Published online Apr 21, 2019. doi: 10.3748/wjg.v25.i15.1890
Peer-review started: December 27, 2018
First decision: January 18, 2019
Revised: February 17, 2019
Accepted: February 22, 2019
Article in press: February 23, 2019
Published online: April 21, 2019
Processing time: 112 Days and 21.1 Hours
Exosomes contain proteins, lipids, and biological molecules such as DNA and RNA. Nucleic acids in exosomes are a group of molecules that can act as biomarkers. Currently, there are many reports on exosomal microRNAs, which are ideal biomarkers for the early diagnosis of cancer. However, there are few reports on the role of exosomal microRNAs in the diagnosis and prognosis of hepatocellular carcinoma (HCC).
To understand the mechanism of exosomal microRNA-224 (miR-224) in the development of HCC and evaluate its diagnostic and prognostic value.
Cell culture and transfection of exosomal miRNA-224, real-time quantitative PCR, luciferase reporter assay, and other methods were used to find new biomarkers related to the development of HCC that can be used to diagnose HCC and predict HCC prognosis.
By targeting glycine N-methyltransferase, incubating exosomes with miR-224 mimic resulted in a significant increase in cell proliferation compared to that of the control group, while incubation with the miR-224 inhibitor significantly reduced cell proliferation. The same results were obtained for the cell invasion assay. Serum exosomal miR-224 did have some ability to differentiate patients with HCC from healthy controls, with an area under the curve of 0.910, and HCC patients with higher serum exosomal miR-224 expression had lower overall survival.
Exosomal miR-224 is a tumor promotor and can be a marker of diagnosis and prognosis of HCC patients, however, its ability to distinguish liver diseases needs further verification.
Core tip: We aimed to understand the mechanism of exosomal microRNA-224 (miR-224) in the development of hepatocellular carcinoma (HCC) and to evaluate its diagnostic and prognostic value for HCC patients. By directly targeting the 3'-untranslated region of glycine N-methyltransferase, exosomal miR-224 inhibits its expression to promote proliferation and invasion. In addition, serum exosomal miR-224 also has potential diagnostic and prognostic value for HCC.
- Citation: Cui Y, Xu HF, Liu MY, Xu YJ, He JC, Zhou Y, Cang SD. Mechanism of exosomal microRNA-224 in development of hepatocellular carcinoma and its diagnostic and prognostic value. World J Gastroenterol 2019; 25(15): 1890-1898
- URL: https://www.wjgnet.com/1007-9327/full/v25/i15/1890.htm
- DOI: https://dx.doi.org/10.3748/wjg.v25.i15.1890
Liver cancer is a malignant tumor with a high mortality rate in China[1]. Liver cancer has the fourth-highest incidence rate of all tumors, and the fatality rate ranks third[1]. A total of 25% of the cancer cases are caused by carcinogenic infections such as hepatitis virus and human papillomavirus, and this figure includes a high proportion of patients in low-income and middle-income countries[2]. Therefore, early detection, diagnosis, and treatment are of great significance for the prognosis of patients with liver cancer. An early clinical diagnosis can effectively improve the survival of patients with hepatocellular carcinoma (HCC)[3]. However, the diagnostic sensitivity and specificity of current noninvasive indicators such as serum alpha-fetoprotein (AFP) and imaging studies for HCC, especially early HCC, still need to be improved[4]. Therefore, new methods for diagnosing HCC need to be developed.
Exosomes are secreted and released by various types of cells and contain nanovesicles with various active factors[5]. Previous literature has reported that serum microRNAs (miRNAs) are mainly present in the exosomes formed from phospholipid membranes, thus avoiding degradation by RNase in circulation[6]. Exosomes have been shown to play an important role in tumorigenesis, development, metastasis, deterioration, and immune escape[7,8]. For example, Gu et al[9] found that exosomes secreted by mesenchymal stem cells can promote the growth of gastric cancer cells, which indicated that exosomes can promote tumorigenesis and cancer development. Yu et al[10] found that exosomes secreted by breast cancer cells can promote tumor angiogenesis, growth, and proliferation, proving that exosomes can promote tumor cell metastasis. Exosomes secreted by breast cancer cells can also promote the proliferation of surrounding normal breast cells and inhibit their apoptosis. In addition, exosomes may also promote complications in cancer patients; for example, patients with pancreatic cancer often have diabetes[11].
Exosomes contain proteins, lipids, and biological molecules such as DNA and RNA[12]. Nucleic acids in exosomes are a group of molecules that can act as biomarkers. Currently, there are many reports on exosomal miRNAs, which are ideal biomarkers for the early diagnosis of cancer[13-16]. However, there are few reports on the role of exosomal miRNAs in the diagnosis and prognosis of HCC. In this study, cell culture and transfection of exosomal miRNA, real-time quantitative PCR (RT-qPCR), luciferase reporter assay, and other methods were performed to find new biomarkers related to the development of HCC and to determine if the biomarkers can be used for the diagnosis and prognosis of HCC.
