Published online Feb 7, 2019. doi: 10.3748/wjg.v25.i5.584
Peer-review started: October 25, 2018
First decision: November 29, 2018
Revised: January 10, 2019
Accepted: January 18, 2019
Article in press: January 18, 2019
Published online: February 7, 2019
Processing time: 99 Days and 1.3 Hours
Claudin-7, one of the important components of cellular tight junctions, is currently considered to be expressed abnormally in colorectal inflammation and colorectal cancer. However, there is currently no effective animal model to study its specific mechanism. Therefore, we constructed three lines of Claudin-7 knockout mice using the Cre/LoxP system.
To determine the function of the tumor suppressor gene Claudin-7 by generating three lines of Claudin-7 gene knockout mice.
We crossed Claudin-7-floxed mice with CMV-Cre, vil1-Cre, and villin-CreERT2 transgenic mice, and the offspring were self-crossed to obtain conventional Claudin-7 knockout mice, conditional (intestinal specific) Claudin-7 knockout mice, and inducible conditional Claudin-7 knockout mice. Intraperitoneal injection of tamoxifen into the inducible conditional Claudin-7 knockout mice can induce the knockout of Claudin-7. PCR and agarose gel electrophoresis were used to identify mouse genotypes, and Western blot was used to confirm the knockout of Claudin-7. The mental state, body length, and survival time of these mice were observed. The dying mice were sacrificed, and hematoxylin-eosin (HE) staining and immunohistochemical staining were performed to observe changes in intestinal structure and proliferation markers.
We generated Claudin-7-floxed mice and three lines of Claudin-7 gene knockout mice using the Cre/LoxP system successfully. Conventional and intestinal specific Claudin-7 knockout mice were stunted and died during the perinatal period, and intestinal HE staining in these mice revealed mucosal gland structure disappearance and connective tissue hyperplasia with extensive inflammatory cell infiltration. The inducible conditional Claudin-7 knockout mice had a normal phenotype at birth, but after the induction with tamoxifen, they exhibited a dying state. Intestinal HE staining showed significant inflammatory cell infiltration, and atypical hyperplasia and adenoma were also observed. Intestinal immunohistochemistry analysis showed abnormal expression and distribution of Ki67, and the normal intestinal proliferation balance was disrupted. The intestinal crypt size in inducible conditional Claudin-7 knockout mice was increased compared with control mice (small intestine: 54.1 ± 2.96 vs 38.4 ± 1.63; large intestine: 44.7 ± 1.93 vs 27.4 ± 0.60; P < 0.001).
The knockout of Claudin-7 in vivo causes extensive inflammation, atypical hyperplasia, and adenoma in intestinal tissue as well as animal death in mice. Claudin-7 may act as a tumor suppressor gene in the development of colorectal cancer.
Core tip: The intestinal tract of conventional and intestinal specific Claudin-7 knockout mice was characterized by extensive and severe inflammation. The development of inducible conditional knockout mice can control the knockout of Claudin-7 in a temporal and compartment specific manner and prolong the survival time of mice, which exhibited atypical hyperplasia and adenoma in the intestine. This study revealed the inhibitory role that Claudin-7 plays in colorectal inflammation and colorectal cancer.
- Citation: Xu C, Wang K, Ding YH, Li WJ, Ding L. Claudin-7 gene knockout causes destruction of intestinal structure and animal death in mice. World J Gastroenterol 2019; 25(5): 584-599
- URL: https://www.wjgnet.com/1007-9327/full/v25/i5/584.htm
- DOI: https://dx.doi.org/10.3748/wjg.v25.i5.584
Members of the Claudin family serve as important components of cellular tight junctions (TJs), and they mainly function to maintain cell polarity, regulate intercellular small molecule flux, and facilitate cell proliferation and differentiation[1-3]. Claudin-7 (Cldn7), one of the 27 members of the Claudin family, is mainly distributed in the stomach, lung, intestine, bladder, and kidney. Cldn7 was originally found in an extracellular Cl- barrier and Na+ channel and shown to affect extracellular permeability[4]. However, recent studies have shown that Cldn7 is abnormally expressed in different cancer tissues, especially in colon cancer, suggesting that alterations in its expression may affect the normal structure and function of TJs and be related to the occurrence of intestinal tumors[5-8]. Cldn7 is currently considered to play an inhibitory role in colorectal inflammation and colorectal cancer by most scholars[9-11].
The most effective way to study inhibitors in vivo is to knock out the gene in an animal and observe its overall phenotype. In recent years, the Cre/LoxP recombinase system has been widely used in novel gene targeting[12,13]. LoxP was inserted at both ends of the Cldn7 sequence to obtain heterozygous floxed mice. After crossing with CMV-Cre and vil1-Cre mice, the sequence between the two LoxP sites was excised and inherited by daughter cells. Shimizu was the first to report time-specific gene knockout animal models in which the time of gene knockout could be artificially controlled by injection with an inducer[14]. Therefore, we constructed conventional Cldn7 gene knockout (CKO) mice and conditional knockout (cKO) mice using the Cre/LoxP system. We also generated inducible conditional Cldn7 knockout (ICKO) mice and induced Cre expression by injecting tamoxifen. Hematoxylin-eosin (HE) staining showed that the intestinal structures in the CKO and cKO mice were severely damaged, and numerous inflammatory cells infiltrated. By injecting tamoxifen into the ICKO mice, we successfully established atypical hyperplasia and intestinal adenoma models. Immunohistochemistry analysis indicated that the expression and distribution of Ki67 in the intestinal tissues were dysregulated. The successful construction of mouse intestinal inflammation and intestinal adenoma models could provide a basis for further studying the role of Cldn7 in intestinal tumors.
We inserted a LoxP site into the intronic sequence downstream of exon 4 of the Cldn7 gene and inserted the FRT-neo-FRT-LoxP element into the upstream intronic sequence of exon 2 to obtain Cldn7-floxed mice. The CMV-Cre mice were purchased from the National Resource Center for Mutant Mice, the vil1-Cre mice were obtained from the China Pharmaceutical University (from the Jackson Laboratory, USA), and the villin-CreERT2 mice were donated by professor Sylvie Robine. All mice were housed according to specific pathogen-free grade animal feeding standards at an indoor temperature of 20-26 °C and a 12-h day/night cycle. The mice were fed a standard diet after sterilization and had free access to food and water. All animals were euthanized for tissue collection. All animal assay protocols were reviewed and approved by the Medical Ethics Committee of the Capital Medical University Affiliated Beijing Shijitan Hospital Institutional Review Board.
