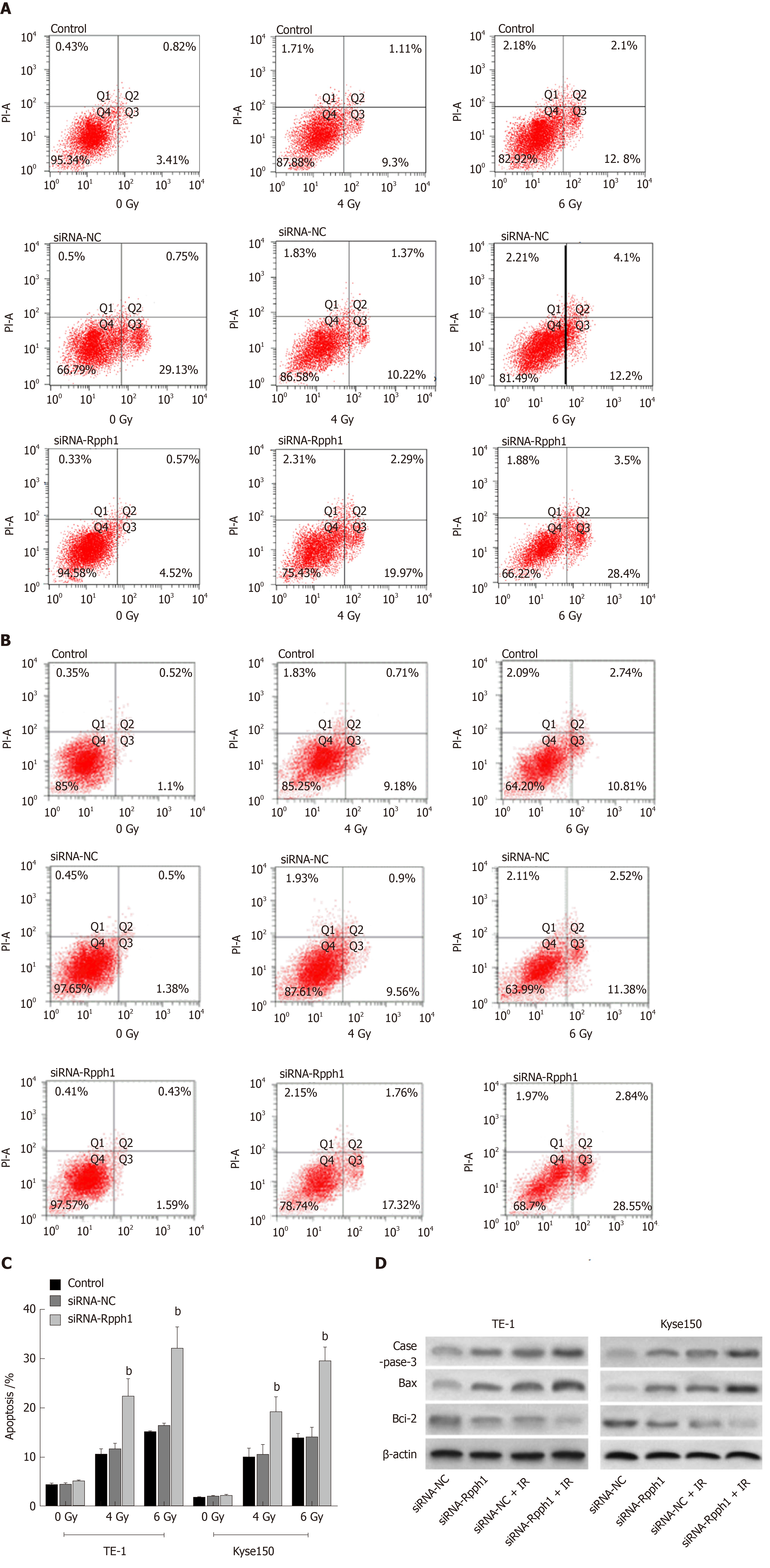
Esophageal cancer cell apoptosis in the blank control group, siRNA-NC group, and siRNA-Rpph1 group under different doses of radiotherapy were tested. Cell apoptosis rates of the three groups were not dramatically different before radiotherapy. After TE-1 and Kyse150 cells were exposed to irradiation at 4 Gy and 6 Gy, the siRNA-Rpph1 group had remarkably higher apoptosis rates than the blank control group and the siRNA-NC group (P < 0.001). Western Blot detection was employed to quantify the expression of apoptosis-related proteins in esophageal cancer cells before and after radiotherapy. As displayed in Figure 4, after cell exposure to 6 Gy irradiation, the siRNA-Rpph1 + IR group and the siRNA-NC group had ramped-up Bax and caspase-3 expression and a reduction in Bcl-2 expression. Cells in the siRNA-Rpph1 + IR group had remarkably higher expression of Bax and caspase-3 proteins, and lower expression of Bcl-2 proteins than cells in the siRNA-NC group.
