Published online Nov 14, 2019. doi: 10.3748/wjg.v25.i42.6311
Peer-review started: August 19, 2019
First decision: September 10, 2019
Revised: October 16, 2019
Accepted: November 1, 2019
Article in press: November 1, 2019
Published online: November 14, 2019
Processing time: 86 Days and 18.9 Hours
Studies have reported that microRNA-30c (miR-30c) has vital functions in the development and progression of multiple cancers.
To investigate the clinical significance and role of miR-30c in pancreatic cancer.
MiR-30c and twinfilin 1 (TWF1) expression levels were analyzed in Gene Expression Omnibus datasets and validated in human pancreatic cancer by quantitative real-time polymerase chain reaction (RT-qPCR). The effects of miR-30c on pancreatic cancer cell growth, apoptosis, and cell cycle were evaluated by CCK-8 and flow cytometry assays. Furthermore, the in vivo effects were investigated using a subcutaneous xenograft experiment. Target gene prediction software and luciferase reporter assays were used to identify TWF1 as a direct target of miR-30c.
The expression of miR-30c was significantly decreased in pancreatic cancer tissues and associated with survival. Gain- and loss-of-function assays showed that miR-30c suppressed pancreatic cancer cell proliferation in vitro and in vivo. RT-qPCR, Western blot, and luciferase reporter assays showed that miR-30c directly targeted TWF1. The expression level of miR-30c was negatively correlated with TWF1 expression in pancreatic cancer tissues. Furthermore, the effects of ectopic miR-30c were rescued by TWF1 overexpression.
Our results identified the role of the miR-30c/TWF1 axis in pancreatic cancer progression and demonstrated that miR-30c might serve as a prognostic biomarker and therapeutic target for pancreatic cancer.
Core tip: Studies have shown that miR-30c exerts vital roles in the oncogenesis of various cancers. However, its expression and role in pancreatic cancer remain unknown. In this study, the expression levels of miR-30c and twinfilin 1 were mined in Gene Expression Omnibus datasets and detected in clinical samples. The relationship of miR-30c expression with clinicopathological factors of pancreatic cancer patients was analyzed. The effect of miR-30c on pancreatic cancer cell proliferation and the underlying regulatory mechanism were investigated. Our study suggested that miR-30c may serve as a prognostic biomarker and therapeutic target for pancreatic cancer.
- Citation: Sun LL, Cheng M, Xu XD. MicroRNA-30c inhibits pancreatic cancer cell proliferation by targeting twinfilin 1 and indicates a poor prognosis. World J Gastroenterol 2019; 25(42): 6311-6321
- URL: https://www.wjgnet.com/1007-9327/full/v25/i42/6311.htm
- DOI: https://dx.doi.org/10.3748/wjg.v25.i42.6311
Pancreatic ductal adenocarcinoma (PDAC) is one of the most malignant gas-trointestinal cancers and the prognosis of PDAC patients is very poor[1]. Although diagnostic methods and therapeutic strategies have been substantially improved, the clinical outcome of pancreatic cancer patients remains poor[2]. Therefore, it is necessary and urgent to understand the molecular mechanisms of pancreatic cancer development and thus identify new precise therapeutic strategies.
MicroRNAs (miRNAs), small noncoding RNAs of 19-22 nucleotides in length, are known to suppress gene expression by binding to their 3’-untranslated regions (3’-UTRs)[3]. Studies have shown that miRNAs are involved in tumor oncogenesis and progression of multiple cancers[4-6]. MiR-30c is one of members of the miR-30 family, which includes miR-30a, miR-30b, miR-30c, miR-30d, miR-30e, and miR-30f[7]. Studies indicate that loss of miR-30c contributes to various malignancies, including gastric cancer, ovarian cancer, and hepatocellular carcinoma[8-10]. However, the expression and role of miR-30c in pancreatic cancer have not been determined.
Twinfilin 1 (TWF1), an actin-binding protein, regulates diverse aspects of actin dynamics[11]. This protein was shown to promote cardiac hypertrophy[12]. The miR-206/TWF1/MKL1-SRF/IL11 signaling pathway inhibited the stemness and metastasis of breast cancer cells[13]. TWF1 regulated breast cancer cell invasion by STAT3 phosphorylation[14]. Furthermore, TWF1 promoted human breast tumor chemotherapy resistance[15]. Overexpression of TWF1 was identified as an inferior prognosis indicator in lung adenocarcinoma[16]. The findings of these studies suggest that TWF1 is a putative driver gene in cancers.
In the present study, we investigated the expression of miR-30c and its relationship with clinical features in pancreatic cancer. In addition, the function of miR-30c in pancreatic cancer was explored, as well as the potential molecular mechanisms. Our findings suggest that miR-30c might be a potential therapeutic target for pancreatic cancer.
Fresh human pancreatic cancer tissues and adjacent normal tissues were collected from 40 patients who received pancreaticoduodenectomy from 2012 to 2013 at the First Affiliated Hospital of Zhengzhou University (ZZU). All tissues were stored in liquid nitrogen. The patients did not receive chemotherapy or radiotherapy before surgery. All patients were independently diagnosed with adenocarcinoma by two experienced pathologists. All samples were collected with informed written consent from all patients, and our study was approved by the Ethical Committee of Zhengzhou University.
