Published online Apr 14, 2019. doi: 10.3748/wjg.v25.i14.1715
Peer-review started: January 7, 2019
First decision: February 13, 2019
Revised: March 6, 2019
Accepted: March 15, 2019
Article in press: March 16, 2019
Published online: April 14, 2019
Processing time: 96 Days and 23.7 Hours
Cellular senescence is a recognized barrier for progression of chronic liver diseases to hepatocellular carcinoma (HCC). The expression of a cluster of genes is altered in response to environmental factors during senescence. However, it is questionable whether these genes could serve as biomarkers for HCC patients.
To develop a signature of senescence-associated genes (SAGs) that predicts patients’ overall survival (OS) to improve prognosis prediction of HCC.
SAGs were identified using two senescent cell models. Univariate COX regression analysis was performed to screen the candidate genes significantly associated with OS of HCC in a discovery cohort (GSE14520) for the least absolute shrinkage and selection operator modelling. Prognostic value of this seven-gene signature was evaluated using two independent cohorts retrieved from the GEO (GSE14520) and the Cancer Genome Atlas datasets, respectively. Time-dependent receiver operating characteristic (ROC) curve analysis was conducted to compare the predictive accuracy of the seven-SAG signature and serum α-fetoprotein (AFP).
A total of 42 SAGs were screened and seven of them, including KIF18B, CEP55, CIT, MCM7, CDC45, EZH2, and MCM5, were used to construct a prognostic formula. All seven genes were significantly downregulated in senescent cells and upregulated in HCC tissues. Survival analysis indicated that our seven-SAG signature was strongly associated with OS, especially in Asian populations, both in discovery and validation cohorts. Moreover, time-dependent ROC curve analysis suggested the seven-gene signature had a better predictive accuracy than serum AFP in predicting HCC patients’ 1-, 3-, and 5-year OS.
We developed a seven-SAG signature, which could predict OS of Asian HCC patients. This risk model provides new clinical evidence for the accurate diagnosis and targeted treatment of HCC.
Core tip: In the present study, we identified a total of 42 senescence-associated genes (SAGs) and found seven of them were significantly downregulated in senescent cells and upregulated in HCC tissues. By using the least absolute shrinkage and selection operator, we constructed a seven-SAG signature that could predict the overall survival (OS) of hepatocellular carcinoma. This seven-SAG signature was further validated and developed in another independent dataset from The Cancer Genome Atlas project. Moreover, our risk score system showed better utility in predicting the OS than classic serum biomarker α-fetoprotein.
- Citation: Xiang XH, Yang L, Zhang X, Ma XH, Miao RC, Gu JX, Fu YN, Yao Q, Zhang JY, Liu C, Lin T, Qu K. Seven-senescence-associated gene signature predicts overall survival for Asian patients with hepatocellular carcinoma. World J Gastroenterol 2019; 25(14): 1715-1728
- URL: https://www.wjgnet.com/1007-9327/full/v25/i14/1715.htm
- DOI: https://dx.doi.org/10.3748/wjg.v25.i14.1715
Hepatocellular carcinoma (HCC) becomes the third leading cause of cancer deaths worldwide, with approximately 80% of mortalities occurring within 5 years[1,2]. During the past decades, great effects have been made to improve the management of HCC. As more and more HCCs are diagnosed at an early stage, treatment efficacy is greatly improved[3]. Whereas, deaths caused by HCC always occur when patients undergo treatment and the clinical outcome of HCC patients is still poor[4]. Moreover, HCC is an extremely heterogeneous disease, which must be monitored for high-risk patients with poor clinical outcomes and adopt effective treatments to improve patient survival[5]. Traditional serum markers, especially alpha-fetoprotein (AFP), have been the most common prognostic indicators in clinic. However, they significantly depend on tumor burden, which limits their value in diagnosing early stage tumors. Therefore, identifying novel prognostic biomarkers contributes to early diagnosis and reducing HCC mortality.
Cellular senescence is considered to be a response of a proliferating somatic cell to stress and damage derived from both exogenous and endogenous sources, and persistent DNA damage is the most common cause[6]. It is characterized as a permanent cell cycle arrest[7]. Cellular senescence has been deemed as a mechanism of limited cell division due to progressive telomere shortening[8]. In cancer cells, there exists telomere-independent senescence due to their activation of telomerase. For instance, numerous studies found that RAS activation could induce cellular senescence in many cancer cell types. Both telomere-dependent and -independent pathways induced senescence by inducing a DNA damage response. Recently, it has been demonstrated that hepatocytes escaping from senescence played a key role in hepatic carcinogenesis and HCC progression[9,10]. During the senescence process, a large number of genes are expressed differentially. Hence, it is reliable to focus on senescence associated genes (SAGs) as a novel approach in cancer diagnosis and monitor.
Bearing this in mind, we first identified the SAGs by analyzing the genome profiling data derived from two types of senescent cells, replicative senescence (RS) and oncogene-induced senescence (OIS). Next, we investigated the prognostic value of several candidate SAGs in predicting survival of HCC by multistep comparisons. Finally, we validated that the candidate SAGs were superior to classic serum biomarker AFP in predicting the OS in HCC cohort retrieved from the Cancer Genome Atlas (TCGA). As far as we know, the study is the first to investigate the expression patterns of SAGs between senescence and HCC and their association with clinical prognosis of Asian patients with HCC.