A total of 89 HCC and 50 normal serum samples were collected from 2014 to 2016, and the clinical information is shown in Table 1. The patients were not treated before the samples were collected, and the diagnosis of HCC was confirmed by pathological analysis of tumor tissue. The analysis of the samples was approved by the patients and the ethics committee.
| Variable | HCC (n = 89) | Normal controls (n = 50) | P-value |
| Average age (yr) | 59.03 (25-90) | 53.58 (25-76) | 0.10 |
| Sex | 0.62 | ||
| Male n (%) | 43 (48.31) | 22 (44.00) | |
| Female n (%) | 46 (51.69) | 28 (56.00) | |
| Cirrhosis | |||
| Yes n (%) | 58 (65.17) | ||
| No n (%) | 31 (34.83) | ||
| T classification | |||
| T1-T2 n (%) | 37 (41.57) | ||
| T3-T4 n (%) | 52 (58.43) | ||
| Tumor size | |||
| > 3 cm n (%) | 35 (39.33) | ||
| < 3 cm n (%) | 54 (60.67) |
The hepatocyte lines WRL68, HepG2, and SKHEP1 were selected and cultured in RPMI-1640 (Sigma) containing 10% fetal bovine serum (FBS; Gibco) at 37 °C in 5% CO2. An miRNA mimic or inhibitor was transfected into cells with preincubated exosomes or Lipofectamine 2000.
The cells were starved in serum-free medium overnight and then centrifuged for 3 min at 2000 rpm, followed by filtration. The exosomes in the cell culture medium and in patient serum were extracted using the Total Exosome Isolation Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. The resulting precipitate was observed under a transmission electron microscope as described previously[17].
Exosomal RNA in serum or cell culture medium was extracted with Trizol (Thermo Fisher Scientific). The expression of exosomal miRNA was detected on the Quant Studio 7 Flex RT PCR System (Applied Biosystems) using a hydrolysis probe according to the manufacturer’s instructions. All experiments were performed in triplicate, and a mixture of let-7i, let-7g, and let-7d (let-7d/g/i) was used as endogenous controls to calculate the relative concentration of miRNA[18-20].
A Cell Counting Kit-8 (CCK8) assay (Dojindo, Japan) followed by measuring the spectrophotometric absorbance at 450 nm was used to estimate cell proliferation. All experiments were performed in triplicate, and data are presented as the mean. A total of 2 × 105 cells were cultured for 48 h in serum-free medium, while the lower chamber was filled with medium containing 10% FBS to analyze the invasion abilities of the cells; the cancer cells in the lower chamber were ultimately counted[21].