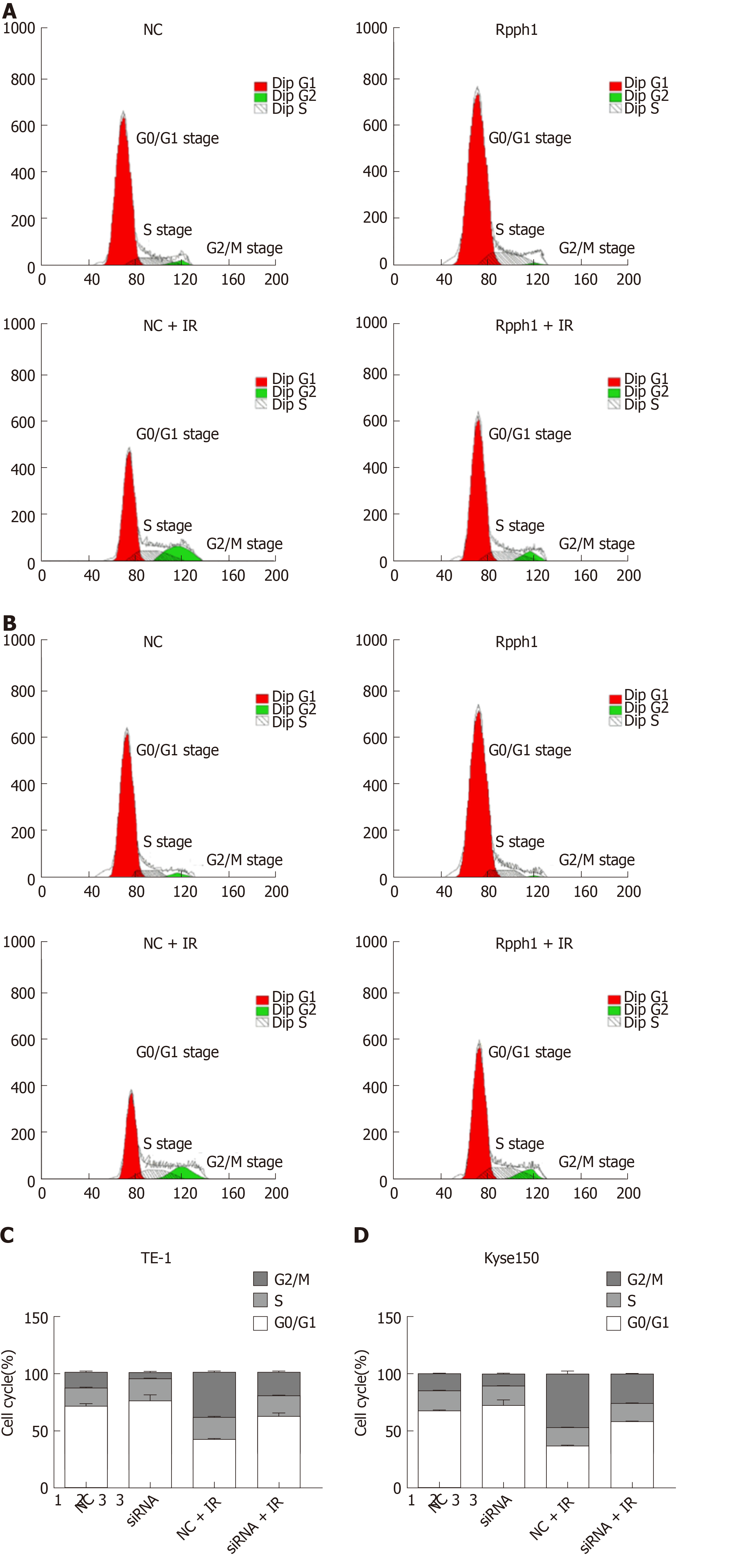
Figure 5 shows that radiotherapy resulted in more G2/M phase cells. The siRNA-Rpph1 + IR group (6 Gy) had markedly fewer G2/M phase cells than the siRNA-NC + IR group (P < 0.05), implying that downregulation of Rpph1 expression can effectively alleviate radiation-induced G2/M phase arrest, thereby enhancing the sensitivity of the esophageal cancer cell lines TE-1 and Kyse150 to radiotherapy.