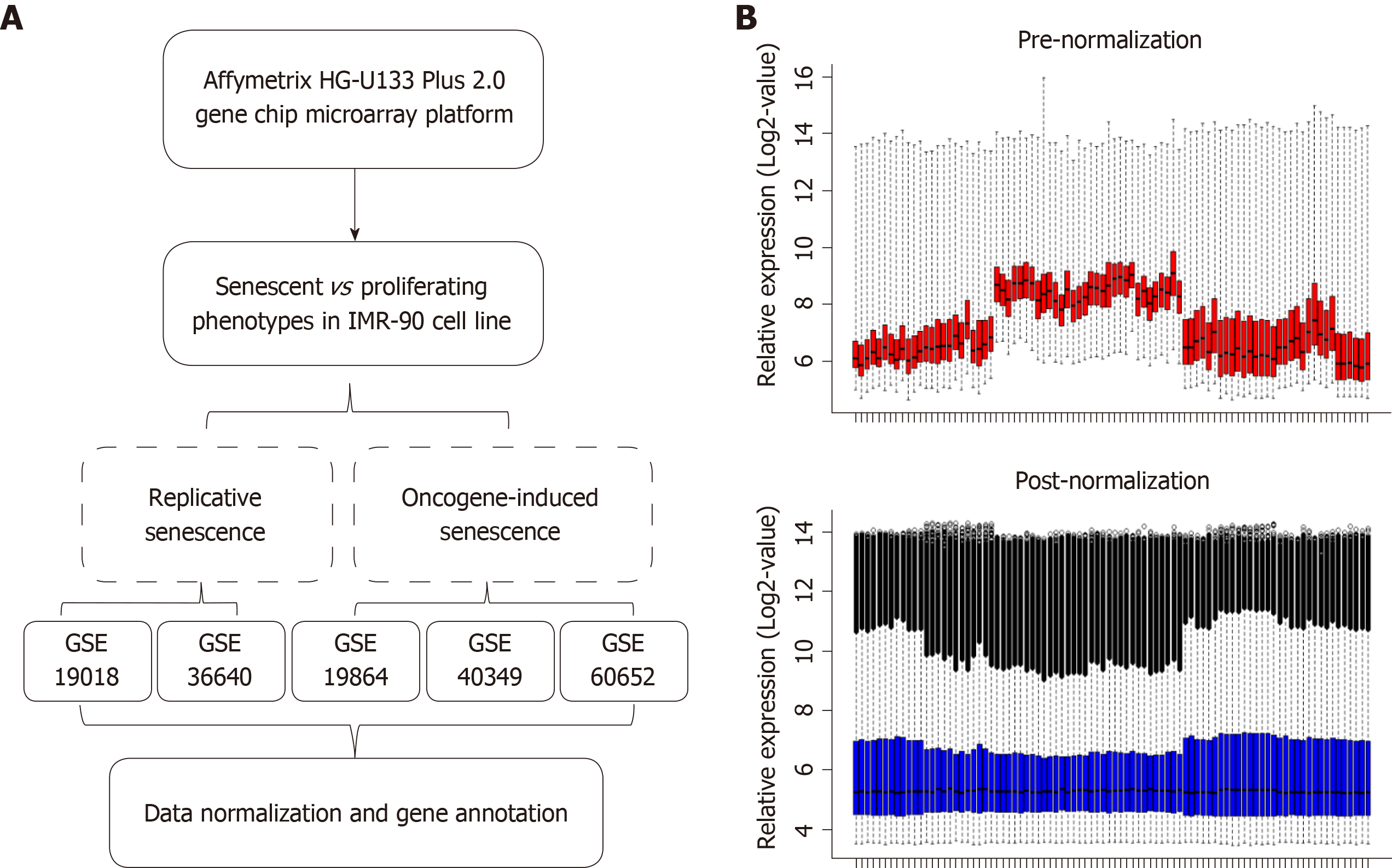
Two genomic profiling datasets of RS cells (GSE19018 and GSE36640), three datasets of OIS cells (GSE19864, GSE40349, and GSE60652), and one HCC dataset (GSE14520) were obtained from the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/). RMA algorithm was performed to normalize and transform all the selected data from GEO to expression values in the R environment (v3.5.1). Among them, GSE14520 (based on GPL3921 platform) enrolled 209 HCC patients with survival data. An independent HCC cohort derived from TCGA database (TCGA-LIHC) was used as a validation group. The level 3 RNAseqv2 data and clinical data were derived from the TCGA data portal. A total of 370 HCC samples were included.
Differentlly expressed genes (DEGs) were calculated in RS and OIS cell models, respectively. Only fold change (FC) ≥ 1.5 and q-value < 0.05 were considered statistically significant. Then, we picked up the overlapped significantly expressed genes in the two senescent models. Venn diagram was carried out using Venny 2.1.0. Moreover, the expression levels of candidate genes were plotted and analyzed by the t-test in GSE14520 and TCGA-LIHC cohorts.
A prognostic model was created by the seven-SAG signature according to least absolute shrinkage and selection operator (LASSO) analysis. LASSO is one of the most popular approaches for sparse linear regression[11]. “Almnet” package was carried out based on a series of λ in the R environment (v3.5.1)[12] and the coefficients of each gene in the risk score system were generated. We got a risk score for every patient based on their own expression levels of the seven genes after the LASSO regression analysis.
Univariate and multivariate survival analyses were carried out and further multivariate COX regression analysis only included variables with P < 0.05. All tests were carried out using SPSS (version 24.0; Chicago, United States). Kaplan-Meier curves were generated using GraphPad Prism 7.0. Comparisons between different subgroups were performed by the Log-Rank test. Patients are divided into high- and low- risk groups by the median.
ROC curve is extended to evaluate biomarker’s accuracy of discriminating binary outcomes[13]. Individuals with a high risk of developing the disease later may be disease-free in earlier life and their markers’ value may change from baseline during follow-up. Therefore, time-dependent ROC curve analysis is more appropriate and outperforms the conventional method adopted for handling censored biomarker data. In this study, the time-dependent ROC curve analysis was performed with “survival ROC” package (R version 3.5.1). The prognostic performance was evaluated at 1, 3, and 5 years to compare the predictive accuracy and sensitivity of different prognostic models.