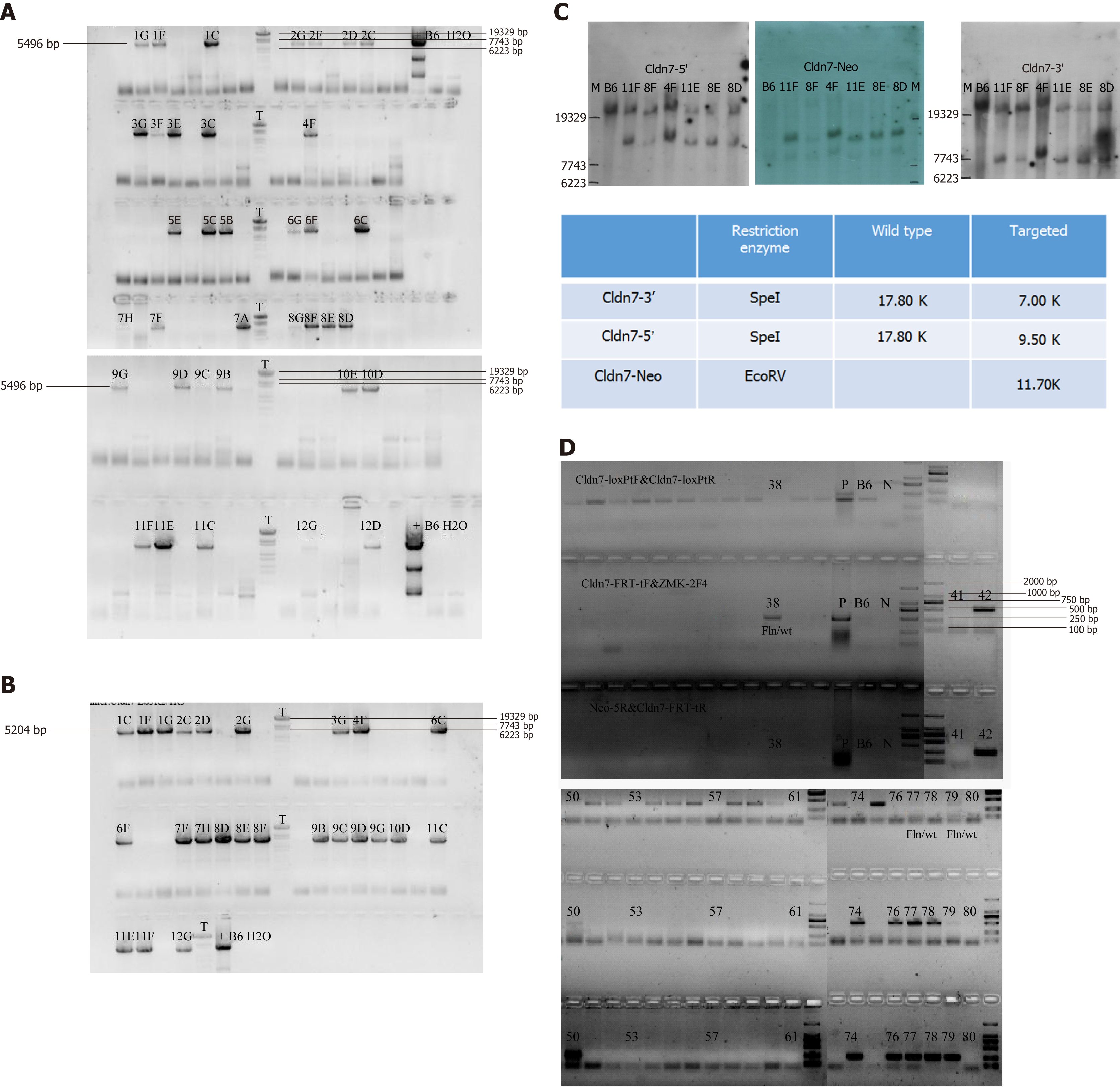
We constructed a targeting vector as shown in Figure 1A. After the final vector was sequenced for validation, it was transfected into embryonic stem (ES) cells by electroporation. From the 8th-10th day, ES cell clones were picked, and genomic DNA was extracted, amplified, digested with the EcoRV enzyme overnight, and slowly electrophoresed for 36 h. The target clones were screened by long range PCR and Southern blot.
Approximately 4-wk-old C57BL/6N female mice were selected and injected with pregnant mare serum gonadotropin and human chorionic gonadotropin to promote ovulation. Embryos were harvested on the 2nd day after cohousing the female mice with the male mice, and 12-15 ES cells were injected into each blastocyst after culturing overnight. After the injection, the blastocysts were cultured for 3 h in an incubator, and those with a normal morphology and intact transparent bands were selected for transplantation. After 8-10 wk of sexual maturation, female C57BL/6N mice were selected for uterine blastocyst transplantation. The mice born after successful transplantation were identified by PCR, and those with the fln/wt genotype were deemed to be chimeric mice. Chimeric mice were crossed with Flper mice and then backcrossed with wild-type C57BL/6N mice to obtain Cldn7-floxed mice (genotype: fl/wt), which were missing the entire Neo resistance gene.
Cldn7-floxed mice were crossed with CMV-Cre mice, vil1-Cre mice, and villin-CreERT2 mice to obtain Cldn7 CKO mice, Cldn7 cKO mice, and Cldn7 ICKO mice, respectively. Next, 50 mg of tamoxifen was dissolved in 5 mL of sterilized sunflower oil and mixed for 30 min to obtain the tamoxifen dilution. Six- to eight-wk-old ICKO mice were intraperitoneally injected with 100 μL of the tamoxifen dilution every 5 d to induce the Cldn7 knockout.
Various Cldn7 knockout mouse tissues were minced on ice and mixed with appropriate grinding beads and total protein extraction reagents containing different protease inhibitors; a tissue homogenizer was then used to extract total protein. Proteins were separated by SDS-PAGE and transferred onto nitrocellulose membranes. The membranes were incubated with a diluted rabbit polyclonal anti-Cldn7 antibody (1:1000, ab27487, Abcam, United States) at 4 °C overnight and then with a donkey anti-rabbit IgG antibody (1:10000, ab175780, Abcam). After blotting, the signals were detected with a Western blot scanner. GAPDH was used as the internal reference.
The intestinal tissues of Cldn7 knockout mice and control mice were washed in PBS and then placed in 10% formalin/PBS at 4 °C. After dehydration and clearing, the tissues were immersed in wax and then cut into 5-8-micron-thick sections. The sections were then dewaxed and stained with HE.
All tissues were embedded in wax blocks and cut into paraffin sections. After dewaxing, hydration, and antigen retrieval, the tissue sections were incubated for 10 min in 3% H2O2 and washed with 0.01 mol/L PBS. The sections were then incubated with a diluted rabbit polyclonal anti-Cldn7 antibody (ab27487, 1:200, Abcam, United States) and a rabbit monoclonal anti-Ki67 antibody (ab16667, 1:1000, Abcam, United States), followed by incubation with the corresponding horseradish peroxidase-labelled secondary antibody. The proteins were then developed in 3,3′-diaminobenzidine for coloration and assessment.
Statistical analyses were performed using IBM SPSS version 17.0 and GraphPad Prism version 6.0. All data are expressed as the mean ± SD. Differences between two groups were analysed by Student’s t-test and considered significant at P < 0.05.
We constructed the Cldn-7 gene knockout targeting vector as shown in Figure 1A, and sequenced the final vector using a unidirectional primer (LoxPtF: GTACGAGTTTGGACCTGCCA) to detect whether the 34 bp LoxP site was inserted correctly (Figure 1B). The LoxP site is shown in yellow, and the 3’-untranslated region is shown in green. FRT sequencing from the 5’ end was performed using a unidirectional primer (Cldn7-FRT-tF: CTGATCTGGGTGTCCCACGT), as the FRT site serves as a screening marker for Neo. By removing the Neo resistance gene, the FRT site was also removed. The second LoxP site is shown in green, the FRT site is shown in yellow, and Neo is shown in purple (Figure 1C). FRT 3’ sequencing was then performed (FRT-tR: CGATGAAACCGTTCCAGGTA), and the presence of another FRT site is shown in pink font (Figure 1D). Therefore, the final gene targeting vector was correct.
