Published online Jun 14, 2019. doi: 10.3748/wjg.v25.i22.2763
Peer-review started: March 28, 2019
First decision: April 11, 2019
Revised: April 15, 2019
Accepted: May 3, 2019
Article in press: May 3, 2019
Published online: June 14, 2019
Processing time: 79 Days and 0.6 Hours
Gastric cancer (GC) is the fourth most frequent malignancy all over the world. The diagnosis of GC is challenging and the prognosis of GC is very unfavorable. Accumulating evidence reveals that serum long noncoding RNAs (lncRNAs) can function as biomarkers in various types of cancers, including GC.
To explore the level and molecular mechanism of the lncRNA HOXA11-AS in GC and the diagnostic and prognostic significance of serum HOXA11-AS in GC.
HOXA11-AS levels in GC tissue, cell lines, and serum samples were measured. The correlation between HOXA11-AS expression and clinicopathological characteristics was analyzed. The role of HOXA11-AS in the diagnosis and prognosis of GC was evaluated. Cell function assays were performed for exploration of the roles of HOXA11-AS in GC cells. Moreover, Western blot was performed to explore the target regulated by HOXA11-AS in GC cells.
Up-regulation of HOXA11-AS was found in GC tissues, cell lines, and serum samples. In GC patients, decreased serum HOXA11-AS levels were negatively related with tumor size, TNM stage, and lymph node metastasis. The area under the receiver operating characteristic curve of serum HOXA11-AS in the diagnosis of GC was 0.924 (95%CI: 0.881-0.967; sensitivity, 0.787; specificity 0.978). Results of the Kaplan-Meier survival curves suggested the GC patients with a lower HOXA11-AS level having a better overall survival rate. HOXA11-AS promoted GC cell proliferation and invasion. SRSF1 may be the target regulated by HOXA11-AS in GC cells.
HOXA11-AS promotes GC cell proliferation and invasion via SRSF1 and may function as a promising marker in GC.
Core tip: The long noncoding RNA (lncRNA) HOXA11-AS is up-regulated in gastric cancer (GC) tissues, cell lines, and serum samples. In GC patients, increased serum HOXA11-AS levels were positively related with tumor size, TNM stage, and lymph node metastasis. HOXA11-AS functions as a diagnostic and prognostic biomarker in GC, and promotes GC cell proliferation and invasion. SRSF1 may be the target regulated by HOXA11-AS in gastric cells.
- Citation: Liu Y, Zhang YM, Ma FB, Pan SR, Liu BZ. Long noncoding RNA HOXA11-AS promotes gastric cancer cell proliferation and invasion via SRSF1 and functions as a biomarker in gastric cancer. World J Gastroenterol 2019; 25(22): 2763-2775
- URL: https://www.wjgnet.com/1007-9327/full/v25/i22/2763.htm
- DOI: https://dx.doi.org/10.3748/wjg.v25.i22.2763
Gastric cancer (GC) is a global health problem, holding the fourth most frequent malignancy and the second most common cause of death from cancer all over the world[1]. Estimated 27510 cases diagnosed with GC and 11140 deaths from GC will occur in the United States in 2019[2]. The diagnosis of GC is challenging owing to the fact that patients often present with vague and non-specific symptoms[3], which causes many patients being at the progressive stage at first diagnosis. However, patients with metastatic GC carry a very poor prognosis and their median survival ranges from 4 mo to around 12 mo[4,5]. Although GC is associated with Helicobacter pylori infection, the molecular mechanisms of its occurrence and progression remain unclear. Consequently, it is ineluctably necessary to probe into the molecular mechanism of the occurrence as well as progression of GC and finding a new marker applying to the detection, therapy, and prognosis of GC.
Long noncoding RNAs (lncRNAs), a class of transcripts > 200 nucleotides (nts) in length, exert their significant functions in the progression and metastasis of malignancy[6]. LncRNAs typically exhibit tissue-specific expression patterns and are readily detectable in body fluids because of their high stability, in comparison with other protein biomarkers expressed in various types of tissues, making them ideal biomarkers[7]. Accumulating evidence reveals that serum lncRNAs function as biomarkers in various types of cancers, such as HOTTIP in GC[8], MALAT1 in epithelial ovarian cancer[9], GIHCG in cervical cancer[10], and LRB1 in hepatocellular carcinoma[11].
Previous studies suggested that aberrantly expressed lncRNA HOXA11-AS plays significant roles in the development and progression of malignancies[12]. HOXA11-AS functions as an oncogene and promotes cell proliferation, invasion, and metastasis in GC[13,14]. However, the molecular mechanism of HOXA11-AS in GC is far from fully elucidated and the diagnostic and prognostic role of HOXA11-AS in GC is still unclear. In this study, we thoroughly investigated the molecular mechanism of HOXA11-AS and the diagnostic and prognostic roles of serum HOXA11-AS in GC.
GC tissue specimens and corresponding paracancerous gastric tissue specimens were obtained from 25 GC patients from Binzhou People's Hospital. Each patient was diagnosed with GC pathologically and none of the patients received any treatment before operation. The tissue samples were processed within 1 hour and submerged in RNAlater reagent from Qiagen GmbH (Hilden, Germany) for half an hour. After that, the samples were stored at -80 ˚C till RNA extraction. All tissue samples were examined and classified under the management of experienced pathologists.
As for blood samples, the present study totaled 134 participants, consisting of 94 patients with GC and 40 healthy controls. Postoperative blood specimens were also obtained from 25 patients with GC. All the blood samples were disposed within 2 h. Serum was disposed at 4 °C and recruited by centrifugation (1200 × g , 10 min). Another centrifugation (10000 × g, 10 mins, at 4 °C) was performed for completely removing residual cellular debris. The serum specimens were kept in liquid nitrogen till RNA extraction.
Our research was managed by the ethics committee of Binzhou People's Hospital. Consent statement was obtained from each participant. Table 1 lists the clinical characteristics of the GC patients.
| Tissue samples | Serum samples | Healthy controls | |
| (n = 25) | |||
| Gender | |||
| Male | 15 | 57 | 26 |
| Female | 10 | 37 | 14 |
| Age | |||
| Mean ± SD | 60 ± 7 | 59 ± 7 | 59 ± 8 |
| Median (range) | 61 (45-72) | 60 (40-74) | 60 (41-74) |
| Tumor size (cm) | |||
| <5 | 19 | 59 | |
| ≥5 | 6 | 35 | |
| TNM stage | |||
| I | 13 | 18 | |
| II | 9 | 21 | |
| III | 3 | 42 | |
| IV | 0 | 13 | |
| Differentiation | |||
| Well and moderate | 13 | 50 | |
| Poor | 12 | 44 | |
| Invasion depth | |||
| T1 | 3 | 9 | |
| T2 | 7 | 32 | |
| T3 | 12 | 35 | |
| T4 | 3 | 18 | |
| Lymph node metastasis | |||
| Positive | 18 | 50 | |
| Negative | 7 | 44 |
A culture medium consisting of 88% of Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific. Inc., Waltham, MA, United States), 10% of fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.), 1% of antibiotics (100 μL/mL penicillin and 100 mg/mL streptomycin sulfate), and 1% of glutamine was used to cultivate normal gastric mucosal epithelial cells (GES-1) and GC cells (MKN-45). MKN-45 and GES-1 cells were maintained in an incubator (37 °C, 5% CO2). MKN-45 cells were pre-seeded in the 6-well plates (3.0 × 105 cells/per well) and maintained until the confluence of 60%. HOXA11-AS primers, siRNA, and negative control (si-NC) were provided by Ribobio (Guangzhou, China); pcDNA-HOXA11-AS and pcDNA3.1 were constructed by Generay (Shanghai, China). siRNAs or pcDNAs were transfected into GES-1 cells with Lipofectamine RNAiMAX Reagent (Life Technologies, Carlsbad, CA, United States).
