Published online Mar 28, 2020. doi: 10.3748/wjg.v26.i12.1329
Peer-review started: December 5, 2019
First decision: January 12, 2020
Revised: February 7, 2020
Accepted: February 28, 2020
Article in press: February 28, 2020
Published online: March 28, 2020
Processing time: 114 Days and 2.7 Hours
Polygonum multiflorum is one of the leading causes of herb-induced liver injury in China. HLA-B*35:01 is reported to be a potential biomarker of Polygonum multiflorum-induced liver injury (PM-DILI). However, little is known about the relationship between single-nucleotide polymorphisms (SNPs) and PM-DILI.
To identify SNPs that indicate susceptibility to PM-DILI.
We conducted a systematic study enrolling 382 participants from four independent hospitals, including 73 PM-DILI patients, 118 patients with other drug-induced liver injury (other-DILI) and 191 healthy controls. Whole-exome sequencing was performed for 8 PM-DILI patients and 8 healthy controls who were randomly selected from the above subjects. Nineteen SNPs that showed high frequencies in the 8 PM-DILI patients were selected as candidate SNPs and then screened in 65 PM-DILI patients, 118 other-DILI patients and 183 healthy controls using the MassARRAY system. HLA-B high-resolution genotyping was performed for the 73 PM-DILI and 118 other-DILI patients. The Han-MHC database was selected as a population control for HLA-B analysis. P < 6.25 × 10-3 after Bonferroni correction was considered significant.
The frequencies of rs111686806 in the HLA-A gene, rs1055348 in the HLA-B gene, and rs202047044 in the HLA-DRB1 gene were significantly higher in the PM-DILI group than in the control group [27.2% vs 11.6%, P = 1.72 × 10-5, odds ratio (OR) = 3.96, 95% confidence interval (CI): 2.21-7.14; 42.5% vs 8.6%, P = 1.72 × 10-19, OR = 13.62, 95%CI: 7.16-25.9; 22.9% vs 8.1%, P = 4.64 × 10-6, OR = 4.1, 95%CI: 2.25-7.47]. Only rs1055348 showed a significantly higher frequency in the PM-DILI group than in the other-DILI group (42.5% vs 13.6%, P = 1.84 × 10-10, OR = 10.06, 95%CI: 5.06-20.0), which suggested that it is a specific risk factor for PM-DILI. rs1055348 may become a tag for HLA-B*35:01 with 100% sensitivity and 97.7% specificity in the PM-DILI group and 100% sensitivity and 98.1% specificity in the other-DILI group. Furthermore, HLA-B*35:01 was confirmed to be associated with PM-DILI with a frequency of 41.1% in the PM-DILI group compared with 11.9% (P = 4.30 × 10-11, OR = 11.11, 95%CI: 5.57-22.19) in the other-DILI group and 2.7% (P = 6.22 × 10-166, OR = 62.62, 95%CI: 35.91-109.20) in the Han-MHC database.
rs111686806, rs1055348, and rs202047044 are associated with PM-DILI, of which, rs1055348 is specific to PM-DILI. As a tag for HLA-B*35:01, rs1055348 may become an alternative predictive biomarker of PM-DILI.
Core tip: We aimed to identify single-nucleotide polymorphisms that indicate susceptibility to Polygonum multiflorum-induced liver injury (PM-DILI). We successfully identified three single-nucleotide polymorphisms, rs111686806 in the HLA-A gene, rs1055348 in the HLA-B gene and rs202047044 in the HLA-DRB1 gene that were associated with PM-DILI. Of which, rs1055348 had the strongest effect and was a specific risk factor for PM-DILI. We also validated the association between HLA-B*35:01 and PM-DILI. Furthermore, we analyzed the correlation between rs1055348 and HLA-B*35:01. With 100% sensitivity and > 95% specificity, rs1055348 may serve as a tag for HLA-B*35:01 and an alternative predictive biomarker of PM-DILI.
- Citation: Yang WN, Pang LL, Zhou JY, Qiu YW, Miao L, Wang SY, Liu XZ, Tan KA, Shi WW, Wang GQ, Hou FQ. Single-nucleotide polymorphisms of HLA and Polygonum multiflorum-induced liver injury in the Han Chinese population. World J Gastroenterol 2020; 26(12): 1329-1339
- URL: https://www.wjgnet.com/1007-9327/full/v26/i12/1329.htm
- DOI: https://dx.doi.org/10.3748/wjg.v26.i12.1329
Herbs are usually considered to be natural and safe. However, herbs are often major causes of drug-induced liver injury (DILI)[1]. A previous retrospective study estimated the annual incidence of DILI in the general population at 23.80 per 100000 persons in mainland China, with traditional Chinese medicines, herbal and dietary supplements, and antituberculosis drugs being the leading causes[2]. Among herbal supplements, Polygonum multiflorum (PM) has been reported in several studies to be one of the leading causative agents of DILI[3-5].
PM is a well-known traditional Chinese herbal medicine called He Shou Wu in China and East Asia and Fo-ti in North America. PM has been used since ancient times as a medical prescription or food supplement for gray hair, hair loss, constipation, and hyperlipemia[6]. Nonetheless, reports of PM-induced liver injury (PM-DILI) have been increasing in recent years. Indeed, several cases of hepatitis associated with PM have been reported in patients from Australia, China, Italy, Japan, the Netherlands, the United States and Slovakia[7-11]. In addition, experiments both in vivo and in vitro have indicated that PM is toxic to the liver[12-14]. The diagnosis of DILI can be difficult because it relies largely on the exclusion of other potential causes and requires a high index of suspicion, as patients are often asymptomatic[15]. To improve the safe use of PM, the identification of biomarkers to prevent and diagnose PM-DILI is essential.
Susceptibility to DILI is considered to be genetically determined[16]. The HLA genotype as well as drug metabolism genes affect susceptibility to DILI due to a range of drugs and correlate with the underlying mechanisms[16,17]. For example, associations have been found between HLA-DRB1*15:01 and amoxicillin-clavulanate-DILI[18], HLA-A*33:01 and fenofibrate-DILI[19], and NAT2, GST and CYP2E1 polymorphisms and antituberculosis-DILI[20]. Recently, Li et al[17] reported an association between HLA-B*35:01 and PM-DILI.
However, HLA genotyping is time consuming and expensive compared with the detection of single-nucleotide polymorphisms (SNPs). Here, we present a systematic multicenter study that aimed to identify SNP risk factors for PM-DILI.