The OD values of TE-1 and Kyse150 cells in the three groups (siRNA-Rpph1, siRNA-NC, and blank control group) were measured by MTT assay at 24, 48, 72 and 96 h after cell culture. Comparison between the three groups in the OD value at 24 h showed no dramatic differences. The OD values of each group underwent a gradual ramp-up. The siRNA-Rpph1 group had slightly lower OD values than the other two groups at 48 h. No great differences in OD values were observed between the blank control group and the siRNA-NC group at each time point, while the OD values at 72 and 96 h were much lower in the siRNA-Rpph1 group than in the other two groups (P < 0.05). All three groups were exposed to different doses of irradiation (0, 2, 4, 6, and 8 Gy). The cell proliferation was weaker in the siRNA-Rpph1 group than in the blank control group and siRNA-NC group after each dose of irradiation (P < 0.05), while the blank control group and the siRNA-NC group were not different in cell proliferation rate. Cell migration was significantly weaker in the siRNA-Rpph1 group than in the siRNA-NC group (P < 0.05). After exposure to irradiation at 6 Gy, the cell migration ability of TE-1 and Kyse150 cells was poorer in the siRNA-Rpph1 group than in the siRNA-NC group (P < 0.05). Expression of EMT-related proteins: Cytoskeletal protein (Vimentin), epithelial cadherin (E-cadherin), and neuro-cadherin (N-cadherin) expression levels were determined. The siRNA-Rpph1 group had markedly lower expression of Vimentin and N-cadherin, and higher expression of E-cadherin than the siRNA-NC group (P < 0.05). After radiotherapy, the expression of Vimentin and N-cadherin in the siRNA-Rpph1 group significantly decreased, while showing much higher E-cadherin expression than the siRNA-NC group (P < 0.05) (Figure 6).
As one of the most prevalent malignant tumors, esophageal cancer has an incidence that increases with age[21]. Postoperative radiotherapy is an effective method to improve survival rates and reduce recurrence rates[22]. Surgery and radiotherapy are effective in esophageal cancer patients, but the 5-year survival rate is poor[21]. LncRNA, involved in many complex regulatory networks, has become a research hotspot in recent years. Rpph1 is known as the subunit of RNase P RNA that regulates tRNA maturation in all life domains[23], and it often works as an internal reference for RNA quantification[24-26]. A previous study suggested that Rpph1 may play a part in disease progression in animals and humans[27].