The extraction of total RNA from tissue and cell samples was done using TRIzol reagent from Invitrogen (Thermo Fisher Scientific) following the manufacturer’s procedures. The isolation of total RNA from serum samples was done with Qiagen miRNeasy Serum/Plasma Kit from Qiagen GmbH (Hilden, Germany) following the manufacturers’ procedures. The quantity and purity of the extracted RNA samples were detected with NanoDrop 2000c (Thermo Fisher Scientific, Inc.). The RNA samples with an optical density ratio (260/280) of 1.8-2.0 were accepted into later study. RNA samples were either kept at -80 °C or used for direct synthesis of cDNA.
The PrimerScript RT Master from Takara Biotechnology (Dalian, China) was used for cDNA synthesis. The level of HOXA11-AS was measured by RT-qPCR with SYBR-Green PCR Master mix (Roche, Mannheim, Germany) on the Roche Lightcycler 480 Real-Time PCR system (Roche Diagnostics, Basel, Switzerland). The qRT-PCR reactions were performed in triplicate. Changes in HOXA11-AS level were calculated using the 2−ΔCt method. Gene expression values were normalized to the expression of GADPH. The sequences used in qRT-PCR are as follows: HOXA11-AS-F: 5′-GATTTCTCCAGCCTCCCTTC-3′ and HOXA11-AS-R: 5′-AGAAATTG GACGAGACTGCG-3′; GAPDH-F: 5′-TGGTATCGTGGAAGGACTCAT-3′ and GAPDH-R: 5′- GTGGGTGTCGCTGTTGAAGTC-3′.
CCK-8 assay was conducted to explore cell proliferation ability. In this assay, a 96-well plate was used and transfected MKN-45 cells were seeded in each well at a density of 3.0 × 103 cells/well. The cells were maintained in a cell incubator. Ten microliters of CCK-8 reagent (US Everbright Inc.) were added to each well at 24, 48, and 72 h after the cells were incubated, respectively. The optical density of each well of 96-well plates was detected using an enzyme immunoassay instrument from Bio Rad Laboratories (Hercules, CA, USA).
Transwell assay was conducted to explore cell invasive capability. Transfected MKN-45 cells (4 × 104) with 200 μL of DMEM medium without serum were added into a transwell chamber (BD Biosciences, New York, NJ, United States) with Matrigel, which was inserted into a 24-well plate. Each well of the 24-well plate was added with 450 μL of DMEM medium and 50 μL of FBS. MKN-45 cells were cultured for another 36 hours. The MKN-45 cells that did not invade through the membrane were washed and cleaned. Crystal violet (1%) was used and the invaded MKN-45 cells invading through the membrane were stained for 25 min. After washing cells with PBS three times, the number of MKN-45 cells invading through the membrane was calculated in three random visual places using a light microscope.
After rinsing with pre-cooling PBS, MKN-45 cells were dissociated with RIPA buffer (Thermo Fisher Scientific) mixed with protease and phosphatase inhibitor cocktail (Roche). The total protein of MKN-45 cells was isolated on the basis of instructions. The Bio-Rad assay system (Bio-Rad Laboratories, Hercules, CA, United States) was used to detect the protein concentration. In Western blot analysis, 12% sodium dodecyl sulfate-polyacrylamide gels were prepared and the same amounts of protein extract were separated by electrophoresis and then transferred to polyvinylidene difluoride (PVDF) membranes (Millipore, Billerica, MA, USA). After blocking in 5% skim milk, the blotted membranes were immersed in primary antibodies (SRSF1, 1:1000; GAPDH, 1:2000, Proteintech, Wuhan, China) for 24 h. The membranes were rinsed with TBST three times (10 min/time). Subsequently, the membranes were immersed in a secondary antibody (HRP conjugated goat anti-rabbit antibody, 1:1000, Sigma) at room temperature for 1 h. GAPDH was used as the internal control. Kodak film (Kodak, Rochester, NY, United States) was used to detect the blots with an enhanced chemiluminescence kit (Merck Millipore).
SPSS 21.0 software and Graphpad prism 7 software were used for statistical analyses. The Student’s t-test was conducted to evaluate the differences between subgroups. The Chi-square test and Fisher’s exact test was conducted to explore the associations between HOXA11-AS expression and clinicopathological features. ROC curve analysis was performed for the assessment of the possibility of serum HOXA11-AS for GC detection. The significance of serum HOXA11-AS for prognosis evaluation of GC was evaluated by the Kaplan-Meier method. Cox regression analysis was conducted for evaluating the possible factors that can predict the prognosis of GC. P < 0.05 was considered statistically significant.