MicroRNA-224 (miR-224) mimic, glycine N-methyltransferase (GNMT) wild- or 3’-untranslated region (UTR) mutant-type, and controls were cotransfected into SKHEP1 cells in 24-well plates for 24 h. The harvested cells were analyzed for fluorescence intensity using a dual luciferase reporter assay kit as indicated.
Differences between groups were analyzed by t-tests using GraphPad Prism 6.0. The receiver operating characteristic (ROC) curve was constructed using SPSS 22.0 software, and the area under the curve (AUC) was calculated to assess the specificity and sensitivity of the prediction of HCC cases and controls. A P-value < 0.05 was considered significant. The Kaplan-Meier survival curve was used to analyze the survival of the patients.
The morphological characteristics of the exosomes in the cell culture medium were observed under a transmission electron microscope. The exosomes showed a vesicular structure with a diameter of approximately 50-150 nm, which is consistent with the literature[22].
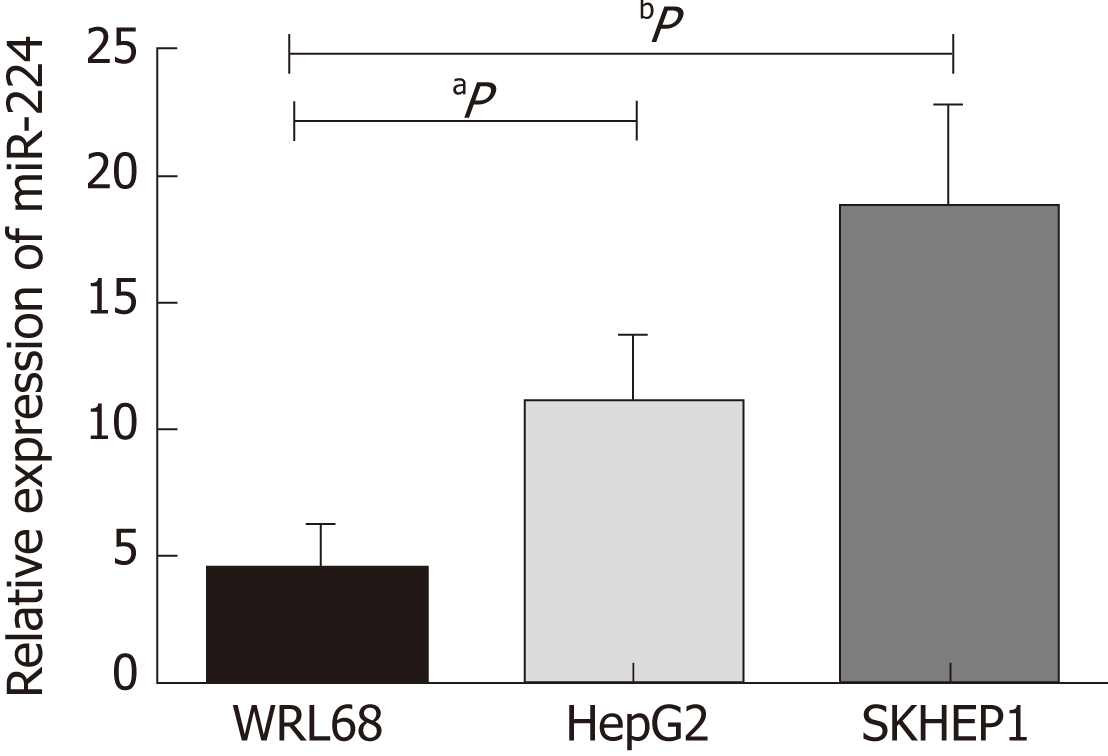
Based on previous experimental results, we found that the expression level of miR-224 in the HCC tissues was significantly higher than that in the normal controls. In this study, we verified the expression of miR-224 in the exosomes of WRL68, HepG2, and SKHEP1 cell lines by RT-qPCR. The expression level of exosomal miR-224 was significantly increased in the two liver cancer cell lines, HepG2 and SKHEP1, compared to that of the normal hepatic cell line, WRL68 (Figure 1).
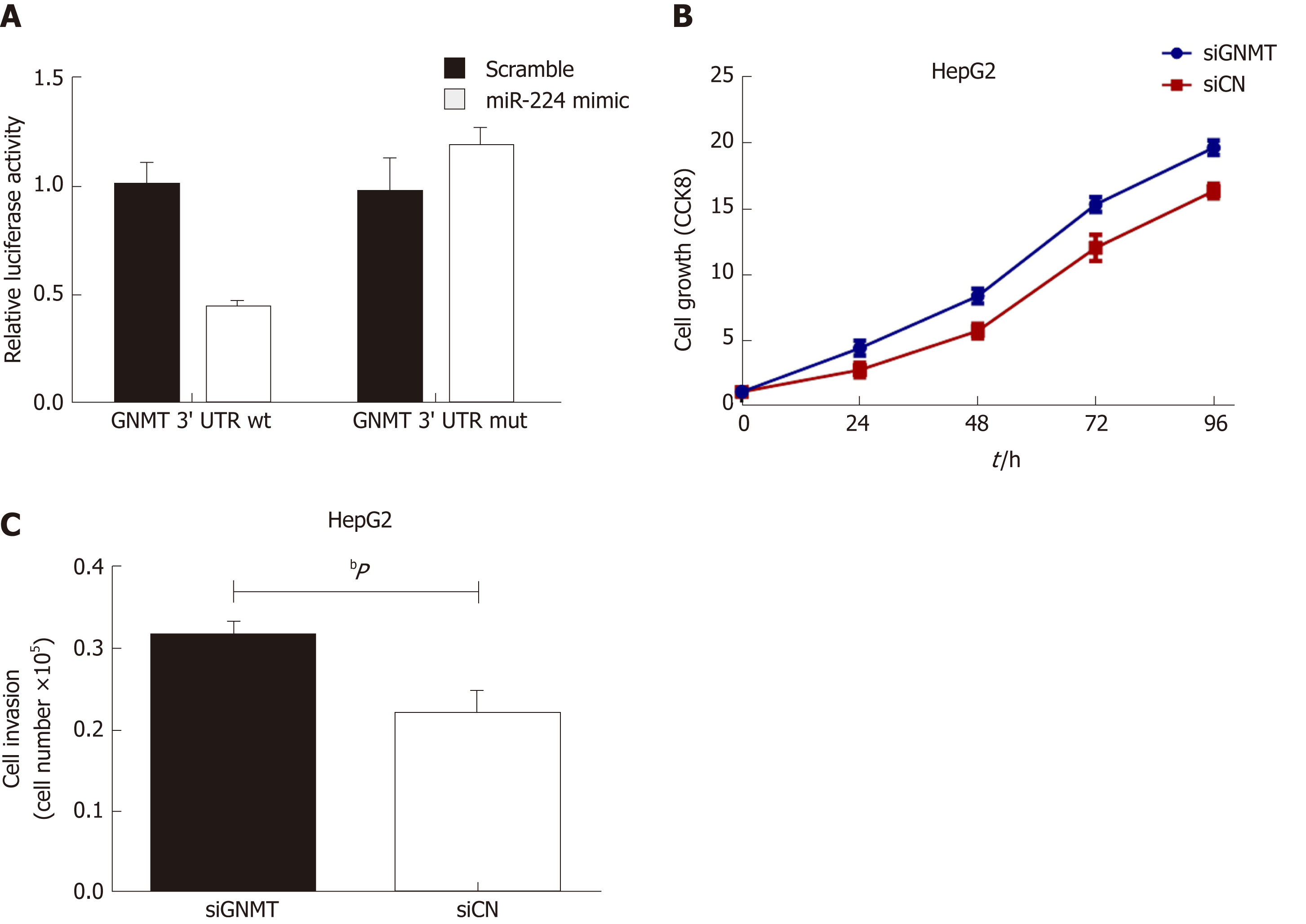
Exosomes incubated with miR-224 mimic or inhibitor were added to HepG2 and SKHEP1 cells to measure cell proliferation. The results showed that exosomes incubated with the miR-224 mimic resulted in a significant increase in cell proliferation compared to the proliferation in the control group, while the exosomes incubated with the miR-224 inhibitor exhibited significantly reduced cell proliferation (Figure 2A and B). These results indicated that exosomal miR-224 can promote the proliferation of liver cancer cells. The same results were obtained for the cell invasion assay (Figure 2C and D). Exosomes incubated with the miR-224 mimic resulted in more cells passing through the insert membranes to the lower chamber, indicating that the exosomal miR-224 can also promote liver cancer cell invasion.