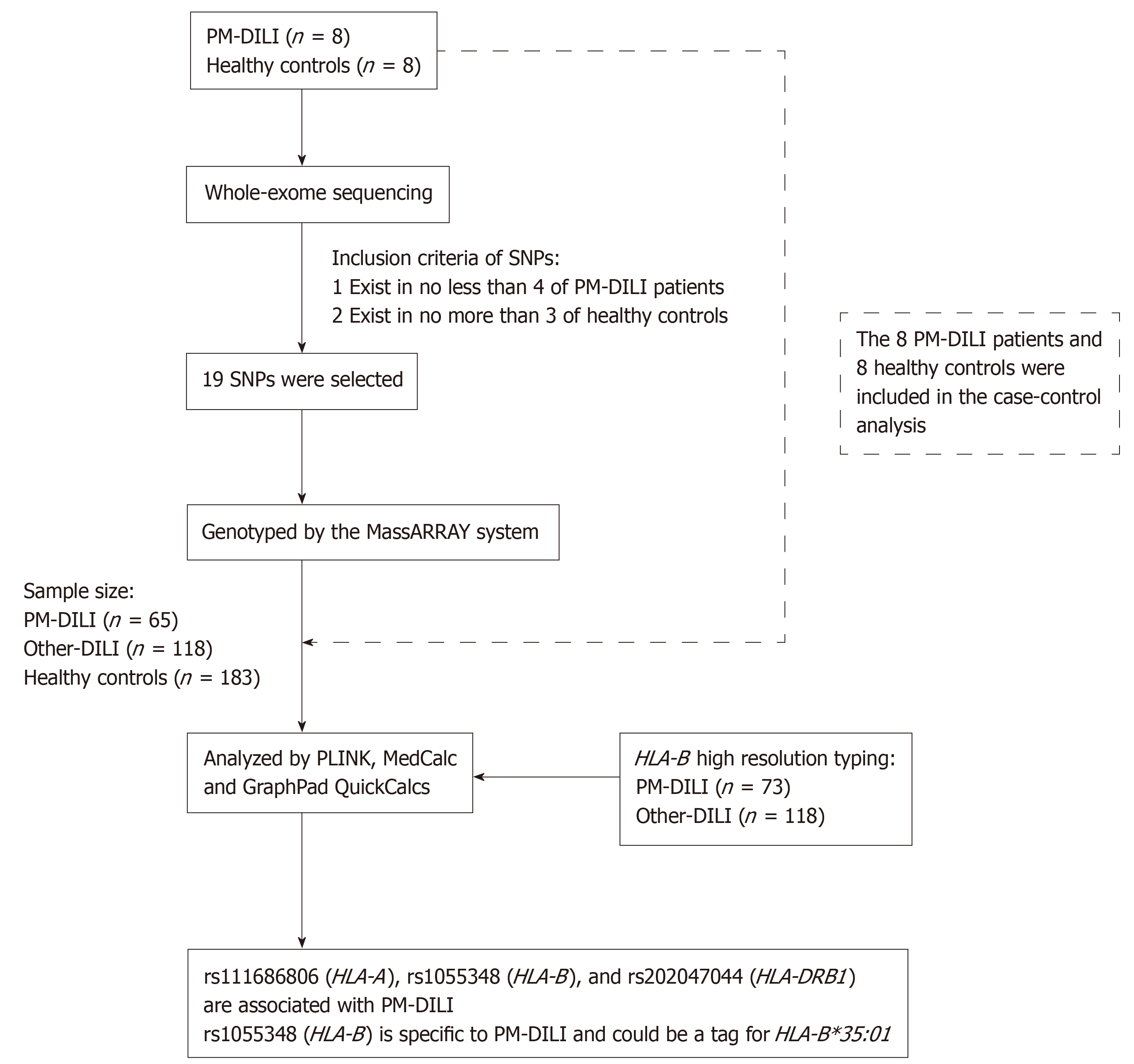
The 73 adult PM-DILI patients involved in our study were recruited at Peking University First Hospital (Beijing, China), the Fifth People's Hospital of Wuxi (Jiangsu, China), the Third Hospital of Qinhuangdao (Hebei, China), and Yantai City Hospital for Infectious Diseases (Shandong, China) from January 2012 to October 2017. An additional 118 adult other drug-induced liver injury (other-DILI) patients and 191 adult healthy controls were recruited at Peking University First Hospital (Beijing, China). The DILI patients met one of the following criteria: Alanine aminotransferase (ALT) levels ≥ 5 × the upper limit of normal (ULN), ALT ≥ 3 × ULN and total bilirubin level (TBil) ≥ 2 × ULN, alkaline phosphatase (ALP) level ≥ 2 × ULN, or pathological diagnosis of DILI. Patients who took both other drugs and single PM or PM preparations were excluded. The study protocol conformed to the ethical guidelines of the 1975 Declaration of Helsinki and was approved by the Human Research Committee of Peking University First Hospital (approval number, 2015[892]). The participants were Han Chinese and provided written informed consent before donating blood samples. Subjects with viral hepatitis, autoimmune liver diseases, alcoholic liver disease and non-alcoholic fatty liver disease were excluded. The overall study design is shown in Figure 1.
The causality of DILI cases was evaluated by a panel of two hepatologists, and all patients had a Roussel Uclaf Causality Assessment Method (RUCAM) score of 3 or greater. RUCAM scores were grouped into likelihood levels of “highly probable” (≥ 9), “probable” (6-8), “possible” (3-5), “unlikely” (1-2), and “excluded” (≤ 0)[21].
According to the Council for International Organizations of Medical Sciences criteria[22], the clinical pattern of DILI was based on the earliest identified liver biochemistry abnormalities and was defined using the R value. R values > 5 were classified as hepatocellular injury, < 2 as cholestatic injury, and 2-5 as mixed injury, where R = (ALT / ULN) / (ALP / ULN).
Genomic DNA was extracted from lymphocytes using DNA Blood Mini Kits (QIAGEN, Hilden, Germany) according to the manufacturer’s instructions and stored at -80 °C in Peking University First Hospital.