The targeting vector was electroporated into B6/BLU ES cells for targeting, and some drug-resistant ES cell clones were obtained. Two methods were used to prevent false-positive results and detect target clones, long range PCR and Southern blot. First, we tested whether the 5' homologous arm was correct (Figure 2A). The 5496 bp product was a positive clone containing LoxP. The 3’ end was also detected (Figure 2B), and the 5204 bp product was a positive clone. The Southern blot results are shown in Figure 2C. Genomic DNA extracted from the transfected ES cells was digested with Spe I restriction endonuclease. Gene fragments of 17.8 kb and 9.5 kb were obtained from the Cldn7 5’ end of the wild-type and target clones, respectively, and gene fragments of 17.8 kb and 7 kb were obtained from the Cldn7 3’ end. When using the EcoRV restriction enzyme (probe on Neo), an 11.7 kb gene fragment was obtained from the target clone. The Southern blot results showed that clones 8D, 8E, 4F, 11E, and 11F were the final positive clones.
The positive clone 8D was selected for blastocyst injection, and the newborn mice after blastocyst transfer were genotyped by PCR. The primer information is shown in Table 1. The mice numbered 50 and 77 were deemed chimeric mice with the genotype fln/wt (Figure 2D). The chimeric mice were crossed with Flper mice to remove the Neo resistance gene, and the resulting mice were mated with wild-type C57BL/6N mice to completely delete the Neo resistance gene, successfully yielding Cldn7-floxed mice.
| Sequence (5’-3’) | Results |
| Cldn7 loxP-tF primer : GTACGAGTTTGGACCT GCCA | Fln/Fln = 493 bp; Fln/wt = 493/375 bp; wt/wt = 375 bp |
| Cldn7-LoxP-tR primer : TGTGCAA GGATCTGGGTCTG | |
| Cldn7-FRT-tF primer : CTGATCTGGGTGTCCCACGT | Fln/Fln = 507 bp; Fln/wt = 507/0 bp; wt/wt = 0bp |
| ZMK-2F4 primer: GCATCGCATTGTCTGAG TAGGTG | |
| Neo-5R primer : GGCTGG ACGTAAACTCCTC | Fln/Fln = 259 bp; Fln/wt = 259/0bp; wt/wt = 0 bp |
| Cldn7-FRT-tR primer: CGATGAAACC GTTCCAGGTA |
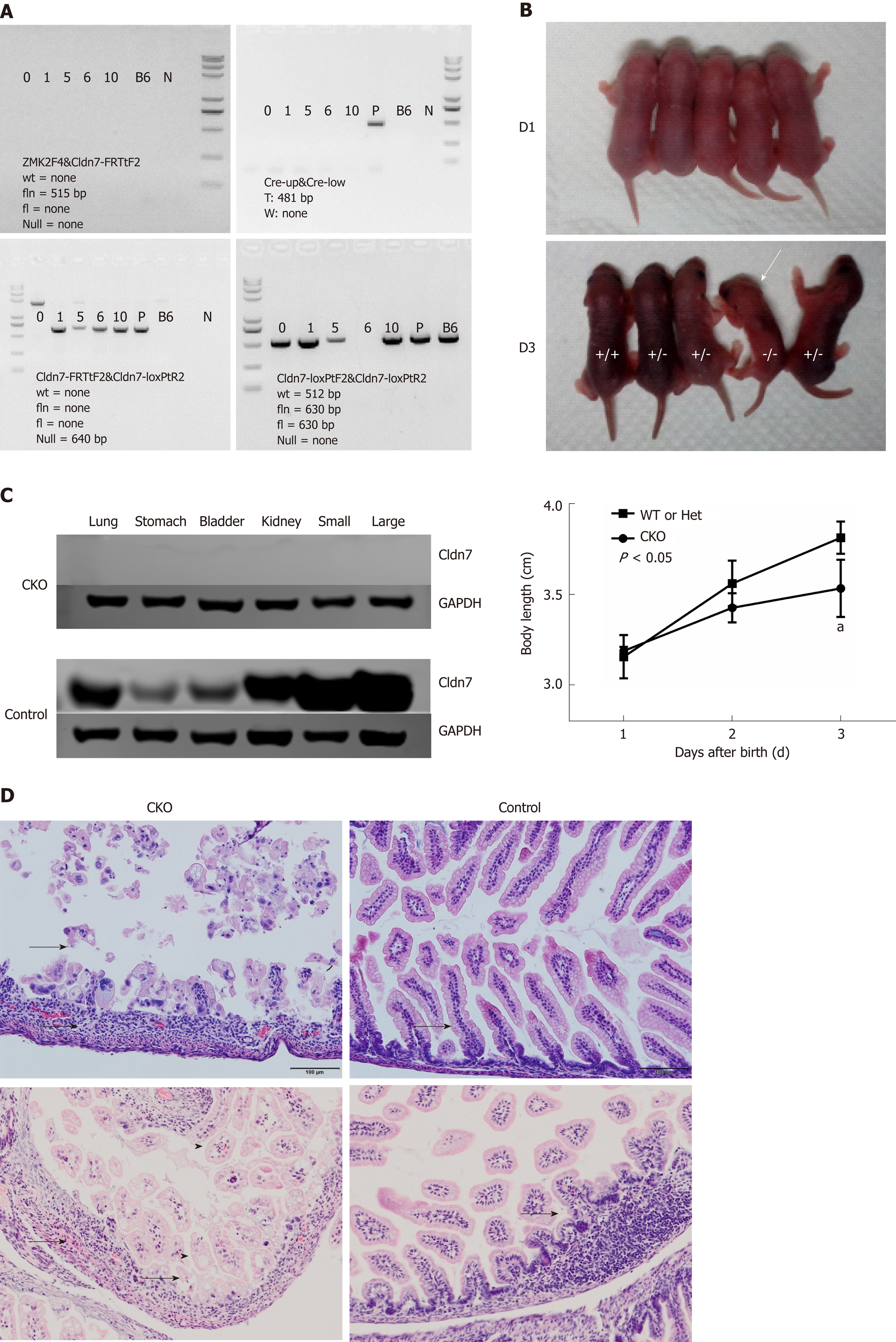
Cldn7-floxed mice were crossed with CMV-Cre mice, and the offspring were then self-crossed. Genomic DNA was isolated from the tails for genotyping. The primer information is shown in Table 2. The mouse with the Null/Null CreW genotype was considered the Cldn7 CKO mouse. We considered newborn mice from the same litter as an example. We first evaluated whether Neo was completely deleted. None of the samples showed an fln band at 515 bp, indicating that Neo had been deleted completely. Furthermore, none of the samples showed the Cre band at 481 bp, suggesting that the genotype of all samples was CreW. Next, the banding results showed that mice numbered 1, 5, 6, and 10 had a null band at 640 bp, suggesting the presence of Cre-mediated recombination. The final step was to identify whether the mice were homozygous, and mouse 6 was determined to be homozygous for the Null/Null genotype. Therefore, mouse 6 was deemed the Cldn7 CKO mouse with the Null/Null CreW genotype (Figure 3A).