Pancreatic cancer cell lines (BxPC-3, Capan-2, Mia PaCa-2, Panc-1, and SW1990) were purchased from the American Type Culture Collection (ATCC, Manassas, VA, United States). An immortalized human pancreatic ductal epithelial cell line (HPDE) was obtained from the Cell Repository, Chinese Academy of Sciences (Shanghai, China). All cell lines in this study were authenticated by short tandem repeat DNA profiling and cultured according to the manufacturer’s instructions. MiR-30c mimics, miR-30c inhibitors, and negative control sequences were purchased from GeneChem (Shanghai, China). Lentiviruses overexpressing miR-30c and the lentivirus control were produced by GeneChem (Shanghai, China). Transfection of the recombinant lentiviruses was performed according to the supplier’s instructions. Transfection efficiency was monitored by quantitative real-time polymerase chain reaction (RT-qPCR), which was independently repeated at least three times.
After treatment, cells were seeded into 96-well plates (1 × 103 cells per well). Cell viability was detected by Cell Counting Kit-8 assays (Dojindo Laboratories, Kumamoto, Japan) according to the manufacturer’s instructions.
Cells were transfected with control, miR-30c mimics, or miR-30c inhibitors. After transfection for 48 h, cells were harvested and stained with an Annexin V/FITC and propidium iodide Apoptosis Detection Kit (MultiSciences, Hangzhou, China) according to the manufacturer’s protocol. The stained cells were then detected using the fluorescence-activated cell sorting (FACS) Caliber system (BD Immunocytometry Systems, San Jose, CA, United States).
Cells were transfected with control, miR-30c mimics, or miR-30c inhibitors in six-well plates. After treatment for 48 h, cells were washed, harvested, and stained according to the instructions of the Cell Cycle Analysis Kit (Multi Sciences, Hangzhou, China). Next, the cells were analyzed using the FACS Caliber system. The percentages of the cells in each phase were determined.
Total RNA from each group of cells was extracted using TRIzol reagent (Invitrogen, United States) and synthesized into cDNA via a reverse transcription kit (TaKaRa, Dalian, China) according to the manufacturer’s instructions. The primers used are listed below: MiR-30c: forward 5'-GCCGCTGTAAACATCCTACACT-3' and reverse 5'-GTGCAGGGTCCGAGGT-3'; U6: forward 5’-CTCGCTTCGGCAGCACA-3’ and reverse 5’-AACGCTTCACGAATTTGCGT-30; TWF1: forward 5’-ACGTG GGTGTGGACACT AAG-3’ and reverse 5’-GGGAATCCTCTTTGGC AAATCTT-3’; and GAPDH: forward 5’-CGTGGGCCGCCCTAGGCACCA-3’ and reverse 5’-TTG GCTTAGGGTTCAGGGGGG-3’. U6 was used as the miRNA internal control and the housekeeping gene GAPDH was used as the mRNA internal control. RT-qPCR was performed with an ABI 7500 system (Applied Biosystems, United States) according to the manufacturer’s instructions.
Western blot analysis was carried out as previously described (Lai et al[5], 2017). Antibodies against human TWF1 were purchased from Cell Signaling Technology (Beverly, MA, United States). Antibodies against human GAPDH and secondary antibodies were purchased from Boster (Wuhan, China).
IHC analysis was performed as previously described (Lai et al[5], 2017). Antibodies for IHC against human TWF1 were purchased from Abcam (Cambridge, MA, United States). Antibodies against human Ki67 were purchased from Boster (Wuhan, China). Semi-quantitative scoring of immunohistochemical staining was performed using the H-score method, and stain score was calculated as intensity × positive rate.
Bioinformatics analysis was performed to predict target genes of miR-30c with TargetScan, miRDB, and miRTarBase. The results indicated that TWF1 is the strongest potential target of miR-30c. MiR-30c expression in Gene Expression Omnibus (GEO) datasets was analyzed with GEO2R. Expression levels were log2-transformed and assessed by an unpaired t test between the tumor and control groups.
Wild-type and mutant 3’-UTRs of TWF1 luciferase reporter vectors were purchased from Promega (Madison, WI, United States). After incubation for 48 h, a dual-luciferase reporter assay system (Promega) was used to measure the luciferase activity. Relative luciferase activity was normalized by the ratio of firefly and Renilla luciferase signals.
Ten 4-6-week-old male nude mice were purchased from HFK Bioscience (Beijing, China) and bred in specific pathogen-free conditions. After treatment, 2 × 106 pancreatic cancer cells were injected in the axilla subcutaneously in each group. Tumor volume was measured using calipers every week and calculated as length × width2 × 0.5. Five weeks later, mice were sacrificed and tumors were removed, weighed, and further analyzed. The animal study was conducted in accordance with NIH animal use guidelines and approved by the Animal Care Committee of Zhengzhou University.
A two-tailed paired t-test was used to analyze the expression difference of miR-30c and TWF1 between cancerous tissues and adjacent noncancerous tissues. Paired or unpaired t-test was used to analyze the expression difference between two groups. The association of miR-30c expression with clinicopathological parameters was analyzed using chi-square tests. Data analyses were performed with SPSS 17.0 (SPSS Inc., Chicago, IL, United States) and presented with GraphPad Prism 6.0 (GraphPad Software, Inc., La Jolla, CA, United States). All experiments were carried out at least three times. P < 0.05 was defined as statistically significant.