Whole-exome sequencing was carried out for 8 PM-DILI patients and 8 healthy controls who were randomly selected from the 73 PM-DILI patients and 191 healthy controls. Whole-exome sequencing was performed by GENEWIZ, Inc. with a depth of 50-fold and a coverage of 90%. All sequencing data are available through the NCBI Sequence Read Archive under accession number PRJNA564440. A total of 19 candidate SNPs that were found in no less than 4 of the PM-DILI patients and no more than 3 of the healthy controls were selected. Detailed information on the SNPs is shown in Table 1 and can be obtained from the SNP database system of the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/snp).
| SNP ID | Chromosome | Gene | Number of PM-DILI patients with candidate SNP | Number of healthy controls with candidate SNP |
| rs1042597 | 2 | UGT1A8 | 6 | 0 |
| rs37959583 | 2 | DRC1 | 6 | 3 |
| rs111686806 | 6 | HLA-A | 5 | 0 |
| rs2231119 | 6 | HLA-A | 8 | 0 |
| rs11369033 | 6 | HLA-A | 6 | 0 |
| rs1156624013 | 6 | HLA-A | 6 | 0 |
| rs1171629793 | 6 | HLA-A | 5 | 2 |
| rs10595163 | 6 | HLA-A | 5 | 0 |
| rs10503412 | 6 | HLA-B | 5 | 0 |
| rs1055348 | 6 | HLA-B | 5 | 0 |
| rs23086553 | 6 | HLA-B | 6 | 0 |
| rs10504581 | 6 | HLA-B | 5 | 0 |
| rs10514881 | 6 | HLA-B | 6 | 0 |
| rs202047044 | 6 | HLA-DRB1 | 4 | 0 |
| rs111534875 | 6 | HLA-DRB1 | 5 | 3 |
| rs78246137 | 6 | HLA-DRB1 | 7 | 0 |
| rs23087683 | 6 | HLA-DRB1 | 5 | 0 |
| rs17999313 | 8 | NAT2 | 4 | 0 |
| rs5747933 | 22 | PRODH | 7 | 3 |
The 19 candidate SNPs were then screened in 65 PM-DILI patients, 118 other-DILI patients and 183 healthy controls using the MassARRAY system which can test up to 40 target-specific DNA fragments in a single reaction. Quality control of the detection data was performed using PLINK software (version 1.07). SNPs with genotype failure rates larger than 5% (2 SNPs), a minor allele frequency less than 5% (1 SNP), or Hardy-Weinberg equilibrium test P values < 0.05 (8 SNPs) were excluded. As a result, 8 SNPs were included in the analysis. Statistical analysis was performed on the 8 SNPs among 73 PM-DILI patients, 118 other-DILI patients and 191 healthy controls.
To investigate the relationship between rs1055348 and HLA-B*35:01, high-resolution genotyping of HLA-B in the 73 PM-DILI patients and 118 other-DILI patients was performed by BFR Gene Diagnostics Co., Ltd. The 10689 healthy individuals from the Han-MHC database were selected as study controls[23]. Sensitivity, specificity, positive predictive value, negative predictive value and Cohen’s kappa coefficient were calculated.
A comparison of the clinical features between the PM-DILI group and other-DILI group was performed using SPSS software, version 20.0 (SPSS Inc., Chicago, IL, United States). Quantitative variables were analyzed by Student’s t test. Categorical variables were compared using the chi-square or Fisher’s exact test, and the results are shown as a number and percentage. Results were considered statistically significant at P < 0.05 (2-tailed).
PLINK (version 1.07) software was used to compare allele frequencies between groups. We used the chi-square or Fisher’s exact test to compare allele frequencies between groups. Logistic regression was applied to investigate independent risk factors for PM-DILI. We set a threshold of P < 6.25 × 10-3 by Bonferroni correction for SNPs and HLA-B alleles.
Sensitivity, specificity, and positive and negative predictive values were analyzed by MedCalc for Windows, version 15.2 (MedCalc Software, Ostend, Belgium). Cohen’s kappa coefficient was calculated using GraphPad QuickCalcs (GraphPad Software Inc., La Jolla, CA, United States).
Of the 73 PM-DILI patients recruited, disease was caused in 11 patients by PM alone and in 58 patients by PM preparations; 4 patients were rechallenged with PM preparations. Of the 118 other-DILI patients, 65 cases were mainly induced by Chinese herbal medicines and 53 by Western medicines. The top 6 causative agents in the other-DILI group were antibiotics (18), Radix Bupleuri (16), statins (11), Fructus Psoraleae (10), Cortex Dictamni (8), and methimazole (4). There were more female and younger patients in the PM-DILI group (P < 0.05). ALT, aspartate aminotransferase (AST) and TBil levels were higher in the PM-DILI group (P < 0.05), although body mass index, ALP level, distribution of injury type and RUCAM scores were not significantly different between the two groups (Table 2).
| Characteristics | PM-DILI (n = 73) | Other-DILI (n = 118) | P |
| Mean age, yr | 47.7 ± 15.8 | 52.3 ± 14.2 | 0.045 |
| Female, n (%) | 56 (76.7) | 74 (62.7) | 0.044 |
| BMI | 23.6 ± 3.2 | 24.0 ± 3.7 | 0.447 |
| Medication time1, d | 30 (45) | 30 (60.8) | 0.097 |
| ALT (U/L) | 1105.0 ± 679.9 | 626.3 ± 670.1 | < 0.001 |
| AST (U/L), n | 797.7 ± 557.3 (65) | 485.6 ± 596.5 (118) | 0.001 |
| ALP (U/L), n | 148.5 ± 58.4 (59) | 160.5 ± 212.3 (105) | 0.673 |
| TBil (mg/dL), n | 112.9 ± 107.2 (61) | 76.9 ± 92.5 (99) | 0.026 |
| Injury type | n = 59 | n = 105 | 0.917 |
| Hepatocellular | 57 | 82 | |
| Cholestatic | 1 | 12 | |
| Mixed | 1 | 11 | |
| RUCAM score | 0.319 | ||
| 3-5 (Possible) | 7 | 20 | |
| 6-8 (Probable) | 53 | 82 | |
| ≥ 9 (Highly probable) | 13 | 16 |
The frequencies of rs111686806 in the HLA-A gene, rs1055348 in the HLA-B gene and rs202047044 in the HLA-DRB1 gene were significantly higher in the PM-DILI group than in the control group (P < 6.25 × 10-3); no significant difference was found for the other 5 SNPs (Table 3). Logistic regression analysis showed that rs111686806, rs1055348, and rs202047044 were risk factors for PM-DILI compared with the control group.