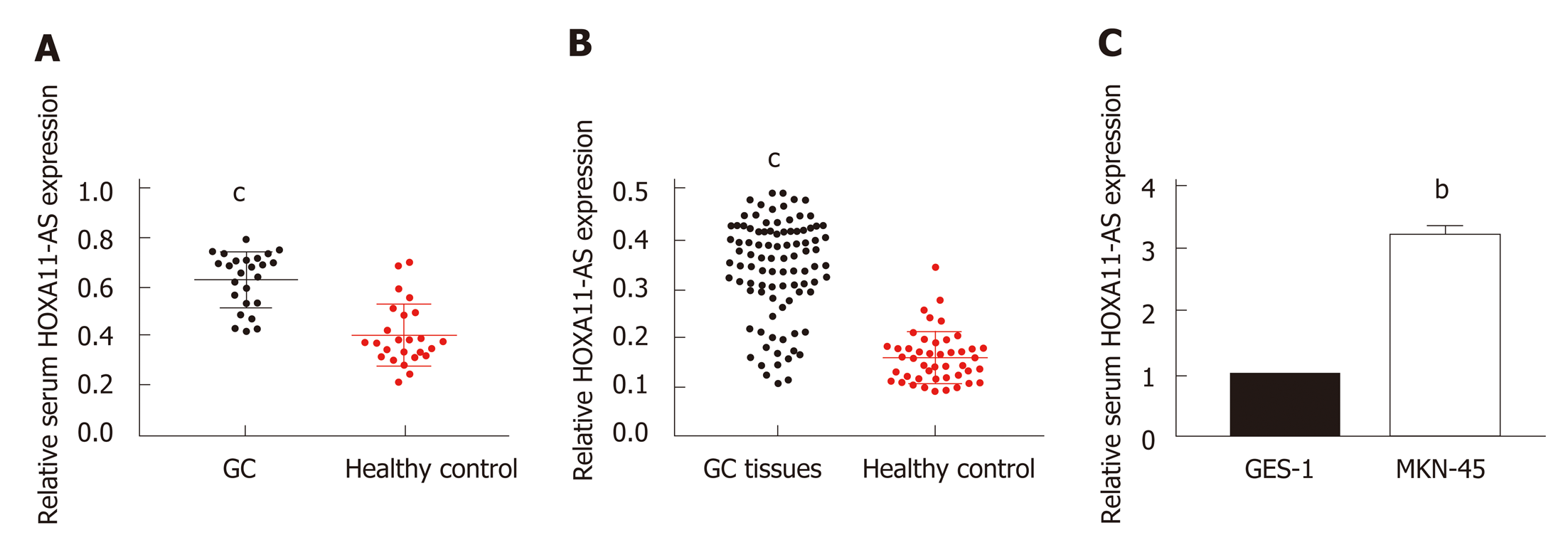
We first explored the levels of HOXA11-AS in tissue and serum samples. As presented in Table 1, there was no significant difference in age or gender between GC patients and healthy controls. A significantly increased level of HOXA11-AS was found in GC tissues vs the paired normal gastric tissues (Figure 1A, P < 0.001). Furthermore, a similar result was obtained in serum samples. Significant up-regulation of HOXA11-AS was found in the serum of GC patients vs healthy individuals (Figure 1B, P < 0.001). As expected, the same result was obtained in GC cell lines, which revealed that HOXA11-AS was up-regulated in MKN-45 cells compared with GES-1 cells (Figure 1C, P < 0.01).
We next thoroughly investigated the association between HOXA11-AS level and GC patients’ clinicopathologic features. The 94 GC patients were split into two parts using median serum HOXA11-AS l as the cutoff (47 with a high serum HOXA11-AS level and 47 with a low serum HOXA11-AS level). As shown in Table 2, there was no significant correlation between serum HOXA11-AS and gender, age, differentiation, or invasion depth (P > 0.05), while a remarkable association was obtained between serum HOXA11-AS and TNM stage (P = 0.001), tumor size (P = 0.001), as well as lymph node metastasis (P = 0.011). We found that the GC patients with decreased serum HOXA11-AS more probably had smaller tumor, earlier TNM stage, and negative lymph node metastasis.
| Clinical feature | Cases | Serum HOXA11-AS | P-value |
| Gender | 0.824 | ||
| Male | 57 | 0.345 ± 0.104 | |
| Female | 37 | 0.349 ± 0.091 | |
| Age (yr) | 0.304 | ||
| <60 | 38 | 0.337 ± 0.111 | |
| ≥60 | 56 | 0.355 ± 0.089 | |
| Tumor size (cm) | 0.001 | ||
| <5 | 59 | 0.320 ± 0.094 | |
| ≥5 | 35 | 0.391 ± 0.092 | |
| TNM stage | 0.001 | ||
| I and II | 39 | 0.250 ± 0.073 | |
| III and IV | 55 | 0.415 ± 0.040 | |
| Differentiation | 0.934 | ||
| Well and moderate | 50 | 0.346 ± 0.112 | |
| Poor | 44 | 0.347 ± 0.084 | |
| Invasion depth | 0.867 | ||
| T1 and T2 | 41 | 0.348 ± 0.100 | |
| T3 and T4 | 53 | 0.345 ± 0.099 | |
| Lymph node metastasis | 0.011 | ||
| Positive | 50 | 0.377 ± 0.088 | |
| Negative | 44 | 0.312 ± 0.100 |
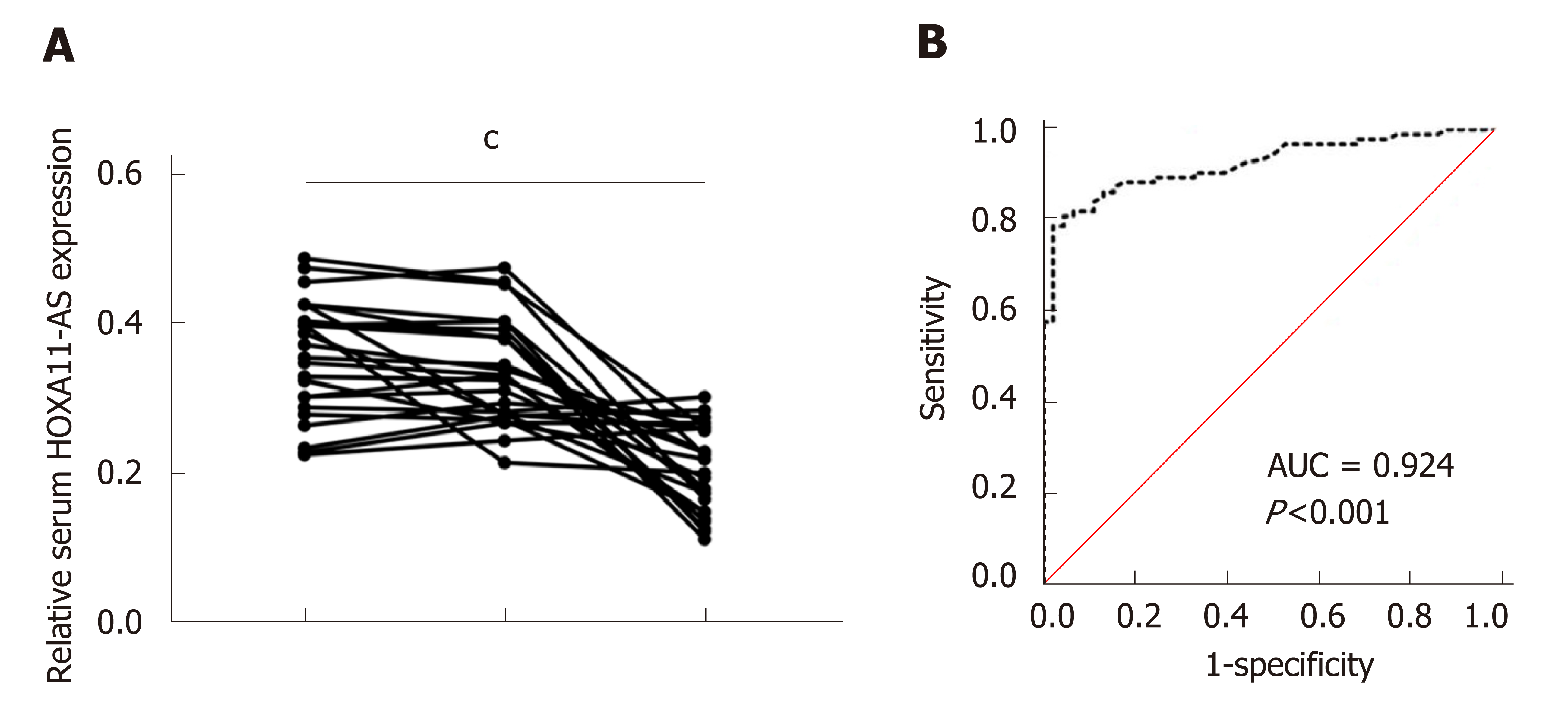
We further tested HOXA11-AS expression in serum samples from postoperative GC patients. The data demonstrated that compared with preoperative samples, serum HOXA11-A levels showed no significant difference in the samples obtained 1 d after surgery (P > 0.05), but they were greatly decreased in the samples obtained 1 mo after surgery (P = 0.001) (Figure 2A). Furthermore, the area under the ROC curve (AUC) of serum HOXA11-AS in the diagnosis of GC was 0.924 (95%CI: 0.881-0.967, sensitivity 0.787, specificity 0.978) (Figure 2A). These data demonstrated that HOXA11-AS possibly acts as a potential marker in the diagnosis of GC.