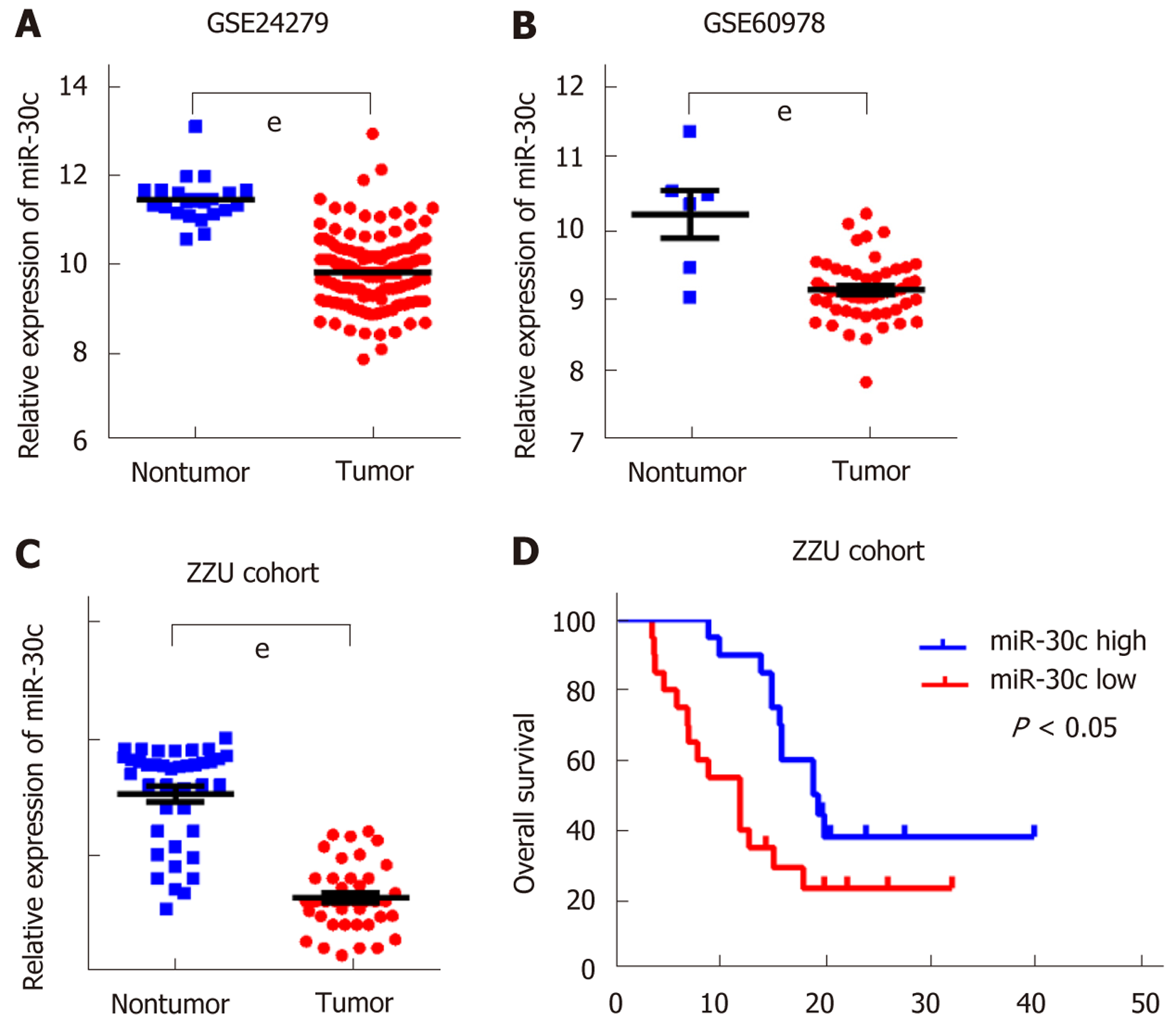
To study the expression of miR-30c in human pancreatic cancer, we first analyzed two GEO datasets GSE24279 and GSE60978. The results showed that miR-30c levels were frequently downregulated in pancreatic cancer tissue samples compared with nontumor tissues (Figure 1A and B). Then, we validated the expression of miR-30c in 40 matched pancreatic cancer patient samples and the corresponding adjacent nontumor tissues by RT-qPCR. MiR-30c was also downregulated in pancreatic cancer (Figure 1C). Then, we stratified all patients into miR-30c high and miR-30c low groups by the median of miR-30c expression and analyzed the clinical information. Pancreatic cancer patients with low miR-30c expression had poorer survival status than those with high miR-30c expression (median survival: 12 mo vs 19.2 mo; log-rank test, P < 0.05; Figure 1D). Meanwhile, miR-30c expression levels in tumors were significantly correlated with tumor stage (American Joint Committee on Cancer 7th edition) but not with sex, age, or tumor grade of pancreatic cancer (Table 1). Collectively, these data indicate that miR-30c is downregulated in pancreatic cancer and correlates with a poor prognosis.