One related study has reported the upregulation of Rpph1 in human gastric cancer tissues[18]. Thus far, however, no studies on Rpph1 in esophageal cancer have been published. This study revealed high Rpph1 expression in esophageal cancer tissues and serum, implying the possibility that Rpph1 has an effect on the occurrence and development of esophageal cancer. Due to its accessibility and the limited invasiveness required for its collection, serum samples for the diagnosis of esophageal cancer are currently popular. The study by Wang et al[28] found that HOTAIR, a lncRNA transcribed from the HOXC locus, could be used as a novel diagnostic marker for esophageal squamous cell carcinoma by detecting its serum expression. This study reveals that serum lncRNA Rpph1 is associated with short-term responses, T stages, N stages, and clinical stages in patients with esophageal cancer, and can help diagnose esophageal cancer. Some studies reported that T stages, N stages, and clinical stages were factors influencing the prognosis of esophageal cancer patients[29,30]. This study conducted regular follow-up for patients and found that esophageal cancer patients receiving radiotherapy had an overall survival rate of 42.17%, and that the 3-year overall survival rate was markedly lower in patients with high serum Rpph1 expression than in those with low serum Rpph1 expression. Such findings suggest that Rpph1 is related to the prognosis of esophageal cancer patients. Rpph1 can therefore be used as a biomarker to diagnose esophageal cancer and evaluate prognosis.
An extensive amount of literature has revealed that radiotherapy depends on ionizing radiation to induce tumor cell apoptosis, the occurrence of which is regulated by multiple genes[9,31,32]. Therefore, changes in apoptosis-related genes can accurately reflect cell sensitivity to radiotherapy. Previous studies have revealed that the esophageal cancer cell lines TE-1 and Kyse150 can effectively reflect radio-sensitivity. We found no contaminations in TE-1 and Kyse150 cell lines, so the detection of these two cell lines can provide a certain theoretical basis for the experiment[33,34]. To validate the role of Rpph1 on the radio-sensitivity of esophageal cancer cells, this study employed a colony formation assay to assess cell viability. The results of the colony formation assay demonstrated that Rpph1 silencing did not affect the colony sizes of TE-1 and Kyse150 cells. The cell-surviving fractions decreased with increasing irradiation doses in all transfection groups, with a stronger decrease in the siRNA-Rpph1 group, suggesting that silencing Rpph1 expression can significantly enhance cell sensitivity to radiotherapy. Colony formation assays conducted by Jiang et al[35] showed that downregulation of the lncRNA TUG1 increased the radio-sensitivity of bladder cancer cells by suppressing the expression of HMGB1. Whether Rpph1 uses the same mechanism of action requires further studies. One former study believed that radiation-induced apoptosis is one form of radiation-induced cell death[36]. The apoptosis experiments in this study revealed that apoptosis of the siRNA-Rpph1 group was much stronger than that of the blank control group and the siRNA-NC group when TE-1 and Kyse150 cells were exposed to irradiation at a dose of 4 Gy and 6 Gy, indicating that Rpph1 silencing can increase radiation-induced cell apoptosis. After radiotherapy, the siRNA-Rpph1+IR group exhibited ramped-up Bax and caspase-3 expression, and a reduction in Bcl-2 expression (anti-apoptotic protein), suggesting that Rpph1 silencing can intensify the radio-sensitivity of esophageal cancer cells by promoting cell apoptosis.
Cell cycle regulation may be critical for the radio-sensitivity of tumor cells[37]. Radiation can induce DNA double-strand breaks and then lead to G2/M phase arrest and DNA damage repair, while G2/M phase arrest is a process of tumor cell self-repair[38,39]. Zhang et al[20] claimed that Rpph1 may influence the progression of breast cancer by regulating cell proliferation and cell cycle. In this study, radiotherapy led to more G2/M phase cells. The siRNA-Rpph1+IR group (6 Gy) had significantly more G2/M phase cells than the siRNA-NC + IR group (P < 0.05), indicating that downregulation of Rpph1 expression can effectively alleviate radiation-induced G2/M phase arrest, thereby enhancing the radio-sensitivity of TE-1 and Kyse150 cells. To figure out the possible biological potential of Rpph1 in esophageal cancer, we silenced Rpph1 expression and discovered that Rpph1 silencing significantly inhibited the cell proliferation and migration of esophageal cancer. A recent study revealed that Rpph1 had a regulatory effect on the inflammation and proliferation of mesangial cells by stimulating the Gal-3/Mek/Erk pathway in diabetic nephropathy[40]. However, the way Rpph1 regulates the biological behavior of esophageal cancer cells requires further studies. EMT is a basic biological process by which epithelial cells lose their polarity and transform into a mesenchymal phenotype[41]. EMT is associated with radio-sensitivity in esophageal cancer[41]. In this study, the expression levels of EMT-related proteins Vimentin and N-cadherin were greatly reduced, E-cadherin expression increased in the siRNA-Rpph1 group, and the decrease or increase of EMT-related proteins was sharper after radiotherapy. While we speculate that Rpph1 may be involved in the regulation of EMT, the specific mechanism needs to be confirmed by further research.