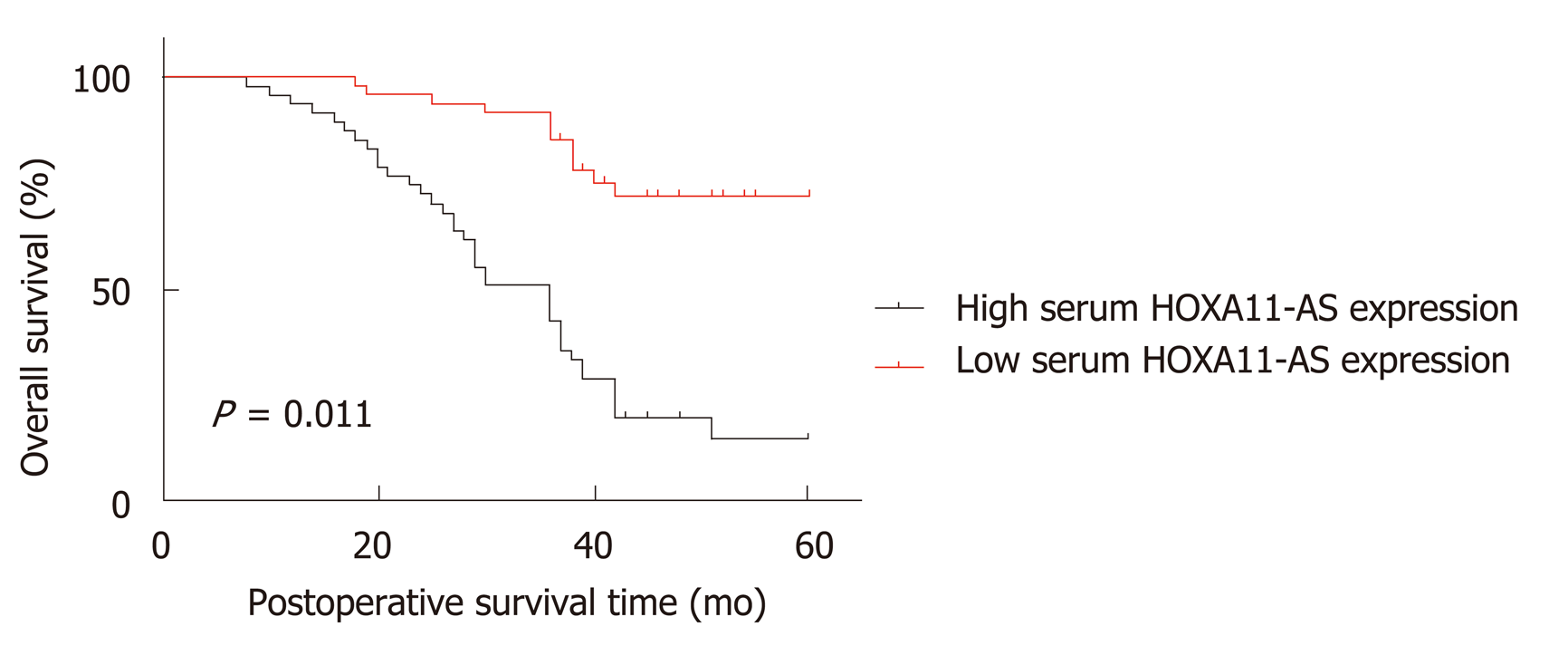
Subsequently, we analyzed the role of HOXA11-AS expression in the prognosis evaluation of GC patients. As shown in Table 3, the univariate Cox proportional hazard regression analysis revealed that tumor size (HR = 0.527, 95%CI: 0.301-0.924, P = 0.025), TNM stage (HR = 0.195, 95%CI: 0.092-0.417, P = 0.001), and serum HOXA11-AS (HR = 0.201, 95%CI: 0.104-0.0.835, P = 0.001) were potential factors affecting the overall survival of GC patients, and the multivariate Cox proportional hazard regression analysis demonstrated that serum HOXA11-AS (HR = 0.338, 95%CI: 0.115-0.989, P = 0.048) was a potential factor affecting the overall survival of GC patients. What's more, the Kaplan-Meier survival curve was drawn, which demonstrated that GC individuals with lower expression of HOXA11-AS had a better overall survival rate (P = 0.001, Figure 3). These data demonstrated that HOXA11-AS may act as a potential marker for the prognosis of GC.