| Characteristic | n | High | Low | P value |
| Sex | 40 | 20 | 20 | 0.386 |
| Male | 26 | 13 | 13 | |
| Female | 14 | 5 | 9 | |
| Age, yr | 0. 972 | |||
| < 50 | 29 | 13 | 16 | |
| ≥ 50 | 11 | 5 | 6 | |
| Grade | < 0.05 | |||
| Low (I and II) | 31 | 20 | 11 | |
| High (III and IV) | 9 | 6 | 3 | |
| Stage | 0.842 | |||
| Early (I and II) | 14 | 8 | 6 | |
| Late (III and IV) | 26 | 14 | 12 |
To investigate the biological role of miR-30c in vitro, we examined miR-30c expression in five pancreatic cancer cell lines (BxPC-3, Capan-2, Mia PaCa-2, Pan-1, and SW1990) and HPDE cell line by RT-qPCR (Figure 2A). Then, BxPC-3 and Mia PaCa-2 cells were transfected with control, miR-30c mimics, or miR-30c inhibitors. The transfection efficiency in the two cell lines was validated by RT-qPCR (Figure 2B). The CCK-8 assays showed that the proliferation of BxPC-3 and Mia PaCa-2 cells was markedly decreased after transfection with miR-30c mimics compared with the control group (Figure 2C and D). The proliferation ability of both cell lines was markedly increased after transfection with miR-30c inhibitors (Figure 2C and D). The above results revealed that miR-30c suppressed the proliferation of pancreatic cancer cells, which is associated with cell apoptosis and cell cycle processes. Therefore, we studied whether miR-30c could regulate cell apoptosis and the cell cycle by flow cytometry. Flow cytometry analysis revealed that gain of miR-30c markedly increased the cell apoptosis rate, whereas loss of miR-30c decreased the apoptosis rate (Figure 2E). Cell cycle results showed that gain of miR-30c significantly increased the percentage of G1 phase cells, and loss of miR-30c decreased the proportion of cells in G1 phase (Figure 2F). Taken together, these results indicate that miR-30c represses pancreatic cancer cell proliferation by inducing apoptosis and cell cycle arrest in vitro.
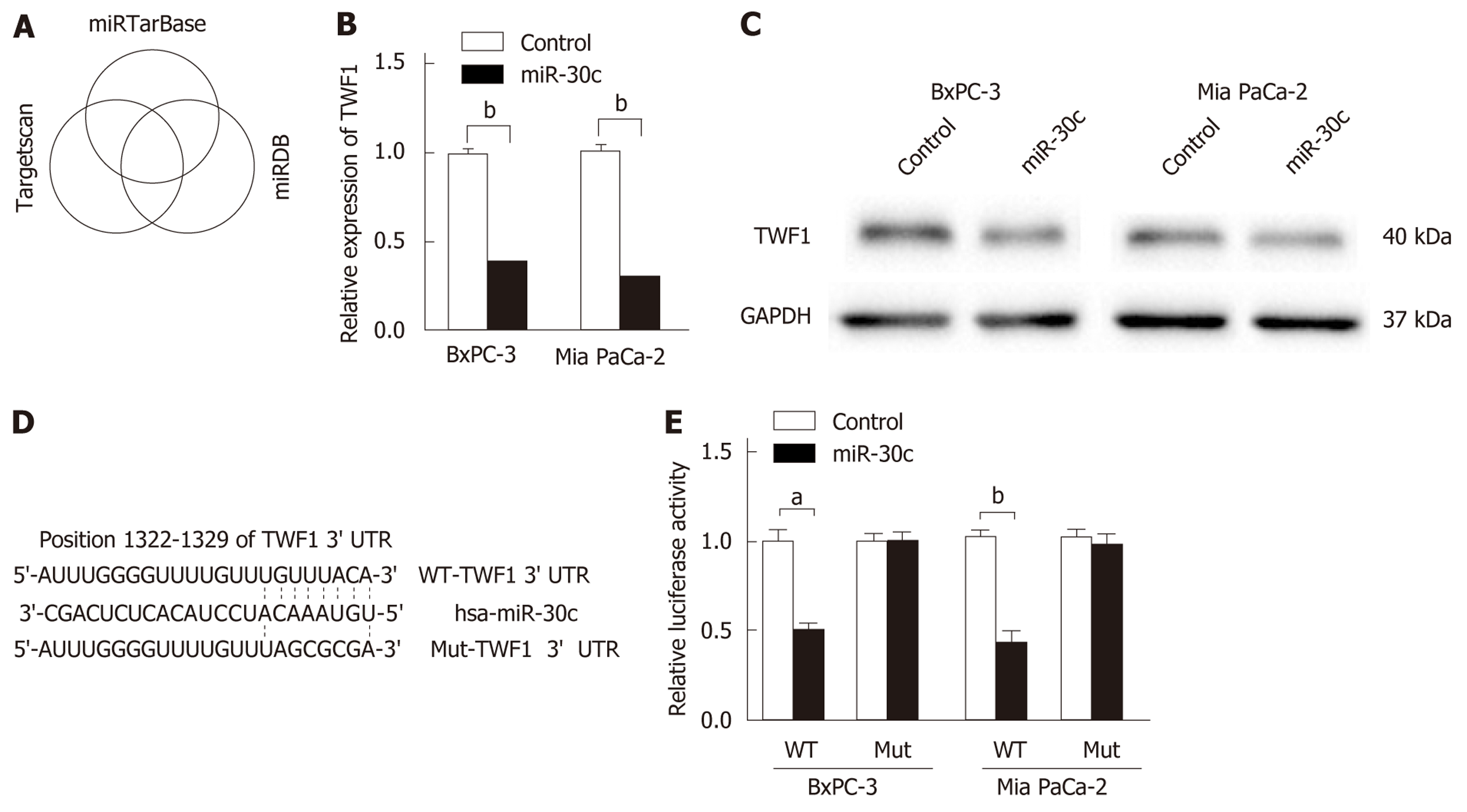
To further explore the potential downstream targets of miR-30c, three online bioinformatics tools (TargetScan, miRDB, and miRTarBase) were used, and the prediction results were comprehensively analyzed (Figure 3A). There were 55 predicted targets for TargetScan, 849 for miRDB, and 521 for miRDB. Five predicted genes (TWF1, RAD23B, S100PBP, MIA3, and VPS33A) in common were identified. We focused on the actin-binding protein TWF1, which regulates diverse aspects of actin dynamics. We first transfected control or miR-30c mimics into pancreatic cancer cells, and then, TWF1 expression levels were detected by RT-qPCR and Western blot analyses. The results showed that re-expression of miR-30c inhibited the mRNA and protein expression of TWF1 (Figure 3B and C). Then, wild-type or mutant TWF1 luciferase reporter vector was constructed (Figure 3D). After transfection, we found that miR-30c mimics dramatically inhibited the luciferase activity of wild-type TWF1, whereas the luciferase activity of mutant TWF1 showed no significant difference (Figure 3E). Collectively, these results demonstrate that TWF1 is a direct target of miR-30c.