The chi-square test was carried out to discover the relationship between gene expression and clinical parameters. Unpaired student’s t-test was used to analyze the difference of gene expression in HCC patients of different features. P < 0.05 was considered statistically different. Statistical analyses were performed using IBM SPSS Statistics software program version 24.0 (IBM Corp, NY, United States).
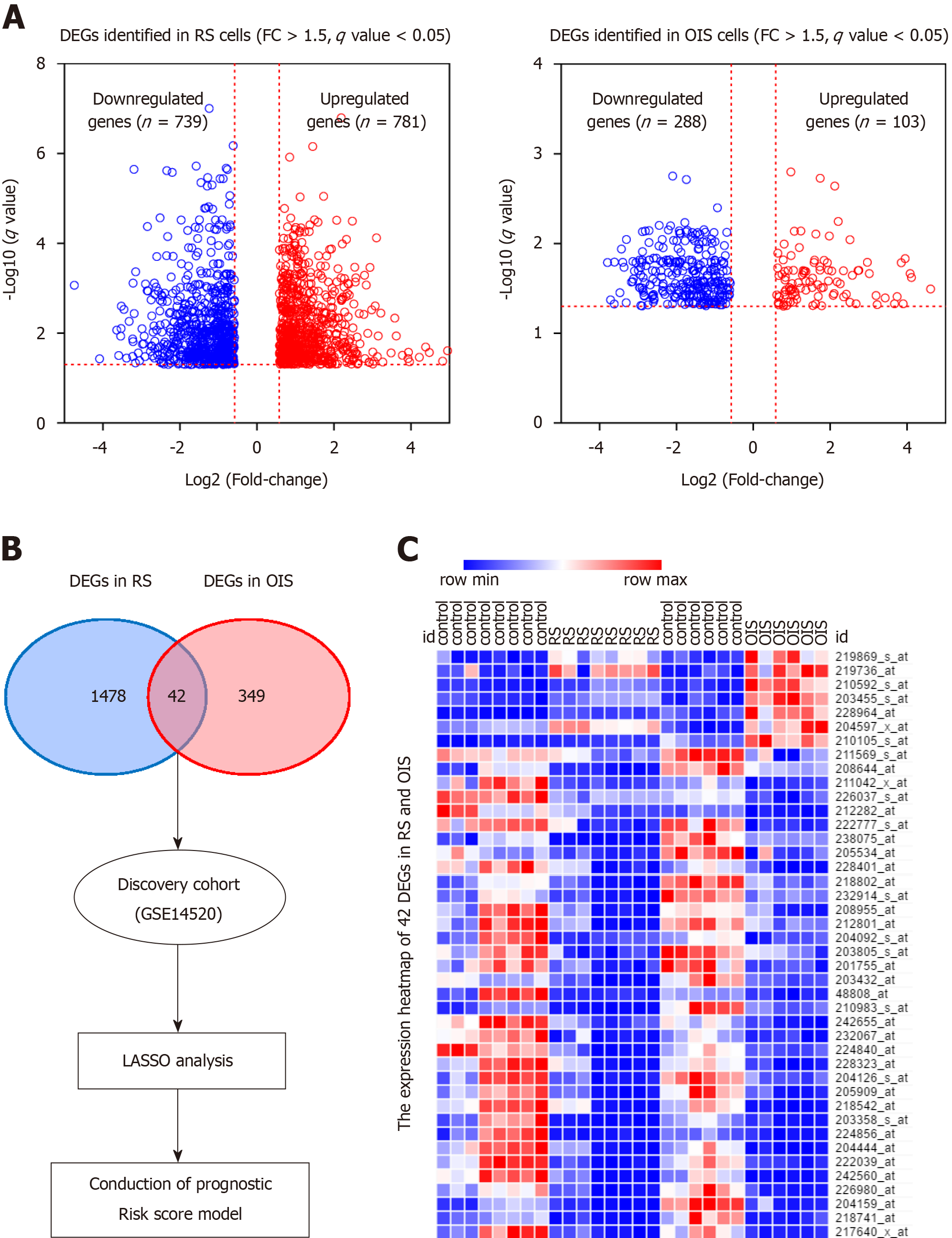
The overall workflow of the data processing is presented in Figure 1A. To identify SAGs, we first integrated five different microarray profiles (GSE19018, GSE36640, GSE19864, GSE40349, and GSE60652). All datasets used in the present study were normalized before analysis. The relative expression of all samples pre- and post-normalization is shown in Figure 1B. Next, we screened the DEGs, which were identified as FC ≥ 1.5 and q-value < 0.05 (senescent vs proliferating cells), in RS and OIS models, respectively. A total of 781 up-regulated and 739 down-regulated genes were selected in the RS model, and 103 up-regulated and 288 down-regulated genes in the OIS model (Figure 2A). By overlapping the two DEG lists, 42 common differentially expressed genes (35 downregulated and only 7 upregulated genes) were selected as SAGs (Figure 2B) and the expression levels of these genes in RS and OIS models are presented as a heat map in Figure 2C.
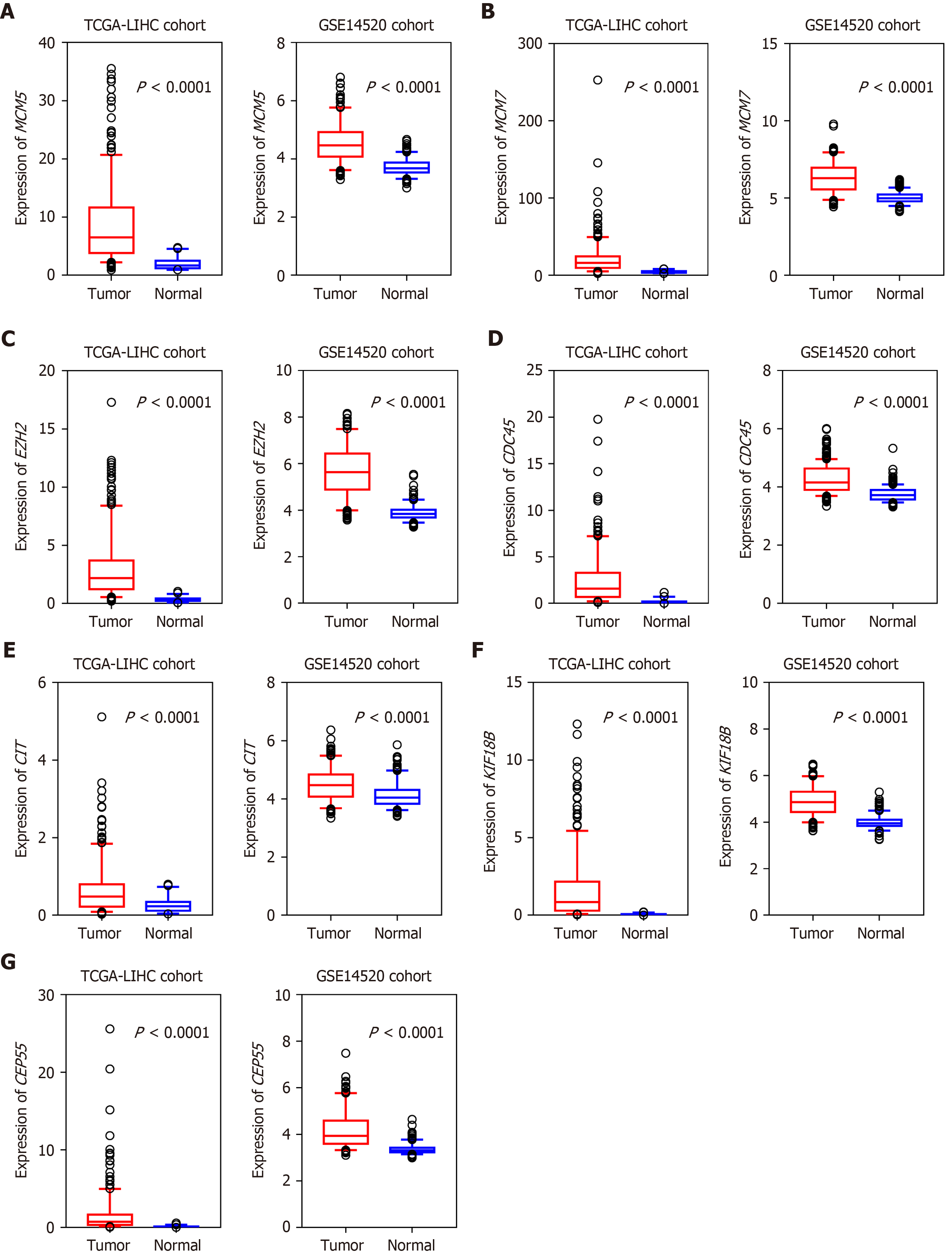
To obtain a prognostic SAG signature for HCC survival prediction, we first performed univariate COX regression analysis to evaluate the prognostic value of each candidate gene. And we found that all seven genes (CEP55, MCM7, CDC45, MCM5, KIF18B, CIT, and EZH2) were proved to be risk factors for HCC patients. Next, we screened the expression pattern of the above 7 candidate genes in HCC cohorts. Intriguingly, seven downregulated genes in senescent cells, were significantly upregulated in HCC tissues in both discovery and validation groups, with a P-value < 0.0001 (Figure 3). We then developed a risk score formula based on these seven SAGs using the LASSO method: Risk score = (0.243 × relative expression value of KIF18B) + (0.274 × relative expression value of CEP55) + (0.282 × relative expression value of CIT) + (0.266 × relative expression value of MCM7) + (0.678 × relative expression value of CDC45) + (0.175 × relative expression value of EZH2) + (0.536 × relative expression value of MCM5). In this risk score system, the contribution of every gene to the risk score model was weighted by absolute value of coefficients. Every patient would get a risk score based on the expression of the seven SAGs of themselves.