| Variable | Univariate analysis | Multivariate analysis | ||||
| HR | 95%CI | P-value | HR | 95%CI | P-value | |
| Gender (Male vs Female) | 1.310 | 0.749-2.291 | 0.344 | - - - | - - - | - - - |
| Age (yr) (<60 vs ≥60) | 0.987 | 0.560-1.739 | 0.963 | - - - | - - - | - - - |
| Tumor size (cm) (<5 vs ≥5) | 0.527 | 0.301-0.924 | 0.025 | 0.816 | 0.442-1.508 | 0.516 |
| TNM stage (I + II vs III + IV) | 0.195 | 0.092-0.417 | 0.001 | 0.437 | 0.125-1.522 | 0.193 |
| Differentiation (Well and moderate vs Poor) | 0.923 | 0.529-1.61 | 0.779 | - - - | - - - | - - - |
| Invasion depth (T1 + T2 vs T3 vs T4) | 1.050 | 0.601-1.836 | 0.863 | - - - | - - - | - - - |
| Lymph node metastasis (Positive vs Negative) | 0.667 | 0.379-1.176 | 0.162 | - - - | - - - | - - - |
| Serum HOXA11-AS (High vs Low) | 0.201 | 0.104-0.385 | 0.001 | 0.338 | 0.115-0.989 | 0.048 |
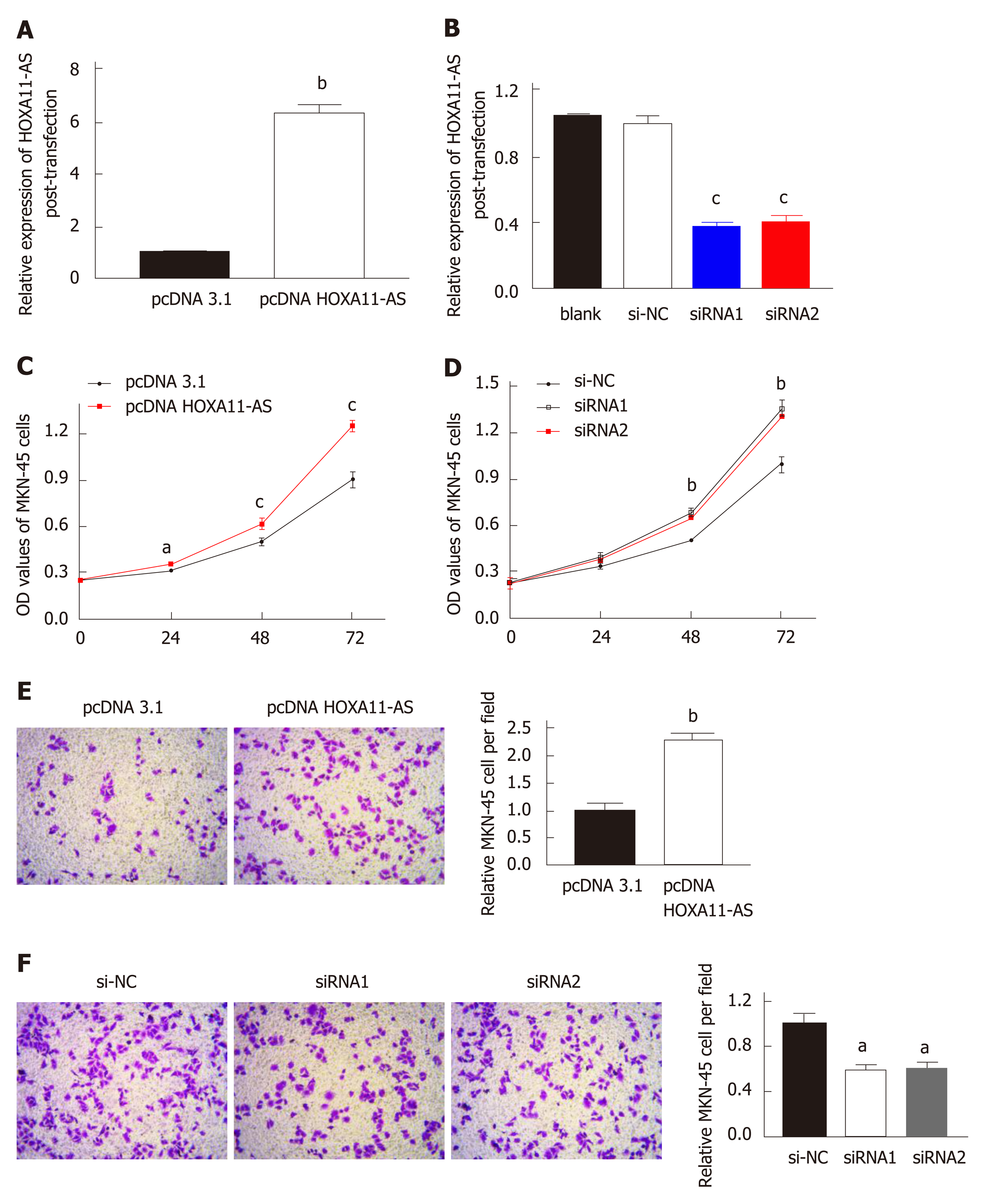
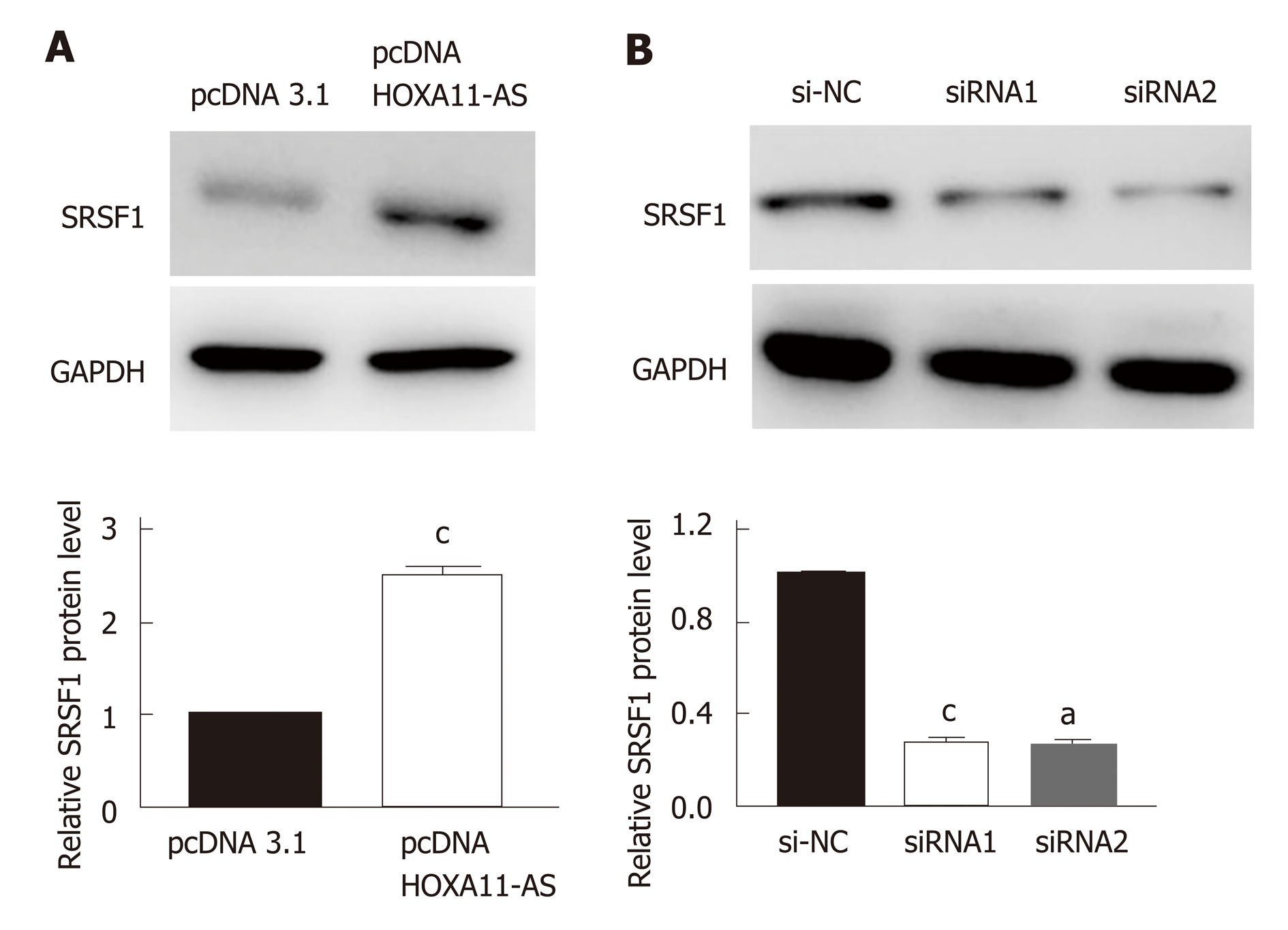
The down-regulation or up-regulation of HOXA11-AS in MKN-45 cells transfected with pcDNA HOXA11-AS or siRNA was authenticated by RT-qPCR. The amount of HOXA11-AS in MKN-45 cells transfected with pc-DNA-HOXA11-AS was noteworthily upregulated vs those transfected with pcDNA3.1 (P < 0.01, Figure 4A). Moreover, MKN-45 cells transfected with HOXA11-AS-siRNA1 (P < 0.001) or HOXA11-AS -siRNA2 (P < 0.01) showed a higher level of HOXA11-AS than those transfected with si-NC (Figure 4B).