| Primer sequence (5’-3’) | Results | Description |
| Cldn7-FRTtF2: CCTGGGATCTGATCTGGGTG | wt = 600 bp, fln = none; fl = 756 bp, Null = none | Check if flper is reorganized |
| Cldn7-FRTtR2: GGCAGGTAGCCTTAGGATGG | ||
| ZMK2F4: GCATCGCATTGTCTGAGTAGGTG | wt = none, fln = 515 bp; fl = none, Null = none | Check if Neo is deleted completely |
| Cldn7-FRTtF2: CCTGGGATCTGATCTGGGTG | ||
| Cldn7-loxPtF2: CTTGGGAGACATCAGGTCCG | wt = 512 bp, fln = 630 bp; fl = 630 bp, Null = none | Homozygous identification |
| Cldn7-loxPtR2: GAGGCAATAGGCCCAAGGAG | ||
| Cldn7-FRTtF2: CCTGGGATCTGATCTGGGTG | wt = none, fln = none; fl = none, Null = 640 bp | Check if Cre mediated recombination occurs |
| Cldn7-loxPtR2: GAGGCAATAGGCCCAAGGAG | ||
| Cre-up: GCCTGCATTACCGGTCGATGC | T: 481 bp; W: none | Detect Cre |
| Cre-low: CAGGGTGTTATAAGCAATCCC |
Cldn7 CKO mice were born similarly to heterozygous and wild-type mice. However, the lengths of the Cldn7 CKO mice increased significantly slower than those of the control mice (P < 0.05).
Beginning on the third day, CKO mice were thin, lacked energy, showed signs of lethargy, exhibited decreased body temperature, and had reduced activities, all of which suggested a state of dying (Figure 3B).
The dying mice were sacrificed, and their lungs, stomachs, bladders, kidneys, small intestines, and large intestines were collected. Western blot analysis showed that Cldn7 was not expressed in any of the tissues analyzed in CKO mice, while the control mice expressed Cldn7 in all of these tissues (Figure 3C). Intestinal HE staining showed obvious atrophy, thinning or loss of intestinal mucosa, connective tissue hyperplasia with inflammatory cell infiltration, residual intestinal mucosal epithelial vacuolar degeneration, villus shortening, and lymphatic expansion. Intestinal HE staining of the control mice showed no obvious histopathological changes (Figure 3D). Therefore, Cldn7 CKO mice showed significantly slow growth and appeared to be dying on the third day. HE staining showed severe intestinal destruction, loss of intestinal mucosal structure, and infiltration of numerous inflammatory cells.
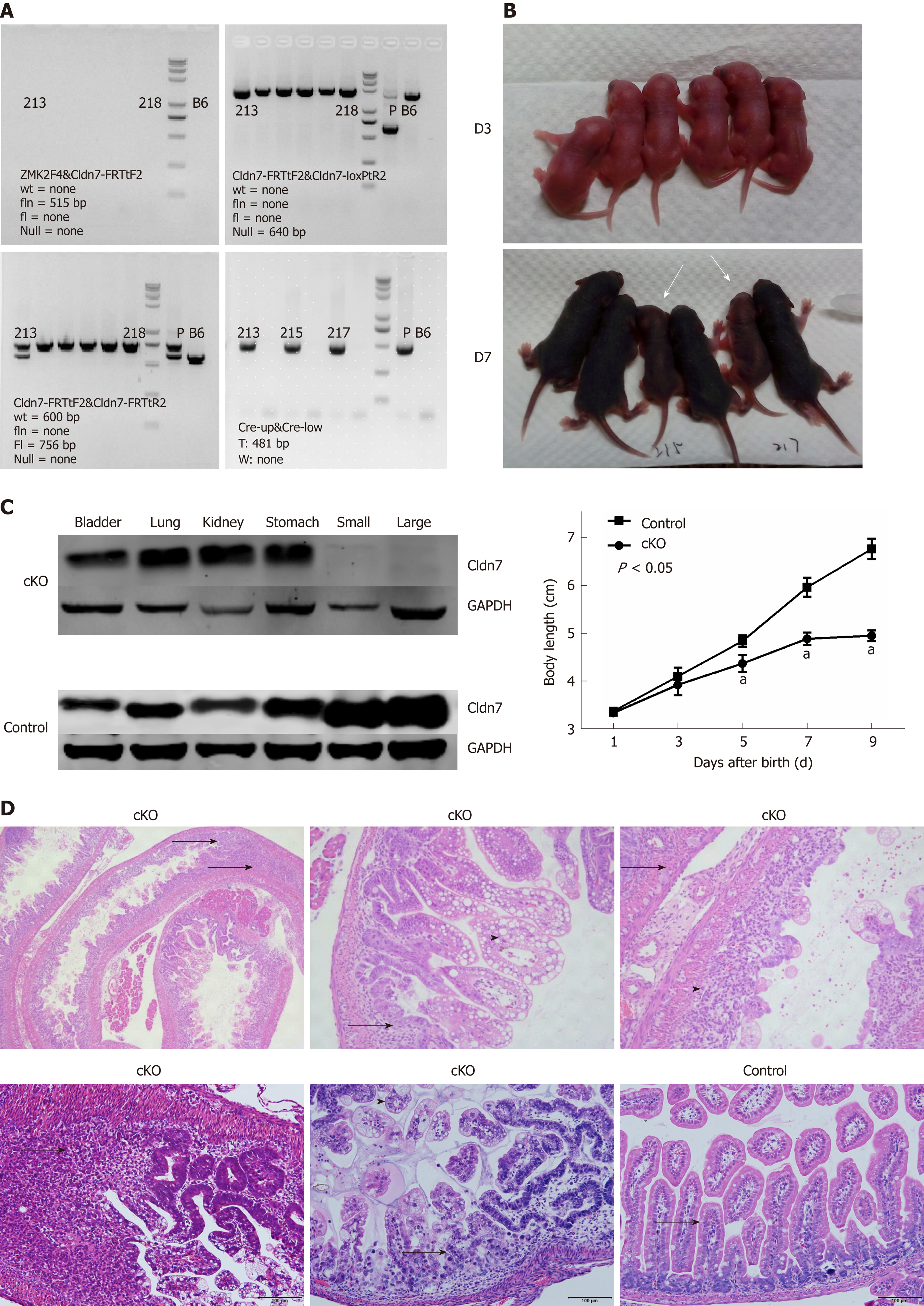
Cldn7-floxed mice were crossed with vil1-Cre mice, and the offspring were then self-crossed. Genomic DNA was isolated for genotyping analysis. The mouse with the Cldn7fl/fl; villin-CreT (fl/fl CreT) genotype was deemed the Cldn7 cKO mouse. None of the samples showed a fln band at 515 bp or a null band at 640 bp, indicating that Neo had been deleted completely and that no Cre recombination occurred. We next detected whether flper recombination occurred by the detection of an fl band at 756 bp. The results showed that No. 213 mouse had the fl/wt genotype, while all other mice had the fl/fl genotype. Finally, Cre was detected, and a band at 481 bp, which corresponded to CreT, was observed in mice numbered 213, 215, and 217, while all other mice displayed a band corresponding to CreW. Therefore, the mice numbered 215 and 217 were deemed Cldn7 cKO mice (Figure 4A).