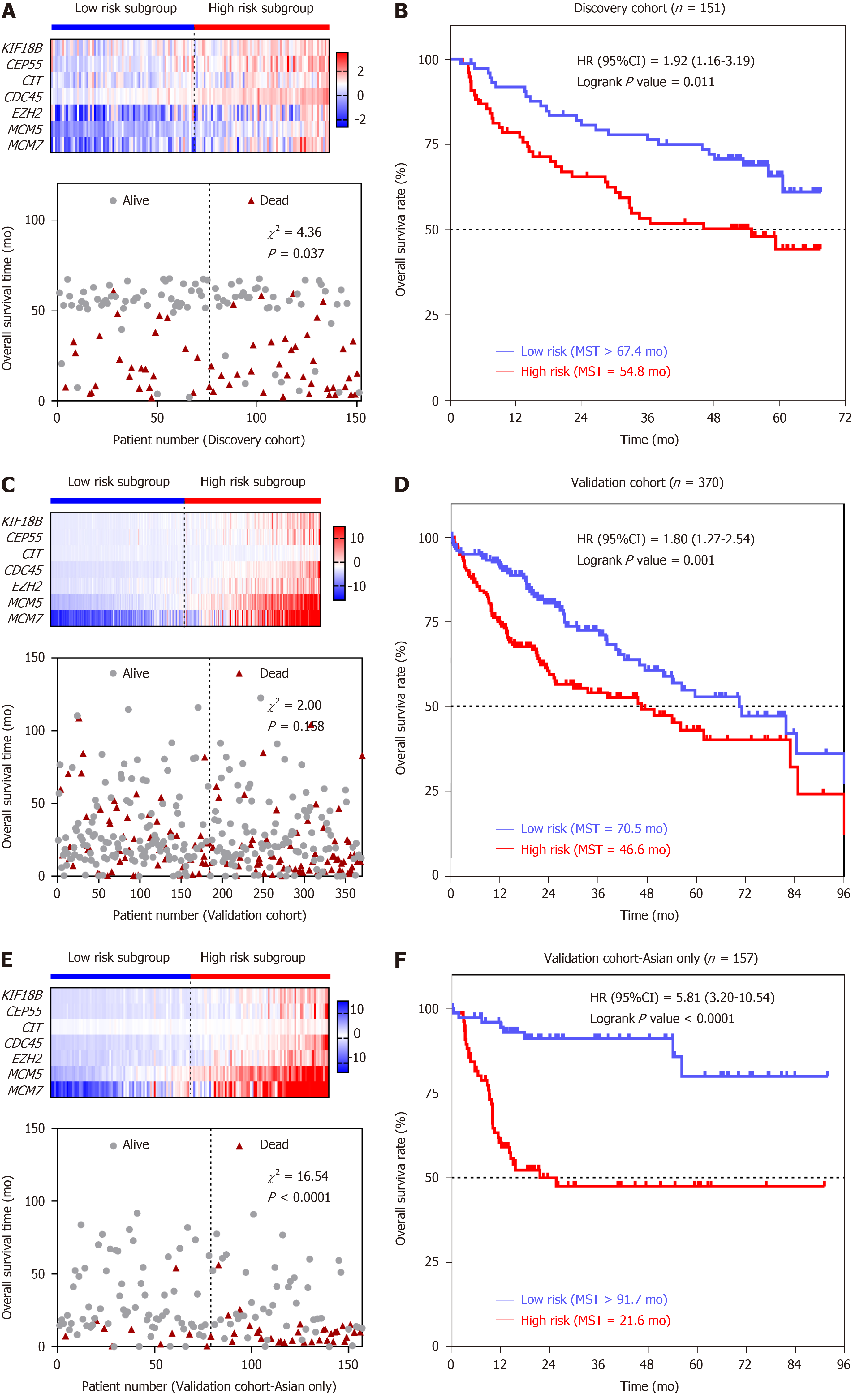
To confirm the potentiality of the seven-SAG prognostic model, Kaplan-Meier curve was carried out to evaluate the association between the overall survival (OS) and our gene signature in discovery (GSE14520) and validation (TCGA-LIHC) cohorts. The whole group was divided into the high- and low-risk subgroups according to the median of all patients’ risk scores. In the discovery cohort, with the increase in the risk score, the expression of all the seven genes was increasing, and the death events accumulated (Figure 4A). The patients in the high-risk subgroup had a 1.92-fold higher death risk than the low subgroup [hazard ratio (HR), 95% confidence interval (CI) = 1.92, 1.16-3.19; log-rank P value = 0.011] (Figure 4B). We then tempted to test these findings in the validation cohort (TCGA-LIHC) (Figure 4C). Similar to the findings obtained from the discovery cohort, patients in the high-risk group [median survival time (MST) = 46.6 m] had significantly shorter OS time than patients with a low-risk score (MST = 70.5 m) [HR (95%CI) = 1.80 (1.27-2.54), log-rank P value = 0.001] (Figure 4D). Interestingly, when we analyzed the data in the Asian population, we observed a highly significant association between the seven-SAG signature and OS. The majority of death events occurred in the high-risk group (Figure 4E). Asian HCC patients with a high-risk score were shown to have a > 5-fold increased death risk than low-risk patients [HR (95%CI) = 5.81 (3.20-10.54), log-rank P value < 0.0001]. The MST of the high-risk subgroup was only 60% of that of the low-risk group (MST = 21.6 m vs 91.7 m) (Figure 4F). In order to investigate the prognostic value of the risk score system in different patient groups with different characteristics, we performed univariate/multivariate Cox regression analysis of clinicopathologic factors associated with OS in the discovery and validation cohorts. From the Cox regression results, both the seven-SAG signature and serum AFP level were confirmed to be independent risk factors of OS in the two cohorts (Table 1).