In our study, CCK‑8 assay was conducted to evaluate cell proliferative capability. As expected, cell proliferation was faster in MKN-45 cells transfected with pcDNA HOXA11-AS compared with those transfected with pcDNA 3.1 (Figure 4C). On the contrary, cell proliferation was slower in MKN-45 cell transfected with siRNA1 or siRNA2 in comparison with those transfected with si-NC (Figure 4D).
Transwell assay was conducted to evaluate cell invasion capability, and the results are presented in Figure 4E and F, which revealed that the quantity of invaded cells was significantly higher in MKN-45 cells transfected with pcDNA HOXA11-AS than in those transfected with pcDNA 3.1 (P < 0.01, Figure 4E). However, the quantity of invaded cells was remarkably lower in MKN-45 cells transfected with siRNA1 (P < 0.05) or siRNA2 (P < 0.05) than in those transfected with si-NC (Figure 4F).
The result of Western blot indicated that the protein expression of SRSF1 in MKN-45 cells was upregulated when the expression of HOXA11-AS was increased (P < 0.001, Figure 5A). On the contrary, the protein expression of SRSF1 in MKN-45 cells was downregulated when the expression of HOXA11-AS was decreased (P < 0.01, Figure 5B).
GC is one of the most frequent and fatal malignancies, seriously threatening human health. Because of the position of the stomach and atypical clinical features of GC, it is quite difficult to identify GC patients from the healthy control at an early curable stage. Therefore, over half of GC patients are already in the advanced stage when they are initially diagnosed with GC and the 5-year overall survival (OS) rate of GC is less than 30%[15]. However, there are no biomarkers that are accurate and easy to detect for the diagnosis, prognosis evaluation, and treatment of GC clinically, which may be attributed to the mystery of the molecular mechanism about the genesis and progression of GC.
HOXA11-AS, located at the 5′ region of the HOXA cluster, is one of frequently studied lncRNAs[12]. The level of HOXA11-AS was significantly increased in lung cancer[16], breast cancer[17], renal cancer[18], and osteosarcoma[19]. Previous studies have found that HOXA11-AS served as a biomarker in head and head-neck carcinoma[20] and esophageal carcinoma[21]. Moreover, a meta-analysis revealed that HOXA11-AS functioned as a prognostic marker in human solid tumors[22]. As for non-small cell lung cancer (NSCLC), an increased level of HOXA11-AS had a promotive role in cell epithelial–mesenchymal transition, which was achieved through suppressing miR-200b[23]. Moreover, the levels of HOXA11-AS in hepatocellular carcinoma tissues and cell lines were upregulated. HOXA11-AS promoted cell proliferation by regulating cell cycle and apoptosis via DUSP5 in hepatocellular carcinoma[24]. These results demonstrated that HOXA11-AS may play a significant role in various types of cancers, including GC. In GC, HOXA11-AS was reported to be act as an oncogene and promote GC cell proliferation, invasion, and metastasis[13,14]. However, the roles of HOXA11-AS in the diagnosis and prognosis of GC have yet to be fully elucidated.
In our study, the levels of HOXA11-AS in GC tissue samples, cell lines, and serum samples were detected, which revealed increased levels of HOXA11-AS in these samples and cell lines. Moreover, in GC patients, serum HOXA11-AS was negatively related to tumor size, TNM stage, and lymph node metastasis. The role of HOXA11-AS in the diagnosis of GC was explored, which revealed that the AUC of serum HOXA11-AS in the diagnosis of GC was 0.924 (95%CI: 0.881-0.967, sensitivity 0.787, specificity 0.978). The Kaplan-Meier survival curve was drawn to evaluate HOXA11-AS in the prognosis of GC and the data revealed that the GC patients with low HOXA11-AS expression has a better overall survival rate. Further functional assays were conducted to probe into how HOXA11-AS affects the biological behavior of GC cells, which revealed that HOXA11-AS promoted GC cell proliferation and invasion.
Serine/arginine splicing factor 1 (SRSF1), an important family member of SR proteins, was reported to participate in multiple biological functions, including splicing regulation, translation control, RNA transport, cell proliferation, invasion, and senescence[25-29]. SRSF1 has emerged as a key oncodriver in numerous solid tumors, such as GC. In our research, SRSF1 was found to be positively co-expressed with HOXA11-AS in GC by StarBase 3.0 (http://starbase.sysu.edu.cn/index.php). Therefore, we assume that SRSF1 may be a target of HOXA11-AS in GC cells. Western blot analysis was performed, and as expected, the results showed that SRSF1 may be the target of HOXA11-AS in GC cells.
LncRNAs play a significant role in the genesis and progression of malignant tumors. Previous research found that downregulation of HOXA11-AS caused positive changes in NSCLC cell proliferation and migration, reversed the epithelial mesenchymal transition process, and induced cell apoptosis[30]. HOXA11-AS was increased in breast cancer and promoted cell invasion and metastasis via EMT, providing a theoretical basis and important molecular target for the therapy of breast cancer[17]. Results of previous research verified that HOXA11-AS promoted liver metastasis in colorectal cancer by acting as a miR-125a-5p sponge[31]. HOXA11-AS also exerted its functions in glioblastoma, uveal melanoma, renal cancer, and cervical cancer[18,32-34].
Previous studies revealed that lncRNAs can function as diagnostic biomarkers in GC. It was found that serum lncRNA CTC-497E21.4 may function as a potentially diagnostic marker for GC[35]. A lower level of circulating LINC00086 expression was identified in GC patients than in normal individuals. LINC00086 distinguished GC patients from healthy controls with a high sensitivity and specificity, suggested that LINC00086 is a potential biomarker for the diagnosis of GC[36]. Moreover, circulating H19 was upregulated in GC patients in comparison with healthy individuals and H19 expression declined significantly upon surgical removal of the tumors, suggesting that H19 can act as a diagnostic biomarker of GC[37].
Previous studies also revealed that lncRNAs can function as prognostic biomarkers in GC. It was found that serum lncRNA HULC was upregulated in GC patients and Kaplan-Meier curve analysis suggested that HULC was a good predictor of the prognosis of GC[7]. Circulating GACAT2 level was noteworthily up-regulated in preoperative GC patients than in the postoperative group. Moreover, the expression of GACAT2 were associated with the clinicopathologic features (lymphatic metastasis, distal metastasis, and perineural invasion) of patients with GC, which suggested that GACAT2 may be a potential tumor marker for the prediction of GC patient prognosis[16]. It was reported that up-regulation of plasma MALAT1 was inde-pendently correlated with a poor prognosis in GC, demonstrating MALAT1 as a novel marker for the distant metastasis of GC patients[38].