| SNP ID | Chromose (Gene) | Minor allele | Minor allele frequency (%) | PM-DILI vs control | PM-DILI vs Other-DILI | ||||
| PM-DILI (n = 73) | Other-DILI (n = 118) | Control (n = 191) | P value | OR (95%CI) | P value | OR (95%CI) | |||
| rs1055348 | 6 (HLA-B) | G | 42.5% | 13.6% | 8.6% | 1.72 × 10-19a | 13.62 (7.16-25.9) | 1.84 × 10-10a | 10.06 (5.06-20) |
| rs202047044 | 6 (HLA-DRB1) | C | 22.9% | 17.5% | 8.1% | 4.64 × 10-6a | 4.1 (2.25-7.47) | 0.212 | 1.53 (0.84-2.76) |
| rs111686806 | 6 (HLA-A) | T | 27.2% | 27.6% | 11.6% | 1.72 × 10-5a | 3.96 (2.21-7.1) | 0.937 | 0.97 (0.53-1.77) |
| rs2231119 | 6 (HLA-A) | A | 26.0% | 24.4% | 36.8% | 0.020 | 0.57 (0.36-0.90) | 0.714 | 1.11 (0.66-1.85) |
| rs111534875 | 6 (HLA-DRB1) | A | 29.6% | 32.0% | 20.1% | 0.022 | 1.64 (1.06-2.55) | 0.622 | 0.89 (0.57-1.40) |
| rs1042597 | 2 (UGT1A8) | G | 42.1% | 44.0% | 51.0% | 0.071 | 0.71 (0.48-1.04) | 0.723 | 0.93 (0.61-1.41) |
| rs78246137 | 6 (HLA-DRB1) | T | 29.5% | 31.6% | 22.5% | 0.095 | 1.45 (0.94-2.25) | 0.656 | 0.9 (0.57-1.43) |
| rs5747933 | 22 (PRODH) | T | 13.6% | 16.7% | 10.2% | 0.279 | 1.35 (0.77-2.39) | 0.424 | 0.8 (0.45-1.42) |
Notably, only rs1055348 in the HLA-B gene exhibited a significantly different distribution between the PM-DILI group and the other-DILI group. The frequency of rs1055348 was 42.5% in the PM-DILI group and 13.6% in the other-DILI group, a difference that was significant (P = 1.84 × 10-10). A logistic regression model showed that rs1055348 was a risk factor for increased susceptibility to PM-DILI compared with other-DILI [Odds ratio (OR) = 10.06, 95% confidence interval (CI): 5.06-20.0] and healthy controls (OR = 13.62, 95%CI: 7.16-25.9).
Moreover, in the PM-DILI group, 10 of 11 (90.91%) PM-DILI patients whose disease was caused by PM alone, 4 of 4 (100%) PM-DILI patients who were rechallenged with PM preparations and 44 of 58 (75.86%) PM-DILI patients whose disease was caused by PM preparations carried the rs1055348 G allele. No significant difference was found between the three subgroups (P = 0.305).