This study is subject to certain limitations. The specific target genes or signaling pathways on which Rpph1 acts to regulate the biological function of esophageal cancer cells remains unknown. The function of tumor suppression by Rpph1 in esophageal cancer animal models needs to be further explored. In addition, the possibility of Rpph1 functioning as a target for treating esophageal cancer should be confirmed by abundant future studies.
In summary, Rpph1 is highly expressed in esophageal cancer. To a certain extent, silencing Rpph1 expression can promote cell apoptosis, inhibit cell proliferation and migration, relieve radiation-induced G2/M phase arrest, and enhance radio-sensitivity.
As a common digestive tract tumor, esophageal cancer is typically treated by radiotherapy. Poor responses to radiotherapy in most patients are prone to causing local radiotherapy failure. It is therefore essential to find new radiosensitizers to enhance the response of cancer cells to radiotherapy and increase the survival rate of esophageal cancer patients with radiation resistance. The long non-coding RNA (lncRNA) Rpph1 is highly expressed in human gastric cancer tissues, which also decreases breast cancer cell proliferation as well as tumorigenesis. In fact, the expression of lncRNA Rpph1 in esophageal cancer and its relationship with radio-sensitivity have not been thoroughly investigated.
LncRNA Rpph1 is found abnormally expressed in a variety of cancers. The possibility that Rpph1 impacts the radio-sensitivity of esophageal cancer cells requires more research.
This study was intended to reveal the value of lncRNA Rpph1 in esophageal cancer as well as its effect on cell sensitivity to radiotherapy.
We initially detected the expression of lncRNA Rpph1 in esophageal cancer tissue samples obtained surgically. Subsequently, siRNA-NC or siRNA-Rpph1 was transfected into esophageal cancer cell lines, and a blank control group was set where cells were not transfected with anything. Consequently, we analyzed the effect of Rpph1 on the biological behavior of esophageal cancer cells exposed to irradiation.
We found that Rpph1 is highly expressed in esophageal cancer. Rpph1 can be applied for the diagnosis of esophageal cancer and identify the pathological characteristics of patients. The results of 3-year survival supported patients with low Rpph1 expression over those with high Rpph1 expression (P < 0.05). Cytological studies indicated that silencing the expression of Rpph1 contributed to the enhancement of radio-sensitivity of esophageal cancer cells and cell apoptosis via regulating apoptosis-linked proteins, thus relieving the radiation-induced G2/M phase cell cycle arrest. It also helped restrain cell proliferation and migration and regulate the expression of epithelial-mesenchymal transition (EMT)-related proteins.
Rpph1 is highly expressed in esophageal cancer. Silencing Rpph1 expression has an influence on the biological behavior of tumor cells and the enhancement of radio-sensitivity.