In conclusion, serum HOXA11-AS significantly increases in GC patients. HOXA11-AS promotes GC cell proliferation and invasion via SRSF1 and may function as a diagnostic and prognostic biomarker in GC.
As one of the most frequent cancers, gastric cancer (GC) caused more than 700000 deaths in just 2012 worldwide. Many recent studies have demonstrated the molecular mechanisms involved in transcriptional regulation in GC, and long noncoding RNAs (lncRNAs) play an irreplaceable role in the initiation and progression of GC, such as maintaining cell growth, evasion of apoptosis, promotion of invasion and metastasis, stemness maintenance, and EMT.
To identify more biomarkers for the diagnosis and treatment of GC.
This study aimed to investigate the underlying mechanisms of HOXA11-AS in GC.
HOXA11-AS expression was detected by qRT-PCR assay in GC tissues, cell lines, and serum samples. Clinicopathological characteristics were collected and expression analysis of HOXA11-AS was performed to evaluate the role of HOXA11-AS. Cell function assays were performed to explore the functions of HOXA11-AS in GC cell lines. Moreover, Western blot was performed to explore the target regulated by HOXA11-AS in GC cell lines.
We found that HOXA11-AS was upregulated in GC tissues, cell lines, and serum samples, and exhibited a significant negative correlation with tumor size, TNM stage, and lymph node metastasis. Cell experiments showed that HOXA11-AS promoted the proliferation and invasion capacity of GC cell lines, and SRSF1 may be the target regulated by HOXA11-AS in GC cells. Especially, GC patients with a lower HOXA11-AS level had a better overall survival rate.
Our study demonstrated that HOXA11-AS can significantly promote GC cell growth, migration, and invasion. Furthermore, it can work through SRSF1. Therefore, our study provides the possible molecular mechanism and two new biomarkers for GC.
In the future, research may reveal the important role of HOXA11-AS that enhances the sensitivity of GC detection and facilitate its application in anti-cancer treatments. The identification of the HOXA11-AS/SRSF1 molecular axis may further explain the underlying mechanism.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Lazăr DC, Tanabe S S-Editor: Gong ZM L-Editor: Wang TQ E-Editor: Zhang YL
| 1. | Van Cutsem E, Sagaert X, Topal B, Haustermans K, Prenen H. Gastric cancer. Lancet. 2016;388:2654-2664. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1282] [Cited by in RCA: 1468] [Article Influence: 163.1] [Reference Citation Analysis (0)] |
| 2. | Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7-34. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13300] [Cited by in RCA: 15475] [Article Influence: 2579.2] [Reference Citation Analysis (2)] |
| 3. | Thrumurthy SG, Chaudry MA, Hochhauser D, Mughal M. The diagnosis and management of gastric cancer. BMJ. 2013;347:f6367. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 109] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 4. | Murad AM, Santiago FF, Petroianu A, Rocha PR, Rodrigues MA, Rausch M. Modified therapy with 5-fluorouracil, doxorubicin, and methotrexate in advanced gastric cancer. Cancer. 1993;72:37-41. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 5. | Wagner AD, Unverzagt S, Grothe W, Kleber G, Grothey A, Haerting J, Fleig WE. Chemotherapy for advanced gastric cancer. Cochrane Database Syst Rev. 2010;CD004064. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 231] [Cited by in RCA: 350] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 6. | Lee YR, Kim G, Tak WY, Jang SY, Kweon YO, Park JG, Lee HW, Han YS, Chun JM, Park SY, Hur K. Circulating exosomal noncoding RNAs as prognostic biomarkers in human hepatocellular carcinoma. Int J Cancer. 2019;144:1444-1452. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 202] [Article Influence: 28.9] [Reference Citation Analysis (0)] |
| 7. | Jin C, Shi W, Wang F, Shen X, Qi J, Cong H, Yuan J, Shi L, Zhu B, Luo X, Zhang Y, Ju S. Long non-coding RNA HULC as a novel serum biomarker for diagnosis and prognosis prediction of gastric cancer. Oncotarget. 2016;7:51763-51772. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 70] [Cited by in RCA: 91] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 8. | Zhao R, Zhang Y, Zhang X, Yang Y, Zheng X, Li X, Liu Y, Zhang Y. Exosomal long noncoding RNA HOTTIP as potential novel diagnostic and prognostic biomarker test for gastric cancer. Mol Cancer. 2018;17:68. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 140] [Cited by in RCA: 235] [Article Influence: 33.6] [Reference Citation Analysis (0)] |
| 9. | Chen Q, Su Y, He X, Zhao W, Wu C, Zhang W, Si X, Dong B, Zhao L, Gao Y, Yang X, Chen J, Lu J, Qiao X, Zhang Y. Plasma long non-coding RNA MALAT1 is associated with distant metastasis in patients with epithelial ovarian cancer. Oncol Lett. 2016;12:1361-1366. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 49] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 10. | Zhang X, Mao L, Li L, He Z, Wang N, Song Y. Long noncoding RNA GIHCG functions as an oncogene and serves as a serum diagnostic biomarker for cervical cancer. J Cancer. 2019;10:672-681. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 23] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 11. | Wang ZF, Hu R, Pang JM, Zhang GZ, Yan W, Li ZN. Serum long noncoding RNA LRB1 as a potential biomarker for predicting the diagnosis and prognosis of human hepatocellular carcinoma. Oncol Lett. 2018;16:1593-1601. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 20] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 12. | Lu CW, Zhou DD, Xie T, Hao JL, Pant OP, Lu CB, Liu XF. HOXA11 antisense long noncoding RNA (HOXA11-AS): A promising lncRNA in human cancers. Cancer Med. 2018;7:3792-3799. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 60] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 13. | Liu Z, Chen Z, Fan R, Jiang B, Chen X, Chen Q, Nie F, Lu K, Sun M. Over-expressed long noncoding RNA HOXA11-AS promotes cell cycle progression and metastasis in gastric cancer. Mol Cancer. 2017;16:82. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 97] [Cited by in RCA: 136] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 14. | Sun M, Nie F, Wang Y, Zhang Z, Hou J, He D, Xie M, Xu L, De W, Wang Z, Wang J. LncRNA HOXA11-AS Promotes Proliferation and Invasion of Gastric Cancer by Scaffolding the Chromatin Modification Factors PRC2, LSD1, and DNMT1. Cancer Res. 2016;76:6299-6310. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 328] [Cited by in RCA: 405] [Article Influence: 45.0] [Reference Citation Analysis (0)] |
| 15. | Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5-29. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9172] [Cited by in RCA: 9958] [Article Influence: 995.8] [Reference Citation Analysis (0)] |
| 16. | Tan L, Yang Y, Shao Y, Zhang H, Guo J. Plasma lncRNA-GACAT2 is a valuable marker for the screening of gastric cancer. Oncol Lett. 2016;12:4845-4849. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 29] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 17. | Li W, Jia G, Qu Y, Du Q, Liu B, Liu B. Long Non-Coding RNA (LncRNA) HOXA11-AS Promotes Breast Cancer Invasion and Metastasis by Regulating Epithelial-Mesenchymal Transition. Med Sci Monit. 2017;23:3393-3403. [PubMed] |