Furthermore, we compared biochemical characteristics between patients with the rs1055348 G allele and those without the rs1055348 G allele (Table 4). In both the PM-DILI and other-DILI groups, patients with the rs1055348 G allele had higher ALT and AST levels than did patients without the rs1055348 G allele (P < 0.05). No significant difference in the TBil level was found.
| Variables | PM-DILI | Other-DILI | |||||
| With the rs1055348 G allele (n = 58) | Without the rs1055348 G allele (n = 15) | P value | With the rs1055348 G allele (n = 32) | Without the rs1055348 G allele (n = 86) | P value | ||
| Age, mean ± SD, yr | 46.7 ± 15.1 | 51.2 ± 16.2 | 0.317 | 49.2 ± 14.4 | 53.4 ± 14.0 | 0.152 | |
| Female, n (%) | 42 (72.4) | 14 (93.3) | 0.167 | 22 (68.8) | 52 (60.5) | 0.408 | |
| BMI | 23.7 ± 2.9 | 23.0 ± 3.0 | 0.407 | 23.8 ± 3.0 | 24.0 ± 3.9 | 0.774 | |
| Medication time1, d | 30 (40.8) | 30 (22) | 0.533 | 46.5 (63.5) | 30 (54) | 0.269 | |
| ALT (U/L) | 1234.8 ± 609.5 | 603.2 ± 517.0 | < 0.0001 | 936.7 ± 866.7 | 510.8 ± 532.7 | 0.013 | |
| AST (U/L), n | 876.9 ± 537.8 (51) | 509.2 ± 507.6 (14) | 0.025 | 712.8 ± 691.5 (32) | 401.0 ± 513.4 (86) | 0.009 | |
| ALP (U/L), n | 155.8 ± 59.8 (48) | 116.7 ± 40.0 (11) | 0.044 | 140.3 ± 99.4 (28) | 167.8 ± 240.8 (77) | 0.559 | |
| TBil (mg/dL), n | 123.9 ± 110.5 (48) | 70.7 ± 84.9 (13) | 0.113 | 85.7 ± 104.8 (29) | 73.3 ± 87.4 (70) | 0.547 | |
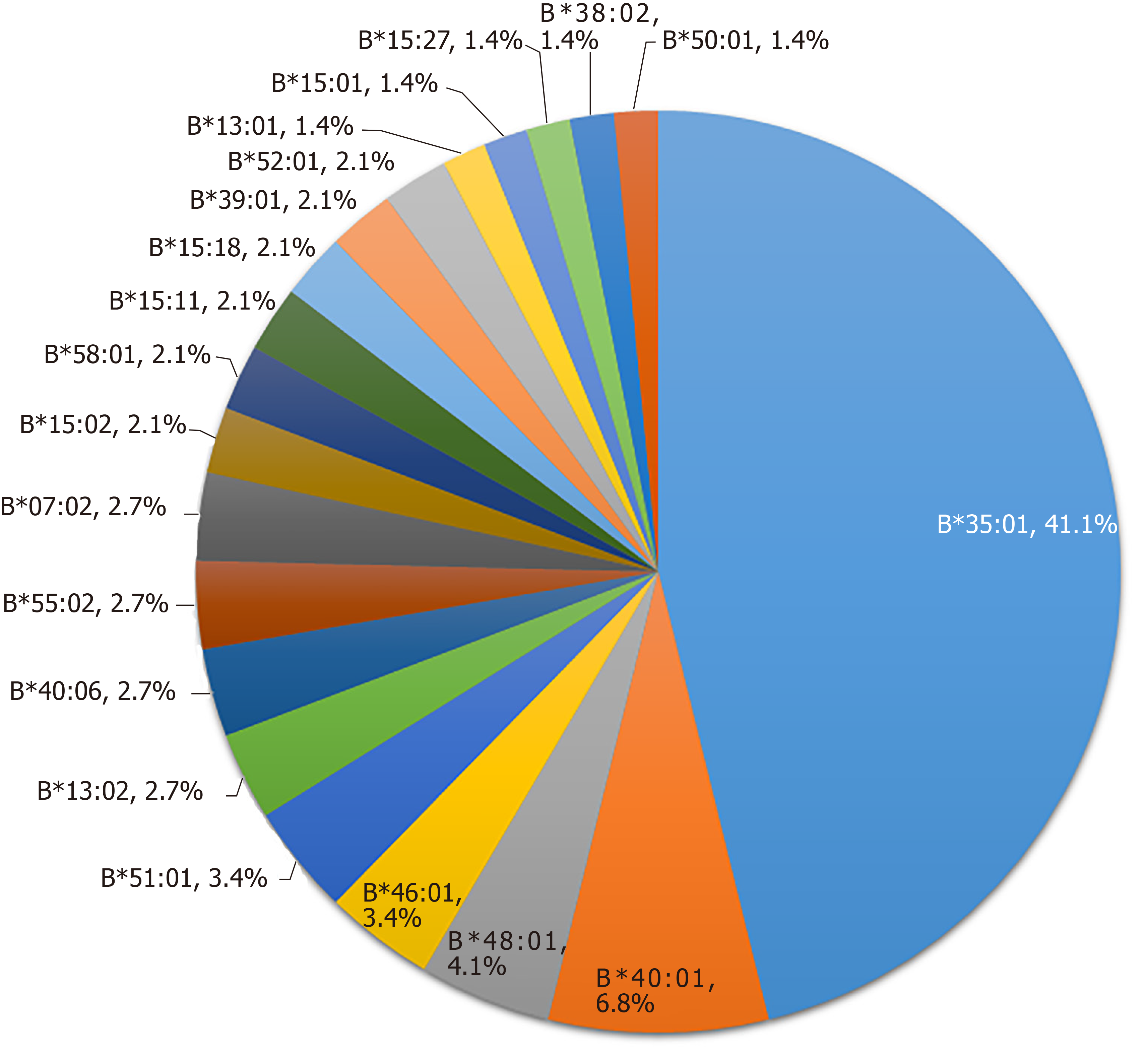
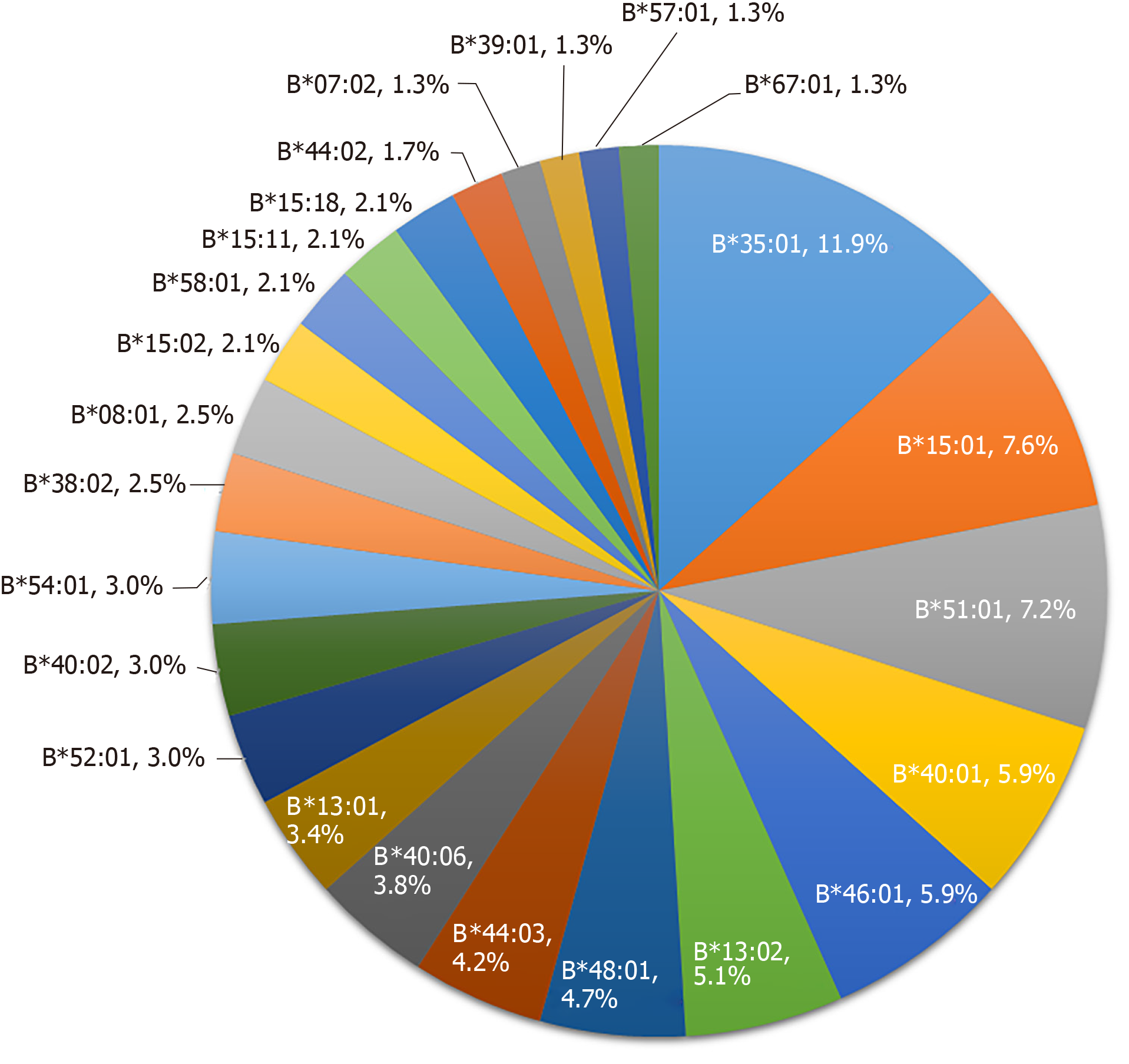
HLA-B alleles showed considerable variability. A total of 48 alleles were identified in the 73 PM-DILI patients and 118 other-DILI patients, and the distribution of alleles with frequencies > 1% is shown in Figure 2 and Figure 3. The frequency of HLA-B*35:01 was 41.1% in the PM-DILI group compared with 11.9% (P = 4.30×10-11, OR = 11.11, 95%CI: 5.57-22.19) in the other-DILI group and 2.7% (P = 6.22 × 10-166, OR = 62.62, 95%CI: 35.91-109.20) in the Han-MHC database of 10689 healthy control subjects[23]. These results validated the previous conclusion that HLA-B*35:01 is a genetic risk factor for PM-DILI[17].