This study revealed the value of Rpph1 in the diagnosis of esophageal cancer and concluded that silencing Rpph1 expression can enhance the radiotherapy of tumor cells, illuminating a new target for the future treatment of esophageal cancer.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Jeong KJ, Surucu E S-Editor: Gong ZM L-Editor: Filipodia E-Editor: Ma YJ
| 1. | Giri S, Pathak R, Aryal MR, Karmacharya P, Bhatt VR, Martin MG. Incidence trend of esophageal squamous cell carcinoma: an analysis of Surveillance Epidemiology, and End Results (SEER) database. Cancer Causes Control. 2015;26:159-161. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 16] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 2. | Yamamoto M, Weber JM, Karl RC, Meredith KL. Minimally invasive surgery for esophageal cancer: review of the literature and institutional experience. Cancer Control. 2013;20:130-137. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 34] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 3. | Funakawa K, Uto H, Sasaki F, Nasu Y, Mawatari S, Arima S, Nakazawa J, Taguchi H, Hashimoto S, Kanmura S, Setoyama H, Numata M, Tsubouchi H, Ido A. Effect of endoscopic submucosal dissection for superficial esophageal neoplasms and risk factors for postoperative stricture. Medicine (Baltimore). 2015;94:e373. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 40] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 4. | Arnold M, Soerjomataram I, Ferlay J, Forman D. Global incidence of oesophageal cancer by histological subtype in 2012. Gut. 2015;64:381-387. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 944] [Cited by in RCA: 1027] [Article Influence: 102.7] [Reference Citation Analysis (0)] |
| 5. | Zheng H, Ren W, Pan X, Zhang Q, Liu B, Liu S, He J, Zhou Z. Role of intravoxel incoherent motion MRI in early assessment of the response of esophageal squamous cell carcinoma to chemoradiotherapy: A pilot study. J Magn Reson Imaging. 2018;48:349-358. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 25] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 6. | Parisi E, Genestreti G, Sarnelli A, Ghigi G, Arpa D, Burgio MA, Gavelli G, Rossi A, Scarpi E, Monti M, Tesei A, Polico R, Romeo A. Accelerated hypofractionated radiotherapy plus chemotherapy for inoperable locally advanced non-small-cell lung cancer: final results of a prospective phase-II trial with a long-term follow-up. Radiat Oncol. 2019;14:112. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 7. | Wu CC, Chen CJ. Esophageal carcinoma. N Engl J Med. 2015;372:1472. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 52] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 8. | Luo LL, Zhao L, Wang YX, Tian XP, Xi M, Shen JX, He LR, Li QQ, Liu SL, Zhang P, Xie D, Liu MZ. Insulin-like growth factor binding protein-3 is a new predictor of radiosensitivity on esophageal squamous cell carcinoma. Sci Rep. 2015;5:17336. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 16] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 9. | Park M, Yoon HJ, Kang MC, Kwon J, Lee HW. MiR-338-5p enhances the radiosensitivity of esophageal squamous cell carcinoma by inducing apoptosis through targeting survivin. Sci Rep. 2017;7:10932. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 41] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 10. | Wakita A, Motoyama S, Sato Y, Koyota S, Usami S, Yoshino K, Sasaki T, Imai K, Saito H, Minamiya Y. REG Iα activates c-Jun through MAPK pathways to enhance the radiosensitivity of squamous esophageal cancer cells. Tumour Biol. 2015;36:5249-5254. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 13] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 11. | Cheng J, Liu W, Zeng X, Zhang B, Guo Y, Qiu M, Jiang C, Wang H, Wu Z, Meng M, Zhuang H, Zhao L, Hao J, Cai Q, Xie D, Pang Q, Wang P, Yuan Z, Qian D. XRCC3 is a promising target to improve the radiotherapy effect of esophageal squamous cell carcinoma. Cancer Sci. 2015;106:1678-1686. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 18] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 12. | Beermann J, Piccoli MT, Viereck J, Thum T. Non-coding RNAs in Development and Disease: Background, Mechanisms, and Therapeutic Approaches. Physiol Rev. 2016;96:1297-1325. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1378] [Cited by in RCA: 1329] [Article Influence: 147.7] [Reference Citation Analysis (1)] |