Cldn7 cKO mice were normal at birth compared to control mice. But Cldn7 cKO mice were obviously thin, their body length increased slowly from the 5th day after birth, and their growth rate was significantly slower than that of control mice (P < 0.05). On the 9th day, Cldn7 cKO mice appeared to be languid, with reduced or even inactive activities, leaving only a slight breath. The body temperature of the mouse was reduced and it was in a state of dying (Figure 4B).
The dying mice were sacrificed, and we found that the expression levels of Cldn7 in the lung, stomach, bladder, and kidney tissues of cKO mice were normal, while Cldn7 expression was absent in the small and large intestines. All control mouse tissues expressed Cldn7 (Figure 4C). Intestinal HE staining showed obvious mucosal atrophy, mucosal gland structure disappearance, and connective tissue hyperplasia with extensive inflammatory cell infiltration. Inflammatory lesions were observed everywhere (Figure 4D), and mucosal epithelial vacuolar degeneration was observable after magnification. No obvious histopathological changes were observed in the mucosal glands, submucosa, or muscular layers of the intestines of fl/fl CreW mice. Cldn7 cKO mice excluded the influence of other organs lacking Cldn7 expression on mice and prolonged the survival time. However, because the cKO mice still showed intestinal inflammation, we generated Cldn7 ICKO mice on this basis to try to obtain an adenoma model.
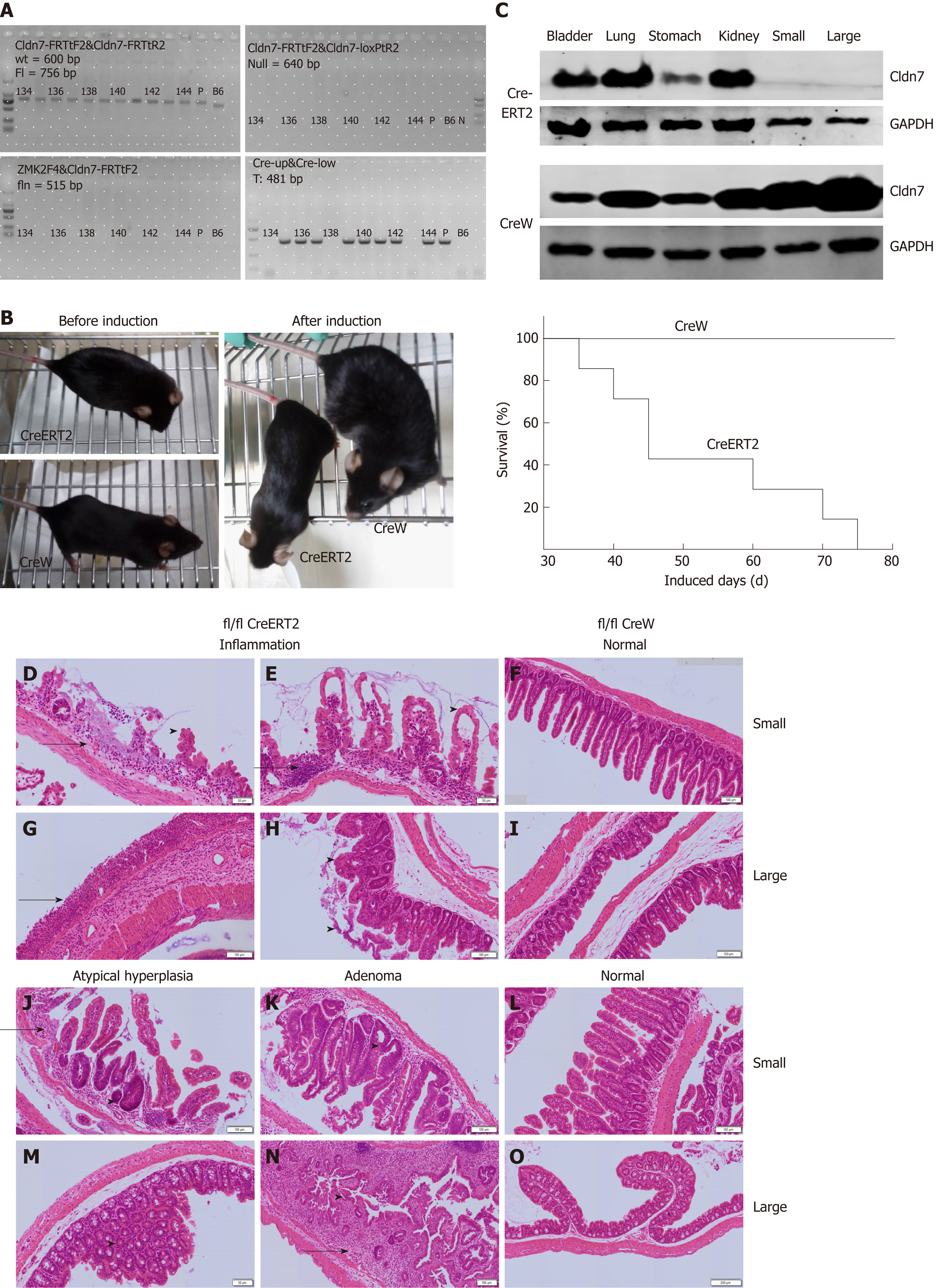
Cldn7-floxed mice were crossed with villin-CreERT2 mice, and the offspring were then self-crossed. PCR and agarose gel electrophoresis results showed the mice numbered 135-137, 139-142, and 144 were Cldn7 ICKO mice with the genotype of Cldn7fl/fl; villin-CreERT2 (fl/fl CreERT2) (Figure 5A).
Cldn7 ICKO mice were normal at birth and developed smoothly, unlike Cldn7fl/fl; villin-CreW (fl/fl CreW) mice (Figure 5B left). A tamoxifen solution (10 mg/mL, 100 μL) was intraperitoneally injected into 6 to 8-wk-old ICKO mice every 5 d. Beginning at the 7th injection, the ICKO mice were lethargic and lack of activity, appeared thin, and exhibited a dying state, while the control mice showed no abnormalities (Figure 5B left). All ICKO mice died within 75 d (15 tamoxifen injections, Figure 5B right).