It has been reported that miR-224 can affect cancer development by targeting glycine N-methyltransferase (GNMT)[23], so we used a luciferase reporter assay to verify whether miR-224 can directly interact with GNMT. As shown in Figure 3A, the wild-type GNMT reporter gene combined with the miR-224 mimic exhibited lower luciferase activity in the HepG2 cell line than that of the control group. However, when the 3'-UTR of the GNMT gene was mutated, this reduction could be eliminated. The siGNMT was added to HepG2 cells to knock out GNMT mRNA, which can reduce the expression of GNMT. The results showed that the proliferation and invasion of cells increased notably (Figure 3B and C). It is suggested that miR-224 may directly target GNMT to promote the proliferation and invasion of liver cancer cells.
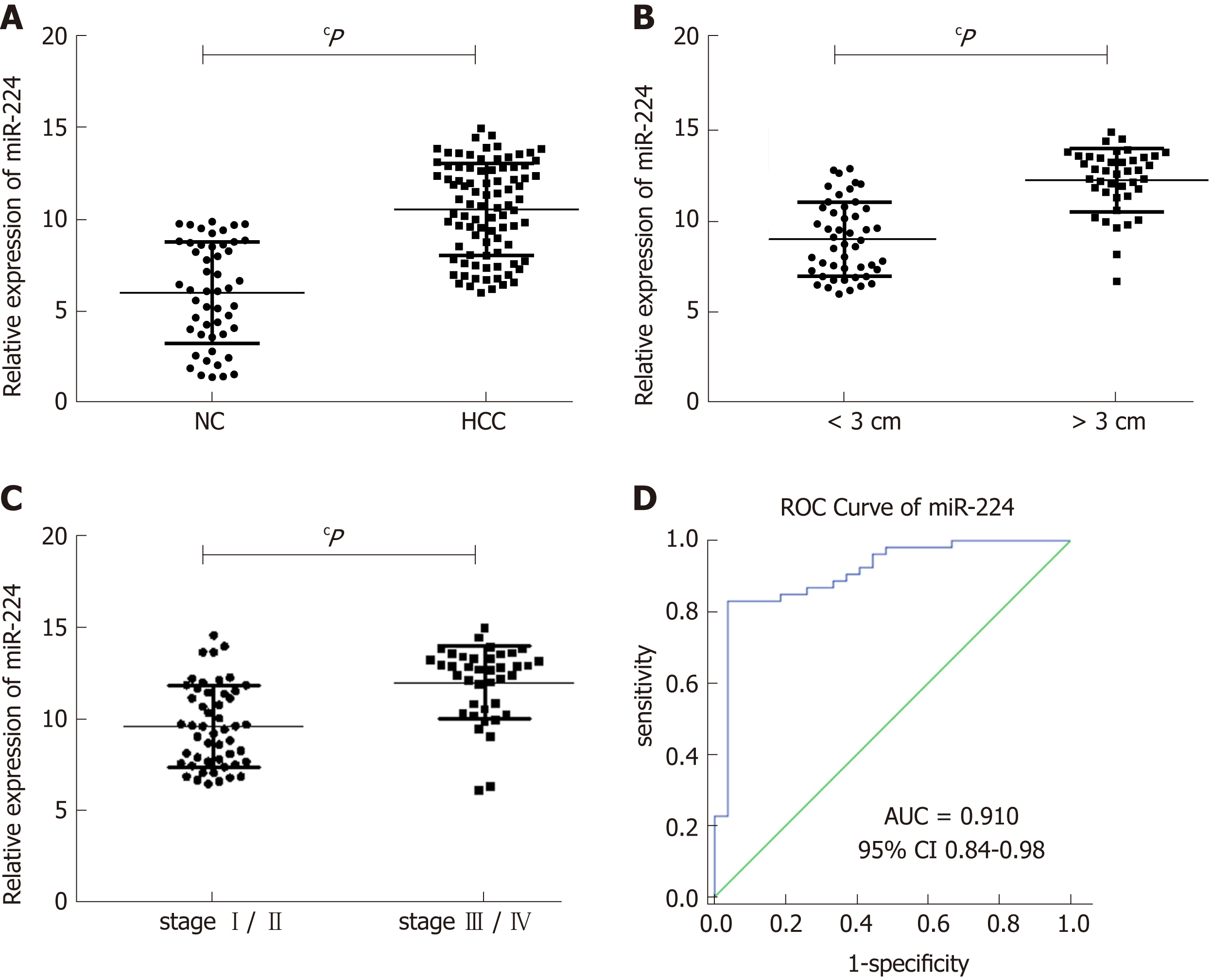
Using RT-qPCR, we determined that the expression levels of exosomal miR-224 in the 89 HCC samples were significantly higher than those in the 50 healthy controls (Figure 4A). In addition, according to the clinical characteristics of the sample group, the levels of exosomal miR-224 in the serum of patients with large tumors or late-stage tumors were significantly higher (Figure 4B and C). In the ROC curve analysis, serum exosomal miR-224 showed an ability to differentiate HCC patients from healthy controls, with an AUC of 0.910 (Figure 4D). These results indicated that serum exosomal miR-224 can be used as a potential biomarker for the diagnosis of HCC.