| 13. | Neumann P, Jaé N, Knau A, Glaser SF, Fouani Y, Rossbach O, Krüger M, John D, Bindereif A, Grote P, Boon RA, Dimmeler S. The lncRNA GATA6-AS epigenetically regulates endothelial gene expression via interaction with LOXL2. Nat Commun. 2018;9:237. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 120] [Cited by in RCA: 156] [Article Influence: 22.3] [Reference Citation Analysis (0)] |
| 14. | García-Padilla C, Aránega A, Franco D. The role of long non-coding RNAs in cardiac development and disease. AIMS Genet. 2018;5:124-140. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 21] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 15. | Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17:47-62. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2868] [Cited by in RCA: 2719] [Article Influence: 302.1] [Reference Citation Analysis (0)] |
| 16. | Bhan A, Mandal SS. Long noncoding RNAs: emerging stars in gene regulation, epigenetics and human disease. ChemMedChem. 2014;9:1932-1956. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 177] [Cited by in RCA: 214] [Article Influence: 19.5] [Reference Citation Analysis (0)] |
| 17. | Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354-361. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1424] [Cited by in RCA: 1565] [Article Influence: 111.8] [Reference Citation Analysis (0)] |
| 18. | Xia T, Liao Q, Jiang X, Shao Y, Xiao B, Xi Y, Guo J. Long noncoding RNA associated-competing endogenous RNAs in gastric cancer. Sci Rep. 2014;4:6088. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 246] [Cited by in RCA: 331] [Article Influence: 30.1] [Reference Citation Analysis (0)] |
| 19. | Lipovich L, Dachet F, Cai J, Bagla S, Balan K, Jia H, Loeb JA. Activity-dependent human brain coding/noncoding gene regulatory networks. Genetics. 2012;192:1133-1148. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 143] [Cited by in RCA: 162] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 20. | Zhang Y, Tang L. Inhibition of breast cancer cell proliferation and tumorigenesis by long non-coding RNA RPPH1 down-regulation of miR-122 expression. Cancer Cell Int. 2017;17:109. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 35] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 21. | Wen X, Han XR, Fan SH, Zhang ZF, Chen L, Yi G, Wu DM, Lu J, Zheng YL. Down-regulation of XIAP enhances the radiosensitivity of esophageal cancer cells in vivo and in vitro. Biosci Rep. 2017;37. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 8] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 22. | Li CL, Zhang FL, Wang YD, Han C, Sun GG, Liu Q, Cheng YJ, Jing SW, Yang CR. Characteristics of recurrence after radical esophagectomy with two-field lymph node dissection for thoracic esophageal cancer. Oncol Lett. 2013;5:355-359. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 40] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 23. | Evans D, Marquez SM, Pace NR. RNase P: interface of the RNA and protein worlds. Trends Biochem Sci. 2006;31:333-341. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 159] [Cited by in RCA: 163] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 24. | Page K, Hava N, Ward B, Brown J, Guttery DS, Ruangpratheep C, Blighe K, Sharma A, Walker RA, Coombes RC, Shaw JA. Detection of HER2 amplification in circulating free DNA in patients with breast cancer. Br J Cancer. 2011;104:1342-1348. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 62] [Cited by in RCA: 71] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 25. | Raoul C, Abbas-Terki T, Bensadoun JC, Guillot S, Haase G, Szulc J, Henderson CE, Aebischer P. Lentiviral-mediated silencing of SOD1 through RNA interference retards disease onset and progression in a mouse model of ALS. Nat Med. 2005;11:423-428. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 333] [Cited by in RCA: 337] [Article Influence: 16.9] [Reference Citation Analysis (0)] |