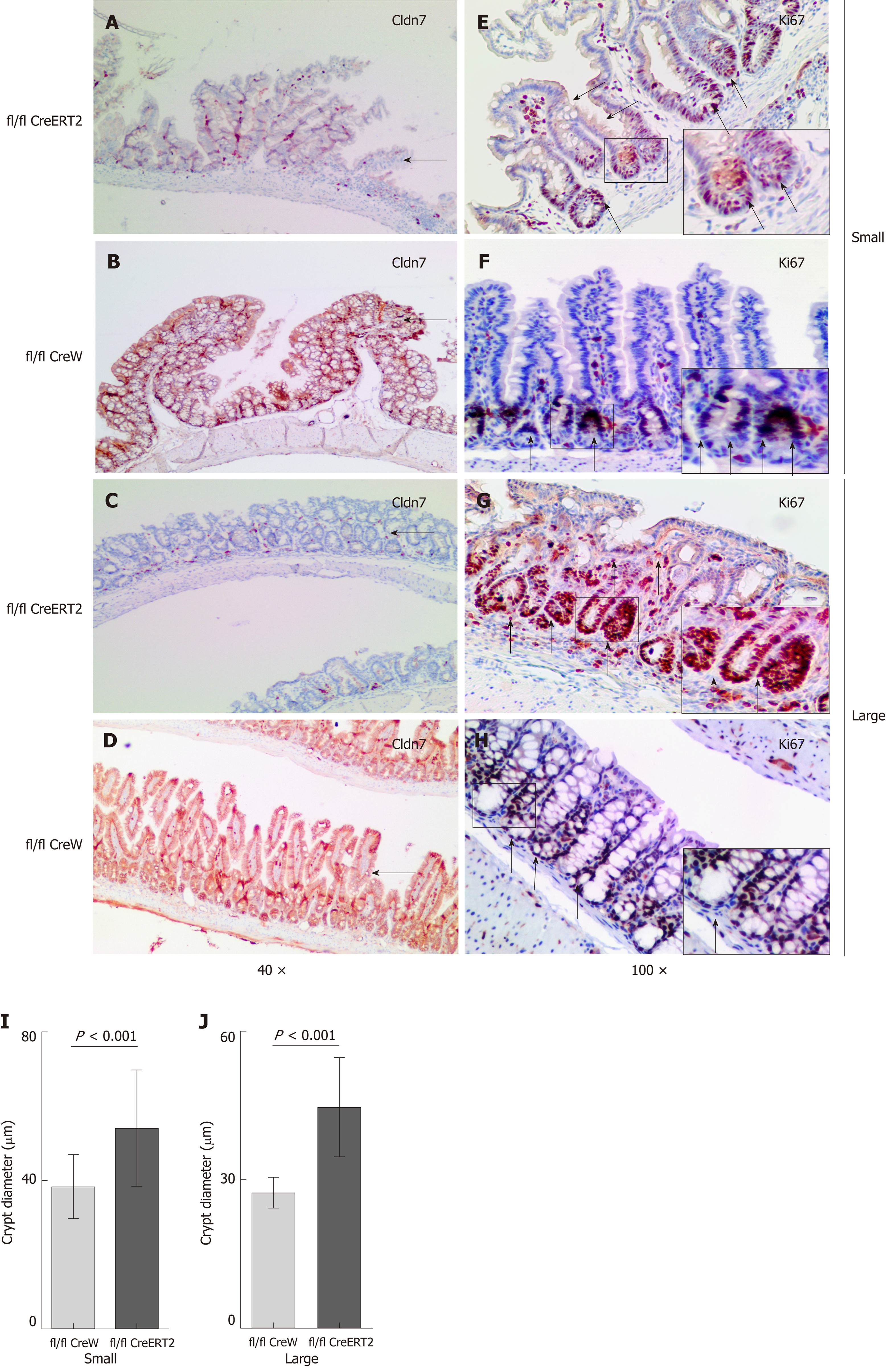
Both the dying mice and control mice were sacrificed. The expression levels of Cldn7 in the lung, stomach, bladder, and kidney tissues of ICKO mice were normal, but Cldn7 expression was absent in their small and large intestines. All control mouse tissues expressed Cldn7 (Figure 5C). Intestinal HE staining showed obvious inflammatory manifestations (Figure 5D, E, G, and H), numerous infiltrated inflammatory cells, abnormal or absent intestinal villi and intestinal gland structure, mucosal epithelial cell shedding, and disordered residual intestinal villus mucosal epithelial cells that lacked polarity. Atypical hyperplasia (Figure 5J and M) and intestinal adenoma (Figure 5K and N) were also observed. The intestinal structure was normal and intact in the control mice (Figure 5F, I, L, and O), with no obvious pathological changes observed.
Immunohistochemistry analysis was also used to detect the expression of intestinal Cldn7 and the nuclear proliferation marker Ki67 in ICKO and control mice, revealing that Cldn7 was strongly expressed in the intestinal epithelial junction of control mice, but was significantly weakened in the intestines of ICKO mice (Figure 6A-D). In addition, the expression of Ki67 was also altered. In control mice, Ki67 was mainly distributed in portions of the crypt, while Ki67-positive cells occupied the entire intestinal crypt in ICKO mice (Figure 6E-H). The intestinal crypt size in ICKO mice was increased compared with that in control mice (Figure 6I and J). Furthermore, Ki67 was mostly expressed in the crypt area in control mice, while Ki67 in the intestinal tract of the ICKO mice was no longer confined to the crypt area. Cells expressing Ki67 were observed throughout the entire intestinal villi, and this manifestation was more pronounced in the large intestine.
Abnormal expression of Cldn7 can lead to the destruction of TJ structure and function as well as cell proliferation and migration abnormalities[15], which are closely related to the occurrence and development of malignant tumors, such as lung cancer, ovarian cancer, and gastric cancer[16-18]. However, the specific mechanisms underlying these phenomena remain elucidated.
Cldn7 is widely considered to be a tumor-suppressor gene[19-21], and one of the best methods for researching tumor-suppressor genes is to knock out the gene in vivo and then observe the phenotype of the entire animal[22]. Tamura et al[23] generated Cldn15-/- mice using the conventional gene targeting strategy. Cldn15 is similar to Cldn7 and strongly expressed in the duodenum, jejunum, ileum, and colon, while other Claudin family proteins, such as Cldn6, 9, 10, 11, and 14, are not expressed in the intestine[20]. Cldn15-deficient mice formed mega-intestines, in which the upper small intestine was two times larger than the normal intestine. Moreover, because Cldn15 deletion is not lethal, researchers can observe intestinal development at different time points, such as at 1 wk, 4 wk, and 10 wk after birth[22]. Similarly, we constructed Cldn7 CKO mice using the same strategy, and this mouse model showed severe intestinal defects that included mucosal ulcerations, epithelial cell sloughing, and inflammation. However, Cldn7 CKO mice died beginning on the third day, which was not conducive to long-term observation or subsequent experiments. Additionally, the effects of Cldn7 deletion in other organs on survival time and morphological changes could not be excluded. Therefore, prolonging the survival time of Cldn7 knockout mice is necessary for further studying the function of Cldn7.