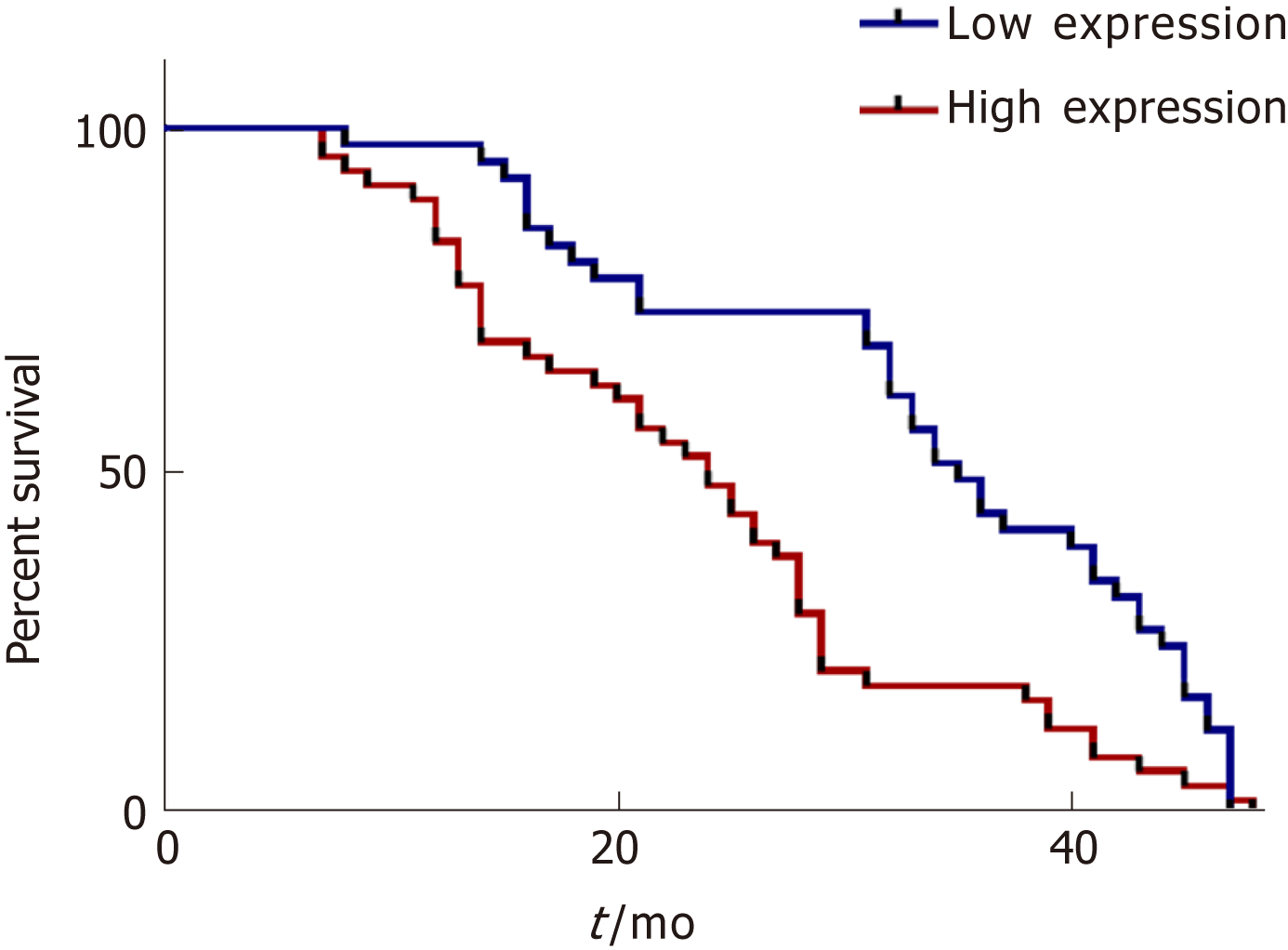
In addition, we analyzed the overall survival of the followed patients. Kaplan-Meier survival curves showed that HCC patients with higher serum exosomal miR-224 expression had lower overall survival (Figure 5), suggesting that serum exosomal miR-224 can be an independent prognostic factor for HCC.