| 26. | Soler-Alfonso C, Carvalho CM, Ge J, Roney EK, Bader PI, Kolodziejska KE, Miller RM, Lupski JR, Stankiewicz P, Cheung SW, Bi W, Schaaf CP. CHRNA7 triplication associated with cognitive impairment and neuropsychiatric phenotypes in a three-generation pedigree. Eur J Hum Genet. 2014;22:1071-1076. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 34] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 27. | Cai Y, Sun Z, Jia H, Luo H, Ye X, Wu Q, Xiong Y, Zhang W, Wan J. Rpph1 Upregulates CDC42 Expression and Promotes Hippocampal Neuron Dendritic Spine Formation by Competing with miR-330-5p. Front Mol Neurosci. 2017;10:27. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 58] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 28. | Wang W, He X, Zheng Z, Ma X, Hu X, Wu D, Wang M. Serum HOTAIR as a novel diagnostic biomarker for esophageal squamous cell carcinoma. Mol Cancer. 2017;16:75. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 81] [Cited by in RCA: 98] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 29. | Yin H, Li D, Zhu C, Wang M, Wei N. Factors relevant to the prognosis of patients with esophageal cancer who received intensity-modulated radiotherapy. Thorac Cancer. 2018;9:1215-1219. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 6] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 30. | Chen J, Lin Y, Cai W, Su T, Wang B, Li J, Wu J, Pan J, Chen C. A new clinical staging system for esophageal cancer to predict survival after definitive chemoradiation or radiotherapy. Dis Esophagus. 2018;31. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 20] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 31. | Xu G, Zhu L, Wang Y, Shi Y, Gong A, Wu C. Stattic Enhances Radiosensitivity and Reduces Radio-Induced Migration and Invasion in HCC Cell Lines through an Apoptosis Pathway. Biomed Res Int. 2017;2017:1832494. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 22] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 32. | Qian D, Zhang B, Zeng XL, Le Blanc JM, Guo YH, Xue C, Jiang C, Wang HH, Zhao TS, Meng MB, Zhao LJ, Hao JH, Wang P, Xie D, Lu B, Yuan ZY. Inhibition of human positive cofactor 4 radiosensitizes human esophageal squmaous cell carcinoma cells by suppressing XLF-mediated nonhomologous end joining. Cell Death Dis. 2014;5:e1461. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 37] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 33. | Zhang DJ, Xiang J, Wang X, Wang J, Xiao JC, Xu W, Xu H, Xin Y, Zhang LZ, Pei DS, Zheng JN, Gu YM. RPA1 expression in esophageal carcinoma and its influence on radiosensitivity of esophageal carcinoma TE-1 cells. Panminerva Med. 2015;57:183-189. [PubMed] |
| 34. | Su H, Wu Y, Fang Y, Shen L, Fei Z, Xie C, Chen M. MicroRNA‑301a targets WNT1 to suppress cell proliferation and migration and enhance radiosensitivity in esophageal cancer cells. Oncol Rep. 2019;41:599-607. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 19] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 35. | Jiang H, Hu X, Zhang H, Li W. Down-regulation of LncRNA TUG1 enhances radiosensitivity in bladder cancer via suppressing HMGB1 expression. Radiat Oncol. 2017;12:65. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 78] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 36. | Eriksson D, Stigbrand T. Radiation-induced cell death mechanisms. Tumour Biol. 2010;31:363-372. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 416] [Cited by in RCA: 473] [Article Influence: 31.5] [Reference Citation Analysis (0)] |
| 37. | Cao R, Ding Q, Li P, Xue J, Zou Z, Huang J, Peng G. SHP1-mediated cell cycle redistribution inhibits radiosensitivity of non-small cell lung cancer. Radiat Oncol. 2013;8:178. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 18] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 38. | Shibata A, Jeggo P. A historical reflection on our understanding of radiation-induced DNA double strand break repair in somatic mammalian cells; interfacing the past with the present. Int J Radiat Biol. 2019;95:945-956. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 31] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 39. | Liu X, Sun C, Jin X, Li P, Ye F, Zhao T, Gong L, Li Q. Genistein enhances the radiosensitivity of breast cancer cells via G₂/M cell cycle arrest and apoptosis. Molecules. 2013;18:13200-13217. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 63] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 40. | Zhang P, Sun Y, Peng R, Chen W, Fu X, Zhang L, Peng H, Zhang Z. Long non-coding RNA Rpph1 promotes inflammation and proliferation of mesangial cells in diabetic nephropathy via an interaction with Gal-3. Cell Death Dis. 2019;10:526. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 75] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 41. | Su H, Jin X, Zhang X, Zhao L, Lin B, Li L, Fei Z, Shen L, Fang Y, Pan H, Xie C. FH535 increases the radiosensitivity and reverses epithelial-to-mesenchymal transition of radioresistant esophageal cancer cell line KYSE-150R. J Transl Med. 2015;13:104. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 47] [Article Influence: 4.7] [Reference Citation Analysis (0)] |