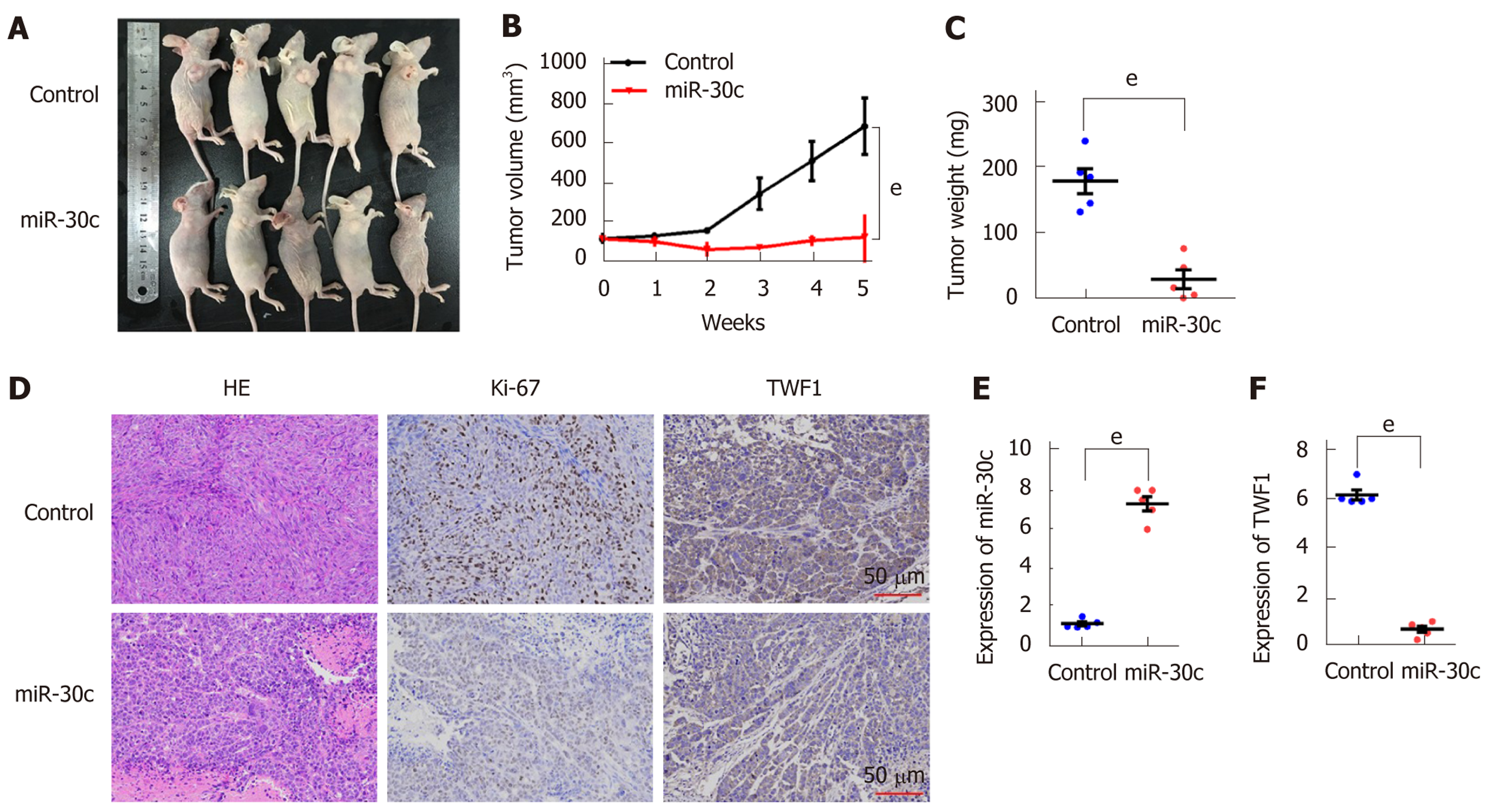
To further evaluate the oncogenic role of miR-30c in vivo, xenograft tumor models were established in BALB/C nude mice using BxPC-3 cells transfected with lentivirus-control or lentivirus-miR-30c vector. As shown in Figure 4A, all the nude mice developed xenograft tumors 5 wk after injection. Furthermore, the average tumor volume and weight of the miR-30c overexpression group were significantly smaller than those in the control group (Figure 4B and C). IHC analysis showed that tumors derived from lentivirus-miR-30c group showed weaker staining of Ki-67 than those in the control group (Figure 4D). Interestingly, tumors derived from the lentivirus-miR-30c overexpression group also showed weaker staining for the target gene TWF1 than those in the control group (Figure 4D). The xenograft tumor tissues were analyzed to verify miR-30c and TWF1 expression using RT-qPCR, which showed similar results to the IHC results (Figure 4E and F). These data suggest that re-expression of miR-30c inhibits tumor growth in vivo.