| Variable | Discovery cohort | Validation cohort-Asian only | ||||||
| Univariate analysis | Multivariate analysis | Univariate analysis | Multivariate analysis | |||||
| HR (95%CI) | P-value | HR (95%CI) | P-value | HR (95%CI) | P-value | HR (95%CI) | P-value | |
| Gender (male/female) | 1.36 (0.55-3.40) | 0.507 | - | - | 0.89 (0.43-1.86) | 0.755 | - | - |
| Age (> 60/≤ 60 yr) | 1.06 (0.57-2.00) | 0.851 | - | - | 1.21 (0.66-2.24) | 0.541 | - | - |
| Cirrhosis (yes/no) | 3.09 (0.76-12.65) | 0.117 | - | - | 2.44 (1.56-3.85) | < 0.001a | 1.67 (1.04-2.70) | 0.034a |
| AFP (> 300/≤ 300 ng/mL) | 2.30 (1.37-3.86) | 0.002a | 2.26 (1.38-3.69) | 0.001a | 3.70 (2.13-6.25) | < 0.001a | 2.13 (1.30-3.57) | 0.003a |
| Risk score (high/low) | 1.93 (1.15-3.23) | 0.012a | 1.99 (1.19-3.34) | 0.009a | 5.91 (2.74-12.76) | < 0.001a | 4.22 (1.89-9.41) | < 0.001a |
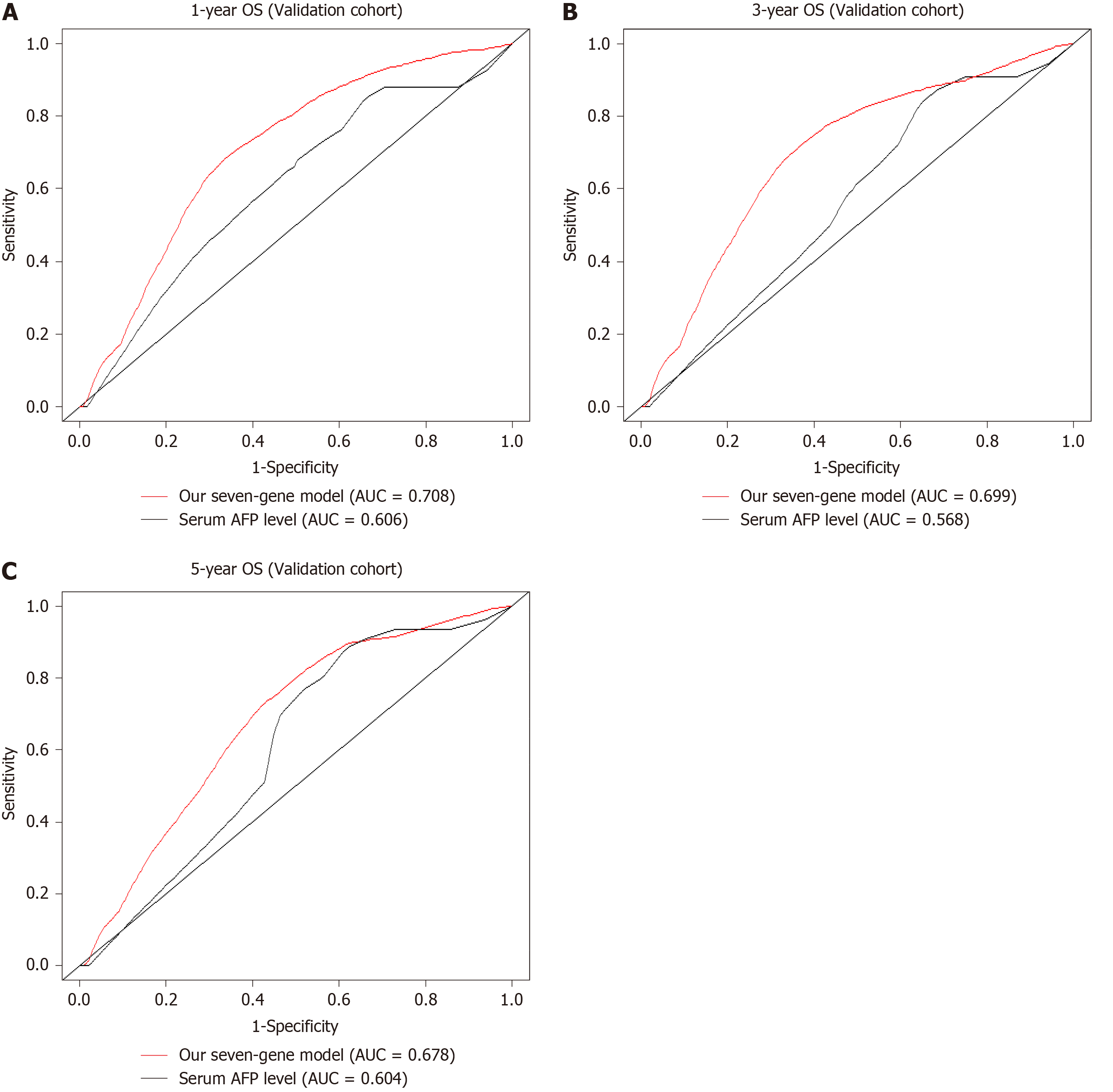
To assess the prognostic accuracy of the seven-SAG signature, we performed time-dependent ROC analysis of 1-, 3-, and 5-year OS of the validation cohort. The area under the curve (AUC) of the seven-SAG signature model indicated an acceptable predictive accuracy, which is superior to AFP, a widely used traditional serum marker, at 1 year (our seven-gene model AUC = 0.708, serum AFP level AUC = 0.606) (Figure 5A), 3 years (our seven-gene model AUC = 0.699, serum AFP level AUC = 0.568) (Figure 5B) as well as 5 years (our seven-gene model AUC = 0.678, serum AFP level AUC = 0.604) (Figure 5C). These results indicated the validation of the prognostic signature.