In this study, we identified that the expression level of exosomal miR-224 in the liver cancer cell lines HepG2 and SKHEP1 was significantly higher than that in the healthy liver cell line WRL68 using gene chip and RT-qPCR. We also determined that the high expression of exosomal miR-224 can promote the proliferation and invasion of liver cancer cells. In addition, lower luciferase activity was observed when the miR-224 mimic and the GNMT wild type were cotransfected into the HepG2 cell line, while this phenomenon did not occur when the GNMT 3’-UTR was mutated, indicating that miR-224 can directly target the 3'-UTR of GNMT mRNA. In addition, siGNMT was added to the HepG2 cell line, and as a result, the proliferation and invasion of the cancer cells increased. Therefore, miR-224 can directly target GNMT to promote the proliferation and invasion of cancer cells.
We verified that miR-224 can directly target GNMT to increase the proliferation and invasion of liver cancer cells. In colorectal cancer, miR-224 targets caspase-3 and caspase-7, and this inverse relationship was evident from the earliest phases of transformation in the intestinal mucosa[24]. In addition, miR-224-5p inhibited autophagy by targeting Smad4 in breast cancer cells, suggesting a novel regulatory network contributing to the metastasis of breast cancer[25]. Both miR-224 overexpression and PTX3 silencing promoted cell proliferation, migration, and invasion, whereas the aforementioned properties were reduced when miR-224 was inhibited, indicating that miR-224 inhibition may significantly prevent cervical carcinoma progression by targeting the PTX3 gene[26]. MiR-224 inversely regulated thioredoxin-interacting protein (TXNIP) by binding directly to its 3'-UTR, which resulted in the activation of hypoxia-inducible factor 1α (HIF-1α), while either TXNIP re-expression or HIF-1α depletion abolished the effects of miR-224 on the proliferation and migration of PDAC cells in vitro and in vivo, suggesting that TXNIP is a target of miR-224[27]. HIF-1α inhibits the NCR1/NKp46 pathway by upregulating miR-224, which affects the killing capability of natural killer cells in prostate cancer, thus inducing the immune escape of tumor cells[28]. In summary, miR-224 may play an important role in the development and occurrence of tumors. Furthermore, miR-224 has many target genes, suggesting that miR-224 may participate in different pathways for cancer regulation, which requires further exploration.
We collected preoperative serum from HCC patients and healthy controls and isolated exosomes with high and stable purity. The expression level of serum exosomal miR-224 in HCC patients was significantly higher than that in healthy controls, as determined by RT-qPCR. The ability of serum exosomal miR-224 as a biomarker to distinguish HCC patients from healthy controls was confirmed by the ROC curve analysis. In addition, the relationship between tumor size and stage and miR-224 expression in HCC patients was analyzed, and we found that the expression of serum exosomal miR-224 was higher in patients with larger tumors and later stages. In addition, we also analyzed the relationship between the expression of exosomal miR-224 and the overall survival of patients. The Kaplan-Meier survival curve showed that the higher the expression level of serum exosomal miR-224, the shorter the patient's overall survival, suggesting that serum exosomal miR-224 can be used as a prognostic factor in patients with HCC. However, in this study we only compared the expression of exosomal miR-224 in the serum of HCC patients and healthy controls. It is not clear whether the expression of exosomal miR-224 in cirrhosis, hepatitis, or other liver cancer type is different, so the ability of miR-224 to distinguish between HCC and other liver diseases needs further verification.
In conclusion, exosomal miR-224 can decrease the expression of GNMT by directly targeting the 3'-UTR of GNMT mRNA to promote the proliferation and invasion of HCC cells, which may provide a new method for HCC treatment. In addition, serum exosomal miR-224 may be used as a biomarker for the diagnosis of HCC and a prognostic factor for patients with HCC.
Hepatocellular carcinoma (HCC) is a malignant tumor with a high mortality rate. Exosomes have been shown to play an important role in tumorigenesis, cancer development, metastasis, deterioration, and immune escape.
We aimed to research the mechanism of exosomal microRNA-224 (miR-224) and its target in the development and invasion of HCC, and we also evaluated the diagnostic and prognostic value of miR-224 for patients with HCC.
Cell culture and transfection of exosomal miR-224, its mimics, and its inhibitor; real-time quantitative PCR; and luciferase reporter assay were used to explore the mechanism of exosomal miR-224. HCC patients and healthy controls were used to assess the value of exosomal miR-224 in diagnosing HCC and predicting HCC prognosis.
Serum exosomes incubated with the miR-224 mimic showed a significant increase in cell proliferation and invasion when compared to the control group, while those incubated with the inhibitor showed a significant reduction. For discriminating HCC from healthy controls, serum exosomal miR-224 showed an area under the ROC curve of 0.910. Higher serum exosomal miR-224 expression levels in HCC patients were associated with lower overall survival.