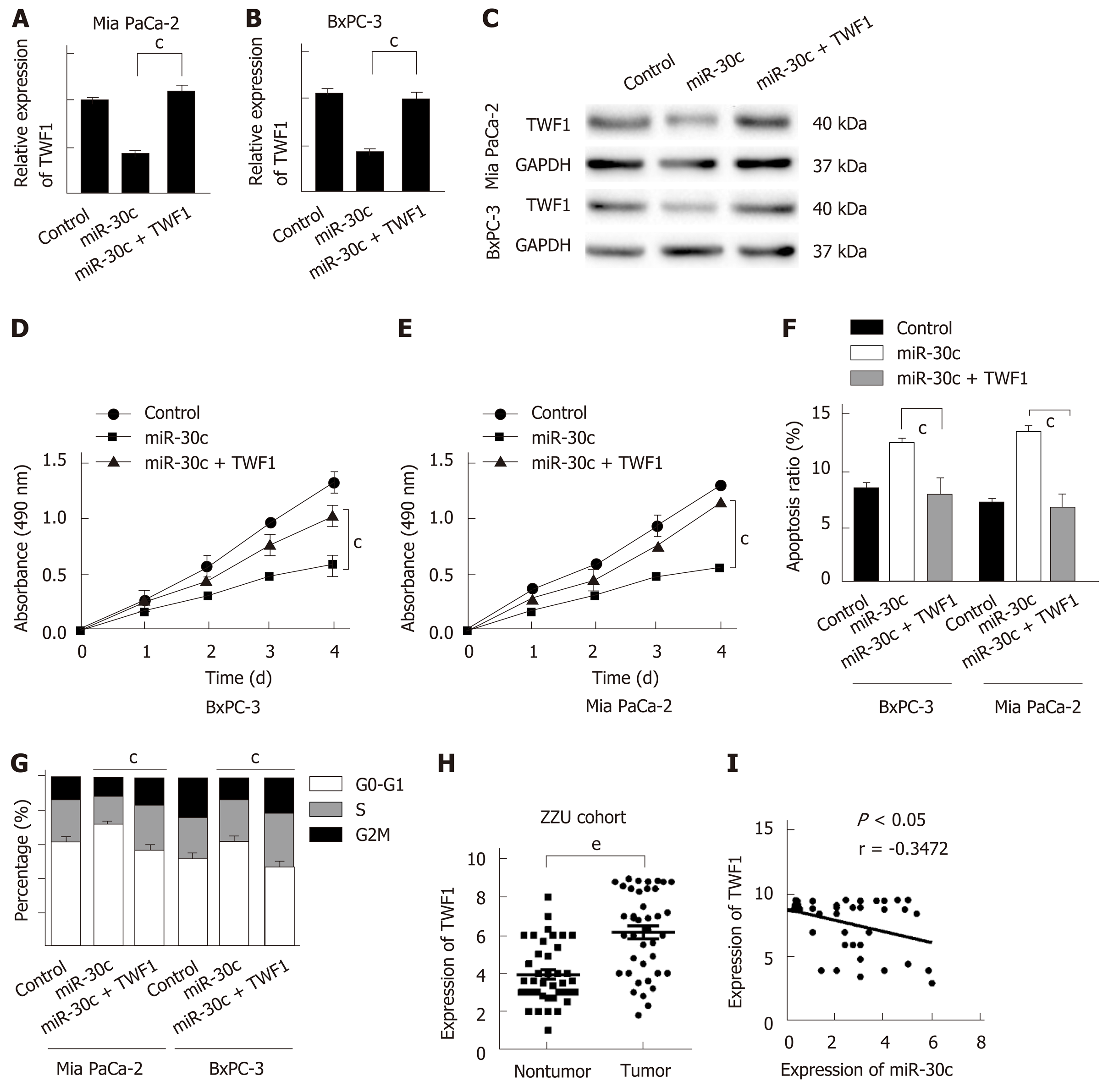
We demonstrated that the re-expression of miR-30c suppressed the proliferation of pancreatic cancer cells and inhibited TWF1 expression. To further confirm whether the effect of miR-30c in PDAC cells is mediated by regulation of TWF1, we overexpressed TWF1 in BxPC-3 and Mia PaCa-2 cells transfected with stable lentivirus-miR-30c. Compared with controls, cells transfected with lentivirus-TWF1 showed significantly higher expression of TWF1 at both the mRNA and protein levels (Figure 5A-C). CCK-8 assays revealed that ectopic TWF1 expression effectively reversed the inhibition of proliferation induced by miR-30c overexpression (Figure 5D and E). Apoptosis analysis showed that ectopic TWF1 expression effectively reversed the promotion of apoptosis induced by miR-30c overexpression (Figure 5F). Similarly, TWF1 upregulation significantly reversed the inhibitory effects of miR-30c induced cell cycle progression (Figure 5G). TWF1 expression was detected in 40 paired pancreatic specimens by RT-qPCR. TWF1 was increased in pancreatic cancer tissues (Figure 5H). Moreover, the mRNA levels of TWF1 and miR-30c exhibited a significant inverse correlation as shown by the Pearson correlation test in pancreatic cancer tissues (Figure 5I). Taken together, these results reveal that miR-30c acts as a negative regulator in the growth of pancreatic cancer cells, which is at least partly dependent on the modulation of TWF1.