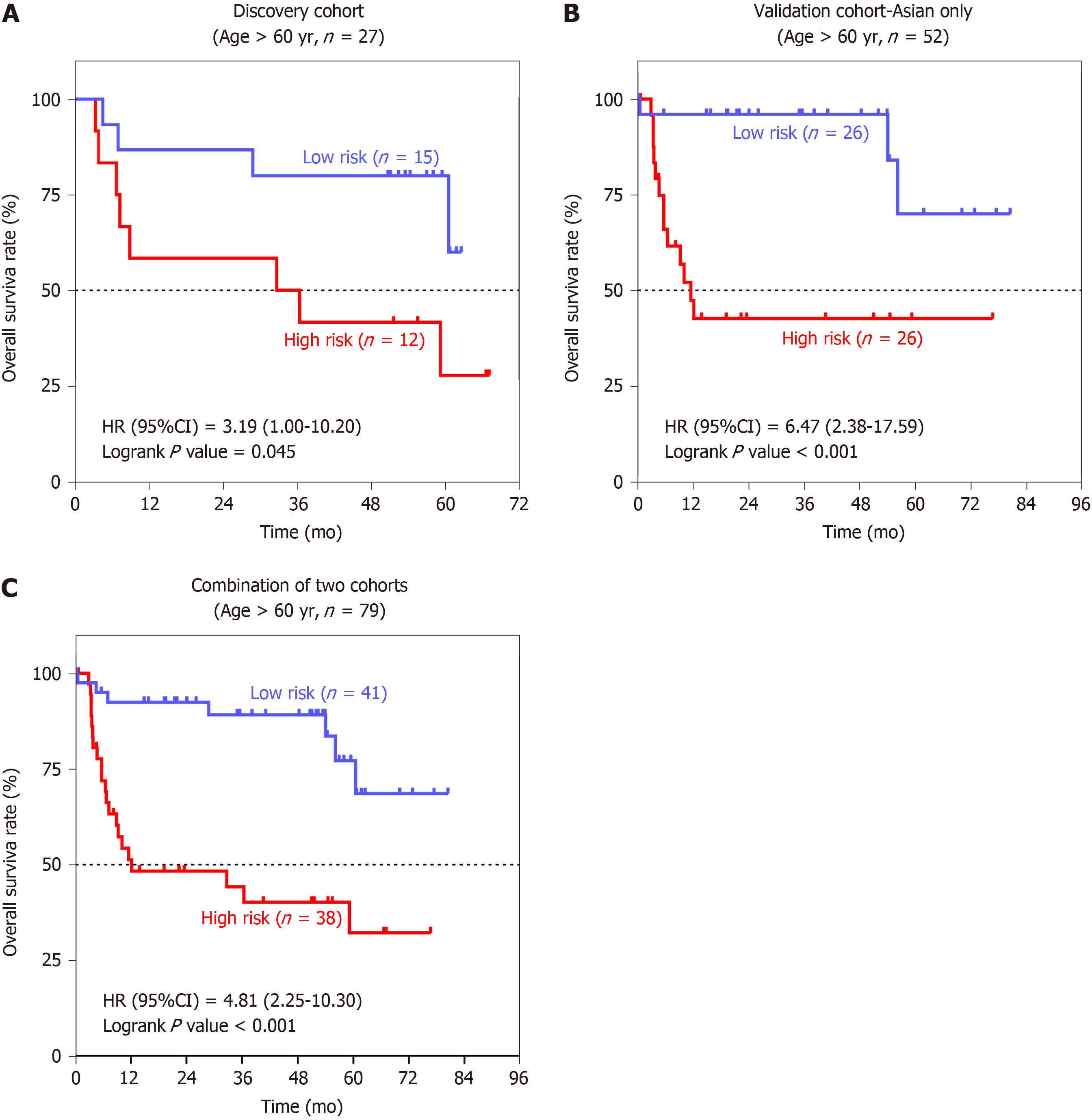
Stratified analyses based on the clinical characteristics were carried out to identify the suitable Asian patient groups for the seven-SAG signature (Table 2). In the elderly population, patients with a high-risk score had a more than 3-fold increased risk of death than the low-risk group (Figure 6A-C). These results suggested that our seven-SAG signature was more applicable to the HCC patients with older age in predicting OS.
| Variable | Discovery cohort | Validation cohort-Asian only | ||||
| High-risk/low-risk | HR (95%CI) | P-value | High-risk/low-risk | HR (95%CI) | P-value | |
| Overall | 76/75 | 1.92 (1.16-3.19) | 0.011a | 79/78 | 5.81 (3.20-10.54) | < 0.001a |
| Gender | ||||||
| Male | 69/67 | 2.15 (1.27-3.64) | 0.004a | 62/61 | 5.18 (2.66-10.10) | < 0.001a |
| Female | 7/8 | 0.60 (0.10-3.47) | 0.569 | 17/17 | 9.63 (2.59-35.77) | 0.009a |
| Age (yr) | ||||||
| ≤ 60 | 64/60 | 1.70 (0.97-2.98) | 0.064 | 53/52 | 5.22 (2.49-10.97) | < 0.001a |
| > 60 | 12/15 | 3.19 (1.00-10.20) | 0.045a | 26/26 | 6.47 (2.38-17.59) | < 0.001a |
| Cirrhosis | ||||||
| Yes | 70/69 | 1.93 (1.15-3.22) | 0.012a | 17/24 | 3.50 (0.66-18.47) | 0.122 |
| No | 6/6 | 1.73 (0.10-30.65) | 0.695 | 11/28 | 0.98 (0.19-5.00) | 0.979 |
| AFP (ng/mL) | ||||||
| ≤ 300 | 40/39 | 1.73 (0.77-3.87) | 0.179 | 34/55 | 2.94 (1.01-8.57) | 0.036a |
| > 300 | 34/34 | 2.11 (1.09-4.08) | 0.025a | 16/16 | 2.65 (0.60-11.66) | 0.226 |
In this study, we first identified 42 overlapped DEGs using the RS and OIS models. Among them, seven downregulated genes in senescent cells, KIF18B, CEP55, CIT, MCM7, CDC45, EZH2, and MCM5, were shown to be upregulated in HCC tissues and selected to construct a prognostic model. The seven-SAG signature was shown to be associated with OS in both discovery and validation cohorts. Stratified analysis showed that our seven-SAG signature was significantly associated with OS in elderly Asian patients. Moreover, time-dependent ROC analysis showed a favorable prognostic value of our seven-SAG signature when compared with serum AFP.
The cellular senescence is considered an aging hallmark. With the increase in age, the number of senescent cells is increasing. Cellular senescence is widely considered to be an anti-tumor mechanism. Studies have shown that a source of stress that triggers liver senescence is chronic inflammation, which causes damage to liver cell regeneration. Importantly, abrogation of senescence leads to aggressive HCC development[14]. In the present study, we found that the HCC patients carrying high expression of seven SAGs had a shorter OS time. Stratified results further suggested the seven-SAG signature was more applicable to the elderly HCC patients. The potential explanation might be that due to the increasing number of senescent cells, the expression of the seven-SAG signature is decreased, while its high expression indicates a higher proliferation rate and poorer OS.
It has been widely accepted that senescence pathways are collectively at the level of activation of CDKIs, which play pivotal roles in regulating the cell cycle progression. Of the seven genes, KIF18B, a member of the kinesin-8 subfamily, is involved in cell cycle process[15] and acts as an oncogene in cervical cancer[16]. CEP55, also known as c10orf3 and FLJ10540, promotes tumorigenesis and regulates stemness in various cancers, such as lung adenocarcinoma[17-19]. CIT (Serine/threonine kinase 21) encoding a serine/threonine protein kinase, is a downstream effector of Rho family GTPases and participates in cell cycle regulation. Liu et al[20] and Xu et al[21] have demonstrated that CIT is up-regulated in HCC and regulates the G2/M transition in rat hepatocytes. EZH2 is a subunit of polycomb repressive complex 2 (PRC2), a protein complex that induces epigenetically silencing of genes[22]. And it has been reported that several lncRNAs are able to regulate gene transcription by binding to PRC2[23,24].
In most eukaryotes, the MCM complex consists of six highly conserved MCM proteins, namely MCM2–7, which functions as a replicative DNA helicase to unwind the DNA duplex template during DNA replication[25]. Recent evidence has demonstrated that several MCM proteins are tightly associated with tumorigenesis[26-28]. The MCM2-7 hexamer complexes with CDC45 and the hetero-tetrameric GINS complex, the Cdc45-Mcm2-7-GINS (CMG) complex, function as a potential target for cancer treatment and CDC45 interacts with MCM2[29]. Furthermore, our previous study showed that MCM7 promotes cancer progression through cyclin D1-dependent signaling in HCC[30]. Three members of CMG complex, MCM5, MCM7, and CDC45, were included in the risk formula. This evidence indicated that more attention should be focused on the pro-oncogenic mechanisms of senescence escape of HCC cells.