The results showed that exosomal miR-224 directly targets the 3'-UTR of glycine N-methyltransferaseto and impacts the proliferation and invasion of HCC, and exosomal miR-224 may be used as a potential diagnostic and prognostic factor for patients with HCC.
Our study provides novel insight into the mechanism of exosomal miR-224 in the development and invasion of HCC and may provide a potential biomarker for the diagnosis and prognosis of HCC.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Gordon LG, Jin M, Shimizu Y S-Editor: Ma RY L-Editor: Wang TQ E-Editor: Ma YJ
| 1. | Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115-132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11444] [Cited by in RCA: 13210] [Article Influence: 1467.8] [Reference Citation Analysis (3)] |
| 2. | Plummer M, de Martel C, Vignat J, Ferlay J, Bray F, Franceschi S. Global burden of cancers attributable to infections in 2012: a synthetic analysis. Lancet Glob Health. 2016;4:e609-e616. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1068] [Cited by in RCA: 1018] [Article Influence: 113.1] [Reference Citation Analysis (0)] |
| 3. | Santi V, Trevisani F, Gramenzi A, Grignaschi A, Mirici-Cappa F, Del Poggio P, Di Nolfo MA, Benvegnù L, Farinati F, Zoli M, Giannini EG, Borzio F, Caturelli E, Chiaramonte M, Bernardi M; Italian Liver Cancer (ITA. LI.CA) Group. Semiannual surveillance is superior to annual surveillance for the detection of early hepatocellular carcinoma and patient survival. J Hepatol. 2010;53:291-297. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 210] [Cited by in RCA: 237] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 4. | Sun W, Liu Y, Shou D, Sun Q, Shi J, Chen L, Liang T, Gong W. AFP (alpha fetoprotein): who are you in gastrology? Cancer Lett. 2015;357:43-46. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 38] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 5. | Huang Z, Huang D, Ni S, Peng Z, Sheng W, Du X. Plasma microRNAs are promising novel biomarkers for early detection of colorectal cancer. Int J Cancer. 2010;127:118-126. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 750] [Cited by in RCA: 750] [Article Influence: 50.0] [Reference Citation Analysis (0)] |
| 6. | Lima LG, Chammas R, Monteiro RQ, Moreira ME, Barcinski MA. Tumor-derived microvesicles modulate the establishment of metastatic melanoma in a phosphatidylserine-dependent manner. Cancer Lett. 2009;283:168-175. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 169] [Cited by in RCA: 201] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 7. | Hood JL, San RS, Wickline SA. Exosomes released by melanoma cells prepare sentinel lymph nodes for tumor metastasis. Cancer Res. 2011;71:3792-3801. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 719] [Cited by in RCA: 785] [Article Influence: 56.1] [Reference Citation Analysis (0)] |
| 8. | Wolfers J, Lozier A, Raposo G, Regnault A, Théry C, Masurier C, Flament C, Pouzieux S, Faure F, Tursz T, Angevin E, Amigorena S, Zitvogel L. Tumor-derived exosomes are a source of shared tumor rejection antigens for CTL cross-priming. Nat Med. 2001;7:297-303. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1117] [Cited by in RCA: 1263] [Article Influence: 52.6] [Reference Citation Analysis (0)] |
| 9. | Gu H, Ji R, Zhang X, Wang M, Zhu W, Qian H, Chen Y, Jiang P, Xu W. Exosomes derived from human mesenchymal stem cells promote gastric cancer cell growth and migration via the activation of the Akt pathway. Mol Med Rep. 2016;14:3452-3458. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 85] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 10. | Yu DD, Wu Y, Shen HY, Lv MM, Chen WX, Zhang XH, Zhong SL, Tang JH, Zhao JH. Exosomes in development, metastasis and drug resistance of breast cancer. Cancer Sci. 2015;106:959-964. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 122] [Cited by in RCA: 165] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 11. | Javeed N, Sagar G, Dutta SK, Smyrk TC, Lau JS, Bhattacharya S, Truty M, Petersen GM, Kaufman RJ, Chari ST, Mukhopadhyay D. Pancreatic Cancer-Derived Exosomes Cause Paraneoplastic β-cell Dysfunction. Clin Cancer Res. 2015;21:1722-1733. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 136] [Cited by in RCA: 144] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 12. | Kosaka N, Yoshioka Y, Fujita Y, Ochiya T. Versatile roles of extracellular vesicles in cancer. J Clin Invest. 2016;126:1163-1172. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 206] [Cited by in RCA: 269] [Article Influence: 29.9] [Reference Citation Analysis (0)] |
| 13. | Joyce DP, Kerin MJ, Dwyer RM. Exosome-encapsulated microRNAs as circulating biomarkers for breast cancer. Int J Cancer. 2016;139:1443-1448. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117] [Cited by in RCA: 147] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 14. | Tanaka Y, Kamohara H, Kinoshita K, Kurashige J, Ishimoto T, Iwatsuki M, Watanabe M, Baba H. Clinical impact of serum exosomal microRNA-21 as a clinical biomarker in human esophageal squamous cell carcinoma. Cancer. 2013;119:1159-1167. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 303] [Cited by in RCA: 359] [Article Influence: 27.6] [Reference Citation Analysis (0)] |