Sustained proliferation is a hallmark of cancer and is regulated by multiple molecules, including miRNAs[17]. The roles and molecular mechanisms of miRNAs in tumorigenesis have attracted increased attention. MiR-30c was proven to be a critical regulator in the malignant progression of various cancers. However, the clinical significance and biological role of miR-30c in pancreatic cancer remain unknown. Our results showed that TWF1 is a direct target of miR-30c in pancreatic cancer. Furthermore, ectopic overexpression of miR-30c blocked pancreatic cancer cell proliferation in vitro and in vivo.
MiR-30c has been identified to be tumor suppressive and downregulated in various cancers. In esophageal squamous cell carcinoma, downregulated miR-30c inhibited biological behaviors and epithelial-mesenchymal transition of ESCC by directly targeting SNAI1[18]. In breast cancer, micRNA-30c negatively regulated collagen triple helix repeat containing-1 and suppressed cell proliferation and metastasis[5]. MiRNA-30c inhibited proliferation of non-small cell lung cancer cells by targeting Rab18[19]. In our study, we found that downregulation of miR-30c occurred widely in pancreatic cancer, and predicted a poor prognosis. Similarly, miR-30c was downregulated in five pancreatic cancer cell lines compared with HPDE. Consistent with these findings, experiments in vitro and in vivo showed that re-expression of miR-30c significantly inhibited cell proliferation by inducing apoptosis and G1-phase arrest. Bioinformatics prediction analysis was carried out to search for potential targets of miR-30c and identified TWF1 as a promising target. TWF1 was shown to be a target of miR-30c. Furthermore, rescue experiments showed that enforced overexpression of TWF1 could strongly restore the proliferation of miR-30c-overexpressing cells. The TWF1 mRNA level exhibited an inverse correlation with the level of miRNA-30c in both pancreatic cancer patient tissues and subcutaneous tumors derived from nude mice. We concluded that miR-30c might exert its effect by influencing TWF1 expression to inhibit pancreatic cancer proliferation.
In this study, we provide evidence that miR-30c functions as a tumor-suppressive gene through direct inhibition of TWF1 in pancreatic cancer. Our results suggest that miR-30c might represent a potential therapeutic target for the treatment of human pancreatic cancer.
Pancreatic ductal adenocarcinoma (PDAC) is one of the most malignant gastrointestinal cancers worldwide. Current diagnostic methods and therapeutic strategies are very limited, and the prognosis of pancreatic cancer patients remains poor. To understand the molecular mechanisms of pancreatic cancer development is necessary and urgent. Little is known regarding miR-30c expression and its role in the progression of PDAC.
Our study will provide a new therapeutic target for pancreatic cancer.
To study the expression, role, and target gene of miR-30c in pancreatic cancer.
We detected the expression levels of miR-30c and twinfilin 1 (TWF1) in Gene Expression Omnibus datasets and validated in clinical samples by quantitative real-time polymerase chain reaction. The relationship of miR-30c expression with clinicopathological factors of pancreatic cancer patients was analyzed. The effect and mechanism miR-30c on pancreatic cancer cell proliferation were investigated in vitro and in vivo. Assays were performed to explore potential target gene TWF1 of miR-30c in pancreatic cancer.
In the present study, we found that miR-30c was downregulated and associated with a poor prognosis in pancreatic cancer patients. We showed that re-expression of miR-30c reduced pancreatic cancer cell proliferation in vitro and in vivo by targeting TWF1. Meanwhile, overexpression of TWF1 abolished the effects of miR-30c in pancreatic cancer.
MiR-30c is downregulated and promotes the proliferation of pancreatic cancer cells by targeting TWF1. Overexpression of TWF1 abolishes the effects of miR-30c.