| 18. | Yang FQ, Zhang JQ, Jin JJ, Yang CY, Zhang WJ, Zhang HM, Zheng JH, Weng ZM. HOXA11-AS promotes the growth and invasion of renal cancer by sponging miR-146b-5p to upregulate MMP16 expression. J Cell Physiol. 2018;233:9611-9619. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 48] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 19. | Cui M, Wang J, Li Q, Zhang J, Jia J, Zhan X. Long non-coding RNA HOXA11-AS functions as a competing endogenous RNA to regulate ROCK1 expression by sponging miR-124-3p in osteosarcoma. Biomed Pharmacother. 2017;92:437-444. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 63] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 20. | Yao Y, Chen X, Lu S, Zhou C, Xu G, Yan Z, Yang J, Yu T, Chen W, Qian Y, Ding S, Tang J, Chen Y, Zhang Y. Circulating Long Noncoding RNAs as Biomarkers for Predicting Head and Neck Squamous Cell Carcinoma. Cell Physiol Biochem. 2018;50:1429-1440. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 30] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 21. | Sun XY, Wang XF, Cui YB, Cao XG, Zhao RH, Wei HY, Cao W, Wu W. [Expression level and clinical significance of LncRNA HOXA11-AS in esophageal squamous cell carcinoma patients]. Zhonghua Zhong Liu Za Zhi. 2018;40:186-190. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 22. | Mu S, Ai L, Fan F, Sun C, Hu Y. Prognostic and clinicopathological significance of long noncoding RNA HOXA11-AS expression in human solid tumors: a meta-analysis. Cancer Cell Int. 2018;18:1. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 44] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 23. | Chen JH, Zhou LY, Xu S, Zheng YL, Wan YF, Hu CP. Overexpression of lncRNA HOXA11-AS promotes cell epithelial-mesenchymal transition by repressing miR-200b in non-small cell lung cancer. Cancer Cell Int. 2017;17:64. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51] [Cited by in RCA: 72] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 24. | Liu B, Li J, Liu X, Zheng M, Yang Y, Lyu Q, Jin L. Long non-coding RNA HOXA11-AS promotes the proliferation HCC cells by epigenetically silencing DUSP5. Oncotarget. 2017;8:109509-109521. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 25] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 25. | Erkelenz S, Mueller WF, Evans MS, Busch A, Schöneweis K, Hertel KJ, Schaal H. Position-dependent splicing activation and repression by SR and hnRNP proteins rely on common mechanisms. RNA. 2013;19:96-102. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 150] [Cited by in RCA: 174] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 26. | Wang Y, Xiao X, Zhang J, Choudhury R, Robertson A, Li K, Ma M, Burge CB, Wang Z. A complex network of factors with overlapping affinities represses splicing through intronic elements. Nat Struct Mol Biol. 2013;20:36-45. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 75] [Cited by in RCA: 82] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 27. | Das R, Yu J, Zhang Z, Gygi MP, Krainer AR, Gygi SP, Reed R. SR proteins function in coupling RNAP II transcription to pre-mRNA splicing. Mol Cell. 2007;26:867-881. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 261] [Cited by in RCA: 260] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 28. | Fregoso OI, Das S, Akerman M, Krainer AR. Splicing-factor oncoprotein SRSF1 stabilizes p53 via RPL5 and induces cellular senescence. Mol Cell. 2013;50:56-66. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 93] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 29. | Zhou X, Wang R, Li X, Yu L, Hua D, Sun C, Shi C, Luo W, Rao C, Jiang Z, Feng Y, Wang Q, Yu S. Splicing factor SRSF1 promotes gliomagenesis via oncogenic splice-switching of MYO1B. J Clin Invest. 2019;129:676-693. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 114] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 30. | Zhao X, Li X, Zhou L, Ni J, Yan W, Ma R, Wu J, Feng J, Chen P. LncRNA HOXA11-AS drives cisplatin resistance of human LUAD cells via modulating miR-454-3p/Stat3. Cancer Sci. 2018;109:3068-3079. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 101] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 31. | Chen D, Sun Q, Zhang L, Zhou X, Cheng X, Zhou D, Ye F, Lin J, Wang W. The lncRNA HOXA11-AS functions as a competing endogenous RNA to regulate PADI2 expression by sponging miR-125a-5p in liver metastasis of colorectal cancer. Oncotarget. 2017;8:70642-70652. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 47] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 32. | Se YB, Kim SH, Kim JY, Kim JE, Dho YS, Kim JW, Kim YH, Woo HG, Kim SH, Kang SH, Kim HJ, Kim TM, Lee ST, Choi SH, Park SH, Kim IH, Kim DG, Park CK. Underexpression of HOXA11 Is Associated with Treatment Resistance and Poor Prognosis in Glioblastoma. Cancer Res Treat. 2017;49:387-398. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 33] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 33. | Lu Q, Zhao N, Zha G, Wang H, Tong Q, Xin S. LncRNA HOXA11-AS Exerts Oncogenic Functions by Repressing p21 and miR-124 in Uveal Melanoma. DNA Cell Biol. 2017;36:837-844. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 50] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 34. | Kim HJ, Eoh KJ, Kim LK, Nam EJ, Yoon SO, Kim KH, Lee JK, Kim SW, Kim YT. The long noncoding RNA HOXA11 antisense induces tumor progression and stemness maintenance in cervical cancer. Oncotarget. 2016;7:83001-83016. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 69] [Cited by in RCA: 74] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 35. | Zong W, Feng W, Jiang Y, Ju S, Cui M, Jing R. Evaluating the diagnostic and prognostic value of serum long non-coding RNA CTC-497E21.4 in gastric cancer. Clin Chem Lab Med. 2019; Epub ahead of print. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 21] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 36. | Ji B, Huang Y, Gu T, Zhang L, Li G, Zhang C. Potential diagnostic and prognostic value of plasma long noncoding RNA LINC00086 and miR-214 expression in gastric cancer. Cancer Biomark. 2019;24:249-255. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 23] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 37. | Yörüker EE, Keskin M, Kulle CB, Holdenrieder S, Gezer U. Diagnostic and prognostic value of circulating lncRNA H19 in gastric cancer. Biomed Rep. 2018;9:181-186. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 30] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 38. | Xia H, Chen Q, Chen Y, Ge X, Leng W, Tang Q, Ren M, Chen L, Yuan D, Zhang Y, Liu M, Gong Q, Bi F. The lncRNA MALAT1 is a novel biomarker for gastric cancer metastasis. Oncotarget. 2016;7:56209-56218. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 70] [Cited by in RCA: 95] [Article Influence: 13.6] [Reference Citation Analysis (0)] |