Our study showed that AFP was an independent risk factor for HCC patients. AFP is often expressed at high levels in most HCC patients and is considered a reliable clinical tumor biomarker. As a classic serum biomarker, AFP was found to be associated with prognosis in HCC patients[31,32]. Park et al[33] reported that AFP combined with PIVKA-II were useful in predicting survival in the radiological treatment of locally advanced HCC. Jiang et al[34] also found that preoperative AFP and fibrinogen showed a predictive power for recurrence of HCC after liver transplantation. Here, our study demonstrated that the seven-SAG signature model was superior to serum AFP level and more applicable in predicting OS of HCC patients with older age. The above findings suggested a potential clinical application of the seven-SAG signature in HCC patients.
However, there are some limitations in our study. First, the samples for screening SAG were small, which might cause false positive results. Second, we constructed the risk score system merely based on the gene expression levels, rather than the other genetic events that probably have an effect on the initiation and progression of cancer. Third, patients in the discovery cohort were from Asia, thus, the risk score system was established based on an Asian background. And further stratified analysis in the validation cohort also showed that this model was more suitable for Asian patients. Hence, our HCC prognostic signature still needs to be validated in a larger group of patients from various populations.
In conclusion, we constructed and confirmed a prognostic risk score system comprised of seven SAGs. The seven-SAG signature could be a potential predictor for OS, particularly in elderly Asian HCC patients. Our data provide new promising evidence on prediction biomarkers and targeted therapy for HCC.
Hepatocellular carcinoma (HCC) is a common malignancy that remains a serious cause of death worldwide. Recently, molecular markers and prognostic models have been used to improve the diagnosis and treatment of HCC, but few can be applied clinically. Currently, bioinformatics technology has been used for data mining in large public databases. The abundant sample size in the public database can make up for the shortcomings of small samples in real hospitals and help to seek for a more accurate and applicable prognostic model for HCC.
Researchers have been making efforts to find molecular markers or prognostic models that can effectively predict the prognosis of HCC. Senescence is a cell cycle arrest caused by stress in cells, but the cells are still alive. Studies have shown that the proportion of senescent cells in tissues of patients with cirrhosis increases, but a considerable number of patients with cirrhosis can develop liver cancer, and its specific molecular mechanism has rarely been reported.
By analyzing the database of two cellular senescence models from Gene Expression Omnibus, we screened for senescence-associated genes and validated these genes in the liver cancer databases (GSE14520 and TCGA-LIHC). Then, we constructed an HCC prognostic model and evaluate its prognostic accuracy.
Senescence-associated genes (SAGs) were identified using R package “limma”. The latest statistical algorithm-the least absolute shrinkage and selection operator (LASSO) was applied to create our prognostic model. Time-dependent receiver operating characteristic (ROC) curves were used to compare the prognostic accuracy between the seven-SAG signature and serum α-fetoprotein.
The prognostic model for predicting the overall survival (OS) of HCC was constructed by LASSO, consisting of the seven senescence-associated genes (SAGs) (KIF18B, CEP55, CIT, MCM7, CDC45, EZH2, and MCM5). All seven SAGs were highly expressed in HCC and proliferating cells, while lowly expressed in normal tissues and senescent cells. Survival analysis showed that our seven-SAG characteristics are closely related to OS, especially in Asian populations, both in the discovery and validation cohorts. In addition, the time-dependent ROC curve analysis indicated that the seven-gene marker is better than serum alpha-fetoprotein in predicting 1-, 3-, and 5-year OS of HCC patients.
The seven-SAG signature was more applicable to evaluate OS of Asian HCC patients, which may provide new clinical evidence for the diagnosis and treatment of HCC transformed from cirrhosis.
The current study provides clues that the expression changes of senescence-associated gene are the molecular basis for the progression of cirrhosis to liver cancer. Finding effective senescence-associated molecular biomarkers and predictive features of HCC prognosis is necessary.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: El Eneen Khattab MA, Kamimura K, Toriguchi K S-Editor: Yan JP L-Editor: Wang TQ E-Editor: Song H
| 1. | Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115-132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11444] [Cited by in RCA: 13215] [Article Influence: 1468.3] [Reference Citation Analysis (3)] |
| 2. | Reddy SK, Steel JL, Chen HW, DeMateo DJ, Cardinal J, Behari J, Humar A, Marsh JW, Geller DA, Tsung A. Outcomes of curative treatment for hepatocellular cancer in nonalcoholic steatohepatitis versus hepatitis C and alcoholic liver disease. Hepatology. 2012;55:1809-1819. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 155] [Cited by in RCA: 184] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 3. | Bruix J, Gores GJ, Mazzaferro V. Hepatocellular carcinoma: Clinical frontiers and perspectives. Gut. 2014;63:844-855. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 929] [Cited by in RCA: 1108] [Article Influence: 100.7] [Reference Citation Analysis (1)] |
| 4. | Dutkowski P, Linecker M, DeOliveira ML, Müllhaupt B, Clavien PA. Challenges to liver transplantation and strategies to improve outcomes. Gastroenterology. 2015;148:307-323. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 167] [Cited by in RCA: 203] [Article Influence: 20.3] [Reference Citation Analysis (0)] |
| 5. | Llovet JM, Villanueva A, Lachenmayer A, Finn RS. Advances in targeted therapies for hepatocellular carcinoma in the genomic era. Nat Rev Clin Oncol. 2015;12:408-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 315] [Cited by in RCA: 392] [Article Influence: 39.2] [Reference Citation Analysis (0)] |
| 6. | Kuilman T, Michaloglou C, Mooi WJ, Peeper DS. The essence of senescence. Genes Dev. 2010;24:2463-2479. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1416] [Cited by in RCA: 1623] [Article Influence: 108.2] [Reference Citation Analysis (0)] |
| 7. | Demidenko ZN, Korotchkina LG, Gudkov AV, Blagosklonny MV. Paradoxical suppression of cellular senescence by p53. Proc Natl Acad Sci U S A. 2010;107:9660-9664. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 177] [Cited by in RCA: 230] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 8. | Wiemann SU, Satyanarayana A, Tsahuridu M, Tillmann HL, Zender L, Klempnauer J, Flemming P, Franco S, Blasco MA, Manns MP, Rudolph KL. Hepatocyte telomere shortening and senescence are general markers of human liver cirrhosis. FASEB J. 2002;16:935-942. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 374] [Cited by in RCA: 382] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 9. | Wei W, Ji S. Cellular senescence: Molecular mechanisms and pathogenicity. J Cell Physiol. 2018;233:9121-9135. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 146] [Article Influence: 20.9] [Reference Citation Analysis (0)] |