The sensitivity, specificity, positive predictive value, negative predictive value and Cohen’s kappa coefficient of the rs1055348 G allele for HLA-B*35:01 are shown in Table 5. Both in the PM-DILI group and the other-DILI group, the rs1055348 G allele had 100% sensitivity and > 95% specificity for HLA-B*35:01, which indicated that HLA-B*35:01 could be tagged by rs1055348. And a nearly perfect agreement was found between the rs1055348 G allele and HLA-B*35:01 with κ = 0.959 in the PM-DILI group and κ = 0.911 in the other-DILI group.
| Group | TP | TN | FP | FN | Sensitivity (95%CI) | Specificity (95%CI) | PPV (95%CI) | NPV (95%CI) | κ (95%CI) |
| PM-DILI | 60 | 84 | 2 | 0 | 100.0 (94.0-100.0) | 97.7 (91.9-99.7) | 96.8 (88.8-99.6) | 100.0 (95.7-100.0) | 0.959 (0.879-1.000) |
| Other-DILI | 28 | 204 | 4 | 0 | 100.0 (87.7-100.0) | 98.1 (95.1-99.5) | 87.5 (71.0-96.5) | 100.0 (98.2-100.0) | 0.911 (0.825-0.996) |
It is difficult to assess the relationship between PM and liver injury or to predict PM-DILI. In China, the use of PM is often combined with Western drugs. Identification of a specific genetic biomarker of PM-DILI is helpful for the diagnosis or prediction of PM-DILI. We successfully identified three SNPs, rs111686806 in the HLA-A gene, rs1055348 in the HLA-B gene and rs202047044 in the HLA-DRB1 gene, to be risk factors for PM-DILI compared with the control group. HLA-A and HLA-B are HLA class I alleles associated with cytotoxic CD8+ T cell-mediated livery injury, while HLA-DRB1 is an HLA class II allele involved in the interaction between antigen-presenting cells and CD4+ T cells, which leads to liver injury[24]. These results indicate that the adaptive immunity response may play an important role in the pathogenesis of PM-DILI.
Notably, when compared with that in the other-DILI group, rs1055348 in the HLA-B gene was the only potential genetic risk factor, which suggests that rs1055348 in the HLA-B gene is specific to PM-DILI and may help strengthen the diagnosis of PM-DILI, particularly in distinguishing PM-DILI from other-DILI.
rs1055348 is a T > G or T > C mutation in the 3’-untranslated region of the HLA-B gene. The 3’-untranslated region of a gene is best known for regulating mRNA expression, including localization, translation, and degradation[25]. Thus, rs1055348 in the HLA-B gene may increase susceptibility to PM-DILI by regulating expression of the HLA-B gene.
On the other hand, HLA-associated DILI is normally correlated with HLA genotyping which has different amino acid sequences, but not the untranslated region. Therefore, another mechanism may be that rs1055348 in the HLA-B gene may be a tag for one HLA-B genotype. HLA-B*35:01 was reported to be a genetic risk factor for PM-DILI and a potential biomarker for predicting PM-DILI[17]. The clinical performance of rs1055348 in the HLA-B gene to become a tag for HLA-B*35:01 has been evaluated in African, Ad Mixed American, East Asian and European populations from the 1000 Genomes Dataset[26], and it was concluded that rs1055348 may not be suitable due to a low specificity of 80.22%.
Regardless, in the current study, we evaluated the potential for the rs1055348 G allele in the HLA-B gene to become a tag for HLA-B*35:01. In both the PM-DILI group and the other-DILI group, the sensitivity was 100%, and the specificity was > 95%. The 100% sensitivity and > 95% specificity were considered sufficient for the clinical utility of tagging SNPs in a previous study[27]. Thus, the rs1055348 G allele in the HLA-B gene may serve as a tag for HLA-B*35:01 in PM-DILI patients and other-DILI patients in the Han Chinese population.
Furthermore, compared with HLA-B*35:01 typing, testing of rs1055348, a tag SNP, is cost-effective and time efficient. In fact, real-world experience demonstrated that using tag SNP rs116488202 to screen for axial spondyloarthritis can yield a 94% reduction in costs relative to cases in which all patients were assessed with HLA-B*27[28], which suggests that screening for tag SNP rs1055348 before consuming PM would produce considerable economic benefits.
Studies on the biochemical characteristics of traditional Chinese medicines and Western medicines are controversial[4,5,29]. In the current study, the distribution of gender, age, ALT level, AST level and TBil level were significantly different between the PM-DILI and other-DILI groups (P < 0.05). Patients with the rs1055348 G allele showed higher levels of ALT and AST than did those without the rs1055348 G allele in both groups, which indicates that immune factors are involved not only in the onset of DILI but also in the process of inflammation. In addition, the higher frequency of rs1055348 may be another reason for the higher levels of ALT and AST in the PM-DILI group. However, the causative agents in the other DILI group consisted of multiple types of drugs, including both Chinese herbal medicines and Western medicines. More cases are needed to further compare the clinical features of PM-DILI with other-DILI.
The limitations of this study include the small sample size and number of patients taking PM only. A prospective study with a larger sample size of patients taking PM only is needed to further evaluate the role of rs1055348 in the HLA-B gene in PM-DILI.
In summary, our systematic study identified rs111686806 in the HLA-A gene, rs1055348 in the HLA-B gene, and rs202047044 in the HLA-DRB1 gene as genetic risk factors for PM-DILI, of which, rs1055348 in the HLA-B gene was specific to PM-DILI compared to other drugs. As a tag for HLA-B*35:01, rs1055348 in the HLA-B gene may become an alternative predictive biomarker of PM-DILI. Further study to explore the mechanisms of rs1055348 in PM-DILI is needed.
Polygonum multiflorum (PM) is a well-known traditional Chinese herbal medicine. However, reports of PM-induced liver injury (PM-DILI) have been increasing in recent years. To increase the safe use of PM, the identification of biomarkers to prevent and diagnose PM-DILI is essential.
Susceptibility to DILI is considered to be genetically determined. Recently, an association between HLA-B*35:01 and PM-DILI was reported. However, HLA genotyping is time consuming and expensive compared with the detection of single-nucleotide polymorphisms (SNPs). The identification of SNPs which could serve as biomarkers of PM-DILI would improve the application of PM and produce considerable economic benefits.