This study provides insight into the role of miR-30c in promoting pancreatic cancer development by targeting TWF1. MiR-30c might be a new therapeutic target for pancreatic cancer.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
Published online: November 14, 2019
P-Reviewer: Dambrauskas Z, Kleeff J S-Editor: Tang JZ L-Editor: Wang TQ E-Editor: Zhang YL
| 1. | Miller KD, Siegel RL, Lin CC, Mariotto AB, Kramer JL, Rowland JH, Stein KD, Alteri R, Jemal A. Cancer treatment and survivorship statistics, 2016. CA Cancer J Clin. 2016;66:271-289. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3134] [Cited by in RCA: 3542] [Article Influence: 393.6] [Reference Citation Analysis (0)] |
| 2. | McGuigan A, Kelly P, Turkington RC, Jones C, Coleman HG, McCain RS. Pancreatic cancer: A review of clinical diagnosis, epidemiology, treatment and outcomes. World J Gastroenterol. 2018;24:4846-4861. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 1338] [Cited by in RCA: 1262] [Article Influence: 180.3] [Reference Citation Analysis (39)] |
| 3. | Chen Y, Zhao ZX, Huang F, Yuan XW, Deng L, Tang D. MicroRNA-1271 functions as a potential tumor suppressor in hepatitis B virus-associated hepatocellular carcinoma through the AMPK signaling pathway by binding to CCNA1. J Cell Physiol. 2019;234:3555-3569. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 21] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 4. | Fu Y, Liu X, Chen Q, Liu T, Lu C, Yu J, Miao Y, Wei J. Downregulated miR-98-5p promotes PDAC proliferation and metastasis by reversely regulating MAP4K4. J Exp Clin Cancer Res. 2018;37:130. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 64] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 5. | Lai YH, Chen J, Wang XP, Wu YQ, Peng HT, Lin XH, Wang WJ. Collagen triple helix repeat containing-1 negatively regulated by microRNA-30c promotes cell proliferation and metastasis and indicates poor prognosis in breast cancer. J Exp Clin Cancer Res. 2017;36:92. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 34] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 6. | Liu Y, Wu L, Li K, Liu F, Wang L, Zhang D, Zhou J, Ma X, Wang S, Yang S. Ornithine aminotransferase promoted the proliferation and metastasis of non-small cell lung cancer via upregulation of miR-21. J Cell Physiol. 2019;234:12828-12838. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 19] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 7. | Mao L, Liu S, Hu L, Jia L, Wang H, Guo M, Chen C, Liu Y, Xu L. miR-30 Family: A Promising Regulator in Development and Disease. Biomed Res Int. 2018;2018:9623412. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 63] [Cited by in RCA: 106] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 8. | Cao JM, Li GZ, Han M, Xu HL, Huang KM. MiR-30c-5p suppresses migration, invasion and epithelial to mesenchymal transition of gastric cancer via targeting MTA1. Biomed Pharmacother. 2017;93:554-560. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 56] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 9. | Han X, Zhen S, Ye Z, Lu J, Wang L, Li P, Li J, Zheng X, Li H, Chen W, Li X, Zhao L. A Feedback Loop Between miR-30a/c-5p and DNMT1 Mediates Cisplatin Resistance in Ovarian Cancer Cells. Cell Physiol Biochem. 2017;41:973-986. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 62] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 10. | Liu D, Wu J, Liu M, Yin H, He J, Zhang B. Downregulation of miRNA-30c and miR-203a is associated with hepatitis C virus core protein-induced epithelial-mesenchymal transition in normal hepatocytes and hepatocellular carcinoma cells. Biochem Biophys Res Commun. 2015;464:1215-1221. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 36] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 11. | Ydenberg CA, Johnston A, Weinstein J, Bellavance D, Jansen S, Goode BL. Combinatorial genetic analysis of a network of actin disassembly-promoting factors. Cytoskeleton (Hoboken). 2015;72:349-361. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 18] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 12. | Li Q, Song XW, Zou J, Wang GK, Kremneva E, Li XQ, Zhu N, Sun T, Lappalainen P, Yuan WJ, Qin YW, Jing Q. Attenuation of microRNA-1 derepresses the cytoskeleton regulatory protein twinfilin-1 to provoke cardiac hypertrophy. J Cell Sci. 2010;123:2444-2452. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 119] [Cited by in RCA: 116] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 13. | Samaeekia R, Adorno-Cruz V, Bockhorn J, Chang YF, Huang S, Prat A, Ha N, Kibria G, Huo D, Zheng H, Dalton R, Wang Y, Moskalenko GY, Liu H. miR-206 Inhibits Stemness and Metastasis of Breast Cancer by Targeting MKL1/IL11 Pathway. Clin Cancer Res. 2017;23:1091-1103. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 98] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 14. | Bockhorn J, Yee K, Chang YF, Prat A, Huo D, Nwachukwu C, Dalton R, Huang S, Swanson KE, Perou CM, Olopade OI, Clarke MF, Greene GL, Liu H. MicroRNA-30c targets cytoskeleton genes involved in breast cancer cell invasion. Breast Cancer Res Treat. 2013;137:373-382. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 85] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 15. | Bockhorn J, Dalton R, Nwachukwu C, Huang S, Prat A, Yee K, Chang YF, Huo D, Wen Y, Swanson KE, Qiu T, Lu J, Park SY, Dolan ME, Perou CM, Olopade OI, Clarke MF, Greene GL, Liu H. MicroRNA-30c inhibits human breast tumour chemotherapy resistance by regulating TWF1 and IL-11. Nat Commun. 2013;4:1393. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 183] [Cited by in RCA: 199] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 16. | Kaishang Z, Xue P, Shaozhong Z, Yingying F, Yan Z, Chanjun S, Zhenzhen L, Xiangnan L. Elevated expression of Twinfilin-1 is correlated with inferior prognosis of lung adenocarcinoma. Life Sci. 2018;215:159-169. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 17] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 17. | Wang J, Guo XJ, Ding YM, Jiang JX. miR-1181 inhibits invasion and proliferation via STAT3 in pancreatic cancer. World J Gastroenterol. 2017;23:1594-1601. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 26] [Cited by in RCA: 31] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 18. | Ma T, Zhao Y, Lu Q, Lu Y, Liu Z, Xue T, Shao Y. MicroRNA-30c functions as a tumor suppressor via targeting SNAI1 in esophageal squamous cell carcinoma. Biomed Pharmacother. 2018;98:680-686. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 20] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 19. | Zhong K, Chen K, Han L, Li B. MicroRNA-30b/c inhibits non-small cell lung cancer cell proliferation by targeting Rab18. BMC Cancer. 2014;14:703. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 74] [Cited by in RCA: 94] [Article Influence: 8.5] [Reference Citation Analysis (0)] |