The Cre/LoxP technology makes it possible to knock out a gene in a site- or time-specific manner[22]. Cre recombinase can be expressed in specific cell types, and the crossing between floxed mice and Cre mice can then be used to remove the sequence between two LoxP sites in specific tissues. When Cre recombinase is fused to a ligand-binding domain of a mutant human estrogen receptor (ER), it becomes a tamoxifen-dependent Cre recombinase (Cre-ERT)[24]. Time-specific gene knockout can be achieved by injecting tamoxifen into transgenic mice at different growth stages. Using this method, we constructed intestinal specific Cldn7 cKO mice. A Western blot assay confirmed that only intestinal Cldn7 was ablated. While Cldn7 cKO mice had a normal phenotype after birth, by the fifth day, the mice grew slowly and lacked energy. On the 9th day, the dying mice were sacrificed; their intestinal tracts showed severe inflammation, and their mucosal or glands appeared abnormal. Tanaka et al also constructed intestinal specific Cldn7 cKO mice[25], which had a longer survival period of 28 d due to the use of a different Cre enzyme mouse model. They found that intestinal Cldn7 knockout changed only the paracellular flux of small molecule solutes and did not completely destroy the TJ structure. However, both knockout mice are intestinal inflammation models not yet showing adenomas or tumors. We further constructed Cldn7 ICKO mice and controlled their survival time and intestinal morphological changes by changing the dose and frequency of tamoxifen. When the mice received 1 mg of tamoxifen every day, they began dying after the 5th injection, and severe inflammation was observed in the intestine (data not shown). When the mice received 1 mg of tamoxifen every 5 d, they began to show signs of dying after the 7th injection, and all mice died within 15 tamoxifen injections. Approximately 71.4% of the mice developed adenomas in different regions, including the duodenum, jejunum, ileum, and colon.
Immunohistochemical staining showed that compared with that in control mice, the intestinal crypt size in ICKO mice was increased, and cells positively expressing Ki67 covered the entire crypt. Ki67 expression was no longer limited to the crypt but rather to the crypt-villus axis. These phenomena suggested that the loss of Cldn7 led to both expansion of the intestinal crypt and proliferation of cells at the crypt, and the normal proliferation-differentiation balance of intestinal cells along the crypt-villus axis was disrupted.
In the intestine, proliferating epithelial cells are specifically confined to the crypts[26]. Recent studies have shown that the crypt microenvironment is important for the generation and maintenance of proliferating cells[27]. Because TJs play a critical role in maintaining intestinal homeostasis, they may be important for formation of the niche, the microenvironment of the crypt for stem and transit-amplifying cells[24]. Therefore, the construction of Cldn7 ICKO mice is of great significance for studying the relationship among intestinal barrier destruction, dysregulation of intestinal stem cell proliferation and differentiation along the crypt-villus axis, changes in the intestinal crypt microenvironment, and intestinal tumor formation.
In conclusion, we report the novel findings that Cldn7 knockout caused extensive inflammation, atypical hyperplasia, and adenoma in intestinal tissue as well as animal death in three lines of knockout mice. Therefore, Cldn7 may act as a tumor suppressor gene in the development of colorectal cancer, and the specific mechanism of Cldn7 can be elucidated by performing further research on the ICKO mouse model.
Claudin-7, one of the important components of cellular tight junctions, is currently considered to be expressed abnormally in colorectal inflammation and colorectal cancer. However, there is currently no effective animal model to study its specific mechanisms. Therefore, we constructed three lines of Claudin-7 knockout mice using the Cre/LoxP system to provide a basis for further studying the role of Claudin-7 in intestinal tumors.
Claudin-7 is currently considered to play an inhibitory role in colorectal inflammation and colorectal cancer. The most effective way to study inhibitors in vivo is to knock out the gene in an animal and observe its overall phenotype. Therefore, we constructed conventional Claudin-7 gene knockout (CKO) mice and conditional Claudin-7 gene knockout (cKO) mice using Cre/LoxP system, and we also generated inducible conditional Claudin-7 gene knockout (ICKO) mice and induced Cre expression by injecting tamoxifen. The successful construction of these mouse lines as intestinal inflammation and intestinal adenoma models could provide a basis for further studying the role of Claudin-7 in intestinal tumors.
The main objective was to construct three lines of Claudin-7 gene knockout mice to achieve space- and time-specific knockout of Claudin-7 and prolong the survival time of mice. Due to the prolonged growth time of mice, the organs matured when Claudin-7 was knocked out. So this animal model can provide a basis for further study of Claudin-7.
We constructed three lines of Claudin-7 knockout mice using the Cre/LoxP system. First, chimeric mice were constructed by transfecting the target vector into embryonic stem cells, screening the hybrid clones and injecting them into the female mouse blastocysts, and transplanting the blastocysts into the female mouse uterus. Chimeric mice were then purified to obtain Claudin-7-floxed mice. Second, Claudin-7-floxed mice were crossed with CMV-Cre mice, vil1-Cre mice, and villin-CreERT2 mice ot obtain Claudin-7 CKO mice, Claudin-7 cKO mice, and Claudin-7 ICKO mice, respectively. ICKO mice were induced by intraperitoneal injection of tamoxifen to knockout Claudin-7 in intestinal tissue. Finally, Western blot was used to verify the knockout efficiency of Claudin-7. Hematoxylin-eosin (HE) staining was used to confirm the structural changes and pathological changes of the intestinal tract in Claudin-7 knockout mice. Immunohistochemical staining was used to observe the proliferation markers. The construction of cKO mice prolonged the lifespan of CKO mice, and the ICKO mouse was the first animal model to specifically knock out Claudin-7 in a spatial and temporal manner.
We generated Claudin-7-floxed mice and three lines of Claudin-7 gene knockout mice successfully. Claudin-7 CKO mice and Claudin-7 cKO mice were stunted and died during the perinatal period, and intestinal HE staining revealed mucosal gland structure disappearance and connective tissue hyperplasia with extensive inflammatory cell infiltration. Claudin-7 ICKO mice had a normal phenotype at birth, but after the induction with tamoxifen, the mice exhibited a dying state. Intestinal HE staining showed significant inflammatory cell infiltration, and atypical hyperplasia and adenoma were also observed. Intestinal immunohistochemistry analysis showed abnormal expression and distribution of Ki67, and the normal intestinal proliferation balance was disrupted.
This study innovatively constructed three lines of Claudin-7 gene knockout mice, which clarified that Claudin-7 plays an inhibitory role in colon inflammation and colon adenoma, and initially found that Claudin-7 may promote the development of colon adenomas by affecting proliferation. This study successfully simulated intestinal inflammation and intestinal adenoma, and proposed new animal models. This study clarified the role of Claudin-7 in colonic inflammation and tumors, laying the groundwork for finding early clinical diagnosis and potential therapeutic targets.
This article describes the construction of Claudin-7-floxed mice and the process of crossing with three Cre mice. Based on this experience, we can construct ICKO mice in which Claudin-7 is ablated in the kidney, skin, and some other organs, and then observe changes in mice before and after Claudin-7 deletion. In the next step of the study, the dose of tamoxifen should be clarified, and the tumor should be induced in the intestinal tract of ICKO mice. The direction of the future research is to clarify the specific mechanism of Claudin-7 in inflammatory bowel disease and intestinal tumorigenesis, invasion, and metastasis; and to explore the relationship between Claudin-7 and stem cells as well as its role in intestinal development.