| 10. | Guo M. Cellular senescence and liver disease: Mechanisms and therapeutic strategies. Biomed Pharmacother. 2017;96:1527-1537. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 12] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 11. | Arbet J, McGue M, Chatterjee S, Basu S. Resampling-based tests for Lasso in genome-wide association studies. BMC Genet. 2017;18:70. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 14] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 12. | Friedman J, Hastie T, Tibshirani R. Regularization Paths for Generalized Linear Models via Coordinate Descent. J Stat Softw. 2010;33:1-22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 9] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 13. | Kim Y, Kong L. Time-dependent ROC analysis for censored biomarker data due to limit of detection. J Biopharm Stat. 2018;28:612-621. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 14. | Zeng S, Shen WH, Liu L. Senescence and Cancer. Cancer Transl Med. 2018;4:70-74. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 66] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 15. | Lee YM, Kim E, Park M, Moon E, Ahn SM, Kim W, Hwang KB, Kim YK, Choi W, Kim W. Cell cycle-regulated expression and subcellular localization of a kinesin-8 member human KIF18B. Gene. 2010;466:16-25. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 36] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 16. | Wu Y, Wang A, Zhu B, Huang J, Lu E, Xu H, Xia W, Dong G, Jiang F, Xu L. KIF18B promotes tumor progression through activating the Wnt/β-catenin pathway in cervical cancer. Onco Targets Ther. 2018;11:1707-1720. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 53] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 17. | Tao J, Zhi X, Tian Y, Li Z, Zhu Y, Wang W, Xie K, Tang J, Zhang X, Wang L, Xu Z. CEP55 contributes to human gastric carcinoma by regulating cell proliferation. Tumour Biol. 2014;35:4389-4399. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 62] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 18. | Chen CH, Lu PJ, Chen YC, Fu SL, Wu KJ, Tsou AP, Lee YC, Lin TC, Hsu SL, Lin WJ, Huang CY, Chou CK. FLJ10540-elicited cell transformation is through the activation of PI3-kinase/AKT pathway. Oncogene. 2007;26:4272-4283. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 77] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 19. | Martinez-Garay I, Rustom A, Gerdes HH, Kutsche K. The novel centrosomal associated protein CEP55 is present in the spindle midzone and the midbody. Genomics. 2006;87:243-253. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 69] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 20. | Liu H, Di Cunto F, Imarisio S, Reid LM. Citron kinase is a cell cycle-dependent, nuclear protein required for G2/M transition of hepatocytes. J Biol Chem. 2003;278:2541-2548. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 37] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 21. | Xu XR, Huang J, Xu ZG, Qian BZ, Zhu ZD, Yan Q, Cai T, Zhang X, Xiao HS, Qu J, Liu F, Huang QH, Cheng ZH, Li NG, Du JJ, Hu W, Shen KT, Lu G, Fu G, Zhong M, Xu SH, Gu WY, Huang W, Zhao XT, Hu GX, Gu JR, Chen Z, Han ZG. Insight into hepatocellular carcinogenesis at transcriptome level by comparing gene expression profiles of hepatocellular carcinoma with those of corresponding noncancerous liver. Proc Natl Acad Sci U S A. 2001;98:15089-15094. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 254] [Cited by in RCA: 268] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 22. | Kim KH, Roberts CW. Targeting EZH2 in cancer. Nat Med. 2016;22:128-134. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1059] [Cited by in RCA: 1156] [Article Influence: 128.4] [Reference Citation Analysis (0)] |
| 23. | Su M, Xiao Y, Tang J, Wu J, Ma J, Tian B, Zhou Y, Wang H, Yang D, Liao QJ, Wang W. Role of lncRNA and EZH2 Interaction/Regulatory Network in Lung Cancer. J Cancer. 2018;9:4156-4165. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 51] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 24. | Dong P, Xiong Y, Yue J, Hanley SJB, Kobayashi N, Todo Y, Watari H. Long Non-coding RNA NEAT1: A Novel Target for Diagnosis and Therapy in Human Tumors. Front Genet. 2018;9:471. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 122] [Cited by in RCA: 186] [Article Influence: 26.6] [Reference Citation Analysis (0)] |
| 25. | Ishimi Y. Regulation of MCM2-7 function. Genes Genet Syst. 2018;93:125-133. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 63] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 26. | Zhou YM, Zhang XF, Cao L, Li B, Sui CJ, Li YM, Yin ZF. MCM7 expression predicts post-operative prognosis for hepatocellular carcinoma. Liver Int. 2012;32:1505-1509. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 44] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 27. | Toyokawa G, Masuda K, Daigo Y, Cho HS, Yoshimatsu M, Takawa M, Hayami S, Maejima K, Chino M, Field HI, Neal DE, Tsuchiya E, Ponder BA, Maehara Y, Nakamura Y, Hamamoto R. Minichromosome Maintenance Protein 7 is a potential therapeutic target in human cancer and a novel prognostic marker of non-small cell lung cancer. Mol Cancer. 2011;10:65. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 85] [Cited by in RCA: 96] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 28. | You Z, Ishimi Y, Masai H, Hanaoka F. Roles of Mcm7 and Mcm4 subunits in the DNA helicase activity of the mouse Mcm4/6/7 complex. J Biol Chem. 2002;277:42471-42479. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 61] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 29. | Abid Ali F, Renault L, Gannon J, Gahlon HL, Kotecha A, Zhou JC, Rueda D, Costa A. Cryo-EM structures of the eukaryotic replicative helicase bound to a translocation substrate. Nat Commun. 2016;7:10708. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 106] [Cited by in RCA: 105] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 30. | Qu K, Wang Z, Fan H, Li J, Liu J, Li P, Liang Z, An H, Jiang Y, Lin Q, Dong X, Liu P, Liu C. MCM7 promotes cancer progression through cyclin D1-dependent signaling and serves as a prognostic marker for patients with hepatocellular carcinoma. Cell Death Dis. 2017;8:e2603. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 89] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 31. | Lee SH, Lee JS, Na GH, You YK, Kim DG. Immunohistochemical markers for hepatocellular carcinoma prognosis after liver resection and liver transplantation. Clin Transplant. 2017;31. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 23] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 32. | Liu G, Wang K, Li J, Xia Y, Lu L, Wan X, Yan Z, Shi L, Lau WY, Wu M, Shen F. Changes in serum alpha fetoprotein in patients with recurrent hepatocellular carcinoma following hepatectomy. J Gastroenterol Hepatol. 2015;30:1405-1411. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 14] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 33. | Park H, Kim SU, Park JY, Kim DY, Ahn SH, Chon CY, Han KH, Seong J. Clinical usefulness of double biomarkers AFP and PIVKA-II for subdividing prognostic groups in locally advanced hepatocellular carcinoma. Liver Int. 2014;34:313-321. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 29] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 34. | Jiang N, Zeng KN, Dou KF, Lv Y, Zhou J, Li HB, Tang JX, Li JJ, Wang GY, Yi SH, Yi HM, Li H, Chen GH, Yang Y. Preoperative Alfa-Fetoprotein and Fibrinogen Predict Hepatocellular Carcinoma Recurrence After Liver Transplantation Regardless of the Milan Criteria: Model Development with External Validation. Cell Physiol Biochem. 2018;48:317-327. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 10] [Article Influence: 1.4] [Reference Citation Analysis (0)] |