The objective of this study was to identify SNPs that indicate susceptibility to PM-DILI.
The study enrolled 73 PM-DILI patients, 118 other drug-induced liver injury (other-DILI) patients and 191 healthy controls. Whole-exome sequencing was performed on 8 PM-DILI patients and 8 healthy controls who were randomly selected from 73 PM-DILI patients and 191 healthy controls. Nineteen SNPs which were selected from previous whole-exome sequencing were screened in the remaining subjects using the MassARRAY system. HLA-B high-resolution genotyping was performed for 73 PM-DILI patients and 118 other-DILI patients. The Han-MHC database was selected as a population control for HLA-B analysis. SPSS (version 20.0, SPSS Inc., Chicago, IL, United States), PLINK software (version 1.07), MedCalc for Windows (version 15.2, MedCalc Software, Ostend, Belgium), and GraphPad QuickCalcs (GraphPad Software Inc., La Jolla, CA, United States) were used for statistical analyses.
Compared with the control group, three SNPs, rs111686806 in the HLA-B gene, rs1055348 in the HLA-B gene, and rs202047044 in the HLA-DRB1 gene were associated with PM-DILI. Only rs1055348 had a significantly higher frequency in the PM-DILI group than in the other-DILI group (P = 1.84 × 10-10), which suggested that rs1055348 was a specific risk factor for PM-DILI. HLA-B*35:01 was confirmed to be associated with PM-DILI. Furthermore, rs1055348 may serve as a tag for HLA-B*35:01 with 100% sensitivity and > 95% specificity.
rs111686806, rs1055348 and rs202047044 are associated with PM-DILI, and rs1055348 is specific to PM-DILI. As a tag for HLA-B*35:01, rs1055348 may be used as an alternative predictive biomarker of PM-DILI.
Screening for tag rs1055348 before consuming PM would improve the safe use of PM and produce considerable economic benefits. A prospective study with a larger sample size of patients taking PM only is needed to further evaluate the role of rs1055348 in the HLA-B gene in PM-DILI.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Ng QX, Kagawa T, Shimizu Y, Tarantino G S-Editor: Tang JZ L-Editor: Webster JR E-Editor: Ma YJ
| 1. | Tarantino G, Pezzullo MG, di Minno MN, Milone F, Pezzullo LS, Milone M, Capone D. Drug-induced liver injury due to "natural products" used for weight loss: a case report. World J Gastroenterol. 2009;15:2414-2417. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 47] [Cited by in RCA: 36] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 2. | Shen T, Liu Y, Shang J, Xie Q, Li J, Yan M, Xu J, Niu J, Liu J, Watkins PB, Aithal GP, Andrade RJ, Dou X, Yao L, Lv F, Wang Q, Li Y, Zhou X, Zhang Y, Zong P, Wan B, Zou Z, Yang D, Nie Y, Li D, Wang Y, Han X, Zhuang H, Mao Y, Chen C. Incidence and Etiology of Drug-Induced Liver Injury in Mainland China. Gastroenterology. 2019;156:2230-2241.e11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 245] [Cited by in RCA: 384] [Article Influence: 64.0] [Reference Citation Analysis (0)] |
| 3. | Byeon JH, Kil JH, Ahn YC, Son CG. Systematic review of published data on herb induced liver injury. J Ethnopharmacol. 2019;233:190-196. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 62] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 4. | Lu RJ, Zhang Y, Tang FL, Zheng ZW, Fan ZD, Zhu SM, Qian XF, Liu NN. Clinical characteristics of drug-induced liver injury and related risk factors. Exp Ther Med. 2016;12:2606-2616. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 44] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 5. | Zhu Y, Niu M, Chen J, Zou ZS, Ma ZJ, Liu SH, Wang RL, He TT, Song HB, Wang ZX, Pu SB, Ma X, Wang LF, Bai ZF, Zhao YL, Li YG, Wang JB, Xiao XH. Specialized Committee for Drug-Induced Liver Diseases, Division of Drug-Induced Diseases, Chinese Pharmacological Society. Hepatobiliary and pancreatic: Comparison between Chinese herbal medicine and Western medicine-induced liver injury of 1985 patients. J Gastroenterol Hepatol. 2016;31:1476-1482. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 79] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 6. | Chinese Pharmacopoeia Commission. Pharmacopoeia of the people’s republic of China (Part one). 1st ed. Beijing: China Medical Science Press. 2015;175-176. |
| 7. | Zhu Y, Liu SH, Wang JB, Song HB, Li YG, He TT, Ma X, Wang ZX, Wang-Li-ping, Zhou K, Bai YF, Zou ZS, Xiao XH. Clinical Analysis of Drug-induced Liver Injury Caused by Polygonum multiflorum and its Preparations. Zhongguo Zhong Xi Yi Jie He Za Zhi. 2015;35:1442-1447. [PubMed] |
| 8. | Wang Y, Wang L, Saxena R, Wee A, Yang R, Tian Q, Zhang J, Zhao X, Jia J. Clinicopathological features of He Shou Wu-induced liver injury: This ancient anti-aging therapy is not liver-friendly. Liver Int. 2019;39:389-400. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 19] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 9. | Laird AR, Ramchandani N, deGoma EM, Avula B, Khan IA, Gesundheit N. Acute hepatitis associated with the use of an herbal supplement (Polygonum multiflorum) mimicking iron-overload syndrome. J Clin Gastroenterol. 2008;42:861-862. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 32] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 10. | Jung KA, Min HJ, Yoo SS, Kim HJ, Choi SN, Ha CY, Kim HJ, Kim TH, Jung WT, Lee OJ, Lee JS, Shim SG. Drug-Induced Liver Injury: Twenty Five Cases of Acute Hepatitis Following Ingestion of Polygonum multiflorum Thunb. Gut Liver. 2011;5:493-499. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 101] [Cited by in RCA: 118] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 11. | Bounda GA, Feng YU. Review of clinical studies of Polygonum multiflorum Thunb. and its isolated bioactive compounds. Pharmacognosy Res. 2015;7:225-236. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 72] [Cited by in RCA: 70] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 12. | Wu X, Chen X, Huang Q, Fang D, Li G, Zhang G. Toxicity of raw and processed roots of Polygonum multiflorum. Fitoterapia. 2012;83:469-475. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 83] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 13. | Meng YK, Li CY, Li RY, He LZ, Cui HR, Yin P, Zhang CE, Li PY, Sang XX, Wang Y, Niu M, Zhang YM, Guo YM, Sun R, Wang JB, Bai ZF, Xiao XH. Cis-stilbene glucoside in Polygonum multiflorum induces immunological idiosyncratic hepatotoxicity in LPS-treated rats by suppressing PPAR-γ. Acta Pharmacol Sin. 2017;38:1340-1352. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 51] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 14. | Lin L, Liu Y, Fu S, Qu C, Li H, Ni J. Inhibition of Mitochondrial Complex Function-The Hepatotoxicity Mechanism of Emodin Based on Quantitative Proteomic Analyses. Cells. 2019;3:pii: E263. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 52] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 15. | Ng QX, Yong CSK, Loke W, Yeo WS, Soh AYS. Escitalopram-induced liver injury: A case report and review of literature. World J Hepatol. 2019;11:719-724. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 6] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 16. | Urban TJ, Daly AK, Aithal GP. Genetic basis of drug-induced liver injury: present and future. Semin Liver Dis. 2014;34:123-133. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 83] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 17. | Li C, Rao T, Chen X, Zou Z, Wei A, Tang J, Xiong P, Li P, Jing J, He T, Bai Z, Yin J, Tan Z, Yu P, Zhou H, Wang J, Xiao X, Ouyang D. HLA-B*35:01 Allele Is a Potential Biomarker for Predicting Polygonum multiflorum-Induced Liver Injury in Humans. Hepatology. 2019;70:346-357. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 71] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 18. | Lucena MI, Molokhia M, Shen Y, Urban TJ, Aithal GP, Andrade RJ, Day CP, Ruiz-Cabello F, Donaldson PT, Stephens C, Pirmohamed M, Romero-Gomez M, Navarro JM, Fontana RJ, Miller M, Groome M, Bondon-Guitton E, Conforti A, Stricker BH, Carvajal A, Ibanez L, Yue QY, Eichelbaum M, Floratos A, Pe'er I, Daly MJ, Goldstein DB, Dillon JF, Nelson MR, Watkins PB, Daly AK; Spanish DILI Registry; EUDRAGENE; DILIN; DILIGEN; International SAEC. Susceptibility to amoxicillin-clavulanate-induced liver injury is influenced by multiple HLA class I and II alleles. Gastroenterology. 2011;141:338-347. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 408] [Cited by in RCA: 350] [Article Influence: 25.0] [Reference Citation Analysis (0)] |
| 19. | Ahmad J, Odin JA, Hayashi PH, Chalasani N, Fontana RJ, Barnhart H, Cirulli ET, Kleiner DE, Hoofnagle JH. Identification and Characterization of Fenofibrate-Induced Liver Injury. Dig Dis Sci. 2017;62:3596-3604. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 40] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 20. | Singla N, Gupta D, Birbian N, Singh J. Association of NAT2, GST and CYP2E1 polymorphisms and anti-tuberculosis drug-induced hepatotoxicity. Tuberculosis (Edinb). 2014;94:293-298. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 52] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 21. | Danan G, Benichou C. Causality assessment of adverse reactions to drugs--I. A novel method based on the conclusions of international consensus meetings: application to drug-induced liver injuries. J Clin Epidemiol. 1993;46:1323-1330. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1015] [Cited by in RCA: 1071] [Article Influence: 33.5] [Reference Citation Analysis (0)] |
| 22. | Bénichou C. Criteria of drug-induced liver disorders. Report of an international consensus meeting. J Hepatol. 1990;11:272-276. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 780] [Cited by in RCA: 773] [Article Influence: 22.1] [Reference Citation Analysis (0)] |
| 23. | Zhou F, Cao H, Zuo X, Zhang T, Zhang X, Liu X, Xu R, Chen G, Zhang Y, Zheng X, Jin X, Gao J, Mei J, Sheng Y, Li Q, Liang B, Shen J, Shen C, Jiang H, Zhu C, Fan X, Xu F, Yue M, Yin X, Ye C, Zhang C, Liu X, Yu L, Wu J, Chen M, Zhuang X, Tang L, Shao H, Wu L, Li J, Xu Y, Zhang Y, Zhao S, Wang Y, Li G, Xu H, Zeng L, Wang J, Bai M, Chen Y, Chen W, Kang T, Wu Y, Xu X, Zhu Z, Cui Y, Wang Z, Yang C, Wang P, Xiang L, Chen X, Zhang A, Gao X, Zhang F, Xu J, Zheng M, Zheng J, Zhang J, Yu X, Li Y, Yang S, Yang H, Wang J, Liu J, Hammarström L, Sun L, Wang J, Zhang X. Deep sequencing of the MHC region in the Chinese population contributes to studies of complex disease. Nat Genet. 2016;48:740-746. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 146] [Cited by in RCA: 171] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 24. | Chalasani N, Bonkovsky HL, Fontana R, Lee W, Stolz A, Talwalkar J, Reddy KR, Watkins PB, Navarro V, Barnhart H, Gu J, Serrano J; United States Drug Induced Liver Injury Network. Features and Outcomes of 899 Patients With Drug-Induced Liver Injury: The DILIN Prospective Study. Gastroenterology. 2015;148:1340-52.e7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 635] [Cited by in RCA: 643] [Article Influence: 64.3] [Reference Citation Analysis (0)] |
| 25. | Mayr C. Regulation by 3'-Untranslated Regions. Annu Rev Genet. 2017;51:171-194. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 312] [Cited by in RCA: 403] [Article Influence: 50.4] [Reference Citation Analysis (0)] |
| 26. | Erlichster M. Improving Application of Human Leukocyte Antigen Genotyping in Precision Medicine. PhD Thesis, University of Melbourne. 2018. Available from: https://minerva-access.unimelb.edu.au/handle/11343/218155. |
| 27. | Liu X, Sun J, Yu H, Chen H, Wang J, Zou H, Lu D, Xu J, Zheng SL. Tag SNPs for HLA-B alleles that are associated with drug response and disease risk in the Chinese Han population. Pharmacogenomics J. 2015;15:467-472. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 10] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 28. | Nguyen HV, O'Rielly DD, Rahman P. Real-world Experience of Using HLA-B*27 Tag-single-nucleotide Polymorphism Assay to Screen for Axial Spondyloarthritis. J Rheumatol. 2018;45:1712. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 29. | Amadi CN, Orisakwe OE. Herb-Induced Liver Injuries in Developing Nations: An Update. Toxics. 2018;6. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 48] [Article Influence: 6.9] [Reference Citation Analysis (0)] |