We thank the National Resource Center for Mutant Mice for their assistance and technical support.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: McMillin MA, Subramanian VS S- Editor: Yan JP L- Editor: Wang TQ E- Editor: Huang Y
| 1. | Van Itallie CM, Anderson JM. Claudins and epithelial paracellular transport. Annu Rev Physiol. 2006;68:403-429. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 837] [Cited by in RCA: 865] [Article Influence: 45.5] [Reference Citation Analysis (0)] |
| 2. | Ding L, Lu Z, Lu Q, Chen YH. The claudin family of proteins in human malignancy: A clinical perspective. Cancer Manag Res. 2013;5:367-375. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 66] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 3. | Farkas AE, Capaldo CT, Nusrat A. Regulation of epithelial proliferation by tight junction proteins. Ann N Y Acad Sci. 2012;1258:115-124. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 45] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 4. | Alexandre MD, Lu Q, Chen YH. Overexpression of claudin-7 decreases the paracellular Cl- conductance and increases the paracellular Na+ conductance in LLC-PK1 cells. J Cell Sci. 2005;118:2683-2693. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 167] [Cited by in RCA: 172] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 5. | Darido C, Buchert M, Pannequin J, Bastide P, Zalzali H, Mantamadiotis T, Bourgaux JF, Garambois V, Jay P, Blache P, Joubert D, Hollande F. Defective claudin-7 regulation by Tcf-4 and Sox-9 disrupts the polarity and increases the tumorigenicity of colorectal cancer cells. Cancer Res. 2008;68:4258-4268. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 96] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 6. | Akizuki R, Shimobaba S, Matsunaga T, Endo S, Ikari A. Claudin-5, -7, and -18 suppress proliferation mediated by inhibition of phosphorylation of Akt in human lung squamous cell carcinoma. Biochim Biophys Acta Mol Cell Res. 2017;1864:293-302. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 39] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 7. | Tsujiwaki M, Murata M, Takasawa A, Hiratsuka Y, Fukuda R, Sugimoto K, Ono Y, Nojima M, Tanaka S, Hirata K, Kojima T, Sawada N. Aberrant expression of claudin-4 and -7 in hepatocytes in the cirrhotic human liver. Med Mol Morphol. 2015;48:33-43. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 19] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 8. | Garcia-Hernandez V, Quiros M, Nusrat A. Intestinal epithelial claudins: Expression and regulation in homeostasis and inflammation. Ann N Y Acad Sci. 2017;1397:66-79. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 184] [Cited by in RCA: 305] [Article Influence: 38.1] [Reference Citation Analysis (0)] |
| 9. | Schölch S, García SA, Iwata N, Niemietz T, Betzler AM, Nanduri LK, Bork U, Kahlert C, Thepkaysone ML, Swiersy A, Büchler MW, Reissfelder C, Weitz J, Rahbari NN. Circulating tumor cells exhibit stem cell characteristics in an orthotopic mouse model of colorectal cancer. Oncotarget. 2016;7:27232-27242. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 41] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 10. | Oshima T, Miwa H, Joh T. Changes in the expression of claudins in active ulcerative colitis. J Gastroenterol Hepatol. 2008;23 Suppl 2:S146-S150. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 146] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 11. | Ding L, Wang L, Sui L, Zhao H, Xu X, Li T, Wang X, Li W, Zhou P, Kong L. Claudin-7 indirectly regulates the integrin/FAK signaling pathway in human colon cancer tissue. J Hum Genet. 2016;61:711-720. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 24] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 12. | Trinh KR, Morrison SL. Site-specific and directional gene replacement mediated by Cre recombinase. J Immunol Methods. 2000;244:185-193. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 15] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 13. | Le Y, Sauer B. Conditional gene knockout using Cre recombinase. Mol Biotechnol. 2001;17:269-275. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 47] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 14. | Shimizu E, Tang YP, Rampon C, Tsien JZ. NMDA receptor-dependent synaptic reinforcement as a crucial process for memory consolidation. Science. 2000;290:1170-1174. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 412] [Cited by in RCA: 405] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 15. | Singh AB, Dhawan P. Claudins and cancer: Fall of the soldiers entrusted to protect the gate and keep the barrier intact. Semin Cell Dev Biol. 2015;42:58-65. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 52] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 16. | Lu Z, Ding L, Hong H, Hoggard J, Lu Q, Chen YH. Claudin-7 inhibits human lung cancer cell migration and invasion through ERK/MAPK signaling pathway. Exp Cell Res. 2011;317:1935-1946. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 84] [Cited by in RCA: 84] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 17. | Dahiya N, Becker KG, Wood WH, Zhang Y, Morin PJ. Claudin-7 is frequently overexpressed in ovarian cancer and promotes invasion. PLoS One. 2011;6:e22119. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 63] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 18. | Johnson AH, Frierson HF, Zaika A, Powell SM, Roche J, Crowe S, Moskaluk CA, El-Rifai W. Expression of tight-junction protein claudin-7 is an early event in gastric tumorigenesis. Am J Pathol. 2005;167:577-584. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 88] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 19. | Usami Y, Chiba H, Nakayama F, Ueda J, Matsuda Y, Sawada N, Komori T, Ito A, Yokozaki H. Reduced expression of claudin-7 correlates with invasion and metastasis in squamous cell carcinoma of the esophagus. Hum Pathol. 2006;37:569-577. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 117] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 20. | Bornholdt J, Friis S, Godiksen S, Poulsen SS, Santoni-Rugiu E, Bisgaard HC, Lothe IM, Ikdahl T, Tveit KM, Johnson E, Kure EH, Vogel LK. The level of claudin-7 is reduced as an early event in colorectal carcinogenesis. BMC Cancer. 2011;11:65. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 39] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 21. | Lu Z, Kim DH, Fan J, Lu Q, Verbanac K, Ding L, Renegar R, Chen YH. A non-tight junction function of claudin-7-Interaction with integrin signaling in suppressing lung cancer cell proliferation and detachment. Mol Cancer. 2015;14:120. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 64] [Cited by in RCA: 65] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 22. | Metzger D, Chambon P. Site- and time-specific gene targeting in the mouse. Methods. 2001;24:71-80. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 288] [Cited by in RCA: 286] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 23. | Tamura A, Kitano Y, Hata M, Katsuno T, Moriwaki K, Sasaki H, Hayashi H, Suzuki Y, Noda T, Furuse M, Tsukita S, Tsukita S. Megaintestine in claudin-15-deficient mice. Gastroenterology. 2008;134:523-534. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 160] [Cited by in RCA: 165] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 24. | el Marjou F, Janssen KP, Chang BH, Li M, Hindie V, Chan L, Louvard D, Chambon P, Metzger D, Robine S. Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis. 2004;39:186-193. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 738] [Cited by in RCA: 860] [Article Influence: 43.0] [Reference Citation Analysis (0)] |
| 25. | Tanaka H, Takechi M, Kiyonari H, Shioi G, Tamura A, Tsukita S. Intestinal deletion of Claudin-7 enhances paracellular organic solute flux and initiates colonic inflammation in mice. Gut. 2015;64:1529-1538. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 144] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 26. | Cheng H, Leblond CP. Origin, differentiation and renewal of the four main epithelial cell types in the mouse small intestine. V. Unitarian Theory of the origin of the four epithelial cell types. Am J Anat. 1974;141:537-561. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1049] [Cited by in RCA: 1037] [Article Influence: 20.3] [Reference Citation Analysis (0)] |
| 27. | Matsuda M, Kubo A, Furuse M, Tsukita S. A peculiar internalization of claudins, tight junction-specific adhesion molecules, during the intercellular movement of epithelial cells. J Cell Sci. 2004;117:1247-1257. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 177] [Cited by in RCA: 186] [Article Influence: 8.9] [Reference Citation Analysis (0)] |