| 15. | Thind A, Wilson C. Exosomal miRNAs as cancer biomarkers and therapeutic targets. J Extracell Vesicles. 2016;5:31292. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 294] [Cited by in RCA: 305] [Article Influence: 33.9] [Reference Citation Analysis (0)] |
| 16. | Fang T, Lv H, Lv G, Li T, Wang C, Han Q, Yu L, Su B, Guo L, Huang S, Cao D, Tang L, Tang S, Wu M, Yang W, Wang H. Tumor-derived exosomal miR-1247-3p induces cancer-associated fibroblast activation to foster lung metastasis of liver cancer. Nat Commun. 2018;9:191. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 407] [Cited by in RCA: 748] [Article Influence: 106.9] [Reference Citation Analysis (0)] |
| 17. | Hu R, Yan H, Fei X, Liu H, Wu J. Modulation of glucose metabolism by a natural compound from Chloranthus japonicus via activation of AMP-activated protein kinase. Sci Rep. 2017;7:778. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 25] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 18. | Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402-408. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149116] [Cited by in RCA: 133837] [Article Influence: 5576.5] [Reference Citation Analysis (1)] |
| 19. | Chen X, Liang H, Guan D, Wang C, Hu X, Cui L, Chen S, Zhang C, Zhang J, Zen K, Zhang CY. A combination of Let-7d, Let-7g and Let-7i serves as a stable reference for normalization of serum microRNAs. PLoS One. 2013;8:e79652. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 76] [Cited by in RCA: 89] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 20. | Wu C, Wang C, Guan X, Liu Y, Li D, Zhou X, Zhang Y, Chen X, Wang J, Zen K, Zhang CY, Zhang C. Diagnostic and prognostic implications of a serum miRNA panel in oesophageal squamous cell carcinoma. PLoS One. 2014;9:e92292. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 78] [Cited by in RCA: 89] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 21. | Hu R, Huffman KE, Chu M, Zhang Y, Minna JD, Yu Y. Quantitative Secretomic Analysis Identifies Extracellular Protein Factors That Modulate the Metastatic Phenotype of Non-Small Cell Lung Cancer. J Proteome Res. 2016;15:477-486. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 45] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 22. | Yuan R, Zhi Q, Zhao H, Han Y, Gao L, Wang B, Kou Z, Guo Z, He S, Xue X, Hu H. Upregulated expression of miR-106a by DNA hypomethylation plays an oncogenic role in hepatocellular carcinoma. Tumour Biol. 2015;36:3093-3100. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 26] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 23. | Hung JH, Li CH, Yeh CH, Huang PC, Fang CC, Chen YF, Lee KJ, Chou CH, Cheng HY, Huang HD, Chen M, Tsai TF, Lin AM, Yen CH, Tsou AP, Tyan YC, Chen YA. MicroRNA-224 down-regulates Glycine N-methyltransferase gene expression in Hepatocellular Carcinoma. Sci Rep. 2018;8:12284. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 17] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 24. | Fassan M, Cui R, Gasparini P, Mescoli C, Guzzardo V, Vicentini C, Munari G, Loupakis F, Lonardi S, Braconi C, Scarpa M, D'Angelo E, Pucciarelli S, Angriman I, Agostini M, D'Incá R, Farinati F, Gafà R, Lanza G, Frankel WL, Croce CM, Valeri N, Rugge M. miR-224 Is Significantly Upregulated and Targets Caspase-3 and Caspase-7 During Colorectal Carcinogenesis. Transl Oncol. 2019;12:282-291. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 13] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 25. | Cheng Y, Li Z, Xie J, Wang P, Zhu J, Li Y, Wang Y. MiRNA-224-5p inhibits autophagy in breast cancer cells via targeting Smad4. Biochem Biophys Res Commun. 2018;506:793-798. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 42] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 26. | Yu LM, Wang WW, Qi R, Leng TG, Zhang XL. MicroRNA-224 inhibition prevents progression of cervical carcinoma by targeting PTX3. J Cell Biochem. 2018;119:10278-10290. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 14] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 27. | Zhu G, Zhou L, Liu H, Shan Y, Zhang X. MicroRNA-224 Promotes Pancreatic Cancer Cell Proliferation and Migration by Targeting the TXNIP-Mediated HIF1α Pathway. Cell Physiol Biochem. 2018;48:1735-1746. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 58] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 28. | Chen CH, Li SX, Xiang LX, Mu HQ, Wang SB, Yu KY. HIF-1α induces immune escape of prostate cancer by regulating NCR1/NKp46 signaling through miR-224. Biochem Biophys Res Commun. 2018;503:228-234. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 16] [Article Influence: 2.3] [Reference Citation Analysis (0)] |